Abstract
Iron metabolism is critical for sustaining life and maintaining human health. Here, we find that iron homeostasis is linked to facultative heterochromatin assembly and regulation of gene expression during adaptive genome control. We show that the fission yeast Clr4/Suv39h histone methyltransferase is part of a rheostat-like mechanism, in which transcriptional up-regulation of mRNAs in response to environmental change provides feedback to prevent their uncontrolled expression through heterochromatin assembly. Interestingly, proper iron homeostasis is required, as depletion of iron or down-regulation of iron transporters caused defects in heterochromatin assembly and unrestrained upregulation of gene expression. Remarkably, an unbiased genetic screen revealed that restoration of iron homeostasis is sufficient to re-establish facultative heterochromatin and proper gene control genome-wide. These results establish a role for iron homeostasis in facultative heterochromatin assembly and reveal a dynamic mechanism for reprogramming the genome in response to environmental changes.
The genome of eukaryotic cells can be reprogrammed to allow adaptation to changes in environmental growth conditions or to respond to developmental signals1–4. Reprogramming requires intricate epigenetic mechanisms that orchestrate dynamic changes in gene expression patterns through factors involved in the assembly of “open” or “closed” chromatin domains, referred to as euchromatin or heterochromatin, respectively5–8. Moreover, posttranscriptional mechanisms that regulate RNA stability/degradation also control gene expression9–12. However, how transcriptional and posttranscriptional factors are coordinated to establish genome-wide patterns of gene expression critical for growth under diverse conditions is poorly understood.
The fission yeast Schizosaccharomyces pombe is an excellent model system to explore adaptive control of the genome. Chromatin modifying factors involved in the regulation of gene expression in higher eukaryotes, such as histone modifying activities involved in the assembly of specialized chromatin domains, are highly conserved in fission yeast5. Clr4, a member of the mammalian SUV39 histone methyltransferase family6, is the sole enzyme that methylates histone H3 lysine 9 (H3K9me) to assemble constitutive heterochromatin domains as well as discrete facultative heterochromatin islands13,14. Additionally, histone deacetylases (HDACs) deacetylate histones to assemble repressive chromatin15. Among the HDACs, Clr6 acts broadly across the genome and exists in at least two distinct protein complexes: complex-I, which contains the essential Sin3 homolog Pst1 and targets gene promoters; and complex-II, which contains the non-essential Sin3 homolog Pst2 and targets open reading frames (ORFs)16.
Gene expression is also regulated by RNA processing/degradation factors, which are also conserved from S. pombe to mammals9. A network of nuclear RNA elimination factors, including a protein complex composed of the Mtr4-like RNA helicase Mtl1 and the zinc-finger protein Red1 (MTREC, Mtl1-Red1 core) prevents the untimely expression of genes12,17–19. MTREC, which is functionally related to mammalian PAXT20, and its associated proteins promote RNA degradation by the 3’→5’ exosome exonuclease (Rrp6)17,19. Additionally, the RNAi machinery including Argonaute (Ago1), Dicer (Dcr1) and the RNA-dependent RNA polymerase (Rdp1), as well as the CCR4-NOT complex that plays diverse roles in RNA metabolism from synthesis to decay21, target various mRNAs for degradation17,18,22–27 (see also Supplementary Fig. 1).
In addition to RNA degradation, RNA processing factors mediate RNA-dependent targeting of heterochromatin5,14,17,22,25,26,28–30. The RNA interference (RNAi) machinery targets facultative heterochromatin assembly at regulated genes17,26,29,31, in addition to constitutive heterochromatin assembly at centromeric repeats27,32. In another pathway, MTREC associated proteins recruit Clr4 to assemble facultative heterochromatin islands at meiotic genes14,17,25,33. These facultative heterochromatin domains can be modulated in response to nutritional conditions14,33. However, whether these pathways directly regulate the fluctuating pool of transcripts as part of cellular adaptation to environmental changes remains to be examined.
In this study, we explore the roles of heterochromatin and RNA processing factors in regulating gene expression patterns at low temperature. We find that transcriptional up-regulation of mRNAs in response to an environmental change provides negative feedback to prevent their uncontrolled expression through a buffering mechanism involving heterochromatin factor Clr4. Surprisingly, we find that iron homeostasis is critical for facultative heterochromatin assembly and regulation of gene expression at low temperature. We discuss the implications of these findings for understanding the cellular strategies employed to fine-tune gene expression, and show that Clr4 limits RNA polymerase II (RNAPII) transcription at target genes to optimize their expression.
Results
Cells grown at low temperature show widespread changes in gene expression
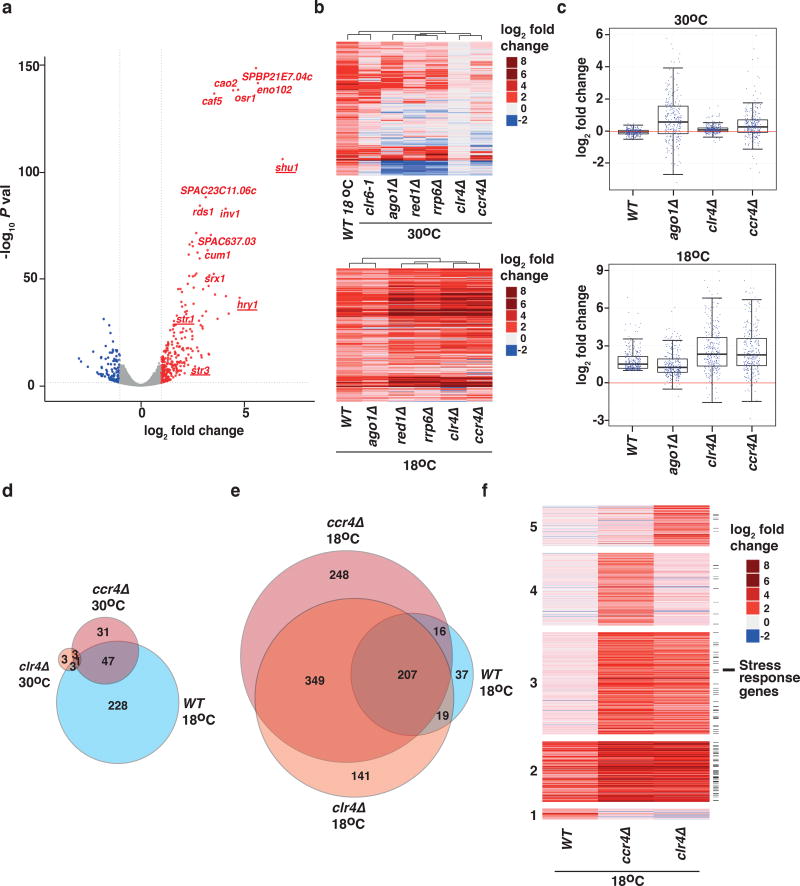
To explore mechanisms of gene control in response to varying growth conditions, we first asked how the transcriptome of S. pombe cells is affected by growth at a suboptimal low temperature (18°C). We analyzed the transcriptome of wild-type cells cultured at 18°C using RNA-sequencing analysis (RNA-seq) and compared our results to those obtained from cells grown under standard laboratory conditions (30°C). We observed widespread transcriptome changes in cells grown at low temperature, including 279 transcripts that were up-regulated and 129 transcripts that were down-regulated at least two-fold when compared to cells grown at 30°C (Fig. 1a). We found that among the loci with increased expression at 18°C, the most highly up-regulated transcripts included stress response genes34, non-coding RNAs, and genes encoding transmembrane transporters (Supplementary Fig. 2). Notably, we found that genes encoding iron transporters were significantly up-regulated at 18°C (Fig. 1a). Thus, cells exposed to low temperature undergo an adaptive response that involves significant changes at the transcript level.
Figure 1. Transcriptome analyses of cells grown at 18°C.
(a) Volcano plot showing statistical significance (−log10 P value) versus fold change (log2 fold change) of RNA-seq data from WT (wild-type) cells grown at 30°C or 18°C (n=2 independent experiments per condition). Genes with increased expression (fold change value ≥ 1 and P value <0.05) are shown in red and genes with decreased expression (fold change value ≤ 1 and P value <0.05) are shown in blue. Iron transporter genes are underlined. (b) Heat map of fold change values relative to WT 30°C cells for the 279 transcripts up-regulated in WT 18°C cells, compared to mutant cells grown at 30°C (upper) or 18°C (lower). Data for clr6-1 temperature-sensitive mutant is shown for 30°C only. (c) Box plots representing fold-change values for the indicated strains (top, 30 °C; bottom, 18 °C) relative to WT cells at 30 °C for the 279 transcripts upregulated in WT cells at 18 °C. Whiskers for box plots are drawn at the 12.5 and 87.5 percentiles. Top and bottom edges of the box approximate the third and first quantiles, respectively, and the central line is drawn at the median. (d, e) Area proportional Venn diagrams representing the numbers of genes with increased expression in WT 18°C cells compared with ccr4Δ and clr4Δ cells grown at 30°C (d) or 18°C (e). (f) Heat map of fold change values for up-regulated transcripts relative to WT 30°C cells in the indicated strains at 18°C. Clusters are grouped according to transcripts up-regulated in WT only (1); WT, ccr4Δ, and clr4Δ (2); ccr4Δ and clr4Δ (3); ccr4Δ only (4); and clr4Δ only (5). Black lines on the right indicate stress response genes. Source data for Fig. 1a–f are available with the paper online.
Transcripts upregulated at 18°C are normally repressed by Clr6
To understand the mechanisms that control the expression of transcripts up-regulated at low temperature in wild-type cells, we first tested whether the loss of heterochromatin or RNA degradation factors affects levels of these RNAs at 30°C. Specifically, we analyzed the transcriptomes of a panel of mutant strains grown at 30°C including cells lacking Clr4, RNAi factor Ago1, CCR4-NOT subunit Ccr4, MTREC component Red1, or nuclear exosome subunit Rrp6, as well as a strain expressing a temperature-sensitive mutant allele of Clr6 HDAC. Interestingly, expression profiling revealed the highest correlation between transcripts up-regulated in wild-type cells grown at 18°C and those in the clr6 HDAC mutant grown at 30°C (Fig. 1b and Supplementary Fig. 3a), suggesting that the loci that are up-regulated at low temperature are repressed by Clr6 under normal growth conditions.
Notably, we found iron transporter genes among the loci repressed by Clr6 (Supplementary Fig. 3b). To address whether Clr6 directly targets iron transporter genes to control their expression we performed chromatin immunoprecipitation coupled to microarray analysis (ChIP-chip) of Clr6 complex subunits. Indeed, we found Clr6 and Pst1 localized to iron transporter genes (Supplementary Fig. 3c), suggesting that Clr6 complex plays a direct role in silencing these genes.
We noted that the loss of Ago1, Red1, or Rrp6 impacted a subset of up-regulated transcripts in wild-type cells grown at 18°C (Fig. 1b). These mutants formed a distinct cluster based on their correlation coefficients (Fig. 1b), which is consistent with our previous findings that RNAi, MTREC, and the exosome share targets across the genome26. Surprisingly, we found that loss of Clr4 or Ccr4 in cells cultured at 30°C had no major impact on gene expression, suggesting that these factors are largely dispensable, at least under standard growth conditions, for the regulation of transcripts that are up-regulated in wild-type cells grown at 18°C (Fig. 1b–d).
Cells lacking Clr4 or Ccr4 show hyper-elevation of transcripts up-regulated at 18°C
We further tested whether loss of heterochromatin or RNA degradation factors affects transcript levels in cells grown at low temperature. Interestingly, loss of Ago1 caused only minimal changes in the levels of transcripts up-regulated in wild-type cells grown at 18°C (Fig. 1b,c), suggesting that RNAi is dispensable for their regulation at low temperature. By contrast, transcripts up-regulated in wild-type cells grown at 18°C were further de-repressed in the absence of the RNA elimination factors Red1 or Rrp6 (Fig. 1b).
Surprisingly, we found that despite Clr4 and Ccr4 having little effect in regulating transcript levels under standard laboratory growth conditions (Fig. 1b–d), they play a critical role in limiting them at 18°C (Fig. 1b,c,e,f). Notably, Clr4 and Ccr4 affected a common set of targets, including transcripts up-regulated in wild-type cells cultured at low temperature, which were further de-repressed in cells lacking Clr4 or Ccr4 (Fig. 1b,c,e,f). In addition, many other coding and non-coding RNAs were de-repressed in the absence of Ccr4 or Clr4 (Fig. 1e,f). Strikingly, we noted that a large fraction of loci that are up-regulated in clr4Δ cells at 18°C are normally repressed by Clr6 HDAC under standard growth conditions (Supplementary Fig. 3a), indicating a global shift in the gene expression control strategy used as cells to respond to a temperature change. These results suggest that loss of Clr4 or Ccr4 disables an important buffering mechanism that normally prevents unrestrained gene expression during adaptation to low temperature growth.
Loss of Clr4 affects meiotic gene silencing at low temperature
Clr4 methylates H3K9 at several meiotic genes that are maintained in a silent state during vegetative growth14. However, deletion of clr4 alone has little or no effect on meiotic gene expression under standard growth conditions (Supplementary Fig. 4a)14. Given the change in gene control strategy that occurs at 18°C, we wondered whether Clr4 becomes required for the repression of meiotic genes under suboptimal growth conditions. Indeed, cells lacking Clr4 exhibited untimely expression of several meiotic genes during vegetative growth at 18°C (Supplementary Fig. 4a), suggesting a critical role for this conserved factor in preventing inappropriate gene expression. Notably, clr4Δ cells displayed a slow growth phenotype at 18°C compared to wild-type cells (Supplementary Fig. 4b).
Cells cultured at low temperature form new heterochromatin islands
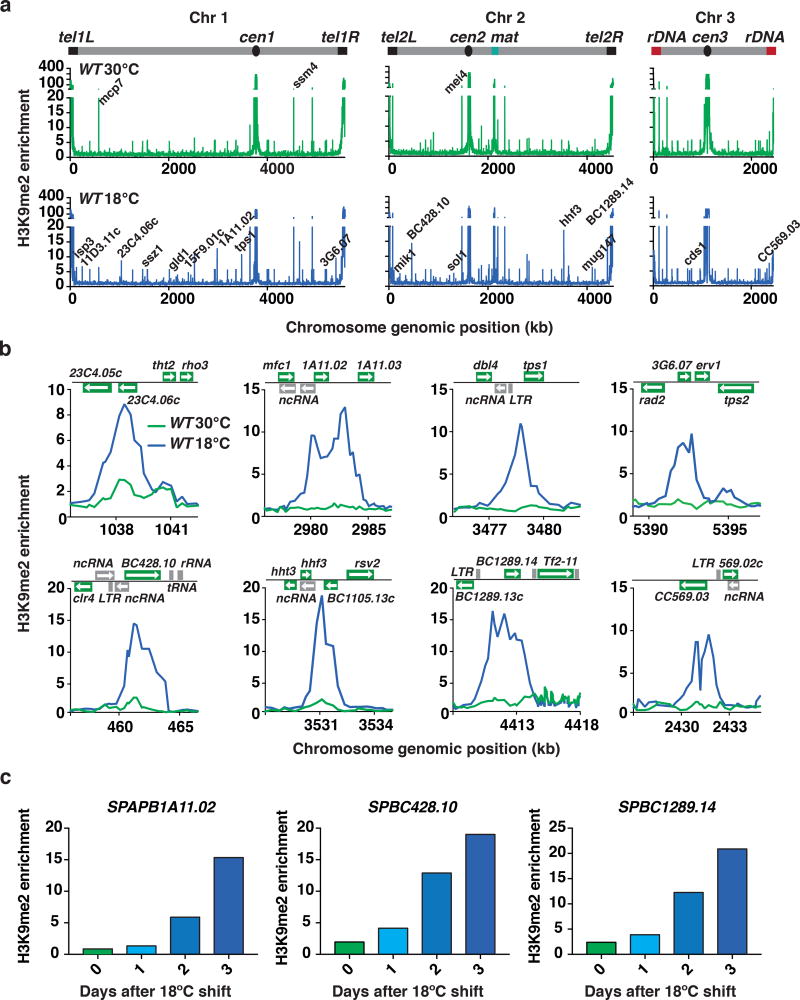
Our finding that loss of Clr4 causes further up-regulation of various transcripts at low temperature suggested that cells grown at 18°C might exhibit a unique heterochromatin profile and display additional facultative heterochromatin islands, besides the previously reported facultative heterochromatin domains14,26. To explore potential changes in heterochromatin distribution across the genome, we performed genome-wide analyses of H3K9me in wild-type cells grown at 18°C. Remarkably, we identified 36 novel facultative heterochromatin islands (Fig. 2a,b). Our quantitative ChIP-PCR (ChIP-qPCR) time course analyses revealed a gradual increase in H3K9me at selected new facultative heterochromatin islands following the shift to low temperature (Fig. 2c). Importantly, the new heterochromatin islands mapped preferentially to stress responsive genes (Supplementary Fig. 2), and are coincident with loci that show up-regulation of transcripts at 18°C (Fig. 2a–c). These transcripts were also further up-regulated in clr4Δ cells grown at 18°C, which was also confirmed in our quantitative RT-PCR (RT-qPCR) analysis of SPAPB1A11.02 (Fig. 3a). Together, these findings suggest that Clr4 is involved in the assembly of new facultative heterochromatin islands at 18°C and regulates the expression of environmentally-controlled transcripts.
Figure 2. New facultative heterochromatin domains form at low temperature.
(a) Genome-wide H3K9me relative fold enrichment in WT cells grown at 30°C or 18°C was determined by chromatin immunoprecipitation followed by microarray hybridization (ChIP-chip). Representative heterochromatin islands are indicated on the 30°C plot and new 18°C facultative heterochromatin peaks are indicated on the 18°C plot. (b) H3K9me relative enrichment at individual loci in WT cells grown at 30°C or 18°C, as determined by ChIP-chip. (c) H3K9me enrichments in WT cells grown at 30°C or 18°C for 1, 2, or 3 days were determined by ChIP-qPCR. H3K9me fold enrichments at SPAPB1A11.02, SPBC428.10, and SPBC1289.14 are shown relative to the control leu1 gene. Data presented is from a representative experiment performed at least twice. Source data for Fig. 2a are available with the paper online.
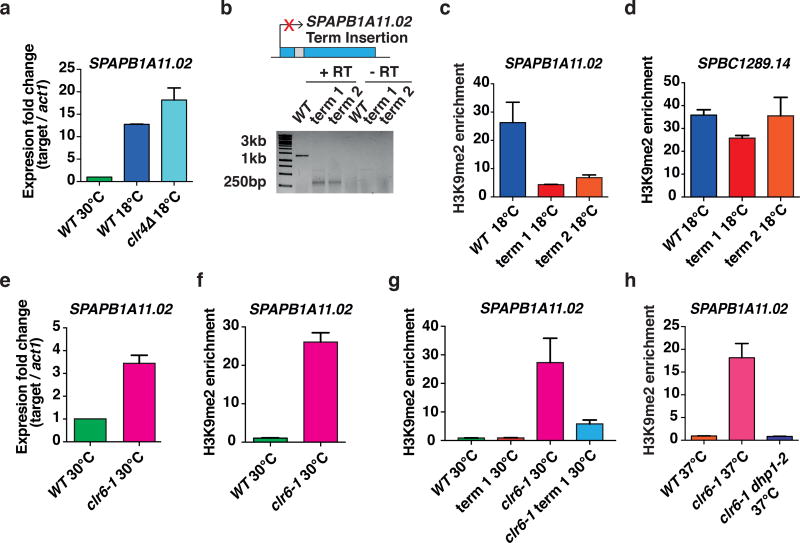
Figure 3. Facultative heterochromatin forms by an RNA-based mechanism at 18°C.
(a) Expression levels of SPAPB1A11.02 relative to act1 were determined by RT-qPCR and are shown as the mean +s.d. (n=2 independent experiments). Expression levels are normalized to WT cells grown at 30°C. (b) Schematic of the ura4 terminator (Term) sequence inserted into SPAPB1A11.02. Term 1 and Term 2 refer to two independent strains containing the ura4 terminator sequence in the SPAPB1A11.02 ORF. The intron is shown as a grey box (upper). Cells were grown at 18°C and termination of SPAPB1A11.02 was determined using 3’RACE (lower). The uncropped image is shown in Supplementary Data Set 1. (c, d) ChIP-qPCR analysis of H3K9me enrichments at 18°C in WT and two strains containing the ura4 terminator sequence in the SPAPB1A11.02 gene. H3K9me fold enrichments at SPAPB1A11.02 (c) or SPBC1289.14 (d) relative to the control leu1 gene are shown as the mean +s.d. (n=3 independent experiments). (e) Expression levels of SPAPB1A11.02 relative to act1 in WT and clr6-1 cells grown at 30°C were determined by performing strand-specific RT-qPCR and are shown as the mean +s.d. (n=2 independent experiments). Expression levels are normalized to WT cells. (f) ChIP-qPCR analysis of H3K9me enrichments at 30°C in WT and clr6-1 cells. H3K9me fold enrichments at SPAPB1A11.02 relative to the control leu1 gene are shown as the mean +s.d. (n=3 independent experiments). (g) ChIP-qPCR analysis of H3K9me enrichments at 30°C in WT, WT containing the ura4 terminator sequence in the SPAPB1A11.02 gene, clr6-1, and clr6-1 containing the ura4 terminator sequence in the SPAPB1A11.02 gene. H3K9me fold enrichments at SPAPB1A11.02 relative to the control leu1 gene are shown as the mean +s.d. (n=3 independent experiments). (h) ChIP-qPCR analysis of H3K9me enrichments relative to the control leu1 gene at SPAPB1A11.02 is shown for the indicated strains. Enrichments are shown as the mean +s.d. (n=3 independent experiments).
An RNA-based mechanism forms new heterochromatin islands
We were intrigued by the finding that new H3K9me peaks form at loci showing up-regulation of mRNAs at 18°C, and that these mRNAs are further de-repressed upon loss of Clr4. Taken together, these results suggested that mRNAs might provide feedback to regulate their own expression through recruitment of Clr4. We tested this possibility by inserting the ura4 terminator sequence into the SPAPB1A11.02 ORF to impair mRNA production, which we confirmed using 3’RACE (Fig. 3b). Disruption of SPAPB1A11.02 mRNA production resulted in defects in heterochromatin assembly specifically at this locus (Fig. 3c), and did not affect H3K9me enrichment at another locus, SPBC1289.14, at 18°C (Fig. 3d). These results suggest that gene transcripts up-regulated at low temperature are likely part of a self-regulatory feedback mechanism that recruits Clr4 in cis to buffer against their uncontrolled expression.
To further confirm this self-regulatory feedback mechanism, we examined whether an increase in SPAPB1A11.02 mRNA could trigger heterochromatin formation under standard growth conditions. We tested this using a clr6 mutant that showed an increase in SPAPB1A11.02 mRNA at 30°C, similar to wild-type cells cultured at 18°C (Fig. 3e). Remarkably, we found that an increase in SPAPB1A11.02 mRNA was linked to an increase in H3K9me enrichment in clr6 mutant cells grown at 30°C (Fig. 3e,f). Moreover, disruption of transcription by the ura4 terminator sequence in the SPAPB1A11.02 ORF caused a strong reduction in H3K9me levels in clr6 mutant cells (Fig. 3g), thus further supporting our model in which mRNAs generated from the target genes themselves recruit Clr4 to modulate their expression.
RNA elimination and termination factors but not RNAi are required for new H3K9me islands at 18°C
Transcription and RNA-mediated assembly of heterochromatin involves RNA processing factors. Whereas RNAi machinery is required for establishing H3K9me at repetitive DNA elements and certain regulated genes26,31, RNAi-independent mechanisms involving MTREC and its associated factors assemble facultative heterochromatin at meiotic genes14,17,25. Our ChIP-chip and ChIP analyses showed that loss of the RNAi component Ago1 did not affect H3K9me peaks that are detected in cells grown at 18°C (Supplementary Fig. 5a,b), indicating that RNAi is dispensable for the assembly of these heterochromatin islands.
We next tested the potential involvement of RNA elimination factors, including MTREC and CCR4-NOT, in heterochromatin island assembly. H3K9me peaks detected at 18°C did not form in the absence of Ccr4 (Supplementary Fig. 5a,c) (see also below). Moreover, cells lacking Red1 were defective in the assembly of these heterochromatin islands (Supplementary Fig. 5c). These data are consistent with our transcriptome analyses showing that RNA elimination factors, but not RNAi machinery, are required for preventing inappropriate gene expression at low temperature (Fig. 1b,c).
RNA elimination factors cooperate with the conserved exoribonuclease Dhp1 (Xrn2 in higher eukaryotes) that, in addition to promoting transcription termination, recruits Clr4 to assemble heterochromatin35,36. We tested whether the formation of heterochromatin islands detected at 18°C requires Dhp1. Since dhp1 is an essential gene, we used a temperature-sensitive dhp1 mutant allele in a clr6 mutant background, and assayed H3K9me enrichment at the restrictive temperature (37°C). Unlike the single clr6 mutant that showed significant H3K9me enrichment at the SPAPB1A11.02 locus, no enrichment could be detected in the clr6 dhp1 double-mutant (Fig. 3h), suggesting that Dhp1 is required for heterochromatin assembly.
Genetic screen connects iron homeostasis to cellular adaptation
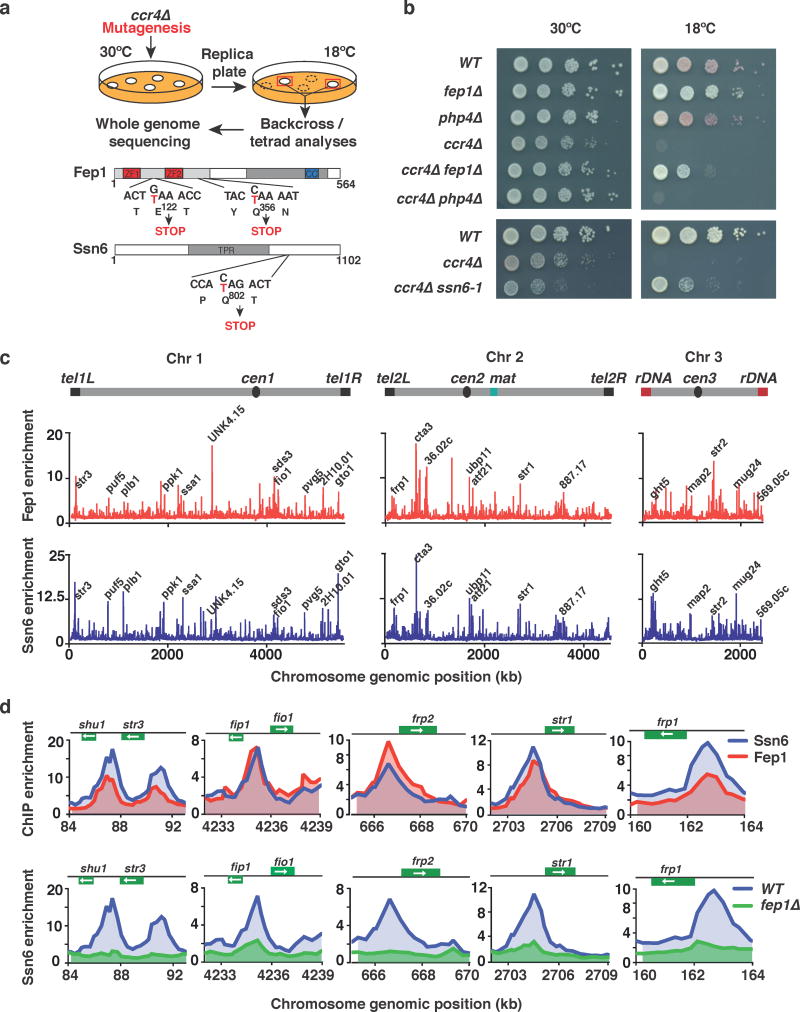
Among the factors affecting facultative heterochromatin at 18°C, ccr4Δ cells showed the most severe cold-sensitivity. We hypothesized that the cold-sensitivity of ccr4Δ cells might be related to misregulated gene expression. To explore this, we performed a genetic screen for suppressors of the ccr4Δ cold-sensitive phenotype (see Fig. 4a and Online Methods), and isolated three suppressor mutants. Whole genome sequencing analyses revealed that two suppressors contained mutations in a gene encoding the iron-sensing transcription factor fep1, which represses iron transporter genes37–40. Both mutations introduced premature stop codons into the fep1 ORF (Fig. 4a). Supporting these results, we found that the deletion of fep1 mimicked the suppression of the ccr4Δ cold-sensitivity that we observed with the mutant alleles (Fig. 4b). This suppression was specific to fep1, as deletion of php4, which is involved in the repression of genes encoding iron-using proteins37, did not suppress the growth defect of ccr4Δ cells at 18°C (Fig. 4b).
Figure 4. Mutation of fep1 or ssn6 restores growth in ccr4Δ cells at 18°C.
(a) Genetic screen for suppressors of the cold-sensitive growth phenotype of ccr4Δ cells. Cells were mutagenized by UV irradiation and suppressors were isolated. Mutants were backcrossed to WT, and the phenotype was tracked in ccr4Δ cells by assaying growth at 18°C. Whole genome sequencing and nucleotide variant analysis identified two independent suppressor mutations in fep1 (fep1-1 E122→STOP and fep1-2 (Q365→STOP), as well as a suppressor mutation in ssn6 (ssn6-1 Q802→STOP). (b) Serial dilutions of the indicated strains were spotted onto rich media (YEA) and incubated at 30°C for 3 days, or 18°C for 11 days. (c) ChIP-chip analysis of genome-wide Fep1-GFP (upper) or Ssn6-Myc (lower) relative enrichment in cells grown at 30°C. Iron transporter loci and other peaks are indicated. (d) ChIP-chip analysis of Fep1-GFP and Ssn6-Myc relative enrichment (upper) or Ssn6-Myc relative enrichment at iron transporter genes in WT or fep1Δ cells (lower) grown at 30°C.
Interestingly, the third suppressor mutation mapped to the gene encoding Ssn6 (Fig. 4a), a component of the conserved Ssn6-Tup11/12 transcriptional corepressor that acts together with HDACs41,42. Tup11 physically associates with Fep143. Loss of Tup11 or Tup12 causes up-regulation of iron transporter genes38, which we found are also repressed by the Clr6 HDAC (Supplementary Fig. 3b). Our genome-wide ChIP-chip analysis revealed widespread co-localization of Fep1 and Ssn6, including at the promoters of genes involved in various physiological processes (Fig. 4c,d). Moreover, their localization coincided with the Pst1-containing Clr6 complex-I at several gene promoters (Supplementary Fig. 6a). We also observed preferential enrichment of these proteins at the promoters of iron transporter genes (Fig. 4d and Supplementary Fig. 6b). Collectively, these results suggest an important connection between factors that regulate iron homeostasis and growth at 18°C.
The remarkable identification of two distinct factors involved in iron homeostasis as suppressors of the ccr4Δ cold-sensitive phenotype prompted us to test iron uptake in ccr4Δ cells, and to explore the connection between Fep1 and Ssn6 in controlling this process. Strikingly, iron transporter genes were down-regulated in ccr4Δ cells at 18°C, as determined by our RNA-seq and RT-qPCR analyses (Fig. 5a). However, loss of Fep1, which abolished the localization of the Ssn6 corepressor at iron transporter gene promoters (Fig. 4d), fully restored the expression of these loci in ccr4Δ cells (Fig. 5a, Supplementary Fig. 6c). Consistently, we found that intracellular iron levels were lower in ccr4Δ cells relative to wild-type cells cultured at 18°C and that loss Fep1 in these cells restored intracellular iron (Supplementary Fig. 6d). Together with our results showing that iron transporters are up-regulated in wild-type cells grown at 18°C (Fig. 1a), these findings suggest that proper control of iron homeostasis is crucial for cellular adaptation to growth at low temperature.
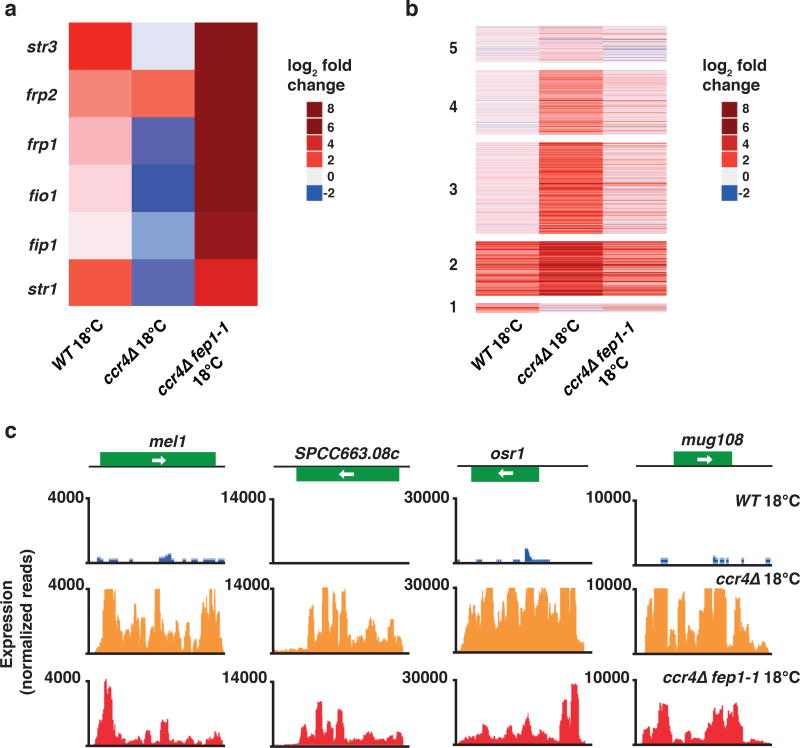
Figure 5. Mutation of fep1 restores gene expression in ccr4Δ cells grown at 18°C.
(a) Heat map of fold change values relative to WT 30°C cells for six representative iron transporter genes in the indicated strains grown at 18°C. (b) Heat map of fold change values relative to WT 30°C cells for up-regulated transcripts in the indicated strains at 18°C. Clusters are grouped according to the analysis presented in Fig. 1f. (c) RNA-seq analysis of the expression levels of four representative genes up-regulated in ccr4Δ cells at 18°C are shown for WT, ccr4Δ, and ccr4Δ fep1-1 cells. Source data for Fig. 5a,b are available with the paper online.
Iron homeostasis affects facultative heterochromatin and global gene control
We next investigated whether the suppression of cold-sensitivity is coupled to restoration of proper global gene expression in ccr4Δ fep1-1 double-mutant cells, as compared to the ccr4Δ single-mutant. Our expression profile comparisons revealed widespread normalization of transcript levels in ccr4Δ fep1-1 cells (Fig. 5b,c). A particularly striking finding was that the expression levels of a wide array of transcripts that were up-regulated in ccr4Δ cells compared to wild-type cells cultured at 18°C were restored to near wild-type levels in the ccr4Δ fep1-1 double-mutants (Fig. 5b,c). This remarkably broad restoring effect on gene expression included not only the highly up-regulated transcripts in wild-type cells grown at 18°C, but also other transcripts that were de-repressed genome-wide in ccr4Δ and clr4Δ cells (Fig. 5b).
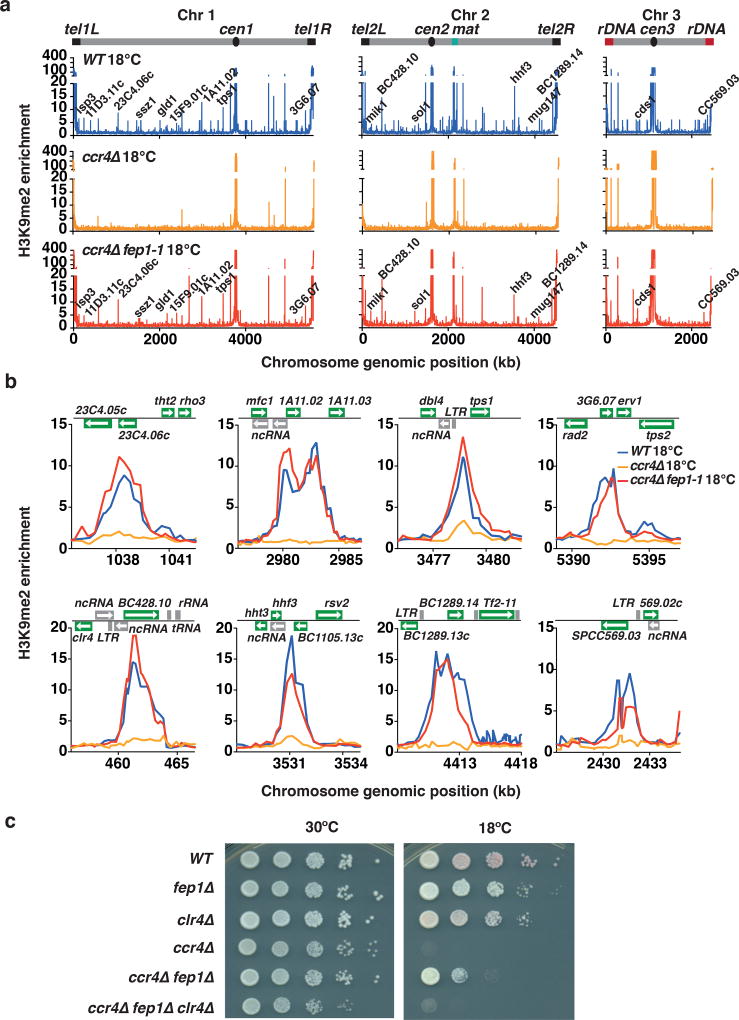
We wondered whether restoration of iron homeostasis upon loss of Fep1 in ccr4Δ cells could also restore facultative heterochromatin assembly. Remarkably, our ChIP-chip analyses showed that H3K9me peaks lost in ccr4Δ cells were restored in ccr4Δ fep1-1 cells to levels comparable to those in wild-type cells cultured at 18°C (Fig. 6a,b). Similarly, a mutation in ssn6 restored H3K9me at loci showing defects in facultative heterochromatin assembly in ccr4Δ cells at 18°C (Supplementary Fig. 6e).
Figure 6. Mutation of fep1 restores heterochromatin assembly in ccr4Δ cells grown at 18°C.
(a) ChIP-chip analysis of genome-wide H3K9me relative enrichment in WT, ccr4Δ, and ccr4Δ fep1-1 cells grown at 18°C. New 18°C facultative heterochromatin peaks are indicated. WT ChIP-chip data is also presented in Fig. 2a. (b) H3K9me relative enrichment at individual loci in WT, ccr4Δ, and ccr4Δ fep1-1 cells grown at 18°C. WT ChIP-chip data is also presented in Fig. 2b. (c) Serial dilutions of the indicated strains were spotted onto rich media (YEA) and incubated at 30°C for 3 days, or at 18°C for 11 days.
Given the restoration of facultative heterochromatin in the ccr4 fep1 double-mutant, we explored whether Clr4 is required by the factors that restore iron homeostasis and suppress the cold-sensitivity of ccr4Δ cells. Indeed, we found that ccr4Δ fep1Δ clr4Δ cells failed to grow at 18°C (Fig. 6c), suggesting that Clr4 is indeed functionally important. Collectively, our analyses of cells lacking Ccr4, which show down-regulation of iron transporter genes, reveal an intimate connection between iron homeostasis and proper gene control via a mechanism involving Clr4, RNA processing factors, and facultative heterochromatin assembly.
Iron depletion disrupts facultative heterochromatin assembly and gene regulation
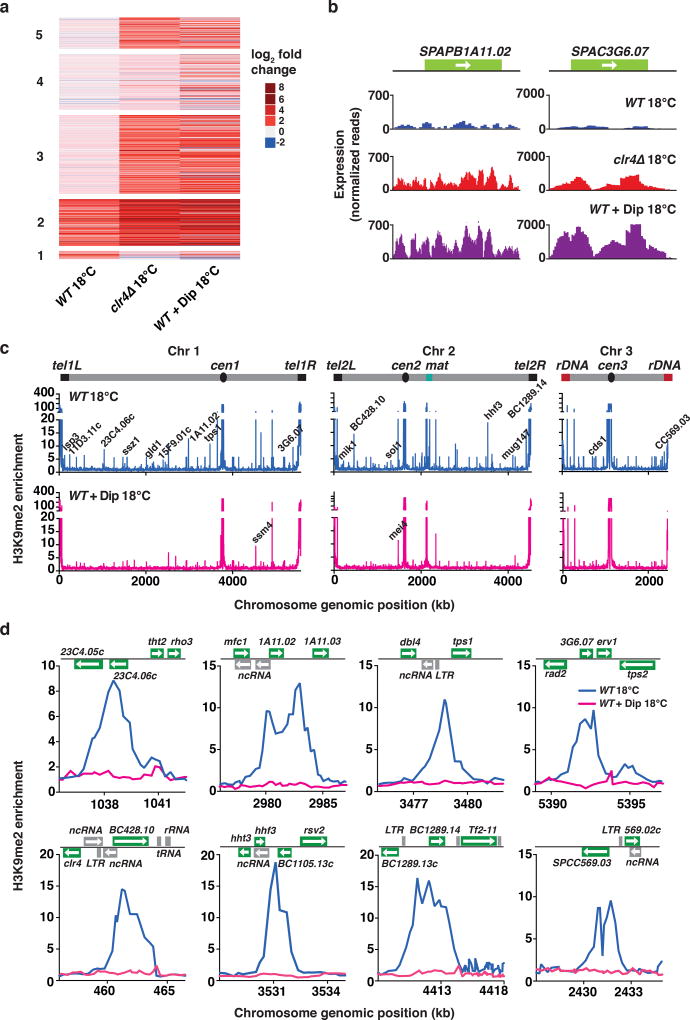
Our genetic analyses suggested that iron homeostasis is critical for regulating gene expression and facultative heterochromatin formation at low temperature. To directly explore the role of iron in this process, we depleted intracellular iron using the membrane-permeable iron chelating agent 2,2’-bipyridyl (Dip). Wild-type cells cultured at 18°C in the presence of Dip showed up-regulation of transcripts that are regulated by Clr4 at low temperature (Fig. 7a,b). In addition to further de-repression of transcripts that are up-regulated in wild-type cells grown at 18°C, iron chelation in wild-type cells affected additional gene clusters that were de-repressed in clr4Δ cells grown at 18°C (Fig. 7a). Transcripts that were up-regulated in iron-depleted cells were enriched in stress response and catabolic genes, as well as ncRNAs, as we observed in clr4Δ cells cultured at 18°C. We obtained similar results when ccr4Δ fep1-1 double-mutant cells were treated with Dip (Supplementary Fig. 7a), suggesting that the normalization of gene expression observed in ccr4Δ fep1-1 requires iron, and therefore is linked to restoration of iron homeostasis.
Figure 7. Depletion of iron from growth medium abolishes heterochromatin assembly at low temperature.
(a) Heat map of fold change values for up-regulated transcripts relative to WT 30°C cells in the indicated strains grown at 18°C. Clusters are grouped according to the analysis presented in Fig. 1f. (b) RNA-seq analysis of the expression levels of two representative heterochromatic loci are shown for the indicated strains grown at 18°C. (c) ChIP-chip analysis of genome-wide H3K9me relative enrichment in WT cells at 18°C, either untreated or treated with 250µM Dip. New 18°C facultative heterochromatin peaks are indicated on the WT 18°C plot. Note that H3K9me at constitutive heterochromatin domains (cen, mat and tel) and meiotic islands (ssm4 and mei4) is largely maintained in Dip-treated cells. WT (untreated) ChIP-chip data is also presented in Fig. 2a. (d) H3K9me relative enrichment at individual loci in WT cells, either untreated or treated with 250µM Dip, at 18°C. WT (untreated) ChIP-chip data is also presented in Fig. 2b. Source data for Fig. 7a are available with the paper online.
We next investigated the effect of iron depletion on facultative heterochromatin assembly. We found that Dip treatment abolished H3K9me peaks corresponding to heterochromatin islands in wild-type cells cultured at 18°C (Fig. 7c,d and Supplementary Fig. 7b). Similar results were observed when ccr4Δ fep1-1 cells were treated with Dip (Supplementary Fig. 7c). Importantly, Dip had no major impact on H3K9me levels at constitutive heterochromatic loci including centromeres and telomeres (Fig. 7c), suggesting that iron depletion selectively affected the assembly of facultative heterochromatin islands at 18°C. Collectively, our findings underscore the importance of iron homeostasis in RNA-mediated targeting of Clr4 to assemble facultative heterochromatin and define gene expression patterns in response to environmental change.
Loss of Clr4 enhances RNAPII transcription of target loci at 18°C
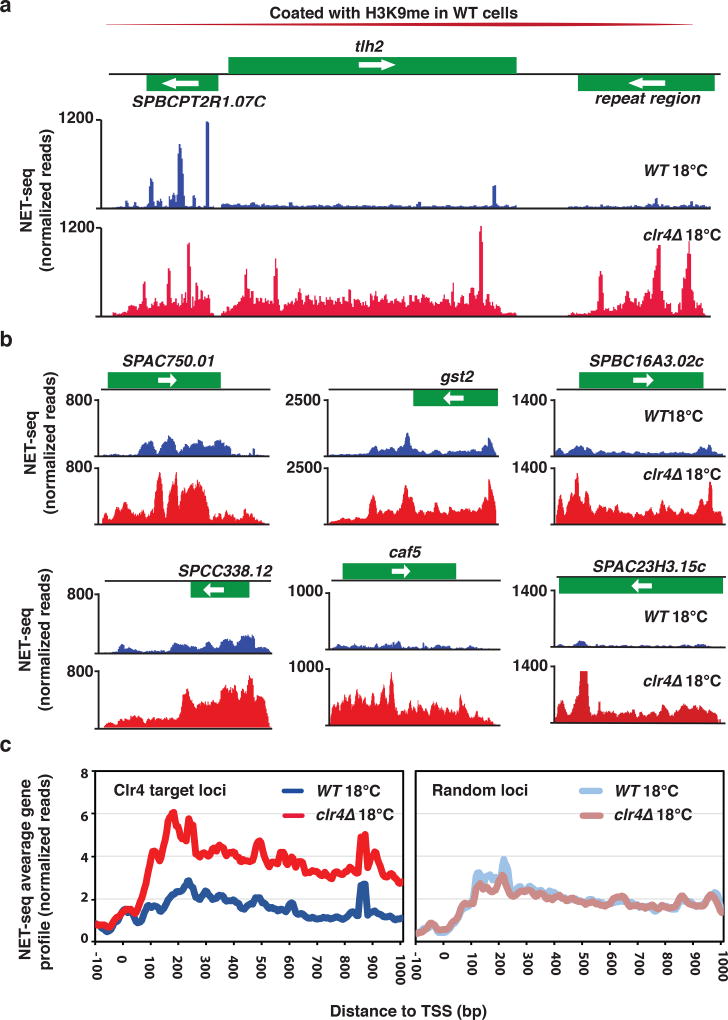
We next addressed how Clr4 could affect transcript levels at low temperature. At constitutive heterochromatin regions, Clr4 affects expression at both transcriptional as well as posttranscriptional levels44. Since the assembly of heterochromatin islands at 18°C involves RNA-mediated targeting of Clr4 (Fig. 3), we postulated that the observed changes in transcript levels in clr4Δ cells might be linked to defects in RNA degradation. However, it was also possible that Clr4 affects RNAPII transcription. To test the latter, we performed native elongating transcript sequencing (NET-seq)45 to assay RNAPII transcription in wild-type and clr4Δ cells grown at 18°C. We found that clr4Δ cells showed an increase in RNAPII transcription at constitutive heterochromatin domains, supporting our previous ChIP analyses46. We also observed an increase in RNAPII transcription at telomere-linked helicase (tlh) genes and several other loci located within subtelomeric domains (Fig. 8a), which are coated with H3K9me in wild-type cells47. However, a surprising finding was that clr4Δ cells cultured at low temperature showed a marked increase in RNAPII transcription specifically at genes that showed elevated transcript levels at 18°C upon loss of Clr4 (Fig. 8b). At some locations, clr4Δ cells showed an increase in RNAPII transcription at genes proximal to the heterochromatin islands detected in wild-type cells (e.g. SPAC23H3.15)(Fig. 8b).
Figure 8. NET-seq reveals enhanced RNAPII transcription in clr4Δ cells.
(a) Active RNAPII distributions at subtelomeric transcripts at the tlh2 locus are shown for WT and clr4Δ cells grown at 18°C. (b) Active RNAPII distributions at six representative loci up-regulated in RNA-seq analysis of clr4Δ 18°C cells are shown for WT and clr4Δ cells grown at 18°C. (c) Average active RNAPII distributions across gene bodies at Clr4 target loci (left) and other loci (right) is plotted for WT and clr4Δ cells grown at 18°C (see Online Methods).
To address the specific effect on RNAPII transcription at loci showing hyper-elevation (>5-fold) of transcripts in clr4Δ cells grown at 18°C, we next systematically compared the NET-seq profiles of wild-type and clr4Δ cells. Remarkably, the NET-seq average gene profile clearly showed higher levels of RNAPII transcription at Clr4 target loci in clr4Δ cells as compared to wild-type cells cultured at 18°C (Fig. 8c). This difference was not observed at other loci which show no detectable changes in expression in clr4Δ cells cultured at 18°C (Fig. 8c). Thus, in addition to constitutive heterochromatic loci, Clr4 affects RNAPII transcription at specific genes. In particular, Clr4 limits RNAPII activity at loci that are up-regulated in cells cultured at low temperature.
Discussion
In this study, we provide evidence that heterochromatin factor Clr4/Suv39h as well as nuclear RNA processing and termination factors are components of a dynamically regulated mechanism that controls gene expression during growth at low temperature. Furthermore, we show that iron homeostasis is critical for facultative heterochromatin assembly and the normalization of up-regulated gene expression that occurs during adaptation to this suboptimal condition.
Our analyses suggest that a rheostat-like mechanism prevents up-regulated transcripts from reaching hyper-elevated levels. Many genes that are repressed by Clr6 HDAC under standard growth conditions are controlled by a Clr4-based mechanism that buffers and fine-tunes their expression at low temperature. Specifically, we find that transcriptional up-regulation of the mRNAs provides feedback and limits gene expression through recruitment of Clr4. Consistently, transcripts that are up-regulated in wild-type cells are considerably further elevated in cells lacking Clr4. Moreover, we show that Clr4 inhibits RNAPII transcription at its target loci.
How cells recruit Clr4 to suppress RNAPII activity remains a key question. Clr4-mediated suppression of RNAPII activity may involve RNAi machinery implicated in transcription-coupled heterochromatin formation. However, we find that RNAi is dispensable for both facultative heterochromatin assembly and gene regulation at 18°C. Instead, this process requires RNA elimination machinery and Dhp1/Xrn2, a RNAPII termination factor linked to heterochromatin modifications35,36,48. Clr4 might recruit silencing effectors by methylating H3K9, as it does at constitutive heterochromatic loci44. Nevertheless, we note that Clr4 exists in a cullin 4 (Cul4)-containing complex49–51, which contains E3 ubiquitin ligase activity that might also contribute to limiting RNAPII transcription, particularly at loci showing little or no H3K9me. Regardless of the mechanism, these findings will help to elucidate the functions of heterochromatin factors at transcribed genes in higher eukaryotes52–54. Interestingly, the localization of heterochromatin factors at heat-shock puffs in Drososphila is contemporaneous with the wave of increased expression53, and in mammals H3K9me is detected in the coding regions of activated genes54. Whether, or even how, heterochromatin machinery regulates gene activity at these loci is unclear. In light of this study, these observations are remarkably consistent with the view that the transcription-coupled recruitment of heterochromatin factors buffers against uncontrolled gene expression in response to developmental and environmental signals.
Another surprising finding from our current study was that iron is required for adaptive gene control at low temperature. Notably, cells depleted of iron, as well as ccr4Δ cells that are defective in iron homeostasis due to down-regulation of iron transporter genes, show defects in facultative heterochromatin assembly and hyper-elevation of Clr4 target genes. Suppressor mutations that restore iron homeostasis in ccr4Δ cells re-establish facultative heterochromatin and proper gene control genome-wide, supporting a critical role for iron in these adaptive processes. In particular, mutations in the iron-sensing factor fep1 de-repress iron transporters silenced by Clr6 HDAC and the Ssn6-Tup11/12 corepressor (this study)41,43, which in turn restores intracellular iron levels required for adaptive genome control. Interestingly, wild-type cells cultured at 18°C show elevated transcription of iron transporters that correlates with the formation of facultative heterochromatin islands across the genome. It is likely that intracellular iron concentrations are carefully regulated as part of adaptive gene control in response to changes in growth conditions.
How does iron regulate facultative heterochromatin assembly and gene expression? We can speculate that iron may affect chromatin modifying activities55. However, given that iron depletion has no impact on constitutive heterochromatin formation, it is unlikely that iron affects Clr4 activity, but rather iron may promote specific RNA-dependent targeting of H3K9me by RNA processing/termination factors. Supporting this, we find that iron depletion causes the accumulation of aberrantly spliced RNAs, which are normally degraded by RNA processing factors (Supplementary Fig. 8a). Furthermore, our hierarchical clustering analysis of expression profiles showed that iron-depleted cells have the highest correlation with mtl1 mutant cells (Supplementary Fig. 8b,c), which are defective in RNA processing and facultative heterochromatin assembly17. Thus, we envision that factors that connect RNA processing to facultative heterochromatin assembly require iron for their function (Supplementary Fig. 9). Indeed, RNA modifying enzymes containing iron-sulfur clusters and radical SAM domains catalyze a variety of RNA modifications that affect RNA stability or function56,57. Determining whether Clr4 recruitment may be facilitated through iron-dependent RNA modification activities will be an important topic for future studies. The conservation of RNA processing and heterochromatin pathways from S. pombe to mammals underscores the importance of these findings for understanding epigenetic reprogramming of higher eukaryotic genomes, and for discovering the molecular underpinnings of human diseases linked to iron deficiencies58,59.
Methods
Strains and media
Yeast cell culture and genetic manipulations were performed using standard methods60. S. pombe strains used in this study are listed in Supplementary Table 1. Yeast extract rich medium supplemented with adenine (YEA) was used to grow cultures. Cells cultured at 18°C were grown to mid-log phase (OD600 = 0.4–0.8) at 30°C and shifted to 18°C for 3 days. Cultures were maintained in early- to mid-log phase (OD600 = 0.1–0.8) during 18°C growth. Cells cultured at 37°C were first grown at 26°C and shifted to 37°C for 5 hours. For iron chelation experiments, cultured cells were treated with 250µM 2,2’-bipyridyl (Dip) (Sigma) immediately before the shift to 18°C.
RNA analyses
RNA-sequencing libraries were constructed as previously described17. Briefly, total RNA was prepared using the MasterPure Yeast RNA Purification Kit (Epicentre) according to the manufacturer’s instructions. rRNA was removed using the Ribo-Zero Gold rRNA Removal Magnetic Kit (Yeast) (Epicentre) prior to library construction using the ScriptSeq v2 RNA-seq Library Preparation Kit (Epicentre). Sequencing was performed on the Illumina MiSeq platform. Adapter trimmed reads were aligned to the S. pombe ASM294v2.30 reference genome61 using Tophat262. Minimum and maximum intron sizes were set to 40 and 900, respectively. Differential expression was analyzed using the Bioconductor DESeq2 R library63–65. Significantly increased or decreased transcript levels were defined as those with log2 fold change values ≥1 or ≤1, respectively, and a P value of <0.05 after a Benjamini and Hochberg adjustment for multiple tests66. Heatmaps were drawn using the ComplexHeatmap R library67, while other plots were drawn using the ggplot268, R library, and standard R plotting functions. Gene categories used in the analysis were derived from a combination of gene descriptions from Pombase, GO Slim terms (http://go.princeton.edu/) compiled for S. pombe using the Web-based GO slimming tool, Kegg pathway annotations, and the list of core environmental stress response genes identified by Chen and colleagues34. Hierarchical clustering of the samples for the plots was performed using the Ward.D2 method as implemented in the standard R 'hclust' function. Scatterplots of gene expression between pairs of samples were generated using R from the log2FC of all detected transcripts. Best fit lines were generated using the standard linear model (lm) R function. Novel introns were identified by comparing the alignment coordinates of split-reads (discontiguous alignments for single reads) found in the RNA-seq data. Split-read alignment coordinates were compared with the coordinates of canonical introns annotated on the Pombase v2.29 genome and those implying splice-sites differing on both ends of the splice by more than 5 bp from canonical introns were designated as novel. RNA-seq data for ago1Δ 30°C, dcr1Δ 30°C, erh1Δ 30°C, mmi1Δ 30°C, red1Δ 30°C, mtl1-1 30°C, cwf10-1 26°C, rrp6Δ 30°C and ccr4Δ 30°C were published previously17,23,25.
ChIP and ChIP-chip
ChIP and ChIP-chip experiments were performed as previously described17,47. In ChIP experiments performed at 18°C, cells were fixed at 18°C with 3% paraformaldehyde for 30 minutes. In ChIP experiments performed at 30°C or 37°C, cells were fixed at room temperature with 3% paraformaldehyde for 30 minutes. Anti-H3K9me2 (ab115159, Abcam), anti-HA (12CA5, Roche), anti-GFP (ab290, Abcam) or anti-Myc (9E10, Santa Cruz) antibodies were used for immunoprecipitation. Antibodies were recovered using a Protein A: Protein G (1:1) bead slurry (Invitrogen). Immunoprecipitated DNA and input DNA were analyzed by performing ChIP-chip analysis or quantitative PCR (qPCR) using iTaq Universal SYBR® Green Supermix (Biorad). In ChIP-chip experiments, immunoprecipitated DNA and input DNA were labeled with Cy5/Cy3 for microarray analyses using a custom 4×44K oligonucleotide array (Agilent). Oligonucleotides used for ChIP-qPCR are listed in Supplementary Table 1.
RT-PCR
Total RNA was prepared using the MasterPure Yeast RNA Purification Kit (Epicentre) according to the manufacturer’s instructions. Gene-specific cDNA was synthesized with random hexamers (Thermo Scientific) and strand-specific cDNA was synthesized with custom DNA oligos using SuperScript III Reverse Transcriptase (Invitrogen). Expression was analyzed by performing quantitative PCR (qPCR) using iTaq Universal SYBR® Green Supermix (Biorad). Oligonucleotides used for cDNA synthesis and RT-qPCR are listed in Supplementary Table 1.
3′ RACE
Total RNA was prepared using the MasterPure Yeast RNA Purification Kit (Epicentre) according to the manufacturer’s instructions. cDNA was synthesized using the 3′-RACE CDS Primer A oligonucleotide using SuperScript III Reverse Transcriptase (Invitrogen). cDNA was then amplified by touchdown PCR with a gene-specific oligonucleotide and the universal primer mix using the Advantage 2 Polymerase mix (Clontech). Gene-specific oligonucleotides are listed in Supplementary Table 1.
Genetic screen
Exponentially growing cells were either plated to identify spontaneous suppressors of the ccr4Δ cold sensitive phenotype or mutagenized with UV (150J/m2) using a UV cross-linker (Thermo Scientific). Colonies were screened for growth on rich media (YEA) at 18°C. Mutants were isolated and backcrossed to a non-mutagenized strain. Genomic DNA isolated from three cold-sensitive (control) and three mutant segregants were subjected to whole genome sequencing. DNA-sequencing libraries were constructed using the Nextera XT Kit (Illumina) and sequenced on the NextSeq 500 platform to an average depth of 40–50×. Mutations were identified as follows: adapter trimmed reads were aligned to the S. pombe ASM294v2.30 reference genome using the BWA69 short read aligner with default parameters. Duplicate reads were marked in the resulting BAM files using picard-tools (http://broadinstitute.github.io/picard/) MarkDuplicates. Mutations were called from the duplicate-marked BAMS using samtools70 mpileup followed by bcftools71 to produce a single VCF file containing mutations called in each of the six genomes. Mutations found in the mutant segregants but not in any of the controls were identified from the VCF file using an in-house Perl script (https://www.perl.org/). Mutation impact was assessed using SnpEff72.
Iron measurement
Iron-55 labeling of S. pombe cells was performed as previously described73 with some modifications. Yeast cells were inoculated in 50ml of YEA rich medium in 125ml flasks and grown overnight in an orbital platform shaker. The cultures were diluted to give an OD600 of 0.2 in 100ml of fresh YEA in 250ml flasks, and grown for 48 hours at 18°C. Cells were transferred to 50ml Falcon tubes and spun down at 2000×g for 5 minutes. Cell pellets were washed with water and resuspended in synthetic iron-free medium at 0.5g of cell pellet per 10ml of medium supplemented with iron-55 (Perkin Elmer, cat # NEZ043002M) at 2µCi/ml of cell suspension. Resuspended cells were incubated at 150 rpm for 3 h at 18°C. Cells were then collected, and washed first with 50mM sodium citrate (50mM citrate, 1mM EDTA, pH 7.0), then with 20mM HEPES (20mM HEPES/KOH pH 7.4) buffer to remove residual iron-55 from the medium and the outside of cells. Washed cells were resuspended at 1g/ml in TENTG (10mM Tris/HCl pH 7.4, 2.5mM EDTA, 150mM NaCl, 10% (vol/vol) glycerol, 0.5% (vol/vol) Triton X-100) supplemented with proteinase inhibitors. Whole cell extracts were prepared and radioactivity was determined by scintillation counting as described 73.
NET-seq
NET-seq libraries were constructed as previously described74 with a few modifications. Briefly, 2 L of cells expressing FLAG epitope tagged Rpb3 subunit of RNAPII (Rpb3-FLAG) were cultured at 18 °C for 3 d. Cells were harvested at early- to mid-log phase (OD600 = 0.3–0.5) by vacuum filtration and flash frozen in liquid nitrogen. Frozen cells were lysed using a CryoMill (Retsch) via 6 cycles of pulverization for 3 minutes at 15 Hz. Nascent RNA was purified by immunoprecipitating RNAPII using anti-FLAG M2 affinity gel (Sigma) in lysis buffer (150mM HEPES pH 7.4, 110mM KOAc, 10mM MnCl2, 0.5% Triton X-100, 0.1% Tween 20, 1× Protease Inhibitors EDTA-free complete- Roche, 50 U/mL SUPERase In (Thermo Scientific). Ligation of the DNA 3’-linker was performed with 2–3µg of purified nascent RNA. For reverse transcription, the following RT primer was used to allow for paired end sequencing: 5’Phos/AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC/iSp18/CACTCA/iSp18/TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG-3’. cDNA was circularized and PCR amplification was performed using minimal amplification cycles with the following primers: 5’ AATGATACGGCGACCACCGAGATCTACAC 3’, 5’ CAAGCAGAAGACGGCATACGAGATACGACTGTGACTGGAGTTCAGACGTGTGCTCTTCCG 3’. Sequencing was performed on the Illumina MiSeq platform. Oligonucleotides used for library construction are listed in Supplementary Table 1. Adapter trimmed reads were aligned to the S. pombe ASM294v2.30 reference genome using Tophat2 and FPKMs were calculated using Cufflinks. The position matching to the 3' end of the nascent RNA fragment was recorded and used for subsequent analysis. To calculate average gene profiles, 175 transcripts that showed >5-fold up-regulation in clr4Δ 18°C cells in RNA-seq analysis or randomly selected loci were aligned with respect to the transcription start site (TSS). RNAPII localization, in the sense strand, at regions from 100bp upstream to the 1000bp downstream with respect to the TSS were determined for both wild-type and clr4Δ cells. Normalized read counts at 10 bp probing windows overlapped by 5bp across this region were used to determine the average gene profile.
Statistical methods
The degree of overlap in sets of upregulated transcripts between strains or conditions or the degree of overrepresentation of upregulated transcripts within gene classes was evaluated using the binomial distribution. Conservative P-values were calculated as the right-hand tail density of the appropriate model distributions constructed for a total of 6380 coding and noncoding transcripts. The Pearson's correlation coefficient for the correspondence of transcript levels between strains or conditions was calculated from the paired log2 fold changes of 6380 transcripts. All transcript fold changes were calculated relative to the levels of wild-type S. pombe grown at 30 °C (n = 2 independent cell cultures).
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data Availability
Genomic datasets are deposited in the Gene Expression Omnibus with accession number GSE104547. Source data for Figures 1, 2, 5, 7 and Supplementary Figure 8 are available with the paper online. Other primary data are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank E. Hidalgo (Universitat Pompeu Fabra, Spain) for strains, J. Zhu, V. Bliskovsky and S. Shema for valuable technical advice, S. Holla for helpful contributions, J. Barrowman for editing the manuscript, and members of the Laboratory of Biochemistry and Molecular Biology, in particular the Grewal laboratory, for discussions. This study used the Helix Systems and Biowulf Linux cluster at the National Institutes of Health. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Author contributions
S.I.S.G and P.S.G. conceived and supervised the project. P.S.G., M.L., J.D., V.B., H.X., C.W. performed experiments and analyzed data, R.C, G.T, and D.W performed bioinformatics analyses of genomic datasets. S.I.S.G. and P.S.G. wrote the manuscript.
Competing interests
The authors declare no competing financial conflicts.
Additional Information
Supplementary information accompanies this paper.
References
- 1.Berger SL, Sassone-Corsi P. Metabolic signaling to chromatin. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry S, Dean C. Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 2015;83:133–48. doi: 10.1111/tpj.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rando OJ, Simmons RA. I'm eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol. Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Jia ST, Jia S. New Insights into the Regulation of Heterochromatin. Trends Genet. 2016;32:284–94. doi: 10.1016/j.tig.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilchert C, Wittmann S, Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016;17:227–39. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- 10.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–77. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 11.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto M. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:788–97. doi: 10.2183/pjab.86.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 14.Zofall M, et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–15. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Nicolas E, et al. Distinct roles of HDAC complexes in promoter silencing, antisense suppression and DNA damage protection. Nat. Struct. Mol. Biol. 2007;14:372–80. doi: 10.1038/nsmb1239. [DOI] [PubMed] [Google Scholar]
- 17.Lee NN, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–74. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama T, Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30:1027–39. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, et al. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat. Commun. 2015;6:7050. doi: 10.1038/ncomms8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meola N, et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell. 2016;64:520–533. doi: 10.1016/j.molcel.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Miller JE, Reese JC. Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 2012;47:315–33. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronner C, Salvi L, Zocco M, Ugolini I, Halic M. Accumulation of RNA on chromatin disrupts heterochromatic silencing. Genome Res. 2017;27:1174–1183. doi: 10.1101/gr.216986.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folco HD, et al. Untimely expression of gametogenic genes in vegetative cells causes uniparental disomy. Nature. 2017;543:126–130. doi: 10.1038/nature21372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiriart E, et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiyama T, et al. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol. Cell. 2016;61:747–759. doi: 10.1016/j.molcel.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamanaka S, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–60. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 28.Cotobal C, et al. Role of Ccr4-Not complex in heterochromatin formation at meiotic genes and subtelomeres in fission yeast. Epigenetics Chromatin. 2015;8:28. doi: 10.1186/s13072-015-0018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28:231–44. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touat-Todeschini L, et al. Selective termination of lncRNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 2017;36:2626–2641. doi: 10.15252/embj.201796571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joh RI, et al. Survival in quiescence requires the euchromatic deployment of Clr4/SUV39H by Argonaute-associated small RNAs. Mol. Cell. 2016;64:1088–1101. doi: 10.1016/j.molcel.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 33.Tashiro S, Asano T, Kanoh J, Ishikawa F. Transcription-induced chromatin association of RNA surveillance factors mediates facultative heterochromatin formation in fission yeast. Genes Cells. 2013;18:327–39. doi: 10.1111/gtc.12038. [DOI] [PubMed] [Google Scholar]
- 34.Chen D, et al. Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell. 2003;14:214–29. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalamcharla VR, Folco HD, Dhakshnamoorthy J, Grewal SI. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl. Acad. Sci. U. S. A. 2015;112:15548–55. doi: 10.1073/pnas.1522127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker JF, et al. A novel epigenetic silencing pathway involving the highly conserved 5'-3' exoribonuclease Dhp1/Rat1/Xrn2 in Schizosaccharomyces pombe. PLoS Genet. 2016;12:e1005873. doi: 10.1371/journal.pgen.1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labbe S, Khan MG, Jacques JF. Iron uptake and regulation in Schizosaccharomyces pombe. Curr. Opin. Microbiol. 2013;16:669–76. doi: 10.1016/j.mib.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Pelletier B, Beaudoin J, Mukai Y, Labbe S. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 2002;277:22950–8. doi: 10.1074/jbc.M202682200. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier B, Beaudoin J, Philpott CC, Labbe S. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 2003;31:4332–44. doi: 10.1093/nar/gkg647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Encinar del Dedo J, Gabrielli N, Carmona M, Ayte J, Hidalgo E. A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast. PLoS Genet. 2015;11:e1005106. doi: 10.1371/journal.pgen.1005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagerstrom-Billai F, Wright AP. Functional comparison of the Tup11 and Tup12 transcriptional corepressors in fission yeast. Mol. Cell. Biol. 2005;25:716–27. doi: 10.1128/MCB.25.2.716-727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson AD, et al. Ssn6-Tup1 interacts with class I histone deacetylases required for repression. Genes Dev. 2000;14:2737–44. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Znaidi S, Pelletier B, Mukai Y, Labbe S. The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J. Biol. Chem. 2004;279:9462–74. doi: 10.1074/jbc.M312787200. [DOI] [PubMed] [Google Scholar]
- 44.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 45.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 47.Cam HP, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 2005;37:809–19. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 48.Wagschal A, et al. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell. 2012;150:1147–57. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005;19:1705–14. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 2005;7:1007–13. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 51.Hong EJ, Villen J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2:106–11. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 52.Greil F, et al. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2825–38. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–14. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–91. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 55.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 56.Kimura S, Suzuki T. Iron-sulfur proteins responsible for RNA modifications. Biochim. Biophys. Acta. 2015;1853:1272–83. doi: 10.1016/j.bbamcr.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 57.Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Andrews NC. Disorders of iron metabolism. N. Engl. J. Med. 1999;341:1986–95. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 59.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 60.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 61.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–80. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Team RCR. A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. [Google Scholar]
- 66.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 67.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 68.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- 69.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pierik AJ, Netz DJ, Lill R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat. Protoc. 2009;4:753–66. doi: 10.1038/nprot.2009.39. [DOI] [PubMed] [Google Scholar]
- 74.Churchman LS, Weissman JS. Native elongating transcript sequencing (NET-seq) Curr. Protoc. Mol. Biol. 2012 doi: 10.1002/0471142727.mb0414s98. Chapter 4, Unit 4 14 1–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic datasets are deposited in the Gene Expression Omnibus with accession number GSE104547. Source data for Figures 1, 2, 5, 7 and Supplementary Figure 8 are available with the paper online. Other primary data are available from the corresponding author upon reasonable request.