Abstract
Evolutionarily conserved diurnal circadian mechanisms maintain oscillating patterns of gene expression based on the day-night cycle. Xiphophorus fish have been used to evaluate transcriptional responses after exposure to various light sources and it was determined that each source incites distinct genetic responses in skin tissue. However, basal expression levels of genes that show oscillating expression patterns in day-night cycle, may affect the outcomes of such experiments, since basal gene expression levels at each point in the circadian path may influence the profile of identified light responsive genes. Lack of knowledge regarding diurnal fluctuations in basal gene expression patterns may confound the understanding of genetic responses to external stimuli (e.g., light) since the dynamic nature of gene expression implies animals subjected to stimuli at different times may be at very different stages within the continuum of genetic homeostasis. We assessed basal gene expression changes over a 24-hour period in 200 select Xiphophorus gene targets known to transcriptionally respond to various types of light exposure. We identified 22 genes in skin, 36 genes in brain and 28 genes in liver that exhibit basal oscillation of expression patterns. These genes, including known circadian regulators, produced the expected expression patterns over a 24-hour cycle when compared to circadian regulatory genes identified in other species, especially human and other vertebrate animal models. Our results suggest the regulatory network governing diurnal oscillating gene expression is similar between Xiphophorus and other vertebrates for the three Xiphophorus organs tested. In addition, we were able to categorize light responsive gene sets in Xiphophorus that do, and do not, exhibit circadian based oscillating expression patterns.
Keywords: circadian rhythm, Xiphophorus, gene expression
Introduction
The diurnal or circadian cycle has been shown to affect the physiology of all organisms where it has been studied (Panda et al., 2002a). These physiological changes are coincide with oscillating gene expression patterns that are coordinated to adapt cellular activity to periods of activity and inactivity. The core mechanisms that lead to endogenous circadian oscillation of gene expression involve a transcription-translation feedback loop controlled by transcription factors CLOCK and brain and muscle ARNT-like protein 1 (BMAL), and their antagonistic transcriptional targets Cryptochrome (CRY) and Period (PER). CRY and PER serve as repressors of CLOCK and BMAL transactivation activity (Mohawk et al., 2012). Oscillating expression patterns of these circadian regulators can be entrained by external environmental cues, such as alteration in light periods, feeding patterns, or changes in temperature (Raible et al., 2017; Migaud et al., 2010; Carr et al., 2006; Rensing and Ruoff, 2002).
Teleost fish represent useful vertebrate model systems to investigate circadian gene expression. The diversity of physiological adaptations to extremely varied environments allows exploration of the plasticity in patterns of basal gene expression. Circadian cycle regulator genes have been characterized in several teleost fish species, including zebrafish, medaka, flounder, amberjack, sea bass and blind cavefish (Beale et al., 2013; Cavallari et al., 2011; Cuesta et al., 2014; Sanchez et al., 2010; Toyama et al., 2009; Wang, 2008; Watanabe et al., 2012). Comparative genetic analyses among fish models have identified different regulatory mechanisms associated with basal gene expression. For example, cave-dwelling populations of blind cavefish Astyanax mexicanus, compared to non-cave living populations, both exhibit robust circadian cycling of the per1 gene, but the cave-dwelling blind cavefish show elevated levels of light-induced genes (e.g., per2; Beale et al., 2013). The understanding of differences in regulation of basal gene expression, as a result of adaptation to an environment niche, may enhance our understanding of chronobiology. Compared to mammals in which the suprachiasmatic nucleus (SCN) serves as a master oscillation regulator, fish appear to utilize a photoreceptive pineal gland as the autonomous clock to drive melatonin synthesis and circadian rhythm (Bailes and Lucas, 2010). Additionally, fish have been shown to possess oscillation centers in peripheral organs (i.e., “peripheral clocks”) that may be directly entrained by light exposure (Vallone et al., 2004; Whitmore et al., 2000). These attributes make fish attractive models to study circadian gene regulation and light-induced genetic effects.
Xiphophorus represent a tractable vertebrate model that has recently been employed to investigate the molecular genetic responses to exposure from varied light sources and select light wavebands (Boswell et al., 2015; Chang et al., 2015; Lu et al., 2015; Walter et al., 2014; Walter et al., 2015; Yang et al., 2014; Boswell et al., 2017a; Boswell et al., 2017b). Genome sequence and assembly for several Xiphophorus species indicates they possess compact genomes retaining remarkable syntenic conservation with mammalian genomes (Amores et al., 2014; Schartl et al., 2013; Shen et al., 2016). Previous studies employing gene expression profiling of Xiphophorus skin reported that ultraviolet light effects the transcription of genes associated with apoptosis, cell cycle, circadian rhythm, fatty acid/lipid biosynthesis, wound healing, and cell differentiation (Boswell et al., 2015; Lu et al., 2015; Yang et al., 2014). In contrast, exposure to 4,100 K (“cool white”) fluorescent light (FL) was shown to incite a genetic response in Xiphophorus skin involving transcriptional suppression of gene sets (<130 genes) associated with cell cycle progression, chromosome segregation, DNA repair and recombination, as well as expected induction of circadian genes (Walter et al., 2015). Our previous reports showing light induced changes in transcriptional profiles were performed at a single time point (i.e., 7 am) in the normal diurnal or circadian cycle. However, due to oscillating expression patterns, light responsive genes are expected to be in different homeostatic activity states over the circadian cycle, perhaps affecting light induced gene expression. To better understand this experimental parameter, we sought to define basal gene expression patterns inherent to genetic homeostasis through the circadian cycle.
Herein, we assessed basal gene expression levels of previously identified light-responsive genes, to determine if they would exhibit oscillating gene expression patterns. We report gene expression patterns for a selected gene set in Xiphophorus skin, brain and liver over a 24-hour period. The three target organs represent external (skin), and internal (brain and liver) organs that play central roles in behavior, physiology, and metabolism. We designed a custom NanoString nCounter panel that contains 65 previously identified light-responsive genes, 60 reference circadian genes, and 10 housekeeping genes as internal controls to measure basal gene expression. Additionally, as tumors are known to have disrupted circadian cycle, for future studies involved with melanoma etiology, we have also assessed diurnal gene expression patterns of 65 genes that known to be associated with the Xiphophorus melanoma development.
Materials and Methods
Fish Utilized and RNA Isolation
Xiphophorus maculatus Jp 163 A in the 105th inbred generation were provided by the Xiphophorus Genetic Stock Center (http://www.xiphophorus.txstate.edu/). Mature male and female (age > 9 months) X. maculatus were maintained in separate 38-liter aquaria filled with filtered aquifer water from San Marcos, TX on a 14 hr light/10 hr dark cycle under 10,000 K FL (Coralife T8 Lamp 10,000 K, 32 W). All fish utilized, including 24 males and 12 females were combined in the same aquaria two weeks prior to the experiment. Fish were fed a supplement of liver paste at precisely 8 a.m. and brine shrimp at 3 p.m. during this period. For all analyses presented herein, 8 a.m. corresponds to Circadian Time (Ct) 0 hr. Two male X. maculatus were dissected for skin and brain at each circadian time point (Ct 0, 3, 6, 9, 12, 15, 18, and 21 hr) and for liver at Ct 3, 9, 15 and 21 hr. Two female fish were dissected for skin at Ct 0, 6, 12 and 18 hr; and liver at Ct 0, 6, 12 and 18 hr. At dissection, fish were anesthetized in an ice bath and upon loss of gill movement were sacrificed by cranial resection. Skin tissue were dissected directly into TRI Reagent (Sigma Inc., St. Louis, MO, USA), and flash frozen in a dry ice-ethanol bath for immediate RNA isolation, brain and liver tissues were dissected into RNAlater (Ambion Inc.) and maintained at −80°C for later use. All fish were maintained and samples taken in accordance with approved protocols (IACUC# 2015107711).
Total RNA was isolated as previously detailed (Lu et al., 2015). Briefly, samples were homogenized in TRI Reagent (Sigma Inc., St. Louis, MO, USA) followed by addition of 200 µL/mL chloroform, vigorously shaken, and subjected to centrifugation at 12,000 × g for 5 min at 4°C. Total RNA was further purified using an RNeasy mini RNA isolation kit (Qiagen, Valencia, CA, USA). Column DNase digestion was performed at 25°C for 15 min to remove residual DNA. Total RNA concentration was determined using a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY, USA), and RNA quality was verified on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) to confirm RIN scores were above 8.0 prior to NanoString analysis of gene expression.
Gene expression analysis by Nanostring nCounter
A 200-gene capture and reporter probe set was custom designed to represent the selected Xiphophorus genes. Of these 200 gene targets, 65 were designed to assess gene targets we have previously shown to transcriptionally respond to FL exposure (4,100 K or 10,000 K), ultraviolet B (UVB, 311 nm), or various 50 nm wavebands between 300–600 nm (Boswell et al., 2015; Chang et al., 2015; Lu et al., 2015; Walter et al., 2014; Walter et al., 2015; Yang et al., 2014). These light responsive genes were identified by comparing post light exposure gene expression profiles to unexposed controls as previously detailed (see: Boswell et al., 2015; Chang et al., 2015; Lu et al., 2015; Walter et al., 2015; Yang et al., 2014). This 200-gene target NanoString nCounter panel also contained 60 Xiphophorus homologs of genes reported in the literature to be circadian regulators or circadian responsive genes (Supplement Table 1). Additionally, 65 probes were selected and designed to assess expression of genes that are associated with spontaneous melanoma that has been documented among select Xiphophorus interspecies hybrids (Supplement Table 1). Differentially expressed genes between melanoma tumors and normal skin samples were identified by comparing RNA-Seq derived gene expression profiles of 16 melanoma tumors isolated from X. hellerii × (X. maculatus × X. hellerii) interspecies hybrids. Tumor and normal skin samples were taken from the same tumor-bearing fish (Log2FC≥ 1 or ≤ −1, FDR < 0.05). Ten genes that are not light responsive, nor differentially expressed between melanoma tumors and normal skin, and do not show circadian gene expression patterns were selected to serve as housekeeping genes to provide internal controls. Design and production of the nCounter probes, was performed by the Nanostring bioinformatics group (Nanostring, Seattle, WA). Transcript sequences corresponding to each gene target were downloaded from Ensembl database by Biomart, and used as templates to design probes. Each probe is 100 nt long, with a melting temperature between 73 and 91°C and will not form secondary structures inhibiting the assay. Probes were tested in silico to avoid cross hybridization to other loci.
NanoString hybridization of RNA samples with the target panel was initiated by mixing 500 ng of RNA (100 ng/µL) with the custom designed capture and reporter probe set. Samples were incubated for 12 hrs at 65°C and then processed the NanoString Prep Station (NanoString Technologies, Seattle, WA, USA) and subsequent nCounter analysis to determine gene expression. Built-in positive control probes were used to control the binding efficiency of each sample as read counts generated by these probes are independent of the RNA samples. A scaling factor is calculated based on the mean value of positive control probe generated read counts. Samples with the scaling factor between 0.3 to 3 are qualified for further analyses. Subsequently, the scaled read counts were normalized to the geometrical mean of the housekeeping genes to normalize potentially different total RNA input. Finally, background signal noise was determined by the read counts of negative control probes and was removed from normalized read counts (Geiss et al., 2008; Malkov et al., 2009; Brumbaugh et al., 2011).
Identification of oscillating gene expression patterns
NanoString nCounter gene expression counts were utilized to calculate the relative expression of each target gene for each time point. Herein, relative expression = (Average gene expression counts of biological replicate of each time point) / (Average expression counts of all time points). To identify genes exhibiting oscillating circadian expression patterns, a RAIN algorithm was applied to skin, brain and liver gene expression profiles (Thaben and Westermark, 2014) where a p-value < 0.05 was applied to identify genes showing oscillating expression patterns over a 24 hr period. Heatmaps, dot plots and line plots were used to represent basal gene expression patterns over a diurnal cycle. All plots were created using R plot function as well as heatmap.2 function of R package gplots.
Results
Identification of genes exhibiting diurnal oscillation of expression in Xiphophorus skin, brain and liver
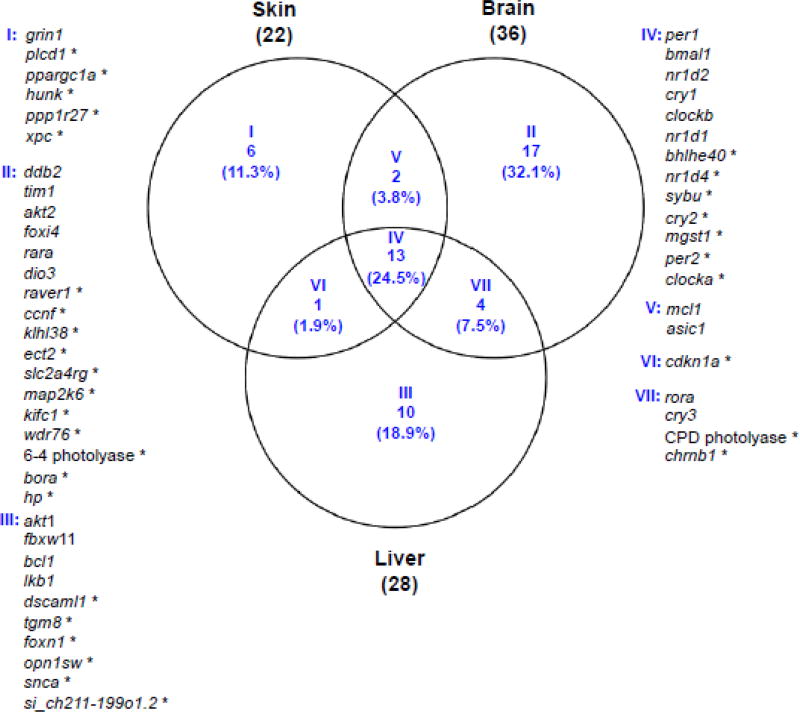
The basal expression patterns over a 24 hr period of 200 genes, including 65 genes previously identified as light-responsive, 65 genes associated with melanomagenesis, 60 genes selected based on circadian expression reported in other animal models, and 10 housekeeping genes, were assessed using the NanoString nCounter platform (Supplement Table 1). In this study, we aimed to identify genes exhibiting circadian expression patterns throughout the 24 hr period. Housekeeping genes, as expected, did not exhibit oscillating expression patterns (p-value > 0.05). In contrast, 22 genes in skin, 36 genes in brain and 28 genes in liver, were identified as exhibiting basal oscillation of their expression patterns over the 24 hr period (Fig. 1; p-value < 0.05). Thirteen genes (bmal1, per1, per2, cry1, cry2, clocka, clockb, nr1d1, nr1d2, nr1d4, bhlhe40, sybu, mgst1) were identified as showing basal oscillating expression patterns in all 3 organs analyzed (Fig. 1, group IV). In addition to these 13 genes, we also identified 2 genes that were shared in skin and brain (Fig. 1, group V), 1 gene shared in skin and liver (Fig. 1, group VI), and 4 genes shared in brain and liver (Fig. 1, group VII) that exhibit diurnal cycling of basal expression.
Figure 1. Basal oscillation of gene expression over 24-hour light-dark cycle in Xiphophorus skin, brain and liver.
Genes exhibiting basal oscillating expression patterns over a 24 hr diurnal cycle were identified using the RAIN algorithm (p<0.05). Among the light responsive and reference circadian gene targets on the NanoString panel, 22 genes were identified in skin, 36 genes were identified in brain and 28 genes were identified in liver that showed circadian oscillation. Genes exhibiting oscillating expression patterns identified from different organs were compared to each other. Organ specific oscillating genes and shared oscillating genes among the three organs are categorized into seven groups. Group I, II and III represent genes that were uniquely identified from skin, brain and liver. Group IV represent genes that are shared by all three organs, and Group V, VI and VII represent genes that are shared by two of the three organs. A NanoString nCounter panel was designed to capture gene expression of known light-responsive genes and known circadian rhythm regulator genes. Asterisk (*) highlights previously identified Xiphophorus light-responsive genes.
Organ specific genes (i.e., genes with p-value < 0.05 in one tissue) showing oscillation of expression for each of the 3 organs were also identified in skin (6 genes; Fig. 1, group I), brain (17 genes; Fig. 1, group II) and liver (10 genes; Fig. 1, group III).
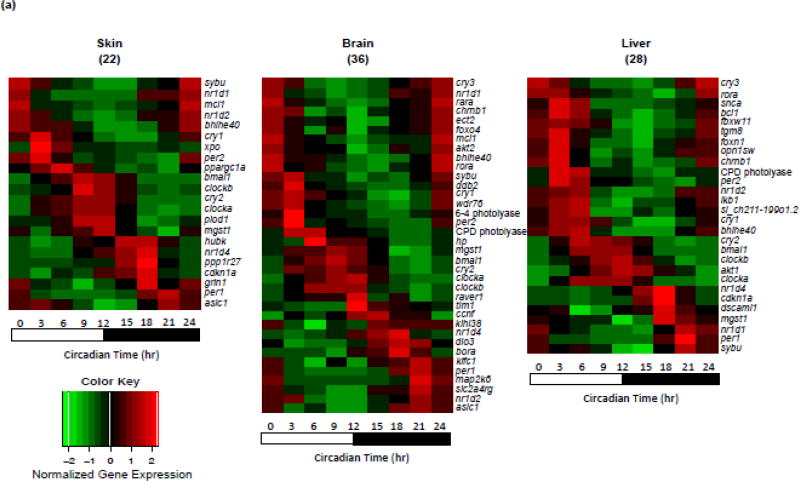
Thirty-two of the previously identified light responsive genes showed oscillating circadian expression patterns in Xiphophorus skin, brain or liver (Fig. 1; p-value < 0.05), while 33 of 65 light-responsive genes incorporated into the NanoString panel did not exhibit oscillation of expression (Fig. 2b, Table 1; Supplement Table 1; p-value > 0.05). These light responsive genes were selected for the NanoString panel based on treatment of fish with 10 kJ/m2 of 4,100 K FL, 10,000 K FL, or 8 and 16 kJ/m2 of UVB (311 nm). The 33 light responsive genes that did not exhibit oscillating expression patterns may represent a new and interesting class of genes regulated outside the normal light/dark circadian cycle.
Figure 2. Expression patterns of basal oscillating genes in skin, brain and liver. Heatmap.
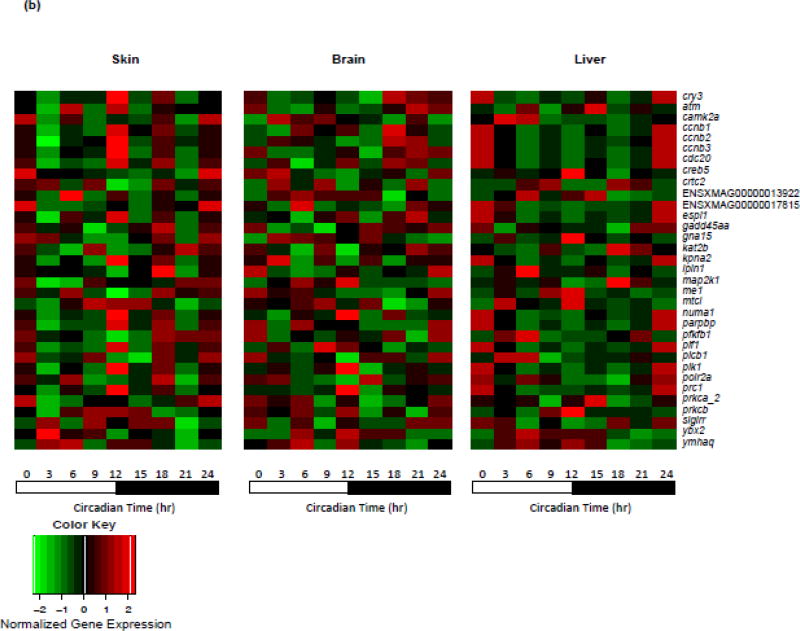
(A) Basal oscillating gene expression patterns identified in skin, brain and liver are represented. Twenty-two genes (13 light responsive genes) were identified in skin, 36 genes (20 light responsive genes) were identified in brain and 28 genes (16 light responsive genes) were identified in liver. Gene expression is normalized to the mean expression level over the entire diurnal cycle and scaled for visualization. Genes are ordered on the Ct time when they exhibited peak expression. (B) Thirty-three genes that are light-responsive and do not exhibit diurnal oscillation of expression in any of the organs tested are represented. Expression of these light-responsive, non-circadian influenced genes (ordered alphabetically) was normalized to the mean expression level over the entire diurnal cycle.
Table 1.
Summary Table of light responsive genes in NanoString nCounter panel
| ID | name | Category | Oscillating pattern | UVB(FC) | 4,100K FL (FC) | 10,000K FL (FC) | 50nm waveband (FC) |
|---|---|---|---|---|---|---|---|
| ENSXMAT00000000218 | RAVER1 | Light responsive gene | B | −2.62 | −3.90 | −3.52 | −4.01 |
| ENSXMAT00000004757 | CCNF | Light responsive gene | B | −4.03 | −3.63 | −3.86 | −2.38 |
| ENSXMAT00000008690 | KLHL38 | Light responsive gene | B | 1.09 | 14.28 | 2.83 | 15.62 |
| ENSXMAT00000014150 | ECT2 | Light responsive gene | B | −7.51 | −5.13 | −4.59 | −1.71 |
| ENSXMAT00000015684 | SLC2A4RG | Light responsive gene | B | −6.75 | |||
| ENSXMAT00000016798 | MAP2K6 | Light responsive gene | B | −4.90 | −3.70 | −1.99 | |
| ENSXMAT00000016886 | KIFC1 | Light responsive gene | B | −3.91 | −3.55 | −2.97 | −2.43 |
| ENSXMAT00000017117 | WDR76 | Light responsive gene | B | 5.28 | |||
| ENSXMAT00000017176 | photolyase ( | Light responsive gene | B | 2.48 | |||
| ENSXMAT00000018276 | BORA | Light responsive gene | B | −3.67 | −3.33 | −2.07 | |
| ENSXMAT00000018403 | HP | Light responsive gene | B | 3.01 | |||
| ENSXMAT00000004084 | DSCAML1 | Light responsive gene | L | −2.85 | 2.70 | ||
| ENSXMAT00000009163 | TGM8 | Light responsive gene | L | 3.22 | 5.36 | 5.36 | |
| ENSXMAT00000011812 | FOXN1 | Light responsive gene | L | −3.68 | |||
| ENSXMAT00000013132 | OPN1SW | Light responsive gene | L | −3.51 | |||
| ENSXMAT00000014172 | SNCA | Light responsive gene | L | 6.54 | 3.58 | 1.70 | 2.83 |
| ENSXMAT00000020266 | CH211-199O | Light responsive gene | L | 4.01 | 2.54 | 7.08 | |
| ENSXMAT00000000265 | PLCD1 | Light responsive gene | S | −3.47 | |||
| ENSXMAT00000007789 | PPARGC1A | Light responsive gene | S | 5.18 | |||
| ENSXMAT00000017652 | HUNK | Light responsive gene | S | −3.32 | |||
| ENSXMAT00000018069 | PPP1R27 | Light responsive gene | S | 1.24 | 5.98 | 5.72 | |
| ENSXMAT00000019435 | XPC | Light responsive gene | S | 3.56 | 2.11 | ||
| ENSXMAT00000004764 | PD photolya | Light responsive gene | B,L | 1.60 | 2.16 | 2.10 | |
| ENSXMAT00000013726 | CHRNB1 | Light responsive gene | B,L | 2.63 | |||
| ENSXMAT00000002257 | BHLHE40 | Light responsive gene | S,B,L | −2.60 | −2.55 | ||
| ENSXMAT00000002359 | NR1D4 | Light responsive gene | S,B,L | −2.43 | 2.31 | 2.32 | |
| ENSXMAT00000007109 | SYBU | Light responsive gene | S,B,L | 2.28 | −2.47 | ||
| ENSXMAT00000009327 | CRY2 | Light responsive gene | S,B,L | 1.80 | 2.62 | 1.71 | 1.03 |
| ENSXMAT00000015774 | MGST1 | Light responsive gene | S,B,L | −2.55 | |||
| ENSXMAT00000016248 | PER2 | Light responsive gene | S,B,L | 8.01 | 3.07 | 1.77 | 2.58 |
| ENSXMAT00000017010 | CLOCKA | Light responsive gene | S,B,L | 1.84 | 3.44 | 2.14 | 2.99 |
| ENSXMAT00000001085 | CDKN1A | Light responsive gene | S,L | 4.02 | |||
| ENSXMAT00000000868 | MTCL | Light responsive gene | na | 5.84 | |||
| ENSXMAT00000001067 | CREB5 | Light responsive gene | na | 3.29 | 2.92 | 4.21 | |
| ENSXMAT00000001124 | CRTC2 | Light responsive gene | na | −2.00 | |||
| ENSXMAT00000001311 | PFKFB1 | Light responsive gene | na | 2.15 | |||
| ENSXMAT00000002159 | ESPL1 | Light responsive gene | na | −2.65 | −3.77 | −3.72 | −1.81 |
| ENSXMAT00000002966 | YWHAQ | Light responsive gene | na | −2.53 | |||
| ENSXMAT00000003274 | KAT2B | Light responsive gene | na | −3.05 | |||
| ENSXMAT00000003526 | MAP2K1 | Light responsive gene | na | −3.20 | |||
| ENSXMAT00000003968 | PIF1 | Light responsive gene | na | −3.83 | −6.39 | ||
| ENSXMAT00000004000 | GADD45AA | Light responsive gene | na | 2.23 | |||
| ENSXMAT00000004686 | LPIN1 | Light responsive gene | na | 3.41 | 2.78 | ||
| ENSXMAT00000005357 | PRKCB | Light responsive gene | na | −3.45 | −2.19 | ||
| ENSXMAT00000006617 | CAMK2A | Light responsive gene | na | −2.90 | |||
| ENSXMAT00000006647 | PRKCA_2 | Light responsive gene | na | −2.29 | −2.35 | ||
| ENSXMAT00000006913 | PRC1 | Light responsive gene | na | −3.32 | −3.41 | −5.15 | −1.82 |
| ENSXMAT00000006917 | ME1 | Light responsive gene | na | 2.91 | |||
| ENSXMAT00000007091 | SIGIRR | Light responsive gene | na | −4.72 | |||
| ENSXMAT00000008371 | KPNA2 | Light responsive gene | na | −4.58 | −4.59 | −5.89 | −2.15 |
| ENSXMAT00000008587 | PLCB1 | Light responsive gene | na | −2.96 | 1.78 | ||
| ENSXMAT00000008618 | CCNB1 | Light responsive gene | na | −3.53 | −4.77 | −6.90 | −2.45 |
| ENSXMAT00000011193 | NUMA1 | Light responsive gene | na | −4.17 | −3.49 | −2.26 | −2.14 |
| ENSXMAT00000012882 | PLK1 | Light responsive gene | na | −4.37 | −4.40 | −5.18 | −1.77 |
| ENSXMAT00000013490 | GNA15 | Light responsive gene | na | 2.66 | 2.74 | 3.22 | |
| ENSXMAT00000013968 | MAG000000 | Light responsive gene | na | 12.30 | |||
| ENSXMAT00000014334 | CCNB2 | Light responsive gene | na | −4.29 | −5.05 | −4.26 | −3.86 |
| ENSXMAT00000014517 | ARHGAP19 | Light responsive gene | na | −3.79 | −4.55 | −3.81 | −2.63 |
| ENSXMAT00000015109 | CCNB3 | Light responsive gene | na | −3.58 | −3.59 | −5.05 | −2.39 |
| ENSXMAT00000015760 | POLR2A | Light responsive gene | na | 1.16 | −2.52 | ||
| ENSXMAT00000015815 | YBX2 | Light responsive gene | na | 2.35 | 2.97 | 1.45 | 1.90 |
| ENSXMAT00000016054 | CDC20 | Light responsive gene | na | −2.78 | −5.49 | −7.61 | −1.65 |
| ENSXMAT00000017002 | ATM | Light responsive gene | na | −2.90 | −2.72 | −2.99 | |
| ENSXMAT00000017412 | PARPBP | Light responsive gene | na | −4.32 | −7.53 | −3.45 | −3.39 |
| ENSXMAT00000017917 | MAG000000 | Light responsive gene | na | −14.02 | −19.31 | −2.95 | |
| ENSXMAT00000000552 | FAM102B | housekeeper gene | na | ENSXMAT00000000653 | TPM4 | housekeeper gene | |
| na ENSXMAT00000003489 | SORT1 | housekeeper gene | na | ENSXMAT00000004652 | WDR37 | housekeeper gene | |
| na ENSXMAT00000007048 | LCP2 | housekeeper gene | na | ENSXMAT00000009755 | FLT4 | housekeeper gene | |
| na ENSXMAT00000010762 | INTS9 | housekeeper gene | na | ENSXMAT00000012666 | H211-114I1 | housekeeper gene | |
| na ENSXMAT00000016883 | PCP4 | housekeeper gene | na | ENSXMAT00000019159 | TMEM204 | housekeeper gene | |
| na | |||||||
na= absence of oscillating expression pattern S=Skin; B=Brain; L=Liver
Our previous studies have reported differences between male and female skin in the gene expression patterns after light exposures (i.e., UVB and fluorescent light, Boswell et al., 2015; 2017b). Therefore, we compared basal gene expression over the diurnal cycle for Xiphophorus in skin from both females and males. Only one light responsive gene, asic1, exhibited a circadian oscillating expression pattern in male skin, and did not also show an oscillating gene expression pattern in female skin (Supplement Fig. 2a).
Expression patterns of genes showing diurnal oscillation of transcription in Xiphophorus skin, brain and liver
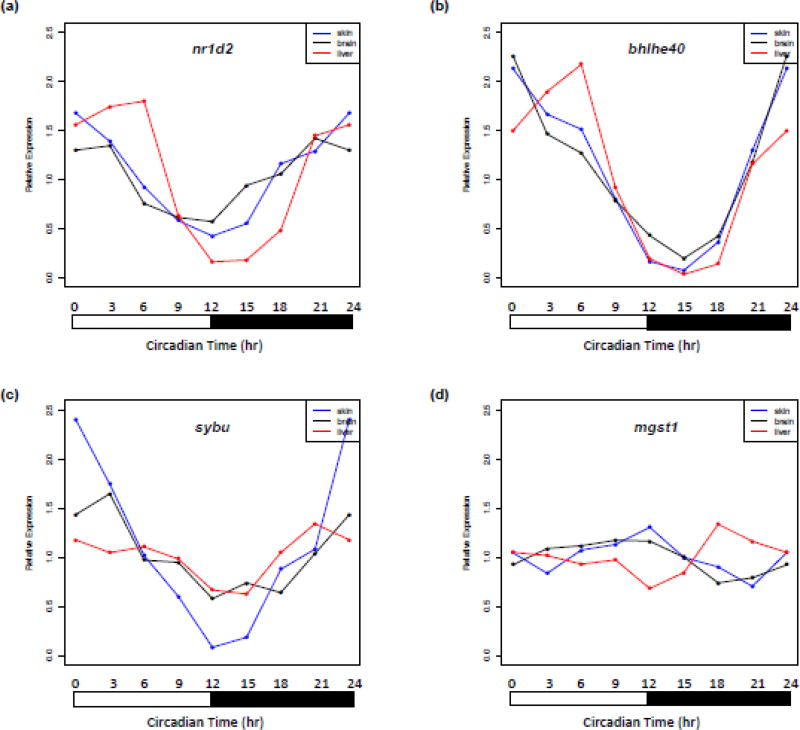
We compared basal oscillation of gene expression patterns between different organs to determine peak expression times, and thus organ specific circadian phasing of gene expression, relative to one another. Oscillating genes shared by two organs (Fig. 1, group V, VI, VII) all showed consistent peak expression at the same circadian time (Fig. 2). In contrast, 4 oscillating genes shared by all three organs (Fig. 1, group IV) showed retardation of peak expression in at least one organ (Fig. 2; Fig. 3). For example, nr1d2 showed peak expression at the dark-light transition (Ct 0 and Ct 21 hr) in skin and brain, but peak expression at Ct 6 hr in liver (Fig. 3a). A similar gene expression pattern shift is also observed for bhlhe40 (Fig. 3b). Circadian times for peak gene expression of sybu are Ct 0, Ct 3 and Ct 21 hr, respectively in skin, brain and liver (Fig. 3c). The mgst1 gene exhibits peak expression at Ct 12 and Ct 9 hr in skin and brain, with peak expression at Ct 18 hr in liver (Fig. 3d).
Figure 3. Shift in peak expression times of select basal oscillating genes in different organs.
(A) nr1d2 shows peak expression at Ct 0 and Ct 21 hr in skin and brain, but peak expression at Ct 6 hr in liver. (B) bhlhe40 shows peak gene expression at Ct 0 hr in both skin and brain, while exhibiting peak expression at Ct 6 hr in liver. (C) Circadian times for peak gene expression of sybu are Ct 0, Ct 3 and Ct 21 hr, respectively in skin, brain and liver. (D) mgst1 exhibits peak expression at Ct 12 and Ct 9 hr in skin and brain, but peak expression at Ct 18 hr in liver.
Xiphophorus melanoma associated genes exhibiting oscillating expression patterns
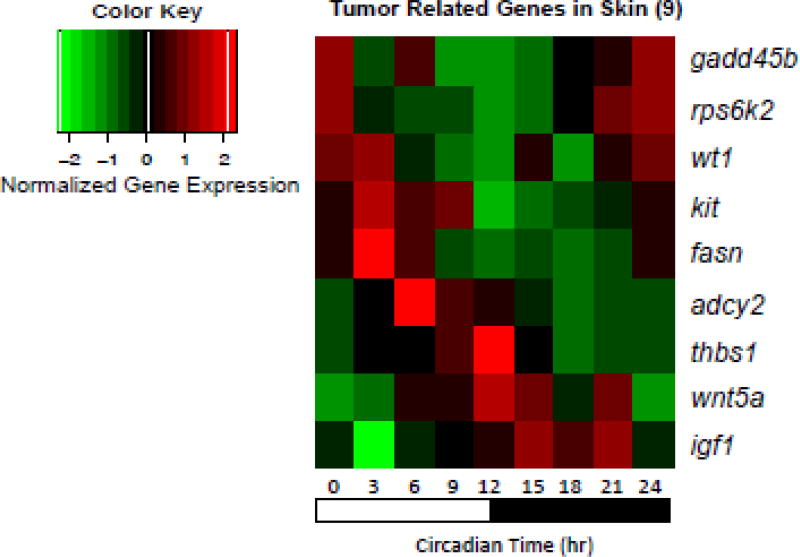
The Xiphophorus melanoma model is a well-established experimental system wherein melanoma develops in progeny produced from an interspecies backcross: [X. hellerii × (X. maculatus × X. hellerii); For review, see Schartl and Walter, 2016]. To determine whether genes associated with melanomagenesis exhibit oscillating expression patterns, we assessed the expression of 65 Xiphophorus melanoma related genes in skin. Nine of the 65 genes represented in the NanoString panel were observed to exhibit oscillation of expression consistent with circadian regulation (Fig. 4). The DNA repair gene gadd45b, and cell growth and differentiation related genes, rps6ka2, both show peak expression at the dark-light transition phase (Ct 0 hr). Transcription factor wt1, melanocyte differentiation marker kit, fatty acid synthase fasn, and cAMP synthase adcy2, show peak expression in the light phase (Ct 3–9 hr). Microenvironment related gene thbs1 and development related gene wnt5a exhibit peak expression at the light-dark transition phase (Ct 12 hr). Insulin growth factor igf1 is the only melanoma related gene to show peak expression in the dark phase of diurnal cycle (Ct 21 hr; Fig. 4). Compared to males, the female fish exhibited the same expression pattern for 8 of these genes, but showed an absence of expression oscillation for thbs1 in skin (Supplement Fig. 2).
Figure 4. Basal oscillating gene expression of Xiphophorus melanoma associated genes.
Nine genes associated with Xiphophorus melanomagenesis showed basal oscillating expression patterns. These genes include DNA repair gene gadd45b, cell growth and differentiation related gene rps6ka2, transcription factor wt1, melanocyte differentiation marker kit, fatty acid synthase fasn, cAMP synthase adcy2, microenvironment related gene thbs1, development related gene wnt5a and insulin growth factor igf1.
Discussion
Organisms possess an endogenous biological clock that dynamically interacts with environmental cues to temporally control biological processes in response to the light-dark cycle (Mohawk et al., 2012; Takahashi et al., 2008). Disruption of the normal gene expression patterns is associated with several physiological disorders and can predispose animals to disease (Coogan and McGowan, 2017; Panda, 2016; Zhang et al., 2016). The relatively recent and ever-increasing use of artificial light (i.e., fluorescent light) and extension of light period has made understanding the effects exposure to various light types may have on gene expression in vertebrates an area of interest. Artificial lighting has been reported to affect several physiological activities (Blask, 2009; Blask et al., 2005; Cajochen et al., 2005; Koo et al., 2016; Lewy et al., 1980; Lucassen et al., 2016; Ohayon and Milesi, 2016; Romeo et al., 2017; Schwimmer et al., 2014; Stevens et al., 2013), and one biological parameter associated with this is established as interference with the normal circadian cycle (Stevens et al., 2013). Our previous studies identified transcriptional responses 6 hrs after exposure to FL (10 kJ/m2), UVB (8 kJ/m2), or 50 nm wavebands of light between 300 to 600 nm (Boswell et al., 2015; Chang et al., 2015; Lu et al., 2015; Walter et al., 2015; Yang et al., 2014). These results suggest each light source may incite very different genetic responses, and thereby alter the genetic homeostasis. However, the circadian position of basal gene expression was not investigated as a potential parameter effecting light driven transcriptional responses. To better understand light-induced gene expression effects, we performed NanoString analyses of 200 light responsive, circadian, and tumor associated gene targets over a 24 hr period in Xiphophorus skin, brain and liver.
The identification of well-known circadian master regulators clocka, clockb, bmal1 (arntl), cry1, cry2, per1, per2, nr1d1, nr1d2, nr1d4, bhlhe40 in skin, brain and liver (Fig. 1; Fig. 2) shows expression patterns for core circadian regulators are similar in Xiphophorus fish compared to other vertebrates. Additionally, the identification of genes in skin, brain and liver that exhibit organ-specific oscillating expression patterns suggests circadian gene expression is hallmarked by organ-specificity as previously reported (Kita et al., 2002; Korencic et al., 2014; Panda et al., 2002b; Reppert and Weaver, 2002; Storch et al., 2002; Ueda et al., 2002).
Global transcriptome analyses are required to comprehensively delineate basal gene expression patterns; however, our use of a custom NanoString panel focused on previously identified light responsive gene targets allowed us to segregate genes into transcriptional response sets as either light responsive and exhibiting circadian expression (i.e., 32 genes in this study; Fig. 1), or light responsive without diurnal oscillating transcription (i.e., 33 genes, Table 1). We believe identification of genes that are light responsive, yet non-circadian influenced, represent a novel class of genes with currently unknown regulatory mechanism(s). Many of these light-responsive, non-circadian oscillating genes encode kinases (i.e., camk2a, map2k1, plk1, prkca, prkcb) that are known to play fundamental roles in regulating cell proliferation, cell cycle regulation, and/or have been suggested to be regulated by circadian cycle. Similarly, other cell cycle regulators (ccnb1, ccnb2, ccnb3, cdc20, creb5, lpin1, plcb1) also showed transcriptional light responsiveness, but non-oscillating expression patterns (Fig. 2b, Table 1; Bieler et al., 2014; Feillet et al., 2014; Feillet et al., 2015; Grechez-Cassiau et al., 2008; Shostak, 2017). These results, collectively, suggest the genetic response to light, outside the light phase of diurnal cycle, may shift the genetic homeostasis of these genes and affect their function in circadian regulation at post-transcriptional/translational level (e.g., phosphorylation; Cao et al., 2015; Reischl and Kramer, 2011; Robles et al., 2017). In future studies, identification of light-responsive but non-circadian influenced genetic regulatory elements may provide useful tools for development of facile expression vectors.
The presence of basal oscillating expression patterns in the light responsive genes suggest that light exposure at different times of the day may lead to different downstream genetic effects. For example, the master cell cycle regulator cdkn1a has been shown to possess a concentration-dependent effect on induction of apoptosis versus initiation of DNA repair via cell cycle arrest (Chen et al., 2015; Esteve-Puig et al., 2014; Hall et al., 2014; Hollmann et al., 2016; Karimian et al., 2016). The cdkn1a gene showed both basal oscillation of gene expression with peak expression at Ct 18 hr, and also shows a UVB response (Boswell et al., 2015). These features suggest UVB exposure at different times of the day may lead to different cdkn1a activated cell fate (DNA repair vs. apoptosis; Fig. 2).
In this study, gene expression assessments were performed on 3 hr intervals. This period defines assessment of peak expression to ± 3 hrs from a peak expression time. With this caveat in mind, we were still able to identify four genes showing shifted peak expression (retardation) of at least 6 hrs in one of the three organs (Fig. 3). Although skin and brain showed consistent expression patterns, liver exhibited uncoupled expression patterns for nr1d2, bhlhe40, sybu and mgst1. Both nr1d2 and bhlhe40 are transcriptional products of the BMAL/CLOCK transcription factor, and also serve as repressors of BMAL/CLOCK transcriptional activity (Honma et al., 2002; Solt et al., 2011). However, unlike the reported use of restricted feeding to uncouple circadian gene expression in peripheral organs (e.g., liver), whereupon expression patterns in all circadian genes are shifted together, we note a shift only in nr1d2 and bhlhe40 expression with no coincident shift of expression of the other core circadian regulators (e.g., bmal1, per1, per2, cry1, cry2, clocka and clockb; (Damiola et al., 2000; Le Minh et al., 2001). Therefore, the differences in expression patterns of nr1d2, bhlhe40, sybu and mgst1 in liver may be a result of different regulatory mechanisms than circadian uncoupling. In other fish models (e.g., zebrafish) peripheral clocks have been shown to be directly light entrainable, suggesting differences in peak oscillating gene expression within different organs may be due to organ specific photoreception as a regulator of gene expression (Whitmore et al., 2000). Recent studies have identified novel opsins expressed within internal organs and these may serve as peripheral photoreceptors to establish organ-specific light-entrainable expression patterns (Bellingham et al., 2006; Cavallari et al., 2011; Moutsaki et al., 2003; Peirson et al., 2009; Pierce et al., 2008). Interestingly, we identified one light responsive opsin gene, opn1sw, that exhibits a basal oscillating expression pattern in Xiphophorus liver, and may be involved in the liver-specific expression patterns. Further work will be required to test this idea.
Comparison of basal gene expression patterns in female and male skin identified one gene, asic1 (acid-sensing ion channel 1), that exhibits gender specific patterns of expression (Supplement Fig. 2). This gene has also been identified to exhibit sex-specific expression patterns in mice (Kobayashi et al., 2009), and has been shown to affect light-dependent locomotion in zebrafish (Liu et al., 2014). Taken together, we may speculate that asic1 plays a role in differentiation of the basal gene expression in males and females.
Xiphophorus melanoma tumors share both phenotypic and genetic features with human melanoma, making this model appropriate for research into cancer etiology (Schartl and Walter, 2016). Cancer cells show dysregulated circadian gene expression patterns, and are capable of changing the normal circadian gene expression patterns of other organs (Masri et al., 2016). Here we found 9 genes that play significant roles (i.e., DNA repair genes, kinases, metabolism enzymes, microenvironment regulators, cell development and differentiation regulators) in melanomagenesis and also exhibit basal oscillating expression patterns (Fig. 4). One may expect from these observations that disruption of the normal circadian phasing may lead to dysregulation of oncogene and tumor suppressor expression and interaction (For review: Eismann et al., 2010; Fu and Lee, 2003; Sahar and Sassone-Corsi, 2007; Shostak, 2017). Interestingly, we also found thbs1, a microenvironment regulator, did not exhibit oscillating gene expression in females. Considering males have been shown to be more susceptible to melanoma than females, this difference may be associated with differences in regulation of cell-cell and cell-microenvironment interactions in the sexes (Supplement Fig. 2; Fernandez and Bowser, 2010; Schartl et al., 1995).
Conclusions
We conclude that a central regulatory network governing oscillating gene expression exist in Xiphophorus as other studied vertebrate animals. Further, we have distinguished previously identified light responsive genes in Xiphophorus into genes that do, or do not, exhibit circadian based oscillating expression patterns. Categorization of light responsive genes further delineates light-induced genetic effects and highlights a new direction for future research.
Supplementary Material
Acknowledgments
The authors would like to thank the staff of the Xiphophorus Genetic Stock Center, Texas State University, for maintaining the pedigreed fish lines and caring for the animals used in this study. Support for this project was provided in part by the NIH, ORIP grants R24OD-011120, R24 OD-018555 and R25-GM-102783.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amores A, Catchen J, Nanda I, Warren W, Walter R, Schartl M, Postlethwait JH. A RAD-tag genetic map for the platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among teleost fish. Genetics. 2014;197:625–641. doi: 10.1534/genetics.114.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes HJ, Lucas RJ. Melanopsin and inner retinal photoreception. Cell Mol Life Sci. 2010;67:99–111. doi: 10.1007/s00018-009-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale A, Guibal C, Tamai TK, Klotz L, Cowen S, Peyric E, Reynoso VH, Yamamoto Y, Whitmore D. Circadian rhythms in Mexican blind cavefish Astyanax mexicanus in the lab and in the field. Nat Commun. 2013;4:2769. doi: 10.1038/ncomms3769. [DOI] [PubMed] [Google Scholar]
- Bellingham J, Chaurasia SS, Melyan Z, Liu C, Cameron MA, Tarttelin EE, Iuvone PM, Hankins MW, Tosini G, Lucas RJ. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler J, Cannavo R, Gustafson K, Gobet C, Gatfield D, Naef F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol. 2014;10:739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blask DE. Melatonin, sleep disturbance and cancer risk. Sleep Med Rev. 2009;13:257–264. doi: 10.1016/j.smrv.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Blask DE, Brainard GC, Dauchy RT, Hanifin JP, Davidson LK, Krause JA, Sauer LA, Rivera-Bermudez MA, Dubocovich ML, Jasser SA, Lynch DT, Rollag MD, Zalatan F. Melatonin-depleted blood from premenopausal women exposed to light at night stimulates growth of human breast cancer xenografts in nude rats. Cancer Res. 2005;65:11174–11184. doi: 10.1158/0008-5472.CAN-05-1945. [DOI] [PubMed] [Google Scholar]
- Boswell M, Boswell W, Lu Y, Savage M, Mazurek Z, Chang J, Muster J, Walter R. The transcriptional response of skin to fluorescent light exposure in viviparous (Xiphophorus) and oviparous (Danio, Oryzias) fishes. Comp Biochem Physiol C Toxicol Pharmacol. 2017a doi: 10.1016/j.cbpc.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell W, Boswell M, Titus J, Savage M, Lu Y, Shen J, Walter RB. Sex-specific molecular genetic response to UVB exposure in Xiphophorus maculatus skin. Comp Biochem Physiol C Toxicol Pharmacol. 2015;178:76–85. doi: 10.1016/j.cbpc.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boswell WT, Boswell M, Walter DJ, Navarro KL, Chang J, Lu Y, Savage MG, Shen J, Walter RB. Exposure to 4100K fluorescent light elicits sex specific transcriptional responses in Xiphophorus maculatus skin. Comp Biochem Physiol C Toxicol Pharmacol. 2017b doi: 10.1016/j.cbpc.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinformatics. 2011;12:479. doi: 10.1186/1471-2105-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajochen C, Munch M, Kobialka S, Krauchi K, Steiner R, Oelhafen P, Orgul S, Wirz-Justice A. High sensitivity of human melatonin, alertness, thermoregulation, and heart rate to short wavelength light. J Clin Endocrinol Metab. 2005;90:1311–1316. doi: 10.1210/jc.2004-0957. [DOI] [PubMed] [Google Scholar]
- Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC, Amir S, Sonenberg N. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci. 2015;18:855–862. doi: 10.1038/nn.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr AJ, Tamai TK, Young LC, Ferrer V, Dekens MP, Whitmore D. Light reaches the very heart of the zebrafish clock. Chronobiol Int. 2006;23:91–100. doi: 10.1080/07420520500464395. [DOI] [PubMed] [Google Scholar]
- Cavallari N, Frigato E, Vallone D, Frohlich N, Lopez-Olmeda JF, Foa A, Berti R, Sanchez-Vazquez FJ, Bertolucci C, Foulkes NS. A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol. 2011;9:e1001142. doi: 10.1371/journal.pbio.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Lu Y, Boswell WT, Boswell M, Caballero KL, Walter RB. Molecular genetic response to varied wavelengths of light in Xiphophorus maculatus skin. Comp Biochem Physiol C Toxicol Pharmacol. 2015;178:104–115. doi: 10.1016/j.cbpc.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Huang X, Xue Z, Cao D, Huang K, Chen J, Pan Y, Gao Y. The Role of p21 in Apoptosis, Proliferation, Cell Cycle Arrest, and Antioxidant Activity in UVB-Irradiated Human HaCaT Keratinocytes. Med Sci Monit Basic Res. 2015;21:86–95. doi: 10.12659/MSMBR.893608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coogan AN, McGowan NM. A systematic review of circadian function, chronotype and chronotherapy in attention deficit hyperactivity disorder. Atten Defic Hyperact Disord. 2017 doi: 10.1007/s12402-016-0214-5. [DOI] [PubMed] [Google Scholar]
- Cuesta IH, Lahiri K, Lopez-Olmeda JF, Loosli F, Foulkes NS, Vallone D. Differential maturation of rhythmic clock gene expression during early development in medaka (Oryzias latipes) Chronobiol Int. 2014;31:468–478. doi: 10.3109/07420528.2013.856316. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eismann EA, Lush E, Sephton SE. Circadian effects in cancer-relevant psychoneuroendocrine and immune pathways. Psychoneuroendocrinology. 2010;35:963–976. doi: 10.1016/j.psyneuen.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Esteve-Puig R, Gil R, Gonzalez-Sanchez E, Bech-Serra JJ, Grueso J, Hernandez-Losa J, Moline T, Canals F, Ferrer B, Cortes J, Bastian B, Ramon YCS, Martin-Caballero J, Flores JM, Vivancos A, Garcia-Patos V, Recio JA. A mouse model uncovers LKB1 as an UVB-induced DNA damage sensor mediating CDKN1A (p21WAF1/CIP1) degradation. PLoS Genet. 2014;10:e1004721. doi: 10.1371/journal.pgen.1004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P, Teboul M, Saito S, Levi FA, Bretschneider T, van der Horst GT, Delaunay F, Rand DA. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci U S A. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet C, van der Horst GT, Levi F, Rand DA, Delaunay F. Coupling between the Circadian Clock and Cell Cycle Oscillators: Implication for Healthy Cells and Malignant Growth. Front Neurol. 2015;6:96. doi: 10.3389/fneur.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AA, Bowser PR. Selection for a dominant oncogene and large male size as a risk factor for melanoma in the Xiphophorus animal model. Mol Ecol. 2010;19:3114–3123. doi: 10.1111/j.1365-294X.2010.04738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- Hall JR, Bereman MS, Nepomuceno AI, Thompson EA, Muddiman DC, Smart RC. C/EBPalpha regulates CRL4(Cdt2)-mediated degradation of p21 in response to UVB-induced DNA damage to control the G1/S checkpoint. Cell Cycle. 2014;13:3602–3610. doi: 10.4161/15384101.2014.962957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann G, Linden R, Giangrande A, Allodi S. Increased p53 and decreased p21 accompany apoptosis induced by ultraviolet radiation in the nervous system of a crustacean. Aquat Toxicol. 2016;173:1–8. doi: 10.1016/j.aquatox.2015.12.025. [DOI] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Kita Y, Shiozawa M, Jin W, Majewski RR, Besharse JC, Greene AS, Jacob HJ. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics. 2002;12:55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Yoshiyama M, Zakoji H, Takeda M, Araki I. Sex differences in the expression profile of acid-sensing ion channels in the mouse urinary bladder: a possible involvement in irritative bladder symptoms. BJU Int. 2009;104:1746–1751. doi: 10.1111/j.1464-410X.2009.08658.x. [DOI] [PubMed] [Google Scholar]
- Koo YS, Song JY, Joo EY, Lee HJ, Lee E, Lee SK, Jung KY. Outdoor artificial light at night, obesity, and sleep health: Cross-sectional analysis in the KoGES study. Chronobiol Int. 2016;33:301–314. doi: 10.3109/07420528.2016.1143480. [DOI] [PubMed] [Google Scholar]
- Korencic A, Kosir R, Bordyugov G, Lehmann R, Rozman D, Herzel H. Timing of circadian genes in mammalian tissues. Sci Rep. 2014;4:5782. doi: 10.1038/srep05782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewy AJ, Wehr TA, Goodwin FK, Newsome DA, Markey SP. Light suppresses melatonin secretion in humans. Science. 1980;210:1267–1269. doi: 10.1126/science.7434030. [DOI] [PubMed] [Google Scholar]
- Liu S, Wang MX, Mao CJ, Cheng XY, Wang CT, Huang J, Zhong ZM, Hu WD, Wang F, Hu LF, Wang H, Liu CF. Expression and functions of ASIC1 in the zebrafish retina. Biochem Biophys Res Commun. 2014;455:353–357. doi: 10.1016/j.bbrc.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Lu Y, Bowswell M, Bowswell W, Yang K, Schartl M, Walter RB. Molecular genetic response of Xiphophorus maculatus-X. couchianus interspecies hybrid skin to UVB exposure. Comp Biochem Physiol C Toxicol Pharmacol. 2015;178:86–92. doi: 10.1016/j.cbpc.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen EA, Coomans CP, van Putten M, de Kreij SR, van Genugten JH, Sutorius RP, de Rooij KE, van der Velde M, Verhoeve SL, Smit JW, Lowik CW, Smits HH, Guigas B, Aartsma-Rus AM, Meijer JH. Environmental 24-hr Cycles Are Essential for Health. Curr Biol. 2016;26:1843–1853. doi: 10.1016/j.cub.2016.05.038. [DOI] [PubMed] [Google Scholar]
- Masri S, Papagiannakopoulos T, Kinouchi K, Liu Y, Cervantes M, Baldi P, Jacks T, Sassone-Corsi P. Lung Adenocarcinoma Distally Rewires Hepatic Circadian Homeostasis. Cell. 2016;165:896–909. doi: 10.1016/j.cell.2016.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, Mashadi-Hossein A, Fare T. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes. 2009;2:80. doi: 10.1186/1756-0500-2-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migaud H, Davie A, Taylor JF. Current knowledge on the photoneuroendocrine regulation of reproduction in temperate fish species. J Fish Biol. 2010;76:27–68. doi: 10.1111/j.1095-8649.2009.02500.x. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsaki P, Whitmore D, Bellingham J, Sakamoto K, David-Gray ZK, Foster RG. Teleost multiple tissue (tmt) opsin: a candidate photopigment regulating the peripheral clocks of zebrafish? Brain Res Mol Brain Res. 2003;112:135–145. doi: 10.1016/s0169-328x(03)00059-7. [DOI] [PubMed] [Google Scholar]
- Ohayon MM, Milesi C. Artificial Outdoor Nighttime Lights Associate with Altered Sleep Behavior in the American General Population. Sleep. 2016;39:1311–1320. doi: 10.5665/sleep.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002b;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature. 2002a;417:329–335. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Peirson SN, Halford S, Foster RG. The evolution of irradiance detection: melanopsin and the non-visual opsins. Philos Trans R Soc Lond B Biol Sci. 2009;364:2849–2865. doi: 10.1098/rstb.2009.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce LX, Noche RR, Ponomareva O, Chang C, Liang JO. Novel functions for Period 3 and Exo-rhodopsin in rhythmic transcription and melatonin biosynthesis within the zebrafish pineal organ. Brain Res. 2008;1223:11–24. doi: 10.1016/j.brainres.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, Takekata H, Tessmar-Raible K. An Overview of Monthly Rhythms and Clocks. Front Neurol. 2017;8:189. doi: 10.3389/fneur.2017.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585:1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- Rensing L, Ruoff P. Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int. 2002;19:807–864. doi: 10.1081/cbi-120014569. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Robles MS, Humphrey SJ, Mann M. Phosphorylation Is a Central Mechanism for Circadian Control of Metabolism and Physiology. Cell Metab. 2017;25:118–127. doi: 10.1016/j.cmet.2016.10.004. [DOI] [PubMed] [Google Scholar]
- Romeo S, Vitale F, Viaggi C, di Marco S, Aloisi G, Fasciani I, Pardini C, Pietrantoni I, Di Paolo M, Riccitelli S, Maccarone R, Mattei C, Capannolo M, Rossi M, Capozzo A, Corsini GU, Scarnati E, Lozzi L, Vaglini F, Maggio R. Fluorescent light induces neurodegeneration in the rodent nigrostriatal system but near infrared LED light does not. Brain Res. 2017;1662:87–101. doi: 10.1016/j.brainres.2017.02.026. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–1331. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Madrid JA, Sanchez-Vazquez FJ. Molecular cloning, tissue distribution, and daily rhythms of expression of per1 gene in European sea bass (Dicentrarchus labrax) Chronobiol Int. 2010;27:19–33. doi: 10.3109/07420520903398633. [DOI] [PubMed] [Google Scholar]
- Schartl A, Malitschek B, Kazianis S, Borowsky R, Schartl M. Spontaneous melanoma formation in nonhybrid Xiphophorus. Cancer Res. 1995;55:159–165. [PubMed] [Google Scholar]
- Schartl M, Walter RB. Xiphophorus and Medaka Cancer Models. Adv Exp Med Biol. 2016;916:531–552. doi: 10.1007/978-3-319-30654-4_23. [DOI] [PubMed] [Google Scholar]
- Schartl M, Walter RB, Shen Y, Garcia T, Catchen J, Amores A, Braasch I, Chalopin D, Volff JN, Lesch KP, Bisazza A, Minx P, Hillier L, Wilson RK, Fuerstenberg S, Boore J, Searle S, Postlethwait JH, Warren WC. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat Genet. 2013;45:567–572. doi: 10.1038/ng.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwimmer H, Metzer A, Pilosof Y, Szyf M, Machnes ZM, Fares F, Harel O, Haim A. Light at night and melatonin have opposite effects on breast cancer tumors in mice assessed by growth rates and global DNA methylation. Chronobiol Int. 2014;31:144–150. doi: 10.3109/07420528.2013.842925. [DOI] [PubMed] [Google Scholar]
- Shen Y, Chalopin D, Garcia T, Boswell M, Boswell W, Shiryev SA, Agarwala R, Volff JN, Postlethwait JH, Schartl M, Minx P, Warren WC, Walter RB. X. couchianus and X. hellerii genome models provide genomic variation insight among Xiphophorus species. BMC Genomics. 2016;17:37. doi: 10.1186/s12864-015-2361-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak A. Circadian Clock, Cell Division, and Cancer: From Molecules to Organism. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18040873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Kojetin DJ, Burris TP. The REV-ERBs and RORs: molecular links between circadian rhythms and lipid homeostasis. Future Med Chem. 2011;3:623–638. doi: 10.4155/fmc.11.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens RG, Brainard GC, Blask DE, Lockley SW, Motta ME. Adverse health effects of nighttime lighting: comments on American Medical Association policy statement. Am J Prev Med. 2013;45:343–346. doi: 10.1016/j.amepre.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben PF, Westermark PO. Detecting rhythms in time series with RAIN. J Biol Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama R, Chen X, Jhawar N, Aamar E, Epstein J, Reany N, Alon S, Gothilf Y, Klein DC, Dawid IB. Transcriptome analysis of the zebrafish pineal gland. Dev Dyn. 2009;238:1813–1826. doi: 10.1002/dvdy.21988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, Iino M, Shigeyoshi Y, Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Vallone D, Gondi SB, Whitmore D, Foulkes NS. E-box function in a period gene repressed by light. Proc Natl Acad Sci U S A. 2004;101:4106–4111. doi: 10.1073/pnas.0305436101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter DJ, Boswell M, Volk de Garcia SM, Walter SM, Breitenfeldt EW, Boswell W, Walter RB. Characterization and differential expression of CPD and 6-4 DNA photolyases in Xiphophorus species and interspecies hybrids. Comp Biochem Physiol C Toxicol Pharmacol. 2014;163:77–85. doi: 10.1016/j.cbpc.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter RB, Walter DJ, Boswell WT, Caballero KL, Boswell M, Lu Y, Chang J, Savage MG. Exposure to fluorescent light triggers down regulation of genes involved with mitotic progression in Xiphophorus skin. Comp Biochem Physiol C Toxicol Pharmacol. 2015;178:93–103. doi: 10.1016/j.cbpc.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Comparative analysis of period genes in teleost fish genomes. J Mol Evol. 2008;67:29–40. doi: 10.1007/s00239-008-9121-5. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Itoh K, Mogi M, Fujinami Y, Shimizu D, Hashimoto H, Uji S, Yokoi H, Suzuki T. Circadian pacemaker in the suprachiasmatic nuclei of teleost fish revealed by rhythmic period2 expression. Gen Comp Endocrinol. 2012;178:400–407. doi: 10.1016/j.ygcen.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404:87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- Yang K, Boswell M, Walter DJ, Downs KP, Gaston-Pravia K, Garcia T, Shen Y, Mitchell DL, Walter RB. UVB-induced gene expression in the skin of Xiphophorus maculatus Jp 163 B. Comp Biochem Physiol C Toxicol Pharmacol. 2014;163:86–94. doi: 10.1016/j.cbpc.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WX, Chen SY, Liu C. Regulation of reproduction by the circadian rhythms. Sheng Li Xue Bao: [Acta Physiologica Sinica] 2016;68:799–808. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.