Abstract
The fibers of cotton (Gossypium hirsutum) are single-cell trichomes that undergo rapid and synchronous elongation. Cortical microtubules provide spatial information necessary for the alignment of cellulose microfibrils that confine and regulate cell elongation. We used gene-specific probes to investigate α-tubulin transcript levels in elongating cotton fibers. Two discrete patterns of transcript accumulation were observed. Whereas transcripts of α-tubulin genes GhTua2/3 and GhTua4 increased in abundance from 10 to 20 d post anthesis (DPA), GhTua1 and GhTua5 transcripts were abundant only through to 14 DPA, and dropped significantly at 16 DPA with the onset of secondary wall synthesis. This is the first report, to our knowledge, of gene-specific changes in tubulin transcript levels during the development of a terminally differentiated plant cell. The decrease in abundance of GhTua1 and GhTua5 transcripts was correlated with pronounced changes in cell wall structure, suggesting that α-tubulin isoforms may be functionally distinct in elongating fiber cells. Although total α-tubulin transcript levels were much higher in fiber than several other tissues, including the hypocotyl and pollen, none of the α-tubulins was specific to fiber cells.
Microtubules are components of the filamentous cytoskeleton of eukaryotic cells and participate in many cell processes, including cell division, intracellular transport, cell motility, and cell morphogenesis. In plants, microtubules have a number of specialized roles, including participation in cell differentiation. Differentiation in plants is directed by regulation of the plane of cell division and the direction of cell elongation. During cell elongation, highly organized microfibrils of cellulose confine turgor-driven cell expansion to a single major axis of growth (Giddings and Staehelin, 1991; Delmer and Amor, 1995). The cellulose microfibrils are oriented at right angles to the major axis of elongation (Gertel and Green, 1977), and the organization of these microfibrils is controlled by cortical microtubules (Giddings and Staehelin, 1991; Cyr and Palevitz, 1995). The cortical microtubule array is thought to provide spatial information to the cellulose-synthesizing machinery by mechanisms that are yet to be identified. Also unknown is how the cortical microtubules become aligned. Recent experiments using inhibitors of cellulose biosynthesis suggest a bidirectional flow of information, whereby biophysical forces generated by the microfibril arrays in the elongating cell are necessary for microtubule alignment (Fisher and Cyr, 1998).
The fibers of cotton (Gossypium hirsutum) are a good experimental system for studying the role of microtubules in cell elongation, since elongation occurs at a fast rate over a relatively long period, uninterrupted by cell division. Furthermore, changes in the cell wall structure of elongating cotton fibers have been well characterized (Basra and Malik, 1984; Seagull, 1986, 1992, 1993). Cotton fibers are single-cell trichomes that result from elongation of epidermal cells of the ovule. Ultrastructural evidence indicates that elongation occurs by a diffuse growing mechanism (Seagull, 1990; Tiwari and Wilkins, 1995). In elongating fibers a thin primary wall is deposited. Secondary wall synthesis is initiated approximately 16 to 18 DPA, overlapping the final stages of elongation. Cotton is unique in that its secondary wall contains nearly pure cellulose and no lignin.
In expanding cotton fibers the patterns of microtubule deposition correlate precisely with the wall microfibril arrays (Seagull, 1986, 1992). During fiber development, microtubules exhibit specific changes in orientation, organization, number, length, and proximity to the plasmalemma. These changes are most apparent in the transition from rapid elongation and primary wall synthesis to the onset of secondary cell wall synthesis and slowing of elongation. We were particularly interested in cytoskeletal changes that occur during this developmental transition at approximately 16 to 18 DPA.
The major structural component of microtubules is tubulin, a heterodimeric protein composed of two highly conserved subunits, α and β. A less abundant form, γ-tubulin (Oakley et al., 1989), is also found in higher plants (Liu et al., 1994). Both α- and β-tubulins are encoded by multigene families in eukaryotes (Cleveland and Sullivan, 1985; Silflow et al., 1987). Tubulin genes have been studied in only a few plant species and have been best characterized in Arabidopsis and maize. In Arabidopsis at least six α-tubulin genes and nine β-tubulin genes are expressed (Kopczak et al., 1992; Snustad et al., 1992). There is evidence of at least seven α-tubulin genes (Montoliu et al., 1989, 1990, 1992; Villemur et al., 1992) and six β-tubulin genes (Villemur et al., 1994) expressed in maize. Tissue-specific preferences in accumulation of tubulin transcripts have been reported in both Arabidopsis (Ludwig et al., 1988; Oppenheimer et al., 1988; Carpenter et al., 1992; Snustad et al., 1992) and maize (Joyce et al., 1992; Villemur et al., 1994; Uribe et al., 1998), and in Arabidopsis, specific β-tubulin genes were shown to be regulated by light (Leu et al., 1995) and temperature (Chu et al., 1993). Therefore, gene-specific expression of tubulins is regulated by both developmental and environmental signals.
In cotton fiber cells, nine α-tubulin and seven β-tubulin isotypes have been identified on immunoblots of two-dimensional gels (Dixon et al., 1994). Two α-tubulin and two β-tubulin isotypes showed preferential accumulation in fibers and appeared to be temporally regulated (Dixon et al., 1994). Since we were interested in comparing the promoter structure of members of this multigene family, we have begun to characterize the α-tubulin genes expressed in developing fibers. Due to the high degree of nucleotide sequence conservation among α-tubulin-coding regions in higher plants, we chose to PCR-amplify the 3′-UTRs of cotton α-tubulins for use as gene-specific probes. Five distinct α-tubulin cDNA fragments from cotton fiber were amplified. We report the results of an examination of α-tubulin transcript levels during fiber elongation using these gene-specific probes.
MATERIALS AND METHODS
Plant Material
Cotton (Gossypium hirsutum var MD51 ne) plants were grown in the field during the summer of 1997. First-position flowers were tagged on the day of anthesis during 6 consecutive days in July. Developing bolls were collected at 2-d intervals from 10 through 20 DPA. Immediately after harvest, cottonseeds were excised from each boll, frozen in liquid nitrogen, and stored at −80°C. Fiber cells were harvested while frozen for RNA extraction. The fibers were carefully scraped from the frozen ovules using a scalpel, ensuring that the fibers were not contaminated with other cell types.
Hypocotyls and cotyledons were harvested from 10-d-old seedlings of the cotton var Texas Marker-1 grown in a greenhouse at 25°C to 32°C. Ungerminated pollen was collected from the flowers of var MD51 ne on the day of anthesis. These plants were also grown in a greenhouse at 25°C to 32°C. Roots were harvested from 10-d-old seedlings of var Texas Marker-1 grown in a controlled-environment chamber (30°C, 240 μE m−2 s−1 incandescent and fluorescent light). The seedlings were grown in 16 h of light d−1 for 7 d, followed by darkness for 3 d, prior to harvest of root tissues. All tissues were frozen in liquid nitrogen and stored at −80°C prior to nucleic acid extraction.
Isolation of Cotton RNA
RNA was isolated from fiber, root, hypocotyl, and cotyledon tissues with a kit (RNeasy Midi Kit, Qiagen, Valencia, CA), using a modification of the manufacturer's protocol. Fiber material (5 g) was ground to a powder in liquid nitrogen using a mortar and pestle. The powder was added to 20 mL of lysis buffer (RLT, Qiagen) containing 1% (v/v) β-mercaptoethanol. The slurry was homogenized in a conventional rotor-stator homogenizer for 2 min at maximum speed, centrifuged at 3,000g for 10 min at room temperature, and filtered through Miracloth (Calbiochem). One-half volume of absolute ethanol was added to the filtered lysate, and mixed by vortexing. The mixture was applied to the spin column (Qiagen) in serial aliquots of 3.8 mL. Each aliquot was centrifuged at 4,000g for 2 min at room temperature. The column was washed and RNA eluted according to the manufacturer's instructions. The yield of RNA was dependent on the stage of fiber development. Typical yields of total RNA were in the range of 40 to 100 μg from 5 g of fiber.
PCR Amplification and Cloning
First-strand cDNA was reverse transcribed from total RNA isolated from 10-DPA cotton fiber. The first-strand cDNA was used as a template for amplification of partial cDNAs encoding 3′ fragments of α-tubulin genes.
GhTua1, GhTua2, and GhTua3 were amplified with the 5′ primer 5′-GGAAAGTACATGGCTTGCTGTTTGATG-3′ and the 3′ primer oligo d(T)16, using an annealing temperature of 65°C. The PCR mixture was 1× PCR buffer (10 mm Tris-HCl, pH 8.3, and 50 mm KCl), 2 mm MgCl2, 0.2 mm dNTPs, 0.15 μm for each primer, and 2.5 units of Taq DNA polymerase. GhTua4 and GhTua5 were amplified using the partially degenerate 5′ primer 5′-TGTTTGATGTACCGWGGWGAYGT-3′ and the 3′ primer oligo d(T)16, using an annealing temperature of 45°C. The PCR mixture was 1× PCR buffer, 3 mm MgCl2, 0.2 mm dNTPs, 0.5 μm 5′ primer, 4 μm 3′ primer, and 2 units of Taq DNA polymerase. Amplification was carried out using either a model 480 or a GeneAmp 9700 thermal cycler (both Perkin-Elmer Cetus). The PCR products were blunt-cloned in the plasmid vectors pCRScript (Stratagene) or pCRII (Invitrogen, Carlsbad, CA). The DNA sequence of clones GhTua1 to GhTua5 was determined for both strands using the dideoxynucleotide chain-termination method with a DNA-sequencing kit (Sequitherm Excel II Long Read, Epicentre Technologies, Madison, WI), a thermal cycler (model 480), and a DNA sequencer (model 4000L, LI-COR). The accession numbers are as follows: GhTua1, AF106567; GhTua2, AF106568; GhTua3, AF106569; GhTua4, AF106570; and GhTua5, AF106571.
A cotton 26S rRNA gene was isolated from a Lambda Zap II (Stratagene) cDNA library. The Bluescript plasmid containing the 26S rRNA gene was recovered from bacteriophage by an in vivo excision protocol according to the manufacturer's instructions.
Northern Analysis
Total RNA (2 μg) was heat denatured and electrophoresed through a 1.2% (w/v) agarose nondenaturing gel, as described by Kevil et al. (1997). The RNA was transferred to a positively charged nylon membrane (BrightStar-Plus, Ambion, Austin, TX) by downward capillary transfer (Ausubel et al., 1987) using a transfer solution of 5× SSC (750 mm NaCl and 75 mm tri-sodium citrate [pH 7.0]), and 10 mm NaOH. After transfer, RNA was cross-linked to the membrane using a 50-mJ UV pulse (GS GeneLinker, Bio-Rad). Blots were prehybridized for 2 h at 48°C to 55°C in 5× SSPE (750 mm NaCl, 50 mm NaH2PO4, and 5 mm EDTA [pH 7.4]), 50% formamide, 7% SDS, and 100 μg mL−1 herring-sperm DNA.
[α-32P]UTP-labeled antisense RNA probes were transcribed from plasmid DNA linearized by digestion with an appropriate restriction enzyme. These probes were transcribed using a kit (Strip-EZ, Ambion) that incorporates a chemically modified CTP, facilitating probe removal for re-use of blots. Approximately 200 μmol of each probe was used, and standard membrane hybridization protocols were followed. All probes were well in excess of the target transcripts in each RNA sample assayed. The blots were hybridized to RNA probes for at least 16 h under the same conditions used for pre-hybridization. After hybridization, the blots were washed twice in 1× SSPE and 0.5% (w/v) SDS for 10 min at hybridization temperature, and twice in 5× SSPE, 50% (v/v) formamide, and 7% (w/v) SDS for 30 min at 53°C to 65°C. The blots were exposed to x-ray film (BioMax, Kodak) with an intensifying screen at −80°C. The amount of radioactive signal on the blots was quantified by phosphor imaging (model GS-525, Bio-Rad). Probes were stripped from the membranes as described by the transcription kit manufacturer, and the membranes were re-exposed to x-ray film for at least 18 h to confirm probe removal.
Physical Properties of Cotton Fiber Samples
Fiber Length
Fiber length was estimated by placing ovules on a watchglass and gently spraying fibers with a stream of distilled water from a wash bottle (Schubert et al., 1973). The distance from the chalazal end of the ovule to the tip of the spread fibers was measured to the nearest 0.1 mm with calipers. Two replicate samples with 20 ovules per replicate were measured.
Fiber Weight per Ovule
Fibers were gently removed from all ovules of each replicate sample, dried, and weighed on an analytical balance (model AE163, Mettler-Toledo, Columbus, OH). Fiber dry weight was divided by the total number of ovules in each sample.
Cellulose Content
Fiber samples were dried, cut into 1-mm sections with scissors, and weighed. Replicate 10-mg fiber samples were placed into 5-mL vials (Reacti-vials, Pierce). Noncellulosic material was hydrolyzed with acetic-nitric reagent (Updegraff, 1969). (Reagent volumes were reduced from the original Updegraff procedure, permitting triplicate assays to be conducted in 2.0-mL microcentrifuge tubes.) Cellulose was hydrolyzed with sulfuric acid, and a colorimetric assay using anthrone was used to detect Glc. After cooling the microcentrifuge tubes on ice, 150 μL of each assay was transferred to a well in a 96-well microtiter plate. A plate reader (ThermoMax, Molecular Devices, Sunnyvale, CA) was used to measure the sample A650, with cellulose (Avicel PH-101, FMC, Rockland, ME) serving as the standard.
Fiber α-Tubulin Content
Fiber scraped from frozen ovules was collected in liquid nitrogen and ground in a mortar. Proteins were extracted by the method of Barent and Elthon (1992) and resuspended in SDS sample buffer containing a protease-inhibitor tablet (Complete, Boehringer Mannheim). Protein content was measured by applying the samples to nitrocellulose, staining with Amido black (Goldring and Ravaioli, 1996), and scanning the membrane with an imaging densitometer (GS-700, Bio-Rad). BSA was used as the standard. Replicate western blots were prepared as described previously (Dixon et al., 1994) using a 1:1,000 dilution of anti-α-tubulin (YOL 1/34) (Accurate Chemical and Scientific Corporation, Westbury, NY) and a 1:1,000 dilution of secondary antibody (sheep anti-rat IgG coupled with horseradish peroxidase, Amersham). A chemiluminescent peroxidase substrate (SuperSignal, Pierce) was used according to the manufacturer's protocol. A phosphor imager (Molecular Analyst, Bio-Rad) was used to detect and quantify chemiluminescent signals from the membrane.
RESULTS
Cellulose and α-Tubulin Levels in Elongating Cotton Fibers
The cellulose content was measured in fiber cell walls isolated from the same field-grown samples used for RNA isolation. These values were used as a reference point for analyses of tubulin expression. A distinct increase in cellulose content was observed between 14 and 16 DPA (Table I), indicating increased cellulose synthetic activity and the onset of secondary wall synthesis. Fiber elongation was continuous from 10 through 20 DPA. An overlap between fiber elongation and secondary wall synthesis in developing cotton fibers has been extensively reported (for review, see Ryser, 1985). Levels of α-tubulin protein were determined by immunoblot analysis (Table I), using an antibody that binds an epitope that is conserved in all known cotton α-tubulin isotypes (Dixon et al., 1994). α-Tubulin protein levels increased in these fiber samples throughout the developmental period between 10 and 20 DPA, which is in agreement with the results of Kloth (1989). This increase in α-tubulin protein is consistent with the increase in microtubule length and number that occurs during these stages of fiber development (Seagull, 1992).
Table I.
Fiber growth characteristics, cellulose content, and α-tubulin protein content
| Fiber Characteristic | 10 DPA | 12 DPA | 14 DPA | 16 DPA | 18 DPA | 20 DPA |
|---|---|---|---|---|---|---|
| Dry mass (mg/ovule) | 0.84 | 1.50 | 14.41 | 41.7 | 69.7 | 71.9 |
| Length (cm) | 1.5 ± 0.27 | 1.9 ± 0.33 | 2.3 ± 0.43 | 2.7 ± 0.48 | 2.9 ± 0.48 | 3.5 ± 0.44 |
| Cellulose content (%) | 11.9 ± 3.69 | 13.5 ± 2.02 | 14.4 ± 1.99 | 41.7 ± 14.3 | 61.7 ± 15.5 | 71.9 ± 15.4 |
| α-Tubulin protein accumulation (%) | 12.7 ± 3.4 | 36.5 ± 9.8 | 63.3 ± 17.1 | 86.5 ± 23.3 | 95.7 ± 12.8 | 100 ± 6.2 |
Fiber length was measured for two sets of 20 ovules randomly selected from the same population of cottonseed used for RNA isolation. After length measurement, fibers were stripped from the seed and lyophilized to constant weight. Fiber samples were stored over phosphorus pentoxide prior to weight determinations and cellulose measurements. Replicate fiber samples were hydrolyzed with acetic-nitric reagent, followed by cellulose determination with anthrone. Avicel cellulose (PH-101, FMC) was used as the standard in the cellulose assays. The relative amount of α-tubulin protein was measured by scanning triplicate western blots of equal amounts of total fiber protein isolated from selected stages of development and probed with an antibody YOL 1/34 to α-tubulin. Protein values are expressed as the percentages of the highest protein content in the series.
Isolation and Characterization of Probes for Northern Analyses
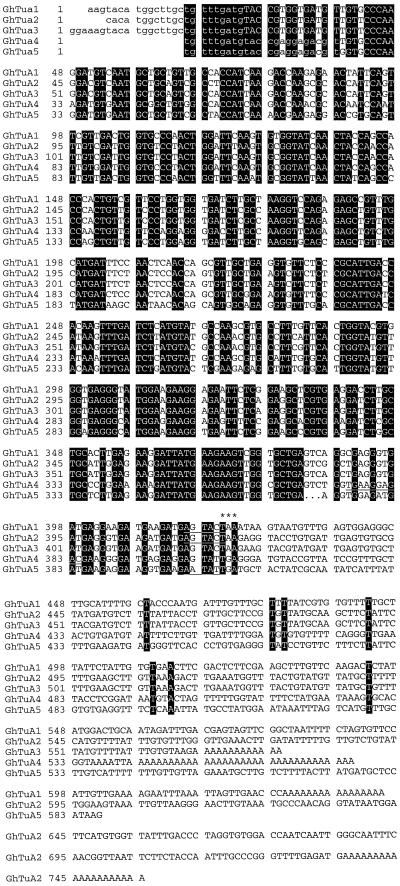
Partial cDNAs encoding 3′ fragments of α-tubulin genes were amplified from a cDNA pool derived from 10-DPA cotton fibers. Primer design was based on a highly conserved region of amino acid identity shared by plant α-tubulin cDNAs located near the 3′ end of the ORF. The cDNAs were cloned in plasmid vectors containing flanking promoters to facilitate transcription of antisense RNA probes for northern analyses. The clones were characterized by restriction analysis and determination of the complete nucleotide sequence of five distinct clones (GhTua1 to GhTua5; Fig. 1). The sequences of clones GhTua1 to GhTua5, excluding PCR primers, were deposited in GenBank and accession numbers are given in Methods. For each of the cDNA clones GhTua1, GhTua2, and GhTua3, one or more homologs differing only by the length of the 3′UTR and the poly(A+) tail were identified, indicating multiple genes and/or polyadenylation sites. GhTua1, GhTua2, and GhTua3 were the longest clones from each group of homologs. Clone GhTua4 was represented by two identical clones; and no homologs of clone GhTua5 were isolated. A 286-bp fragment of an α-tubulin ORF (accession no. AF009565) amplified from cotton cv Acala SJ-2 cDNA (Smart et al., 1998) differed from GhTua2 by only three nucleotides. The deduced amino acid sequences of these gene fragments were completely conserved. It is likely that AF009565 and GhTua2 were amplified from the same gene; the differences in nucleotide sequence may have been due to polymorphism between the two cotton cultivars or to PCR misincorporation during the amplification of one or both cDNAs.
Figure 1.
Alignment of nucleotide sequences of α-tubulin cDNA clones. Sequences were aligned and displayed using the programs PileUp and Pretty (version 9.1, Genetics Computer Group, Madison, WI). Sequences homologous to the oligonucleotide primers used for PCR amplification are shown in lowercase; conserved nucleotides are indicated by solid black boxes; and dots indicate gaps that have been inserted for optimal alignment; the putative translational stop codons are marked by asterisks; and the restriction sites at which probe transcription was terminated are underlined. The restriction sites are cut by ScaI (GhTua1 and GhTua2), BsmAI (GhTua4), and SspI (GhTua5).
A comparison of the deduced amino acid sequences of GhTua1 to GhTua5 and the α-tubulin gene family of Arabidopsis is shown in Figure 2. The amino acid sequences of GhTua1 to GhTua5 are distinct, but show high identity to each other and to the Arabidopsis sequences. Clone GhTua5 was the most divergent of the cotton sequences; a Gly residue that was conserved in clones GhTua1 to GhTua4 was absent in GhTua5. Pairwise comparisons of cotton and Arabidopsis amino acid sequences indicated 87% to 94% identity.
Figure 2.
Alignment of partial deduced amino acid sequences of α-tubulin cDNAs from cotton and Arabidopsis. Sequences were aligned and displayed using the programs PileUp and Pretty (Genetics Computer Group). The amino acids are numbered consecutively from 1 through 128, beginning with the first amino acid represented in all of the partial clones. Sequences homologous to the oligonucleotide primers used for PCR amplification of GhTua1 to GhTua5 have been excluded from the comparison. Solid black boxes indicate conserved amino acid residues; translational stop codons are indicated by asterisks; the dot indicates a gap that has been inserted for optimal alignment. The Arabidopsis α-tubulin sequences are from Ludwig et al. (1987, 1988) and Kopczak et al. (1992).
The 3′-UTRs of clones GhTua1, GhTua4, and GhTua5 were highly divergent (Fig. 1) and were selected as gene-specific probes for northern analyses. Clones GhTua2 and GhTua3 shared high identity across both the ORF and 3′-UTRs (Fig. 1), and therefore we have not attempted to distinguish the transcripts of these two genes. Clones GhTua1, GhTua2, GhTua4, and GhTua5 were cut at restriction sites near the stop codon (Fig. 1) to generate templates for the transcription of [α-32P]UTP-labeled probes to the 3′-UTRs. These probes (tua1UTR, tua2UTR, tua4UTR, and tua5UTR) are described in Table II. A 394-bp subclone of GhTua2 encompassing the ORF fragment was used as a template for the transcription of the antisense probe tua2ORF. This region has high nucleotide sequence identity to all of the cDNA fragments we have amplified (Table II), so is likely to be well conserved in all cotton α-tubulin genes.
Table II.
α-Tubulin probes used for northern analysis
| Probe | Template (Excluding Vector) | Nucleotide Identity
|
||||
|---|---|---|---|---|---|---|
| GhTua1 | GhTua2 | GhTua3 | GhTua4 | GhTua5 | ||
| % | ||||||
| tua1UTR | GhTua1 (419–645) | 100 | 46 | 37 | 45 | 39 |
| tua2UTR | GhTua2 (416–755) | 43 | 100 | 89 | 43 | 40 |
| tua4UTR | GhTua4 (375–575) | 48 | 41 | 48 | 100 | 38 |
| tua5UTR | GhTua5 (404–587) | 38 | 49 | 39 | 41 | 100 |
| tua2ORF | GhTua2 (1–415) | 88 | 100 | 99 | 83 | 78 |
α-32P-labeled antisense RNA probes were transcribed from the cDNA clones (Fig. 1) following linearization with the appropriate restriction enzyme. Probes tua1UTR, tua2UTR, tua4UTR, and tua5UTR were transcribed from the 3′-UTR of the corresponding cDNA, whereas tua2ORF was transcribed from an ORF fragment. The nucleotide identity of the probes to each α-tubulin cDNA are shown. Values in parentheses are the corresponding nucleotide numbers.
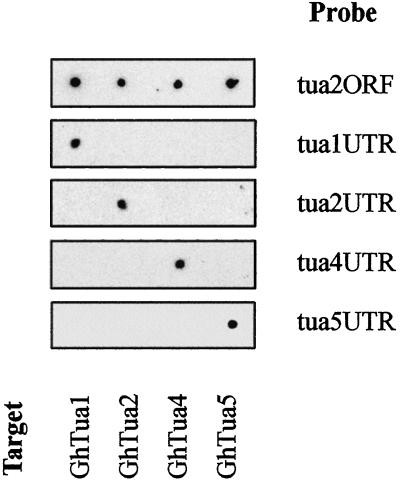
To confirm the specificity of the α-tubulin probes, each was hybridized to sense transcripts of GhTua1, GhTua2, GhTua4, and GhTua5 (Fig. 3). Probes tua1UTR, tua2UTR, tua4UTR, and tua5UTR showed no significant cross-hybridization under high-stringency conditions, while at lower stringency tua2ORF hybridized to all of the sense transcripts, demonstrating cross-hybridization at 78% nucleotide identity (Table II). Due to the high sequence identity of GhTua2 and GhTua3 in the 3′-UTR, it is possible that probe tua2ORF may hybridize to both GhTua2 and GhTua3 transcripts under high-stringency conditions. Length heterogeneity was observed in the 3′-UTRs of GhTua1, GhTua2, and GhTua3 homologs, and clones GhTua2 and GhTua3, which showed high sequence identity across the region of overlap in their 3′-UTRs, also differed in the length of their 3′-UTRs (Fig. 1). Further, it is possible that variable-length homologs of GhTua4 and GhTua5 are also transcribed in fibers. We anticipate that α-tubulin transcripts with more truncated 3′-UTRs may not be assayed by our antisense probes under stringent washing conditions, due to the misalignment of target and/or probe poly(A+) tails. We limited this study to characterizing the expression of four distinct transcripts, GhTua1, GhTua2, GhTua4, and GhTua5, and total α-tubulin transcript levels; we did not investigate the expression of polyadenylation site variants.
Figure 3.
Specificity of α-tubulin probes under assay conditions. Sense transcripts of the cDNA clones GhTua1, GhTua2, GhTua4, and GhTua5 (1 ng) were spotted onto each of five positively charged nylon membranes. The membranes were hybridized to each of the α-tubulin antisense probes listed in Table II and washed in 5× SSPE, 50% (v/v) formamide, and 7% (w/v) SDS at 53°C to 65°C. Probe identities are shown at the right of each panel, and sense transcript positions are noted at the bottom of the figure.
A 26S rRNA probe was used to control for variations in sample loadings. Clone Gh26S (1.6 kb) was isolated from a cotton cDNA library, and its identity as a 26S rDNA fragment was confirmed by partial sequencing (data not shown). The clone was linearized with the restriction enzyme BsmAI, and an antisense probe of 256 bp (gh26S) was transcribed with T7 RNA polymerase.
Northern Analyses of α-Tubulin Transcripts
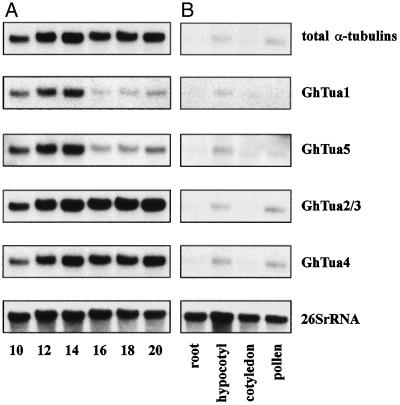
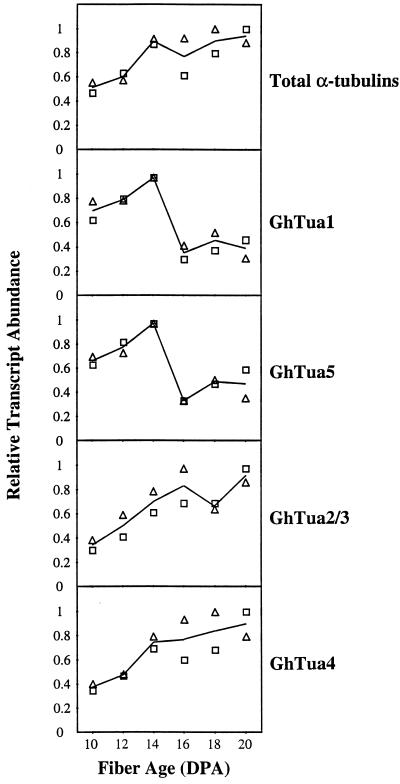
Total RNA was extracted from developing cotton fibers harvested at 2-d intervals from 10 to 20 DPA. This sample series was chosen to investigate the relative transcript levels of specific α-tubulin genes at the onset of secondary wall synthesis (Table I), when significant changes in the organization of the cortical cytoskeleton occur (Seagull, 1986, 1992). RNA was also extracted from root, hypocotyl, cotyledon, and pollen samples to investigate the tissue specificity of each gene. Transcript levels were measured by northern analysis using sequentially hybridized probes (Fig. 4). Since the probes were of slightly different lengths and GC content, and may have been labeled to different specific activities, it was not appropriate to compare the levels of different transcripts within a single tissue or stage of development. Rather, these experiments were designed to investigate relative differences in accumulation for each transcript across the sample series. The quantified results of duplicate northern analyses are shown in Figure 5. Relative transcript levels were consistent in the independently extracted and assayed RNA samples.
Figure 4.
Northern analysis of α-tubulin mRNA levels. A, Developing cotton fiber. Fiber age is shown in DPA. B, Root, hypocotyl, cotyledon, and ungerminated pollen. Approximately 2 μg of total RNA was electrophoresed in each lane. The membrane was stripped and rehybridized to each probe in succession, as described in Methods. The α-tubulin probes (described in Table II) are as follows: total α-tubulins, tua2ORF; GhTua1, tua1UTR; GhTua2/3, tua2UTR; GhTua4, tua4UTR; and GhTua5, tua5UTR. For the gene-specific probes, hybridization and washing were carried out under stringencies sufficient to eliminate cross-hybridization (Fig. 3). The ribosomal probe gh26S was used to quantify differences in RNA loading.
Figure 5.
Quantification of northern analyses of α-tubulin transcript levels in developing cotton fibers. The signals for each α-tubulin probe were quantified by phosphor-imaging the blots. These values were normalized against the 26S rRNA signal and are expressed relative to the highest value in the series. Duplicate northern analyses (not shown) were performed using independently extracted RNA samples. □, Results shown in Figure 4A; ▵, results of duplicate northern analyses.
Levels of total α-tubulin transcripts in fiber assayed with probe tua2ORF (Fig. 4A) were much higher than levels in other tissues (Fig. 4B). Transcript levels in pollen and hypocotyls were approximately 5-fold and 30-fold lower, respectively, than levels in 20-DPA fibers. Levels in roots and cotyledons were still lower and could not be quantified accurately. In developing fibers, two discrete patterns of transcript accumulation were observed (Figs. 4A and 5). The transcripts of GhTua1 to GhTua5 all increased in abundance from 10 through 14 DPA. However, GhTua2/3 and GhTua4 transcripts remained abundant to 20 DPA, while GhTua1 and GhTua5 transcripts dropped significantly after 14 DPA, when secondary wall synthesis began.
Transcripts of GhTua1, GhTua2/3, GhTua4, and GhTua5 were all detected in hypocotyls, while in roots and cotyledons, these transcripts were either seen at lower levels or were undetectable. In pollen, GhTua2/3 and GhTua4 transcripts were preferentially expressed. Transcripts of GhTua2/3 and GhTua4 were at least 3-fold higher in abundance in pollen than in hypocotyls, whereas GhTua1 and GhTua5 transcript levels in pollen were too low to quantify.
DISCUSSION
We investigated steady-state transcript levels for five distinct α-tubulin genes in developing cotton fibers from 10 through 20 DPA, focusing on the onset of secondary wall synthesis, which occurred at approximately 16 DPA (Table I). PCR-based strategies were used to isolate cDNA fragments corresponding to the variable 3′-UTR of α-tubulins to serve as probes for these studies.
Levels of total α-tubulin transcripts in fibers (Fig. 4A) were much higher than levels in other tissues (Fig. 4B), reflecting the rapid cell elongation occurring in developing fibers. These levels are consistent with the high relative abundance of tubulin proteins observed in cotton fibers by Dixon et al. (1994). This result strengthens our working hypothesis that characterization of fiber α-tubulin promoters may be a useful step in determining which signals specify the high levels of tubulin expression seen in this cell type. Accordingly, we have isolated genomic clones of fiber α-tubulin genes and have begun characterization and sequence analysis of these clones (D.J. Whittaker and B.A. Triplett, unpublished data).
Our most important finding was that α-tubulins show gene-specific differences in transcript accumulation in developing fibers. While all assayed transcripts were abundant in fibers from 10 to 14 DPA, only GhTua2/3 and GhTua4 remained abundant following the onset of secondary wall synthesis (Figs. 4A and 5). Immunoblot analyses indicate greater isotype diversity in elongating fibers prior to secondary wall synthesis (Dixon et al., 1994). Two distinct groups of isoforms were seen in 10-DPA fibers and in hypocotyls; the more acidic group of α-tubulins was not expressed at 20 DPA. The differences in transcript accumulation we observed indicate that differential transcription of distinct genes contributes to the change in isotype populations.
A total of nine α-tubulin isotypes have been distinguished in 10-DPA cotton fibers using immunoblot analysis (Dixon et al., 1994). We identified fragments of genes encoding only five distinct α-tubulins, and therefore it is possible that another novel α-tubulin gene or genes is transcribed in this tissue. However, a de-tyrosinated subset of α-tubulins has been identified in fibers (D.C. Dixon and B.A. Triplett, unpublished data), indicating that posttranslational modifications contribute to the observed number of isotypes.
At the onset of secondary wall synthesis there is a dramatic change in cortical microtubule orientation, from transverse to steeply pitched helices (Seagull, 1992). The microtubules also increase in number, length, and proximity to the plasma membrane. Re-orientation of the microtubules is mirrored by microfibrils in the cell wall, supporting the hypothesis that microtubules control microfibril alignment (Williamson, 1991; Cyr and Palevitz, 1995). We observed a steady increase in abundance of total α-tubulin transcripts in fiber cells from 10 through to 20 DPA (Figs. 4A and 5), by which time secondary wall synthesis was active. The onset of secondary wall synthesis, which occurred between 14 and 16 DPA (Table I), was not marked by a change in abundance of total α-tubulin transcripts but, rather, by a change in the relative abundance of two distinct transcripts. This correlation suggests that transcriptional control of specific tubulins may influence microtubule organization in elongating fiber cells and consequently affect patterns of microfibril deposition and cell form.
Transcripts of the α-tubulins GhTua1 and GhTua5 appear to accumulate preferentially in rapidly elongating tissues, i.e. in hypocotyls and in fiber cells prior to secondary wall synthesis. Transcripts of several genes have been shown to accumulate at high levels before and during rapid expansion in cotton fibers, including genes encoding expansin and endo-1,4-β-glucanase (Shimizu et al., 1997) and genes involved in the regulation of cell turgor and extensibility (Smart et al., 1998). The coordinated regulation of GhTua1 and GhTua5 with these elongation-related genes suggests that the tubulins encoded by GhTua1 and GhTua5 may contribute to the specialized cytoskeletal architecture of elongating cells. Common regulatory elements might facilitate coupling of cytoskeletal structure with the changes in turgor regulation and cell wall structure necessary for elongation. Smart et al. (1998) propose that transcripts of another α-tubulin gene (accession no. AF009565) peaked during rapid expansion and declined during the onset of secondary wall synthesis. However, these transcript levels are not consistent with our data, which indicated that transcription of GhTua2 (a homolog of AF009565) is sustained until 20 DPA, when secondary wall synthesis is well under way.
Developmentally regulated patterns of tubulin transcription have been observed in several plant species, and have been most widely studied in Arabidopsis (Ludwig et al., 1988; Oppenheimer et al., 1988; Carpenter et al., 1992; Kopczak et al., 1992; Chu et al., 1998) and maize (Montoliu et al., 1989, 1990; Hussey et al., 1990; Joyce et al., 1992; Villemur et al., 1994; Uribe et al., 1998). Recent in situ hybridization studies in maize indicate transcription of different tubulin genes in distinct groups of differentiating cells (Uribe et al., 1998). Furthermore, tubulin promoters from maize (Uribe et al., 1998) and Arabidopsis (Chu et al., 1998) directed preferential reporter gene expression in specialized organs and cell types. We have shown that in cotton, gene-specific changes in transcript abundance also occur during the development of a terminally differentiated cell. As yet, there is no evidence for functionally distinct tubulin isoforms in plants like there is in animals, where it has been demonstrated that microtubule architecture can be intrinsic to the tubulin primary sequence (Raff et al., 1997). We believe that the clearly defined cytoskeletal changes seen in cotton fibers make this cell type a good system for investigating a possible functional role for specific tubulin isotypes.
ACKNOWLEDGMENTS
The authors wish to acknowledge the contribution of Dave Dixon (Michigan Technological University, Houghton), who amplified the α-tubulin cDNA fragments GhTua1, GhTua2, and GhTua3. We also thank Reiner Kloth (U.S. Department of Agriculture-Agricultural Research Service, Stoneville, MS) and Thomas Pesacreta (University of Southwest Louisiana, Lafayette) for critical reading of the manuscript.
Footnotes
This work was supported by the U.S. Department of Agriculture, Agricultural Research Service, Current Research Information System project no. 6435–21440–0001–00D.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Stuhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1987. [Google Scholar]
- Barent RL, Elthon TE. Two-dimensional gels: an easy method for large quantities of proteins. Plant Mol Biol Rep. 1992;10:338–344. [Google Scholar]
- Basra AS, Malik CP. Development of the cotton fiber. Int Rev Cytol. 1984;89:65–113. [Google Scholar]
- Carpenter JL, Ploense SE, Snustad DP, Silflow CD. Preferential expression of an α-tubulin gene of Arabidopsis in pollen. Plant Cell. 1992;4:557–571. doi: 10.1105/tpc.4.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B, Snustad DP, Carter JV. Alteration of β-tubulin gene expression during low-temperature exposure in leaves of Arabidopsis thaliana. Plant Physiol. 1993;103:371–377. doi: 10.1104/pp.103.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B, Wilson TJ, McCune-Zierath C, Snustad DP, Carter JV. Two β-tubulin genes, TUB1 and TUB8, of Arabidopsis exhibit largely nonoverlapping patterns of expression. Plant Mol Biol. 1998;37:785–790. doi: 10.1023/a:1006047129410. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Sullivan KF. Molecular biology and genetics of tubulin. Annu Rev Biochem. 1985;54:331–365. doi: 10.1146/annurev.bi.54.070185.001555. [DOI] [PubMed] [Google Scholar]
- Cyr RJ, Palevitz BA. Organization of cortical microtubules in plant cells. Curr Opin Cell Biol. 1995;7:65–71. doi: 10.1016/0955-0674(95)80046-8. [DOI] [PubMed] [Google Scholar]
- Delmer DP, Amor Y. Cellulose biosynthesis. Plant Cell. 1995;7:987–1000. doi: 10.1105/tpc.7.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DC, Seagull RW, Triplett BA. Changes in the accumulation of α- and β-tubulin isotypes during cotton fiber development. Plant Physiol. 1994;105:1347–1353. doi: 10.1104/pp.105.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DD, Cyr RJ. Extending the microtubule/microfibril paradigm. Plant Physiol. 1998;116:1043–1051. doi: 10.1104/pp.116.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertel ET, Green PB. Cell growth pattern and wall microfibrillar arrangement. Plant Physiol. 1977;60:247–254. doi: 10.1104/pp.60.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Staehelin LA. Microtubule-mediated control of microfibril deposition: a re-examination of the hypothesis. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. London: Academic Press; 1991. pp. 85–99. [Google Scholar]
- Goldring JPD, Ravaioli L. Solubilization of protein-dye complexes on nitrocellulose to quantify proteins spectrophotometrically. Anal Biochem. 1996;242:197–201. doi: 10.1006/abio.1996.0453. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Haas N, Hunsperger J, Larkin J, Snustad DP, Silflow CD. The β-tubulin gene family in Zea mays: two differentially expressed β-tubulin genes. Plant Mol Biol. 1990;15:957–972. doi: 10.1007/BF00039438. [DOI] [PubMed] [Google Scholar]
- Joyce CM, Villemur R, Snustad DP, Silflow CD. Tubulin gene expression in maize (Zea mays L.): change in isotype expression along the developmental axis of seedling root. J Mol Biol. 1992;227:97–107. doi: 10.1016/0022-2836(92)90684-c. [DOI] [PubMed] [Google Scholar]
- Kevil CG, Walsh L, Laroux FS, Kalogeris T, Grisham MB, Alexander JS. An improved, rapid northern protocol. Biochem Biophys Res Commun. 1997;238:277–279. doi: 10.1006/bbrc.1997.7284. [DOI] [PubMed] [Google Scholar]
- Kloth RH. Changes in the level of tubulin subunits during development of cotton (Gossypium hisutum) fiber. Physiol Plant. 1989;76:37–41. [Google Scholar]
- Kopczak SD, Haas NA, Hussey PJ, Silflow CD, Snustad DP. The small genome of Arabidopsis thaliana contains at least six expressed α-tubulin genes. Plant Cell. 1992;4:539–547. doi: 10.1105/tpc.4.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu W-M, Cao X-L, Wilson TJ, Snustad DP, Chua N-H. Phytochrome A and phytochrome B mediate the hypocotyl-specific down-regulation of TUB1 by light in Arabidopsis. Plant Cell. 1995;7:2187–2196. doi: 10.1105/tpc.7.12.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Joshi HC, Wilson TJ, Silflow CD, Palevitz BA, Snustad DP. γ-Tubulin in Arabidopsis: gene sequence, immunoblot, and immunofluorescence studies. Plant Cell. 1994;6:303–314. doi: 10.1105/tpc.6.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP. Characterization of the α-tubulin gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1987;84:5833–5837. doi: 10.1073/pnas.84.16.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Oppenheimer DG, Silflow CD, Snustad DP. The α-1 tubulin gene family of Arabidopsis thaliana: primary structure and preferential expression in flowers. Plant Mol Biol. 1988;120:311–321. doi: 10.1007/BF00029881. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Puigdomènech P, Rigau J. The Tubα3 gene from Zea mays: structure and expression in dividing plant tissues. Gene. 1990;94:201–207. doi: 10.1016/0378-1119(90)90388-8. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènech P. A tandem of α-tubulin genes preferentially expressed in radicular tissues from Zea mays. Plant Mol Biol. 1989;193:427–438. doi: 10.1007/BF00015650. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Rigau J, Puigdomènech P. Analysis by PCR of the number of homologous genomic sequences to α-tubulin in maize. Plant Sci. 1992;84:179–185. [Google Scholar]
- Oakley CE, Oakley BR. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Oppenheimer DG, Haas N, Silflow CD, Snustad DP. The β-tubulin family of Arabidopsis thaliana: preferential accumulation of the β1 transcript in roots. Gene. 1988;63:87–102. doi: 10.1016/0378-1119(88)90548-3. [DOI] [PubMed] [Google Scholar]
- Raff EC, Fackenthal JD, Hutchens JA, Hoyle HD, Turner FR. Microtubule architecture specified by a β-tubulin isoform. Science. 1997;275:70–73. doi: 10.1126/science.275.5296.70. [DOI] [PubMed] [Google Scholar]
- Ryser U. Cell wall biosynthesis in differentiating cotton fibers. Eur J Cell Biol. 1985;39:236–256. [Google Scholar]
- Schubert AM, Benedict CR, Berlin JD, Kohel RJ. Cotton fiber developmental kinetics of cell elongation and secondary wall thickening. Crop Sci. 1973;13:704–709. [Google Scholar]
- Seagull RW. Changes in microtubule organization and wall microfibril orientation during in vitro cotton fiber development: an immunofluorescent study. Can J Bot. 1986;64:1373–1381. [Google Scholar]
- Seagull RW. Tip growth and transition to secondary wall synthesis in developing cotton hairs. In: Heath IB, editor. Tip Growth in Plant and Fungal Cells. San Diego: Academic Press; 1990. pp. 261–284. [Google Scholar]
- Seagull RW. A quantitative electron microscopic study of changes in microtubule arrays and wall microfibril orientation during in vitro cotton fiber development. J Cell Sci. 1992;101:561–577. [Google Scholar]
- Seagull RW. Cytoskeletal involvement in cotton fiber growth and development. Micron. 1993;24:643–660. [Google Scholar]
- Shimizu Y, Aotsuka S, Hasegawa O, Kawada T, Sakuno T, Sakai F, Hayashi T. Changes in levels of mRNAs for cell wall-related enzymes in growing cotton fiber cells. Plant Cell Physiol. 1997;38:375–378. doi: 10.1093/oxfordjournals.pcp.a029178. [DOI] [PubMed] [Google Scholar]
- Silflow CD, Oppenheimer DG, Kopczak SD, Ploense SE, Ludwig SR, Haas NA, Snustad DP. Plant tubulin genes: structure and differential expression during development. Dev Genet. 1987;8:435–460. [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA. Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol. 1998;116:1539–1549. doi: 10.1104/pp.116.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis thaliana contains at least nine expressed β-tubulin genes. Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SC, Wilkins TA. Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Bot. 1995;73:746–757. [Google Scholar]
- Updegraff DM. Semi-micro determination of cellulose in biological materials. Anal Biochem. 1969;32:420–424. doi: 10.1016/s0003-2697(69)80009-6. [DOI] [PubMed] [Google Scholar]
- Uribe X, Torres MA, Capellades M, Puigdomènech P, Rigau J. Maize α-tubulin genes are expressed according to specific patterns of cell differentiation. Plant Mol Biol. 1998;37:1067–1078. doi: 10.1023/a:1006067710312. [DOI] [PubMed] [Google Scholar]
- Villemur R, Joyce CM, Haas NA, Goddard RH, Kopczak SD, Hussey PJ, Snustad DP, Silflow CD. α-Tubulin gene family of maize (Zea mays L.): evidence for two ancient α-tubulin genes in plants. J Mol Biol. 1992;227:81–96. doi: 10.1016/0022-2836(92)90683-b. [DOI] [PubMed] [Google Scholar]
- Villemur R, Haas NA, Joyce CM, Snustad DP, Silflow CD. Characterization of four new β-tubulin genes and their expression during male flower development in maize (Zea mays L.) Plant Mol Biol. 1994;24:295–315. doi: 10.1007/BF00020169. [DOI] [PubMed] [Google Scholar]
- Williamson RE. Orientation of cortical microtubules in interphase plant cells. Int Rev Cytol. 1991;129:135–206. [Google Scholar]