Abstract
A new class of peptidomimetics termed “γ-AApeptides” was recently developed by our group. Similar to other peptidomimetics, γ-AApeptides are resistant to proteolytic degradation, and possess limitless potential to introduce chemically diverse functional groups. γ-AApeptides have shown great promise in biomedical applications. In this article, we will review a few examples of γ-AApeptides with biological potential. Certain γ-AApeptides can permeate cell membranes and therefore they can be used as potential drug carrier. γ-AApeptides can also bind to HIV RNA with high specificity and affinity, suggesting their potential application as anti-HIV agents. Moreover, they can mimic host-defense peptides and display potent and broad-spectrum activity towards a range of drug-resistant bacterial pathogens. They are also potential anti-cancer agents. For instance, they have shown great promise in targeted imaging of tumor in mouse model, and they are also capable of disrupting p53/DNA interactions, and thus antagonize STAT3 signaling pathway. Recently, from combinatorial screening, γ-AApeptides are identified to inhibit Aβ peptide aggregation, and thus they can be developed into potential anti-Alzheimer’s Disease agent.
Keywords: γ-AApeptides, Peptidomimetics, Structures, Anticancer activity, Antimicrobial activity, Anti-HIV activity, Anti-Aβ aggregation
1. INTRODUCTION
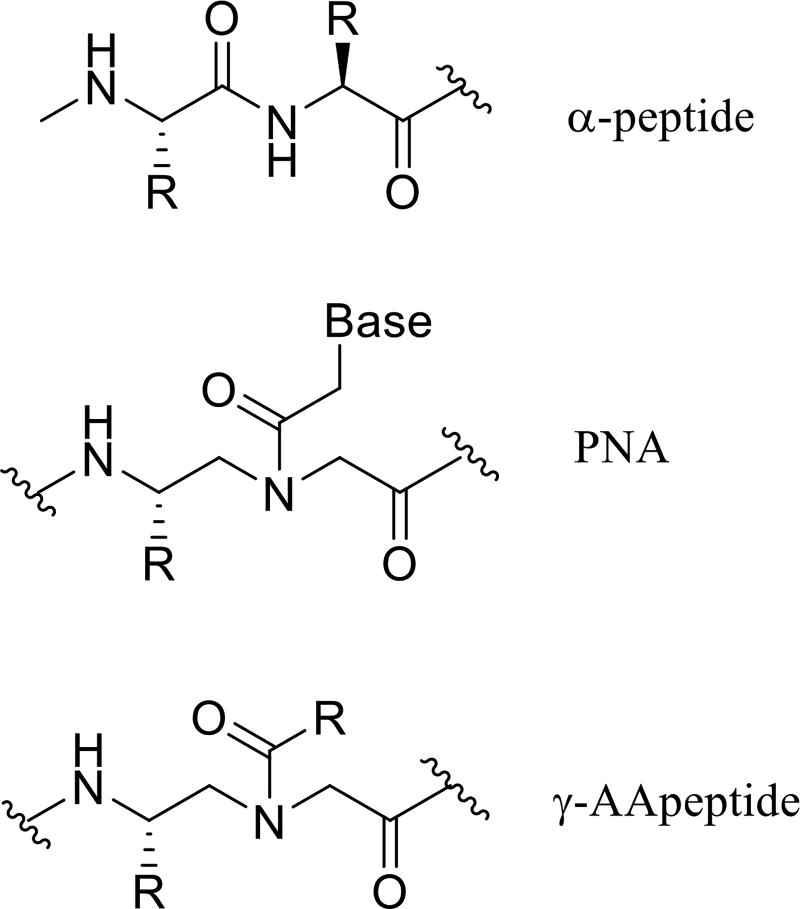
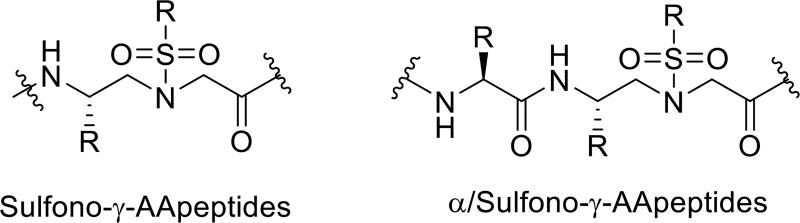
The peptidomimetic oligomers have started to find important applications in chemical biology and biomedical sciences in the past two decades [1,2]. Consisting of unnatural backbones, peptidomimetics could overcome obstacles associated with canonical peptides, including low bioavailability, susceptibility to proteolytic degradation, and limitation in chemodiversity [3]. Thus, they could lead to the development of promising lead compounds and drug candidates. Indeed, peptidomimetics, such as β-peptides [4], α/β-peptides [5], peptoids [6–7], oligoureas [8], show capability to mimic hierarchical structures of proteins, and are extensively studied for therapeutic applications. In the meantime, they are also resistant to proteolytic degradation. To expand the family of peptidomimetics and to facilitate their application in biomedical sciences, we recently developed a new class of peptidomimetics - “γ-AApeptides” [9–10]. They are termed “γ-AApeptides”, as they are oligomers of γ-substituted-N-acylated-N-aminoethyl amino acids (Figure 1), similar to γ-PNAs [11–13]. Our research endeavors have manifested that γ-AApeptides are resistant to enzymatic degradation, and are amendable to chemical modification. In this review we highlight some therapeutic applications of this class of peptidomimetics.
Figure 1.
The chemical structure of α-peptide and γ-AApeptide.
2. Synthesis of γ-AApeptides
2.1 Synthesis of linear γ-AApeptides
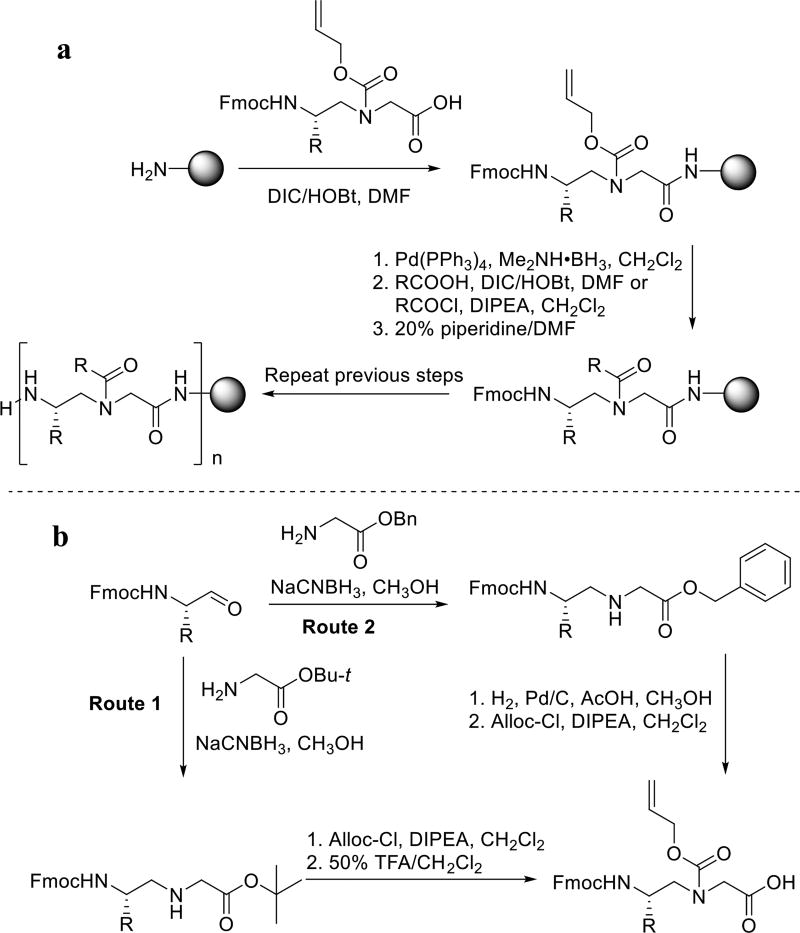
The linear γ-AApeptides (Scheme 1) are synthesized on the solid phase combining both building-block [14] and submonomeric approaches [15–16]. In this method, N-alloc γ-AApeptide building blocks are prepared (Scheme 1b) via route 1 or route 2 depending on different R groups, and are used subsequently for solid phase synthesis in Scheme 1a. The alloc group can be removed effectively, allowing introduction of a variety of side chains through acylation. One advantage of this synthetic approach is that N-alloc γ-AApeptide building blocks are very stable and thus they can be prepared in large batches. Moreover, the same N-alloc γ-AApeptide building block can be functioned with virtually limitless type of side chains, which significantly enhances the chemical diversity.
Scheme 1.
Synthesis of linear γ-AApeptides (a) and γ-AApeptide building blocks (b).
2.2 Synthesis of cyclic γ-AApeptides
It is known that cyclic peptides generally exhibit improved biological activity and stability compared to their linear counterpart due to the rigidification of backbone and functional groups. Although γ-AApeptides are resistant to enzymatic degradation, we envision that cyclization of γ-AApeptides may enhance binding affinity and cell permeability, and therefore we have also developed effective methods for the synthesis of cyclic γ-AApeptides, through either head-to-tail or head-to-side chain cyclization.
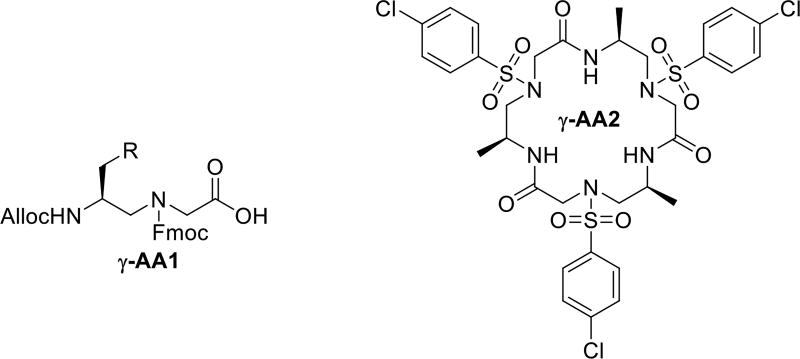
The head-to-tail cyclization is achieved in solution by cyclizing N-terminal NH2 group and C-terminal COOH group of the protected γ-AApeptide synthesized on the CTC (2-chlorotrityl chloride) resin [17]. The Alloc-N’-Fmoc-N γ-AApeptide building block (Figure 2) is used as the first building block attached to the solid support, because regular Fmoc protected building block is easy to self-cleave from the solid support due to the formation of ketopiperazine side products [18]. Such side reactions can be avoided by using the Alloc protected building block because it’s removal is carried out under neutral condition. One example of head-to-tail γ-AApeptide γ-AA2 is shown in Figure 2.
Figure 2.
The structure of γ-AA1 and γ-AA2.
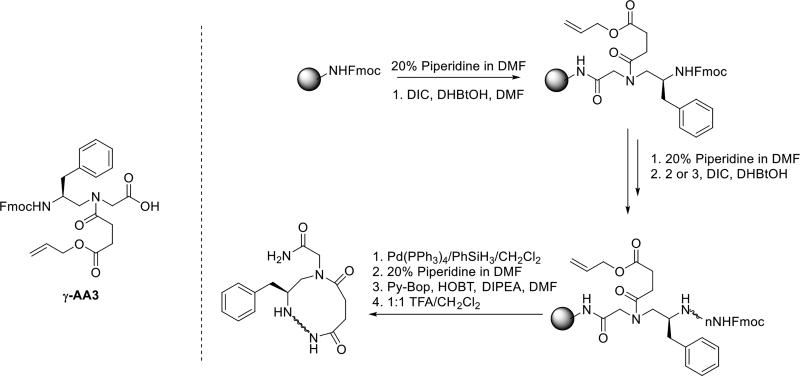
The synthesis of cyclic γ-AApeptides through the head-to-side chain cyclization can be accomplished on the solid support (Scheme 2) [19]. To facilitate on-resin cyclization, a γ-AApeptide building block bearing the mono-allyl succinate group (γ-AA3) is introduced as the first building block attached to the solid support. After the desired sequence is synthesized on the resin, the allyl ester is removed by Pd(PPh3)4/PhSiH3. The cyclization is then conducted by reacting the free carboxylate group with the amino group of the N-terminal building block.
Scheme 2.
The strategy of the head to side chain cyclization.
3. The structure of γ-AApeptides
Three-dimensional structures of natural biopolymers including proteins and nucleic acids determine their function [20]. Understanding the folding of peptidomimetics could lead to rational design of molecules for therapeutic applications [21–22]. We recently have set out to study the secondary structure of γ-AApeptides. To date, we have investigated folding propensity of sulfono-γ-AApeptides and 1:1 α/sulfono-γ-AApeptides (Scheme 3) by X-ray, CD, and 2D-NMR. Akin to the α-peptide, the protons of the secondary amide moieties can potentially form intramolecular hydrogen bonding. Meanwhile, bulky nature of sulfonamido moieties could contribute to the secondary folding by inducing backbone curvature.
Scheme 3.
The sulfono-γ-AApeptide and 1:1 α/sulfono-γ-AA hybrid.
3.1 Sulfono- γ-AApeptides
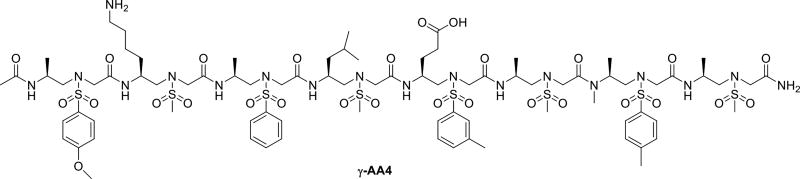
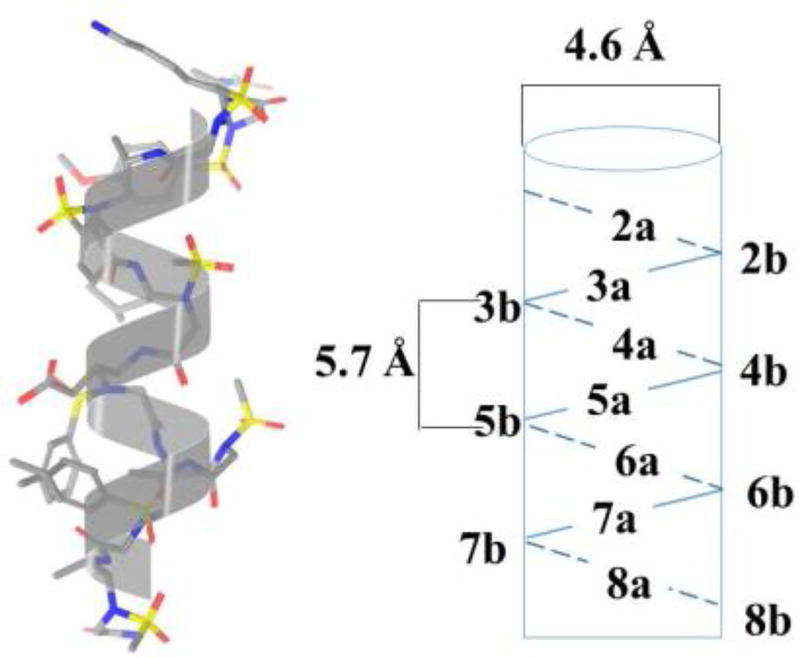
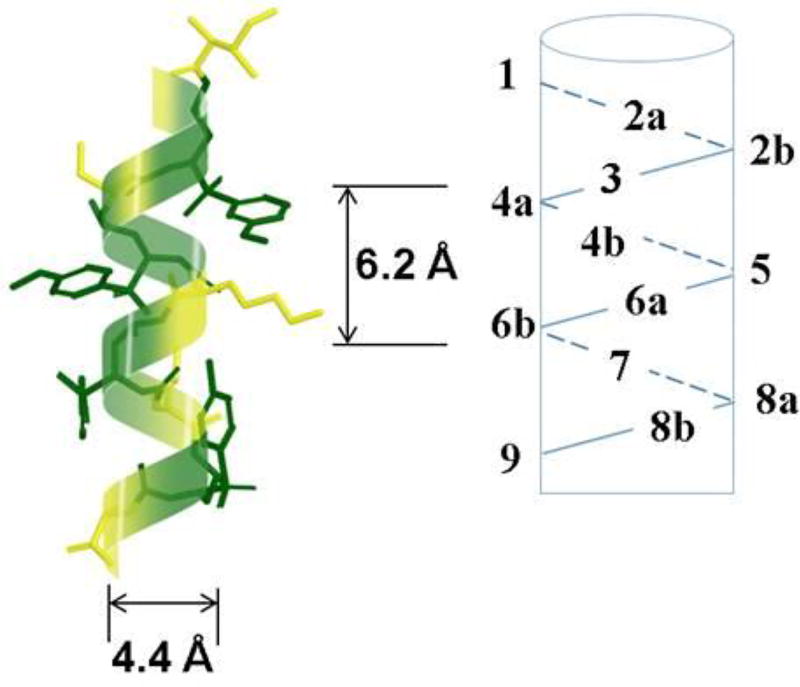
2D-NMR studies suggest that the sequence γ-AA4 (Scheme 4) comparable to a 16-mer peptide in length adopts a helical conformation in solution, and interestingly, its helical parameters including helical radius and pitch are similar to the α-helix (Figure 3) [23]. The subsequent Circular Dichroism (CD) studies indicate longer sequences may possess more pronounced helicity than shorter sequences. It should be noted that these helical structures are reasonably stable over a wide range of temperature change.
Scheme 4.
The chemical structure of the sulfono-γ-AApeptides.
Figure 3.
The helical structure of γ-AA4. Reproduced with permission from ref 23. Copyright 2015 John Wiley & Sons, Inc.
3.2 1:1 α/sulfono- γ-AA peptides
Encouraged by the abovementioned findings that sulfono-γ-AApeptides exhibit helical propensity, we further investigated the folding conformation of a new class of heterogeneous peptides comprised of α and sulfono-γ-AApeptide residues in a 1:1 ratio (Scheme 5) [24]. In contrast to homogeneous sulfono-γ-AApeptides, we hypothesized that inclusion of α-amino acid residues may lead to the sequences with more stable folding structure due to the introduction of more hydrogen bonding.
Scheme 5.
The chemical structure of the α/sulfono-γ-AA peptides.
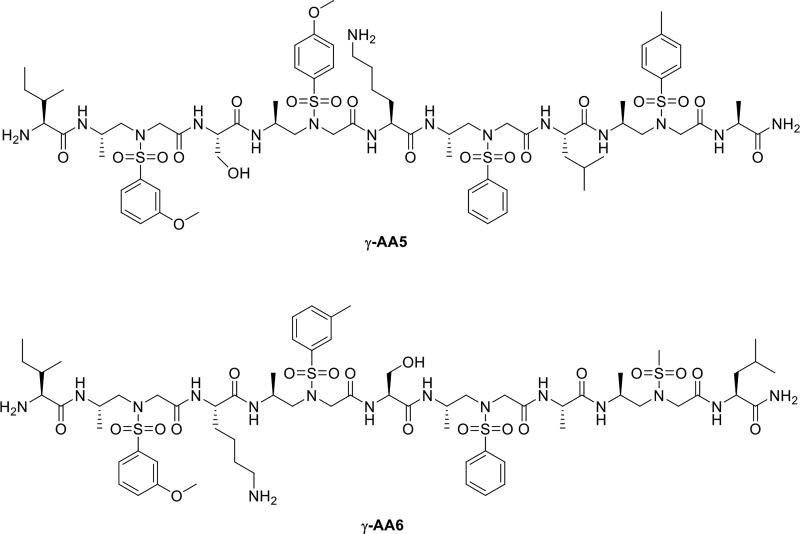
As anticipated, 2D-NMR of a heterogeneous sequence γ-AA5 containing alternative α and sulfono-γ-AApeptide residues strongly manifests the presence of a defined helical folding structure (Figure 4). The availability of hydrogen bonds was confirmed by the existence of most backbone NH resonances even more than 24 h in the H/D exchange studies. The folding propensity seems to be universal in this class of oligomer as another 1:1 α/sulfono-γ-AA peptide γ-AA6 with different side chains also show similar right-handed helical structure in solution.
Figure 4.
The helical structure of γ-AA5. Reproduced with permission from ref 24. Copyright 2015 American Chemical Society.
While we are still excavating three-dimensional folding structures of γ-AApeptides, we have also started to explore biological potential of γ-AApeptides. Through either rational design or combinatorial screening, some γ-AApeptides have been shown to hold promise for therapeutic applications.
4. Anti-cancer
4.1 Mimicking RGD peptides
Integrin αvβ3, a cell surface receptor, plays a vital role in biological processes such as angiogenesis and has been found to be overexpressed in many cancers [25]. As such, it is an attractive target for anticancer therapeutic development. It is known that integrin αvβ3 binds Arg-Gly-Asp (RGD) tripeptide motif in proteins and peptides, therefore, a wide variety of peptides/peptidomimetics bearing RGD epitope have been developed.
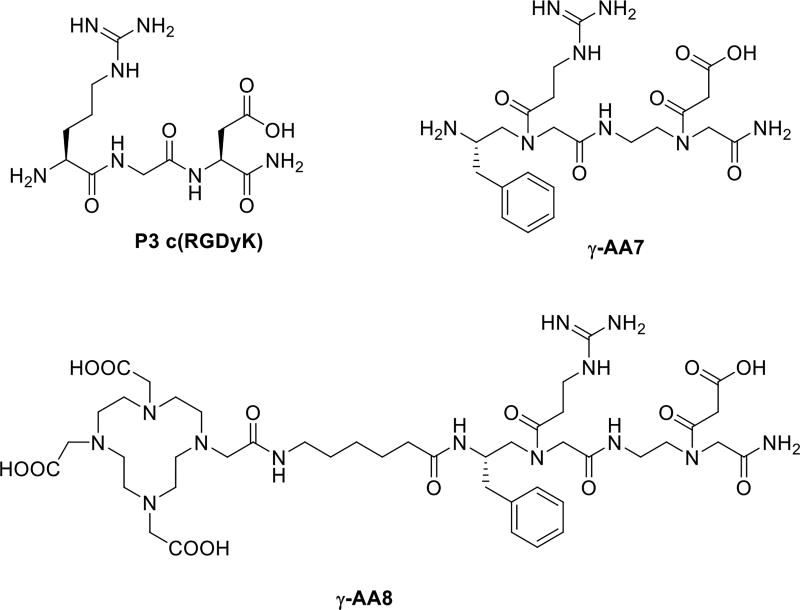
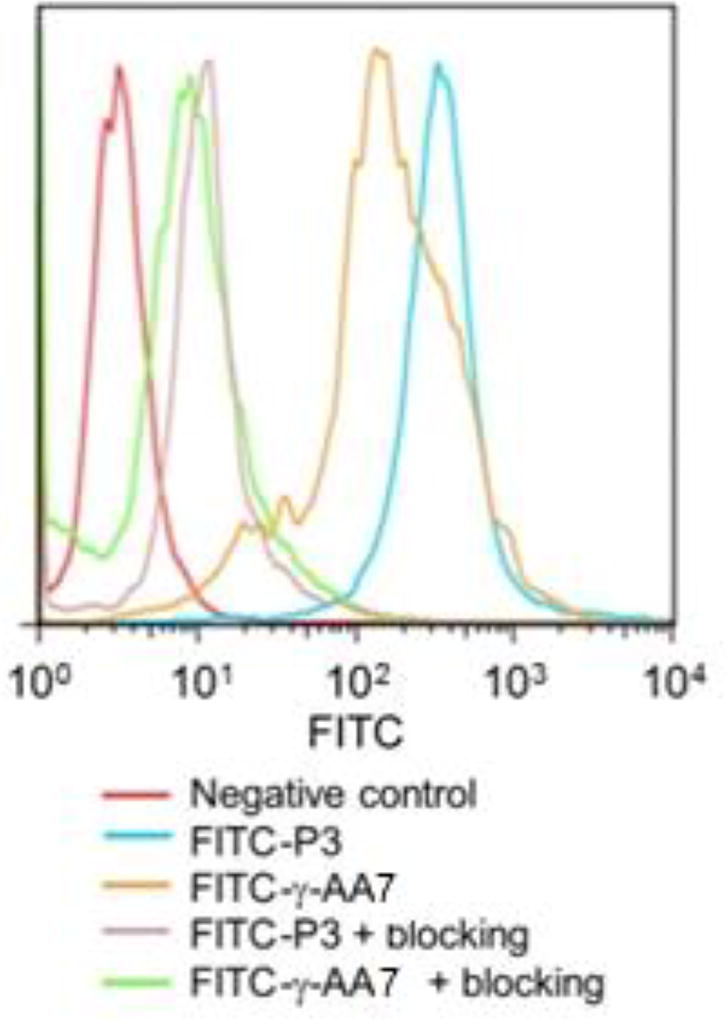
To specifically recognize integrin αvβ3, we designed a γ-AApeptide γ-AA7 bearing guanidino and carboxyl groups, which were introduced to mimic the RGD motif (Scheme 6) [26]. Interestingly, the binding affinity of FITC conjugated γ-AA7 towards integrin αvβ3 on U87MG human glioblastoma cells was similar to the FITC labeled c(RGDyK) peptide P3 (Figure 5). The binding specificity was confirmed by the blocking assay in which pre-incubation of the cells with 2 µM of P3 largely prevented the binding of FITC-γ-AA7, analogous to what occurred when FITC-P3 was used. It should be noted that although γ-AA7 and P3 exhibited comparable affinity to αvβ3, 64Cu-γ-AA8 was more resistant to proteolytic degradation than 64Cu-DOTA-P3 [26], augmenting its therapeutic potential.
Scheme 6.
The chemical structure γ-AA7, γ-AA8 and control c(RGDyK) P3.
Figure 5.
Flow cytometry analysis of FITC-conjugated γ-AApeptides. Reproduced with permission from ref 26. Copyright 2012 Royal Society of Chemistry.
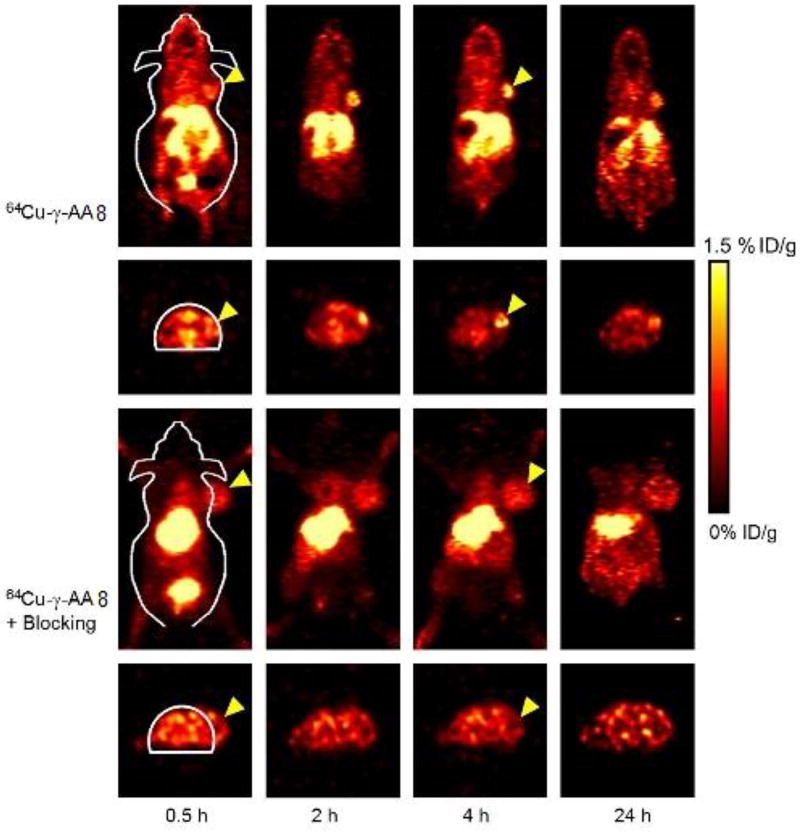
We thus conducted PET imaging on an U87MG tumor-bearing mouse model. After administering the tracer 64Cu-γ-AA8 intravenous (Figure 6), its uptake in tumor could be detected as early as 0.5 h p.i., and remained persistent during the experimental period. Similar to in vitro studies, administering the P3 significantly reduced the tumor uptake (Figure 6), which suggests that the γ-AApeptide tracer targets integrin αvβ3 specifically in vivo.
Figure 6.
Serial PET imaging and biodistribution studies of 64Cu-γ-AA8. Reproduced with permission from ref 26. Copyright 2012 Royal Society of Chemistry.
4.2 Mimicking fMLF peptide
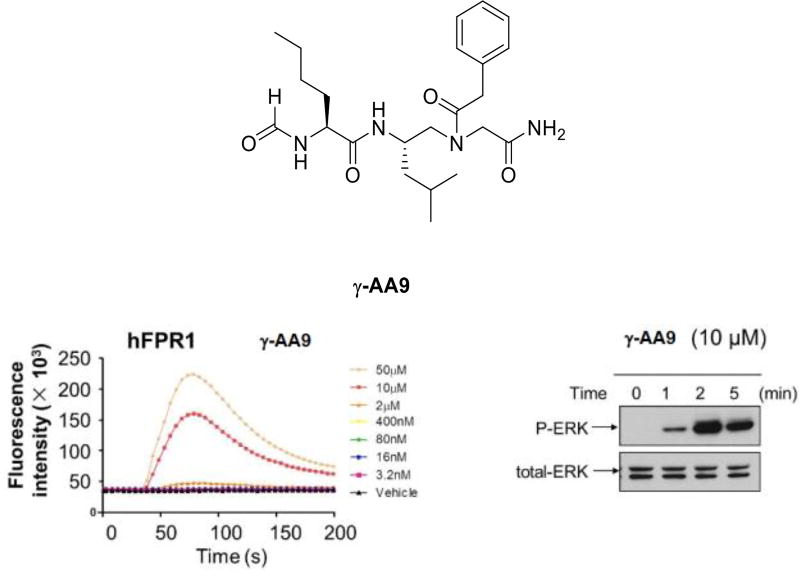
The formyl peptide, N-formyl-Met-Leu-Phe (fMLF), is chemoattractant of neutrophils [27]. It binds to the G protein-coupled N-formylpeptide receptor (FPR) and activates a series of biological events [28]. Its dysfunction has been linked to cancer and other diseases, and thus there is considerable interest in the development of fMLF mimetics for therapeutic applications [29]. To this end, we designed a number of γ-AApeptides to mimic fMLF, and investigated their biological function (Figure 7) [30].
Figure 7.
The induction of calcium mobilization of γ-AA9 and activation of ERKs. Reproduced with permission from ref 30. Copyright 2014 John Wiley & Sons, Inc.
It is known that fMLF can effectively activate calcium mobilization in human FPR1-transfected rat basophilic leukemic (RBL)-2H3 cells, these fMLF-mimicking γ-AApeptides were tested for their ability to induce calcium mobilization in the same cell line (Figure 7). Interestingly, even though the lead compound γ-AA9 was less potent than fMLF at lower concentrations, it was more effective at high concentrations. In addition to calcium mobilization, it is also common that the fMLF peptide can activate the MAP kinases ERK1 and ERK2 in human neutrophils through recognition of FPR. Indeed, γ-AA9 can rapidly activate the phosphorylation of ERK in just a few minutes (Figure 7). It should also be mentioned that our sequent studies indicate that γ-AA9 could also trigger ROS generation and induce chemotaxis.
4.3 Disruption of p53/MDM2 protein-protein interaction
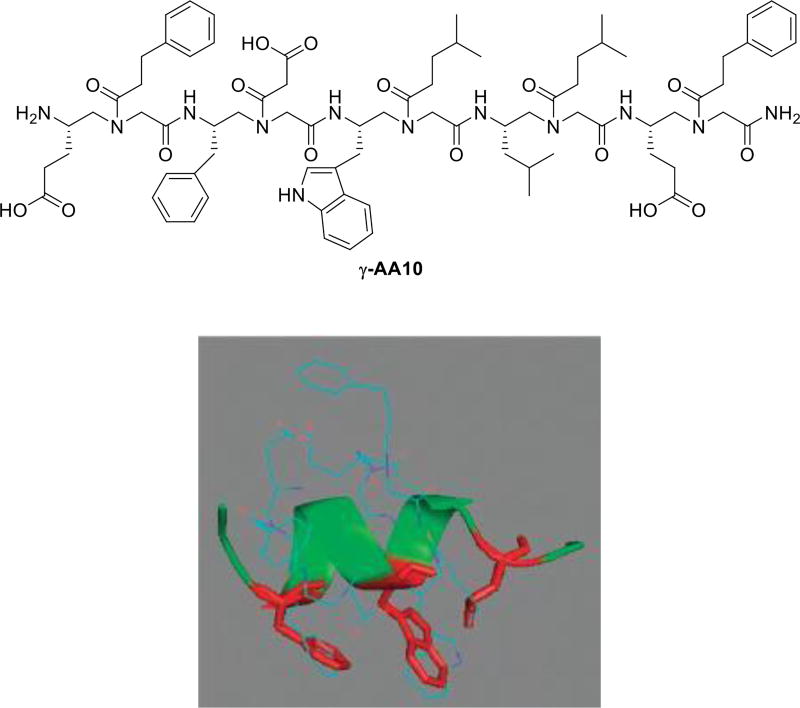
The p53/MDM2 interaction is a viable cancer target, and also serves as a testing ground for the design of helical peptidomimetics [31]. We have synthesized a number of γ-AApeptides and tested these sequences for their ability to modulate p53/MDM2 interaction, in the hope to identify potential anticancer therapeutics [14]. Interestingly, even though the tested sequences were not designed to form helical structures due to the short lengths, a few γ-AApeptides exhibited reasonable activity in the inhibition of p53/MDM2. It is known that Phe, Trp, Leu residues of the p53 helical domain are largely responsible for the strong association with MDM2. Our energy-minimization for the most potent sequence γ-AA10 (Figure 8) suggested the side groups of γ-AA10 may indeed overlap well with those residues in the p53 helical domain.
Figure 8.
The chemical structure of γ-AA10. Energy minimized structure (MM2) of γ-AA10. Reproduced with permission from ref 14. Copyright 2011 Royal Society of Chemistry.
4.4 γ-AApeptides that disrupt STAT3/DNA interaction
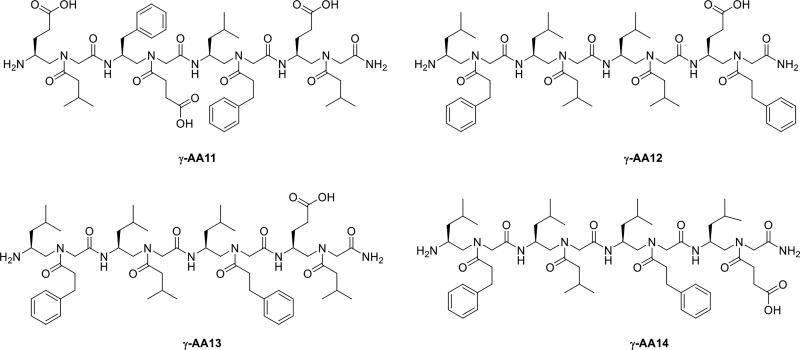
STAT3, the signal transducer and activator of transcription 3, plays a critical role in the regulation of a number of biological processes [32]. The activation of STAT3 is generally transient and is tightly regulated. Upon phosphorylation and activation, STAT3 relocates to cell nucleus, and mediates the expression of a variety of genes. The aberrant activation of STAT3 is linked to many cancers including both solid tumors and hematological cancers [33]. Thus, there is significant interest in the identification of inhibitors of STAT signaling, so as to develop potential anti-cancer therapeutics. Inhibition of STAT3 signaling can be achieved by inhibiting STAT3/DNA interaction or inhibition of STAT3 dimerization [34]. Using the one-bead-one-compound (OBOC) screening method, a library of γ-AApeptides was developed, from which a few sequences were identified to potentially target STAT3-DNA binding (Scheme 7) [35].
Scheme 7.
The chemical structure of γ-AA11, 12, 13, 14.
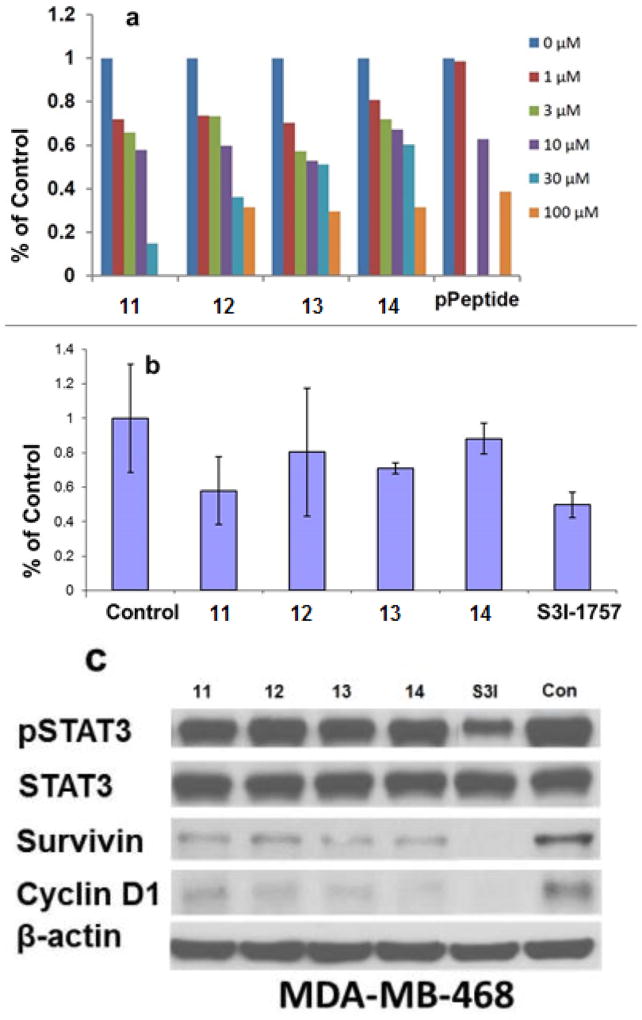
Interestingly, none of these molecules showed any inhibitory activity to inhibit the binding of the fluorescein-labelled GpYLPQTV phosphotyrosine peptide, suggesting that these γ-AApeptides did not prevent STAT3 dimerization. In contrast, all the sequences could effectively disrupt STAT3-DNA interaction (Figure 9a and 9b). Moreover, all γ-AApeptides showed capability to cross cell membranes and inhibit STAT3-DNA interaction. (Figure 9c).
Figure 9.
a and b is the DNA–STAT3 binding assays. c is the effect of the γ-AApeptides on cell signaling. Reproduced with permission from ref 35. Copyright 2014 Royal Society of Chemistry.
5. Antimicrobial
The World Health Organization (WHO) reported that one major threat faced by public in 21st century is the development of resistance to conventional antibiotics by pathogenic bacteria [36]. Recent studies have focused on the development of new antibiotics with novel mechanisms to combat the bacterial resistance [37]. Host-defense peptides (HDPs) may be an alternative strategy to the conventional antibiotics as these peptides show their mode of action by bacterial membrane disruption. It is known that HDPs are diverse in structure and can form helices, β-sheets, and extended structures. However, they frequently attain globally amphipathic nature when approaching the bacterial surface. The hydrophilic cationic groups direct the initial association of HDPs with the negatively charged bacterial membranes, whereas the hydrophobic groups assist in the penetration of HDPs into the bacteria, leading to rupture of the bacterial cell membranes. This kind of mechanism makes it difficult for bacteria to develop resistance. However, HDPs are prone to proteolytic degradation and are only moderately active.
Thus, there has been considerable interest in developing peptidomimetics that mimic HDPs to overcome the disadvantages [38–39]. We recently focused on developing γ-AApeptides that mimic the mechanism of action of HDPs. γ-AApeptides are proved for their stability against enzymatic degradation and capability of forming secondary structures. Hence, we investigated the potency and antimicrobial activity of different classes of γ-AApeptides. The findings suggested that γ-AApeptides could be an alternative approach of HDPs for antibacterial development.
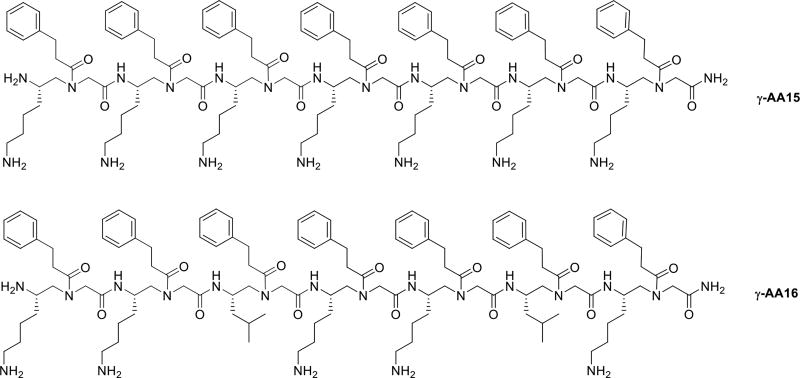
Our initial attempt was made by synthesizing the linear γ-AA15 sequence which has 7 amphipathic building blocks. (Each building block has one hydrophobic group and one cationic group.) (Scheme 8, Table 1) [40]. γ-AA15 showed potent activity against Gram-positive bacteria. We hypothesized that γ-AA15 could adopt a global amphipathic conformation when interacting bacterial membranes. In an attempt to analyze the impact of the hydrophobicity on the antimicrobial activity, γ-AA16 was synthesized by replacing the two amphiphilic building blocks in γ-AA15 with two hydrophobic building blocks. To our delight, γ-AA16 was found to be potent and had broad spectrum antimicrobial activity against Gram-positive and Gram-negative bacteria. Florescence microscopy data suggested that γ-AA16 acts by membrane disruption [41].
Scheme 8.
The chemical structures of γ-AA15, 16.
Table 1.
The antimicrobial and hemolytic activities of γ-AApeptides.
| γ-AA peptides |
MIC (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram negative | Gram positive | |||||||
| E. coli | K.pneumoniae | P.aeruginosa | B.subtilis |
S.epidermidis (MRSE) |
E.faecalis (VRE) |
S.aureus (MRSA) |
Hemolysis (HC50) |
|
| γ-AA15 | 5 | - | >50 | - | 5 | 15 | 5 | >500 |
| γ-AA16 | 5 | - | >50 | - | 5 | 5 | 5 | 300 |
| γ-AA17 | 4 | - | 6 | - | 2 | 2 | 4 | 75 |
| γ-AA18 | 4 | - | 4 | - | 2 | 2 | 2 | 100 |
| γ-AA19 | - | 8 | 8 | 1 | 2 | 5 | 1 | 100 |
| γ-AA20 | 2.5 | - | 5 | - | 4 | 5 | 4 | >500 |
| γ-AA21 | 3 | - | 3 | - | 3 | 4 | 3 | >500 |
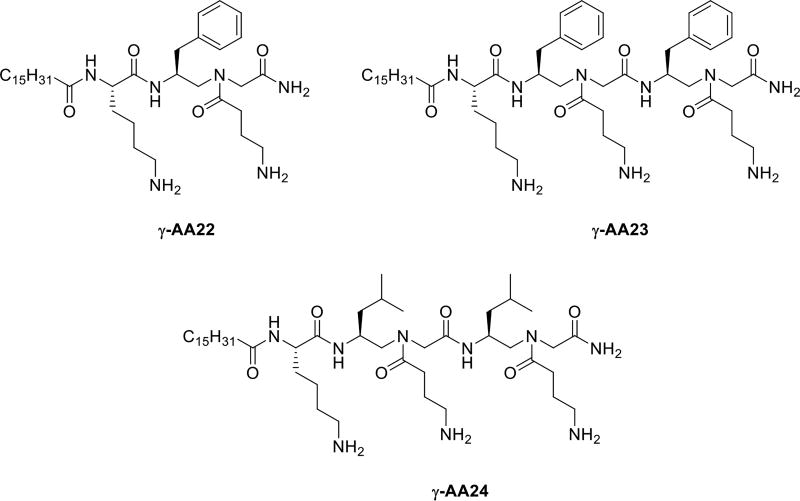
| γ-AA22 | 4 | - | 2 | - | 2 | 2 | 2 | 150 |
| γ-AA23 | 4 | - | 2 | - | 2 | 2 | 2 | 250 |
| γ-AA24 | 4 | - | 2 | - | 2 | 2 | 2 | 350 |
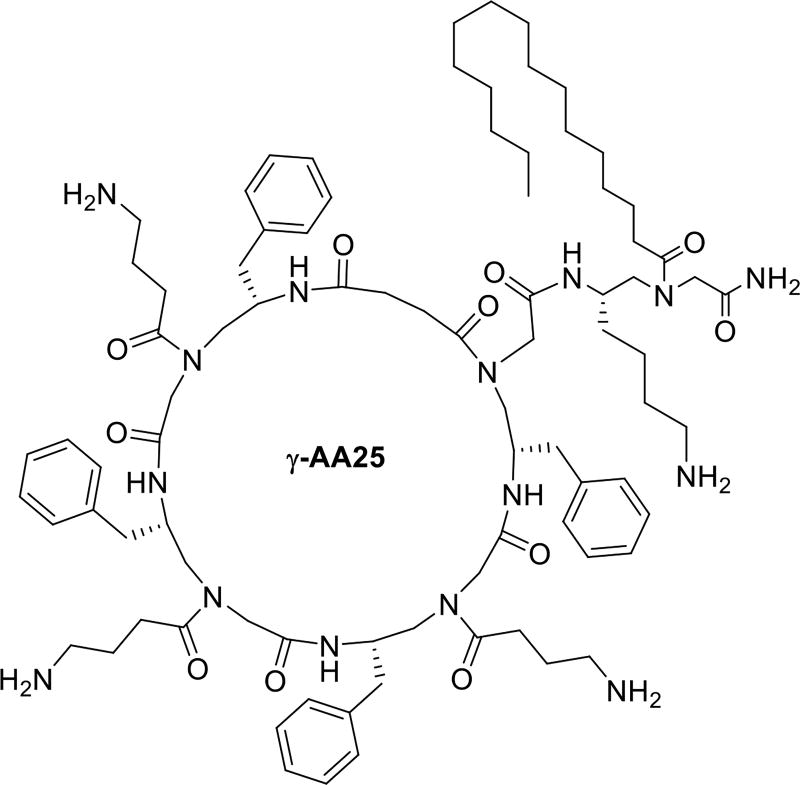
| γ-AA25 | 2 | 3 | 5 | - | 1 | 3 | 2 | 100 |
5.1 Linear helical γ-AApeptides
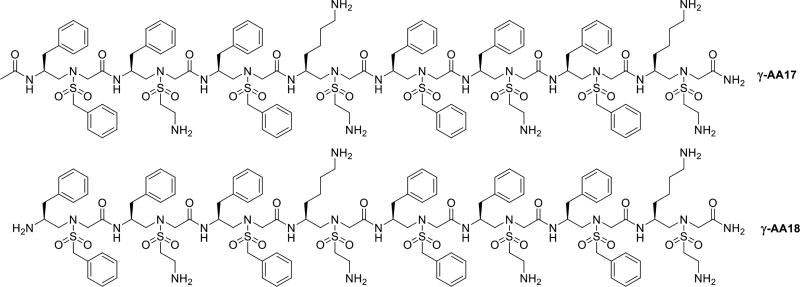
Host-defense peptides, such as magainin [42], can adopt amphipathic helical structure. We previously reported that sulfono-γ-AApeptides can form helical structures [23]. It was intriguing to investigate if antimicrobial sulfono-γ-AApeptides can mimic magainin in terms of helical amphipathic structure and mode of action. We thus designed a series of sulfono-γ-AApeptides. Among all synthesized sequences, γ-AA17 and γ-AA18 were found to be potent against Gram-positive and Gram-negative bacteria (Scheme 9, Table 1) [43]. In fact, γ-AA17 and γ-AA18 were comprised of same γ-AApeptide building blocks. The only difference is that γ-AA18 lacks an acetyl group at the N-terminus when compared to γ-AA17. The SAXS (Small-Angle X-ray Scattering) studies reveals that γ-AA17 adopted more defined folding structure than γ-AA18 in solution. However, γ-AA18 was found to be highly selective and less cytotoxic against human cells in comparison with γ-AA17, which is consistent to the findings that strong helical forming sequences are more hemolytic and less active [44]. Florescence microscopy suggested that γ-AA18 could damage bacterial membranes. In addition to strong antimicrobial activity, γ-AA18 was also proved to be highly stable in pronase for 18h, thus overcoming the drawbacks of HDPs which are susceptible to enzymatic degradation.
Scheme 9.
The chemical structure of γ-AA17, γ-AA18.
5.2 Cyclic γ-AApeptides
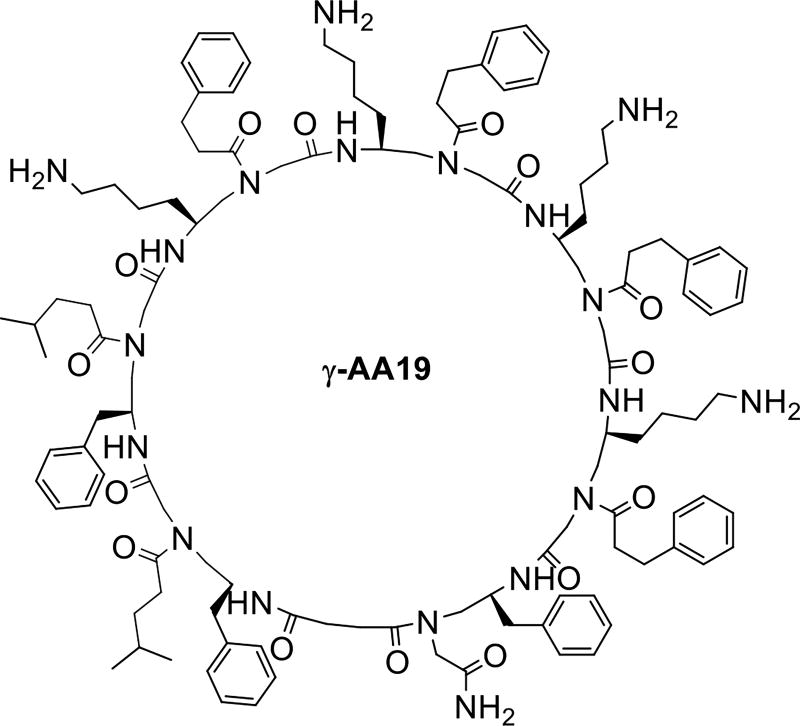
Cyclic peptide antibiotics are widely used and found to have more potent activity than the linear peptide antibiotics due to their more rigid conformation and less susceptibility to proteolytic degradation [45]. Although stability is not an issue for γ-AApeptides, cyclization may lead to sequences of more potent activity. We synthesized different amphipathic cyclic γ-AApeptides and the activity was tuned by changing the number of cationic and hydrophobic groups in the amphipathic building blocks. Of all synthesized cyclic γ-AApeptides γ-AA19 was found to have broad spectrum antimicrobial activity (Table1, Scheme 10) [19].
Scheme 10.
The chemical structure of γ-AA19.
5.3 Lipo γ-AApeptides
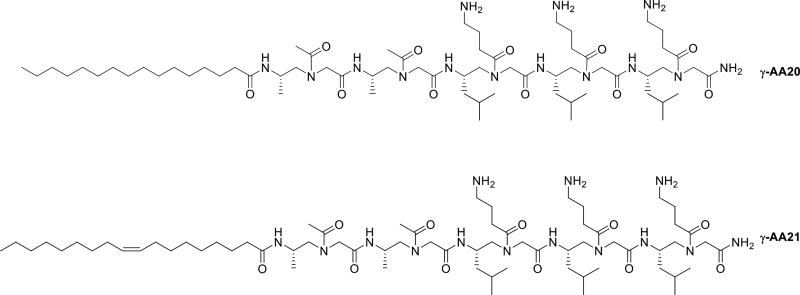
Another class of antibiotic peptides are lipopeptide whose mechanism was found to be different from the HDPs. Lipopeptides are composed of peptides with varying length of aliphatic acid chains [46]. The lipid chains are believed to contribute to the overall lipophilicity of the peptide, which helps in bacterial membrane penetration [47]. Based on the conception, two lipo-linear peptides γ-AA20 and γ-AA21 with saturated and unsaturated alkyl tails were synthesized respectively, and were found to be active against Gram-positive and Gram-negative bacteria (Table 1, Scheme 11) [48]. γ-AA21 showed potent antimicrobial activity and high selectivity to bacteria, suggesting that the peptides with the unsaturated tail are more sensitive to bacteria than the mammalian cells.
Scheme 11.
The chemical structure γ-AA20 and 21.
Moving forward, we developed a series of lipo-linear peptides which had a hybrid back bone fused with γ-AApeptides and lipidated α-peptides (γ-AA22, γ-AA23, γ-AA24, Table 1, Scheme 12). Surprisingly, these fused lipo-linear peptides had demonstrated broad spectrum anti-bacterial activity. These findings can augment the chemo diversity of lipo-linear γ-AApeptides for optimization and enhancement as a new class of antibiotic agents [49].
Scheme 12.
The chemical structure of γ-AA22, γ-AA23, γ-AA24.
Lipo-Cyclic antibiotic peptides such as Daptomycin and Polymyxin are used to treat Gram-positive and Gram-negative bacterial infections, respectively [50]. We assumed a combination of lipidation and cyclization in γ-AApeptides could therefore show enhanced antimicrobial activity than the previously reported lipo and cyclic γ-AApeptides. Initially, lipo-cyclic γ-AApeptides were synthesized with the C16 tail attached to the ring directly. Interestingly, they were not active at all. Subsequently more lipo-cyclic γ-AApeptides were synthesized in which the C16 tail was attached outside of the cyclic ring. Among them, γ-AA25 (Table 1, Scheme 10), which had a small amphipathic ring and a C16 tail, displayed most broad spectrum antimicrobial activity in comparison with other lipo-cyclic γ-AApeptides. The lipo-cyclic γ-AApeptides were also proved to be more potent against Gram-negative bacteria than the non-alkylated cyclic γ-AApeptides. Lastly, γ-AA25 was found to kill the bacteria by membrane disruption mechanism by florescence microscopy study [50].
Remarkably, lipo-cyclic γ-AApeptides displayed dual roles as antimicrobial and inflammatory agents as γ-AA25 was showed to inhibit the lipopolysaccharide (LPS) activated Toll-like receptor 4 (TLR4) signaling. A more recent study also reveals that lipo-cyclic γ-AApeptides could prevent the growth of biofilms efficiently than conventional antibiotics, which might be due to the presence of the lipid tail [51]
6. γ-AApeptides that disrupt Aβ aggregation
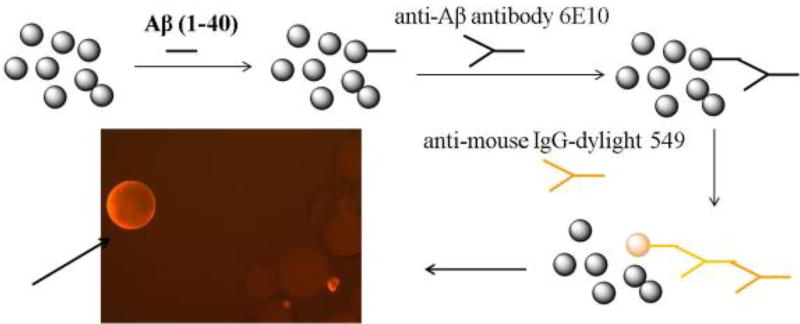
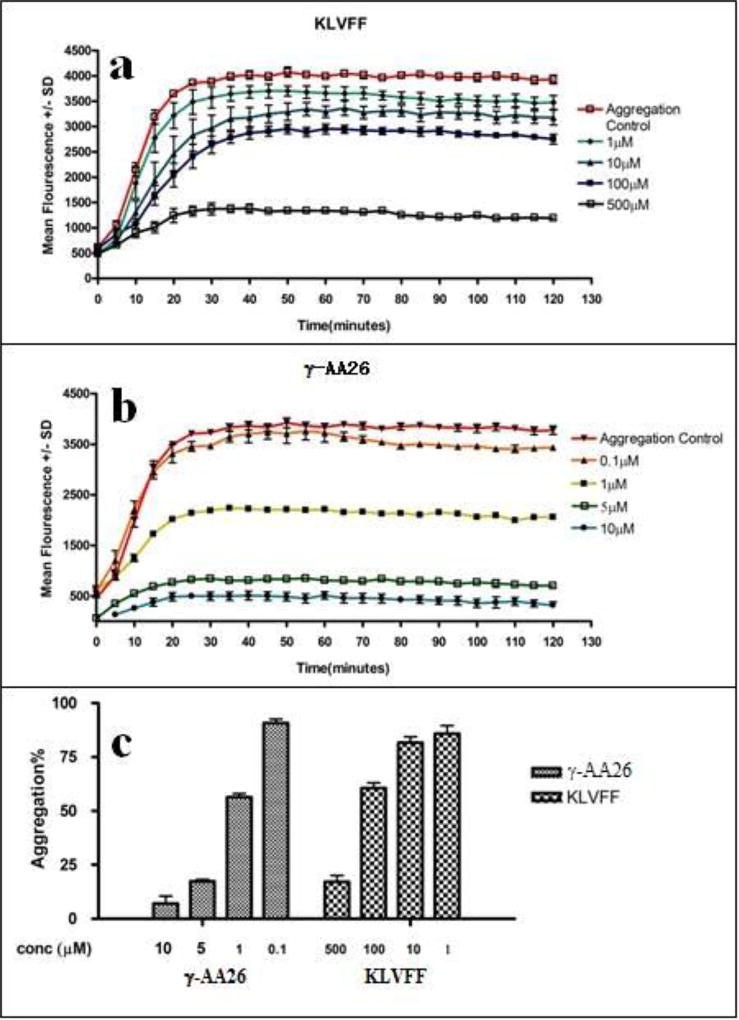
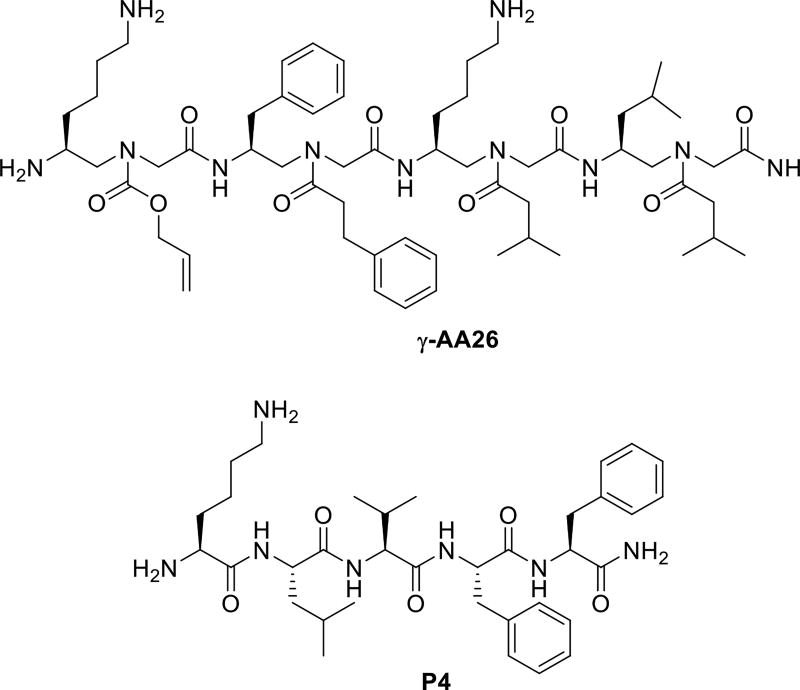
The Aβ40 peptide is recognized as a major etiological factor for Alzheimer’s disease (AD) and plays a key role in AD pathogenesis. Using the split-and-pool method, we prepared a γ-AApeptide library containing 192000 compounds [52], which were screened for their binding to Aβ40 peptide. One hit compound γ-AA26 was identified (Figure 10 and Scheme 13) and tested for its binding activity towards Aβ40 peptide. Surprisingly, γ-AA26 is almost 100-fold as potent as the hydrophobic core of the Aβ peptide P4, KLVFF, in the inhibition of Aβ aggregation (Figure 11).
Figure 10.
Schematic representation of the on-bead screening of the γ-AApeptide library against the Aβ40 peptide. Reproduced with permission from ref 52. Copyright 2014 Royal Society of Chemistry
Scheme 13.
The chemical structure of γ-AA25.
Figure 11.
The ThT assay of compounds against Aβ40. Reproduced with permission from ref 52. Copyright 2014 Royal Society of Chemistry.
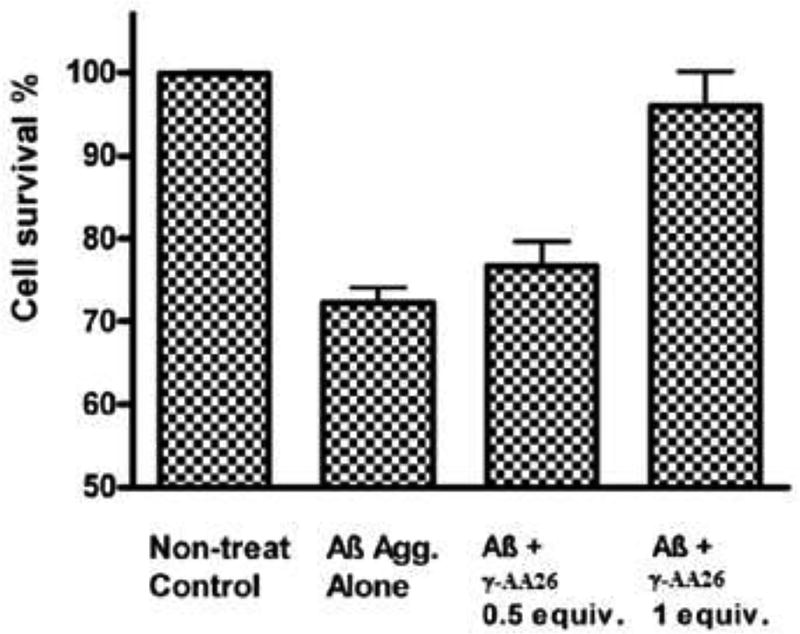
The subsequent TEM studies confirmed the ability of γ-AA26 for the inhibition of Aβ aggregation [52]. In addition, one equivalent of γ-AA26 could remove the toxicity of Aβ aggregates towards N2a neuroblastoma cells. (Figure 12).
Figure 12.
Detoxification of Aβ42 aggregates by γ-AA26. Reproduced with permission from ref 52. Copyright 2014 Royal Society of Chemistry.
7. Anti HIV
Mimicking Tat peptides
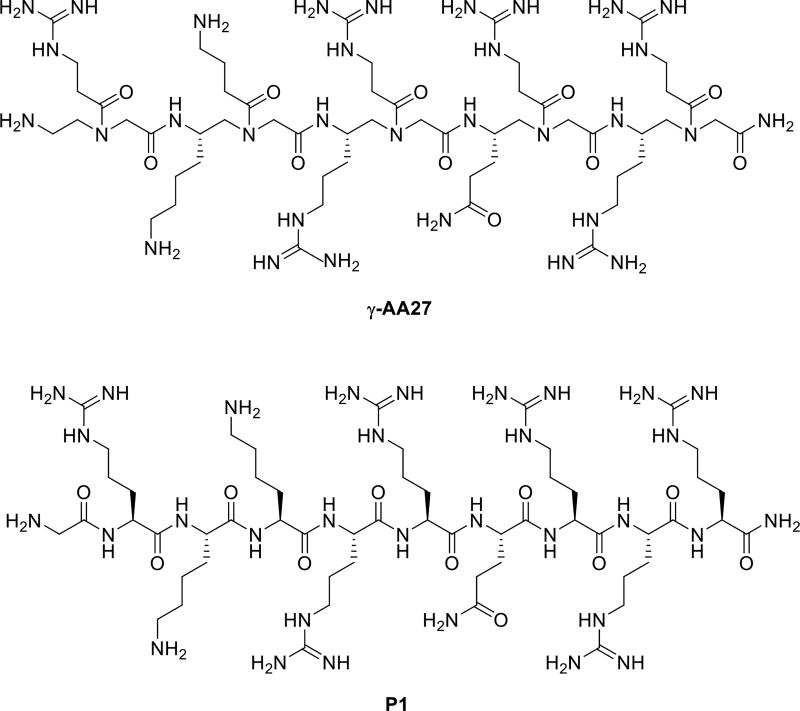
It is of importance in the biological and medicinal chemistry to identify novel molecules that specifically target RNA [53–54]. The Tat peptide is a known peptide which tightly binds to HIV TAR RNA. To the end, we designed a γ-AApeptide γ-AA27 which contains same side chains as Tat 48–57 (Scheme 14). Interestingly, this sequence could bind to HIV TAR RNA and BIV TAR RNA with affinity and specificity akin to the Tat peptide [55].
Scheme 14.
The chemical structure of γ-AA26 and the KLVFF P4.
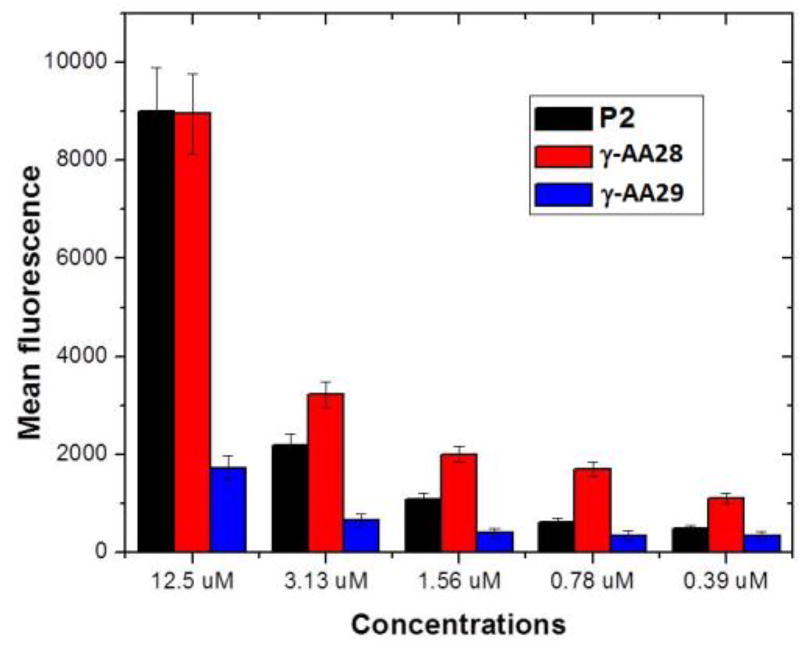
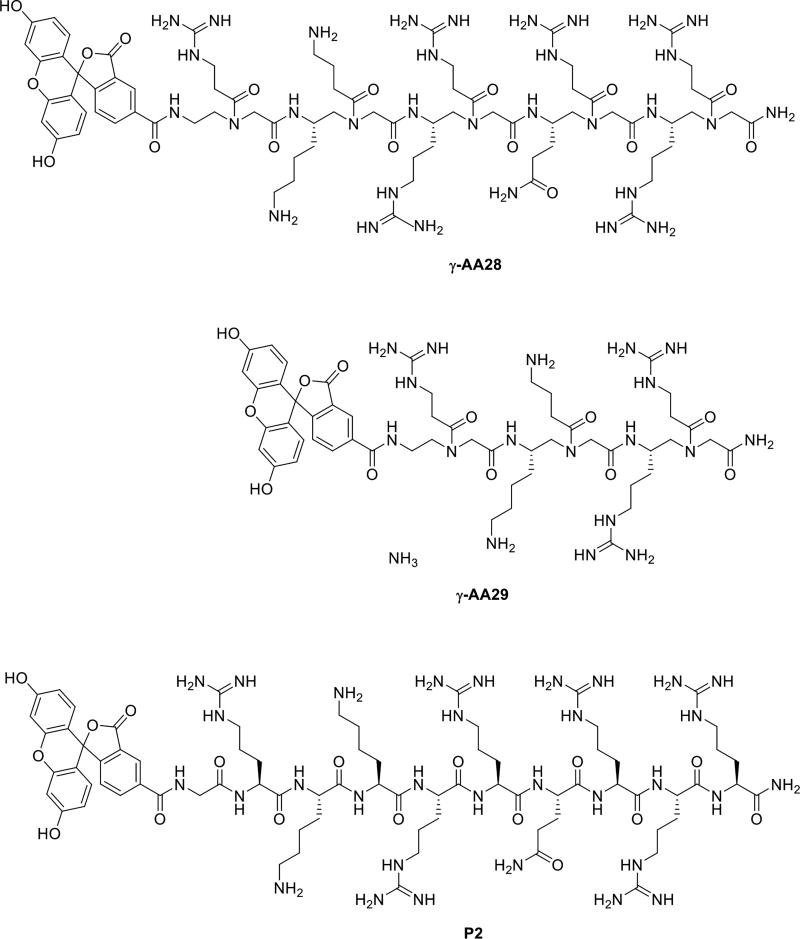
In addition to HIV RNA recognition, it is known that the Tat peptide also exhibits excellent cell permeability [56], and as a result it has been widely used as a carrier to deliver drugs/molecular probes into intact cells [57]. We have also investigated the cell permeability of γ-AApeptides by attaching the fluorescein moiety to γ-AA27 (Scheme 14) [58]. Flow cytometry (Figure 13) showed that the cellular uptake of γ-AA28 (fluorescent γ-AA27, red bar) is similar to that of P2 (fluorescent P1, black bar) at the same concentration. It is interesting that γ-AA28 displayed a higher cellular uptake at low concentrations, possibly because γ-AApeptide backbone contains tertiary amide bonds [59]. In agreement with the previous studies, even at high concentrations the short γ-AA10 (Figure 13, Scheme 16) didn’t display any cellular uptake.
Figure 13.
Flow cytometry study of the Tat peptide analogue. Reproduced with permission from ref 58. Copyright 2012 American Chemical Society.
Scheme 16.
The chemical structures γ-AA28, γ-AA29 and control Tat 48–57.
Future
In this review, we have highlighted a few examples of γ-AApeptides for their therapeutic applications. Our research findings indicate that γ-AApeptides can form well-defined structures, which makes it possible to rationally design novel agents that specially target proteins of medicinal interest. Meanwhile, their chemodiversity and stability allow them to be rich source of ligands for the identification of potential drug candidates. A few research areas in γ-AApeptides could be expanded in the future to strengthen their potential in biomedical sciences. First, although solution structures of sulfono-γ-AApeptides were obtained, crystal structures are still necessary to gain insight into their folding principles, which could guide the design of bioactive peptides. In addition, in vivo animal studies should be conducted as the next step to manifest the biological potential of γ-AApeptides. Moreover, combinatorial libraries could be further developed by introducing functional group diversity and structural constraints, e.g. cyclic γ-AApeptide library. Such development may lead to new γ-AApeptide-based drug candidates or leads.
Scheme 15.
The chemical structure γ-AA27 and control Tat 48–57.
Acknowledgments
We thank financial support from NSF 1351265 and NIH 1R01GM112652-01A1.
Biography

References
- 1.Wu Y-D, Gellman S. Peptidomimetics. Acc. Chem. Res. 2008;41:1231–1232. doi: 10.1021/ar800216e. [DOI] [PubMed] [Google Scholar]
- 2.Goodman CM, Choi S, Shandler S, DeGrado WF. Foldamers as versatile frameworks for the design and evolution of function. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patch JA, Barron AE. Mimicry of bioactive peptides via non-natural, sequence-specific peptidomimetic oligomers. Curr. Opin. Chem. Biol. 2002;6:872–877. doi: 10.1016/s1367-5931(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 4.Cheng RP, Gellman SH, DeGrado WF. β-Peptides: From Structure to Function. Chem. Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 5.Horne WS, Johnson LM, Ketas TJ, Klasse PJ, Lu M, Moore JP, Gellman SH. Structural and biological mimicry of protein surface recognition by α/β-peptide foldamers. Proc. Natl. Acad. Sci. 2009;106:14751–14756. doi: 10.1073/pnas.0902663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laursen JS, Engel-Andreasen J, Olsen CA. β-Peptoid Foldamers at Last. Acc. Chem. Res. 2015;48:2696–2704. doi: 10.1021/acs.accounts.5b00257. [DOI] [PubMed] [Google Scholar]
- 7.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK. Peptoids: a modular approach to drug discovery. Proc. Natl. Acad. Sci. U. S. A. 1992;89:9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fremaux J, Kauffmann B, Guichard G. Synthesis and Folding Propensity of Aliphatic Oligoureas Containing Repeats of Proline-Type Units. J. Org. Chem. 2014;79:5494–5502. doi: 10.1021/jo5006075. [DOI] [PubMed] [Google Scholar]
- 9.Teng P, Shi Y, Sang P, Cai J. γ-AApeptides as a New Class of Peptidomimetics. Chem. Eur. J. 2016;22:5458–5466. doi: 10.1002/chem.201504936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Teng P, Sang P, She F, Wei L, Cai J. γ-AApeptides: Design, Structure, and Applications. Acc. Chem. Res. 2016;49:428–441. doi: 10.1021/acs.accounts.5b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. A Simple γ-Backbone Modification Preorganizes Peptide Nucleic Acid into a Helical Structure. J. Am. Chem. Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 12.Winssinger N, Damoiseaux R, Tully DC, Geierstanger BH, Burdick K, Harris JL. PNA-Encoded Protease Substrate Microarrays. Chem. Biol. 2004;11:1351–1360. doi: 10.1016/j.chembiol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Debaene F, Da Silva JA, Pianowski Z, Duran FJ, Winssinger N. Expanding the scope of PNA-encoded libraries: divergent synthesis of libraries targeting cysteine, serine and metallo-proteases as well as tyrosine phosphatases. Tetrahedron. 2007;63:6577–6586. [Google Scholar]
- 14.Niu Y, Hu Y, Li X, Chen J, Cai J. [gamma]-AApeptides: design, synthesis and evaluation. New J. Chem. 2011;35:542–545. [Google Scholar]
- 15.Wu H, Amin MN, Niu Y, Qiao Q, Harfouch N, Nimer A, Cai J. Solid-Phase Synthesis of γ-AApeptides Using a Submonomeric Approach. Org. Lett. 2012;14(13):3446–3449. doi: 10.1021/ol301406a. [DOI] [PubMed] [Google Scholar]
- 16.Wu H, Teng P, Cai J. Rapid Access to Multiple Classes of Peptidomimetics from Common γ-AApeptide Building Blocks. Eur. J. Org. Chem. 2014;2014:1760–1765. [Google Scholar]
- 17.Wu H, She F, Gao W, Prince A, Li Y, Wei L, Mercer A, Wojtas L, Ma S, Cai J. The synthesis of head-to-tail cyclic sulfono-[gamma]-AApeptides. Org. Biomol. Chem. 2015;13(3):672–676. doi: 10.1039/c4ob02232g. [DOI] [PubMed] [Google Scholar]
- 18.Schwergold C, Depecker G, Giorgio CD, Patino N, Jossinet F, Ehresmann B, Terreux R, Cabrol-Bass D, Condom R. Cyclic PNA hexamer-based compound: modelling, synthesis and inhibition of the HIV-1 RNA dimerization process. Tetrahedron. 2002;58:5675–5687. [Google Scholar]
- 19.Wu H, Niu Y, Padhee S, Wang RE, Li Y, Qiao Q, Bai G, Cao C, Cai J. Design and synthesis of unprecedented cyclic [gamma]-AApeptides for antimicrobial development. Chem. Sci. 2012;3:2570–2575. [Google Scholar]
- 20.Gellman SH. Foldamers: A Manifesto. Acc. Chem. Res. 1998;31:173–180. [Google Scholar]
- 21.Gennari C, Gude M, Potenza D, Piarulli U. Hydrogen-Bonding Donor/Acceptor Scales in β-Sulfonamidopeptides. Chem. Eur. J. 1998;4:1924–1931. [Google Scholar]
- 22.Karlsson AJ, Pomerantz WC, Weisblum B, Gellman SH, Palecek SP. Antifungal Activity from 14-Helical β-Peptides. J. Am. Chem. Soc. 2006;128:12630–12631. doi: 10.1021/ja064630y. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Qiao Q, Hu Y, Teng P, Gao W, Zuo X, Wojtas L, Larsen RW, Ma S, Cai J. Sulfono-γ-AApeptides as a New Class of Nonnatural Helical Foldamer. Chem. Eur. J. 2015;21:2501–2507. doi: 10.1002/chem.201406112. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, Qiao Q, Teng P, Hu Y, Antoniadis D, Zuo X, Cai J. New Class of Heterogeneous Helical Peptidomimetics. Org. Lett. 2015;17:3524–3527. doi: 10.1021/acs.orglett.5b01608. [DOI] [PubMed] [Google Scholar]
- 25.Cai W, Chen X. Anti-Angiogenic Cancer Therapy Based on Integrin αvβ3 Antagonism. Anti-Cancer Agents Med. Chem. 2006;6:407–428. doi: 10.2174/187152006778226530. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Niu Y, Hong H, Wu H, Zhang Y, Engle JW, Barnhart TE, Cai J, Cai W. Radiolabeled [gamma]-AApeptides: a new class of tracers for positron emission tomography. Chem. Commun. 2012;48:7850–7852. doi: 10.1039/c2cc33620k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He R, Tan L, Browning DD, Wang JM, Ye RD. The Synthetic Peptide Trp-Lys-Tyr-Met-Val-d-Met Is a Potent Chemotactic Agonist for Mouse Formyl Peptide Receptor. J. Immunol. 2000;165:4598–4605. doi: 10.4049/jimmunol.165.8.4598. [DOI] [PubMed] [Google Scholar]
- 28.Giordano C, Lucente G, Masi A, Paradisi MP, Sansone A, Spisani S. α-Peptide/β-sulfonamidopeptide hybrids: Analogs of the chemotactic agent for-Met-Leu-Phe-OMe. Bioorg. Med. Chem. 2006;14:2642–2652. doi: 10.1016/j.bmc.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 29.Torino D, Mollica A, Pinnen F, Feliciani F, Spisani S, Lucente G. Novel chemotactic For-Met-Leu-Phe-OMe (fMLF-OMe) analogues based on Met residue replacement by 4-amino-proline scaffold: Synthesis and bioactivity. Bioorg. Med. Chem. 2009;17:251–259. doi: 10.1016/j.bmc.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, Cheng N, Wu H, Kang S, Ye RD, Cai J. Design, Synthesis and Characterization of fMLF-Mimicking AApeptides. ChemBioChem. 2014;15:2420–2426. doi: 10.1002/cbic.201402396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray JK, Gellman SH. Targeting protein-protein interactions: Lessons from p53/MDM2. Peptide Science. 2007;88:657–686. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 32.Debnath B, Xu S, Neamati N. Small Molecule Inhibitors of Signal Transducer and Activator of Transcription 3 (Stat3) Protein. J. Med. Chem. 2012;55:6645–6668. doi: 10.1021/jm300207s. [DOI] [PubMed] [Google Scholar]
- 33.Siddiquee KAZ, Gunning PT, Glenn M, Katt WP, Zhang S, Schrock C, Sebti SM, Jove R, Hamilton AD, Turkson J. An Oxazole-Based Small-Molecule Stat3 Inhibitor Modulates Stat3 Stability and Processing and Induces Antitumor Cell Effects. ACS Chem. Biol. 2009;4:309–309. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher S, Page BDG, Zhang X, Yue P, Li ZH, Sharmeen S, Singh J, Zhao W, Schimmer AD, Trudel S, Turkson J, Gunning PT. Antagonism of the Stat3-Stat3 Protein Dimer with Salicylic Acid Based Small Molecules. ChemMedChem. 2011;6:1459–1470. doi: 10.1002/cmdc.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng P, Zhang X, Wu H, Qiao Q, Sebti SM, Cai J. Identification of novel inhibitors that disrupt STAT3-DNA interaction from a [gamma]-AApeptide OBOC combinatorial library. Chem. Commun. 2014;50:8739–8742. doi: 10.1039/c4cc03909b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu Y, Wang RE, Wu H, Cai J. Recent development of small antimicrobial peptidomimetics. Future Med. Chem. 2012;4:1853–1862. doi: 10.4155/fmc.12.111. [DOI] [PubMed] [Google Scholar]
- 37.Marr AK, Gooderham WJ, Hancock REW. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006;6:468–472. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 38.Karlsson AJ, Pomerantz WC, Weisblum B, Gellman SH, Palecek SP. Antifungal activity from 14-helical beta-peptides. J. Am. Chem. Soc. 2006;128:12630–12631. doi: 10.1021/ja064630y. [DOI] [PubMed] [Google Scholar]
- 39.Patch JA, Barron AE. Helical peptoid mimics of magainin-2 amide. J. Am. Chem. Soc. 2003;125:12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 40.Niu Y, Padhee S, Wu H, Bai G, Harrington L, Burda WN, Shaw LN, Cao C, Cai J. Identification of [gamma]-AApeptides with potent and broad-spectrum antimicrobial activity. Chem. Commun. 2011;47:12197–12199. doi: 10.1039/c1cc14476f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C, Pan F, Zhang S, Hu J, Cao M, Wang J, Xu H, Zhao X, Lu JR. Antibacterial Activities of Short Designer Peptides: a Link between Propensity for Nanostructuring and Capacity for Membrane Destabilization. Biomacromolecules. 2010;11:402–411. doi: 10.1021/bm901130u. [DOI] [PubMed] [Google Scholar]
- 42.Matsuzaki K. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Wu H, Teng P, Bai G, Lin X, Zuo X, Cao C, Cai J. Helical Antimicrobial Sulfono-γ-AApeptides. J. Med. Chem. 2015;58:4802–4811. doi: 10.1021/acs.jmedchem.5b00537. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, Smith C, Wu H, Teng P, Shi Y, Padhee S, Jones T, Nguyen A-M, Cao C, Yin H, Cai J. Short Antimicrobial Lipo-α/γ-AA Hybrid Peptides. ChemBioChem. 2014;15:2275–2280. doi: 10.1002/cbic.201402264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obrecht D, Robinson JA, Bernardini F, Bisang C, DeMarco SJ, Moehle K, Gombert FO. Recent Progress in the Discovery of Macrocyclic Compounds as Potential Anti-Infective Therapeutics. Curr. Med. Chem. 2009;16:42–65. doi: 10.2174/092986709787002844. [DOI] [PubMed] [Google Scholar]
- 46.Makovitzki A, Avrahami D, Shai Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15997–6002. doi: 10.1073/pnas.0606129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makovitzki A, Baram J, Shai Y. Antimicrobial Lipopolypeptides Composed of Palmitoyl Di- and Tricationic Peptides: In Vitro and in Vivo Activities, Self-Assembly to Nanostructures, and a Plausible Mode of Action. Biochemistry. 2008;47:10630–10636. doi: 10.1021/bi8011675. [DOI] [PubMed] [Google Scholar]
- 48.Niu Y, Padhee S, Wu H, Bai G, Qiao Q, Hu Y, Harrington L, Burda WN, Shaw LN, Cao C, Cai J. Lipo-γ-AApeptides as a New Class of Potent and Broad-Spectrum Antimicrobial Agents. J. Med. Chem. 2012;55:4003–4009. doi: 10.1021/jm300274p. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Smith C, Wu H, Teng P, Shi Y, Padhee S, Jones T, Nguyen A-M, Cao C, Yin H, Cai J. Short Antimicrobial Lipo-α/γ-AA Hybrid Peptides. ChemBioChem. 2014;15:2275–2280. doi: 10.1002/cbic.201402264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Smith C, Wu H, Padhee S, Manoj N, Cardiello J, Qiao Q, Cao C, Yin H, Cai J. Lipidated Cyclic γ-AApeptides Display Both Antimicrobial and Anti-inflammatory Activity. ACS Chem. Biol. 2014;9:211–217. doi: 10.1021/cb4006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Padhee S, Li Y, Cai J. Activity of lipo-cyclic γ-AApeptides against biofilms of Staphylococcus epidermidis and Pseudomonas aeruginosa. Bioorg. Med. Chem. Lett. 2015;25:2565–2569. doi: 10.1016/j.bmcl.2015.04.039. [DOI] [PubMed] [Google Scholar]
- 52.Wu H, Li Y, Bai G, Niu Y, Qiao Q, Tipton JD, Cao C, Cai J. [gamma]-AApeptide-based small-molecule ligands that inhibit A[small beta] aggregation. Chem. Commun. 2014;50:5206–5208. doi: 10.1039/c3cc46685j. [DOI] [PubMed] [Google Scholar]
- 53.Leulliot N, Varani G. Current Topics in RNA−Protein Recognition: Control of Specificity and Biological Function through Induced Fit and Conformational Capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 54.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255–270. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 55.Niu Y, Jones AJ, Wu H, Varani G, Cai J. gamma-AApeptides bind to RNA by mimicking RNA-binding proteins. Org. Biomol. Chem. 2011;9:6604–6609. doi: 10.1039/c1ob05738c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potocky TB, Menon AK, Gellman SH. Cytoplasmic and Nuclear Delivery of a TAT-derived Peptide and a β-Peptide after Endocytic Uptake into HeLa Cells. J. Biol. Chem. 2003;278:50188–50194. doi: 10.1074/jbc.M308719200. [DOI] [PubMed] [Google Scholar]
- 57.Pepinsky RB, Androphy EJ, Corina K, Brown R, Barsoum J. Specific Inhibition of a Human Papillomavirus E2 Trans-Activator by Intracellular Delivery of Its Repressor. J. DNA Cell Biol. 1994;13:1011–1019. doi: 10.1089/dna.1994.13.1011. [DOI] [PubMed] [Google Scholar]
- 58.Niu Y, Bai G, Wu H, Wang RE, Qiao Q, Padhee S, Buzzeo R, Cao C, Cai J. Cellular Translocation of a γ-AApeptide Mimetic of Tat Peptide. Mol. Pharmaceutics. 2012;9:1529–1534. doi: 10.1021/mp300070w. [DOI] [PubMed] [Google Scholar]
- 59.Umezawa N, Gelman MA, Haigis MC, Raines RT, Gellman SH. Translocation of a β-Peptide Across Cell Membranes. J. Am. Chem. Soc. 2002;124:368–369. doi: 10.1021/ja017283v. [DOI] [PubMed] [Google Scholar]