Abstract
G protein-coupled receptors (GPCRs) are the largest class of receptors in the human genome and one of the most common drug targets. It is now well-established that GPCRs can signal through multiple transducers, including heterotrimeric G proteins, G protein receptor kinases and βarrestins. While these signaling pathways can be activated or blocked by “balanced” agonists or antagonists, they can also be selectively activated in a “biased” response. Biased responses can be induced by biased ligands, biased receptors, or system bias, any of which can result in preferential signaling through G proteins or βarrestins. At many GPCRs, G protein- and βarrestin-mediated signaling have been shown to have distinct biochemical and physiological actions from one another and an accurate evaluation of biased signaling from pharmacology through physiology is critical for preclinical drug development. Recent structural studies have provided snapshots of GPCR-transducer complexes, which should aid in the structure-based design of novel biased therapies. Our understanding of GPCRs from two-state, on-and-off switches has evolved to that of multistate allosteric microprocessors, in which biased ligands transmit distinct structural information that is processed into distinct biological outputs. The development of biased ligands as therapeutics heralds an era of increased drug efficacy with reduced drug side effects.
Introduction
G protein-coupled receptors (GPCRs) are the most common receptors encoded in the genome, comprising greater than 1% of the coding human genome with approximately 800 members and expressed within every organ system1. All GPCRs share a common architecture consisting of an extracellular N-terminal sequence, seven transmembrane-spanning (TM) domains (TM1–TM7) that are connected by three extracellular and three intracellular loops, and an intracellular C-terminal domain. As they regulate virtually every aspect of physiology, it is unsurprising that GPCRs are also the target of > 30% of all prescription drug sales2,3. GPCRs are sensors of a wide array of extracellular stimuli, including proteins, hormones, small molecules, neurotransmitters, ions, and light. GPCR signaling is primarily controlled by interactions with three protein families: G proteins, G protein receptor kinases (GRKs) and βarrestins which perform distinct functions at the receptor4. Upon stimulation, GPCRs activate heterotrimeric G proteins. Classically, agonist binding causes a conformational change in a GPCR, inducing guanine exchange factor (GEF) activity that catalyzes the exchange of GTP for GDP on Gα subunits of the heterotrimeric G-protein. This, in turn, leads to the dissociation of the heterotrimeric complex into Gα and Gβγ subunits. The dissociated subunits promote the formation of second messenger effectors such as cyclic adenosine monophosphate (cAMP), inositol triphosphate (IP3), diacylglycerol (DAG), as well as modulation of other receptors and channels, such as activation of inward rectifying potassium channels.
Similar to most biological systems, negative feedback loops have evolved to quench sustained second messenger signaling following receptor stimulation for maintenance of biological homeostasis. After ligand binding and G protein activation, the receptor is phosphorylated on its cytoplasmic loops and C-terminus, primarily by GRKs5, which enhance βarrestin binding to the receptor. βarrestins were first discovered for their role in mediating receptor desensitization6, the process whereby repeated stimulation decreases the signaling response over seconds to minutes, through steric hindrance of GPCR interaction with the G proteins. βarrestins also mediate receptor internalization via interactions with clathrin coated pits7–9. This can result in downregulation, a sustained decrease in receptor number over minutes to hours due to trafficking of these internalized receptors to proteasomes or lysosomes. Internalized receptors that are not degraded also can be recycled to the plasma membrane10. It is now established that in addition to acting as negative regulators of G protein signaling, βarrestins also couple to numerous signaling mediators including mitogen-activated protein kinases (MAPKs), AKT, SRC, nuclear factor-κB (NF-κB) and phosphoinositide 3-kinase (PI3K) by acting as adaptors and scaffolds11–17. These pathways are separate from classical G protein signaling, but can involve similar signaling cascades that are often temporally distinct. More recently, it has also been appreciated that some receptors tightly interacting with βarrestins maintain catalytic GEF activity on endosomes, continuing to promote G protein signaling after internalization18,19. Thus, βarrestins regulate nearly all aspects of receptor activity, including desensitization, downregulation, trafficking and signaling.
Most drugs that activate or block GPCRs are thought to “equally” target distinct signaling pathways mediated by different G proteins and βarrestins. These agonists are thought to amplify downstream signaling pathways in a similar fashion to that of the endogenous reference agonist (“balanced agonists”), while most antagonists are believed to inhibit all second messenger systems activated by those agonists. However, it was appreciated three decades earlier that selective agonists or antagonists could specifically target particular receptor-linked effector systems20. Indeed, over the past two decades, a number of ligands have been described that selectively activate some pathways while blocking others downstream of the receptor21,22. Compared to the aforementioned balanced agonists, these “functionally selective”23 or “biased” agonists can selectively activate G proteins while blocking arrestins, or vice versa24. This behavior was initially identified in a number of GPCR systems, including PACAP receptor ligands that differentially activated different G proteins as measured by a reversal in potencies25–27. Biased agonism has become an increasingly active area of research since the discovery of βarrestin-mediated signaling28, with a plethora of biased ligands identified for multiple GPCRs29,30.
The discovery of biased agonism has had important implications for our understanding of GPCR biology. First, biased agonism is not consistent with two-state models for receptor signaling, and therefore it alters our concept of efficacy31. Second, biased signaling suggests that GPCRs should not be modeled as binary switches, but instead as allosteric microprocessors that generate a multitude of conformations in response to different ligands. There are also important clinical implications for these ligands, as selectively activating or inhibiting specific signaling cascades could yield more targeted drugs with reduced side effects32. In this review, we primarily focus on advances in the field of biased agonism within the past five years that have changed our fundamental understanding of receptor biology, from the theoretical and structural bases for receptor signaling to the design of novel agents with unique therapeutic profiles.
Theoretical basis of biased signalling
What factors result in the development of a biased response? Using the ternary complex as a model for receptor activity, agonist activation of a receptor requires three principal components to initiate signaling: ligand, receptor, and transducer(s)33. These three components interact allosterically34: A ligand can increase the affinity of a receptor for transducer, such as a G protein or βarrestin, while transducer binding to intracellular receptor domains can stabilize a conformation that increases the affinity for a specific ligand35,36. Allostery is a widespread biological phenomenon that describes the ability of interactions occurring at one site of a macromolecule to modulate interactions at a spatially distinct binding site on the same macromolecule in a reciprocal manner37. In two state models, there are only binary conditions for the receptor: the inactive state, which is incapable of signaling, and the active state, which can bind and activate transducers35. The receptor is modeled as a switch, with agonists stabilizing the “on” state and antagonists stabilizing the “off” state. Agonist efficacy is defined as the ability of a ligand to modify the signaling state of the receptor by stabilizing the active receptor conformation38. The phenomenon of biased agonism demonstrates that receptors are not acting as simple switches that merely encode states of activity across a binary spectrum, i.e., either agonists or antagonists that equally activate or inhibit all signaling pathways downstream of a receptor. Rather, ligand binding results in the activation or inhibition of multiple GPCR-mediated effectors. These effectors often rely on distinct phases of G protein, GRK and βarrestin signaling. Instead of encoding binary ‘on’ or ‘off’ signals, a more appropriate model is one where a GPCR acts as an allosteric microprocessor with pluridimensional efficacy, responding to different molecules with different transducer coupling efficiencies39,40. Any site on the receptor surface that binds a molecule can, in theory, stabilize a distinct receptor conformation and induce a particular pharmacological output. Therefore, physiological activity of a drug need not be linked to interaction at the orthosteric binding site.
Indeed, biased signaling is not limited to GPCRs. For example, nuclear hormone receptors can display distinct conformational dynamics when bound to different ligands41–43. Selective estrogen receptor modulators (SERMs) display tissue-selective pharmacology, and highlight the clinical relevance of differential signaling. As estrogen antagonists, SERMs oppose the action of estrogens in certain tissues, while mimicking the action of endogenous estrogens (agonists) in other tissue types44. Ligands for receptor tyrosine kinases (RTKs) have been discovered that dissociate kinase domain phosphorylation and receptor dimerization45. Interestingly, βarrestin1 regulates ERK signaling downstream of the RTK insulin-like growth factor-1 receptor (IGF-1R)46, suggesting that βarrestin-biased ligands might be discovered in the RTK family as well. Like GPCRs, selectively modulating the signaling networks of these receptor classes holds immense therapeutic promise. It is likely that we will begin to appreciate biased signaling in other cellular components charged with transducing extracellular stimuli into intracellular responses.
Ligand Bias, Receptor Bias and System Bias
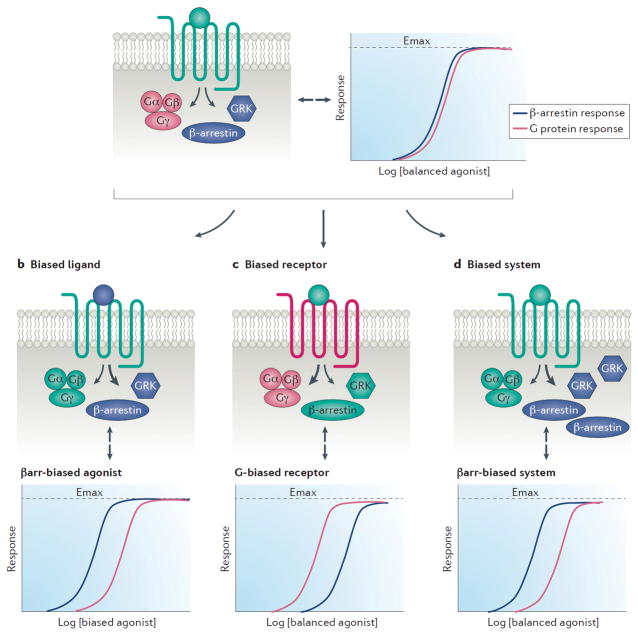
Any of the three components of a ternary complex, ligand, receptor and transducers, can contribute to a biased response (Figure 1). “Ligand bias” or ‘druggable’ biased agonism, refers to the situation in which the ligand induces a unique receptor conformation that results in differential coupling to the signal transduction cascade and a biased response. Distinct agonists, as well as peptides with disparate post translational modifications, can alter intracellular transduction. For example, either differential glycosylation patterning of the endogenous follicle stimulating hormone (FSH)47 or different thiazolidinone small molecule allosteric modulators48,49 can cause divergent FSH-receptor signaling patterns. Biased agonists can alter the properties of this core ternary complex by binding orthosterically, allosterically, or both in a bitopic fashion. Ligand bias should generate a biased response relatively independent of the cell system tested, although if the transducers required for a biased response are either expressed at low levels or absent, then no change in signaling would be observed.
Figure 1. Biased signaling can be encoded through three general mechanisms.
(A) A balanced agonist binding to a balanced receptor in an unbiased system may display equivalent potencies for two different pathways, such as G protein and βarrestin, under assay conditions with similar amplification levels. (B) Biased agonism, or ‘druggable’ biased signaling, is encoded through the ligand. The ligand-receptor-effector complex generates distinct conformation(s) that preferentially signal through certain pathways relative to other pathways (βarrestin biased relative to G protein in this example). Unlike the balanced agonist in panel (A), a βarrestin biased agonist may display a left shift in potency relative to the G protein pathway under the same assay conditions. (C) Biased receptors, such as G protein-biased receptors that lack C-terminal phosphorylation sites necessary for βarrestin recruitment, signal preferentially through one pathway relative to another (G protein biased signaling in this example) despite being stimulated by a balanced agonist. Similar to biased ligands, biased receptors will also display a left shift of one pathway relative to another that may not be observed at an unbiased receptor under the same assay conditions. (D) System bias may be due to differential expression of signaling effectors or other cofactors. For example, higher expression of certain GRK and/or βarrestin isoforms can bias signaling towards the βarrestin pathway (shown here). Alternatively, lack of GRKs or βarrestins can bias signaling towards the G protein pathway (not shown).
Biased receptors can be generated by modifying the receptor to change its ability to bind specific ligands or transducers. This can occur through mutation or through differential splicing, both of which can alter coupling to G proteins and βarrestins. For example, all 19 possible amino acid substitutions at Ala293 of the α1 adrenergic receptor result in constitutive G protein activity50, while a single serine to alanine mutation in the C-terminus of the apelin receptor (APJ) inactivates GRK phosphorylation and blunts βarrestin signaling51. In the C-terminus of neuropeptide Y4 receptor, mutation of glutamic acid, serine or threonine residues disrupts agonist induced recruitment of βarrestin-2 and receptor endocytosis52. The chemokine receptor CXCR7 was initially classified as a non-signaling ‘decoy’ receptor, although it was later shown that CXCR7 engages ligand dependent βarrestin recruitment and signaling while lacking appreciable G-protein signaling53. CXCR7 has also been shown to heterodimerize with CXCR4 to alter CXCR4 signaling54,55, while also having distinct and independent functions mediated by βarrestin56. The chemokine receptor CXCR3 has splice variants, CXCR3-A and CXCR3-B, that differentially activate βarrestins despite only differing in their N-terminal residues 57,58. GPCR designer receptors exclusively activated by a designer drug (DREADDs) utilize a chemical-genetic approach for selective activation of a designer GPCR with an otherwise pharmacologically inert compound59. Biased DREADDs have recently been designed that selectively activate G protein or βarrestin signaling in specific cell types60,61. In addition, biased optogenetic GPCRs for both G protein signaling and βarrestin signaling have been generated and these chimeric light-sensitive receptors allow for precise temporal and spatial pathway dissection62,63.
“System bias”, or “apparent” biased agonism, can be modulated through the differential expression of transducer elements proximal to the receptor, such as the receptor itself, βarrestins or GRKs, as well as expression of amplification cascades distal to the receptor38,64,65 (Figure 1). Similar to the nuclear hormone receptor system, where a ligand may act as an estrogen receptor agonist in one tissue but as an antagonist in another66, GPCRs also have different signaling properties depending on the tissue or system probed. Such systems bias can result in different signaling properties depending on the cell type; for example, certain agonists targeting the dopamine-2 receptor (D2R) have different effects in the striatum and prefrontal cortex, which could be related to the differential expression of βarrestins and GRKs across brain regions67. System bias may also differ between species68, and is important to consider in translational studies. Studies of biased ligands with apparently contradictory results may be due to differences in the experimental system. Part of system bias is “observation bias”, as all measurements are viewed through the lens of a specific assay that is associated with amplification or other properties. To identify ‘druggable’ ligand bias, it is critical to remove the effects of system bias and observation bias from the cellular response38.
Transmission of Ligand Bias
How is bias encoded in the ligand transmitted through the receptor to downstream transducers? Our current understanding is that this is primarily accomplished through the ligand-induced generation of distinct conformations of the receptor allosteric microprocessor via multiple mechanisms. First, there are changes in receptor secondary and tertiary structure. Then, these conformational changes result in the recruitment of proteins that post-translationally modify the intracellular loops and C-terminus of the receptor. Together, these changes in conformation and post-translational modifications contribute to differential transducer coupling. Phosphorylation and ubiquitination of GPCRs, by altering the conformational architecture of the receptor, are the most well described post translational modifications that can bias signaling69. GPCRs require phosphorylation of serine and threonine residues within the C-terminus and intracellular loops for tight binding with βarrestin. Multiple studies have now demonstrated that differential receptor phosphorylation patterns, or receptor “barcodes” 70–73, lead to distinct receptor conformations or transitions that differentially couple to signaling transducers. For example, stimulation of the β2-adrenergic receptor (β2AR) with different biased ligands results in discrete phosphorylation patterns of intracellular residues as assessed by both quantitative phosphoproteomics and antibodies directed against phospho-specific residues70. With this receptor “barcode”, biased ligands that activate βarrestin signaling induce conformations and trafficking patterns of βarrestin that are distinct from those induced by unbiased ligands70,74–78. Disrupting this barcode, through mutations in the C-terminus of GPCRs that remove putative GPCR C-terminal phosphorylation sites can have serious physiological consequences. For example, truncation or mutation of serine or threonine residues in the C-terminus of CXCR4 cause WHIM syndrome by disrupting CXCR4 internalization and sequestering neutrophils within the bone marrow79,80. Mutations that potentiate βarrestin-receptor interactions also cause disease. A constitutively active mutation in the conserved E/DRY “ionic lock” motif of the vasopressin 2 receptor (V2R) leads to constitutive receptor phosphorylation, continual βarrestin recruitment and receptor internalization that results in nephrogenic diabetes insipidus through diminished surface expression of V2R and reduced aquaporin channels in renal collecting ducts81 (Figure 2). Mutating highly conserved polar residues near the transmembrane helical boundaries and core of the GLP1-receptor results in differential regulation of cAMP, calcium, and phospho-ERK signaling and can be a trigger for biased agonism82–84. However, it is still largely unclear how these motifs contribute at the structural level to the transmission of biased information encoded in the ligand to the receptor and then to the transducer.
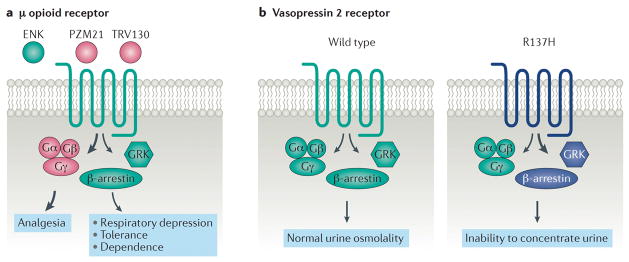
Figure 2. Drug discovery strategies and physiological consequences of biased signalling.
(A) The μ opioid receptor is a common drug target for analgesia. The endogenous ligand enkphalin (ENK) is unbiased (‘balanced’) with respect to G protein and βarrestin signaling. However, current μopioid selective agonists, such as morphine, that provide pain relief also cause adverse effects including respiratory depression, constipation, tolerance, and dependence. Both animal and human studies suggest that G protein signaling primarily mediates the analgesic efficacy, while βarrestin signaling mediates many of the adverse effects. Strongly G protein biased agonists, such as TRV130 and PZM21, may therefore provide clinical superiority to currently available agonists. (B) Relative to the wild-type receptor, the naturally occurring human vasopressin 2 receptor mutation R137H results in βarrestin biased signaling and is associated with familial nephrogenic diabetes insipidus. The vasopression R137H receptor variant is constitutively phosphorylated. This constitutive phosphorylation promotes greater βarrestin recruitment and internalization, even in the absence of arginine vasopressin (AVP, also known as antidiuretic hormone), compared to the wild-type receptor. Presumably, a significant reduction in vasopressin mediated Gαs signaling is sufficient to diminish aquaporin channel insertion into the renal collecting ducts, impeding urine concentration through reduced water resorption and leading to the clinical diagnosis of nephrogenic diabetes insipidus.
Structural Basis for Biased Agonism
Since the first initial crystal structure of a non-rhodopsin GPCR, the β2-adrenergic receptor, was solved in 200785, over 30 high resolution crystal structures of GPCRs have been determined. This structural revolution has provided invaluable insights into GPCR signaling mechanisms. These structures display a common seven transmembrane architecture, but with significant heterogeneity in the orthosteric ligand binding pockets86. Receptors that bind the same ligand have increased conservation within the binding pocket, but still most commonly share only 50–60% of residues87. The production of homogenous complexes necessary for crystallization often requires modification of the flexible C-terminus and other intracellular loops; therefore, many GPCR structures lack detailed information of these regions. While active GPCR structures are critical for structure-based drug design for agonists, inactive GPCR structures provide high resolution insight into the development of therapeutic antagonists and inverse agonists. This is true for, for example, for structures of the angiotensin II type 1 receptor (AT1R) in complex with AT1R blockers (ARBs)88,89, a common therapeutic target for anti-hypertensive medications. Inactive structures also provide important insights into signaling mediated by the active state, as a comparison point for conformational differences between ligand bound and unbound receptor states. There are many receptors for which both ‘inactive’ as well either active or partially active intermediate crystal structures are available, including rhodopsin90,91, the A2A receptor (A2AR)92–94, the M2 muscarinic receptor (M2R)95, and the μOR96,97. Structures of receptors in complex with biologically potent allosteric antagonists or negative allosteric modulators include CC Chemokine receptor 2 (CCR2)98, CC Chemokine receptor 9 (CCR9)99, corticotrophin releasing factor receptor (CRHR1)100, and the glucagon receptor101. These offer high resolution insight into druggable surfaces outside the orthosteric ligand pocket, most notably rearrangements of amino acid contacts between transmembrane regions 3, 6, and 7 that expose residues that could lead to GEF activity and subsequent G protein activation. However, the changes observed in these structures can be quite limited.
These structures have shown that agonists alone are often not sufficient to stabilize an active GPCR conformation. Transducer coupling, or use of a stabilizing modulator such as a nanobody, is often required for trapping the receptor in its active state. To date, only two GPCR structures have been solved in complex with a bona fide G protein102,103. The β2AR was the first such receptor-G protein complex, and the co-crystalization structure reveals that both the N-terminal and C-terminal domains of the Gαs subunit interact with the β2AR intracellular loop 2, transmembrane domain 5 (TM5), and TM6. Recently, the structure of the calcitonin receptor (CTR) in complex with a Gαsβγ heterotrimeric complex was solved by cryo-EM103. Like the β2AR, significant outward movement of TM6 is observed. Despite the β2AR belonging to the class A GPCR sub-family and the CTR belonging to the class B sub-family (comprising of a larger extracellular N-terminal domain when compared with class A GPCRs), an overlay of the β2AR and the CTR G protein complex structures reveal only minor differences in G protein conformation between the two receptors. However, in contrast to the β2AR, helix VIII of the CTR appears to play a more important role in receptor stability at the cell surface and in interactions with Gβ. Indeed, growing evidence suggests an important role of helix VIII in signaling. At the enigmatic angiotensin II type 2 receptor (AT2R), helix VIII lies parallel to the membrane in the active-like state apparently sterically inhibiting G protein and βarrestin interactions with the AT2R, which is in agreement with an unusual lack of observed G protein or βarrestin signaling for this receptor.104.
Other GPCRs have been crystalized with G-protein “mimics,” such as nanobodies or an engineered mini-G protein model, that hold the receptor in a conformation thought to mirror the G protein activated state. These structures include the M2 muscarinic95, the μ-opioid receptor (μOR)97, the viral US28 receptor105, and the adenosine A2A receptor (A2AR)106. Two distinct conformations of the β2AR stabilized by different nanobodies have also been recently reported107.
Structural information on how βarrestins can interact with GPCRs is also beginning to emerge. Truncation of the arrestin1 (visual arrestin) C-terminus mimics an activated arrestin, and releases a critical central-crest ‘finger loop’ by disrupting the polar core108. The structure of this finger loop bound to rhodopsin, as well as of the structure of rhodopsin bound to arrestin1, were recently solved, providing a high resolution view of an additional GPCR:transducer complex109,110. Additional structures of GPCRs in complex with βarrestins, as well as GRKs, will be necessary to determine if conserved receptor:arrestin interaction patterns are also present. Such studies will reveal if G proteins, GRKs, and arrestins have preferences of specific receptor conformations. Comparing structures of the ligand:receptor:transducer ternary activated complex to the inactive receptor state are the cartographical methods of rational GPCR drug design, and represent a powerful tool beginning to be harnessed for increasing the efficiency of drug discovery.
A range of other biophysical studies are now being used to provide high resolution information to enable drug discovery for more challenging targets111. Electron microscopy techniques (EM), notably cryo-EM and negative stain EM, have emerged as powerful tools for determining GPCR-transducer structures. An advantage of EM technology is the ability to study a receptor in its wild-type form, as x-ray crystallography frequently requires receptor modifications to obtain sufficient homogeneity and stability for crystal formation. In addition to high resolution structures such as the CTR-G protein complex described earlier, low resolution structures from negative stain EM have been used to reveal agonist-occupied β2AR in complex with G protein as well as to capture transient intermediate G protein and β2AR complexes 112. Furthermore, EM of a β2AR and vasopressin type 2 receptor (V2R) chimera receptor (utilized to increase βarrestin affinity) demonstrates that βarrestin can adopt at least two distinct conformational states when interacting with a GPCR. One conformation reveals βarrestin bound only to the phosphorylated C-terminus of the receptor, while a separate conformation demonstrates the flexible “finger loop” of βarrestin inserting into the receptor transmembrane core in addition to the C-terminus interaction 113. The functional significance these distinct βarrestin receptor interactions (C-terminus only and C-terminus+core) has also been studied. Partial engagement of βarrestin with the C-terminus appears sufficient for receptor endocytosis and ERK activation114. Contact of the βarrestin finger loop with the receptor transmembrane core appears necessary for desensitization, with negative stain EM providing support that a finger loop-core interaction sterically blocks the apparent G protein binding site115. Negative stain EM studies have also revealed that internalized receptor complexes can consist of a GPCR, βarrestin, and G protein116. Such internalized ‘megaplexes’ demonstrate that a single GPCR can simultaneously interact with both a βarrestin and a G-protein. Visualization of these ‘megaplexes’ provides a molecular basis for sustained GPCR catalytic GEF activity and subsequent G protein signaling following GPCR internalization into endosomes. Taken together, the existing EM studies lend further structural support for the presence of multiple GPCR ‘active’ conformational states.
Additional experimental approaches, including a high-resolution mass spectrometry labeling strategy, NMR, molecular dynamic simulations, conformationally selective RNA aptamers, and single domain camelid antibodies (nanobodies)107,117–119, have shown that functionally similar ligands can induce distinct receptor conformational changes. For example, 19F NMR spectroscopic analysis of the β2AR demonstrated distinct receptor conformational states when bound to balanced or biased ligands. The FDA approved beta-blocker Carvedilol, proven to be particularly effective for the treatment of heart failure, is a moderately βarrestin-biased agonist at the β1AR120,121 and β2AR122. A comparison of β2AR conformations induced by either the balanced agonist isoproterenol or the βarrestin-biased agonist carvedilol revealed that that G protein activity correlates with movement of transmembrane helix VI, whereas carvedilol chiefly alters the conformation of helix VII123. RNA aptamer binding also revealed specific β2AR conformations induced by different ligands124, and 13C-labeled β2ARs further validate distinct receptor states not captured by previous crystal structures125. Double electron electron resonance (DEER) spectroscopy of a trifluoro-methyl labeled cysteine at TM6 of the β2AR has also shown significant conformational heterogeneity and rapid interconversion of multiple receptor states126. In addition to the β2AR, a multitude of active receptor conformations are observed at the ghrelin127 and serotonin 2B128 receptors, demonstrating that a heterogeneous ensemble of active receptor states is a conserved property. βarrestin biosensors have also recently been employed to correlate the conformational signature of arrestins to predict signaling and trafficking functions following drug stimulation76. βarrestin NMR probes show different βarrestin-phosphopeptide interactions encode distinct structures that correlate with specific βarrestin mediated functions129. In summary, most structural studies support a model of the receptor as an allosteric microprocessor, with biased ligands inducing an array of distinct receptor conformations that differentially recruit and activate transducers.
Quantifying Bias
Overview
In pharmacological models of receptor signaling, such as those of Ariens130, Stephenson131, Furchgott132, and Black and Leff133, a ligand is considered to have two primary properties in interactions with a receptor: affinity (the ability of a ligand to form a ligand–receptor complex) and efficacy (the ability of the ligand–receptor complex to increase or decrease downstream signaling response(s)). In mechanistic models of receptor signaling, such as the ternary complex of model, efficacy is a measure of the ability of a ligand to effect the transition between active and inactive receptor conformations (such as the alpha parameter in the ternary complex model), while in pharmacological models, efficacy relates the pharmacological stimulus to the observed response131. These pharmacological and mechanistic estimates of efficacy are closely related mathematically38. As noted above, the concept of biased agonism requires that a receptor display multiple efficacies and determination of these biased efficacies requires a deconvolution of ligand bias from system bias. Multiple approaches have now been proposed to quantify ligand bias134–136, although identifying biased ligands requires multiple steps, from the choice of assays used to assess different signaling pathways to the computational approaches used to quantify ligand bias.
Assays for detecting G protein and βarrestin signaling
Table 1 provides a summary of different experimental techniques that can be utilized to measure signal transduction effects. Broadly speaking, these assays are based on different aspects of transducer activity, which include transducer redistribution, receptor:transducer proximity, transducer conformation, receptor internalization, and transducer signaling. Redistribution assays are based on the movement of transducer either to or away from the receptor or downstream effector with ligand stimulation. Receptor proximity assays quantify changes in the distance of a population of transducers, such as βarrestins, to a GPCR upon agonist binding. Assays of transducer conformation and signaling both require a clear understanding of the relationship between the measured effect and transducer activity. For example, while βarrestin activation is associated with conformational changes, only certain conformational signatures are associated with specific signaling function, such as receptor internalization76. Indeed, receptor internalization does not always require βarrestin recruitment, or vice-versa137. Some signaling assays may have inputs from multiple upstream transducers, such as MAP kinase phosphorylation, which has distinct inputs from both G proteins and βarrestins.
Table 1.
Selected assays for assessing ligand bias.
| Assay | Technology | Strengths | Weaknesses |
|---|---|---|---|
| Proximity | Intermolecular BRET or FRET between receptor and βarrestin214 |
|
|
| Enzyme complementation (e.g. cAMP dependent firefly luciferase) |
|
|
|
| Reporter based assays |
|
|
|
| Conformation | Intramolecular BRET or FRET of βarrestin75,76 or seven transmembrane receptor biosensors216 |
|
|
| RNA aptamers124 |
|
|
|
| Redistribution | Labeling of βarrestin or receptor217 |
|
|
| Labeling of receptor/Receptor internalization |
|
|
|
| Signalling | Multiple outputs, including phosphorylation (e.g. MAPK 1/2, AKT), ubiquitination, chemotaxis, apoptosis, stress fibre formation |
|
|
| TGF alpha ectodomain shedding assay218 |
|
|
|
| Cyclase accumulation |
|
|
|
| Calcium/potassium indicator dyes |
|
|
|
| Calcium influx (Aequorin) |
|
|
When designing experiments to identify biased agonists, it is important to eliminate sources of system bias that can confound data interpretation. Comparisons of efficacies and potencies from assays are often limited by differences in receptor reserve (also known as ‘spare receptors’) or amplification between assays. In assays with significant amplification, such as G protein second-messenger assays, both full and partial agonists can reach the same maximal response. In assays with negligible signal amplification, such as BRET-based recruitment or dissociation assays, a balanced partial agonist will display less efficacy than a full agonist. With a simple comparison of potencies or maximal responses, a partial balanced agonist that yields half maximal response in assay B and a maximal response in the amplified assay A could incorrectly be labeled as a biased ligand relative to the full balanced agonist that that achieved maximal responses in both assays. Potencies are also affected by amplification. In assays with high amplification, a full agonist will have a greater leftward shift in potency (EC50) from its dissociation constant (KD) than a partial agonist. However, in assays with no or minimal receptor reserve, the EC50 will approximate the KD for a partial agonist, but not for a full agonist. It can be difficult to compare proximal and distal signaling effectors due to varied amplification and system bias138. Thus, it is not straightforward to compare Emax and EC50 alone to identify biased agonists. To address this issue, both qualitative and quantitative approaches have been developed to attempt to remove the effects of system bias and identify truly biased ligands.
Identifying Biased Ligands
Qualitative assessment
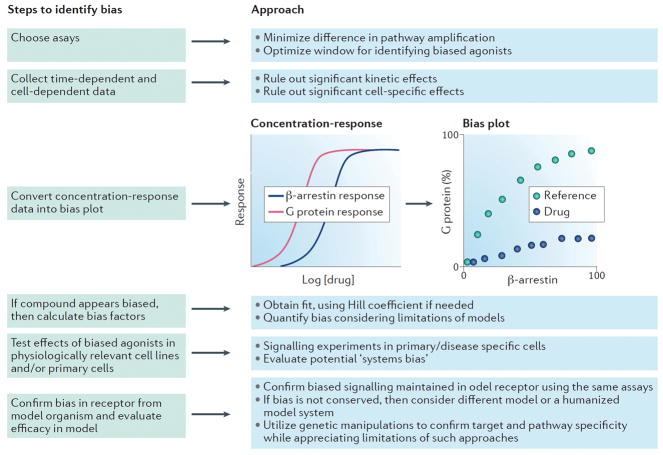
To increase the sensitivity and specificity of identifying biased compounds, both quantitative and qualitative methods should be used to identify potentially biased ligands. The effects of system bias can be assessed qualitatively by a ‘bias plot’139 (Figure 3A). Bias plots are generated by converting dose-response data for two signaling pathways of interest, e.g., G protein and βarrestin, into plots comparing responses in pathway A to responses in pathway B at identical concentrations of ligand138,139. An advantage of the bias plot is it allows an assessment of assay amplification effects that can confound efforts to identify biased ligands. It also provides a way to identify the best assays to quantify ligand bias, as it provides information on the windows for identifying G protein- or βarrestin-biased agonists. When choosing assays to assess ligand bias, it is important to select assays that have similar levels of amplification. Similar levels of amplification provide the largest windows for identifying ligands that could be biased towards either pathway tested. However, if one is interested in identifying ligands biased towards pathway A relative to pathway B, it can make sense to choose an assay with somewhat more amplification in pathway B relative to A. If a ligand does not appear to be biased using a bias plot, it is unlikely to be biased, even if a “bias factor” from a quantitative approach is significant. This is because the bias plot is not prone to errors introduced from different fitting approaches38. However, even bias plots cannot account for other aspects of system bias, such as differential expression of GRKs and βarrestins, which can qualitatively change the relative difference between downstream pathways.
Figure 3. General approach to characterizing biased ligands.
First, assays for different pathways should be chosen with the goal of minimizing differences in signal amplification. Such assay selection optimizes the window for identifying biased agonists (see example of a bias plot below). Time- and cell-dependent data should be obtained to ensure no significant kinetic or cell-specific effects. To qualitatively identify biased agonists, construct a ‘bias plot’ by, for example, graphing βarrestin activity on the x axis and G protein activity on the y axis (each normalized to pathway specific maximal signals in this example). Deviations from the reference agonist suggest the presence of ligand bias. If no biased signaling is observed on a bias plot, then it is unlikely that true biased signaling is present (even if calculated bias factors are statistically significant). Multiple approaches can be used to calculate bias factors (see text), such as a method based on intrinsic relative activities (calculator available online, at https://biasedcalculator.shinyapps.io/calc/). After identification and quantification of ligand bias, the physiological implications of such signaling can be tested in relevant cell lines or primary cells. Biased signaling properties can then be confirmed in the receptor from a relevant model organism and subsequently evaluated in the animal model for safety and efficacy.
Common Questions Regarding Ligand Bias
What assays should I use to identify biased agonists?
In general, it is best to choose assays with similar levels of amplification. However, if one is interested in specifically identifying G protein or βarrestin-biased agonists, assays can be chosen to maximize the “window” for identifying such ligands (see box 1, Qualitative assessment). For example, if screening is being performed for a G protein-biased agonist, the window for identifying such agonists could be maximized by using an assay for G proteins that has less amplification than that for βarrestins (Figure 3). While increased amplification allows the identification of weak partial agonists, it can result in artificial stoichiometric relationships between interacting proteins and a risk for artifacts. If strong partial or full agonists are studied, assays with low or endogenous receptor expression may be better. Ideally, the assays used should be in a cell type that is physiologically relevant, thereby limiting the effects of system bias on the in vitro and in vivo drug profiles.
BOX 1. Quantitative assessment.
Intrinsic relative activity values
Relative activity (RAi) values are calculated from the maximal effect produced by a ligand (Emax value, or maximal response) and the concentration of ligand that produces half-maximal response (EC50 value, or potency) compared to a reference compound from concentration–response curves136. For example, for the signaling of ligand 1 compared to a reference compound in pathway A, the intrinsic relative activity is:
A clear indication of biased signaling between two ligands is if the rank order of RAi changes from one pathway to another within the same system. These ratios are also mathematically identical to transduction coefficients (τ/KA) when the Hill slope is unity136; thus, this method is mathematically identical to the “transduction coefficient” method for most concentration-response data (see below). Bias can also be quantified through the calculation of a bias factor, β, when comparing signaling through two pathways134:
This bias factor quantifies the degree of ligand bias on a logarithmic scale, e.g., a bias factor of 1 corresponds to ten-fold higher signaling through pathway A compared to pathway B. The strength of this approach is that it can provide estimates of bias relatively free of any underlying receptor model from simple fits of concentration-response data (unlike bias factors from the operational model, which require either a dissociation constant or a more complex fitting routine, see below). Limitations of calculating bias through intrinsic relative activity values include that A) it can only be applied to data with a Hill coefficient of 1, and B) it does not provide an estimate of the underlying efficacy of the ligands tested (it only provides an estimate of bias, not of the underlying efficacies). An online calculator for intrinsic relative activity values is available at https://biasedcalculator.shinyapps.io/calc/.
Approaches for quantifying bias using the operational model
The operational model developed by Black and Leff is a pharmacological model that accounts for efficacy (quantified by the factor τ) and affinity (KA) by considering the agonist-receptor complex as the functional unit that leads to a pharmacologic effect:
where the variable n accounts for dose-response curve with a non-unity Hill coefficient (although the curve obtained from an operational model fit is different than a typical dose-response curve with a Hill coefficient); τ accounts for efficacy and system amplification and is equal to the receptor density (Rt) divided by the receptor-transducer coupling efficiency (KE)210; and KA is the ligand dissociation constant. While the operational model assumes that signaling is directly related to the agonist:receptor complex and accounts for receptor concentration and intracellular amplification, mathematically it is nearly identical to other pharmacologic receptor models38.
Transduction coefficients: Assessing bias through functional affinity
In the approach using “transduction coefficients”, which are defined as τ/KA, a “functional affinity”, KA, as opposed to one obtained from a separate binding study, is used to fit the data. As noted earlier, for dose-response data with a Hill coefficient of 1, the transduction coefficient should be mathematically identical to the intrinsic relative activity. Therefore, a similar approach can be used to yield a bias factor. Namely, a reference transduction coefficient is utilized to compare values for a given agonist between the pathways of interest, providing a Δlog(τ/KA). A measure of relative bias (ΔΔlog(τ/KA)) is then calculated through comparing a ligand of interest to a reference ligand:
| 135. |
A strength of this approach is that it does not require separate experiments to obtain dissociation constants for ligands. For partial agonists, this can result in an excellent fit of the data, as there is a clear relationship between potency and KA. Unlike the intrinsic relative activities, this approach can also be used to fit data with a Hill coefficient of non-unity. There are some limitations of this approach as well. First, there can be ambiguity of the KA for full agonists where there is not a clear relationship between potency and functional affinity. Second, only information on bias can be obtained from this analysis and not on the underlying efficacies for signaling through different pathways. These advantages and limitations of this model have been noted elsewhere.211–213
Effective Signaling: Estimates using the Operational Model
If dissociation constants from an independent binding study are available, such data can be used in the operational model to yield not only estimates of bias, βlig, but also of the effective signaling for each ligand, σlig = τlig − τref134. A strength of this approach is that it provides both estimates for bias and efficacy for each ligand. This is an advantage, as a strongly biased weak partial agonist will have significantly different effects than a strongly biased full agonist. This approach can also be used to fit data with a non-unity Hill coefficient. However, there is a major limitation to this approach because of the requirement for having dissociation constants for each ligand. These dissociation constants must be obtained in conditions similar to the signaling assay with decoupling from potential transducers (as shifts in binding associated with transducer coupling are directly related to efficacy140). Even with those conditions, partial agonists may display significant differences between EC50 and KD that cannot be accounted for in a pharmacologic model. In such a situation, either intrinsic relative activities or transduction coefficients should be used.
What is the best way to calculate ligand bias?
All of the approaches discussed (see Box 1) have strengths and weaknesses. A bias plot should always be used to assess for bias qualitatively – if bias is not visualized on a bias plot, it is unlikely to be present. All of the quantitative approaches to assess bias are based on essentially the same framework of the operational model, although with different assumptions. If data is fit well without Hill coefficients and no binding data is available, using the approach of intrinsic relative activities is straightforward and can provide estimates for bias. If the Hill coefficient is non-unity and no binding data is available, transduction coefficients should be used. If binding data is available, the approach of effective signaling can also be used.
My ligand signals through both G proteins and βarrestins? How could it be biased?
Biased and balanced signaling are relative terms. First, it is nearly impossible to state that a receptor signals “equally” to different transducers, unless an assay is performed to assess changes in binding of transducers at the receptor140, which would place signaling through different transducers on the same scale. Rather, nearly all measurements are made through the lens of observation bias, and reflect amplification at different levels. For that reason, accepted approaches to quantifying bias are in comparison to a reference agonist, which, by convention, is usually referred to as “balanced.” It is possible that the endogenous reference agonist may have a significant difference in Emax in two assays while a synthetic ligand reaches higher Emax in both assays. This would not make the synthetic ligand “balanced”; rather, it is biased relative to the reference agonist.
How should I calculate maximal response and potency?
A typical first step in concentration-response analysis is to fit the measured efficacy to varied concentrations of a drug, typically with a standard 3-parameter curve fit utilizing the minimum, maximum, and EC50. Introduction of additional variables, such as a Hill coefficient, may improve the fit, but should only be used if it provides a significant improvement. Poor curve fits that result in a suboptimal definition of the minima, maxima, and/or EC50 will introduce significant error into a bias calculation. Baseline correcting the minima to zero is sometimes necessary in high throughput assays, especially if the lowest concentration of ligand is on the outer wells of 96 or 384 well plates that are at increased risk of suffering from small concentration deviations due to increased evaporation of solvent relative to the inner wells.
Can I still calculate bias with a poor fit?
Curve fits that have poorly defined minima, or more commonly maxima, introduce significant error into bias calculations. If the maximal response is poorly defined, and increasing the concentration of the ligand is not feasible, then how can a model system be altered to produce an improved fit? One method is to increase expression of receptor or transducers, such as G proteins, GRKs, and/or βarrestins, however, one must be very cautious of this approach. Artificial stoichiometric relationships between interacting proteins, such as the receptor: transducer ratio, can produce misleading results. In circumstances where receptors are promiscuous with respect to the G protein isoforms they interact with, high receptor numbers may recruit G protein isoforms that have an opposing signal or activate alternative pathways that would not be found in the tissue of interest. In addition, high levels of βarrestin overexpression may dramatically alter the kinetics of the assay through potentiated desensitization and/or increased receptor internalization.
Translating in vitro bias into in vivo utility
Many of the strategies used to bring a promising biased ligand from the bench to bedside are similar to those for typical drug candidates and have been reviewed elsewhere141–144. Given the significant costs in late phase drug discovery, accurately quantifying the relative signaling properties of biased agonists early in the drug discovery process, as well as evaluating effects in suitable preclinical models of disease, is necessary. A number of approaches for biased ligand drug discovery can build confidence that the observed physiological effects are due to bias at the intended target and pathway (Figure 3). Often, the first step in testing a biased drug is in heterologous expression systems. At this stage, it is important to use pharmacological assays that allow for an accurate assessment of bias, which in practice most often means selecting signaling assays with similar levels of amplification. If biased signaling is noted in a heterologous expression system, the candidate can then be tested in primary cells or a cell line that best models the tissue being targeted to minimize any potential system bias due to different levels of transducer expression from the heterologous expression system. Testing the ligand in human cells and/or tissue prior to direct human trials provides further validation. Before studying the compounds in model organism(s) to test safety and efficacy, it is critical to confirm that the drug binds to and that bias is conserved at the receptor of a model organism. For example, a mouse receptor should be evaluated in heterologous expression systems in the same assays that a human receptor was tested. After confirming binding and biased signaling at the receptor of a model organism, the ligand can be moved ahead with more confidence. Frequently it is helpful to perform experiments with a biased candidate in parallel with balanced agonists and antagonists, to confirm that the effects of the candidate are due to bias and not to simple agonism or antagonism. The use of genetically altered animal models can provide further data about the mechanism of action. For assessing biased ligands, knocking out the receptor target can confirm that it mediates the biological effect, and removing or altering a presumed critical transducer (such as a G protein or βarrestin isoform) that the candidate is biased towards can provide strong support about the specific pathway(s) involved. Caution is warranted when interpreting a phenotype from models lacking a critical transducer, even from conditional knockouts, as these transducers almost always couple to multiple receptors and signaling pathways. Some transducers have overlapping roles, such as the two isoforms of βarrestins, and knocking out the dominant isoform in the tissue of interest is often necessary when dual or multiple knockouts are lethal or not feasible. As in all genetic manipulations, compensation and alternative pathway selection are additional confounding factors to consider.
Biased Physiology
Overview
Biased agonism has the potential to revolutionize GPCR drug discovery. For this reason, groups in academia and industry have active research programs in this area. It is impossible to provide a full description of all the biased agonists whose effects have been tested in preclinical or clinical studies. Rather, we will focus on a few examples that demonstrate the promise of these agents as novel therapeutics. Table 2 provides an abbreviated and incomplete list of therapeutically promising biased ligands under active investigation.
Table 2.
Selected biased agonists under different phases of evaluation for therapeutic use.
| Ligand | Receptor | Signalling bias | Indication | Development status |
|---|---|---|---|---|
| Carvedilol | Beta 1 AR Beta 2 AR |
Mild βarrestin | Congestive heart failure | Approved |
| Oliceridine (TRV130) | Mu opioid | G protein | Moderate to severe pain | Phase III |
| TRV734 | Mu opioid | G protein | Moderate to severe pain | Phase I |
| Triazole 1.1 | Kappa opioid | G protein | Pruritus | Preclinical |
| RB-64 | Kappa opioid | G protein | Pain | Preclinical |
| PZM21 | Mu opioid | G protein | Pain | Preclinical |
| UNC9994, UNC9975 | Dopamine 2 | βarrestin | Schizophrenia and mood disorders | Preclinical |
| TRV250 | Delta opioid | G protein | Migraine | Preclinical |
Mu opioid receptor (μOR)
The μ-opioid receptor (μOR), with its endogenous agonists of enkephalin peptides, is the target of multiple blockbuster drugs. Exogenous opioid agonists of the μOR, such as morphine and fentanyl, provide analgesia while antagonists, such as naloxone and its derivatives, are used in the treatment of opioid overdose and substance abuse. However, current μOR agonists are limited by side effects that include addictive potential, respiratory depression, constipation, and tolerance. Compared to the endogenous agonists of μOR, the enkephalins, morphine induces considerably less receptor phosphorylation and internalization (consistent with bias towards G protein-mediated signaling), while etorphine, fentanyl, and methadone cause robust receptor internalization in cells without high levels of GRKs145,146. Intrinsic efficacies of G protein signaling for several μOR agonists do not correlate with the rank order of agonist induced internalization efficacy, consistent with ligand bias147. Early studies of βarrestin signaling at the μOR suggested that a G protein biased agonist might display increased analgesia with a reduced side effect profile148,149. Such small molecules have since been developed: herkinorin150 (derived from the naturally occurring plant product, salvinorin A), TRV130, and PZM21. The G protein biased μOR agonist, TRV130, increased both analgesia and pain relief while reducing on target adverse effects when compared to morphine in a randomized, double blind control trial151,152. PZM21, which is structurally distinct compared to previously explored μOR ligands, was discovered utilizing computational modeling and structure based optimization strategies153. Like TRV130, PZM21 displayed a potent Gαi signaling profile, limited βarrestin recruitment, selective μOR activity, and provided enhanced analgesia with fewer side effects compared to morphine in a preclinical pain model (Figure 2).
Kappa opioid Receptor (KOR)
Like the μ-opioid receptor, kappa opioid receptor (KOR) signaling can produce analgesia, but, unlike drugs targeting μOR, KOR agonists have a low risk of dependence and abuse. However, the distinct side effect profile of KOR activation by the receptor selective, high efficacy agonists initially developed included psychotomimesis, dysphoria, and sedation154,155. Recent studies have established that the analgesic effects are Gβγ mediated, whereas the aversive effects require βarrestin mediated activation of p38 MAPK which regulates serotonin transporter and inward rectifying potassium channel function in neurons of reward processing centers such as dorsal raphe nucleus and ventral tegmental area156–158. A G protein biased KOR ligand triazole 1.1 retained the antinociceptive and antipruritic efficacies of a conventional KOR agonist, yet did not induce sedation, reduce dopamine signaling, or produce dysphoric-like behaviors in rodent models159. Additional biased KOR agonists are currently being developed, and may provide the precision necessary to successfully drug a very promising clinical target68,160–165. The KOR system also shows interesting ligand bias in antagonist drug action: some are conventional competitive ligands with low efficacy, and some are orthosteric ligands that initiate a different mode of signaling which results in long-lasting receptor inactivation166–168. The implications of this ligand bias are clinically significant because KOR antagonists show therapeutic promise in the treatment of stress disorders underlying depression and addiction risks169,170.
Dopamine 2 Receptor (D2R)
The Gαi-coupled D2R is most common target of antipsychotic drugs. The dopamine hypothesis of schizophrenia proposes that the molecular aetiology of this disease is manifested in part by cortical hypodopaminergia and subcortical hyperdoaminergia171. βarrestins and GRKs are differentially expressed in cortex and subcortical regions, with GRK2 and GRK3, and to a lesser extent GRK6, reduced in the striatum compared to the prefrontal cortex in both rodents and humans67. Due to systems bias in these tissues, it is theoretically possible for a balanced ligand to exhibit differential βarrestin and G protein signalling between the striatum and cortex. Interestingly, using genetically engineered mice lacking βarrestin2 in select population of neurons, Urs, Caron, and colleagues have demonstrated both electrophysiologically and behaviourally that while D2R signalling is inhibitory in the striatum, it is excitatory in the cortex but depends on both GRK2 and βarrestin267. A βarrestin biased agonist could therefore theoretically further exploit the observed systems bias by reducing dopaminergic signalling in the striatum while mitigating cortical hypodopaminergia and increasing excitatory signalling. D2R biased ligands have been identified172,173, including βarrestin biased agonists through structure function studies based on derivations of the FDA approved antipsychotic aripiprazole174,175. In animal models, the D2R selective βarrestin biased ligands UNC9975 or UNC9994 improve animal behaviors that are used to model schizophrenia phenotypes, including reduction of dopamine dependent hyperlocomotion, restoration of pre-pulse inhibition, and normalization of social behavior67,176. However, whether these ligands will be effective at correcting cortically associated memory and executive dysfunction remains to be established177. Interestingly, these βarrestin D2R biased ligands appear to have an improved side effect profile compared to typical antipsychotics, with significantly lower levels of catalepsy compared to haloperidol176.
Calcitonin Receptor (CTR)
Similar to other Gαs-coupled GPCRs, calcitonin binding to the calcitonin receptor increases cAMP production, βarrestin recruitment, calcium mobilization, and ERK activation. Calcitonin signaling through CTR reduces serum calcium, primarily by inhibiting osteoclast activity and reducing renal tubular cell reabsorption of calcium phosphate178. Salmon calcitonin (sCT), FDA approved for treatment of postmenopausal osteoporosis, provides a modest increase in bone density by reducing the rate at which osteoclasts degrade bone tissue. However, its peptide formulation and modest clinical efficacy limit the current clinical benefits of sCT. Recent studies demonstrate that sCT has decreased rate of dissociation from the human CTR and increased βarrestin recruitment compared to hCT179. Beyond differences in βarrestin signaling, additional granularity of ligand bias is apparent at the CTR, as hCT and sCT appear to induce unique receptor-transducer conformations. Interestingly, the hCT-occupied ternary complex was disrupted by GTP at ~10-fold lower GTP concentration than for sCT-occupied ternary complexes. Together, these findings demonstrate that ligand dependent G protein ternary complexes mediate GTP affinity by distinct changes in G protein conformation180. This suggests that, similar to the different signaling effects promoted by distinct βarrestin conformations, G proteins can adopt discrete active states with distinguishable signaling characteristics.
Chemokine receptors
Until recently, it was unclear whether biased agonism was a byproduct of GPCR complexity that can only be exploited by synthetic drugs or whether it is a property that has evolved within GPCR systems as an additional layer of signaling specificity. We now appreciate that some endogenous ligands are biased agonists. One group of GPCRs where endogenous bias is critical is the chemokine system, consisting of over 50 ligands and 20 chemokine receptors that bind one another with significant redundancy and promiscuity181,182. For example, the chemokine receptor CXCR3-A, which plays significant roles in inflammation, vascular disease, and cancer, has four known endogenous ligands: CXCL4, CXCL9, CXCL10, and CXCL11183–185. CXCL10 and CXCL11 signal through Gαi to inhibit cAMP generation with equivalent potency and efficacy that exceeds that of CXCL4 and CXCL957. In contrast, CXCL11 is more potent and efficacious in recruiting βarrestin to CXCR3-A than the other three endogenous ligands, and the rank order of efficacy for CXCR3-A internalization is the same as for βarrestin recruitment186. In addition, CXCL11 promotes CD4+ T cell polarization into FOXP3-negative regulatory T cells via STAT3 and STAT6 dependent pathways, while CXCL9 and CXCL10 promote CD4+ polarization into effector Th1/Th17 cells via STAT1, STAT4, and STAT5 dependent pathways187. βarrestin- and GRK-biased chemokines have also been described at other chemokine receptors, including CXCR2, CCR1, CCR5, and CCR765,186,188. Similar to many chemokines, the endogenous CXCR4 ligand CXCL12 (stromal cell-derived factor 1) can exist in monomeric or dimeric forms. Monomeric CXCL12 signals through both G protein and βarrestin pathways at CXCR4, while dimeric CXCL12 signals through G proteins with minimal to absent βarrestin recruitment and signaling189. Demonstrating further granularity of biased signaling, different chemokines can exhibit G protein subunit bias, as endogenous chemokines for CCR5 and CCR7 signal through overlapping but distinct G protein subtypes190. In addition to endogenous biased signaling, biased synthetic small molecules with affinity for chemokine receptors have also been identified191. Biasing chemokine receptor signaling may provide an avenue for drugging a GPCR family that has been notoriously difficult to therapeutically target.
Angiotensin receptor
The angiotensin-II (AngII) type 1 receptor (AT1R) signaling regulates multiple functions controlling blood pressure and serum osmolality, including vasoconstriction and aldosterone secretion. AT1R antagonists, including the sartan class of small molecules, are used in the treatment of hypertension, heart failure, and diabetic nephropathy. AngII binding to AT1R activates both Gαq and Gαi as well as βarrestin transducers. The first example of a βarrestin-biased ligand was SII angiotensin, a synthetically modified form of AngII that binds AT1R28. More strongly βarrestin biased AT1R agonists, such as the peptide TRV027, have recently been explored as drugs to treat decompensated heart failure. At AT1R, Gαq protein signaling mediates vasoconstriction and cardiac hypertrophy, whereas βarrestin signaling results in modest positive inotropy, desensitization of G protein signaling, receptor internalization, activation of antiapoptotic signals, and volume dependent enhancement of cardiac contractility192,193. In addition, AT1R mediated βarrestin1 signaling promotes increases in intracellular calcium concentration in primary chromaffin cells distinct from Gαq-PLCb mediated calcium flux by directly coupling to ion channels such as the transient receptor potential cation channel subfamily C3 (TRPC3)194. In animal models of heart failure, TRV027 and other AT1R βarrestin biased ligands are cardioprotective, and are thought to act by reducing afterload while increasing cardiac performance and maintaining stroke volume. Like other first in class drugs, development of biased compounds has been met with hurdles: TRV027 recently failed to meet established endpoints for decompensated heart failure in a phase IIb clinical trial192.
Adenosine receptors
Adenosine is a purine nucleoside with cardioprotective properties195. Four isotypes of the adenosine receptor have been identified, all of which are expressed in cardiac tissue, among other regions. The Adenosine A1 receptor couples primarily to Gαi/o, the A2aR primarily to Gas, the A2bR to both Gαs and Gαq, and A3R to Gαi/o196,197. It is currently unclear which receptor(s) mediate the cardioprotective effects of adenosine. On-target adenosine mediated side effects, including bradycardia, atrioventricular conduction blockade, and hypotension, limit the clinical utility of adenosine following cardiac ischemia. Therefore, a ligand that recapitulated adenosine’s cardioprotective effects without heart rate reduction would be superior to adenosine. A screen for biased adenosine orthosteric ligands at adenosine receptors was unsuccessful, however, a 2-amino-3-benzoylthiophene allosteric modulator was identified that promotes biased G protein and ERK signaling in the presence of an orthosteric agonist198. Using this allosteric modulator as a backbone, the rational design of a bitopic ligand for the adenosine receptor was undertaken, producing a compound VCP746 with a mixed pharmacological profile consisting of both competitive (at low concentrations of an antagonist) and noncompetitive (at high concentrations of an antagonist) activity. This signaling profile would be predicted for a bitopic ligand binding with high affinity to the orthosteric site and lower affinity to an allosteric site199. In proof of concept studies, VCP746 provided a cytoprotective benefit to rat cardiomyocytes, as well as stimulated anti-fibrotic signaling pathways in both cardiac and renal derived cell lines without concomitant reduction in rat heart rate197,200. Additional biased ligands at adenosine A3 receptor have been identified that promote differential activation of pAKT, pERK, Gαi, calcium influx, and cell survival201.
Other promising leads
Exploration of additional GPCR biased allosteric modulators has identified promising preclinical leads, including ML314, a neurotensin receptor βarrestin biased allosteric small molecule to treat methamphetamine abuse202,203. Peptide based biased ligands are also being developed, including the short lipidated peptide (known as a pepducin) ATI-2341, which targets CXCR4 and acts as a Gαi-biased allosteric ligand that mobilizes bone marrow polymorphonuclear neutrophils204. The serotonin receptor 5-HT2B was recently crystalized with the psychoactive small molecule lysergic acid diethylamide (LSD)205, providing structural insight into the βarrestin biased signaling properties of LSD and the chemically related ergolines at 5-HT2BR128. In addition, biased ligands have been identified at both the cannabinoid receptor 1 and 2 (CB1R, CB2R) receptors206, including the βarrestin/ERK biased allosteric modulator ORG27569 that stabilizes distinct conformations of the CB1R compared to traditional agonists and antagonists. This compound may provide antinociceptive and/or appetite stimulatory signals that are distinct from effects on psychosis or cognitive function207,208. Furthermore, βarrestin biased agonists at Free Fatty Acid Receptor 1 (GPR140) may provide useful in stimulating insulin secretion209. Further screens for biased ligands are likely to identify a trove of promising drug candidates at multiple receptors.
Conclusions
The paradigm of biased agonism, that different ligands can generate discrete receptor conformations that lead to distinct biological processes, is supported by numerous structure-function and pharmacological studies. These studies suggest that GPCRs act as allosteric microprocessors as opposed to binary ‘switches’ or ‘lock and keys.’ Basic and translational studies conducted within the last five years have led to the explosion of promising compounds with putative biased signaling, and demonstrate that the therapeutic potential for biased GPCR ligands is profound. The discovery of alternative GPCR signaling pathways, such as those mediated by βarrestin, warrants application of drug screening techniques beyond technologies that focus solely on proximal signaling responses mediated by G proteins. In addition, screening methods that are unable to identify allosteric ligands are likely to overlook potentially useful drugs. It is imperative to note the limitations of screening assays, especially when identifying potentially biased ligands. Bias plots, combined with quantification methods based on intrinsic relatively activity, functional affinity, and/or the operational model are all reasonable depending on the context and physiology of the system of interest. The available preclinical data suggest that selectively targeting G protein, βarrestin, or other non-canonical signaling pathways, depending on the physiological response desired, could improve current GPCR-based therapies through enhanced efficacy and reduced side effect profiles. The true therapeutic potential will not be realized until more biased ligands are tested in preclinical and clinical trials. Given the significant costs in late phase drug discovery, accurately quantifying the relative signaling properties of biased agonists early in the drug discovery process and their effects in suitable preclinical models of disease is necessary. Beyond their potential therapeutic superiority, biased ligands can also be utilized as tool compounds which, when combined with advances in signaling pathway analysis, be used to dissect fundamental biological processes. Such use of biased ligands as tools will help to advance our basic understanding of intracellular signaling.
Acknowledgments
The authors thank Justin Silverman for helpful discussion on error propagation and for design and implementation of the online biased calculator resource. We thank Thomas Pack, Dr. Marc Caron, and Dr. Charles Chavkin for comments on sections of the manuscript. This work is supported by NIH Grants T32GM7171 (J.S.S.); Duke Medical Scientist Training Program (J.S.S.); HL16037 (R.J.L.), HL114643 (S.R.); a Burroughs Wellcome Career Award for Medical Scientists (S.R.); R.J.L. is a HHMI Investigator.
Footnotes
Conflict of interest statement
R.J.L. is a cofounder and shareholder of Trevena.
References
- 1.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 2.Luttrell LM, Maudsley S, Bohn LM. Fulfilling the Promise of “Biased” G Protein-Coupled Receptor Agonism. Molecular pharmacology. 2015;88:579–588. doi: 10.1124/mol.115.099630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos R, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith JS, Rajagopal S. The beta-Arrestins: Multifunctional Regulators of G Protein-coupled Receptors. The Journal of biological chemistry. 2016;291:8969–8977. doi: 10.1074/jbc.R115.713313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benovic JL, Strasser RH, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: identification of a novel protein kinase that phosphorylates the agonist-occupied form of the receptor. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2797–2801. doi: 10.1073/pnas.83.9.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 7.Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Association of beta-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization. The Journal of biological chemistry. 1999;274:32248–32257. doi: 10.1074/jbc.274.45.32248. [DOI] [PubMed] [Google Scholar]
- 8.Goodman OB, Jr, et al. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 9.Laporte SA, Oakley RH, Holt JA, Barak LS, Caron MG. The interaction of beta-arrestin with the AP-2 adaptor is required for the clustering of beta 2-adrenergic receptor into clathrin-coated pits. The Journal of biological chemistry. 2000;275:23120–23126. doi: 10.1074/jbc.M002581200. References 7–9 are seminal papers describing the role beta-arrestin in GPCR endocytosis. [DOI] [PubMed] [Google Scholar]
- 10.Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 11.Gao H, et al. Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol Cell. 2004;14:303–317. doi: 10.1016/s1097-2765(04)00216-3. [DOI] [PubMed] [Google Scholar]
- 12.Shenoy SK, et al. beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta2 adrenergic receptor. The Journal of biological chemistry. 2006;281:1261–1273. doi: 10.1074/jbc.M506576200. [DOI] [PubMed] [Google Scholar]
- 13.Beaulieu JM, et al. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell. 2005;122:261–273. doi: 10.1016/j.cell.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Luttrell LM, et al. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. One of the first studies delinating signalling roles of the beta-arrestins. [DOI] [PubMed] [Google Scholar]
- 15.Ahn S, Kim J, Hara MR, Ren XR, Lefkowitz RJ. {beta}-Arrestin-2 Mediates Anti-apoptotic Signaling through Regulation of BAD Phosphorylation. The Journal of biological chemistry. 2009;284:8855–8865. doi: 10.1074/jbc.M808463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kendall RT, et al. Arrestin-dependent angiotensin AT1 receptor signaling regulates Akt and mTor-mediated protein synthesis. The Journal of biological chemistry. 2014;289:26155–26166. doi: 10.1074/jbc.M114.595728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichel K, Jullie D, von Zastrow M. beta-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat Cell Biol. 2016;18:303–310. doi: 10.1038/ncb3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irannejad R, et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature. 2013;495:534–538. doi: 10.1038/nature12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Werthmann RC, Volpe S, Lohse MJ, Calebiro D. Persistent cAMP signaling by internalized TSH receptors occurs in thyroid but not in HEK293 cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26:2043–2048. doi: 10.1096/fj.11-195248. [DOI] [PubMed] [Google Scholar]
- 20.Roth BL, Chuang DM. Multiple mechanisms of serotonergic signal transduction. Life Sci. 1987;41:1051–1064. doi: 10.1016/0024-3205(87)90621-7. One of the first papers to experimentally describe biased signalling. [DOI] [PubMed] [Google Scholar]
- 21.Luttrell LM. Minireview: More than just a hammer: ligand “bias” and pharmaceutical discovery. Mol Endocrinol. 2014;28:281–294. doi: 10.1210/me.2013-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarpe MB, et al. [D-Arg1,D-Phe5,D-Trp7,9,Leu11]Substance P acts as a biased agonist toward neuropeptide and chemokine receptors. The Journal of biological chemistry. 1998;273:3097–3104. doi: 10.1074/jbc.273.5.3097. [DOI] [PubMed] [Google Scholar]
- 23.Urban JD, et al. Functional selectivity and classical concepts of quantitative pharmacology. The Journal of pharmacology and experimental therapeutics. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 24.Violin JD, Lefkowitz RJ. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol Sci. 2007;28:416–422. doi: 10.1016/j.tips.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Spengler D, et al. Differential signal transduction by five splice variants of the PACAP receptor. Nature. 1993;365:170–175. doi: 10.1038/365170a0. [DOI] [PubMed] [Google Scholar]
- 26.Kenakin T. Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol Sci. 1995;16:232–238. doi: 10.1016/s0165-6147(00)89032-x. [DOI] [PubMed] [Google Scholar]
- 27.Gurwitz D, et al. Discrete activation of transduction pathways associated with acetylcholine m1 receptor by several muscarinic ligands. Eur J Pharmacol. 1994;267:21–31. doi: 10.1016/0922-4106(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 28.Wei H, et al. Independent beta-arrestin 2 and G protein-mediated pathways for angiotensin II activation of extracellular signal-regulated kinases 1 and 2. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10782–10787. doi: 10.1073/pnas.1834556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Violin JD, Crombie AL, Soergel DG, Lark MW. Biased ligands at G-protein-coupled receptors: promise and progress. Trends Pharmacol Sci. 2014;35:308–316. doi: 10.1016/j.tips.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Wisler JW, Xiao K, Thomsen AR, Lefkowitz RJ. Recent developments in biased agonism. Curr Opin Cell Biol. 2014;27:18–24. doi: 10.1016/j.ceb.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samama P, Cotecchia S, Costa T, Lefkowitz RJ. A mutation-induced activated state of the beta 2-adrenergic receptor. Extending the ternary complex model. The Journal of biological chemistry. 1993;268:4625–4636. [PubMed] [Google Scholar]
- 34.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 35.Kenakin TP. Biased signalling and allosteric machines: new vistas and challenges for drug discovery. British journal of pharmacology. 2012;165:1659–1669. doi: 10.1111/j.1476-5381.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. The Journal of biological chemistry. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 37.Gentry PR, Sexton PM, Christopoulos A. Novel Allosteric Modulators of G Protein-coupled Receptors. The Journal of biological chemistry. 2015;290:19478–19488. doi: 10.1074/jbc.R115.662759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onaran HO, Rajagopal S, Costa T. What is biased efficacy? Defining the relationship between intrinsic efficacy and free energy coupling. Trends Pharmacol Sci. 2014;35:639–647. doi: 10.1016/j.tips.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Kenakin T. Gaddum Memorial Lecture 2014: receptors as an evolving concept: from switches to biased microprocessors. British journal of pharmacology. 2015;172:4238–4253. doi: 10.1111/bph.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galandrin S, Oligny-Longpre G, Bouvier M. The evasive nature of drug efficacy: implications for drug discovery. Trends Pharmacol Sci. 2007;28:423–430. doi: 10.1016/j.tips.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Wardell SE, Marks JR, McDonnell DP. The turnover of estrogen receptor alpha by the selective estrogen receptor degrader (SERD) fulvestrant is a saturable process that is not required for antagonist efficacy. Biochemical pharmacology. 2011;82:122–130. doi: 10.1016/j.bcp.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norris JD, et al. Peptide antagonists of the human estrogen receptor. Science. 1999;285:744–746. doi: 10.1126/science.285.5428.744. [DOI] [PubMed] [Google Scholar]
- 43.Paige LA, et al. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heldring N, et al. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 45.Kovalenko M, et al. Phosphorylation site-specific inhibition of platelet-derived growth factor beta-receptor autophosphorylation by the receptor blocking tyrphostin AG1296. Biochemistry. 1997;36:6260–6269. doi: 10.1021/bi962553l. [DOI] [PubMed] [Google Scholar]
- 46.Girnita L, et al. Beta-arrestin and Mdm2 mediate IGF-1 receptor-stimulated ERK activation and cell cycle progression. The Journal of biological chemistry. 2007;282:11329–11338. doi: 10.1074/jbc.M611526200. [DOI] [PubMed] [Google Scholar]
- 47.Arey BJ, et al. Induction of promiscuous G protein coupling of the follicle-stimulating hormone (FSH) receptor: a novel mechanism for transducing pleiotropic actions of FSH isoforms. Mol Endocrinol. 1997;11:517–526. doi: 10.1210/mend.11.5.9928. [DOI] [PubMed] [Google Scholar]
- 48.Arey BJ, et al. Differing pharmacological activities of thiazolidinone analogs at the FSH receptor. Biochem Biophys Res Commun. 2008;368:723–728. doi: 10.1016/j.bbrc.2008.01.119. [DOI] [PubMed] [Google Scholar]
- 49.Yanofsky SD, et al. Allosteric activation of the follicle-stimulating hormone (FSH) receptor by selective, nonpeptide agonists. The Journal of biological chemistry. 2006;281:13226–13233. doi: 10.1074/jbc.M600601200. [DOI] [PubMed] [Google Scholar]
- 50.Kjelsberg MA, Cotecchia S, Ostrowski J, Caron MG, Lefkowitz RJ. Constitutive activation of the alpha 1B-adrenergic receptor by all amino acid substitutions at a single site. Evidence for a region which constrains receptor activation. The Journal of biological chemistry. 1992;267:1430–1433. [PubMed] [Google Scholar]
- 51.Chen X, Bai B, Tian Y, Du H, Chen J. Identification of serine 348 on the apelin receptor as a novel regulatory phosphorylation site in apelin-13-induced G protein-independent biased signaling. The Journal of biological chemistry. 2014;289:31173–31187. doi: 10.1074/jbc.M114.574020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wanka L, et al. C-terminal motif of human neuropeptide Y4 receptor determines internalization and arrestin recruitment. Cellular signalling. 2017;29:233–239. doi: 10.1016/j.cellsig.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajagopal S, et al. Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:628–632. doi: 10.1073/pnas.0912852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levoye A, Balabanian K, Baleux F, Bachelerie F, Lagane B. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 2009;113:6085–6093. doi: 10.1182/blood-2008-12-196618. [DOI] [PubMed] [Google Scholar]
- 55.Decaillot FM, et al. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. The Journal of biological chemistry. 2011;286:32188–32197. doi: 10.1074/jbc.M111.277038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith JS, et al. C-X-C Motif Chemokine Receptor 3 Splice Variants Differentially Activate Beta-Arrestins to Regulate Downstream Signaling Pathways. Molecular pharmacology. 2017;92:136–150. doi: 10.1124/mol.117.108522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berchiche YA, Sakmar TP. CXC Chemokine Receptor 3 Alternative Splice Variants Selectively Activate Different Signaling Pathways. Molecular pharmacology. 2016;90:483–495. doi: 10.1124/mol.116.105502. [DOI] [PubMed] [Google Scholar]
- 59.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu J, et al. A G Protein-biased Designer G Protein-coupled Receptor Useful for Studying the Physiological Relevance of Gq/11-dependent Signaling Pathways. The Journal of biological chemistry. 2016;291:7809–7820. doi: 10.1074/jbc.M115.702282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakajima K, Wess J. Design and functional characterization of a novel, arrestin-biased designer G protein-coupled receptor. Molecular pharmacology. 2012;82:575–582. doi: 10.1124/mol.112.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siuda ER, et al. Optodynamic simulation of beta-adrenergic receptor signalling. Nat Commun. 2015;6:8480. doi: 10.1038/ncomms9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spangler SM, Bruchas MR. Optogenetic approaches for dissecting neuromodulation and GPCR signaling in neural circuits. Current opinion in pharmacology. 2017;32:56–70. doi: 10.1016/j.coph.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reiter E, Lefkowitz RJ. GRKs and beta-arrestins: roles in receptor silencing, trafficking and signaling. Trends Endocrinol Metab. 2006;17:159–165. doi: 10.1016/j.tem.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 65.Zidar DA, Violin JD, Whalen EJ, Lefkowitz RJ. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9649–9654. doi: 10.1073/pnas.0904361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Riggs BL, Hartmann LC. Selective estrogen-receptor modulators -- mechanisms of action and application to clinical practice. The New England journal of medicine. 2003;348:618–629. doi: 10.1056/NEJMra022219. [DOI] [PubMed] [Google Scholar]
- 67.Urs NM, et al. Distinct cortical and striatal actions of a beta-arrestin-biased dopamine D2 receptor ligand reveal unique antipsychotic-like properties. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E8178–E8186. doi: 10.1073/pnas.1614347113. A physiological example of how systems bias can influence beta-arrestin signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schattauer SS, Kuhar JR, Song A, Chavkin C. Nalfurafine is a G-protein biased agonist having significantly greater bias at the human than rodent form of the kappa opioid receptor. Cellular signalling. 2017;32:59–65. doi: 10.1016/j.cellsig.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shenoy SK, Lefkowitz RJ. beta-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nobles KN, et al. Distinct phosphorylation sites on the beta(2)-adrenergic receptor establish a barcode that encodes differential functions of beta-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Busillo JM, et al. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. The Journal of biological chemistry. 2010;285:7805–7817. doi: 10.1074/jbc.M109.091173. One of the first studies mechanistically demonstrating how site-specific phosphorylation impacts GPCR signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poulin B, et al. The M3-muscarinic receptor regulates learning and memory in a receptor phosphorylation/arrestin-dependent manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9440–9445. doi: 10.1073/pnas.0914801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo J, Busillo JM, Stumm R, Benovic JL. G Protein-Coupled Receptor Kinase 3 and Protein Kinase C Phosphorylate the Distal C-Terminal Tail of the Chemokine Receptor CXCR4 and Mediate Recruitment of beta-Arrestin. Molecular pharmacology. 2017;91:554–566. doi: 10.1124/mol.116.106468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shukla AK, et al. Distinct conformational changes in beta-arrestin report biased agonism at seven-transmembrane receptors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9988–9993. doi: 10.1073/pnas.0804246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Charest PG, Terrillon S, Bouvier M. Monitoring agonist-promoted conformational changes of beta-arrestin in living cells by intramolecular BRET. EMBO Rep. 2005;6:334–340. doi: 10.1038/sj.embor.7400373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee MH, et al. The conformational signature of beta-arrestin2 predicts its trafficking and signalling functions. Nature. 2016;531:665–668. doi: 10.1038/nature17154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Namkung Y, et al. Monitoring G protein-coupled receptor and beta-arrestin trafficking in live cells using enhanced bystander BRET. Nat Commun. 2016;7:12178. doi: 10.1038/ncomms12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nuber S, et al. beta-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle. Nature. 2016;531:661–664. doi: 10.1038/nature17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hernandez PA, et al. Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet. 2003;34:70–74. doi: 10.1038/ng1149. [DOI] [PubMed] [Google Scholar]
- 80.Balabanian K, et al. WHIM syndromes with different genetic anomalies are accounted for by impaired CXCR4 desensitization to CXCL12. Blood. 2005;105:2449–2457. doi: 10.1182/blood-2004-06-2289. [DOI] [PubMed] [Google Scholar]
- 81.Barak LS, Oakley RH, Laporte SA, Caron MG. Constitutive arrestin-mediated desensitization of a human vasopressin receptor mutant associated with nephrogenic diabetes insipidus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:93–98. doi: 10.1073/pnas.011303698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wootten D, et al. The Extracellular Surface of the GLP-1 Receptor Is a Molecular Trigger for Biased Agonism. Cell. 2016;165:1632–1643. doi: 10.1016/j.cell.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wootten D, et al. Key interactions by conserved polar amino acids located at the transmembrane helical boundaries in Class B GPCRs modulate activation, effector specificity and biased signalling in the glucagon-like peptide-1 receptor. Biochemical pharmacology. 2016;118:68–87. doi: 10.1016/j.bcp.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wootten D, et al. A Hydrogen-Bonded Polar Network in the Core of the Glucagon-Like Peptide-1 Receptor Is a Fulcrum for Biased Agonism: Lessons from Class B Crystal Structures. Molecular pharmacology. 2016;89:335–347. doi: 10.1124/mol.115.101246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Venkatakrishnan AJ, et al. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature. 2016;536:484–487. doi: 10.1038/nature19107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang H, et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–844. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang H, et al. Structural Basis for Ligand Recognition and Functional Selectivity at Angiotensin Receptor. The Journal of biological chemistry. 2015;290:29127–29139. doi: 10.1074/jbc.M115.689000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 91.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang W, et al. Structural insights into micro-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng Y, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oswald C, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 100.Hollenstein K, et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 2013;499:438–443. doi: 10.1038/nature12357. [DOI] [PubMed] [Google Scholar]
- 101.Jazayeri A, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 102.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. The first GPCR structure solved in complex with a bona fide G protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang YL, et al. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017 doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H, et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burg JS, et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science. 2015;347:1113–1117. doi: 10.1126/science.aaa5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carpenter B, Nehme R, Warne T, Leslie AG, Tate CG. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 2016;536:104–107. doi: 10.1038/nature18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim YJ, et al. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–146. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- 109.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Szczepek M, et al. Crystal structure of a common GPCR-binding interface for G protein and arrestin. Nat Commun. 2014;5:4801. doi: 10.1038/ncomms5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Renaud JP, et al. Biophysics in drug discovery: impact, challenges and opportunities. Nat Rev Drug Discov. 2016;15:679–698. doi: 10.1038/nrd.2016.123. [DOI] [PubMed] [Google Scholar]
- 112.Westfield GH, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shukla AK, et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kumari P, et al. Functional competence of a partially engaged GPCR-beta-arrestin complex. Nat Commun. 2016;7:13416. doi: 10.1038/ncomms13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cahill TJ, 3rd, et al. Distinct conformations of GPCR-beta-arrestin complexes mediate desensitization, signaling, and endocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2017 doi: 10.1073/pnas.1701529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thomsen AR, et al. GPCR-G Protein-beta-Arrestin Super-Complex Mediates Sustained G Protein Signaling. Cell. 2016;166:907–919. doi: 10.1016/j.cell.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kahsai AW, et al. Multiple ligand-specific conformations of the beta2-adrenergic receptor. Nat Chem Biol. 2011;7:692–700. doi: 10.1038/nchembio.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Isogai S, et al. Backbone NMR reveals allosteric signal transduction networks in the beta1-adrenergic receptor. Nature. 2016;530:237–241. doi: 10.1038/nature16577. [DOI] [PubMed] [Google Scholar]
- 119.Perez-Aguilar JM, Shan J, LeVine MV, Khelashvili G, Weinstein H. A functional selectivity mechanism at the serotonin-2A GPCR involves ligand-dependent conformations of intracellular loop 2. J Am Chem Soc. 2014;136:16044–16054. doi: 10.1021/ja508394x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim IM, et al. Beta-blockers alprenolol and carvedilol stimulate beta-arrestin-mediated EGFR transactivation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14555–14560. doi: 10.1073/pnas.0804745105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim IM, et al. beta-arrestin1-biased beta1-adrenergic receptor signaling regulates microRNA processing. Circ Res. 2014;114:833–844. doi: 10.1161/CIRCRESAHA.114.302766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wisler JW, et al. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kahsai AW, et al. Conformationally selective RNA aptamers allosterically modulate the beta2-adrenoceptor. Nat Chem Biol. 2016;12:709–716. doi: 10.1038/nchembio.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Manglik A, et al. Structural Insights into the Dynamic Process of beta2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mary S, et al. Ligands and signaling proteins govern the conformational landscape explored by a G protein-coupled receptor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8304–8309. doi: 10.1073/pnas.1119881109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wacker D, et al. Structural features for functional selectivity at serotonin receptors. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yang F, et al. Phospho-selective mechanisms of arrestin conformations and functions revealed by unnatural amino acid incorporation and (19)F-NMR. Nat Commun. 2015;6:8202. doi: 10.1038/ncomms9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ariens EJ. Affinity and intrinsic activity in the theory of competitive inhibition. I. Problems and theory. Archives internationales de pharmacodynamie et de therapie. 1954;99:32–49. [PubMed] [Google Scholar]
- 131.Stephenson RP. A modification of receptor theory. Br J Pharmacol Chemother. 1956;11:379–393. doi: 10.1111/j.1476-5381.1956.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Furchgott RF. In: Advances in Drug Research. Harper NJ, Simmonds AB, editors. Vol. 3. Academic Press; 1966. pp. 21–55. [Google Scholar]
- 133.Black JW, Leff P. Operational models of pharmacological agonism. Proc R Soc Lond B Biol Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 134.Rajagopal S, et al. Quantifying Ligand Bias at Seven-Transmembrane Receptors. Mol Pharmacol. 2011;80:367–377. doi: 10.1124/mol.111.072801. mol.111.072801 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kenakin T, Watson C, Muniz-Medina V, Christopoulos A, Novick S. A simple method for quantifying functional selectivity and agonist bias. ACS Chem Neurosci. 2012;3:193–203. doi: 10.1021/cn200111m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Griffin MT, Figueroa KW, Liller S, Ehlert FJ. Estimation of agonist activity at G protein-coupled receptors: analysis of M2 muscarinic receptor signaling through Gi/o,Gs, and G15. J Pharmacol Exp Ther. 2007;321:1193–1207. doi: 10.1124/jpet.107.120857. jpet.107.120857 [pii] [DOI] [PubMed] [Google Scholar]
- 137.Snyder JC, Rochelle LK, Lyerly HK, Caron MG, Barak LS. Constitutive internalization of the leucine-rich G protein-coupled receptor-5 (LGR5) to the trans-Golgi network. The Journal of biological chemistry. 2013;288:10286–10297. doi: 10.1074/jbc.M112.447540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Onaran HO, et al. Systematic errors in detecting biased agonism: Analysis of current methods and development of a new model-free approach. Sci Rep. 2017;7:44247. doi: 10.1038/srep44247. A detailed comparison of different methods to quantify biased signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gregory KJ, Hall NE, Tobin AB, Sexton PM, Christopoulos A. Identification of orthosteric and allosteric site mutations in M2 muscarinic acetylcholine receptors that contribute to ligand-selective signaling bias. The Journal of biological chemistry. 2010;285:7459–7474. doi: 10.1074/jbc.M109.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Strachan RT, et al. Divergent Transducer-Specific Molecular Efficacies Generate Biased Agonism At A G Protein-Coupled Receptor (GPCR) J Biol Chem. 2014 doi: 10.1074/jbc.M114.548131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 142.Jones LH, Bunnage ME. Applications of chemogenomic library screening in drug discovery. Nat Rev Drug Discov. 2017;16:285–296. doi: 10.1038/nrd.2016.244. [DOI] [PubMed] [Google Scholar]
- 143.Horvath P, et al. Screening out irrelevant cell-based models of disease. Nat Rev Drug Discov. 2016;15:751–769. doi: 10.1038/nrd.2016.175. [DOI] [PubMed] [Google Scholar]
- 144.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 145.Whistler JL, Chuang HH, Chu P, Jan LY, von Zastrow M. Functional dissociation of mu opioid receptor signaling and endocytosis: implications for the biology of opiate tolerance and addiction. Neuron. 1999;23:737–746. doi: 10.1016/s0896-6273(01)80032-5. [DOI] [PubMed] [Google Scholar]
- 146.Zhang J, et al. Role for G protein-coupled receptor kinase in agonist-specific regulation of mu-opioid receptor responsiveness. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7157–7162. doi: 10.1073/pnas.95.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Melief EJ, Miyatake M, Bruchas MR, Chavkin C. Ligand-directed c-Jun N-terminal kinase activation disrupts opioid receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11608–11613. doi: 10.1073/pnas.1000751107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bohn LM, Gainetdinov RR, Lin FT, Lefkowitz RJ, Caron MG. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature. 2000;408:720–723. doi: 10.1038/35047086. [DOI] [PubMed] [Google Scholar]
- 149.Bohn LM, et al. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science. 1999;286:2495–2498. doi: 10.1126/science.286.5449.2495. One of the first examples of a βarrestin-mediated physiological response in an animal model. [DOI] [PubMed] [Google Scholar]
- 150.Groer CE, et al. An opioid agonist that does not induce mu-opioid receptor--arrestin interactions or receptor internalization. Molecular pharmacology. 2007;71:549–557. doi: 10.1124/mol.106.028258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soergel DG, et al. Biased agonism of the mu-opioid receptor by TRV130 increases analgesia and reduces on-target adverse effects versus morphine: A randomized, double-blind, placebo-controlled, crossover study in healthy volunteers. Pain. 2014;155:1829–1835. doi: 10.1016/j.pain.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 152.Viscusi ER, et al. A randomized, phase 2 study investigating TRV130, a biased ligand of the mu-opioid receptor, for the intravenous treatment of acute pain. Pain. 2016;157:264–272. doi: 10.1097/j.pain.0000000000000363. [DOI] [PubMed] [Google Scholar]
- 153.Manglik A, et al. Structure-based discovery of opioid analgesics with reduced side effects. Nature. 2016;537:185–190. doi: 10.1038/nature19112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Land BB, et al. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J Neurosci. 2008;28:407–414. doi: 10.1523/JNEUROSCI.4458-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.White KL, Roth BL. Psychotomimetic effects of kappa opioid receptor agonists. Biol Psychiatry. 2012;72:797–798. doi: 10.1016/j.biopsych.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 156.Bruchas MR, et al. Selective p38alpha MAPK deletion in serotonergic neurons produces stress resilience in models of depression and addiction. Neuron. 2011;71:498–511. doi: 10.1016/j.neuron.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Land BB, et al. Activation of the kappa opioid receptor in the dorsal raphe nucleus mediates the aversive effects of stress and reinstates drug seeking. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19168–19173. doi: 10.1073/pnas.0910705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ehrich JM, et al. Kappa Opioid Receptor-Induced Aversion Requires p38 MAPK Activation in VTA Dopamine Neurons. J Neurosci. 2015;35:12917–12931. doi: 10.1523/JNEUROSCI.2444-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Brust TF, et al. Biased agonists of the kappa opioid receptor suppress pain and itch without causing sedation or dysphoria. Sci Signal. 2016;9:ra117. doi: 10.1126/scisignal.aai8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.White KL, et al. The G protein-biased kappa-opioid receptor agonist RB-64 is analgesic with a unique spectrum of activities in vivo. The Journal of pharmacology and experimental therapeutics. 2015;352:98–109. doi: 10.1124/jpet.114.216820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maillet EL, et al. Noribogaine is a G-protein biased kappa-opioid receptor agonist. Neuropharmacology. 2015;99:675–688. doi: 10.1016/j.neuropharm.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 162.White KL, et al. Identification of novel functionally selective kappa-opioid receptor scaffolds. Molecular pharmacology. 2014;85:83–90. doi: 10.1124/mol.113.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rives ML, Rossillo M, Liu-Chen LY, Javitch JA. 6′-Guanidinonaltrindole (6′-GNTI) is a G protein-biased kappa-opioid receptor agonist that inhibits arrestin recruitment. The Journal of biological chemistry. 2012;287:27050–27054. doi: 10.1074/jbc.C112.387332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lovell KM, et al. Structure-activity relationship studies of functionally selective kappa opioid receptor agonists that modulate ERK 1/2 phosphorylation while preserving G protein over betaarrestin2 signaling bias. ACS Chem Neurosci. 2015;6:1411–1419. doi: 10.1021/acschemneuro.5b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou L, et al. Development of functionally selective, small molecule agonists at kappa opioid receptors. The Journal of biological chemistry. 2013;288:36703–36716. doi: 10.1074/jbc.M113.504381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Melief EJ, et al. Duration of action of a broad range of selective kappa-opioid receptor antagonists is positively correlated with c-Jun N-terminal kinase-1 activation. Molecular pharmacology. 2011;80:920–929. doi: 10.1124/mol.111.074195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Munro TA, et al. Long-acting kappa opioid antagonists nor-BNI, GNTI and JDTic: pharmacokinetics in mice and lipophilicity. BMC Pharmacol. 2012;12:5. doi: 10.1186/1471-2210-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carroll FI, Carlezon WA., Jr Development of kappa opioid receptor antagonists. J Med Chem. 2013;56:2178–2195. doi: 10.1021/jm301783x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chavkin C, Koob GF. Dynorphin, Dysphoria, and Dependence: the Stress of Addiction. Neuropsychopharmacology. 2016;41:373–374. doi: 10.1038/npp.2015.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Van’t Veer A, Carlezon WA., Jr Role of kappa-opioid receptors in stress and anxiety-related behavior. Psychopharmacology (Berl) 2013;229:435–452. doi: 10.1007/s00213-013-3195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III--the final common pathway. Schizophr Bull. 2009;35:549–562. doi: 10.1093/schbul/sbp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Free RB, et al. Discovery and characterization of a G protein-biased agonist that inhibits beta-arrestin recruitment to the D2 dopamine receptor. Molecular pharmacology. 2014;86:96–105. doi: 10.1124/mol.113.090563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shonberg J, et al. A structure-activity analysis of biased agonism at the dopamine D2 receptor. J Med Chem. 2013;56:9199–9221. doi: 10.1021/jm401318w. [DOI] [PubMed] [Google Scholar]
- 174.Chen X, et al. Structure-functional selectivity relationship studies of beta-arrestin-biased dopamine D(2) receptor agonists. J Med Chem. 2012;55:7141–7153. doi: 10.1021/jm300603y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Allen JA, et al. Discovery of beta-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18488–18493. doi: 10.1073/pnas.1104807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park SM, et al. Effects of beta-Arrestin-Biased Dopamine D2 Receptor Ligands on Schizophrenia-Like Behavior in Hypoglutamatergic Mice. Neuropsychopharmacology. 2016;41:704–715. doi: 10.1038/npp.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Urs NM, Peterson SM, Caron MG. New Concepts in Dopamine D2 Receptor Biased Signaling and Implications for Schizophrenia Therapy. Biol Psychiatry. 2016 doi: 10.1016/j.biopsych.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Sexton PM, Findlay DM, Martin TJ. Calcitonin. Curr Med Chem. 1999;6:1067–1093. [PubMed] [Google Scholar]
- 179.Andreassen KV, et al. Prolonged calcitonin receptor signaling by salmon, but not human calcitonin, reveals ligand bias. PloS one. 2014;9:e92042. doi: 10.1371/journal.pone.0092042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Furness SG, et al. Ligand-Dependent Modulation of G Protein Conformation Alters Drug Efficacy. Cell. 2016;167:739–749 e711. doi: 10.1016/j.cell.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 181.Bachelerie F, et al. International Union of Basic and Clinical Pharmacology. [corrected]. LXXXIX. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol Rev. 2014;66:1–79. doi: 10.1124/pr.113.007724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Steen A, Larsen O, Thiele S, Rosenkilde MM. Biased and g protein-independent signaling of chemokine receptors. Front Immunol. 2014;5:277. doi: 10.3389/fimmu.2014.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Muller M, Carter S, Hofer MJ, Campbell IL. Review: The chemokine receptor CXCR3 and its ligands CXCL9, CXCL10 and CXCL11 in neuroimmunity--a tale of conflict and conundrum. Neuropathol Appl Neurobiol. 2010;36:368–387. doi: 10.1111/j.1365-2990.2010.01089.x. [DOI] [PubMed] [Google Scholar]
- 184.Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunology and cell biology. 2011;89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Rashighi M, et al. CXCL10 is critical for the progression and maintenance of depigmentation in a mouse model of vitiligo. Sci Transl Med. 2014;6:223ra223. doi: 10.1126/scitranslmed.3007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rajagopal S, et al. Biased agonism as a mechanism for differential signaling by chemokine receptors. The Journal of biological chemistry. 2013;288:35039–35048. doi: 10.1074/jbc.M113.479113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zohar Y, et al. CXCL11-dependent induction of FOXP3-negative regulatory T cells suppresses autoimmune encephalomyelitis. The Journal of clinical investigation. 2014;124:2009–2022. doi: 10.1172/JCI71951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kohout TA, et al. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. The Journal of biological chemistry. 2004;279:23214–23222. doi: 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- 189.Drury LJ, et al. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17655–17660. doi: 10.1073/pnas.1101133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Corbisier J, Gales C, Huszagh A, Parmentier M, Springael JY. Biased signaling at chemokine receptors. The Journal of biological chemistry. 2015;290:9542–9554. doi: 10.1074/jbc.M114.596098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Corbisier J, Huszagh A, Gales C, Parmentier M, Springael JY. Partial agonist and biased signaling properties of the synthetic enantiomers J113863/UCB35625 at chemokine receptors CCR2 and CCR5. The Journal of biological chemistry. 2016 doi: 10.1074/jbc.M116.757559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Felker GM, et al. Heart failure therapeutics on the basis of a biased ligand of the angiotensin-2 type 1 receptor. Rationale and design of the BLAST-AHF study (Biased Ligand of the Angiotensin Receptor Study in Acute Heart Failure) JACC Heart Fail. 2015;3:193–201. doi: 10.1016/j.jchf.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 193.Abraham DM, et al. beta-Arrestin mediates the Frank-Starling mechanism of cardiac contractility. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:14426–14431. doi: 10.1073/pnas.1609308113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu CH, et al. Arrestin-biased AT1R agonism induces acute catecholamine secretion through TRPC3 coupling. Nat Commun. 2017;8:14335. doi: 10.1038/ncomms14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Donato M, Gelpi RJ. Adenosine and cardioprotection during reperfusion--an overview. Mol Cell Biochem. 2003;251:153–159. [PubMed] [Google Scholar]
- 196.Welihinda AA, Kaur M, Greene K, Zhai Y, Amento EP. The adenosine metabolite inosine is a functional agonist of the adenosine A2A receptor with a unique signaling bias. Cellular signalling. 2016;28:552–560. doi: 10.1016/j.cellsig.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vecchio EA, et al. The hybrid molecule, VCP746, is a potent adenosine A2B receptor agonist that stimulates anti-fibrotic signalling. Biochemical pharmacology. 2016;117:46–56. doi: 10.1016/j.bcp.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 198.Aurelio L, et al. Allosteric modulators of the adenosine A1 receptor: synthesis and pharmacological evaluation of 4-substituted 2-amino-3-benzoylthiophenes. J Med Chem. 2009;52:4543–4547. doi: 10.1021/jm9002582. [DOI] [PubMed] [Google Scholar]
- 199.Valant C, et al. Separation of on-target efficacy from adverse effects through rational design of a bitopic adenosine receptor agonist. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4614–4619. doi: 10.1073/pnas.1320962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chuo CH, et al. VCP746, a novel A1 adenosine receptor biased agonist, reduces hypertrophy in a rat neonatal cardiac myocyte model. Clin Exp Pharmacol Physiol. 2016;43:976–982. doi: 10.1111/1440-1681.12616. [DOI] [PubMed] [Google Scholar]
- 201.Baltos JA, et al. Structure-Activity Analysis of Biased Agonism at the Human Adenosine A3 Receptor. Molecular pharmacology. 2016;90:12–22. doi: 10.1124/mol.116.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Barak LS, et al. ML314: A Biased Neurotensin Receptor Ligand for Methamphetamine Abuse. ACS Chem Biol. 2016;11:1880–1890. doi: 10.1021/acschembio.6b00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peddibhotla S, et al. Discovery of ML314, a Brain Penetrant Non-Peptidic beta-Arrestin Biased Agonist of the Neurotensin NTR1 Receptor. ACS Med Chem Lett. 2013;4:846–851. doi: 10.1021/ml400176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Quoyer J, et al. Pepducin targeting the C-X-C chemokine receptor type 4 acts as a biased agonist favoring activation of the inhibitory G protein. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E5088–5097. doi: 10.1073/pnas.1312515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wacker D, et al. Crystal Structure of an LSD-Bound Human Serotonin Receptor. Cell. 2017;168:377–389 e312. doi: 10.1016/j.cell.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mallipeddi S, Janero DR, Zvonok N, Makriyannis A. Functional selectivity at G-protein coupled receptors: Advancing cannabinoid receptors as drug targets. Biochemical pharmacology. 2016 doi: 10.1016/j.bcp.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ahn KH, Mahmoud MM, Shim JY, Kendall DA. Distinct roles of beta-arrestin 1 and beta-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1) The Journal of biological chemistry. 2013;288:9790–9800. doi: 10.1074/jbc.M112.438804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Fay JF, Farrens DL. Structural dynamics and energetics underlying allosteric inactivation of the cannabinoid receptor CB1. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:8469–8474. doi: 10.1073/pnas.1500895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mancini AD, et al. beta-Arrestin Recruitment and Biased Agonism at Free Fatty Acid Receptor 1. The Journal of biological chemistry. 2015;290:21131–21140. doi: 10.1074/jbc.M115.644450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Molecular pharmacology. 2014;86:463–478. doi: 10.1124/mol.114.094342. [DOI] [PubMed] [Google Scholar]
- 211.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 212.Rajagopal S. Quantifying biased agonism: understanding the links between affinity and efficacy. Nat Rev Drug Discov. 2013;12:483. doi: 10.1038/nrd3954-c1. [DOI] [PubMed] [Google Scholar]
- 213.Kenakin T, Christopoulos A. Measurements of ligand bias and functional affinity. Nat Rev Drug Discov. 2013;12:483. doi: 10.1038/nrd3954-c2. [DOI] [PubMed] [Google Scholar]
- 214.Breton B, et al. Multiplexing of multicolor bioluminescence resonance energy transfer. Biophys J. 2010;99:4037–4046. doi: 10.1016/j.bpj.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kroeze WK, et al. PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat Struct Mol Biol. 2015;22:362–369. doi: 10.1038/nsmb.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Devost D, et al. Conformational Profiling of the AT1 Angiotensin II Receptor Reflects Biased Agonism, G Protein Coupling, and Cellular Context. The Journal of biological chemistry. 2017;292:5443–5456. doi: 10.1074/jbc.M116.763854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. The Journal of biological chemistry. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 218.Inoue A, et al. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods. 2012;9:1021–1029. doi: 10.1038/nmeth.2172. [DOI] [PubMed] [Google Scholar]