Abstract
Ryanodine receptors (RyRs) are ubiquitous intracellular calcium (Ca2+) release channels required for the function of many organs including heart and skeletal muscle, synaptic transmission in the brain, pancreatic beta cell function, and vascular tone. In disease, defective function of RyRs due either to stress (hyperadrenergic and/or oxidative overload) or genetic mutations can render the channels leaky to Ca2+ and promote defective disease-causing signals as observed in heat failure, muscular dystrophy, diabetes mellitus, and neurodegerative disease. RyRs are massive structures comprising the largest known ion channel-bearing macromolecular complex and exceeding 3 million Daltons in molecular weight. RyRs mediate the rapid release of Ca2+ from the endoplasmic/sarcoplasmic reticulum (ER/SR) to stimulate cellular functions through Ca2+-dependent processes. Recent advances in single-particle cryogenic electron microscopy (cryo-EM) have enabled the determination of atomic-level structures for RyR for the first time. These structures have illuminated the mechanisms by which these critical ion channels function and interact with regulatory ligands. In the present chapter we discuss the structure, functional elements, gating and activation mechanisms of RyRs in normal and disease states.
Keywords: Ryanodine receptor (RyR), Calcium release channel, Cryo-EM, Endoplasmic reticulum, Sarcoplasmic reticulum
11.1 Ryanodine Receptors: Physiology and Function
Ryanodine receptors (RyRs) control calcium (Ca2+) release from intracellular endoplasmic/sarcoplasmic reticulum (ER/SR) stores (Santulli et al. 2017b). RyRs are found ubiquitously in mammals, and three different variants (RyR1, RyR2, and RyR3) are known. RyR1 is the major form in skeletal muscle (Inui et al. 1987; Takeshima et al. 1989; Marks et al. 1989); RyR2 is the major form in cardiac muscle (Nakai et al. 1990; Otsu et al. 1990; Brillantes et al. 1992), and is also involved in cognitive function (Liu et al. 2012) and in insulin secretion from pancreatic beta cells (Santulli et al. 2015a); RyR3, originally identified in the brain (Nakashima et al. 1997; Hakamata et al. 1992), is widely expressed (Zhang et al. 2011; Lanner et al. 2010) but its function is less well understood.
In muscle cells RyRs are expressed early in development (Rosemblit et al. 1999; Brillantes et al. 1994b), and their expression is regulated by stress signals (Maki et al. 1996). They are required for excitation-contraction coupling, which translates an electrical signal into a mechanical output via the second messenger Ca2+ (Santulli and Marks 2015). The closely related inositol 1,4,5-trisphosphate receptors (IP3Rs) are also found in most cell types and require the second messenger IP3 for activation (Harnick et al. 1995; Yuan et al. 2016; Santulli et al. 2017a). In skeletal muscle there is a mechanical interaction between RyR1 on the sarcoplasmic reticulum (SR) membrane and the dihydropyridine receptor (Cav1.1) on specialized invaginations of the sarcolemma, called transverse tubules, leading to rapid Ca2+ release (Rios and Brum 1987; Nelson et al. 2013; Santulli et al. 2017b). In cardiac muscle the depolarization of the plasma membrane activates Ca2+ influx via the L-type Ca2+ channel (LCC, Cav1.2), which in turn activates RyR2 via Ca2+-induced Ca2+ release (Fabiato and Fabiato 1975; Santulli et al. 2017b). At low cytosolic Ca2+ (∼100–200 nM), RyR channels are closed, with extremely low open probability (Po). The Po increases at submicromolar levels of cytosolic Ca2+, reaching a maximal Po at ∼10 μM. Cytosolic Ca2+ concentrations above ∼10 μM lower the Po (Bezprozvanny et al. 1991). This behavior points to the presence of two Ca2+ binding sites with different affinities, a moderate-affinity (μM KD) Ca2+ activation binding site (des Georges et al. 2016) and a lower-affinity (∼100 μM KD) Ca2+-dependent inhibition site.
As Ca2+ is involved in many cell signaling pathways of critical importance, mutations affecting the proper function and regulation of the Ca2+-release channel RyR can have severe impact on cellular function. In particular, RyRs are crucial for muscle function, and mutations in the receptor can lead to a number of heart and skeletal muscle pathologies. For instance, mutations in RyR1 are implicated in malignant hyperthermia (MH) and central core disease (CCD) (Quane et al. 1993; Santulli et al. 2017b), while mutations in RyR2 can lead to cardiac arrhythmias (Vest et al. 2005; Xie et al. 2015; Lehnart et al. 2004; Marks et al. 2002; Xie et al. 2013). Understanding the structure of these channels and the mechanism by which they function may enable the design of therapeutics to stabilize damaged RyRs in order to restore normal function.
11.2 Early Attempts to Determine RyR Structure
The gigantic size and large variability of the RyR structure have hindered attempts to obtain highly ordered crystals amenable to X-ray crystallography. Only the structures of individual domains were solved to high resolution: a 559-residue fraction of the N-terminal domain (Lobo et al. 2009), representing 11% of the sequence of the entire channel, and the SPRY1 and tandem-repeat domains (Yuchi et al. 2015). However, the best crystals of the channel in its entirety, obtained in a collaboration between the Marks and Hendrickson labs, did not diffract beyond a resolution of 7 Å (Clarke and Hendrickson, personal communication).
For very large molecules such as RyR, several advantages of the single-particle cryogenic electron microscopy (cryo-EM) approach (Frank 2016) over X-ray crystallography come to bear (also see separate section below): first, high-resolution 3D information can be obtained from the 2D projection images of the molecule flash-frozen in solution, obviating the need for ordered crystals; and second, structural heterogeneity inherent to the function of the molecule may be studied as multiple conformers can be retrieved through the use of image classification.
These aspects prompted us early on, in collaboration with Sidney Fleischer at Vanderbilt University, to try using the single-particle electron microscopy technique on detergent-solubilized RyR, resulting in first image averages of the negatively stained channel in the top view (Saito et al. 1988). Soon after that, first three-dimensional reconstructions of the stained (Wagenknecht et al. 1989) and unstained, frozen-hydrated RyR1 (Radermacher et al. 1992 Radermacher et al. 1994; Wagenknecht and Radermacher 1995) (Fig. 11.1) and RyR2 (Sharma et al. 1998) channels were obtained. The resolution of these first reconstructions was quite limited, but they revealed the overall fourfold symmetric (C4) mushroom-shaped architecture, with the four copies of a large cytosolic domain forming an umbrella, and a small cylindric transmembrane domain containing the pore. Each monomer, or “wing” of the cytosolic umbrella domain, was seen to be perforated by large cavities.
Fig. 11.1. First cryo-EM reconstruction of the RyR1 Ca2+-release channel (top and side view.
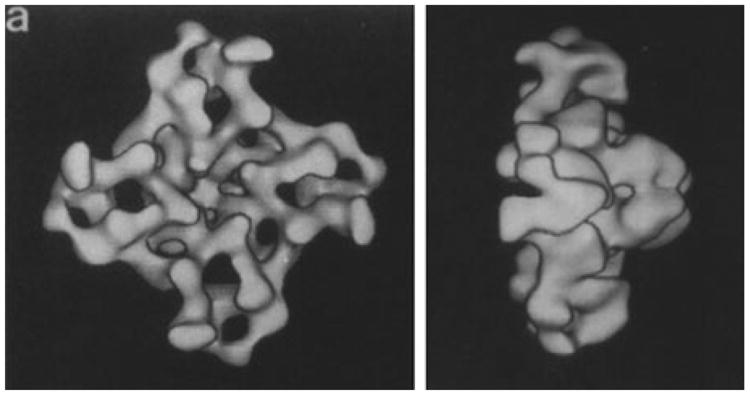
The RyR1 channel was reconstructed from ∼500 particles at 30 Å resolution (Reproduced with permission from Radermacher et al. 1994)
Even as the performance of electron microscopes and the sophistication of image processing programs improved, the resolution of RyR density maps did not keep pace for several years, for an important technical reason: reconstructions were limited by the severe orientation bias as RyR molecules prefer to lie on the carbon-coated grid in their C4-symmetry view, face-up or face-down. In such a situation, the reconstruction is strongly distorted as the resolution is anisotropic, producing biased and misleading representations of the structure. Still, significant progress was achieved in identifying locations of critical effector binding sites on the molecule (see review by Hamilton and Serysheva 2009). The problem of strong view preference of the molecule was finally resolved by preparing the EM grids with the water layer freely suspended over holes, yielding first reconstructions of the closed RyR1 channel with isotropic resolution (Samsò et al. 2005). Details regarding the issues related to view preference of RyR have been reviewed in Baker et al. (2015).
However, until recently, there still existed a fundamental obstacle preventing resolutions better than 5 Å to be reached by single-particle cryo-EM for any structures except for those with very high symmetry, such as icosahedral viruses. The obstacle was presented by the suboptimal performance of recording media in the electron microscope. Density maps of RyR1 with best resolutions in the 10 Å range were interpreted with “helix hunter” programs (Jiang et al. 2007; Baker et al. 2007) that attempt to place helices and other secondary structure elements into the reconstructed density map, based on sequence information and low-resolution localizations of binding sites. On this basis, first tentative all-atomic structures were built for the RyR1 channel in its entirety or for the functionally most relevant, pore-forming region (Ludtke et al. 2005; Samsò et al. 2009). In order to map the low-resolution density map to the sequence of the molecule, Liu and co-workers developed a method of genetic GFP insertion and cryo-EM difference mapping, first applied to RyR2 (Liu et al. 2004). Even at these low resolutions, the masses of density due to insertion of GFP become visible in the difference map as a signal with high statistical significance. This strategy allowed the approximate positions of divergent regions of RyR to be determined.
In 2012 the first direct electron detectors came on the market, which have signal-to- noise ratios and modulation transfer functions superior to those of film and much superior to the only online recording medium then available, the CCD camera. The introduction of these new cameras into electron microscopy spurred an avalanche of molecular structures with resolutions ranging from 2 to 4 Å, allowing chain tracing, sequence-guided modeling and, in the 2–3 Å range, even ab initio modeling. In 2015, the first atomic structures of RyR1 in its closed state were published by three groups, all taking advantage of this new data collection technology (Efremov et al. 2015; Yan et al. 2015; Zalk et al. 2015). Meanwhile several additional structures in different states have been described for RyR1 (des Georges et al. 2016; Wei et al. 2016; Bai et al. 2016) and for RyR2 (Peng et al. 2016). The vast increase of knowledge presented by the cryo-EM studies in the past 2 years will be illuminated in the following sections of this review, and its implications will be discussed in the context of human disease. In particular, the newest structures offer a first glimpse on the dynamic mechanism of channel activation and gating (see Van Petegem 2016; Clarke and Hendrickson 2016).
11.3 A Primer on Single-Particle Cryo-EM
11.3.1 Method of Sample Preparation and Reconstruction
Cryo-EM is a technique of imaging in which a frozen-hydrated sample is visualized in the transmission electron microscope (Dubochet et al. 1981, 1986). Contrast is produced by the difference in elastic scattering (i.e., scattering without loss of energy) between protein (or nucleic acids when applicable) and the surrounding matrix of ice. A frozen-hydrated sample is obtained by quick freeze-plunging of the EM grid (a round 3 mm-diameter copper or gold grid), on which the liquid sample has been deposited, into liquid ethane that is kept at liquid nitrogen temperature. Blotting of excess liquid ensures that the thickness of the aqueous sample will be in the range of thicknesses (∼100 nm) that can be penetrated by the 200-kV or 300-kV electron beam of commercial transmission electron microscopes. Both the high speed of freeze-plunging and the low heat capacity of the grid ensure rapid temperature change of the sample (estimated as 105 degrees Celsius/s). Under these conditions water turns into vitreous (non-crystalline) ice, that is, ice possessing very similar density and amorphous structure as liquid water. The resulting embedment of the molecule in vitreous ice is essential for preserving its native structure. Preparation of samples with consistent ice thickness in the correct range is made possible by “vitrification robots,”, which contain an environmental chamber for control of temperature and humidity and have provisions for automated blotting and rapid plunging of the grid.
The single-particle approach in cryo-EM (see Frank et al. 2002; Frank 2016; Nogales 2016) is distinct from approaches involving crystals (with two-dimensional or helical symmetry) in that the molecules to be visualized are suspended freely in the aqueous buffer, without contacts with neighboring molecules. Thus, there are no constraints on the conformations they can assume, and, provided that the processes of purification and freezing themselves do not significantly alter their structure, the molecules are depicted in their native range of conformations and binding states.
The molecules are visualized as 2D projection images, which carry both amplitude and phase information in the Fourier domain and thus, unlike in X-ray crystallography, obviate the need for phasing. High-resolution 3D information can then be retrieved by alignment of these projections into a common 3D reference frame. The single-particle approach requires, however, that the molecules assume all possible views without major gaps in orientation distribution, as a precondition for artifact-free three-dimensional reconstruction. As pointed out above, many of the early structural studies of the RyR were adversely affected by strong preference of molecules for the top view, rendering the reconstructions difficult to interpret.
After the grid is plunged into the cryogen, it is transferred into the electron microscope, being kept under liquid nitrogen temperature at all times. In the imaging experiment, a beam with low electron dose (in our experiments 25–80e−/Å2) is employed to prevent radiation damage, but at such doses the images are very noisy. The need for collecting large numbers of images, often going in the hundreds of thousands, is due to four reasons: (1) the very low signal-to-noise ratio of the images, (2) beam-induced movement, charging of the specimen and other blurring effects, (3) the need to cover the entire angular range, and (4) the need to sample all major conformations co-existing in the specimen.
The recording of micrographs has been revolutionized recently with the introduction of direct electron detection cameras that possess high signal-to-noise ratio and a modulation transfer function extending to high spatial frequencies (i.e., small spacings in the image) (McMullan et al. 2009; McMullan et al. 2014). Micrographs are recorded as stacks of frames (“movies”), enabling correction of drift and electron beam-induced movement (Brilot et al. 2012; Li et al. 2013) and the weighting of frames according to accumulated electron dose (Grant and Grigorieff 2015). After this pre-processing step the images are sorted, or classified, using maximum-likelihood programs such as RELION (Scheres 2012). Reconstructions from the resulting classes display the whole range of structures co-existing in the sample.
11.4 Structures of the Channel Obtained by High-Resolution Cryo-EM
The above-mentioned advances in single-particle cryo-EM have spurred a revolution in structural biology, enabling the reconstruction of atomic-resolution maps for membrane proteins (Liao et al. 2013). This development has allowed the determination of high-resolution structures of RyRs, for the first time providing a mechanistic understanding of how the channel functions in healthy tissue and giving clues as to how it may be affected in disease states.
As mentioned above, the structure of RyR1 in a closed state, without Ca2+ or other activating ligands, was determined in three recent studies at resolutions ranging from 3.8 Å to 6.1 Å and allowed the building of an atomic-level model of the RyR channel core (Yan et al. 2015) and poly-alanine-level models of the RyR cyto-plasmic shell (Zalk et al. 2015; Yan et al. 2015; Efremov et al. 2015). These structures gave us a better understanding of the overall architecture of RYRs, homotetramer of four 5037-residue protomers with a combined mass of 2.2 mega-Daltons, and divided between pore, activation domain and cytoplasmic shell.
The structures confirmed that the pore domain belongs to the 6TM family of ion channels (Hille 2001) and contains a pseudo-voltage sensor domain (pVSD). RyR has unique features compared to other 6TM-family ion channels, which includes an insertion between S2 and S3 helices (S2S3 domain), a 90 Å long pore helix ending with a C-terminal domain containing a zinc finger and an acidic disordered loop between S1 and S2 (Yan et al. 2015; Fan et al. 2015). A polyproline segment in the S2S3 domain may be a binding site for SH3 domain-containing proteins (des Georges et al. 2016), which by their interaction with this extension of the pVSD may regulate channel function.
A notable feature of the activation domain is that it contains a domain in the shape of a thumb and forefingers (TaF), which clamps the zinc finger-containing C-terminal domain (CTD) (Fig. 11.2), thereby providing the allosteric coupling between the pore and the shell of the receptor. Presence of an EF-hand motif in the activation domain interacting with the pVSD hinted at a putative mechanism of activation by Ca2+ (Wei et al. 2016). This putative mechanism later proved not to be correct (Guo et al. 2016; des Georges 2016), but this interaction between the EF-hand and the pVSD may have an important functional role, which remains to be determined.
Fig. 11.2. Architecture of the ryanodine receptor.
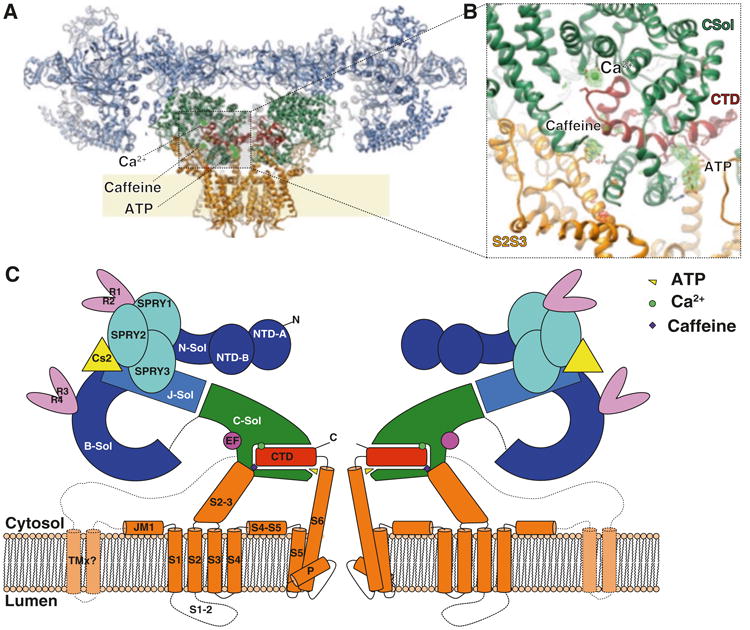
(a) Ribbon representation, with the shell (residues 1–3666; light blue), core solenoid (residues 3667–4253; green), pore domain (residues 4540–4956; orange), and CTD (residues 4957–5037; dark red). (b) Zoom onto the CTD area showing ligands and locally aligned difference density map calculated between the EGTA-only map and Ca2+, ATP, and caffeine map (green/red mesh), contoured at 5s. (c) Schematic representation of RyR1. J-sol, junctional solenoid; B-sol, bridge solenoid; C-sol, core solenoid; N-sol, N-terminus solenoid. TMx, putative additional transmembrane helices (Reproduced with permission from Zalk et al. 2015)
The shell of RyR is principally composed of three α-solenoid repeats. The core solenoid (CSol), which is part of the activation domain, links the pore domain to the shell. The other α-solenoid repeats are the bridging (BSol), and N-terminal (NTD-C), that are joined by a fourth junctional solenoid (JSol) which provides a bridge between all the other ones (des Georges et al. 2016). These solenoids incorporate small folds of other domains, yielding numerous interdomain interfaces in the cytosolic shell. Other domains in the cytosolic shell include the SPRY domains (SPRY1-SPRY3) (Lau and Van Petegem 2014), EF-hands (EF1&2) (Xiong et al. 2006), RyR repeat pairs (RY1&2 and RY3&4) (Yuchi et al. 2015; Sharma et al. 2012), and the N-terminal domains (NTD-A and NTD-B) (Amador et al. 2009).
Three major interfaces provide inter-protomer contacts: SPRY2 with Bsol, NTD-A with NTD-B, and NTD-A with BSol (where the second domain is on the adjacent protomer). These interdomain and interprotomer contacts have been implicated in channel gating (Tung et al. 2010), linking the control of shell dynamics with pore opening. The picture emerging is of the RyR as a large complex network of interconnected helices (Fig. 11.2), which enables allosteric coupling of conformational changes over the span of the entire channel. Many proteins interact with the large cytoplasmic shell and take advantage of this allosteric coupling to allow a range of stimuli to exert control over the channel function (Lanner et al. 2010; Van Petegem 2015).
Important keys for understanding this allosteric coupling between the shell and pore of the channel comes from a most recent study showing a series of structures, and their conformational variabilities, which change upon binding with different combinations of ligands (des Georges et al. 2016). These results have provided the important insight that there are in fact different modes of mechanical coupling, and that these modes are switched upon binding of activating ligands (Clarke and Hendrickson 2016).
11.5 RyR Activation: A Close Look
RyR contains multiple domains forming binding sites for functional ligands, including ions (primarily Ca2+ and Mg2+), proteins including calstabin, calmodulin, the voltage-gated Ca2+ channel Cav and small molecules such as adenine nucleotides, and caffeine (des Georges et al. 2016; Hwang et al. 2012; Choi et al. 2017; Santulli et al. 2017b). These ligands can bias the channel towards specific functional states and Po. Most importantly, RyR gating is regulated by Ca2+ with a characteristic bell-shaped curve (Bezprozvanny et al. 1991), suggesting that RyR must have two Ca2+ binding sites of differing activities. ATP is also a well characterized physiological activating ligand likely to be constitutively bound in physiological conditions since cellular ATP concentration in skeletal muscles is very high (∼5 mM; Kushmerick et al. 1992). Ca2+ and ATP are known to have synergistic effect on RyR gating, with the highest Po when both are present. Although probably not physiological, addition of caffeine increases the Po to an even greater extent (Po = 0.91; des Georges et al. 2016).
It is a striking observation that those three synergistic activating ligands bind at the CTD interdomain interface (des Georges et al. 2016; Fig. 11.2), which is itself directly linked to the pore helix via its Zinc-finger domain. This finding together with the observation that both Ca2+ and ATP/caffeine produce the same or very similar changes in the position of the core solenoid with respect to the CTD (des Georges et al. 2016) can explain their synergistic effect. It also explains why caffeine potentiates RyR Ca2+ binding. As caffeine and Ca2+ binding produce a similar conformational change in the region of the CTD, caffeine acts as an allosteric agonist of Ca2+, thereby shifting the sensitivity to Ca2+ towards lower concentrations (Porta et al. 2011).
The ATP binding site is at the interface formed by the C-terminal portion of the TaF and the S6c-CTD junction (Fig. 11.2). The adenine moiety binds in the T intersection formed by the pore helix and the CTD, next to the Zinc finger motif, and may play a similar structural role, maintaining the geometry of the junction. This would explain why adenine nucleotides are competitive partial agonists of RyR1 while adenosine is an antagonist (Chan et al. 2000; Laver et al. 2001). Other nucleotide phosphates have been shown to not activate RyR.
The triphosphate tail of ATP binds to positively charged residues on the TaF domain of the core solenoid. ATP thereby acts as a bridge between the CTD and the TaF domain, triggering the conformational change between those two domains necessary for gating. As the number of phosphate groups is reduced, the interaction with the TaF positive charges is weakened or abolished and no longer bridges the TaF to the CTD. This observation explains the difference in affinity and channel Po between ATP, ADP and AMP, and may provide a mechanism contributing to muscle fatigue (Rullman et al. 2013; Bellinger et al. 2008a; Chan et al. 2000).
This change in the conformation of the core solenoid with respect to the CTD upon Ca2+ and ATP/caffeine binding activates the channel and allows the lateral displacement of the whole protomer as a rigid unit, resulting in opening of the pore. The fact that ligand binding restricts the range of observed positions of the cytoplasmic shell (des Georges et al. 2016; Fig. 11.3) implies that this observed ligand-dependent conformational change of the core solenoid renders the whole protomer assembly more rigid. This change appears to allow pore opening, but the exact gating mechanism is still unknown. We can postulate two possible and non-exclusive mechanisms: (1) the more rigid assembly allows coupling of the thermal movement of the shell to bending of the S6 pore helix; (2) the conformational change releases a still unknown locking interaction, which allows the outward movement of the protomer and pore opening.
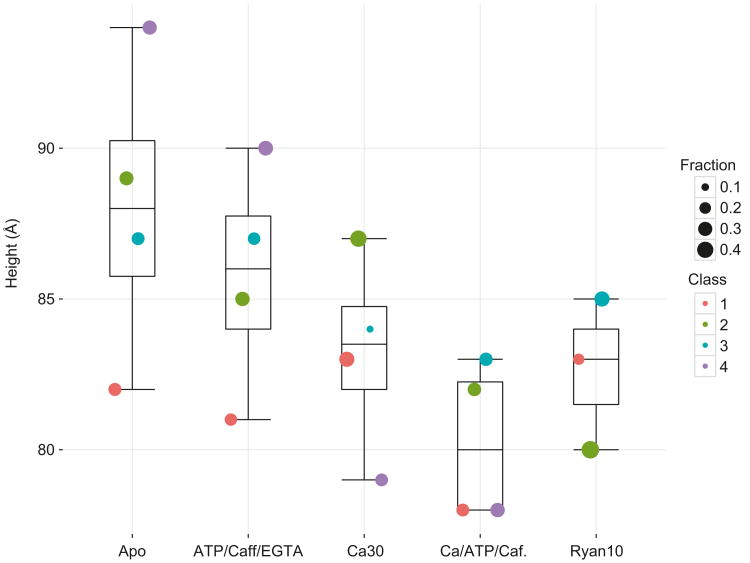
Fig. 11.3. Ligand-dependent change in cytoplasmic shell motions amplitude.
Box and whisker plot of the height measurement of the RY1&2 domain with respect to the SR membrane plane for each class of each dataset. The fraction of particles of each class is represented by the size of the circular marker for each dataset; the class number is given by the color of the marker (Reproduced with permission from des Georges et al. 2016)
The key to the gating mechanism may potentially lie in the pseudo-voltage sensor (pVSD), a domain whose function in RyR is still not well understood. It has been shown that voltage potential on the SR membrane has an effect on RyR gating (Endo 2009), which would imply that the pVSD is playing a role in gating, despite the domain lacking most of the voltage-sensing charges on S4. The S2 helix and RyR-specific S2-S3 domain of the pVSD, together with the EF1/EF2 domain have also been implicated in Ca2+-dependent inactivation of RyR (Gomez and Yamaguchi 2014; Gomez et al. 2016). It has been observed that the EF hand domain comes closer to the S2-S3 domain upon channel opening (des Georges et al. 2016; Wei et al. 2016). A tentative hypothesis could be that low affinity Ca2+-binding on one of those domains triggers a repulsion, pushing those two domains apart and thereby preventing channel opening.
RyRs have a high degree of sequence and structure homology with another intracellular Ca2+ release channel, the IP3R (Santulli et al. 2017a). IP3Rs are found in the ER and mediate the release of intracellular Ca2+ in response to external stimuli. RyRs and IP3Rs have ∼40% homology in the core regions, indicating a common origin for Ca2+ release channels (Santulli et al. 2017a). However, the IP3R does not include much of the cytoplasmic shell and, most notably, lacks the large Bsol (Santulli et al. 2017a). Of particular interest, the extended pore helix, CTD and activation domain are conserved between these two channels, meaning that they have the same or very similar mechanisms of activation by Ca2+ (Santulli et al. 2017a). In IP3R, the CTD is further extended and directly links to the NTD and the IP3 binding site (Fan et al. 2015). Conformational changes induced by IP3 can therefore be directly linked or relayed to Ca2+ binding and gating (Fig. 11.4). In the case of RyR, gating is linked to mechanical coupling of the shell and pore, in IP3R, gating is probably coupled to IP3 binding by a very similar mechanism.
Fig. 11.4. Similarities in allosteric coupling and gating in RyR and IP3R.
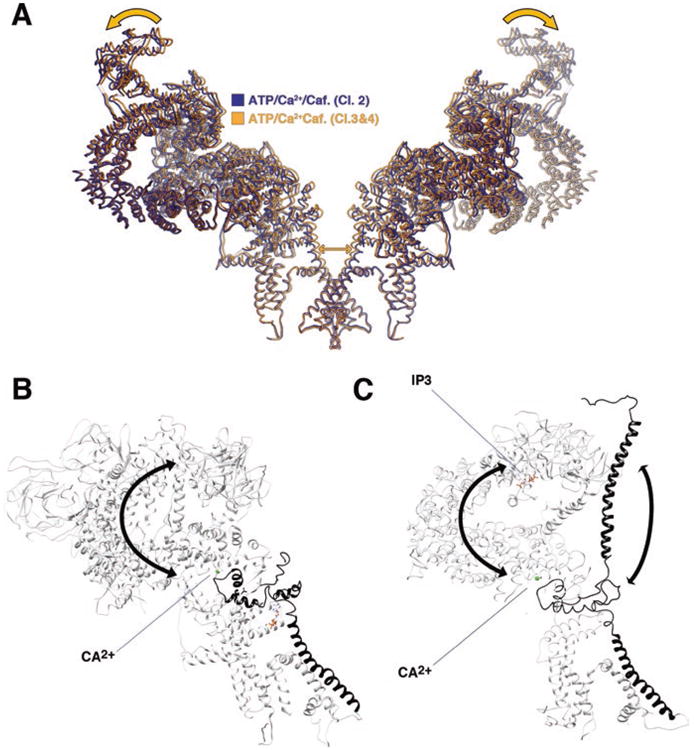
(a) RyR gating is linked to an outwards rigid body motion of opposing protomers resulting in opening of the pore (orange arrows). The ligand-bound closed and open classes are shown in yellow and blue. (b, c) Allosteric coupling between RyR (b) and IP3R (c) cytoplasmic shell and pore helix via the C-terminal domain. Pore helix and CTD are in black. The rest of the molecule is white. Ca2+ (green dot) and IP3 (represented in sticks) are shown in their know binding site. Arrows show possible paths of allosteric coupling
11.6 Mapping of Channel Dynamics by Cryo-EM
RyR gating is triggered by Ca2+-induced conformational changes in the core solenoid and CTD that couple cytoplasmic shell movement to pore opening (des Georges et al. 2016). The cytoplasmic shell dynamics therefore plays a direct role in channel gating by Ca2+, and must play an integral role in Ca2+ release triggered by mechanical coupling with the dihydropyridine receptor. The channel dynamics is also tightly regulated by numerous factors that influence gating including calstabin and calmodulin. It is therefore important to study the complex dynamics of this very large ion channel in order to better understand its function and regulation. Cryo-EM is uniquely placed to do this, as the single-particle method allows the sorting of individual macromolecules based on their conformational state.
In order to appreciate the cryo-EM data analysis for a molecule exhibiting dynamic behavior in the sample, it is important to understand that maximum-likelihood classification by programs such as RELION (Scheres 2012) entails a user decision on how many discrete classes (K) the algorithm should distinguish. In the case of RyR1, for any choice of K, multiple classes with different conformations are found, distinguished by the position of the cytosolic wing domains (des Georges et al. 2016), suggesting that the structure of the isolated receptor is fluctuating as a result of Brownian motion. Binding of ligands (Ca2+, caffeine/ATP, or Ca2+ + caffeine/ATP) changes the range and distribution of cytosolic domain positions found and, as pointed out before, the preference for closed versus open channel. Since increasing K, the number of classes, leads to an ever-increasing number of reconstructions with ever-finer increments in conformational changes, it is safe to say that the discrete classes found are samples from an underlying continuum – reflecting variability of the large molecule that results from being exposed to thermal agitation in the surrounding solvent. (It should be pointed out in this context that the movies showing transitions between the open and closed channel in the work by des Georges et al. 2016 were obtained by morphing between two to four structures, without support by data in between).
The analysis using discrete classes described in des Georges et al. (2016) (classification voluntarily limited to K = 4 with fourfold symmetry imposed in order to reduce the complexity of the analysis), unveiled the conformational changes necessary for gating and the conformation of the open state of the channel. It also showed that the height of the cytoplasmic shell with respect to the membrane plane both lowers and reduces its range of attainable conformations with ligand binding (Fig. 11.3). Activating ligands therefore reinforce the mechanical coupling between the pore and the shell, which must be an integral part of the gating mechanism. Still, even with caffeine/ATP and Ca2+ bound, the analysis shows that the cytoplasmic shell has an important level of intrinsic flexibility not well depicted by the small number of discrete classes used in the analysis.
The continuous character of the distribution of states is in fact borne out by the application of a new kind of analysis that allows continuous ordering of data due to conformational changes to be tracked (Dashti et al. 2014; Frank and Ourmazd 2016). When this method is applied to the data collected in the study of RyR1 bound with various combinations of ligands (des Georges et al. 2016), we indeed see that the channel in each sample assumes an entire continuum of states – albeit with different distributions – exhibiting a combination of wing motions, motions of the activation domain, and channel openings and closings (Dashti et al. 2017). This analysis promises to lead to more profound insights into the nature and dynamics of channel activation and gating than obtainable from single reconstructions in open and closed states (see Van Petegem 2016). It may also be a well-suited tool to study how the numerous regulatory factors shift the equilibrium of states and modulate gating.
11.7 Modulation of RyR Dynamics and Gating
A number of factors affect RyR function in the cell, and a better understanding of their effect on RyR dynamics may be of crucial importance in understanding disease states of the channel.
The Ca2+ channel-binding and -stabilizing proteins calstabin1 and calstabin2 (also known as FKBP12 and FKBP12.6, respectively) are of critical importance for preventing RyRs from leaking Ca2+ and for coupled gating (Marx et al. 1998, 2000) between RyRs (Brillantes et al. 1994a; Timerman et al. 1993; Santulli and Marks 2015; Yuan et al. 2014). Calstabin1 and calstabin2 preferentially bind RyR1 and RyR2, respectively, and help to stabilize the closed state of the channel (Santulli and Marks 2015). Calstabins are the cytosolic targets for the immunosuppressant drugs rapamycin and FK506 (Lombardi et al. 2017a, 2017b). Rapamycin (or sirolimus) coated on coronary artery stents is used to prevent restenosis by inhibiting vascular smooth muscle migration and proliferation (Santulli 2015; Marks 2003; Santulli and Totary-Jain 2013; Santulli et al. 2014). The interaction between calstabins and RyR can regulate the channel and prevent intracellular Ca2+ leak that leads to a variety of disease states (Santulli and Marks 2015; Huang et al. 2006; Brillantes et al. 1994a). These proteins are highly specific, with one calstabin bound per RyR protomer, and have been used to isolate RyR from microsomes (Zalk et al. 2015; Xin et al. 1995). Calstabin binds at the N-terminus of the bridging solenoid, possibly rigidifying the interface between BSol and SPRY1/SPRY2, thus stabilizing the link between the pore and cytoplasmic region (des Georges et al. 2016; Zalk et al. 2015; Yan et al. 2015). Reinforcing such hypothesis, a cryo-EM study of RyR2 in the absence of calstabin shows a much greater level of flexibility of the BSol (Peng et al. 2016). This greater flexibility of the BSol may be due to the absence of calstabin, but may also be intrinsic to RyR2.
Additional modulatory proteins associate directly and indirectly with RyR, including calsequestrin, sorcin, triadin, homer, histidine-rich Ca2+ binding protein, calmodulin, S100A1, and junctin (Santulli et al. 2017b). RyR function is also regulated by post-translational modifications, including phosphorylation, oxidation and nitrosylation (Santulli et al. 2015a, b, 2017a, b; Marx et al. 2000, 2001b; Bellinger et al. 2008b, 2009; Shan et al. 2010a, b; Andersson et al. 2011; Umanskaya et al. 2014).
The majority of the known post-translational modifications and modulatory protein binding sites are situated on the cytoplasmic shell and influence the cytoplasmic shell dynamics and therefore, indirectly, gating (Gambardella et al. 2017). The shell dynamics could be influenced in several ways: The rigidity of the protomer assembly could be altered, as suggested with calstabins; the strength of interprotomer contacts could be modified, thereby influencing shell movement and gating; the shell movement could be directly constrained, which would either promote (e.g., DHPR) or prevent the outward motion necessary for pore opening (Bai et al. 2016; des Georges et al. 2016). It is also possible that other regulatory proteins influence more directly the priming of the core solenoid, which is necessary for gating to occur (des Georges et al. 2016), or influence gating more directly by interacting with the pVSD. For instance, an RyR-specific polyproline segment in the pVSD (des Georges et al. 2016) is a potential binding site for SH3 domain-containing proteins that could have a direct effect on gating.
As discussed above, the movement of the cytoplasmic shell is tightly interconnected with gating. RyRs are known to form paracrystalline arrays in checkerboard patterns (Franzini-Armstrong et al. 1999; Cabra et al. 2016). This feature provides a mechanism by which opening of a single channel can mechanically trigger neighboring channels and greatly amplify Ca2+ release via coupled gating (Marx et al. 1998). RyR repeats 1&2, located in the corners of the cytoplasmic shell, is most likely involved in mediating coupled-gating (Lehnart et al. 2003; Marx et al. 1998, 2001a). By triggering neighboring channels both physically and chemically, RyRs act as both signal amplifiers and signal integrators (Fabiato 1983; Endo et al. 1970). This supramolecular allostery and thus the whole dynamics of excitation-contraction coupling is highly sensitive to the binding of protein factors to the shell assembly.
11.8 Involvement of RyR in Human Diseases
Over 300 mutations causing human diseases have been mapped to RyRs (Santulli et al. 2017a). Mutations in the cardiac RyR2 are primarily linked to cardiac arrhythmias, including catecholaminergic polymorphic ventricular tachycardia (CPVT) (Lehnart et al. 2008; Santulli et al. 2015a), in which exercise-induced sympathetic activation in conjunction with the mutation causes ventricular tachycardia and sudden cardiac death. Importantly, patients typically do not exhibit prior symptoms or structural modification of the heart, but can experience ventricular arrhythmia and syncope during physical exercise, following stress or acute emotions. The majority of mutations linked to CPVT are located in the pore, pVSD and central domain regions (Santulli et al. 2017b), domains directly implicated in activation and gating (des Georges et al. 2016). Other hotspots are located in the NTD and BSol domains, two domains that form interprotomer contacts and therefore influence the cytoplasmic shell dynamics. The majority of those mutations link to malfunction in the gating mechanism which may be exacerbated under stress and lead to spurious Ca2+ sparks and arrhythmia.
Mutations in RyR1 can cause malignant hyperthermia (MH), and RyR1 myopathies previously classified as central core disease (CCD) and multiminicore disease (Santulli et al. 2017a). Additionally, stress-induced post-translational modifications of RyR1 including phosphorylation, oxidation, nitrosylation and depletion of calstabin1 can lead to Ca2+ leak, resulting in deleterious effects on muscle function (Matecki et al. 2016; Shan et al. 2010a, b; Lehnart et al. 2005; Bellinger et al. 2009; Fauconnier et al. 2010; Marx et al. 2000; Umanskaya et al. 2014; Santulli et al. 2017a, b). These myopathies include Duchenne muscular dystrophy (Bellinger et al. 2009), sarcoglycanopathies (Andersson et al. 2012) and sarcopenia (Andersson et al. 2011; Santulli et al. 2017a).
MH involves activation of RyR1 induced by a volatile halogenated anesthetic, which causes a rapid rise in core body temperature and can result in organ failure and death if not promptly treated (Rosenberg et al. 2007; Santulli et al. 2017a). MH is a pharmacogenetic disorder, in which the mutation is latent until it is triggered by a drug. In this condition, Ca2+ is rapidly released by RyR1, causing uncontrolled muscle contraction. Sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) hydrolyzes ATP to pump it back into the SR/ER, consuming ATP and generating the characteristic excessive heat. Although dantrolene is an established therapeutic that resolves MH episodes, mortality from this event remains at approximately 5% (Larach et al. 2014). Despite years of use and studies showing interaction with RyR1, a validated mechanism of action for dantrolene has yet to be reported (Paul-Pletzer et al. 2002; Zhao et al. 2001). A recent study demonstrated that dantrolene efficacy is also affected by the level of Mg2+ (Choi et al. 2017). Mutations causing MH are autosomal dominant and typically seen in the central and N-terminal clusters. Another MH mutation hotspot is at the interprotomer contacts between NTD-A and NTD-B, which is disrupted during channel opening (Kimlicka et al. 2013; Santulli et al. 2017a). The location of those mutations, mostly around the N-terminal region and around the Bsol, points to a misregulation of the shell dynamics that would cause uncontrolled pore opening upon the anesthetic binding to RyR1.
CCD is a congenital myopathy characterized by the presence of metabolically inactive tissue cores in muscle fibers, resulting in progressive muscle weakness (Sewry et al. 2002; Santulli et al. 2017a). CCD presents during infancy as delayed motor development and hypotonia. There are no known therapeutics for CCD and it can result in death. Intriguingly, as RyR1 is also found in the brain, one CCD mutation was also discovered to have a neuronal phenotype (De Crescenzo et al. 2012). Mutations that lead to CCD are clustered in the C-terminal region (Quane et al. 1993; Zhang et al. 1993; Santulli et al. 2017b), region directly responsible for Ca2+ activation (des Georges et al. 2016; Santulli et al. 2017a).
We can see that in each of these disorders, mutations are clustered in similar hotspots, mostly the interface between protomers (Van Petegem 2015; Tung et al. 2010; Santulli et al. 2017a), but also in the activation domain. Mutations at the interface between protomers are likely to influence gating by affecting the outward movement of the shell necessary for pore opening. Weakening of those interfaces would facilitate pore opening and therefore spurious Ca2+ leak (Tateishi et al. 2009; Suetomi et al. 2011). Mutations in the C-terminal region or the core solenoid are more likely to affect the activation mechanism directly, either by facilitating the activated conformation, the binding of the activating ligands Ca2+ and ATP, or by preventing inactivation of the channel.
One hotspot is localized in the phosphorylation domain, near the phosphorylation loop, indicating that disease mutations and phosphorylation can affect the same process (Yuchi et al. 2012; Santulli et al. 2017b). Still, the exact role of post-translational modifications on the channel are poorly understood. Cryo-EM structures of phosphorylated vs dephosphorylated channels showed no differences in the structure and dynamics of the purified receptor (Zalk et al. 2015). These modifications may only be relevant in the context of their interactions with other partners, including DHPR.
11.9 Conclusions
The recently published cryo-EM structures of RyR channels in closed and open states represent a breakthrough after years of slow progress in the structural and molecular understanding of disease-causing mutations in RyRs. The new structures have opened the door toward a detailed mechanistic understanding of their function in health and their dysfunction in disease. The improved understanding of the complex mechanisms of ligand binding and activation of the channel also presents opportunities for targeted drug developments addressing many of the known human diseases implicating RyR1 and RyR2.
Acknowledgments
This work was supported by HHMI (to J.F.) and grants from the National Institutes of Health (R01AR060037 and R01HL061503 to A.R.M., R01GM29169 to J.F., R00DK107895 to G.S.).
Footnotes
Disclosure: ARM is a consultant and board member and owns shares in ARMGO Pharma, Inc. a biotech startup targeting RyR channels for therapeutic purposes.
Contributor Information
Gaetano Santulli, The Wu Center for Molecular Cardiology, Department of Physiology and Cellular biophysics, Columbia University Medical Center, New York, NY, USA; The Wilf Family Cardiovascular Research Institute and the Einstein-Mount Sinai Diabetes Research Center, Department of Medicine, Albert Einstein College of Medicine – Montefiore University Hospital, New York, NY, USA.
Daniel Lewis, Department of Physiology and Cellular Biophysics, Columbia University, New York, NY, USA.
Amedee des Georges, Advanced Science Research Center at the Graduate Center of the City University of New York, New York, NY, USA; Department of Chemistry & Biochemistry, City College of New York, New York, NY, USA; Ph.D. Program in Biochemistry, The Graduate Center of the City University of New York, New York, NY, USA.
Andrew R. Marks, Department of Medicine, Columbia University, New York, NY, USA; Department of Physiology and Cellular Biophysics, Columbia University, New York, NY, USA
Joachim Frank, Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA; Department of Biological Sciences, Columbia University, New York, NY, USA.
References
- Amador FJ, Liu S, Ishiyama N, Plevin MJ, Wilson A, MacLennan DH, Ikura M. Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation “hot spot” loop. Proc Natl Acad Sci U S A. 2009;106(27):11040–11044. doi: 10.1073/pnas.0905186106. https://doi.org/10.1073/pnas.0905186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DC, Betzenhauser MJ, Reiken S, Meli AC, Umanskaya A, Xie W, Shiomi T, Zalk R, Lacampagne A, Marks AR. Ryanodine receptor oxidation causes intracellular calcium leak and muscle weakness in aging. Cell Metab. 2011;14(2):196–207. doi: 10.1016/j.cmet.2011.05.014. https://doi.org/10.1016/j.cmet.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson DC, Meli AC, Reiken S, Betzenhauser MJ, Umanskaya A, Shiomi T, D'Armiento J, Marks AR. Leaky ryanodine receptors in beta-sarcoglycan deficient mice: a potential common defect in muscular dystrophy. Skelet Muscle. 2012;2(1):9. doi: 10.1186/2044-5040-2-9. https://doi.org/10.1186/2044-5040-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Yan Z, Wu J, Li Z, Yan N. The central domain of RyR1 is the transducer for long-range allosteric gating of channel opening. Cell Res. 2016 doi: 10.1038/cr.2016.89. https://doi.org/10.1038/cr.2016.89. [DOI] [PMC free article] [PubMed]
- Baker MR, Fan G, Serysheva II. Single-particle cryo-EM of the ryanodine receptor channel in an aqueous environment. Eur J Transl Myol. 2015;25(1):35–48. doi: 10.4081/ejtm.2015.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007 May 15;104(20):8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger AM, Mongillo M, Marks AR. Stressed out: the skeletal muscle ryanodine receptor as a target of stress. J Clin Invest. 2008a;118(2):445–453. doi: 10.1172/JCI34006. https://doi.org/10.1172/JCI34006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger AM, Reiken S, Dura M, Murphy PW, Deng SX, Landry DW, Nieman D, Lehnart SE, Samaru M, LaCampagne A, Marks AR. Remodeling of ryanodine receptor complex causes “leaky” channels: a molecular mechanism for decreased exercise capacity. Proc Natl Acad Sci U S A. 2008b;105(6):2198–2202. doi: 10.1073/pnas.0711074105. https://doi.org/10.1073/pnas.0711074105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinger AM, Reiken S, Carlson C, Mongillo M, Liu X, Rothman L, Matecki S, Lacampagne A, Marks AR. Hypernitrosylated ryanodine receptor calcium release channels are leaky in dystrophic muscle. Nat Med. 2009;15(3):325–330. doi: 10.1038/nm.1916. https://doi.org/10.1038/nm.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5) P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351(6329):751–754. doi: 10.1038/351751a0. https://doi.org/10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- Brillantes AM, Allen P, Takahashi T, Izumo S, Marks AR. Differences in cardiac calcium release channel (ryanodine receptor) expression in myocardium from patients with end-stage heart failure caused by ischemic versus dilated cardiomyopathy. Circ Res. 1992;71(1):18–26. doi: 10.1161/01.res.71.1.18. [DOI] [PubMed] [Google Scholar]
- Brillantes AB, Ondrias K, Scott A, Kobrinsky E, Ondriasova E, Moschella MC, Jayaraman T, Landers M, Ehrlich BE, Marks AR. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994a;77(4):513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Brillantes AM, Bezprozvannaya S, Marks AR. Developmental and tissue-specific regulation of rabbit skeletal and cardiac muscle calcium channels involved in excitation-contraction coupling. Circ Res. 1994b;75(3):503–510. doi: 10.1161/01.res.75.3.503. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, Potter CS, Carragher B, Henderson R, Grigorieff N. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012 Mar;177(3):630–637. doi: 10.1016/j.jsb.2012.02.003. https://doi.org/10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabra V, Murayama T, Samsò M. Ultrastructural analysis of self-associated RyR2s. Biophys J. 2016;110(12):2651–2662. doi: 10.1016/j.bpj.2016.05.013. https://doi.org/10.1016/j.bpj.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WM, Welch W, Sitsapesan R. Structural factors that determine the ability of adenosine and related compounds to activate the cardiac ryanodine receptor. Br J Pharm. 2000;130(7):1618–1626. doi: 10.1038/sj.bjp.0703459. https://doi.org/10.1038/sj.bjp.0703459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi RH, Koenig X, Launikonis BS. Dantrolene requires Mg2+ to arrest malignant hyper-thermia. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1619835114. https://doi.org/10.1073/pnas.1619835114. [DOI] [PMC free article] [PubMed]
- Clarke OB, Hendrickson WA. Structures of the colossal RyR1 calcium release channel. Curr Opin Struct Biol. 2016 Aug;39:144–152. doi: 10.1016/j.sbi.2016.09.002. https://doi.org/10.1016/j.sbi.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A, Schwander P, Langlois R, Fung R, Li W, Hosseinizadeh A, Liao HY, Pallesen J, Sharma G, Stupina VA, Simon AE, Dinman JD, Frank J, Ourmazd A. Trajectories of the ribosome as a Brownian nanomachine. Proc Natl Acad Sci U S A. 2014 Dec 9;111(49):17492–17497. doi: 10.1073/pnas.1419276111. https://doi.org/10.1073/pnas.1419276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A, Hail DB, Mashayekhi G, Schwander P, des Georges A, Frank J, Ourmazd A. Conformational Dynamics and Energy Landscapes of Ligand Binding in RyR1. bioRxiv. 2017:167080. [Google Scholar]
- des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, Frank J. Structural basis for gating and activation of RyR1. Cell. 2016 Sep 22;167(1):145–157.e17. doi: 10.1016/j.cell.2016.08.075. https://doi.org/10.1016/j.cell.2016.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Crescenzo V, Fogarty KE, Lefkowitz JJ, Bellve KD, Zvaritch E, MacLennan DH, Walsh JV., Jr Type 1 ryanodine receptor knock-in mutation causing central core disease of skeletal muscle also displays a neuronal phenotype. Proc Natl Acad Sci U S A. 2012;109(2):610–615. doi: 10.1073/pnas.1115111108. https://doi.org/10.1073/pnas.1115111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubochet J, Booy FP, Freeman R, Jones AV, Walter CA. Low temperature electron microscopy. Annu Rev Biophys Bioeng. 1981;10:133–149. doi: 10.1146/annurev.bb.10.060181.001025. [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Schultz P, Oudet P. Cryo-electron microscopy of vitrified SV40 minichromosomes: the liquid drop model. EMBO J. 1986 Mar;5(3):519–528. doi: 10.1002/j.1460-2075.1986.tb04241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015 Jan 1;517(7532):39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- Endo M. Calcium-induced calcium release in skeletal muscle. Physiol Rev. 2009;89(4):1153–1176. doi: 10.1152/physrev.00040.2008. https://doi.org/10.1152/physrev.00040.2008. [DOI] [PubMed] [Google Scholar]
- Endo M, Tanaka M, Ogawa Y. Calcium induced release of calcium from the sarcoplasmic reticulum of skinned skeletal muscle fibres. Nature. 1970;228(5266):34–36. doi: 10.1038/228034a0. [DOI] [PubMed] [Google Scholar]
- Fabiato A. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Phys. 1983;245(1):C1–14. doi: 10.1152/ajpcell.1983.245.1.C1. [DOI] [PubMed] [Google Scholar]
- Fabiato A, Fabiato F. Contractions induced by a calcium-triggered release of calcium from the sarcoplasmic reticulum of single skinned cardiac cells. J Physiol. 1975;249(3):469–495. doi: 10.1113/jphysiol.1975.sp011026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, Ludtke SJ, Serysheva II. Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 2015;527(7578):336–341. doi: 10.1038/nature15249. https://doi.org/10.1038/nature15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauconnier J, Thireau J, Reiken S, Cassan C, Richard S, Matecki S, Marks AR, Lacampagne A. Leaky RyR2 trigger ventricular arrhythmias in Duchenne muscular dystrophy. Proc Natl Acad Sci U S A. 2010;107(4):1559–1564. doi: 10.1073/pnas.0908540107. https://doi.org/10.1073/pnas.0908540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J. Generalized single-particle cryo-EM--a historical perspective. Microscopy (Oxf) 2016;65(1):3–8. doi: 10.1093/jmicro/dfv358. https://doi.org/10.1093/jmicro/dfv358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Ourmazd A. Continuous changes in structure mapped by manifold embedding of single-particle data in cryo-EM. Methods. 2016 May 1;100:61–67. doi: 10.1016/j.ymeth.2016.02.007. https://doi.org/10.1016/j.ymeth.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Wagenknecht T, McEwen BF, Marko M, Hsieh CE, Mannella CA. Three-dimensional imaging of biological complexity. J Struct Biol. 2002 Apr-May;138(1-2):85–91. doi: 10.1016/s1047-8477(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong C, Protasi F, Ramesh V. Shape, size, and distribution of Ca2+ release units and couplons in skeletal and cardiac muscles. Biophys J. 1999;77(3):1528–1539. doi: 10.1016/S0006-3495(99)77000-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambardella J, Trimarco B, Iaccarino G, Santulli G. New insights in cardiac calcium handling and excitation-contraction coupling. Adv Exp Med Biol. 2017 doi: 10.1007/5584_2017_106. in press. https://doi.org/10.1007/5584_2017_106. [DOI] [PMC free article] [PubMed]
- Gomez AC, Yamaguchi N. Two regions of ryanodine receptor calcium channel are involved in Ca(2+)-dependent inactivation. Biochemistry. 2014;53(8):1373–1379. doi: 10.1021/bi401586h. https://doi.org/10.1021/bi401586h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez AC, Holford TW, Yamaguchi N. Malignant hyperthermia-associated mutations in the S2-S3 cytoplasmic loop of type 1 ryanodine receptor calcium channel impair calcium-dependent inactivation. Am J Physiol Cell Physiol. 2016;311(5):C749–C757. doi: 10.1152/ajpcell.00134.2016. https://doi.org/10.1152/ajpcell.00134.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, et al. The EF-hand Ca2+ binding domain is not required for cytosolic Ca2+ activation of the cardiac ryanodine receptor. J Biol Chem. 2016 doi: 10.1074/jbc.M115.693325. https://doi.org/10.1074/jbc.M115.693325. [DOI] [PMC free article] [PubMed]
- Hakamata Y, Nakai J, Takeshima H, Imoto K. Primary structure and distribution of a novel ryanodine receptor/calcium release channel from rabbit brain. FEBS Lett. 1992;312(2–3):229–235. doi: 10.1016/0014-5793(92)80941-9. [DOI] [PubMed] [Google Scholar]
- Hamilton SL, Serysheva II. Ryanodine receptor structure: progress and challenges. J Biol Chem. 2009 Feb 13;284(7):4047–4051. doi: 10.1074/jbc.R800054200. https://doi.org/10.1074/jbc.R800054200. Epub 2008 Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnick DJ, Jayaraman T, Ma Y, Mulieri P, Go LO, Marks AR. The human type 1 inositol 1,4,5-trisphosphate receptor from T lymphocytes. Structure, localization, and tyrosine phosphorylation. J Biol Chem. 1995;270(6):2833–2840. doi: 10.1074/jbc.270.6.2833. [DOI] [PubMed] [Google Scholar]
- Hille B. Ion channels of excitable membranes. Vol. 507. Sinauer; Sunderland: 2001. [Google Scholar]
- Huang F, Shan J, Reiken S, Wehrens XH, Marks AR. Analysis of calstabin2 (FKBP12.6)-ryanodine receptor interactions: rescue of heart failure by calstabin2 in mice. Proc Natl Acad Sci U S A. 2006;103(9):3456–3461. doi: 10.1073/pnas.0511282103. https://doi.org/10.1073/pnas.0511282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JH, Zorzato F, Clarke NF, Treves S. Mapping domains and mutations on the skeletal muscle ryanodine receptor channel. Trends Mol Med. 2012;18(11):644–657. doi: 10.1016/j.molmed.2012.09.006. https://doi.org/10.1016/j.molmed.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Inui M, Saito A, Fleischer S. Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. J Biol Chem. 1987;262(4):1740–1747. [PubMed] [Google Scholar]
- Jiang D, Chen W, Wang R, Zhang L, Chen SR. Loss of luminal Ca2+ activation in the cardiac ryanodine receptor is associated with ventricular fibrillation and sudden death. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18309–18314. doi: 10.1073/pnas.0706573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimlicka L, Lau K, Tung CC, Van Petegem F. Disease mutations in the ryanodine receptor N-terminal region couple to a mobile intersubunit interface. Nat Commun. 2013;4:1506. doi: 10.1038/ncomms2501. https://doi.org/10.1038/ncomms2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmerick MJ, Moerland TS, Wiseman RW. Mammalian skeletal muscle fibers distinguished by contents of phosphocreatine, ATP, and Pi. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7521–7525. doi: 10.1073/pnas.89.16.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanner JT, Georgiou DK, Joshi AD, Hamilton SL. Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harb Perspect Biol. 2010;2(11):a003996. doi: 10.1101/cshperspect.a003996. https://doi.org/10.1101/cshperspect.a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larach MG, Brandom BW, Allen GC, Gronert GA, Lehman EB. Malignant hyperthermia deaths related to inadequate temperature monitoring, 2007–2012: a report from the North American malignant hyperthermia registry of the malignant hyperthermia association of the United States. Anesth Analg. 2014;119(6):1359–1366. doi: 10.1213/ANE.0000000000000421. https://doi.org/10.1213/ane.0000000000000421. [DOI] [PubMed] [Google Scholar]
- Lau K, Van Petegem F. Crystal structures of wild type and disease mutant forms of the ryanodine receptor SPRY2 domain. Nat Commun. 2014;5:5397. doi: 10.1038/ncomms6397. https://doi.org/10.1038/ncomms6397. [DOI] [PubMed] [Google Scholar]
- Laver DR, Lenz GK, Lamb GD. Regulation of the calcium release channel from rabbit skeletal muscle by the nucleotides ATP, AMP, IMP and adenosine. J Physiol. 2001;537(Pt 3):763–778. doi: 10.1111/j.1469-7793.2001.00763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnart SE, Huang F, Marx SO, Marks AR. Immunophilins and coupled gating of ryanodine receptors. Cur Top Med Chem. 2003;3(12):1383–1391. doi: 10.2174/1568026033451907. [DOI] [PubMed] [Google Scholar]
- Lehnart SE, Wehrens XH, Laitinen PJ, Reiken SR, Deng SX, Cheng Z, Landry DW, Kontula K, Swan H, Marks AR. Sudden death in familial polymorphic ventricular tachycardia associated with calcium release channel (ryanodine receptor) leak. Circulation. 2004;109(25):3208–3214. doi: 10.1161/01.CIR.0000132472.98675.EC. https://doi.org/10.1161/01.CIR.0000132472.98675.EC. [DOI] [PubMed] [Google Scholar]
- Lehnart SE, Wehrens XH, Reiken S, Warrier S, Belevych AE, Harvey RD, Richter W, Jin SL, Conti M, Marks AR. Phosphodiesterase 4D deficiency in the ryanodine-receptor complex promotes heart failure and arrhythmias. Cell. 2005;123(1):25–35. doi: 10.1016/j.cell.2005.07.030. https://doi.org/10.1016/j.cell.2005.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnart SE, Mongillo M, Bellinger A, Lindegger N, Chen BX, Hsueh W, Reiken S, Wronska A, Drew LJ, Ward CW, Lederer WJ, Kass RS, Morley G, Marks AR. Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J Clin Invest. 2008;118(6):2230–2245. doi: 10.1172/JCI35346. https://doi.org/10.1172/JCI35346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang J, Wang R, Chen SRW, Wagenknecht T. Location of divergent region 2 on the three-dimensional structure of cardiac muscle ryanodine receptor/calcium release channel. J Mol Biol. 2004;338:533–545. doi: 10.1016/j.jmb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Liu X, Betzenhauser MJ, Reiken S, Meli AC, Xie W, Chen BX, Arancio O, Marks AR. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell. 2012;150(5):1055–1067. doi: 10.1016/j.cell.2012.06.052. https://doi.org/10.1016/j.cell.2012.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo PA, Van Petegem F. Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: insights into disease mutations. Structure. 2009 Nov 11;17(11):1505–1514. doi: 10.1016/j.str.2009.08.016. https://doi.org/10.1016/j.str.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Lombardi A, Gambardella J, XL D, Sorriento D, Mauro M, Iaccarino G, Trimarco B, Santulli G. Sirolimus induces depletion of intracellular calcium stores and mitochondrial dysfunction in pancreatic beta cells. Sci Rep. 2017a Nov 20;7(1):15823. doi: 10.1038/s41598-017-15283-y. https://doi.org/10.1038/s41598-017-15283-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A, Trimarco B, Iaccarino G, Santulli G. Impaired mitochondrial calcium uptake caused by tacrolimus underlies beta-cell failure. Cell Commun Signal. 2017b Nov 13;15(1):47. doi: 10.1186/s12964-017-0203-0. https://doi.org/10.1186/s12964-017-0203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. The pore structure of the closed RyR1 channel. Structure. 2005 Aug;13(8):1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki T, Gruver EJ, Davidoff AJ, Izzo N, Toupin D, Colucci W, Marks AR, Marsh JD. Regulation of calcium channel expression in neonatal myocytes by catecholamines. J Clin Invest. 1996;97(3):656–663. doi: 10.1172/JCI118462. https://doi.org/10.1172/JCI118462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks AR. Sirolimus for the prevention of in-stent restenosis in a coronary artery. New Engl J Med. 2003;349(14):1307–1309. doi: 10.1056/NEJMp038141. https://doi.org/10.1056/NEJMp038141. [DOI] [PubMed] [Google Scholar]
- Marks AR, Tempst P, Hwang KS, Taubman MB, Inui M, Chadwick C, Fleischer S, Nadal-Ginard B. Molecular cloning and characterization of the ryanodine receptor/junctional channel complex cDNA from skeletal muscle sarcoplasmic reticulum. Proc Natl Acad Sci U S A. 1989;86(22):8683–8687. doi: 10.1073/pnas.86.22.8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks AR, Priori S, Memmi M, Kontula K, Laitinen PJ. Involvement of the cardiac ryanodine receptor/calcium release channel in catecholaminergic polymorphic ventricular tachycardia. J Cell Physiol. 2002;190(1):1–6. doi: 10.1002/jcp.10031. https://doi.org/10.1002/jcp.10031. [DOI] [PubMed] [Google Scholar]
- Marx SO, Ondrias K, Marks AR. Coupled gating between individual skeletal muscle Ca2+ release channels (ryanodine receptors) Science. 1998;281(5378):818–821. doi: 10.1126/science.281.5378.818. [DOI] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101(4):365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]
- Marx SO, Gaburjakova J, Gaburjakova M, Henrikson C, Ondrias K, Marks AR. Coupled gating between cardiac calcium release channels (ryanodine receptors) Circ Res. 2001a;88(11):1151–1158. doi: 10.1161/hh1101.091268. [DOI] [PubMed] [Google Scholar]
- Marx SO, Reiken S, Hisamatsu Y, Gaburjakova M, Gaburjakova J, Yang YM, Rosemblit N, Marks AR. Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J Cell Biol. 2001b;153(4):699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matecki S, Dridi H, Jung B, Saint N, Reiken SR, Scheuermann V, Mrozek S, Santulli G, Umanskaya A, Petrof BJ, Jaber S, Marks AR, Lacampagne A. Leaky ryanodine receptors contribute to diaphragmatic weakness during mechanical ventilation. Proc Natl Acad Sci U S A. 2016;113(32):9069–9074. doi: 10.1073/pnas.1609707113. https://doi.org/10.1073/pnas.1609707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Faruqi AR, Clare D, Henderson R. Comparison of optimal performance at 300keV of three direct electron detectors for use in low dose electron microscopy. Ultramicroscopy. 2014;147:156–163. doi: 10.1016/j.ultramic.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai J, Imagawa T, Hakamat Y, Shigekawa M, Takeshima H, Numa S. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 1990;271(1–2):169–177. doi: 10.1016/0014-5793(90)80399-4. [DOI] [PubMed] [Google Scholar]
- Nakashima Y, Nishimura S, Maeda A, Barsoumian EL, Hakamata Y, Nakai J, Allen PD, Imoto K, Kita T. Molecular cloning and characterization of a human brain ryanodine receptor. FEBS Lett. 1997;417(1):157–162. doi: 10.1016/s0014-5793(97)01275-1. [DOI] [PubMed] [Google Scholar]
- Nelson BR, Wu F, Liu Y, Anderson DM, McAnally J, Lin W, Cannon SC, Bassel-Duby R, Olson EN. Skeletal muscle-specific T-tubule protein STAC3 mediates voltage-induced Ca2+ release and contractility. Proc Natl Acad Sci U S A. 2013;110(29):11881–11886. doi: 10.1073/pnas.1310571110. https://doi.org/10.1073/pnas.1310571110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. The development of cryo-EM into a mainstream structural biology technique. Nat Methods. 2016 Jan;13(1):24–27. doi: 10.1038/nmeth.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu K, Willard HF, Khanna VK, Zorzato F, Green NM, MacLennan DH. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J Biol Chem. 1990;265(23):13472–13483. [PubMed] [Google Scholar]
- Paul-Pletzer K, Yamamoto T, Bhat MB, Ma J, Ikemoto N, Jimenez LS, Morimoto H, Williams PG, Parness J. Identification of a dantrolene-binding sequence on the skeletal muscle ryanodine receptor. J Biol Chem. 2002;277(38):34918–34923. doi: 10.1074/jbc.M205487200. https://doi.org/10.1074/jbc.M205487200. [DOI] [PubMed] [Google Scholar]
- Peng W, et al. Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2. Science. 2016;354:6310. doi: 10.1126/science.aah5324. https://doi.org/10.1126/science.aah5324. [DOI] [PubMed] [Google Scholar]
- Porta M, Zima AV, Nani A, Diaz-Sylvester PL, Copello JA, Ramos-Franco J, Blatter LA, Fill M. Single ryanodine receptor channel basis of caffeine's action on Ca2+ sparks. Biophys J. 2011;100(4):931–938. doi: 10.1016/j.bpj.2011.01.017. https://doi.org/10.1016/j.bpj.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quane KA, Healy JM, Keating KE, Manning BM, Couch FJ, Palmucci LM, Doriguzzi C, Fagerlund TH, Berg K, Ording H, et al. Mutations in the ryanodine receptor gene in central core disease and malignant hyperthermia. Nat Gen. 1993;5(1):51–55. doi: 10.1038/ng0993-51. https://doi.org/10.1038/ng0993-51. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Grassucci R, Frank J, Inui M, Chadwick C, Fleischer S. Cryo-EM of the native structure of the calcium release channel/ryanodine receptor from sarcoplasmic reticulum. Biophys J. 1992 Apr;61(4):936–940. doi: 10.1016/S0006-3495(92)81900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermacher M, Rao V, Grassucci R, Frank J, Timerman AP, Fleischer S, Wagenknecht T. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J Cell Biol. 1994a Oct;127(2):411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios E, Brum G. Involvement of dihydropyridine receptors in excitation-contraction coupling in skeletal muscle. Nature. 1987;325(6106):717–720. doi: 10.1038/325717a0. https://doi.org/10.1038/325717a0. [DOI] [PubMed] [Google Scholar]
- Rosemblit N, Moschella MC, Ondriasova E, Gutstein DE, Ondrias K, Marks AR. Intracellular calcium release channel expression during embryogenesis. Dev Biol. 1999;206(2):163–177. doi: 10.1006/dbio.1998.9120. [DOI] [PubMed] [Google Scholar]
- Rosenberg H, Davis M, James D, Pollock N, Stowell K. Malignant hyperthermia. Orphanet J Rare Dis. 2007;2:21–21. doi: 10.1186/1750-1172-2-21. https://doi.org/10.1186/1750-1172-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullman E, Andersson DC, Melin M, Reiken S, Mancini DM, Marks AR, Lund LH, Gustafsson T. Modifications of skeletal muscle ryanodine receptor type 1 and exercise intolerance in heart failure. J Heart Lung Transplant. 2013;32(9):925–929. doi: 10.1016/j.healun.2013.06.026. https://doi.org/10.1016/j.healun.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A, Inui M, Radermacher M, Frank J, Fleischer SJ. Ultrastructure of the calcium release channel of sarcoplasmic reticulum. Cell Biol. 1988 Jul;107(1):211–219. doi: 10.1083/jcb.107.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsò M, Wagenknecht T, Allen PD. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat Struct Mol Biol. 2005 Jun;12(6):539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsó M, Feng W, Pessah IN, Allen PD. Coordinated movement of cytoplasmic and transmembrane domains of RyR1 upon gating. PLoS Biol. 2009 Apr 14;7(4):e85. doi: 10.1371/journal.pbio.1000085. https://doi.org/10.1371/journal.pbio.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G. microRNAs distinctively regulate vascular smooth muscle and endothelial cells: functional implications in angiogenesis, atherosclerosis, and in-stent restenosis. Adv Exp Med Biol. 2015;887:53–77. doi: 10.1007/978-3-319-22380-3_4. https://doi.org/10.1007/978-3-319-22380-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Marks AR. Essential roles of intracellular calcium release channels in muscle, brain, metabolism, and aging. Curr Mol Pharmacol. 2015;8(2):206–222. doi: 10.2174/1874467208666150507105105. [DOI] [PubMed] [Google Scholar]
- Santulli G, Totary-Jain H. Tailoring mTOR-based therapy: molecular evidence and clinical challenges. Pharmacogenomics. 2013;14(12):1517–1526. doi: 10.2217/pgs.13.143. https://doi.org/10.2217/pgs.13.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Wronska A, Uryu K, Diacovo TG, Gao M, Marx SO, Kitajewski J, Chilton JM, Akat KM, Tuschl T, Marks AR, Totary-Jain H. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J Clin Invest. 2014;124(9):4102–4114. doi: 10.1172/JCI76069. https://doi.org/10.1172/JCI76069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Pagano G, Sardu C, Xie W, Reiken S, D'Ascia SL, Cannone M, Marziliano N, Trimarco B, Guise TA, Lacampagne A, Marks AR. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J Clin Invest. 2015a;125(5):1968–1978. doi: 10.1172/JCI79273. https://doi.org/10.1172/JCI79273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Xie W, Reiken SR, Marks AR. Mitochondrial calcium overload is a key determinant in heart failure. Proc Natl Acad Sci U S A. 2015b;112(36):11389–11394. doi: 10.1073/pnas.1513047112. https://doi.org/10.1073/pnas.1513047112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santulli G, Lewis D, Marks AR. Physiology and pathophysiology of excitation–contraction coupling in skeletal muscle: the functional role of ryanodine receptor. J Muscle Res Cell Motil. 2017a doi: 10.1007/s10974-017-9470-z. https://doi.org/10.1007/s10974-017-9470-z. [DOI] [PMC free article] [PubMed]
- Santulli G, Nakashima R, Yuan Q, Marks AR. Intracellular calcium release channels: an update. J Physiol. 2017b doi: 10.1113/JP272781. https://doi.org/10.1113/JP272781. [DOI] [PMC free article] [PubMed]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012 Dec;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. https://doi.org/10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewry CA, Muller C, Davis M, Dwyer JS, Dove J, Evans G, Schroder R, Furst D, Helliwell T, Laing N, Quinlivan RC. The spectrum of pathology in central core disease. Neuromusc Disord NMD. 2002;12(10):930–938. doi: 10.1016/s0960-8966(02)00135-9. [DOI] [PubMed] [Google Scholar]
- Shan J, Betzenhauser MJ, Kushnir A, Reiken S, Meli AC, Wronska A, Dura M, Chen BX, Marks AR. Role of chronic ryanodine receptor phosphorylation in heart failure and beta- adrenergic receptor blockade in mice. J Clin Invest. 2010a;120(12):4375–4387. doi: 10.1172/JCI37649. https://doi.org/10.1172/JCI37649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Kushnir A, Betzenhauser MJ, Reiken S, Li J, Lehnart SE, Lindegger N, Mongillo M, Mohler PJ, Marks AR. Phosphorylation of the ryanodine receptor mediates the cardiac fight or flight response in mice. J Clin Invest. 2010b;120(12):4388–4398. doi: 10.1172/JCI32726. https://doi.org/10.1172/JCI32726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MR, Penczek P, Grassucci R, Xin HB, Fleischer S, Wagenknecht T. Cryoelectron microscopy and image analysis of the cardiac ryanodine receptor. J Biol Chem. 1998 Jul 17;273(29):18429–18434. doi: 10.1074/jbc.273.29.18429. [DOI] [PubMed] [Google Scholar]
- Sharma P, Ishiyama N, Nair U, Li W, Dong A, Miyake T, Wilson A, Ryan T, MacLennan DH, Kislinger T, Ikura M, Dhe-Paganon S, Gramolini AO. Structural determination of the phosphorylation domain of the ryanodine receptor. FEBS J. 2012;279(20):3952–3964. doi: 10.1111/j.1742-4658.2012.08755.x. https://doi.org/10.1111/j.1742-4658.2012.08755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetomi T, Yano M, Uchinoumi H, Fukuda M, Hino A, Ono M, Xu X, Tateishi H, Okuda S, Doi M, Kobayashi S, Ikeda Y, Yamamoto T, Ikemoto N, Matsuzaki M. Mutation-linked defective interdomain interactions within ryanodine receptor cause aberrant Ca(2)(+)release leading to catecholaminergic polymorphic ventricular tachycardia. Circulation. 2011;124(6):682–694. doi: 10.1161/CIRCULATIONAHA.111.023259. https://doi.org/10.1161/circulationaha.111.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, et al. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989;339(6224):439–445. doi: 10.1038/339439a0. https://doi.org/10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- Tateishi H, Yano M, Mochizuki M, Suetomi T, Ono M, Xu X, Uchinoumi H, Okuda S, Oda T, Kobayashi S, Yamamoto T, Ikeda Y, Ohkusa T, Ikemoto N, Matsuzaki M. Defective domain-domain interactions within the ryanodine receptor as a critical cause of diastolic Ca2+ leak in failing hearts. Cardiovasc Res. 2009;81(3):536–545. doi: 10.1093/cvr/cvn303. https://doi.org/10.1093/cvr/cvn303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timerman AP, Ogunbumni E, Freund E, Wiederrecht G, Marks AR, Fleischer S. The calcium release channel of sarcoplasmic reticulum is modulated by FK-506-binding protein. Dissociation and reconstitution of FKBP-12 to the calcium release channel of skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1993;268(31):22992–22999. [PubMed] [Google Scholar]
- Tung CC, Lobo PA, Kimlicka L, Van Petegem F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468(7323):585–588. doi: 10.1038/nature09471. https://doi.org/10.1038/nature09471. [DOI] [PubMed] [Google Scholar]
- Umanskaya A, Santulli G, Xie W, Andersson DC, Reiken SR, Marks AR. Genetically enhancing mitochondrial antioxidant activity improves muscle function in aging. Proc Natl Acad Sci U S A. 2014;11(42):15250–15255. doi: 10.1073/pnas.1412754111. https://doi.org/10.1073/pnas.1412754111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Petegem F. Ryanodine receptors: allosteric Ion channel giants. J Mol Biol. 2015;427(1):31–53. doi: 10.1016/j.jmb.2014.08.004. htpps://doi.org/10.1016/j.jmb.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Van Petegem F. How to open a ryanodine receptor. Cell Res. 2016 Oct;26(10):1073–1074. doi: 10.1038/cr.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest JA, Wehrens XH, Reiken SR, Lehnart SE, Dobrev D, Chandra P, Danilo P, Ravens U, Rosen MR, Marks AR. Defective cardiac ryanodine receptor regulation during atrial fibrillation. Circulation. 2005;111(16):2025–2032. doi: 10.1161/01.CIR.0000162461.67140.4C. https://doi.org/10.1161/01.CIR.0000162461.67140.4C. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, Grassucci R, Frank J, Saito A, Inui M, Fleischer S. Three-dimensional architecture of the calcium channel/foot structure of sarcoplasmic reticulum. Nature. 1989 Mar 9;338(6211):167–170. doi: 10.1038/338167a0. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, Radermacher M. Three-dimensional architecture of the skeletal muscle ryanodine receptor. FEBS Lett. 1995 Aug 1;369(1):43–46. doi: 10.1016/0014-5793(95)00581-s. Review. [DOI] [PubMed] [Google Scholar]
- Wei R, et al. Structural insights into Ca2+-activated long-range allosteric channel gating of RyR1. Cell Res. 2016;26:977–994. doi: 10.1038/cr.2016.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Santulli G, Guo X, Gao M, Chen BX, Marks AR. Imaging atrial arrhythmic intracellular calcium in intact heart. J Mol Cell Cardiol. 2013;64:120–123. doi: 10.1016/j.yjmcc.2013.09.003. https://doi.org/10.1016/j.yjmcc.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Santulli G, Reiken SR, Yuan Q, Osborne BW, Chen BX, Marks AR. Mitochondrial oxidative stress promotes atrial fibrillation. Sci Rep. 2015;5:11427. doi: 10.1038/srep11427. https://doi.org/10.1038/srep11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin HB, Timerman AP, Onoue H, Wiederrecht GJ, Fleischer S. Affinity purification of the ryanodine receptor/calcium release channel from fast twitch skeletal muscle based on its tight association with FKBP12. Biochem Biophys Res Commun. 1995;214(1):263–270. doi: 10.1006/bbrc.1995.2283. https://doi.org/10.1006/bbrc.1995.2283. [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhang JZ, He R, Hamilton SL. A Ca2+-binding domain in RyR1 that interacts with the calmodulin binding site and modulates channel activity. Biophys J. 2006;90(1):173–182. doi: 10.1529/biophysj.105.066092. https://doi.org/10.1529/biophysj.105.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Bai XC, Yan C, Wu J, Li Z, Xie T, Peng W, Yin CC, Li X, Scheres SHW, Shi Y, Yan N. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517(7532):50–55. doi: 10.1038/nature14063. https://doi.org/10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Chen Z, Santulli G, Gu L, Yang ZG, Yuan ZQ, Zhao YT, Xin HB, Deng KY, Wang SQ, Ji G. Functional role of Calstabin2 in age-related cardiac alterations. Sci Rep. 2014;4:7425. doi: 10.1038/srep07425. https://doi.org/10.1038/srep07425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Yang J, Santulli G, Reiken SR, Wronska A, Kim MM, Osborne BW, Lacampagne A, Yin Y, Marks AR. Maintenance of normal blood pressure is dependent on IP3R1-mediated regulation of eNOS. Proc Natl Acad Sci U S A. 2016;113(30):8532–8537. doi: 10.1073/pnas.1608859113. https://doi.org/10.1073/pnas.1608859113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuchi Z, Lau K, Van Petegem F. Disease mutations in the ryanodine receptor central region: crystal structures of a phosphorylation hot spot domain. Structure. 2012;20(7):1201–1211. doi: 10.1016/j.str.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Yuchi Z, Yuen SM, Lau K, Underhill AQ, Cornea RL, Fessenden JD, Van Petegem F. Crystal structures of ryanodine receptor SPRY1 and tandem-repeat domains reveal a critical FKBP12 binding determinant. Nat Commun. 2015;6:7947. doi: 10.1038/ncomms8947. https://doi.org/10.1038/ncomms8947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalk R, Clarke OB, des Georges A, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, Marks AR. Structure of a mammalian ryanodine receptor. Nature. 2015 Jan 1;517(7532):44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen HS, Khanna VK, De Leon S, Phillips MS, Schappert K, Britt BA, Browell AK, MacLennan DH. A mutation in the human ryanodine receptor gene associated with central core disease. Nat Gen. 1993;5(1):46–50. doi: 10.1038/ng0993-46. https://doi.org/10.1038/ng0993-46. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu Y, Song F, Zheng H, Hu L, Lu H, Liu P, Hao X, Zhang W, Chen K. Functional SNP in the microRNA-367 binding site in the 3′UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc Natl Acad Sci U S A. 2011;108(33):13653–13658. doi: 10.1073/pnas.1103360108. https://doi.org/10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Li P, Chen SR, Louis CF, Fruen BR. Dantrolene inhibition of ryanodine receptor Ca2+ release channels molecular mechanism and isoform selectivity. J Pys Chem. 2001;276(17):13810–13816. doi: 10.1074/jbc.M006104200. https://doi.org/10.1074/jbc.M006104200. [DOI] [PubMed] [Google Scholar]