Abstract
In recognition of the importance of zebrafish as a model organism for studying human disease, we have created zebrafish content for a web-based reference atlas of microanatomy for comparing histology and histopathology between model systems and with humans (http://bio-atlas.psu.edu). Fixation, decalcification, embedding, and sectioning of zebrafish were optimized to maximize section quality. A comparison of protocols involving six fixatives showed that 10% Neutral Buffered Formalin at 21 °C for 24 h yielded excellent results. Sectioning of juveniles and adults requires bone decalcification; EDTA at 0.35 M produced effective decalcification in 21-day-old juveniles through adults (≥~3 Months). To improve section plane consistency in sets of larvae, we have developed new array casting molds based on the outside contours of larvae derived from 3D microCT images. Tissue discontinuity in sections, a common barrier to creating quality sections of zebrafish, was minimized by processing and embedding the formalin-fixed zebrafish tissues in plasticized forms of paraffin wax, and by periodic hydration of the block surface in ice water between sets of sections. Optimal H&E (Hematoxylin and Eosin) staining was achieved through refinement of standard protocols. High quality slide scans produced from glass histology slides were digitally processed to maximize image quality, and experimental replicates posted as full slides as part of this publication. Modifications to tissue processing are still needed to eliminate the need for block surface hydration. The further addition of slide collections from other model systems and 3D tools for visualizing tissue architecture would greatly increase the utility of the digital atlas.
Keywords: Atlas, Fixation, Decalcification, Embedding, Sectioning, Staining, Histology, Virtual slide
1. Introduction
The use of model organisms in biology is central to our ability to advance knowledge of both normal and disease states in humans. We have created an online atlas of histology and histopathology of the zebrafish to facilitate comparisons of human and model organism histology and histopathology. Zebrafish are widely used as a model for the study of development and human disorders; some 70% of human disease genes have functional homologs in zebrafish (Beckwith et al., 2000, Santoriello and Zon, 2012, Stewart et al., 2014, Zhao et al., 2015). Additionally, mutants can be readily generated, yielding histologically-apparent phenotypes (Mohideen et al., 2003; Amsterdam et al., 2004; Sadler et al., 2005; Santoriello and Zon, 2012). This also creates the opportunity to perform phenotypic comparisons between chemically-induced phenotypes (Hill et al., 2005) and mutant phenotypes. The small size of zebrafish larvae make them particularly well-suited for chemical (experimental or environmental pollutant) screening because small volumes of chemicals can be used (Görge and Nagel, 1990, Hill et al., 2005, Taylor et al., 2010, Stewart et al., 2014).
Since our website is intended to be a resource for members of the scientific community interested in human and model organism histology, consistency of quality, sectioning, staining, and histological imaging are important. To address common difficulties with various aspects of zebrafish histology, we set out to optimize protocols to produce the best possible slides and images. This paper includes answers to the large number of requests we have received for help with zebrafish histology over two decades. Our foundational work for the atlas included the testing of different protocols for tissue fixation, decalcification, and orientation in paraffin. This work addressed the most common and significant problems in zebrafish histology that makes histological sections of randomly oriented fish virtually uninterpretable: poor sectioning, twisting, and sample malorientation. Past work related to these issues includes (Tsao-Wu et al., 1998, Beckwith et al., 2000; Moore et al., 2002; Sabaliauskas et al., 2006). The work of Tsao-Wu et al. (1998) laid the foundation for agarose embedded larval tissue arrays based on a rectangular array mold. This mold was based on a four by eight agarose array in which 64 seven day old larvae can be positioned, two per well. This facilitated histologic analysis of the larvae since they could be sectioned in the same plane and visualized simultaneously. Moore et al. (2002) focused their study on the fixation and decalcification of adult zebrafish with the focus on both fixation and integrity of extracted DNA. They concluded that 10% Neutral Buffered Formalin or 4% Paraformaldehyde followed by decalcification in 0.5 M EDTA yielded satisfactory histological sections and DNA fragments useful for genotypic analysis of adult zebrafish. In subsequent years we decreased the EDTA concentration for decalcification to 0.35 M, based on the idea that this would diminish the amount of EDTA that could contaminate subsequent molecular biological reactions. Sabaliauskas et al. (2006) modified the agarose array mold to fit 50 larvae facing in the same direction. Their refined design has triangular wells that fit one larva each. This modification allowed for more consistent positioning of the larvae and easier digital imaging of individual fish. They also included descriptions of effective methods for tissue processing, staining and digital image capture. Based on the practices of other fish and comparative pathology laboratories, we have evaluated six commonly used fixatives with a focus on duration of fixation. We determined optimal decalcification durations using 0.35 M EDTA for zebrafish age 21 dpf (days post-fertilization) and older. Additionally, we developed and tested two novel agarose larval array molds designed to improve larval alignment). Finally, we refined details of both H&E staining (Hematoxylin and Eosin) and digital image capture.
2. Materials and methods
2.1. Staging and preparation of zebrafish prior to fixation
Wild-type Connor and Ekkwill strains were used for both the development and production of the zebrafish atlas website (www.zfatlas.psu.edu), which is now renamed http://bio-atlas.psu.edu to better engage its use across model systems. Fish originally obtained from Indonesian wild caught sources were acquired through the commercial distributors Connor and Ekkwill. Fish were reared at an average temperature of 28 °C in a recirculating system with a 14:10 h light to dark cycle. Fish were fed three times a day a diet consisting of brine shrimp and flake food. All fish were staged according to the zebrafish developmental staging series of Kimmel et al. (1995; http://onlinelibrary.wiley.com/doi/10.1002/aja.1002030302/pdf). Euthanasia was performed with 160 mg/L Tricaine-S pH 7.0 (tricaine methanesulfonate, Syndel, USA) in system water. It should be noted that the relative merits of Tricaine-S and/or hypothermal shock through immersion in ice water (2–4 °C) have been recently debated (Wilson et al., 2009; Matthews and Varga, 2012). While each method has value, current data suggests that hypothermal shock results in a more rapid and humane euthanasia in small tropical species such as zebrafish. In keeping with this finding, our laboratory currently combines both methods (cold Tricaine) to produce a rapid euthanasia in our fish, ensuring that the fish do not react to subsequent immersion in fixatives.
Manipulations of the fish before and after euthanasia were kept to a minimum and were performed gently to avoid damage to the fish. Pasteur pipettes used to handle larvae were flamed to round sharp edges that can otherwise damage the fish. Fixation was performed immediately after euthanasia to prevent autolysis and degradation of cell morphology. To enhance the penetration of fixative to internal organs, adult zebrafish were incised ventrally midline from the anal pore to the base of the pectoral fin. Fish were held gently using gloved fingers rather than forceps during this process to minimize damage to the skin, external structures, and internal organs.
2.2. Fixation and decalcification
Immediately following euthanasia, fish were placed in two rinses of fixative and then incubated in at least 20× fish volume of fixative. Samples were fixed in flat bottom glass vials to ensure that the fish were fixed in a straight orientation; fixation in a vertical position tends to result in bending of the fish. Several fixatives (Table 1) and fixation parameters such as time and temperature (Table 2) were tested as described. The merits of movement of the fixation solution by stir bar during fixation were tested with the standard fixative used in human pathology, 10% NBF (Neutral Buffered Formalin, Fisher Scientific Cat# SF100-4). Juvenile (57 dpf WT Ekkwill) zebrafish used for this comparison were placed inside embedding cassettes. Up to six cassettes were placed on top of a tray in a glass staining dish and submerged in approximately 100 ml 10% NBF for 48 h at 21 °C. Gentle stirring by stir bar was used, but we detected no effect.
Table 1.
Fixatives tested.
| Fixative | Formulation |
|---|---|
| Zenker’s | Stock solution |
| Potassium dicromate 2.5% | |
| Mercuric chloride 5.0% | |
| Sodium sulfate 1.0% | |
| Distilled water 92% | |
| Working solution | |
| Zenker stock 95 ml | |
| Glacial acetic acid 5 ml | |
| Zamboni’s | Paraformaldehyde 2% |
| Picric acid, saturated aqueous 15% (final 0.195%) | |
| Sodium phosphate di-basic 3.37% | |
| Sodium phosphate monobasic 0.28% | |
| Distilled water 79% | |
| Zinc-formalin | Formalin 3.7% |
| Zinc sulfate 1% | |
| Distilled water 97% | |
| Bouin’s | Picric acid, saturated aqueous 75% (final 0.975%) |
| Formaldehyde 1% | |
| Acetic acid 5% | |
| Distilled Water 19% | |
| Formaldehyde/glutaraldehyde | Formaldehyde 2% |
| Glutaraldehyde 1% | |
| Calcium acetate 2% | |
| Distilled water 95% | |
| 10% neutral buffered formalin | Formaldehyde 4% (10% of ~40% stock solution) |
| Sodium phosphate monobasic 0.4% | |
| Sodium phosphate di-basic 0.65% | |
| Methanol 1.5% | |
| Distilled water 93% |
Table 2.
Fixation times.
| Fixative | Fixation time and temperature | Picric acid time and temperature |
|---|---|---|
| Zenker’s | 24 h 4 °C | None |
| Zamboni’s | 24 h 4 °C | None |
| Zinc-formalin | 6 h 4 °C | None |
| Bouin’s | 24 h 4 °C | None |
| 2% formaldehyde/1% glutaraldehyde | 48 h 21 °C | None |
| 2% formaldehyde/1% glutaraldehyde; 0.65% picric acid (separate step) | 48 h 21 °C | 48 h 21 °C |
| 10% neutral buffered formalin | 24 h 4 °C | None |
| 10% neutral buffered formalin | 48 h 4 °C | None |
| 10% neutral buffered formalin | 8 day 4 °C | None |
| 10% neutral buffered formalin | 24 h 21 °C | None |
| 10% neutral buffered formalin | 48 h 21 °C | None |
| 10% neutral buffered formalin | 8 day 21 °C | None |
We avoid decalcification in acid because it destroys DNA and causes pale H&E staining. Zebrafish age 21 dpf and older were decalcified in 0.35 M EDTA pH 7.8 (Fisher, Cat# BP121). After fixation, fish were rinsed twice in 20× fish volume of 1× PBS (Sigma, Cat# D1408) before being placed in 20× fish volume of 0.35 M EDTA pH 7.8 and incubated at 21 °C for the times listed in Table 3 prior to the next tissue processing steps. After fixation in fish younger than 21 dpf and after decalcification in fish 21 dpf and older, fish were rinsed twice in 20× fish volume of 70% Ethanol (Pharmco-AAPER, Cat# 1,110,000,200). All fish were then incubated overnight at 21 °C in 70% Ethanol prior to pre-embedding or processing.
Table 3.
EDTA decalcification.
| Days post fertilization | Total body length | EDTA incubation time |
|---|---|---|
| 14–20 | 6.2 mm | 0 days |
| 21–29 | 7.8 mm | 4 days |
| 30–44 | 10 mm | 5 days |
| 45–89 | 14 mm | 6 days |
| Adult (90 dpf to 2 years) | Adult | 7 days |
2.3. Pre-embedding
To facilitate proper orientation during sectioning, all stages of fixed fish except the adult stages were pre-embedded in 1% agarose (Sigma, Cat# A5304) in double distilled water (ddH2O) prior to processing (Figs. 1–3). Embryonic and larval stages between 24 hpf (hours post-fertilization) and 13 dpf were pre-embedded in a 1% agarose (w/v in ddH2O) gel array using a casting mold (“triangle mold” Fig. 1A, B) previously developed in our laboratory (Sabaliauskas et al., 2006). The triangle array has 50 triangular wells in a 5 × 10 format. Cooled casts were trimmed and embedded under a dissecting scope. For the transverse and coronal orientation, fish were embedded with their heads in the wide end of the triangle, ventral side up (Fig. 2D). Fish to be sectioned sagittally were placed in the mold on their right sides with heads in the large end of the triangle. Once the fish were positioned, additional 1% melted agarose in ddH2O, cooled to 55 °C, was gently pi-petted over the wells in such a way as not to disturb the position of the fish or fill the well beyond the surface of the well. Once the agarose is solidified, the molds were trimmed as required by the intended orientation before placing in embedding cassettes (Tissue-Tek Uni-Cassette LWS, Cat# 4155-02) in 70% ethanol for tissue processing. During paraffin embedding the molds are positioned in their final orientation in the block.
Fig. 1. Larval Array Molds.
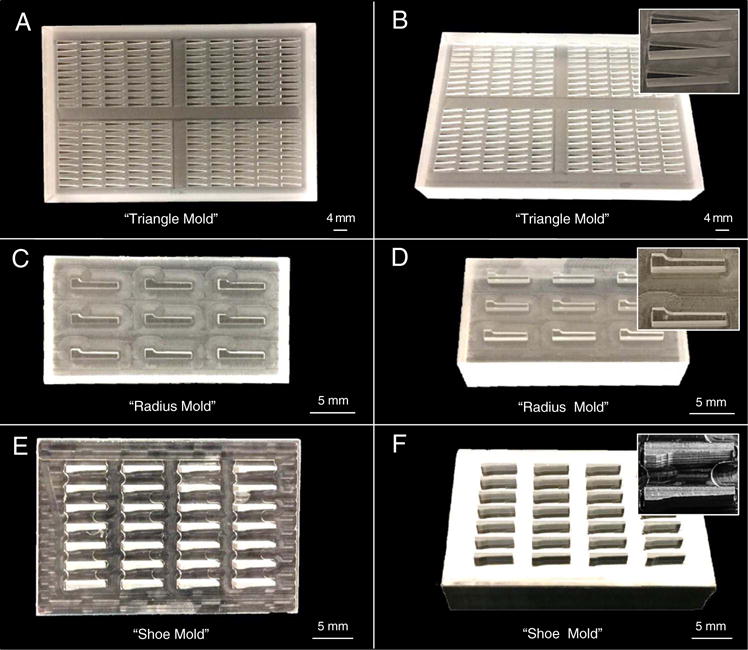
Three different array molds from our laboratory are shown with three different views for each (left: from the top, right: at an angle, right inset: zoomed in view of the positive mold). The original “triangle mold” (A–B), and new “radius mold” (C–D), and “shoe mold” (E–F). Both the “radius mold” and “shoe mold” improved embedding results with the latter showing slightly better fish positioning and reproducibility.
Fig. 3. Juvenile Agarose Embedding.
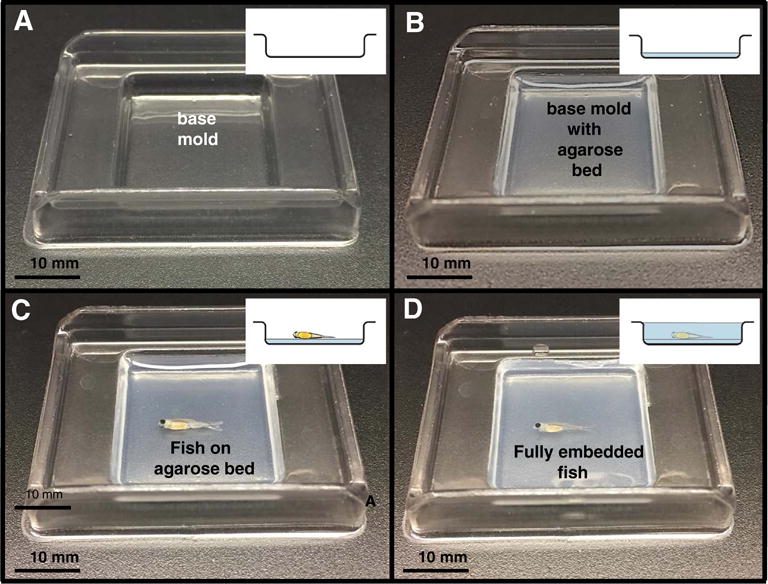
To enhance embedding reproducibility and to facilitate orientation, juvenile zebrafish were embedded in agarose prior to infiltration in paraffin. Figs. A–D show the various embedding stages with inset drawings illustrating the side view. (A) 15 mm × 15 mm base mold. (B) 15 mm × 15 mm base mold filled with agarose. (C) Base mold with fish positioned for embedding. (D) Mold containing an embedded 44 dpf (10 mm) juvenile.
Fig. 2. Agarose Array Embedding.
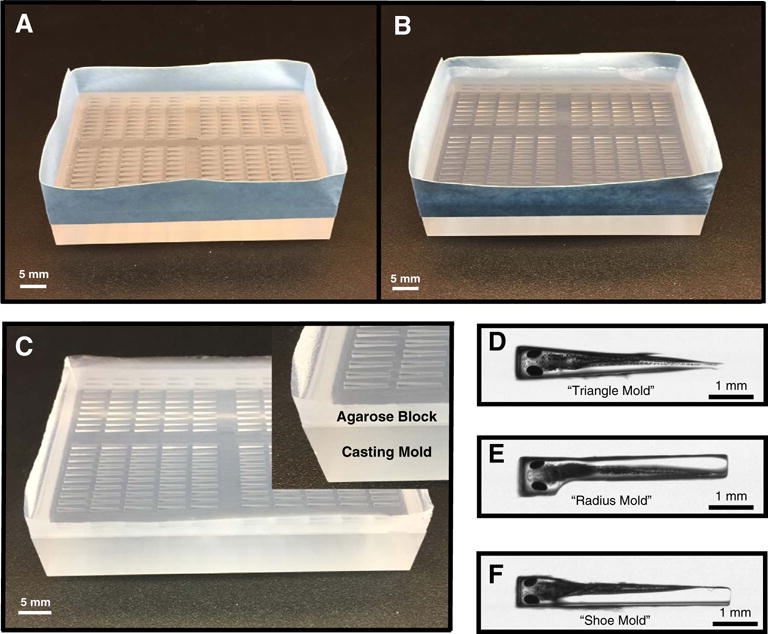
To facilitate larval embedding, casting molds were used to create agarose array blocks. (A) Casting mold taped all the way around the sides before the addition of agarose. (B) Taped casting mold with the agarose added. (C) Agarose block and casting mold after removal of tape (inset: closeup of a corner showing the agarose on top and acrylic mold below it). (D) Closeup view of larvae in the “triangle mold” (E) “radius mold” and (F) “shoe mold” from above to illustrate larval position during embedding. A slight bending apparent for both “triangle” and “radius” molds is not present in the “shoe” mold closeup.
Larval and juvenile stages between the ages 21 dpf to 89 dpf (21–29 dpf, 7.8 mm; 30–44 dpf, 10 mm; 45–89 dpf, 14 mm) were pre-embedded as follows. For each fish, a flat bed of 1% Agarose in ddH2O was poured (about 0.4 ml) into a level disposable 15 mm × 15 mm base mold (Surgipath #03025) just until the bottom of the mold was covered and allowed to solidify (Fig. 3). Fish to be sectioned in the transverse orientation were cut prior to embedding with a Feather double edge blade (Ted Pella Inc. Cat# 121-9). The 7.8 mm and 10 mm fish were cut in half and the 14 mm fish were cut into thirds. Once the molds were solidified they were transferred under a dissecting scope for the positioning and embedding process. Fish designated for sagittal and coronal orientations were laid on their sides on the bed of agarose to enable orthogonal paraffin sections. For the transverse sectioned fish (which were precut in halves or thirds, as described above), the fish sections were laid on their side and on top of each other such that the pieces were in the same rostral-caudal direction. Once the fish were positioned on the bed of agarose, 55 °C 1% agarose in ddH2O was pi-petted over the fish using fire-polished wide-bore Pasteur pipettes, gently enough that the fish position was not disturbed. Some users prefer Pasteur pipettes to be flamed, pulled, and broken to create narrower tips for more controlled agarose distribution. The agarose blocks were trimmed after solidifying and placed in embedding cassettes in 70% Ethanol for tissue processing. During paraffin embedding the fish were positioned in the appropriate final orientation for sectioning.
Embedding larvae using the triangular mold can cause the heads to bend (away from the wall to which the tail adheres by surface tension). Two additional embedding array casting molds were designed by our laboratory to eliminate this bending artifact (Fig. 1C-F and Fig. 2E, F). The “radius mold” is shaped with one side straight and the other side with a notch that fits the head of the fish followed by one of three different curvatures of radius (0.50 mm, 0.55 mm, 0.60 mm) intended to follow the contour of the larvae (Fig. 1C, D). The “shoe mold” is a refinement of the radius mold with one straight side like the radius mold, but follows the actual contour of the larvae as determined by MicroCT on the other side (Fig. 1E, F). Both molds are designed for the larvae to adhere to the contoured side of the well through surface tension.
To test the new mold designs 3 dpf (3.5 mm), 6 dpf (4.2 mm), and 7 dpf (4.5 mm) larvae were staged and fixed overnight at room temperature in 10% NBF before beginning dehydration in 70% ethanol overnight at room temperature. The agarose embedding molds were prepared with 1% agarose in ddH2O for both the radius and shoe casting molds. Solidified agarose molds were placed under a dissecting scope to facilitate embedding. Molds were embedded with the fish ventral side up with their heads in the large end of the well and the sides of their bodies against the contoured side of the well. Warm 55 °C 1% agarose in ddH2O was then pipetted into the well and allowed to harden. Molds were then processed into paraffin blocks, sectioned, mounted and stained before data analysis. Slides were viewed and each larval position was classified into one of the three different categories of orthogonal plane (Fig. 4). Categories were assigned a numerical value and the values within data points were averaged and compared.
Fig. 4. Larval Position Categories.

Larvae were assigned into 3 different categories based on their orthogonal plane alignment from the array mold analysis. For comparison, each coronal section was given one of the following numerical values: 3 (nearly orthogonal), 2 (partially orthogonal), and 1 (not orthogonal), with orthogonal (3) being ideal. Specifically, ‘partially orthogonal’ can be defined by a larva whose eyes are tilted in such a way that the lenses are of unequal size. ‘Not orthogonal’ means that the larva is tilted such that only one eye can be seen in any plane of section.
2.4. Processing, sectioning, and staining
For microtome sectioning, all fish were dehydrated and infiltrated in a RMC Model 1530 automated closed reagent type tissue processor utilizing the distinct programs (Table 4). We used graded alcohols for dehydration, and xylene for tissue clearing before infiltration with Formula R paraffin (Leica Biosystems #3801450), which contains a plastic additive that in turn allows for smoother sectioning and reduced tissue compression. For agarose pre-embedded samples, all ethanol solutions as well as the final xylene were unused to achieve the best possible sections, and to minimize the probability of distortion. For non-pre-embedded samples, solutions are kept fresh and are changed according to the number of blocks processed. Once infiltration was complete, all fish were manually positioned in paraffin blocks in their final orientation in preparation for sectioning. All fish were serially sectioned on a Leica RM2255 automated rotary microtome set to 5 μm thickness. During sectioning, the tissue block face was soaked in ice water for 10 to 15 min between sets of 10–12 section ribbons to minimize cracking of the sections and to facilitate optimal sectioning. Note that leaving out this hydrating manipulation of the block face yields unsatisfactory sections. It is hoped that future modifications to tissue preparation may be found to make this time-consuming step unnecessary. After sectioning and before cover slipping, tissue sections were floated onto slides and stained with H&E as is commonly used in histology (Table 5).
Table 4.
Tissue processing steps.
| Time | Solution | Temp (°C) | Vacuum (inches, Hg) |
|---|---|---|---|
| 45 min | 80% ETOH | 25 | 15 |
| 45 min | 95% ETOH | 25 | 15 |
| 1:00 h | 95% ETOH | 25 | 15 |
| 1:00 h | 100% ETOH | 25 | 15 |
| 1:00 h | 100% ETOH | 25 | 15 |
| 1:00 h | 100% ETOH | 25 | 15 |
| 1:00 h | Xylene | 25 | 15 |
| 1:00 h | Xylene | 25 | 15 |
| 1:30 h | Paraffin | 60 | 15 |
| 1:30 h | Paraffin | 60 | 15 |
| 2:00 h | Paraffin | 60 | 15 |
Table 5.
Automated staining steps.
| Time | Solution |
|---|---|
| 1–3 min | Xylene |
| 2–5 min | Xylene |
| 2 min | 100% ETOH |
| 2 min | 95% ETOH |
| 1 min | Tap H2O |
| 3 min | Hematoxylin |
| 1 min | Tap H2O |
| 1 min | Acidified alcohol |
| 1 min | Tap H2O |
| 0.2 min | Ammoniated H2O |
| 1 min | Tap H2O |
| 0.3 min | Eosin |
| 1 min | 30% ETOH |
| 1 min | 95% ETOH |
| 1 min | 100% ETOH |
| 1 min | Xylene |
2.5. Digital image capture
Over the last 10 years, slide scanner technology has progressed from impractical to highly effective. The first slide-scanner yielding usable scans was made by the dominant vendor, Aperio (Leica Biosystems). The present content of the zebrafish atlas is based on Aperio-based scans of 1 × 3″ glass histology slides made on an Aperio T2 ScanScope (as described by Sabaliauskas et al., 2006) or a Aperio ScanScope XT. The Aperio ScanScope XT has an automated slide loader which can accommodate 120 slides at a time, but has a 20× objective and a “doubler” instead of interchangeable 20× and 40× objectives whose changing tended to cause misalignment issues. The “doubler” mechanism greatly improved scan reproducibility. Additional improvements included moving of the computer (with its vibration and heat) outside of the scanning box, stage orientation, slide handling, actuators, and software, yielding faster scans and improved scan quality. Slide scanning includes a workflow in which the operator selects a scan area and places focus points for each slide prior to automated scanning. To produce the virtual slides displayed on the atlas website, all slides from a given block were preliminarily scanned using an Aperio ScanScope XT slide scanner at 20×. A final subset of slides was selected from these images being careful to represent all structures and to most accurately represent zebrafish histology. Final sets of slides targeted for posting were scanned at the highest resolution (40×) and saved as lossless LZW files to avoid compression artifact.
3. Results and discussion
3.1. Fixation and decalcification
The goal of fixation is to preserve cells and extracellular materials to resemble in vivo tissue architecture as closely as possible (Bancroft and Gamble, 2008). Fixation halts the degradation of tissue by preventing autolysis and by killing microorganisms, while stabilizing the macromolecular structure of the tissue by crosslinking, to allow for downstream tissue processing steps (Bancroft and Gamble, 2008). Fixation without sample distortion is a challenge, since all fixation methods produce artifacts such as tissue shrinkage, swelling, hardening, and color change (Bancroft and Gamble, 2008). Additionally, fixation and fixative choice can have a significant impact on subsequent tissue processing steps, particularly sectioning and staining (Bancroft and Gamble, 2008). With all these factors in mind we set out to determine the most suitable fixative for zebrafish that were ultimately to be stained with H&E, the most commonly used stain for human and veterinary histopathology. Due to the relative importance of fixation as the first step in tissue processing, and a scant literature focusing on tissue processing for zebrafish, we tested an array of commonly used tissue fixatives, including Zenker’s, Zamboni’s, Zinc-Formalin, Bouin’s, Formaldehyde/Glutaraldehyde, and 10% NBF (Table 1).
All of the fixatives were tested on 7 dpf zebrafish larvae (Fig. 5). Variations in fixation length and temperature were determined by the characteristics of individual fixatives and the type of tissues to be fixed. When evaluating the relative quality of the fixatives tested, we considered the quality of tissue preservation, nuclear clarity, rate of fixative penetration, speed of fixation, and staining properties with H&E. While many histological stains are available, we use H&E because it is the most commonly used stain in human pathology. Our goal was to obtain patterns of tissue structure and staining as close as possible to that seen in human histopathology to maximize the utility of zebrafish as a model of human disease. To help quantify our data we ranked all the fixatives tested against each other from best (1) to worst (12) for each of the following (equally weighted): tissue preservation quality, fixative penetration rate, nuclear clarity, staining with H&E and ease of use (Table 6). The rankings were then averaged to give a final rank. When evaluating for tissue preservation quality, we examined the larvae noting the amount of tissue shrinkage or expansion, tearing or displacement of structures and clarity of cells and muscle striations. Fixative penetration rate and fixation were evaluated by looking primarily at the preservation of gut architecture (Fig. 5). In our experience, the intestinal tract, liver and pancreas are the most sensitive to poor fixation, presumably due to the presence of digestive enzymes in these organs. Nuclear clarity was evaluated by looking at the clarity of nuclear edges (primarily for neuronal nuclei), or of nucleoli and chromatin condensations when present (for example, in gut, liver, or cartilage). In determining H&E staining quality we evaluated for color and tissue clarity. Finally, we considered ease of use in terms of fixation time and temperature, fixative ingredient toxicity, especially picric acid, and protocol complexity. Since we would be processing many samples for the Zebrafish Histology Atlas, we sought efficiency and protocol simplicity.
Fig. 5. Comparison of Fixatives in Larvae.

Panels (A–L) show representative sections of the gut for the fixation trials conducted on 7 dpf wild type Connor zebrafish. All fixatives were evaluated for quality of tissue preservation, amount and type of artifact, as well as rate of fixative penetration across the entire fish. (A) Zenker’s, (B) Zamboni’s, (C) Zinc-formalin, (D) Bouin’s, (E) Formaldehyde/Glutaraldehyde with EDTA, (F) Formaldehyde/Glutaraldehyde with Picric Acid, (G) 10% Neutral Buffered Formalin (NBF) 4 °C for 24 h, (H) 10% NBF 4 °C for 48 h, (I) 10% NBF 4 °C for 8 days, (J) 10% NBF 21 °C for 24 h, (K) 10% NBF 21 °C for 48 h, and (L) 10% NBF 21 °C for 8 days. Final results on the trials indicated that fixation with room temperature 10% NBF for 24 h yielded the most optimal results. Because independent high power views would poorly represent the variations in results encountered, we have provided access to full slide scans for each of these conditions at http://bio-atlas.psu.edu
Table 6.
Fixative ranking.
| Fixative | Tissue preservation quality | Fixative penetration rate | Nuclear clarity | Staining with H & E | Ease of use | Overall score average/ranking |
|---|---|---|---|---|---|---|
| NBF 24 h 21 °C | 1 | 1 | 2 | 1 | 1 | 1.2 |
| NBF 48 h 21 °C | 2 | 2 | 3 | 2 | 2 | 2.2 |
| NBF 8 days 21 °C | 3 | 3 | 4 | 3 | 3 | 3.2 |
| NBF 24 h 4 °C | 4 | 4 | 5 | 4 | 4 | 4.2 |
| NBF 48 h 4 °C | 5 | 5 | 6 | 5 | 5 | 5.2 |
| NBF 8 day 4 °C | 6 | 6 | 7 | 6 | 6 | 6.2 |
| Bouin’s | 7 | 7 | 1 | 10 | 10 | 7 |
| Zenker’s | 8 | 8 | 8 | 8 | 8 | 8 |
| Zamboni’s | 9 | 9 | 9 | 9 | 9 | 9 |
| Zinc-formalin | 10 | 10 | 10 | 10 | 7 | 9.4 |
| Formaldehyde/glutaraldehyde | 11 | 11 | 11 | 11 | 11 | 11 |
| Formaldehyde/glutaraldehyde/picric acid | 12 | 12 | 12 | 12 | 12 | 12 |
The first tested fixative, Zenker’s Fluid, contains mercuric chloride and potassium dichromate (both highly toxic). Zenker’s is reputed to fix tissues rapidly, with structural preservation generally occurring within 6 h of fixation (Kiernan, 2008). We found that Zenker’s fixed larvae showed poor fixation, staining, and nuclear clarity (Fig. 5A). For example, poor gut fixation resulted in a hazy appearance of the cells and degradation of the columnar arrangement of the gut epithelial cells.
Zamboni’s fixative contains paraformaldehyde and picric acid. This formulation is used to achieve rapid penetration of large tissue specimens (Sheehan and Hrapchak, 1980). As with the Zenker’s fluid we chose to fix our 7 dpf larvae for 24 h at 4 °C with the lower temperature intended to reduce tissue decomposition. While the Zamboni’s fixative yielded good nuclear clarity, tissue preservation was inconsistent, leading to areas of poor fixation within the fish (Fig. 5B). While the fixation time was short, the gut was poorly preserved. Tissues that were adequately fixed showed good staining with this fixative.
Zinc-formalin, which contains zinc-sulfate and formalin, penetrates and fixes tissues quickly, generally fixing within 6 to 8 h (Kiernan, 2008). For our study, we fixed 7 dpf larvae for 6 h at 4 °C. Review of our Zinc-Formalin fixed larvae revealed adequate nuclear clarity with mixed tissue preservation resulting in areas of good fixation mixed with areas of poor fixation. The Zinc-Formalin fixative appeared to have a quick rate of penetration and fixation, but in our hands yielded inconsistent fixation (Fig. 5C). Larvae stained with H&E after fixation with zinc-formalin produced well-stained sections.
Bouin’s fixative contains picric acid, formalin, and acetic acid. Bouin’s is known for preservation of nuclear features, and is reputed to show minimal tissue distortion and to facilitate ease of tissue sectioning (Kiernan, 2008). In this trial, we fixed 7 dpf larvae in Bouin’s for 24 h at 4 °C; again, the lower temperature intended to decrease cellular degradation prior to fixation. Examination of the larvae after processing and staining showed adequate fixation and superior nuclear clarity, colors in H&E staining were very bright but had a yellow cast indicating the need to adjust staining parameters to accommodate this fixative (Fig. 5D). Additionally, larvae showed an extensive amount of tissue shrinkage resulting in tissue distortion. However, it should be noted that the Bouin’s fixed larvae had by far the best nuclear clarity of the fixatives tested. While this fixative is easy to obtain and commonly used in fish laboratories, picric acid is corrosive and stains things yellow; furthermore, special care is required for storage and disposal due to the reputed susceptibility of dried picric acid to explosion (Zhang et al., 2016).
Due to its common use for electron microscopy, we also tested a 2% formaldehyde/1% glutaraldehyde combination. This combination is known to fix intracytoplasmic structures well and combines the superior fixing but slow penetrating properties of glutaraldehyde with the rapidly penetrating characteristics of formaldehyde (J. A. Kiernan, 2008). We fixed 7 dpf larvae in formaldehyde and glutaraldehyde for 48 h at room temperature (21 °C). In addition to the fixative we also added half saturated picric acid for a separate incubation period to half of the samples to test for better staining (Table 1). Picric acid is known to increase the intensity of staining with acid dyes like Eosin (J. A. Kiernan, 2008). While overall tissue preservation was good, the gut nuclei were consistently less distinct with this fixative, with or without the added picric acid (Fig. 5E, F). Nuclear clarity also tended to be lost, particularly in the brain. Poor gut fixation is consistent with a low rate of tissue penetration or fixation. Overall, larvae fixed with the formaldehyde and glutaraldehyde fixative, both the picric acid and non-picric acid treated samples, stained poorly, yielding an overall hazy staining quality.
The most commonly used fixative in human pathology is “10% neutral buffered formalin” (NBF). Actually, 10% NBF is 4% formaldehyde, deriving from 10% of a 37–40% formaldehyde stock solution. NBF fixative is characterized by rapid tissue penetration and fixation through cross-linking (Kiernan, 2008). It is important to note that the protein molecule cross-linking by formaldehyde is much slower than for other fixatives (Kiernan, 2008). In fact, complete fixation can take as long as 1 to 2 weeks for larger tissue specimens (Kiernan, 2008). In practice, adequate fixation of larval zebrafish occurs within 24 h at either room temperature (21 °C) or 4 °C. As a result of these characteristics of NBF, we tested fixation of 7 dpf larvae in this fixative for 24 h, 48 h, and 8 days at both 4 °C and 21 °C. Review of the fixed larvae showed good overall tissue preservation for all time points and both temperatures. However, larvae fixed at 4 °C (Fig. 5G, H, I) consistently displayed poor gut fixation in all time points perhaps due to slower fixation at lower temperatures (Sheehan and Hrapchak, 1980). Nonetheless, nuclear clarity and staining were good in tissues that were adequately fixed at 4 °C. Larvae fixed at 21 °C at all time points exhibited good tissue preservation, nuclear clarity, and staining with the rate of tissue penetration and fixation adequate to prevent significant tissue degradation and autolysis (Fig. 5J, K, L).
We highlight the digestive tract as an excellent means of evaluating fixative penetration and fixation (Fig. 5). Whole larvae virtual slides are posted on the bio-atlas website. Our virtual slides enable visualization of whole larvae at magnifications ranging from 1× through 40× (0.25 μm per pixel resolution), allowing reader assessment of the criteria we used in our evaluation of the fixatives. While still only a representative sample of the data used in this study, these data will allow readers greater insight into our methods and conclusions. We note that this technology raises the possibility of enhancing independent validation and discovery by associating whole virtual slides with future published work that is based on histological data. Table 6 summarizes our findings based on ranked scoring.
Outside of the native ability of fixative molecules to enter tissues, the concentration gradient of fixative to tissue is a key determinant of fixative diffusion into tissue to enable fixation. Since dilution occurs at the tissue/fixative interface, we asked whether movement of the fixative solution during fixation would increase the speed and completeness of fixation. Juvenile (57 dpf) zebrafish were fixed in NBF at 21 °C for 48 h. Review of the Juvenile fish fixed in NBF that was slowly stirred as compared with Juvenile fish fixed in NBF that was still, showed similar fixation efficiency. In sum we found that the use of NBF for fixation at 21 °C for 24 h yielded good results. This fixative had the best overall tissue preservation and staining of the fixatives tested. It is worth noting that the aforementioned survey of fixatives and fixation variables was performed on larvae; therefore, the optimal protocols for juvenile and adult zebrafish remain to be determined. Additionally, we observed some amount of tissue shrinkage with all the fixatives tested. While the amount varied, from the most with Bouins’ to the least with the NBF, this artifact was persistent across all fixatives.
3.2. Larval mold design for pre-embedding
While the “triangle mold” (Fig. 1A, B) we described in Sabaliauskas et al., 2006 greatly improved the throughput of producing embryonic and larval histology slides, there remain additional challenges to address. One challenge is variability in the position of larvae within the mold, caused by two factors, placement of the fish in the well, and movement of the fish during addition of agarose. Furthermore, the size of the eyes led to a bending of the head away from the surface to which the larvae are allowed to stick by surface tension. To address these problems, we developed two additional casting molds designed to position the fish more precisely than the triangle mold.
The first, “radius mold” expanded upon the triangle mold changing the shape of the mold such that one side was straight and the other side had a notch for the head of the fish followed by one of three different radius curvatures (0.50 mm, 0.55 mm, 0.60 mm) intended to follow the contour of the larvae, followed by a slightly slanting edge thinning towards the tail (Fig. 1C, D). The fish was intended to lie against the side with the curve and through surface tension, provide a more precise positioning. The second mold “shoe mold” is a refinement of the radius mold. Since our laboratory has been involved in 3D reconstructions of zebrafish utilizing microCT scanning, we were able to use these images to determine the outside contours of embryos to develop a mold that better fit the fish. The shoe mold has one straight side like the radius mold, but follows the actual contour of the larvae as determined by microCT on the other side (Fig. 1E, F). As with the radius mold the fish was intended to adhere to the contoured side of the mold through surface tension.
Embedding trials performed on the triangle, radius and shoe molds with 3 dpf (3.5 mm), 6 dpf (4.2 mm), and 7 dpf (4.5 mm) larvae were conducted and larval position within each mold was assigned a score ranging from 1 for fish that were not in plane to 3 for fish with near orthogonal plane alignment (Fig. 4). The values within the datasets for each mold design were averaged and then compared across the three types of mold. The average score for the original triangle mold was 1.8, the radius mold was 2.4 (all curvatures), and the shoe mold, 2.6. With respect to analysis of the radius mold, the three different curvatures resulted in no difference in the orthogonal scoring scheme and so were averaged together. Taken together, review of the data determined that both the radius and shoe mold improved fish position and reproducibility compared with the original triangle mold, with the shoe mold yielding perhaps slightly better results. It is worth noting that the straighter the fish the easier it is to obtain near orthogonal plane alignment (ideal coronal plane section). Straighter fish would be ideal, to minimize the potential scoring of bending artifact as phenotype. We speculate that additional improvements are possible that will further increase positioning precision for larval array histology.
3.3. Processing, sectioning and staining
To support the tissues for microtome sectioning, tissue samples are infiltrated with paraffin (Sheehan and Hrapchak, 1980). Paraffin is commonly used in histology due to its ease of use. Greater rigidity is required for plastic sections than for paraffin sections, due to the increased resistance of plastic to cutting. This rigidity allows the cutting of thinner sections (≤3 μm), but cutting plastic sections is more difficult and labor intensive than paraffin sectioning. To make section cutting smoother, we chose paraffin that contains some plastic: Formula R paraffin (Leica Cat# 3801450). Prior to infiltration with paraffin, the tissues must be completely dehydrated in graded Ethanol solutions to 100%. Since paraffin is not miscible with ethanol, a clearing agent must be used to enable the paraffin to fully saturate the tissue. We used xylene as our clearing agent as it is commonly used in standard histology.
Early efforts to improve high throughput histology through pre-embedding in agarose tissue arrays by us and others frequently resulted in severe distortion of agarose blocks after infiltration, where they would “bow tie” (become indented in the middle) or worse, “cornflake” (severely shrink and wrinkle) – both useless for histology. After testing a variety of variables, we determined that these defects occur when old tissue processing solutions are used, presumably because they no longer close to their original concentrations. When samples are not agarose embedded, solutions in tissue processors can be rotated, and the ethanol solutions not completely changed every run, without untoward effect. However, for agarose embedded tissues, fresh solutions are essential. We therefore change out all the ethanol solutions and the second xylene solution every time we run agarose blocks. Consistent use of fresh solutions has eliminated block distortion.
Another factor that we have found important in the production of optimal sections is soaking the block face in ice water prior to sectioning. We have observed, empirically, that zebrafish sections require between 10 and 15 min of soak time for every 10 to 12 section ribbon cut. Additionally, care must be taken when floating the sections in the water bath prior to mounting on slides; agarose embedded samples tend to detach from their agarose if floated more than a minute prior to mounting (personal observation).
Fixative choice can affect staining and we have optimized staining protocols both for the fixative we use and also for the tissues we are staining. We have found the protocol listed in our staining chart (Table 5) produces optimal results for zebrafish. While most of the protocol is standard for regressive hematoxylin, we do alter our protocol slightly. We have found that our samples require more time in the acid alcohol. A longer duration in the acid alcohol allows for the less basophilic structures to release more of their stain. This results in a gradient of staining that allows for better visualization of structures especially within the zebrafish eye.
3.4. Digital image capture
Central to the task of digital imaging for the atlas project is the ability to create virtual slides that will be posted on our website as a comparative resource for researchers. We have used Aperio slide scanners for this purpose. What differentiates a slide scanner from a standard microscope fitted with a camera is the scanners’ ability to capture an entire slide at a specific magnification and then, through the use of computer software, generate a virtual slide of the original slide. This virtual slide can then be shared through computers or posted on the internet and be viewed on the computer as if it were a standard glass slide on a microscope. This effectively turns any computer into a microscope with the ability to adjust magnification and position anywhere on the virtual slide using the computer mouse or touch pad, with additional potential digital benefits provided by a navigator for orientation, cross-referencing, labeling, and comparisons that are not readily accomplished using glass slides. For our atlas web site (http://bio-atlas.psu.edu) we have scanned our slides at 40× (0.25 μm per pixel) resolution, the highest magnification available with the scanner we have, enabling the full range of magnifications to be used. Scanning at this resolution allows for observation of tissue structure, cellular composition, and even characterization of physiological and disease states.
4. Conclusions
The histological preparation of tissues creates a stable sample that will degrade little over time, and can be viewed on a microscope and captured digitally. In either case, tissue and cellular structures anywhere on the full slide can be resolved. Ideally any methods used to achieve this would minimize the addition of artifact. In the real world, any method is associated with artifact, either accidently as in tissue shrinkage from fixation or intentionally as in staining of tissues (e.g. H&E staining), to better understand structure. With this ideal in mind, we set out to improve methods used in the histological preparation of zebrafish. We have determined satisfactory methods for fixation, decalcification, agarose pre-embedding, paraffin embedding, sectioning, staining and digital image capture. While our work has provided a basis for zebrafish histology preparation, three problems remain. The first is that of differential tissue shrinkage resulting in gaps between tissues that do not exist in the live fish. Shrinkage may be associated with differences between solution and tissue osmolality, which will require further experiments. Additionally, we have observed incomplete fixation of internal organs in some juvenile and adult fish, resulting in tissue degradation. A potential solution to both of these problems is fixative perfusion. We have not evaluated this method to date because we wanted to preserve the presence of blood cells in the vasculature to facilitate the identification of blood vessels. Finally, we would like to find a protocol for tissue processing that eliminates the present need for soaking of the block face during sectioning, which would greatly increase throughput. Eliminating these problems would yield both more accurate representations of the fish and increase throughput.
Discussion in model system meetings internationally has indicated a need for cross-referencing of microanatomic data across model systems. We have identified a number of steps towards the goal of an atlas that extends beyond the zebrafish research community to the full range of model system researchers, particularly those who work on models of human disease. Among the desired goals are the additions of slide collections from the full range of models of human disease, as well as human pathology slides. Reference collections will greatly facilitate education, and interactions between model systems communities and physicians. Advances in 3D imaging have also led to the improved ways to visualize and analyze 3D images; pre-computing and cloud resources can be used to make even large data sets web-accessible. Web-based anatomic reference atlases are likely to advance the development and use of animal models of human disease. Such atlases, in conjunction with the broader use of virtual slides rather than simply photos of single fields of view, can be expected to improve rigor, reproducibility and transparency in biomedical and clinical research and reporting.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Stephen Moorman for discussion about fixatives, Dr. Khai Chung Ang and Spencer Katz for their contributions with manuscript preparation and past and current members of the Cheng Laboratory for their suggestions and help. This work was supported by The Jake Gittlen Memorial Golf Tournament, NIH grants RO1-HD40179, R24-RR17441, and R24-OD018559, as well as Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program Grant SAP-4100026343. The Department of Health specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbpc.2017.11.003.
References
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many Ribosomal Protein Genes are Cancer Genes in Zebrafish. PLoS Biol. 2004;2(5):E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft JD, Gamble M. Theory and Practice of Histological Techniques 2008 [Google Scholar]
- Beckwith LG, Moore JL, Tsao-Wu GS, Harshbarger JC, Cheng KC. Ethylnitrosourea induces neoplasia in zebrafish (Danio rerio) Lab Investig. 2000;80(3):379–385. doi: 10.1038/labinvest.3780042. [DOI] [PubMed] [Google Scholar]
- Görge G, Nagel R. Toxicity of lindane atrazine, and deltamethrin to early life stages of zebrafish (Brachydanio rerio) Ecotoxicol Environ Saf. 1990;20:246–255. doi: 10.1016/0147-6513(90)90004-o. [DOI] [PubMed] [Google Scholar]
- Kiernan JA. Histological and Histochemical Methods 2008 [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of Embryonic Development of the Zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Matthews M, Varga ZM. Anesthesia and Euthanasia in Zebrafish. ILAR J. 2012;53(2):192–204. doi: 10.1093/ilar.53.2.192. [DOI] [PubMed] [Google Scholar]
- Mohideen MA, Beckwith LG, Tsao-Wu GS, Moore JL, Wong AC, Chinoy MR, Cheng KC. Histology-based screen for zebrafish mutants with abnormal cell differentiation. Dev Dyn. 2003;228:414–423. doi: 10.1002/dvdy.10407. [DOI] [PubMed] [Google Scholar]
- Moore JL, Aros M, Steudel KG, Cheng KC. Fixation and decalcification of adult zebrafish for histological, immunocytochemical, and genotypic analysis. BioTechniques. 2002;32(2):296–298. doi: 10.2144/02322st03. [DOI] [PubMed] [Google Scholar]
- Sabaliauskas NA, Foutz CA, Mest JR, Budgeon LR, Sigor AT, Gershenson JA, Joshi SB, Cheng KC. High-Throughput Zebrafish Histology. Methods. 2006;39:246–254. doi: 10.1016/j.ymeth.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132(15):3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Zon LI. Hooked! Modeling Human Disease in Zebrafish. J Clin Invest. 2012;122(7):2337–2343. doi: 10.1172/JCI60434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Theory and Practice of Histology 1980 [Google Scholar]
- Stewart AM, Braubach O, Spitsbergen J, Gerlai R, Kalueff AV. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37(5):264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KL, Grant NJ, Temperley ND, Patton EE. Small molecule screening in zebrafish: an in vivo approach to identifying new chemical tools and drug leads. Cell Commun Signal. 2010;8:11. doi: 10.1186/1478-811X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a Model Vertebrate for Investigating Chemical Toxicity. Toxicol Sci. 2005;86(1):6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- Tsao-Wu GS, Weber CH, Budgeon LR, Cheng KC. Agarose-embedded tissue arrays for histologic and genetic analysis. BioTechniques. 1998;25:614–618. doi: 10.2144/98254st02. [DOI] [PubMed] [Google Scholar]
- Wilson JM, Bunte RM, Carty AJ. Evaluation of Rapid Cooling and Tricaine Methanesulfonate (MS222) as Methods of Euthanasia in Zebrafish (Danio rerio) J Am Assoc Lab Anim Sci. 2009;48(6):785–789. [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Yue YY, Luo HQ, Li NB. Supersensitive and selective detection of picric acid explosive by fluorescent Ag nanoclusters. Analyst. 2016;141(3):1091–1097. doi: 10.1039/c5an02251g. [DOI] [PubMed] [Google Scholar]
- Zhao S, Huang J, Ye J. A fresh look at zebrafish from the perspective of cancer research. J Exp Clin Cancer Res. 2015;34:80. doi: 10.1186/s13046-015-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


