Abstract
Epithelial cultures are commonly used for studying gut health. However, due to the absence of mesenchymal cells and gut structure, epithelial culture systems including recently developed three-dimensional organoid culture cannot accurately represent in vivo gut development, which requires intense cross-regulation of the epithelial layer with the underlying mesenchymal tissue. In addition, organoid culture is costly. To overcome this, a new culture system was developed using mouse embryonic small intestine. Cultured intestine showed spontaneous peristalsis, indicating the maintenance of the normal gut physiological structure. During 10 days of ex vivo culture, epithelial cells moved along the gut surface and differentiated into different epithelial cell types, including enterocytes, Paneth cells, goblet cells and enteroendocrine cells. We further used the established ex vivo system to examine the role of AMP-activated protein kinase (AMPK) on gut epithelial health. Tamoxifen-induced AMPKα1 knockout vastly impaired epithelial migration and differentiation of the developing ex vivo gut, showing the crucial regulatory function of AMPK α1 in intestinal health.
Keywords: intestinal epithelium, differentiation, AMPK, LGR5, stem cell, migration
1. Introduction
Gut epithelium is one of the largest barriers in the body through forming an integrated single cell layer. The gut epithelial barrier is selectively permeable to nutrients, minerals, water and selected antigens, while protecting against potentially harmful bacteria, viruses and antigenic materials. Neonatal gut has a relatively high permeability, which is required for transferring of maternal immunoglobulins in colostrum and selected antigens, further facilitating the proper development of the gut immune system [1,2]. However, the neonatal gut epithelium is susceptible to pathophysiological perturbations [3–6]. A ‘leaky’ neonatal gut transmits harmful antigens, altering immune system development/maturation and pre-disposing offspring to immunological disorders [7]. The elevated gut permeability is a central predisposing factor to inflammatory bowel diseases, autoimmune and related allergic diseases [8–13]. However, many questions regarding the epithelial differentiation and barrier function, especially during the early gut development, remain to be addressed.
Up to now, studies on epithelial differentiation and migration were mainly using epithelial cell lines and in vivo studies. Compared with in vivo studies, in vitro cell culture is convenient, low cost and time-saving, suitable for using chemical stimulators and inhibitors, and suitable for genetic manipulation without physiological complications. However, cell lines are immortalized cells and their cellular behaviour may differ significantly from normal epithelial cells. The recently developed epithelial organoid culture uses Lgr5 (leucine-rich-repeat-containing G protein-coupled receptor) as a surface marker for isolating gut epithelial stem cells, which further develops into organoid structures using three-dimensional matrigel as a scaffold [14]. This organoid system requires expensive growth factors to facilitate epithelial proliferation and differentiation, and has a relatively low efficiency (1–2%) for epithelial stem cells developing into organoids [15,16]. Furthermore, epithelial organoids do not truly represent in vivo epithelial differentiation during gut development due to the lack of underlying mesenchymal tissues that intricately interact with the epithelium to regulate its development [13,17]. Likewise, in vivo studies are either costly or impossible to use chemical inhibitors and activators, or RNA interferences, to manipulate epithelial differentiation without inducing physiological complications or other changes. There is a need to establish an ex vivo model, which can mimic in vivo gut epithelial development but has the convenience and flexibility of in vitro cell culture.
Here, we established an ex vivo model for studying gut epithelial differentiation and migration using embryonic (E) 13.5-day fetal small intestine. The model can effectively track the differentiation of gut epithelium real-time, which, in combination with the immunohistochemical and histochemical examination of structures, provides a powerful venue for elucidating key mechanisms regulating gut epithelial development.
AMP-activated protein kinase (AMPK) is recognized as a master regulator of energy metabolism [18,19], and was recently recognized for its important regulatory role in myogenesis, adipogenesis, cardiac differentiation, smooth muscle formation and osteogenesis [20–24]. We recently found that AMPK promotes intestinal differentiation in vitro and in adult mice [25], but were unable to establish the causal relationship due to the absence of a model for studying AMPK without physiological complications. In this study, the regulatory role of AMPK in regulating embryonic epithelial differentiation and migration was demonstrated using this newly established ex vivo gut model.
2. Material and methods
2.1. Coverslip cleaning, coating and preparation for ex vivo gut culture
Coverslips (22 × 22 mm, Thermo Fisher Scientific, Waltham, MA, USA) were dipped into acidic alcohol (0.1% acetic acid in 95% alcohol, V/V), air dried, then submerged into 2% 3-aminoprophytriethoxysilane (#440140, Sigma, St Louis, MO, USA) in acetone for 15 min [26]. Coverslips were washed twice in acetone and once in water, and then sterilized by autoclaving. The prepared coverslips could be stored up to one year at room temperature. The centre of each sterile coverslip was coated with 200 µl of 50 µg ml−1 fibronectin (#610077, BD Biosciences, San Jose, CA, USA). Coverslips were dried under a biosafety cabinet. Before culturing, coverslips were placed into 35 mm cell culture dishes (#150318, Thermo Fisher Scientific), and four sterile 4.7 × 8 mm cloning cylinders (#C7983, Sigma) were placed onto the fibronectin-coated area on coverslips.
2.2. Mice strains
C57BL/6 J mice were purchased from Jackson Laboratory (#000664, Bar Harbor, ME, USA). Mice harbouring an Lgr5-EGFP-IRES-creERT2 (Lgr5) allele (#008875, Jackson Lab) were crossed with ROSAmT/mG (#007576, Jackson Lab) mice to obtain conditional knockout mice (Lgr5mT/mG). Epithelial stem cells in ex vivo gut from Lgr5mT/mG mice will convert cellular fluorescent protein from red to green signal in Lgr5-positive cells and their derived cells upon induction of 4-hydroxytamoxifen. The double-fluorescent ex vivo gut facilitated the tracing of epithelial movement and reorganization of Lgr5-derived cells. Mice with AMPKα1-floxed gene (Prkaa1tm1.1Sjm/J, #014141, Jackson Lab) were cross-bred with tamoxifen-inducible Cre mice (129-Gt(ROSA)26Sortm1(cre/ERT)Nat/J, #004847, Jackson Lab) as previously described to obtain AMPKα1 conditional knockout (KO) mice [27]. In response to 4-hydroxytamoxifen, AMPKα1 can be deleted in all tissues of AMPKα1 KO mice. Embryonic (E) staging was started from the morning when a copulatory plug was observed and counted as embryonic day 0.5 (E0.5). Pregnant mice at E13.5 were euthanized for ex vivo gut preparation.
2.3. Embryonic gut isolation and culture
E13.5 embryos were removed aseptically and transferred into ice-cold culture medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 1% antibiotic–antimycotic solution (Invitrogen). The small intestine was dissected from embryos according to a procedure described previously with modifications [26]. Briefly, small intestine was cut into 2 mm segments using fine scissors and dissecting needle under a stereomicroscope (Nikon SMZ800, Tokyo, Japan). Then, to each 35 mm dish, a fibronectin-coated coverslip was placed and then 2 ml DMEM supplemented with 20% FBS and 1% antibiotic–antimycotic solution was added. After that, cloning cylinders were placed into dishes, and to each cloning cylinder, 60 µl of medium was added, followed by transferring 2–3 gut sections into the centre of each cloning cylinder. Gut sections were gently pushed down using a dissecting needle so that they could settle on fibronectin-coated coverslips and the dishes containing gut sections were carefully transferred to the 37°C, 5% CO2 humidified incubator. The cloning cylinders were carefully removed after 24 h of culture. The gut segments were cultured for 10 days, and the culture medium was changed every 3 days. To determine Gata4 expression in different segments of small intestine, ex vivo guts from embryonic proximal or distal small intestine were cultured and collected at 7 days separately. To evaluate the effects of signalling regulators on the differentiation of ex vivo guts, culture medium containing 100 nM LDN-193189 (a BMP signalling inhibitor, Stemolecule, Cambridge, MA), 10 μM DAPT (a Notch signalling inhibitor, Stemolecule) or 100 ng ml−1 Wnt 3a and 500 ng ml−1 R-spondin1 (Wnt signalling activators, PeproTech, Rocky Hill, NJ) was prepared to treat the guts for 7 days. To induce AMPK knockout, 4-hydroxytamoxifen (250 µM, Sigma) was used during the first three days of culture.
2.4. Gut fixation and cryosection
Following the removal of culture medium, ex vivo guts were fixed in freshly prepared 4% formaldehyde for 2 h at room temperature and then rinsed with PBS for three times. Ex vivo guts were submerged in PBS with 30% sucrose for 1 h at room temperature, which served as a cryoprotectant for gut during freezing. The cultured guts were carefully peeled from coverslips with dissecting blades under a stereo microscope, then vertically embedded into Tissue TekR OCT compounds (Miles Sakura, Torrance, CA, USA) in embedding moulds (Thermo Fisher Scientific), followed by placement into isopentane (Sigma) pre-cooled by liquid nitrogen for 30 s. The frozen ex vivo gut samples were stored at −80°C and sectioned using a cryostat (Leica, Wetzlar, Germany) to 5 µm thickness and mounted directly onto ultra-stick micro slides (Gold Seal, Bedford, MA, USA), then stored at −80°C till staining.
2.5. Immunofluorescence staining
Sections were permeabilized in 1% Triton X-100 (Sigma) in PBS at room temperature for 30 min, boiled in 10 mM sodium citrate (pH 6.0, Sigma) for 20 min, incubated with 5% goat serum (Vector, Burlingame, CA, USA) in PBS (pH 7.4) for 1 h to block non-specific binding sites, then incubated with primary antibodies overnight at 4°C. The following antibodies were used: Smooth muscle actin (smooth muscle cells), Thermo Fisher Scientific #MS-113-B, 1 : 50; E-cadherin (epithelial junctions), Life Technologies #33–4000, 1 : 200; Chromogranin A (enteroendocrine cells), Developmental Studies Hybridoma Bank #CPTC-CHGA-1, 1 : 200; Vimentin (fibroblasts), Cell Signaling Technology #5741, 1 : 200; Ki67 (proliferative cells), Cell Signaling Technology #9449, 1 : 200; CDX2 (intestinal transcription factor), Cell Signaling Technology #12306, 1 : 300; β-catenin (adherens junctions), Cell Signaling Technology #9562, 1 : 300; Cytokeratin 8 (columnar epithelium), Developmental Studies Hybridoma Bank #TROMA-I, 1 : 400; Lysozyme (Paneth cells), Thermo Fisher Scientific #PA5-16668, 1 : 800. Then the slides were incubated with respective fluorescent secondary antibodies (goat anti-mouse antibody-Alexa Fluor 555, Cell Signaling Technology #4409; goat anti-mouse antibody-Alexa Fluor 488, Cell Signaling Technology #4408; goat anti-rabbit antibody-Alexa Fluor 555, Cell Signaling Technology #4413; goat anti-rabbit antibody-Alexa Fluor 488, Cell Signaling Technology #4412; goat anti-rat antibody-Alexa Fluor 488, Cell Signaling Technology #4416) diluted in 5% goat serum (1 : 1000) at room temperature for 2 h. Immunofluorescence was imaged using a fluorescence microscope (EVOS FL, Life Technologies). Negative controls were stained with secondary antibody without primary antibody incubation.
2.6. Histochemical staining
For Alcian blue staining, tissue sections were stained with 1% Alcian blue solution (pH 2.5, Sigma) for 30 min at room temperature as previously described [28]. Haematoxylin–eosin (H&E) staining was conducted following standard procedures [29]. The images were taken using an inverted microscope (AMG EVOS, Life Technologies).
2.7. RT-qPCR
Total RNA was extracted from 10 ex vivo gut samples for each group using TRIzol (Sigma). Five hundred nanograms of cDNA was synthesized using a Reverse Transcription kit (Bio-Rad, Hercules, CA, USA). qPCRs for 5 ng cDNA per reaction were performed on a CFX RT-PCR detection system (Bio-Rad) using SYBR Green (Bio-Rad). The primers were designed to cross two exons to prevent amplification of genomic DNA, and primer sequences are shown in the electronic supplementary material, table S1. Five nanograms of cDNAs per reaction from adult mouse intestine and fat tissue were used as positive control (PC) and negative control (NC), respectively. 18S rRNA was used as an internal control.
2.8. TUNEL staining
In situ apoptosis was determined using TACS TdT-DAB detection kit (Trevigen, Gaithersburg, MD, USA) per the manufacturer's instructions. Briefly, ex vivo gut sections were treated with Cytopore and blocked with 3% H2O2. DNA fragments were labelled with biotin and HRP-conjugated streptavidin, then incubated with DAB substrates. The counterstaining was performed with 1% Methyl Green. The images were taken using Leica DM2000 LED light microscope (Wetzlar, Germany).
2.9. Statistical analysis
Ex vivo gut section obtained from each fetus was considered as an experimental unit. Statistical analysis was performed using Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). Data were presented as means ± standard error of the means (s.e.m.). Differences between means were determined using two-tailed t-test followed by Duncan's multiple test when appropriate. All experiments were replicated in at least three independent experiments. p ≤ 0.05 was considered to be statistically significant.
3. Results
3.1. Establishment and functionality of ex vivo gut
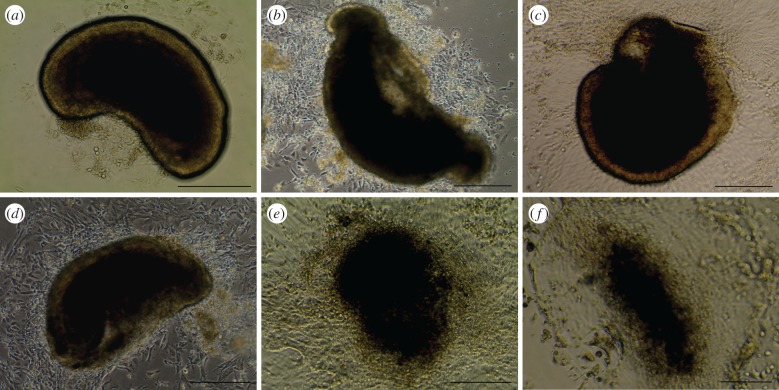
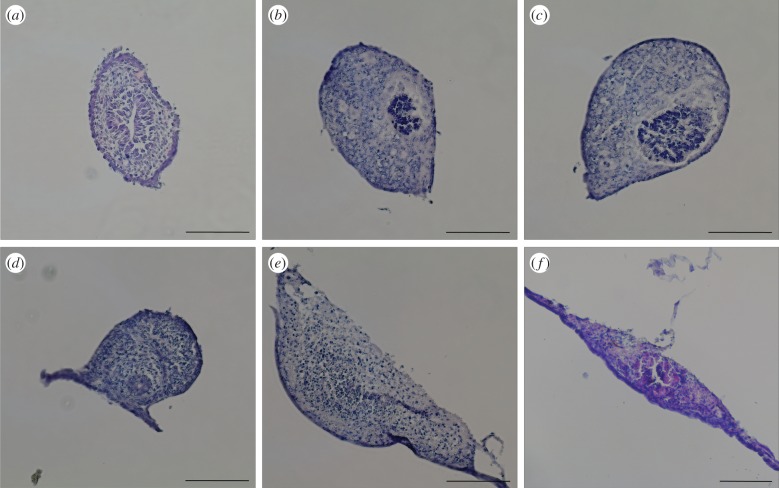
The morphology of ex vivo gut was examined over 10 days. Gut sections adhered to coverslips within 24 h, and gradually expanded and spread out over time (figure 1). Starting day 3, guts demonstrated autonomous contraction with a frequency of about 15 times per minute (electronic supplementary material, movie S1). To examine the overall reorganization of gut structure over time, guts were cross-sectioned and stained with H&E (figure 2). The gut lumen progressively moved closer to the bottom during the initial 5 days of culture (figure 2a–d), and guts gradually flattened out from a tubular structure to a spindle-shaped structure over 10 days (figure 2). To determine cell viability during the 10-day culture, Trypan blue staining was performed (electronic supplementary material, figure S1). Sporadic dead cells were found in ‘spread’ area at 7 or 10 days of culture (electronic supplementary material, figure S1c–f). Addition of Wnt3a and R-spondin1 could extend the culture viability of ex vivo guts to 3 weeks. To further examine the functionality of epithelial cells, the activity of alkaline phosphatase produced by epithelial absorptive cells was tested (electronic supplementary material, figure S1 g). Compared with day 0, the activity increased greatly at 7 days, indicating ex vivo gut remained functional even though cell death slightly occurred.
Figure 1.
Inverted phase contrast images of ex vivo guts. Small intestine from E13.5 embryos were isolated, dissected and cultured to form ex vivo guts at 3 days (a), 4 days (b), 5 days (c), 6 days (d), 7 days (e) and 10 days (f). Scale bar is 250 µm.
Figure 2.
Haematoxylin and eosin staining images of ex vivo gut cross-sections. Ex vivo guts were collected at day 0 (a), and after 3 days (b), 4 days (c), 5 days (d), 7 days (e) and 10 days (f) of culture, fixed in 4% paraformaldehyde and embedded in OCT, cut into 5 µm section and then stained by haematoxylin and eosin. Scale bar is 125 µm.
3.2. Epithelial cell differentiation in cultured gut
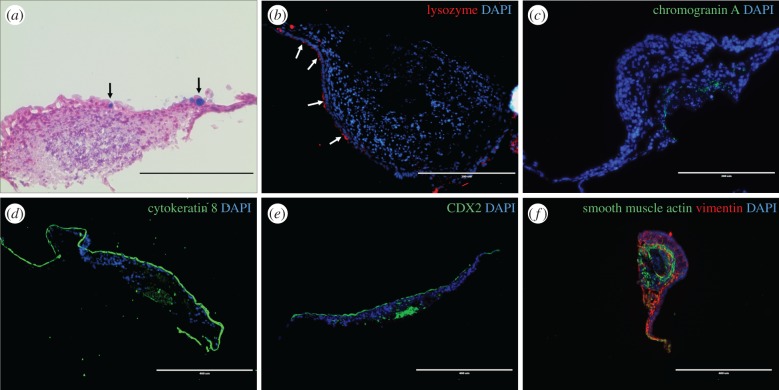
To characterize the differentiation and distribution of intestinal epithelial cells in cultured guts, ex vivo guts at 7 days were stained for the respective differentiated cells. The presence of goblet cells and Paneth cells were visible on the surface of gut section (figure 3a,b). Enteroendocrine cells were identified in the centre of gut lumen, suggested by chromogranin A staining (figure 3c). Absorptive cells, indicated by the columnar epithelial marker cytokeratin 8, formed a tight layer on the outside surface of cultured guts, with a small number concentrated in the centre of the original gut lumen (figure 3d). CDX2, the key epithelial transcriptional factor, had a similar distribution to absorptive cells, which might be due to its crucial role in intestinal differentiation (figure 3e). By contrast, the remaining components were composed of mesenchymal cells, which were concentrated under the epithelium, providing scaffolds for epithelial cells (figure 3f). Fibroblasts formed the first scaffold layer under epithelium, as indicated by vimentin staining (figure 3f). Smooth muscle cells formed the second scaffold layer under fibroblasts based on smooth muscle actin staining (figure 3f). Myofibroblasts were developed in the spreading area (gut ‘tail’), as suggested by co-staining of vimentin and smooth muscle actin (figure 3f). To further test whether interstitial cells of Cajal contribute to spontaneous peristalsis in ex vivo guts [30], mRNA expression of C-kit was tested, which was beyond the detection level (electronic supplementary material, figure S3a). Furthermore, the peristalsis was promoted after treatment with a neurotoxin, botulinum (electronic supplementary material, figure S3c). The expression of Gata4 in ex vivo guts at 7 days from embryonic proximal small intestine was higher than the one from distal small intestine, indicating the segment specificity in cultured gut (electronic supplementary material figure S1h). Additionally, Wnt activators, BMP inhibitor and Notch inhibitor could alter the expression of differentiation markers of ex vivo gut (electronic supplementary material, figure S4), suggesting these signalling pathways regulate the fates of intestinal stem cells during differentiation.
Figure 3.
Localization and distribution of cell differentiation markers in ex vivo guts 7 days post-incubation. Ex vivo guts were cultured for 7 days, fixed and embedded in OCT. The cryosections were examined by Alcian blue staining (a) and immunofluorescence staining (b–f). (a) Goblet cells locate on the surface layer of ex vivo guts shown by Alcian blue staining (black arrows); (b) Paneth cells (white arrows, lysozyme); (c) enteroendocrine cells (chromogranin A staining); (d) columnar epithelium (cytokeratin 8); (e) intestinal transcript factor CDX2; (f) fibroblasts (vimentin), smooth muscle cells (smooth muscle actin) and myofibroblasts (co-immunostaining of smooth muscle actin and vimentin). DAPI was used as nuclei stain. Scale bar is 200 µm in (a–c) and 400 µm in (d–f).
3.3. Migration and polarity of cultured gut
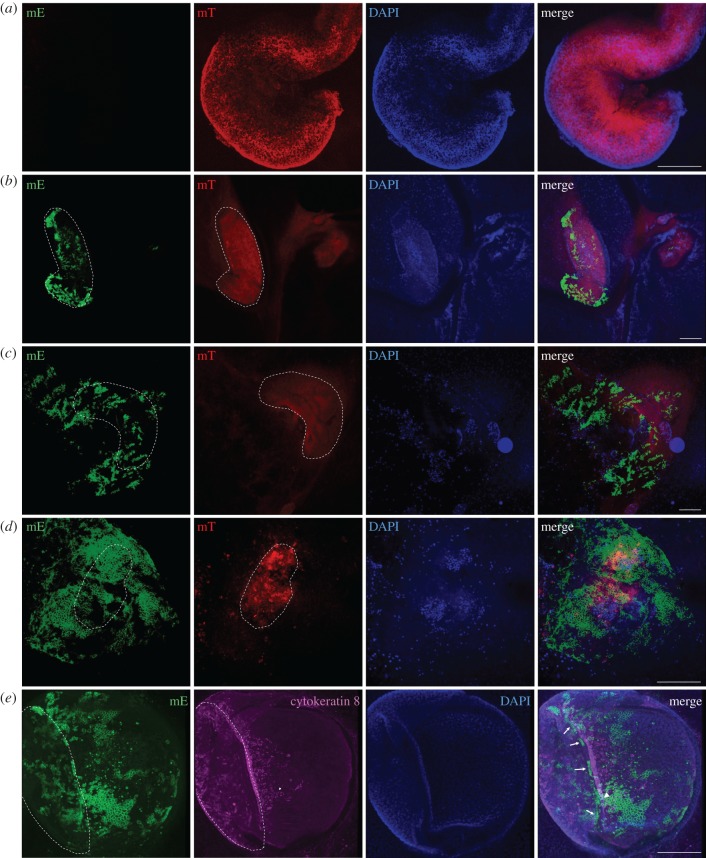
We observed the presence of epithelial layers on the surface of ex vivo gut, which were unexpected, because epithelial cells were anticipated to concentrate inside the gut lumen. To address the originality of the surface epithelial layers, ex vivo guts isolated from intestinal stem cell-derived fluorescent mice (Lgr5mT/mG) were further used for tracing epithelial movement and reorganization.
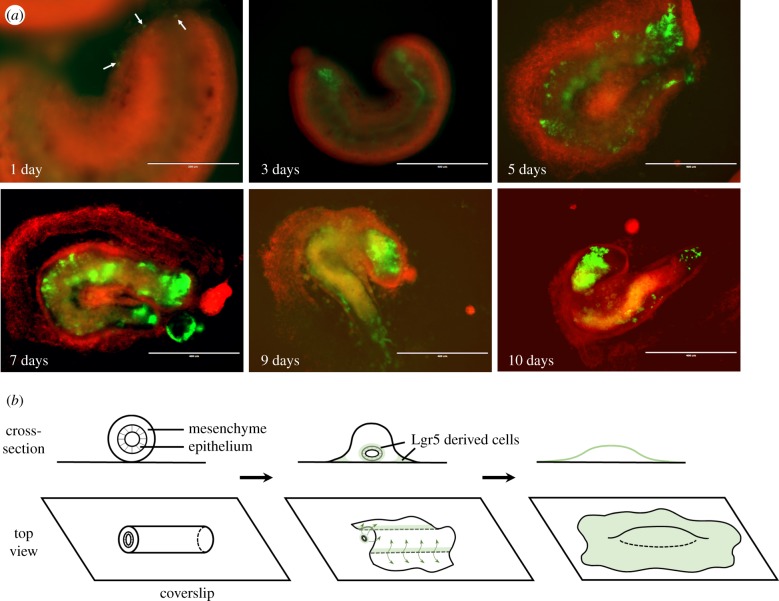
At day 0, ex vivo guts of Lgr5mT/mG showed exclusively red fluorescence (figure 4a). During 9 days of culture, residual cells with red fluorescence remained clearly visible (figure 4a–d). Of note, newly Lgr5-derived epithelial cells with green fluorescence appeared and concentrated at the two ends of gut sections at day 3 (figure 4b; electronic supplementary material, movie S2), and further progressively spread along the surface of the gut at day 6 (figure 4c; electronic supplementary material, movie S3), leading to almost complete coverage of gut at day 9 (figure 4d; electronic supplementary material, movie S4). To identify Lgr5+-clusters, we tested the distribution of columnar epithelial cells using cytokeratin 8 staining. We found that Lgr5+-clusters were localized below the differentiated epithelial layer (figure 4e; electronic supplementary material, movie S5), and reconfirmed that differentiated epithelial cells spread on the surface of the gut (figures 4e and 3d). We further examined the movement of Lgr5-derived cells at the bottom of ex vivo gut (view from the bottom of petri dish; figure 5a). From day 1 to day 10, Lgr5-derived cells started to concentrate at two open ends of ex vivo guts, gradually spread into a large area and formed an epithelial layer on the surface of ex vivo guts (figure 5a,b). mRNA expression of Foxl1 was further detected in ex vivo gut, suggesting Foxl1-expressing mesenchymal cells might facilitate the formation of stem cell niche [31] (electronic supplementary material, figure S3b).
Figure 4.
Migration and reorganization of ex vivo guts. Lgr5mT/mG ex vivo guts were obtained by crossing Lgr5-EGFP-IRES-creERT2 mice with ROSAmT/mG mice. Lgr5mT/mG ex vivo guts were induced by 250 µm 4-hydroxytamoxifen in the first 3 days of culture to mark Lgr5-derived cells into green fluorescence. The fluorescent signal at the surface of ex vivo guts was observed at day 0 (a), and after 3 days (b), 6 days (c) and 9 days (d), by confocal microscope. Columnar epithelium at 7 days is shown by immunostaining for Cytokeratin 8 (e). The dashed lines indicate the contour of the original gut body. Arrows point to Lgr5-derived cell clusters. Arrowhead points to cells across the border between original gut body and the cells spreading from the gut body. DAPI was used as a nuclear stain. Scale bar is 200 µm.
Figure 5.
Development of Lgr5 progeny cells and cell signalling in ex vivo guts. (a) Lgr5mT/mG ex vivo guts were obtained by crossing Lgr5-EGFP-IRES-creERT2 mice with ROSAmT/mG mice. Lgr5mT/mG ex vivo guts were induced by 250 μm 4-hydroxytamoxifen in the first 3 days of culture to mark Lgr5-derived cells by the presence of green fluorescence. The fluorescent signal under ex vivo guts (view from the bottom of Petri dish) was observed at 1, 3, 5, 7, 9 and 10 days by fluorescent inverted microscope. (b) Infographic of morphogenesis and migration in ex vivo guts. Cross-section view is above and the top view is below.
3.4. Application of ex vivo culture to examine the role of AMPK in epithelial development
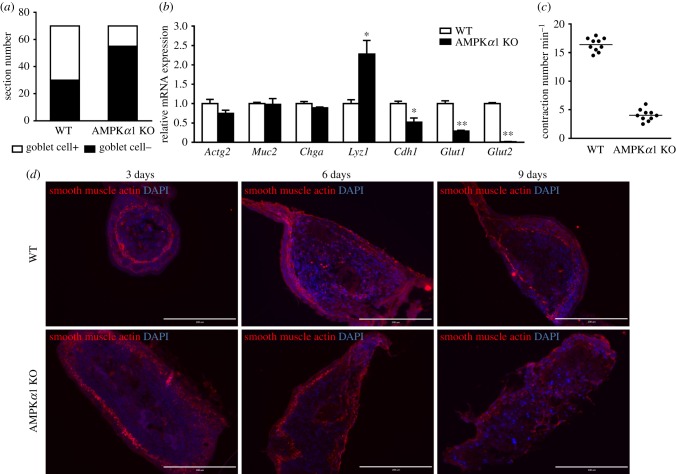
The above established ex vivo culture system was further used to study the regulatory role of AMPKα1 on gut health from wild-type (WT) and AMPKα1 conditional knockout (AMPKα1 KO) mice. Alcian blue staining indicated that the prevalence of goblet cells was remarkably decreased in AMPKα1 KO ex vivo gut compared with that of WT (figure 6a). Lyz1 was higher in AMPK KO group, while Cdh1, Glut1 and Glut2 were higher in WT group at mRNA level (figure 6b).
Figure 6.
Differentiation of WT and AMPKα1 KO ex vivo guts. (a) Goblet cells positive ex vivo guts at 7 days. The ex vivo gut cross-sections were stained with Alcian blue, then goblet-positive and goblet-negative ex vivo guts were counted. A total of 70 ex vivo guts were used. (b) The mRNA expression of gut epithelial differentiation markers. (c) The contraction frequencies of ex vivo guts at 7 days were counted per minute. n = 10. (d) Immunostaining of smooth muscle actin for ex vivo guts at 3, 6 and 9 days to identify mesenchymal cells. DAPI was used as a nuclear stain. Scale bar is 200 µm. Data are represented as mean ± s.e.m. *p < 0.05; **p < 0.01.
Under the normal physiological condition, guts undergo peristalsis. We found that WT ex vivo gut contracted and underwent peristalsis much stronger than that of AMPK KO at 7 days of culture (figure 6c; electronic supplementary material, movies S1 and S6). Because smooth muscle cells contribute to gut peristalsis, we further tested smooth muscle actin. In WT ex vivo guts, smooth muscle actin initiated to localize under the epithelial layer, and gradually spread along the contour of ex vivo gut; while in AMPK KO ex vivo guts, smooth muscle actin became increasingly disorganized over time (figure 6d).
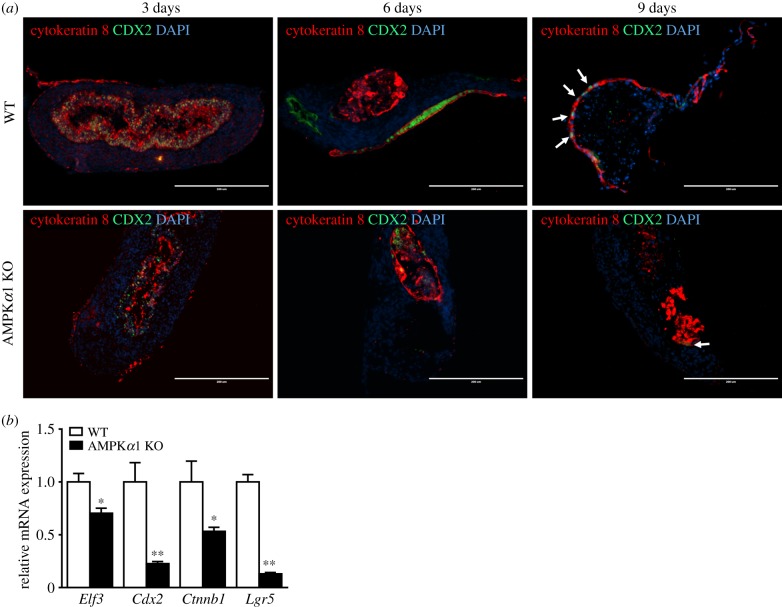
We further tested the effects of AMPK on epithelial cells of ex vivo guts. Both epithelial cells (cytokeratin 8 positive) and intestinal transcription factor (CDX2 positive) progressively moved from gut lumen to surface in WT ex vivo gut (figure 7a). However, this movement was absent in AMPK KO gut where both types of cells remained inside gut lumen (figure 7a), indicating that AMPKα1 deficiency unequivocally impeded the formation and overall arrangement of epithelial layers, which might be due to the downregulated mRNA expression of epithelial transcription factors (figure 7b).
Figure 7.
Expression of transcription factors and epithelial cells in WT and AMPKα1 KO ex vivo guts. (a) Ex vivo guts were cross-sectioned at 3, 6 and 9 days. The CDX2 transcription factor and epithelial cells were identified by immunostaining for Cytokeratin 8 and CDX2. Arrows point to CDX2-positive cells in the epithelial layer. DAPI was used as a nuclear stain. Scale bar is 200 μm. (b) mRNA levels of transcription factors in WT and AMPK KO ex vivo guts. Data are represented as mean ± s.e.m. *p < 0.05; **p < 0.01.
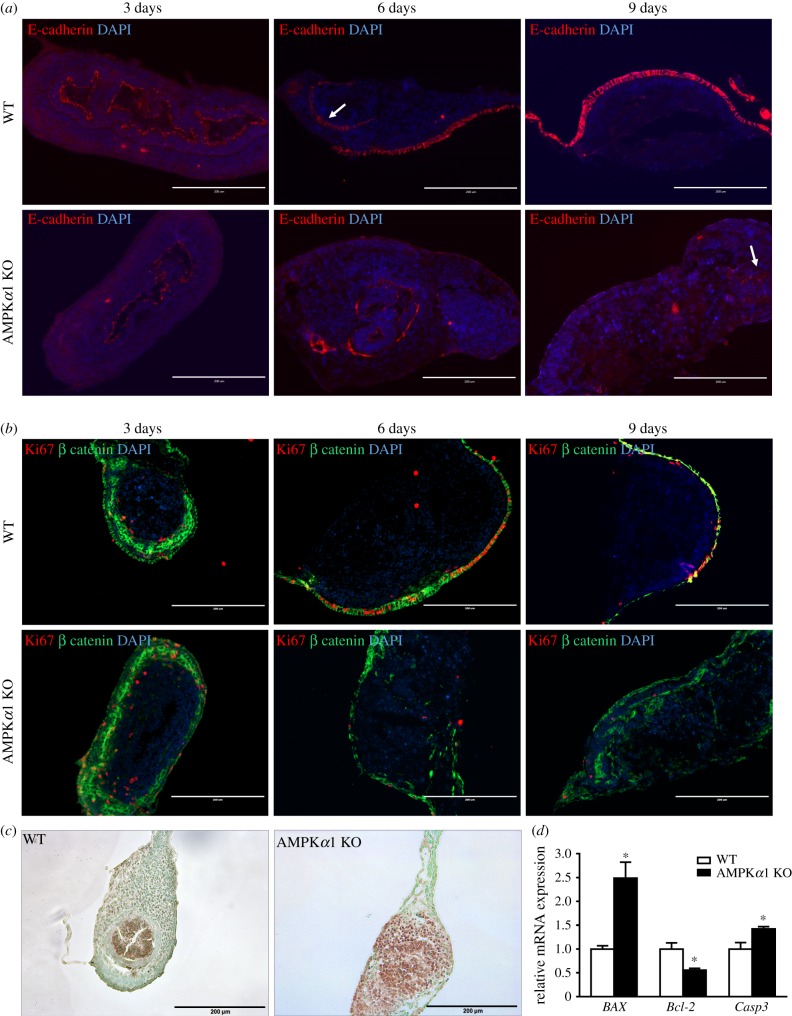
To further determine the role of AMPK in barrier function, E-cadherin and β-catenin, the markers for intestinal paracellular junction, were examined. Consistent with what we observed in figure 7a, E-cadherin and β-catenin in WT ex vivo guts gradually moved towards the exterior layer, and completely covered the surface layer over time; this phenomenon was absent in AMPK KO ex vivo guts (figure 8a,b). Since barrier function is also related to intestinal proliferation, we further tested Ki-67 (figure 8b). Ki67-positive cells were reduced due to AMPKα1 deficiency, which was consistent with the downregulated expression of Ctnnb1 and Lgr5 (figure 7b). Additionally, the apoptosis significantly induced after AMPK deficiency (figure 8c). Pro-apoptotic markers BAX and Casp3 were upregulated in AMPK KO gut, while anti-apoptotic marker Bcl-2 was downregulated (figure 8d). These data collectively demonstrated the necessity of AMPKα1 in proper gut health, and established ex vivo guts as a proper model for examining gut epithelial differentiation and formation.
Figure 8.
Epithelial junctions and proliferating and apoptotic cells of WT and AMPKα1 KO ex vivo guts. (a) Ex vivo guts were cross-sectioned at 3, 6 and 9 days. Junctions are determined by immunostaining for E-cadherin. Arrow points to the staining pattern in the lumen. (b) Adherens junction and proliferative cells were visualized by immunostaining for β-catenin and Ki67. DAPI was used as a nuclear stain. (c) Apoptotic cells of ex vivo guts at 7 days were visualized by TUNEL staining. Scale bar is 200 µm. (d) The mRNA expression of apoptotic markers of ex vivo guts at 7 days. Data are represented as mean ± s.e.m. *p < 0.05.
4. Discussion
Epithelial cell lines and mice are commonly used models for studying epithelial health. An in vivo model is costly, time-consuming and often lacks specific niche for toxicological, pharmacological and nutraceutical studies. The recently developed organoid model from mouse intestinal stem cells [32] or from human-induced pluripotent stem cells [33] allows three-dimensional culture to form crypt-villus structures. These models use expensive growth factors and matrix gel embedding with low efficiency, and the functionality of smooth muscle is missing, making it inaccessible to study motility disorders [34].
The ex vivo gut culture system established in this study allows epithelium developing in a native gut niche without costly growth factors or other promoters. All steps of gut epithelial differentiation and migration can be closely monitored, which is difficult to do with live animals. Furthermore, inhibitors, activators, toxicants or carcinogens can be applied to ex vivo gut to study their biological effects on gut differentiation. This model was successfully applied to one nutraceutical study to determine the effects of purple potato extract on intestinal differentiation [35]. The shortcoming of the ex vivo gut culture is that gut sections are vulnerable to waggle and fall off from the coverslip during the first three days of culturing. Thus, careful handling is needed when moving culture dishes. Also, the defined crypt-villus structure could not be found in ex vivo guts. Thus, intestinal morphogenesis may not be applicable to this model.
During early gut development, the intestinal epithelial structure including crypts and villi starts to form around E14, a critical time window for intestinal development [36]. Thus, the embryonic intestine at E13.5 was used to establish the ex vivo gut culture model. During murine intestinal development, the major differentiated cells, such as absorptive enterocytes, goblet cells and enteroendocrine cells, can be detected [37], while Paneth cells can be detected from postnatal (P) day 14 [38]. CDX2, E-cadherin, smooth muscle actin, vimentin and chromogranin A were detected in the gut of E16.5 and E18.5 mice [33]. Alcian blue and Ki67 were also found in the epithelial layer of P4 mice duodenum [39]. In this study, similar staining in ex vivo guts has been detected. The difference between ex vivo model and in vivo is that ex vivo guts rearrange during culture, with epithelial cells distributing on the surface layer and mesenchymal cells concentrated under the epithelium as scaffolds.
Lgr5mT/mG ex vivo gut was used to trace intestinal stem cells and their derived cells. Lgr5-derived intestinal cells ‘climb’ onto the surface of ex vivo gut, showing the migration of intestinal cells. On the other hand, intestinal stem cells give rise to enterocytes, goblet cells, enteroendocrine cells and Paneth cells, which were also discovered in ex vivo guts at 7 days post-incubation. Of note, Paneth cells, contributing to the niche of intestinal stem cells, normally start to differentiate from P14 in mice [38]. Paneth cells are detected in the organoid derived from a single intestinal stem cell after 14 days in culture [14]. In this study, Paneth cells were observed after 7 days in culture, which possibly differentiate to sustain epithelial stem cell niche [40].
The mesenchymal layers under the epithelial layer are the sources of growth factors and cytokines regulating epithelial differentiation [41,42]. There are several major cell types in the mesenchymal layers. Fibroblasts secrete collagen and synthesize connective tissue as the scaffold for epithelium, smooth muscle cells form contractile apparatus [43], and myofibroblasts possess both features [43]. In addition, smooth muscle cells cooperate with myofibroblasts to regulate contraction, while fibroblasts and myofibroblasts support gut structure. In ex vivo guts, the mesenchymal layers are clearly visible, which located below the epithelial layer; smooth muscle cells, fibroblasts and myofibroblasts were further detected. Importantly, the ex vivo gut demonstrates spontaneous peristalsis, very similar to the gut movement in vivo. Interstitial cells of Cajal (ICC), one type of interstitial cell in the gut, contribute to gut peristalsis, and the absence of ICC is related to motility disorders [30]. However, no ICC was detected, and peristalsis was even enhanced by neurotoxin in ex vivo guts, indicating that peristalsis in ex vivo gut is not due to ICC functionality. Likewise, botulinum toxin could even increase muscular activity in a spinal cord–muscle coculture system [44]. In short, our cultured ex vivo gut has mesenchymal layers which can be useful for studying epithelial motility.
To demonstrate the effectiveness of the model, we evaluated the role of AMPKα1 in embryonic gut epithelial development. We observed an astonishing difference between WT and AMPKα1 KO ex vivo gut. Our results are supported by recent reports showing the positive roles of AMPK in gut epithelial barrier function [25,45], as well as cell differentiation [25,46–48]. A previously published study in adult mice focused on AMPK loss in epithelial cells [25], while this study focused on AMPK loss in the whole guts including mesenchymal cells during development, suggesting the necessity of AMPK α1 in proper peristalsis and overall health of fetal gut. AMPK specific deficiency in intestinal epithelial cells promoted proliferation of crypt cells in adult mice [25], while AMPK deficiency in whole ex vivo gut suppressed the proliferation and induced apoptosis, possibly due to the subsequent apoptosis induced by inadequate or lagging differentiation. Consistently, AMPK mutants in Drosophila cause abnormal epithelial integrity, defective mitotic cell division and even embryonic lethality [49]. The deficient differentiation in AMPKα1 KO ex vivo gut is partially associated with the downregulation of intestinal transcription factors, such as CDX2 [25]. However, the stimulated proliferation in crypts of epithelial AMPK deficient gut could be due to the fully expressed AMPK in mesenchymal cells underneath crypts, which secretes growth factors and cytokines for tissue regeneration to activate crypt cell proliferation [50].
In summary, ex vivo gut culture system not only maintains the physiological structure of the gut but can effectively track intestinal epithelial differentiation and migration. Using the established ex vivo gut model, we further demonstrated a critical regulatory role of AMPKα1 in epithelial differentiation. This model has potential applications in examining impacts of nutrients, microbial metabolites, toxicants and antigenic materials on gut epithelial development, as well as exploring mechanisms regulating differentiation and migration.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Dr Qiyuan Yang for his assistance in primer design, and Dr Xingwei Liang for providing mouse embryos.
Ethics
All procedures involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Washington State University.
Data accessibility
This article has no additional data.
Authors' contributions
X.S. and M.-J.Z. designed the study. X.S. performed the experiments. X.F. helped the immunostaining experiments. X.S., M.-J.Z., X.F. and M.D. analysed and interpreted the data. X.S. and M.-J.Z. wrote and revised the manuscript.
Competing interests
The authors declare no competing or financial interests.
Funding
This work was financially supported partially by National Institutes of Health (NIH) (R15HD073864), USDA National Institute of Food and Agriculture (USDA-NIFA) (2018-67017-27517) and Emerging Research Issues Internal Competitive grant from the Washington State University, Agricultural Research Center (10A-3057-8640).
References
- 1.Calder PC, et al. 2006. Early nutrition and immunity—progress and perspectives. Br. J. Nutr. 96, 774–790. (doi:10.1079/BJN20061881) [PubMed] [Google Scholar]
- 2.Ellis LA, Mastro AM, Picciano MF. 1997. Do milk-borne cytokines and hormones influence neonatal immune cell function? J. Nutr. 127, 985S–988S. (doi:10.1093/jn/127.5.985S) [DOI] [PubMed] [Google Scholar]
- 3.Barreau F, Cartier C, Leveque M, Ferrier L, Moriez R, Laroute V, Rosztoczy A, Fioramonti J, Bueno L. 2007. Pathways involved in gut mucosal barrier dysfunction induced in adult rats by maternal deprivation: corticotrophin-releasing factor and nerve growth factor interplay. J. Physiol. 580, 347–356. (doi:10.1113/jphysiol.2006.120907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Indrio F, Raimondi F, Laforgia N, Riezzo G, Polimeno L, Francavilla R. 2007. Effect of hyperbilirubinemia on intestinal permeability in healthy term newborns. Acta Paediatr. 96, 73–75. (doi:10.1111/j.1651-2227.2006.00007.x) [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Rodenas CL, Bergonzelli GE, Nutten S, Schumann A, Cherbut C, Turini M, Ornstein K, Rochat F, Corthesy-Theulaz I. 2006. Nutritional approach to restore impaired intestinal barrier function and growth after neonatal stress in rats. J. Pediatr. Gastroenterol. Nutr. 43, 16–24. (doi:10.1097/01.mpg.0000226376.95623.9f) [DOI] [PubMed] [Google Scholar]
- 6.Anderson R, Ellis A, Dalziel J, Roy N, Gopal P, Bassett S. 2012. The role of intestinal barrier function in early life in the development of colitis. In Colitis (ed. Fukata M.), pp. 3–30. Rijeka, Croatia: InTechOpen. [Google Scholar]
- 7.Wagner CL, Taylor SN, Johnson D. 2008. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin. Rev. Allergy. Immunol. 34, 191–204. (doi:10.1007/s12016-007-8032-3) [DOI] [PubMed] [Google Scholar]
- 8.Barreau F, Ferrier L, Fioramonti J, Bueno L. 2004. Neonatal maternal deprivation triggers long term alterations in colonic epithelial barrier and mucosal immunity in rats. Gut 53, 501–506. (doi:10.1136/gut.2003.024174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bischoff SC, Kramer S. 2007. Human mast cells, bacteria, and intestinal immunity. Immunol. Rev. 217, 329–337. (doi:10.1111/j.1600-065X.2007.00523.x) [DOI] [PubMed] [Google Scholar]
- 10.Demaude J, Salvador-Cartier C, Fioramonti J, Ferrier L, Bueno L. 2006. Phenotypic changes in colonocytes following acute stress or activation of mast cells in mice: implications for delayed epithelial barrier dysfunction. Gut 55, 655–661. (doi:10.1136/gut.2005.078675) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groschwitz KR, Ahrens R, Osterfeld H, Gurish MF, Han X, Abrink M, Finkelman FD, Pejler G, Hogan SP. 2009. Mast cells regulate homeostatic intestinal epithelial migration and barrier function by a chymase/Mcpt4-dependent mechanism. Proc. Natl Acad. Sci. USA 106, 22 381–22 386. (doi:10.1073/pnas.0906372106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu LC. 2009. The epithelial gatekeeper against food allergy. Pediatr. Neonatol. 50, 247–254. (doi:10.1016/S1875-9572(09)60072-3) [DOI] [PubMed] [Google Scholar]
- 13.Öhman L, Simrén M. 2010. Pathogenesis of IBS: role of inflammation, immunity and neuroimmune interactions. Nat. Rev. Gastroenterol. Hepatol. 7, 163–173. (doi:10.1038/nrgastro.2010.4) [DOI] [PubMed] [Google Scholar]
- 14.Sato T, et al. 2009. Single Lgr5 stem cells build crypt villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. (doi:10.1038/nature07935) [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Clevers H. 2013. Epithelial cell culture protocols, pp. 319–328. Berlin, Germany: Springer. [Google Scholar]
- 16.Sato T, Clevers H. 2013. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194. (doi:10.1126/science.1234852) [DOI] [PubMed] [Google Scholar]
- 17.de Santa Barbara P, van den Brink GR, Roberts DJ. Development and differentiation of the intestinal epithelium. Cell Mol. Life Sci. 2003, 60, 1322–1332. (doi:10.1007/s00018-003-2289-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8, 774–785. (doi:10.1038/nrm2249) [DOI] [PubMed] [Google Scholar]
- 19.Hardie DG. 2007. Biochemistry. Balancing cellular energy. Science 315, 1671–1672. [DOI] [PubMed] [Google Scholar]
- 20.Fu X, Zhao J-X, Liang J, Zhu M-J, Foretz M, Viollet B, Du M. 2013. AMP-activated protein kinase mediates myogenin expression and myogenesis via histone deacetylase 5. Am. J. Physiol. Cell Physiol. 305, C887–C895. (doi:10.1152/ajpcell.00124.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giri S, Rattan R, Haq E, Khan M, Yasmin R, Won J-s, Key L, Singh AK, Singh I. 2006. AICAR inhibits adipocyte differentiation in 3T3L1 and restores metabolic alterations in diet-induced obesity mice model. Nutr. Metab. 3, 31 (doi:10.1186/1743-7075-3-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dzeja PP, Chung S, Faustino RS, Behfar A, Terzic A. 2011. Developmental enhancement of adenylate kinase-AMPK metabolic signaling axis supports stem cell cardiac differentiation. PLoS ONE 6, e19300 (doi:10.1371/journal.pone.0019300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson AM, Martin KA, Rzucidlo EM. 2014. Resveratrol induces vascular smooth muscle cell differentiation through stimulation of SirT1 and AMPK. PLoS ONE 9, e85495 (doi:10.1371/journal.pone.0085495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang WG, Kim EJ, Lee K-N, Son H-J, Koh J-T. 2011. AMP-activated protein kinase (AMPK) positively regulates osteoblast differentiation via induction of Dlx5-dependent Runx2 expression in MC3T3E1 cells. Biochem. Biophys. Res. Commun. 404, 1004–1009. (doi:10.1016/j.bbrc.2010.12.099) [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Yang Q, Rogers CJ, Du M, Zhu M-J. 2017. AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell Death Differ. 24, 819–831. (doi:10.1038/cdd.2017.14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan JM, Yu W-Y, Hornsey MA, Tosh D, Slack JM. 2006. In vitro culture of embryonic mouse intestinal epithelium: cell differentiation and introduction of reporter genes. BMC Dev. Biol. 6, 24 (doi:10.1186/1471-213X-6-24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu X, Zhao J-X, Zhu M-J, Foretz M, Viollet B, Dodson MV, Du M. 2013. AMP-activated protein kinase α1 but not α2 catalytic subunit potentiates myogenin expression and myogenesis. Mol. Cell. Biol. 33, 4517–4525. (doi:10.1128/MCB.01078-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Xue Y, Zhang H, Huang Y, Yang G, Du M, Zhu MJ. 2013. Dietary grape seed extract ameliorates symptoms of inflammatory bowel disease in IL10-deficient mice. Mol. Nutr. Food Res. 57, 2253–2257. (doi:10.1002/mnfr.201300146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu M-J, Ford SP, Nathanielsz PW, Du M. 2004. Effect of maternal nutrient restriction in sheep on the development of fetal skeletal muscle 1. Biol. Reprod. 71, 1968–1973. (doi:10.1095/biolreprod.104.034561) [DOI] [PubMed] [Google Scholar]
- 30.Streutker C, Huizinga J, Driman D, Riddell R. 2007. Interstitial cells of Cajal in health and disease. Part I: normal ICC structure and function with associated motility disorders. Histopathology 50, 176–189. (doi:10.1111/j.1365-2559.2006.02493.x) [DOI] [PubMed] [Google Scholar]
- 31.Aoki R, et al. 2016. Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol. 2, 175–188. (doi:10.1016/j.jcmgh.2015.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyoshi H, Stappenbeck TS. 2013. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8, 2471–2482. (doi:10.1038/nprot.2013.153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spence JR, et al. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. (doi:10.1038/nature09691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells JM, Spence JR. 2014. How to make an intestine. Development 141, 752–760. (doi:10.1242/dev.097386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Du M, Navarre R, Zhu M-J. 2017. Purple potato extract promotes intestinal barrier function and differentiation by activating AMP-activated protein kinase. Mol. Nutr. Food Res. 62, 1700536 (doi:10.1002/mnfr.201700536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson L, Lindahl P, Heath JK, Betsholtz C. 2000. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457–3466. [DOI] [PubMed] [Google Scholar]
- 37.Sinagoga KL, Wells JM. 2015. Generating human intestinal tissues from pluripotent stem cells to study development and disease. EMBO J. 34, 1149–1163. (doi:10.15252/embj.201490686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bry L, Falk P, Huttner K, Ouellette A, Midtvedt T, Gordon JI. 1994. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc. Natl Acad. Sci. USA 91, 10 335–10 339. (doi:10.1073/pnas.91.22.10335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takada Y, Fukuda A, Chiba T, Seno H. 2016. Brg1 plays an essential role in development and homeostasis of the duodenum through regulation of Notch signaling. Development 143, 3532–3539. (doi:10.1242/dev.141549) [DOI] [PubMed] [Google Scholar]
- 40.Clevers HC, Bevins CL. 2013. Paneth cells: maestros of the small intestinal crypts. Ann. Rev. Physiol. 75, 289–311. (doi:10.1146/annurev-physiol-030212-183744) [DOI] [PubMed] [Google Scholar]
- 41.Roman S, Jayewickreme AK, Murtaugh CD, Shivdasani LC, Shivdasani RA. 2014. Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Rep. 2, 127–134. (doi:10.1016/j.stemcr.2013.12.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabiri Z, et al. 2014. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 141, 2206–2215. (doi:10.1242/dev.104976) [DOI] [PubMed] [Google Scholar]
- 43.Roulis M, Flavell RA. 2016. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 92, 116–131. (doi:10.1016/j.diff.2016.05.002) [DOI] [PubMed] [Google Scholar]
- 44.Eckle V-S, Balk M, Thiermann H, Antkowiak B, Grasshoff C. 2016. Botulinum toxin B increases intrinsic muscle activity in organotypic spinal cord–skeletal muscle co-cultures. Toxicol. Lett. 244, 167–171. (doi:10.1016/j.toxlet.2015.08.003) [DOI] [PubMed] [Google Scholar]
- 45.Peng L, Li Z-R, Green RS, Holzman IR, Lin J. 2009. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 139, 1619–1625. (doi:10.3945/jn.109.104638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. 2008. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673. (doi:10.1016/j.devcel.2008.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim EK, Lim S, Park JM, Seo JK, Kim JH, Kim KT, Ryu SH, Suh PG. 2012. Human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by AMP-activated protein kinase. J. Cell. Physiol. 227, 1680–1687. (doi:10.1002/jcp.22892) [DOI] [PubMed] [Google Scholar]
- 48.Nagata D, Mogi M, Walsh K. 2003. AMP-activated protein kinase (AMPK) signaling in endothelial cells is essential for angiogenesis in response to hypoxic stress. J. Biol. Chem. 278, 31 000–31 006. (doi:10.1074/jbc.M300643200) [DOI] [PubMed] [Google Scholar]
- 49.Lee JH, et al. 2007. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature 447, 1017–1020. (doi:10.1038/nature05828) [DOI] [PubMed] [Google Scholar]
- 50.Donnelly JM, Engevik A, Feng R, Xiao C, Boivin GP, Li J, Houghton J, Zavros Y. 2014. Mesenchymal stem cells induce epithelial proliferation within the inflamed stomach. Am. J. Physiol. Gastrointest. Liver Physiol. 36, G1075–G1088. (doi:10.1152/ajpgi.00489.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.