Abstract
Urbanization is considered a global threat to biodiversity; the growth of cities results in an increase in impervious surfaces, soil and air pollution, fragmentation of natural vegetation and invasion of non-native species, along with numerous environmental changes, including the heat island phenomenon. The combination of these effects constitutes a challenge for both the survival and persistence of many native species, while also imposing altered selective regimes. Here, using 110 314 single nucleotide polymorphisms generated by restriction-site-associated DNA sequencing, we investigated the genome-wide effects of urbanization on putative neutral and adaptive genomic diversity in a major insect pollinator, Bombus lapidarius, collected from nine German cities and nine paired rural sites. Overall, genetic differentiation among sites was low and there was no obvious genome-wide genetic structuring, suggesting the absence of strong effects of urbanization on gene flow. We nevertheless identified several loci under directional selection, a subset of which was associated with urban land use, including the percentage of impervious surface surrounding each sampling site. Overall, our results provide evidence of local adaptation to urbanization in the face of gene flow in a highly mobile insect pollinator.
Keywords: RAD-seq, genotype–environment association, local adaptation, landscape genomics, urban evolutionary biology, panmixia
1. Introduction
Urban cover is predicted to increase by 285% across the world from 2000 to 2030, when an extra 5.9 million km2 of land is likely to be converted to urban use [1]. Urbanization is therefore considered a major global threat to biodiversity [1], as well as a driver of evolutionary change [2]. Impacts of urbanization combine both the rapid destruction of natural habitats for infrastructure and their slow degradation through conversion to parks and gardens. The increase in impervious surfaces, fragmentation of natural vegetation and cultivation of non-native species that become invasive, along with long-term environmental changes such as heat island effects, soil and air pollution, have resulted in the creation of ecosystems with novel species composition [3]. Hence, urbanization can directly affect species fitness [4] and may also challenge the persistence of many species, including essential ecosystem service providers like insect pollinators [5] (but see [6,7]).
Recent reports of bee declines in the Northern Hemisphere have focused attention on insect pollinators, including bumblebees (Bombus spp.) [8]. Among wild pollinators, bumblebees have become a model system for studies on behaviour, ecology and evolution, in part because of their ecological and economic importance [9]. The majority of bumblebee species are generalist pollinator, and can therefore facilitate the reproduction of a large number of wild plants and commercial crops in diverse terrestrial habitats in temperate regions [10]. Furthermore, some Bombus species appear to be resilient to land use change and can be found across a gradient of habitat disturbance [10], making them an ideal species group to study the effects of land use on genetic diversity, gene flow [11,12] and local adaptation. Recently, Miller-Struttmann et al. [13] provided evidence of local adaptation and rapid evolution in two alpine bumblebee species in response to a decline in flowering resources caused by increased temperatures; both species seemingly responded to these environmental changes by evolving shorter tongues, which enabled them to use a wider spectrum of floral resources. Adaptive responses of bumblebees to the urban environment have yet to be investigated (for non-adaptive (genetic drift and gene flow) evolutionary processes, see [11,12,14]), despite their potential suitability as models to reveal adaptation to the urban environment.
Understanding the genetic basis of local adaptation is fundamental to predict the evolutionary responses of species to human-mediated global environmental change [15]. The recent development of next-generation DNA sequencing technologies, in particular restriction site-associated DNA sequencing (RAD-seq) [16], has enabled the genome-wide identification of candidate loci involved in local adaptation of several non-model species (e.g. [17,18]). Despite its limitations (e.g. sampling only a small proportion of the genome [19]), RAD-seq has quickly become the method of choice to study genetic differentiation and adaptation in response to ongoing human-induced environmental change, including urbanization [20,21].
Here, we test for genetic evidence of adaptation to the urban environment in a widespread bumblebee, Bombus lapidarius (Linnaeus, 1758), in central European cities using a landscape genomics approach. In addition, our data allowed us to explore patterns of genome-wide genetic variation, differentiation and demographic history of B. lapidarius, factors which need to be taken into account when testing for signatures of selection [22]. To our knowledge, this is the first attempt to use a population genomics approach to uncover the signatures of adaptation to urbanization in an important group of wild pollinators, results of which inform on their adaptive potential as well as on the long-term security of the ecosystem service of pollination.
2. Material and methods
(a). Study species and sampling sites
The red-tailed bumblebee, B. lapidarius, is an important pollinator species for both wild plants and commercial crops [10]. It has a wide distribution throughout Europe and can be found across a range of both semi-natural and managed habitats, including urban areas [23]. Older studies suggest that it has long been abundant within urban environments of temperate Europe, including our study region, since the nineteenth century [24–26]. Though the species exhibits considerable genetic diversity across Europe, populations in the centre of Europe represent a single phylogeographical group [27].
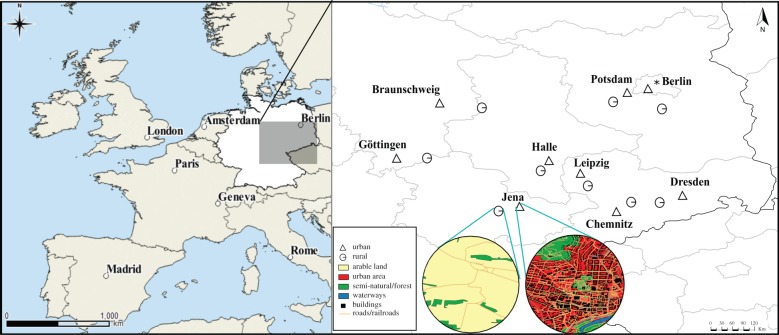
We collected B. lapidarius workers in June–August 2014 from 18 sites located in nine German cities (Berlin, Braunschweig, Chemnitz, Dresden, Göttingen, Halle, Jena, Leipzig and Potsdam) and in nine adjacent rural areas (figure 1; electronic supplementary material, table S1), resulting in a statistically powerful sampling design from multiple (replicated) paired locations. All our urban sites were located either in or in close proximity to the urban core and were surrounded by a high density of roads and human infrastructure (see electronic supplementary material, table S1). Rural sites were selected using land cover maps within a Geographic Information System (quantum GIS), with arable land (agricultural land) and forest/semi-natural cover as the dominant land use types (comprising a mean of 45% and 41% of surrounding (1000 m scale) land use across all rural sites, respectively), typical of the region's rural environment. To do so, we drew a buffer at a circumference of at least 10 km radius from an urban site and then used GIS to identify areas that were largely devoid of ‘residential/industrial’ and dominated by arable land and forest/semi-natural cover within the surrounding 1 km radius, with a low density of roads (see electronic supplementary material, table S1). To ensure independence of sampling, we selected sites at least 10 km distance within each pair, which is beyond the foraging distances of B. lapidarius [10]. Distances between members of different site pairs ranged between 12 and 272 km (figure 1; electronic supplementary material, table S2).
Figure 1.
Location of the 18 study sites in central-eastern Germany, where B. lapidarius workers were sampled. Urban sites are indicated with a triangle, while paired rural sites are shown as a circle. Highlighted are examples of two sites (rural and urban), showing their landscape configuration within a 1000 m radius of a site's geographical centre.
(b). DNA extraction and sequencing
Using hand nets, we collected 198 B. lapidarius workers within 250 m of the centre of our selected sites. In both urban and rural sites, sampling was performed in a single green area (i.e. park or garden for cities and semi-natural area for rural sites). We confirmed species identity by DNA barcoding (sequencing of the mitochondrial cytochrome oxidase I gene), resulting in slight variability in B. lapidarius sample sizes for each site (see electronic supplementary material, table S1). Seventeen of our sampled bees were identified as B. soroeensis proteus (Fabricius, 1777) and excluded from downstream single nucleotide polymorphism (SNP) genotyping, reducing sample sizes at the rural Berlin site, both Göttingen sites and the urban Potsdam site. We generated SNP genotypes for 181 workers using the original RAD-seq protocol [16]. In brief, high-molecular-weight DNA was extracted from thoracic muscle using digestion with DTAB buffer and proteinase K, followed by chloroform : isoamyl alcohol extraction (24 : 1). DNA quality and quantity were assessed using an Epoch spectrophotometer (BioTek, Winooski, USA), agarose gel electrophoresis and a Qubit 3.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). For each individual bee, 2 µg of genomic DNA was normalized to a concentration of 30 ng µl−1. RAD-seq library preparation was carried out by Floragenex, Inc. (Eugene, OR, USA) according to the original RAD protocol, described by, for example, Baird et al. [16] and Etter et al. [28] (for DNA library preparation and sequencing details, see electronic supplementary material, appendix S1).
(c). Data processing, de novo assembly and single nucleotide polymorphism genotyping
An initial quality check of Illumina sequence data was performed using FastQC [29]. Subsequent demultiplexing, removal of barcode sequences, quality filtering (Phred score ≥20) and de novo SNP calling were conducted using Stacks v. 1.42 [30] (for details regarding data processing using Stacks, see electronic supplementary material, appendix S1). We converted the Stacks output to Genepop and BayeScan formats using PGDSpider v. 2.1.0.3 [31].
To check for the potential occurrence of related individuals in the SNP dataset, we used the software KING, with which we calculated kinship coefficients [32]. The kinship analysis did not detect any pairs of individuals coming from the same colony, and thus, all individuals genotyped were assumed to be independent (i.e. from different colonies).
(d). Genetic diversity, inbreeding and genetic structure
Genome-wide measures of genetic diversity (nucleotide diversity (Π), expected heterozygosity (Hexp) and the inbreeding coefficient (FIS)) and overall genetic differentiation, FST, were calculated using the populations program in Stacks. We used 1000 bootstraps to calculate bias-corrected confidence intervals for the FST population pairwise estimates. We used linear mixed models (LMM) to compare heterozygosity, nucleotide diversity and FIS between urban and rural ecosystems. For all mixed models, sampling pair was used as a random effect factor. Mixed models were implemented using the lme4 [33] package in the R statistical software [34]. To test for isolation by distance (IBD) between B. lapidarius populations, we used a Mantel test in the ade4 [35] package in R, using linearized FST values [FST/(1 − FST)], and log-transformed geographical distances separating sampling locations. The data were bootstrapped 10 000 times to generate 95% confidence intervals for the Mantel p-value.
We then used sNMF [36] to examine population structure. sNMF is a fast and efficient method developed for large genomic datasets that uses sparse non-negative matrix factorization to estimate admixture coefficients of individuals. In contrast to other likelihood-based methods such as STRUCTURE, sNMF does not make any model assumptions such as requiring populations to be in Hardy–Weinberg and linkage equilibrium [36]. Prior to running sNMF, we randomly sampled one SNP from each RAD tag to ensure independence of loci. We ran sNMF on 38 695 SNPs and for each value of the number of ancestral populations (K) from 1 to 19 using default parameters, with 10 replicate runs for each value of K. To select the ancestry coefficient with the highest likelihood, sNMF outputs a cross-entropy score; the lowest value with no further decrease in cross-entropy represents the best-supported value of K. As independent support to sNMF results, we additionally conducted a clustering procedure used in discriminant analysis by principal components (DAPC) implemented using the find.clusters function within the adegenet [37] R package. This function first transforms the data using a principal component analysis and then it runs successive K-means clustering with an increasing number of clusters (K). We performed the analysis without prior information on group membership of individuals, and we assessed the optimal number of groups using the Bayesian information criterion (BIC).
We additionally tested whether differentiation across the urban–rural transition was any greater than expected by chance using two methods. Firstly, we used a Mantel spatial correlogram to compare pairwise geographical and genetic distances within distinct distance class bins [38] implemented in the R package vegan [39]. Secondly, to test whether genetic differentiation was greater between habitat types (rural–urban) versus within habitat types (rural–rural, urban–urban), we used a multiple matrix regression with randomization analysis (MMRR [40]) implemented in the ecodist [41] R package with 1000 permutations.
(e). Demographic inference from genome-wide site frequency spectra
The long-term demographic history of B. lapidarius was investigated with δaδi v. 1.7.0 [42]. Our sNMF and find.clusters analyses suggested no genetic structure, and thus, we performed the demographic inference by grouping all 181 individuals into one population representing one evolutionary cluster. Prior to demographic analyses, we converted the Stacks VCF output file to a δaδi folded site frequency spectrum (one-dimensional, i.e. a folded SFS). The folded SFS was then fitted to different demographic scenarios using a diffusion approximation-based method implemented in δaδi [42]. We examined four demographic models: one neutral and three population size change models (i.e. instantaneous expansion model, exponential growth model and bottleneck growth model; see electronic supplementary material, figure S1; for details regarding demographic models and analyses, see electronic supplementary material, appendix S1).
(f). Detection of outlier loci
To identify outlier loci, we applied the frequentist detection method implemented in LOSITAN [43] and the Bayesian approach of BayeScan [44]. Both methods employ an FST-outlier approach, which is based on the idea that genetic differentiation (measured as FST) among loci under selection is higher (directional selection) or lower (balancing selection) than genetic differentiation among neutrally evolving loci, the latter being a mere reflection of neutrally evolving genes' shared demographic history [44]. These two approaches do not explicitly test for urban adaptation; rather, they test for local adaptation among all our tested populations.
LOSITAN is a selection detection workbench that uses a coalescent-based simulation approach to identify outliers in relation to the distributions of heterozygosity and FST [45]. Initially, we ran LOSITAN using all SNPs to estimate the mean neutral FST. We then recalculated the mean neutral FST by removing SNPs outside the confidence intervals (95%) of the first estimate. This allowed us to obtain a better approximation of the mean neutral FST, which was then used as a reference to conduct a final run using all SNPs. We performed 1 000 000 simulations, assuming an infinite allele mutation model with 18 populations and a false discovery rate (FDR) of 0.05 to minimize the number of false positives.
BayeScan also uses FST to detect loci potentially under selection. However, it estimates the probability that a locus is under selection by taking into account population-specific effects of demography and incorporating uncertainties in allele frequencies due to small sample sizes within a Bayesian framework [44]. BayeScan has been shown to be more reliable than alternative methods in detecting outliers and in reducing the number of false positives [46]. In addition, it is relatively robust to confounding demographic processes and hierarchical structure that may inflate FST estimates [47]. We performed the detection of outlier SNPs in BayeScan with default parameters (number of iterations = 5000; thinning interval = 10; number of pilot runs = 20; length of pilot run = 5000; burn-in length = 50 000). Considering the high number of SNPs in our dataset, the prior odds were set to 100 and the FDR was set to 0.05. Convergence of the MCMC chains was confirmed by plotting the log-likelihood trace in R [34].
(g). Detection of associations between genetic differentiation and urbanization
To identify loci for which genetic differentiation is associated with urban versus rural environments, we used BayeScEnv [48]. We did so by means of two approaches. In the first approach, we categorized each sampling site as either rural or urban and tested for an association between genetic differentiation and this binary variable. In a second, more refined analysis, we tested for an association of genetic differentiation with the percentage of impervious surface within a 1000 m buffer zone around each sampling site; impervious surface is a continuous variable which was highly correlated with building cover and road density (see electronic supplementary material, table S3). BayeScEnv aims to identify genetic markers subjected to selection and associated to population-specific covariates. Similar to BayeScan [44], it accounts for population structure by using the F-model of Beaumont & Balding [49]. Prior to the BayeScEnv analysis, all environmental variables were standardized to a mean of zero and standard deviation of one. BayeScEnv was run with several default parameters (number of iterations = 5000; thinning interval = 10; number of pilot runs = 20; length of pilot run = 5000; burn-in length = 50 000) and the following parameter settings: g (upper bound) = 10, α (mean prior) = −1.0, p = 0.5 and π = 0.10. Diagnostics of the log-likelihoods and FST values for the 5000 sampled iterations were checked using R [34].
(h). Single nucleotide polymorphism annotation and gene ontology analysis
To identify candidate genes associated with the 287 detected outlier SNPs (belonging to 176 candidate sequences; see results below), contigs (overlapping set of reads across individuals that together represent a consensus region in the genome) containing outliers were annotated using the Blast2GO pipeline [50], which conducts Blast similarity searches and maps Gene Ontology (GO) terms to homologous sequences. First, we carried out a BLAST search of all candidate sequences containing our 287 candidate SNPs against the complete genome and transcriptome of Bombus terrestris (Linnaeus, 1758) (assembly Bter_1.0, Baylor College of Medicine) to identify candidate genes for GO analysis. As the RAD candidate sequences were only 115 bp in length (125 bp read length minus the 10 bp multiplex identifier (MID) ‘barcode’), this step increased the length of the RAD candidate sequences and hence reduced the number of false positives arising from BLASTing short query sequences against a gene database. Functional annotation steps were performed with Blast2GO default parameters.
Genes under selection were then tested for potential enrichment of specific gene functions using GO enrichment analyses. GO enrichment analyses using Fisher's exact tests were performed in Blast2GO under default parameters and with the entire gene list covered by all of our RAD-Seq data as a reference. Additionally, an enrichment analysis was performed using GOEAST (Gene Ontology Enrichment Analysis Software Toolkit) [51] using the hypergeometric statistical test and corrected p-values for multiple testing using either a Bonferroni correction or the FDR [52] at an overall α of p < 0.05. Both of these enrichment analyses re-classify child terms to higher terms prior to enrichment tests.
3. Results
(a). Restriction site-associated DNA sequencing, genetic diversity and genetic structure
Two lanes of Illumina HiSeq2500 1 × 125 bp SE sequencing produced a total of 354 million reads of 125 bp, of which 285.5 million reads (80.64%) passed initial quality filters. Reads were filtered out due to ambiguous MID barcodes (10.28%), low-quality scores (9.02%) and ambiguous RAD tags (0.06%). After all filtering steps, the catalogue generated by Stacks included a total of 43 568 RAD tags (covering approx. 2% of B. terrestris genome) and 110 314 SNPs with an average of 25 556 (± 6454 s.d.) polymorphic sites in each population. All B. lapidarius populations were well represented in the final dataset, with an average of four to ten individuals genotyped per locus across the 18 populations (see electronic supplementary material, table S4). Expected heterozygosity ranged from 0.0422 to 0.0851 and nucleotide diversity (Π) ranged from 0.049 to 0.090 (see electronic supplementary material, table S4).
The inbreeding coefficient (FIS) was, on average, low and not significantly different from zero (mean 0.055 ± 0.108 s.d.; see electronic supplementary material, table S4). There were no differences in expected heterozygosity (LMM: Hexp; χ2 = 1.058, p = 0.303), nucleotide diversity (LMM: Π; χ2 = 1.044, p = 0.306) and FIS (LMM: χ2 = 0.830, p = 0.362) between rural and urban populations. The Potsdam rural site had an unexpected and exceptionally high FIS value (see electronic supplementary material, table S4); we therefore re-ran all analyses described in this paper after excluding it from the dataset, but results did not change qualitatively or in significance. We therefore retained data from this sampling locality in all analyses presented here.
Genetic differentiation between population pairs was low and ranged from −0.043 to 0.013 (mean FST = 0.003, 95% CI = −0.006–0.009) (see electronic supplementary material, figure S2 and table S5a,b), indicating a lack of significant genetic structure. A Mantel test showed no significant correlation in genetic and geographical distance between pairs of populations (r = 0.055; p = 0.772). sNMF analysis also did not reveal clear population structure across the study area, suggesting a single genetic unit (i.e. K = 1) for the whole dataset. For K > 1, there was no further decrease in the cross-entropy score (Tukey post hoc test: p > 0.05; see electronic supplementary material, figure S4). Similar to sNMF, the find.clusters function indicated the presence of only one genetic cluster based on the BIC (see electronic supplementary material, figure S5). There was also a lack of subtle genetic differentiation between urban and rural members of pairs of sites; a Mantel correlogram (see electronic supplementary material, figure S3) did not reveal any spatial genetic structure (Bonferroni-corrected, p > 0.05) while differentiation across the urban–rural transition was no greater than expected by distance alone (MMR; β = 0.0003, p = 0.680; see electronic supplementary material, table S6).
(i). Bombus lapidarius demographic history
To infer the demographic history of B. lapidarius, we tested four single-population demographic models using the folded SFS and the maximum-likelihood methods implemented in δaδi. All population size change models fitted our data significantly better than the neutral model (LRT; p < 0.001; see electronic supplementary material, figure S1). Based on the AIC, the exponential growth model (AIC = 1089.56) performed better than the instantaneous expansion model (AIC = 1215.39) and marginally outperformed the bottleneck growth model (AIC = 1091.56). Furthermore, the bottleneck growth model gave nearly identical results with the exponential growth model (see electronic supplementary material, figure S1c,d). Results did not change qualitatively when we removed outlier loci (see below).
(b). Outlier Single nucleotide polymorphisms
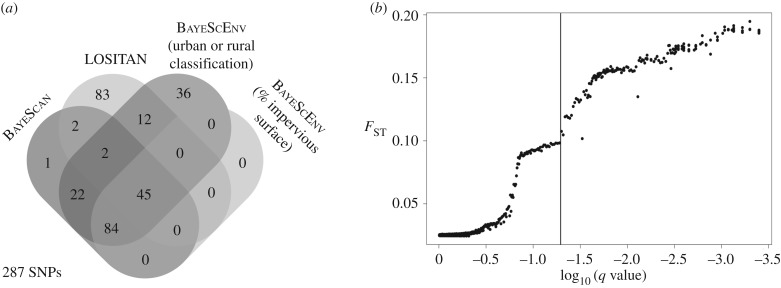
In total, we identified 287 outlier SNPs in 176 contigs, of which 45 (16.3%) were concurrently detected by all programmes we used to detect signatures of selection (figure 2a). LOSITAN and BayeScan shared 49 (19.52%) outlier SNPs, LOSITAN and BayeScEnv shared 59 (20.62%) outlier SNPs, and BayeScan and BayeScEnv shared 153 (75%) outlier SNPs. The LOSITAN approach identified 144 SNPs (figure 2a) and the BayeScan approach 156 SNPs under directional selection (figure 2a,b). The BayeScEnv analysis identified a total of 201 SNPs associated with urbanization. All 201 SNPs were associated with land use type (urban versus rural) and a subset of them (129 SNPs) was associated with the percentage of impervious surface surrounding each sampling site (figure 2a).
Figure 2.
(a) Number of outlier SNPs identified as putatively being under directional selection using two genome scan methods, BayeScan and LOSITAN, as well as BayeScEnv testing for genetic associations with two environmental variables (i.e. ‘urban versus rural landscape type’ and ‘percentage impervious surface’) across all 18 sites. The total number of outlier SNPs (i.e. 287) is reported in the lower left corner of the figure. (b) FST versus log10-transformed q values for the global outlier detection performed in BayeScan. The vertical line represented the FDR threshold of p = 0.05.
(c). Candidate genes for local adaptation
The gene annotation pipeline implemented in Blast2GO allowed us to annotate 61 out of 176 candidate loci (contigs that mapped to the B. terrestris reference genome) for local adaptation. Overall, we could not find a significant BLAST hit for 115 out of 176 contigs. Five of the remaining 61 contigs mapped to the same candidate locus, giving us 56 candidate loci (see electronic supplementary material, table S7). We could not assign GO terms to 15 out of the remaining 56 candidate loci. Of those 41 candidate loci for which we could find a BLAST hit and to which we could assign GO terms, GO functional annotations of the outlier loci yielded 35 GO terms belonging to all the three functional categories: molecular function, biological process and cellular component (see electronic supplementary material, figure S6 and table S7).
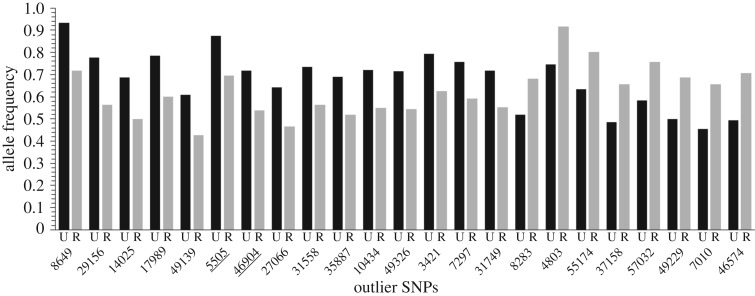
GO term enrichment analysis identified 25 GO terms significantly enriched in functional categories (Blast2GO, p < 0.05; see electronic supplementary material, tables S7 and S8). The enrichment analysis performed with GOEAST identified 18 GO and 11 GO terms significantly enriched when using the FDR and Bonferroni correction, respectively (see electronic supplementary material, table S9). Both of these enrichment analyses of the genes in the candidate regions under divergent selection, as identified by all three outlier methods, highlighted GO terms associated with: metabolic processes, heat stress and oxidative stress (i.e. GO:0043167, GO:0008152 and GO:0044237). The 24 candidate loci under selection identified by at least two methods (LOSITAN, BayeScan and BayeScEnv) and to which we could assign GO terms showed clearly divergent allele frequencies between urban and rural sites (figure 3). Details of all loci containing significant outlier SNPs, along with transcript product names and related GO terms and GO enrichment analyses, are provided in the electronic supplementary material (see electronic supplementary material, tables S7, S8 and S9; note that the GO terms of electronic supplementary material, tables S8 and S9 are hierarchically redundant).
Figure 3.
Allele frequencies of the commonest allele for candidate loci (n = 24) identified as outliers by at least two approaches (LOSITAN, BayeScan and BayeScEnv) and to which we could assign GO terms, ordered left to right from largest positive difference (urban > rural) to largest negative difference (urban < rural). Highlighted are the two loci found in genes coding for calmodulin (5505) and serine/threonine protein kinase (46904). The frequency of the outlier SNP within each land use type, urban (U, black) or rural (R, grey), is plotted on the vertical axis. Each candidate locus is labelled with the contig name on the horizontal axis; see highlighted rows in electronic supplementary material, table S7.
4. Discussion
Our study represents the first high-density SNP-based genome scan for signatures of selection due to urbanization in an ecologically and commercially important pollinator species, Bombus lapidarius. Using a replicated paired sampling design and a RAD-seq approach, we found low levels of genetic differentiation among B. lapidarius populations at the majority of genomic loci screened (approx. 99%). However, a small proportion of loci exhibited a very high degree of genetic differentiation and/or was associated with urbanization, suggesting adaptive divergence among local populations. To this end, our results support the notion that adaptive genetic divergence can occur through urbanization in the face of high gene flow (e.g. [17,18]).
5. Low genetic differentiation in B. lapidarius populations
Although several studies on the population genetics of bumblebees have reported relatively high differentiation across populations, most of them were focused on rare or declining species [53,54]. Our analysis of genetic differentiation and IBD in a common and widely distributed bumblebee, B. lapidarius, suggests panmixia and a lack of marked urban–rural differentiation in central-eastern Germany. This is not surprising as bumblebee reproductive castes (queens and males) have high dispersal abilities. Bombus lapidarius queens can disperse over distances ranging from 3 to 5 km [55] and bumblebee males at least from 2.6 to 9.9 km [56]. In addition, B. lapidarius also exists at high population densities [57]. The low levels of genetic structure we found for B. lapidarius most likely reflect these high dispersal abilities and large population sizes, in accordance with results from other Continental bumblebee populations in Europe (e.g. B. terrestris [58]) and in North America (Bombus impatiens (Cresson, 1863) [59]).
(a). Bombus lapidarius demographic history
From our demographic inference using genome-wide site frequency spectra, we found clear signals of past population growth. Assuming a mutation rate of 3.6 × 10−9 per site per generation (estimated for B. terrestris [60]), the estimated ancestral Ne (effective population size) for B. lapidarius is approximately 35 679 (95% CI = 34 992–36 366) individuals and the contemporary Ne is approximately 423 514 (95% CI = 420 410–426 618), corresponding to mean population growth dating to approximately 67 969 years ago (95% CI = 66 845–69 093). Based on these estimates, we date the population growth of B. lapidarius during the last glaciation period (110–12 ka), not surprising for this cold-tolerant widespread West Palaearctic bumblebee [61]. However, such estimates should be interpreted with caution due to their dependence on an unknown factor: the mutation rate. Nevertheless, from our demographic analyses, we found that B. lapidarius contemporary population size has increased in recent times, as expected given the species' broad and expanding geographical range [10].
(b). Local adaptation to urbanization
Genome scans for SNPs have been widely used to investigate polymorphisms in order to detect and disentangle neutral (demography) and adaptive (selection) evolutionary processes [17,18,21]. The most popular approach to identify potential targets of selection compares allele frequency differentiation, measured as FST between populations [17,18]. Environment association (EA) tests provide fine-scaled analyses of the ecological processes driving selection by identifying loci with allele frequency differentiation that is correlated with environmental factors, for which several EA tools have already been developed [48]. We found differences in the results between the FST-outlier and EA methods implemented in LOSITAN, BayeScan and BayeScEnv, with only 16.3% of loci consistently identified by all three programmes as targets of local selection. However, 98.07% of the outlier SNPs detected with the BayeScan and 40.68% of the outlier SNPs detected with the LOSITAN methods were also classified as candidate loci under selection with the BayeScEnv approach. This is consistent with previous studies, suggesting that the SNP–EA approach may be complementary to FST-outlier methods [62]. Combining population differentiation and environmental association approaches to detect candidate loci related to environmental adaptation not only reduces false positives, but also maximizes the chances of detecting potential signals of selection [63]. In our study, we consistently identified 45 putative targets of local selection that showed both high FST and an association to the environment, including genes associated with metabolism, heat stress and oxidative stress, which may be logically related to urbanization.
In general, organisms living in urban areas face physiological challenges caused by pollution (chemical or from light), urban warming, parasites, diet and costly foraging [64]. Previous studies have documented greater parasite prevalence among bumblebees in urban than rural areas, suggesting that urban habitats may present greater opportunities for parasite transmission [23]. Pathogens are known to reduce colony and individual pollinator fitness and abundance as well as to alter foraging performance [10]. Similarly, foraging habitat, pesticide use and parasite stress were found to be associated with the disruption of metabolic functions related to immunity, energetic resources and antioxidant responses in honeybees [65,66]. In our study, we found that SNPs showing footprints of directional selection were located in genes involved in molecular binding and metabolic processes. For example, we identified footprints of selection in genes coding for serine/threonine protein kinase and calmodulin. Such proteins are involved in regulating downstream kinases in response to environmental stress, such as heat stress and oxidative stress. They also mediate processes, such as apoptosis and the innate immune response, which might be more pronounced in urban than rural environments and might thus be of adaptive relevance for an urban lifestyle. While these associations with environmental parameters and footprints of selection do not necessarily imply causality, they support our interpretation that the signatures of selection we have recovered are likely to be biologically meaningful.
The large number of putative loci under selection that we detected may give cause for concern that some were false positives. From an analytical perspective, the FST-based methods and the EA method used in our study for detecting local adaptation are all correlational; additional lines of evidence (e.g. functional studies on B. lapidarius or comparative studies across other Bombus spp.) are required to determine whether loci are causally involved in local adaptation. Additionally, the lack of a reference genome hindered our annotation of RAD tags. Furthermore, the low genome coverage we achieved with our RAD tags combined with the lack of a genome sequence upon which to link loci meant that we were unable to use the powerful approach of detecting genome-wide signatures of selective sweeps, which can provide a complement to FST-based outlier locus detection and thereby reduce the rate of false positives among candidate loci under selection [67]. In contrast to these caveats, by sequencing only approximately 2% of the B. lapidarius genome, we may well not have detected many additional loci under selection to urbanization.
An open question remains over the establishment and maintenance of allele frequency differences at loci under selection in our study system. We have strong evidence for a lack of differentiation across our study sites at putatively neutral loci, probably because of high Ne and ongoing gene flow. That we consistently detected outlier loci across nine rural–urban transitions suggests that outliers are under selection for the local (urban) environment. We cannot, though, tell if this is due to strong selection each generation (single-generation selection) acting on a panmictic (urban plus rural) gene pool or alternatively due to high Ne coupled to a low migration rate and recent selection on our identified outlier loci (multiple-generation selection). Though B. lapidarius has been recorded since the nineteenth century within German cities [24], arguing against the latter hypothesis, independent lines of evidence are needed to differentiate between single- versus multiple-generation selection. Nevertheless, the outlier loci we detected seem to represent genetic/genomic islands of adaptation in the face of gene flow [68].
6. Conclusion
Rates of urbanization are accelerating globally, with accumulating evidence in support of rapid evolution in response to urbanization [2]. Using a RAD-seq approach, we identified thousands of SNPs in a widespread bumblebee, a subset of which exhibited patterns of genetic differentiation strongly associated with urbanization, despite high ongoing gene flow. To this end, our study demonstrates the power of population genomic approaches to uncover evidence for potential adaptation to human-altered environments. Future work should test whether and how allelic variants are of functional relevance for adaptation to life in the city.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Matthias Seidel, Patricia Landaverde and Lara-Sophie Dey for their assistance with fieldwork, Mike Edward, Joachim Händel, Anselm Kratochwil, Denis Michez and Pierre Rasmont for literature and historical records of B. lapidarius in the urban environment, and the anonymous referees and Special Feature editors for their helpful comments on an earlier version of the manuscript.
Data accessibility
Demultiplexed, raw RAD-Seq reads are available under accession no. SRP099050 at the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra). The variant calling file (VCF) is available on the Dryad Digital Repository at https://doi.org/10.5061/dryad.1hn0951 [69].
Authors' contributions
P.T. participated in the design of the study, collected field data, carried out the molecular lab work, undertook data analysis and drafted the manuscript. R.R. collected field data. B.K., A.S. and I.G. assisted in data interpretation. R.J.P. participated in study design and data interpretation. All authors assisted in drafting the manuscript and gave their final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This research was supported by the German Centre for Integrative Biodiversity Research Halle-Jena-Leipzig (iDiv) Flexible Pool project 50170649, funded by the German Research Foundation (DFG: FZT 118).
References
- 1.Seto KC, Guneralp B, Hutyra LR. 2012. Global forecasts of urban expansion to 2030 and direct impacts on biodiversity and carbon pools. Proc. Natl Acad. Sci. USA 109, 16 083–16 088. ( 10.1073/pnas.1211658109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson MTJ, Munshi-South J. 2017. Evolution of life in urban environments. Science 358, eaam8327 ( 10.1126/science.aam8327) [DOI] [PubMed] [Google Scholar]
- 3.Grimm NB, Faeth SH, Golubiewski NE, Redman CL, Wu J, Bai X, Briggs JM. 2008. Global change and the ecology of cities. Science 319, 756–760. ( 10.1126/science.1150195) [DOI] [PubMed] [Google Scholar]
- 4.Bonier F, Martin PR, Wingfield JC. 2007. Urban birds have broader environmental tolerance. Biol. Lett. 3, 670–673. ( 10.1098/rsbl.2007.0349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates AJ, Sadler JP, Fairbrass AJ, Falk SJ, Hale JD, Matthews TJ. 2011. Changing bee and hoverfly pollinator assemblages along an urban–rural gradient. PLoS ONE 6, e23459 ( 10.1371/journal.pone.0023459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldock KCR, et al. 2015. Where is the UK's pollinator biodiversity? The importance of urban areas for flower-visiting insects. Proc. R. Soc. B 282, 20142849 ( 10.1098/rspb.2014.2849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theodorou P, Albig K, Radzevičiūtė R, Settele J, Schweiger O, Murray TE, Paxton RJ. 2017. The structure of flower visitor networks in relation to pollination across an agricultural to urban gradient. Funct. Ecol. 31, 838–847. ( 10.1111/1365-2435.12803) [DOI] [Google Scholar]
- 8.Vanbergen A, the Insect Polinators Initiative. 2013. Threats to an ecosystem service: pressures on pollinators. Front. Ecol. Environ. 11, 251–259. ( 10.1890/120126) [DOI] [Google Scholar]
- 9.Woodard SH, Lozier JD, Goulson D, Williams PH, Strange JP, Jha S. 2015. Molecular tools and bumble bees: revealing hidden details of ecology and evolution in a model system. Mol. Ecol. 24, 2916–2936. ( 10.1111/mec.13198) [DOI] [PubMed] [Google Scholar]
- 10.Goulson D. 2010. Bumblebees: behaviour, ecology, and conservation. Oxford, UK: Oxford University Press. [Google Scholar]
- 11.Jha S, Kremen C. 2013. Urban land use limits regional bumble bee gene flow. Mol. Ecol. 22, 2483–2495. ( 10.1111/mec.12275) [DOI] [PubMed] [Google Scholar]
- 12.Jha S. 2015. Contemporary human-altered landscapes and oceanic barriers reduce bumble bee gene flow. Mol. Ecol. 24, 993–1006. ( 10.1111/mec.13090) [DOI] [PubMed] [Google Scholar]
- 13.Miller-Struttmann NE, et al. 2015. Functional mismatch in a bumble bee pollination mutualism under climate change. Science 349, 1541–1544. ( 10.1126/science.aab0868) [DOI] [PubMed] [Google Scholar]
- 14.Chapman RE, Wang J, Bourke AFG. 2003. Genetic analysis of spatial foraging patterns and resource sharing in bumble bee pollinators. Mol. Ecol. 12, 2801–2808. ( 10.1046/j.1365-294X.2003.01957.x) [DOI] [PubMed] [Google Scholar]
- 15.Savolainen O, Lascoux M, Merilä J. 2013. Ecological genomics of local adaptation. Nat. Rev. Genet. 14, 807–820. ( 10.1038/nrg3522) [DOI] [PubMed] [Google Scholar]
- 16.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3, e3376 ( 10.1371/journal.pone.0003376) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo B, Li Z, Merilä J. 2016. Population genomic evidence for adaptive differentiation in the Baltic Sea herring. Mol. Ecol. 25, 2833–2852. ( 10.1111/mec.13657) [DOI] [PubMed] [Google Scholar]
- 18.Pujolar JM, et al. 2014. Genome-wide single-generation signatures of local selection in the panmictic European eel. Mol. Ecol. 23, 2514–2528. ( 10.1111/mec.12753) [DOI] [PubMed] [Google Scholar]
- 19.Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, Storfer A. 2016. Breaking RAD: an evaluation of the utility of restriction site associated DNA sequencing for genome scans of adaptation. Mol. Ecol. Resour. 17, 142–152. ( 10.1111/1755-0998.12635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yakub M, Tiffin P. 2016. Living in the city: urban environments shape the evolution of a native annual plant. Glob. Chang. Biol. 23, 2082–2089. ( 10.1111/gcb.13528) [DOI] [PubMed] [Google Scholar]
- 21.Munshi-South J, Zolnik CP, Harris SE. 2016. Population genomics of the Anthropocene: urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evol. Appl. 9, 546–564. ( 10.1111/eva.12357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen R. 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86, 641–647. ( 10.1046/j.1365-2540.2001.00895.x) [DOI] [PubMed] [Google Scholar]
- 23.Theodorou P, Radzevičiūtė R, Settele J, Schweiger O, Murray TE, Paxton RJ. 2016. Pollination services enhanced with urbanization despite increasing pollinator parasitism. Proc. R. Soc. B 283, 20160561 ( 10.1098/rspb.2016.0561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmiedeknecht O. 1882. Apidae Europaeae (Die Bienen Europas) per Genera, Species et Varietates Dispositae atque Descriptae. Berlin, Germany: R. Friedländer & Sons. [Google Scholar]
- 25.Shuckard WE. 1866. British bees: an introduction to the study of the natural history and economy of the bees indigenous to the British Isles. London, UK: Reeve. [Google Scholar]
- 26.Saunders E. 1896. The hymenoptera Aculeata of the British Islands: a descriptive account of the families, genera, and species indigenous to Great Britain and Ireland, with notes as to habits, localities, habitats. London, UK: Reeve. [Google Scholar]
- 27.Lecocq T, Dellicour S, Michez D, Lhomme P, Vanderplanck M, Valterová I, Rasplus JY, Rasmont P. 2013. Scent of a break-up: phylogeography and reproductive trait divergences in the red-tailed bumblebee (Bombus lapidarius). BMC Evol. Biol. 13, 263 ( 10.1186/1471-2148-13-263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etter PD, Bassham S, Hohenlohe PA, Johnson EA, Cresko WA. 2012. SNP Discovery and genotyping for evolutionary genetics using RAD sequencing. In Methods in molecular biology, pp. 157–178. Clifton, NJ: Humana Press; ( 10.1007/978-1-61779-228-1_9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. See http://www.bioinformatics.babraham.ac.uk/projects/fastqc .
- 30.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140. ( 10.1111/mec.12354) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lischer HEL, Excoffier L. 2012. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28, 298–299. ( 10.1093/bioinformatics/btr642) [DOI] [PubMed] [Google Scholar]
- 32.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873. ( 10.1093/bioinformatics/btq559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. ( 10.18637/jss.v067.i01) [DOI] [Google Scholar]
- 34.R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org. [Google Scholar]
- 35.Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20. ( 10.18637/jss.v022.i04) [DOI] [Google Scholar]
- 36.Frichot E, Mathieu F, Trouillon T, Bouchard G, Francois O. 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 196, 973–983. ( 10.1534/genetics.113.160572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jombart T, Ahmed I. 2011. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. ( 10.1093/bioinformatics/btr521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Legendre P, Fortin MJ, Borcard D. 2015. Should the Mantel test be used in spatial analysis? Methods Ecol. Evol. 6, 1239–1247. ( 10.1111/2041-210X.12425) [DOI] [Google Scholar]
- 39.Oksasen J, et al. 2015. vegan: community ecology package. R package version 2.3-1.
- 40.Wang IJ. 2013. Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 67, 3403–3411. ( 10.1111/evo.12134) [DOI] [PubMed] [Google Scholar]
- 41.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19. ( 10.18637/jss.v022.i07) [DOI] [Google Scholar]
- 42.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD, Stephan W. 2009. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 5, e1000695 ( 10.1371/journal.pgen.1000695) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. 2008. LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics 9, 1–5. ( 10.1186/1471-2105-9-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993. ( 10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaumont MA, Nichols RA. 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B 263, 1619–1626. ( 10.1098/rspb.1996.0237) [DOI] [Google Scholar]
- 46.Narum SR, Hess JE. 2011. Comparison of Fst outlier tests for SNP loci under selection. Mol. Ecol. Resour. 11, 184–194. ( 10.1111/j.1755-0998.2011.02987.x) [DOI] [PubMed] [Google Scholar]
- 47.De Villemereuil P, Frichot É, Bazin É, François O, Gaggiotti OE. 2014. Genome scan methods against more complex models: when and how much should we trust them? Mol. Ecol. 23, 2006–2019. ( 10.1111/mec.12705) [DOI] [PubMed] [Google Scholar]
- 48.de Villemereuil P, Gaggiotti OE. 2015. A new Fst-based method to uncover local adaptation using environmental variables. Methods Ecol. Evol. 6, 1248–1258. ( 10.1111/2041-210X.12418) [DOI] [Google Scholar]
- 49.Beaumont MA, Balding DJ. 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13, 969–980. ( 10.1111/j.1365-294X.2004.02125.x) [DOI] [PubMed] [Google Scholar]
- 50.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. ( 10.1093/bioinformatics/bti610) [DOI] [PubMed] [Google Scholar]
- 51.Zheng Q, Wang XJ. 2008. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36, W358–W363. ( 10.1093/nar/gkn276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. ( 10.2307/2346101) [DOI] [Google Scholar]
- 53.Ellis JS, Knight ME, Darvill B, Goulson D. 2006. Extremely low effective population sizes, genetic structuring and reduced genetic diversity in a threatened bumblebee species, Bombus sylvarum (Hymenoptera: Apidae). Mol. Ecol. 15, 4375–4386. ( 10.1111/j.1365-294X.2006.03121.x) [DOI] [PubMed] [Google Scholar]
- 54.Darvill B, Ellis JS, Lye GC, Goulson D. 2006. Population structure and inbreeding in a rare and declining bumblebee, Bombus muscorum (Hymenoptera: Apidae). Mol. Ecol. 15, 601–611. ( 10.1111/j.1365-294X.2006.02797.x) [DOI] [PubMed] [Google Scholar]
- 55.Lepais O, Darvill B, O'connor S, Osborne JL, Sanderson RA, Cussans J, Goffee L, Goulson D. 2010. Estimation of bumblebee queen dispersal distances using sibship reconstruction method. Mol. Ecol. 19, 819–831. ( 10.1111/j.1365-294X.2009.04500.x) [DOI] [PubMed] [Google Scholar]
- 56.Kraus FB, Wolf S, Moritz RFA. 2009. Male flight distance and population substructure in the bumblebee Bombus terrestris. J. Anim. Ecol. 78, 247–252. ( 10.1111/j.1365-2656.2008.01479.x) [DOI] [PubMed] [Google Scholar]
- 57.Dreier S, Redhead JW, Warren IA, Bourke AFG, Heard MS, Jordan WC, Sumner S, Wang J, Carvell C. 2014. Fine-scale spatial genetic structure of common and declining bumble bees across an agricultural landscape. Mol. Ecol. 23, 3384–3395. ( 10.1111/mec.12823) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreira AS, Horgan FG, Murray TE, Kakouli-Duarte T. 2015. Population genetic structure of Bombus terrestris in Europe: isolation and genetic differentiation of Irish and British populations. Mol. Ecol. 24, 3257–3268. ( 10.1111/mec.13235) [DOI] [PubMed] [Google Scholar]
- 59.Lozier JD, Stranfe JP, Stewart IJ, Cameron SA. 2011. Patterns of range-wide genetic variation in six North American bumble bee (Apidae: Bombus) species. Mol. Ecol. 20, 4870–4888. ( 10.1111/j.1365-294X.2011.05314.x) [DOI] [PubMed] [Google Scholar]
- 60.Liu H, Jia Y, Sun X, Tian D, Hurst LD, Yang S. 2017. Direct determination of the mutation rate in the bumblebee reveals evidence for weak recombination-associated mutation and an approximate rate constancy in insects. Mol. Biol. Evol. 34, 119–130. ( 10.1093/molbev/msw226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dellicour S, Kastally C, Varela S, Michez D, Rasmont P, Mardulyn P, Lecocq T. 2017. Ecological niche modelling and coalescent simulations to explore the recent geographical range history of five widespread bumblebee species in Europe. J. Biogeogr. 44, 39–50. ( 10.1111/jbi.12748) [DOI] [Google Scholar]
- 62.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185, 1411–1423. ( 10.1534/genetics.110.114819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.François O, Martins H, Caye K, Schoville SD. 2016. Controlling false discoveries in genome scans for selection. Mol. Ecol. 25, 454–469. ( 10.1111/mec.13513) [DOI] [PubMed] [Google Scholar]
- 64.Isaksson C. 2015. Urbanization, oxidative stress and inflammation: a question of evolving, acclimatizing or coping with urban environmental stress. Funct. Ecol. 29, 913–923. ( 10.1111/1365-2435.12477) [DOI] [Google Scholar]
- 65.Dussaubat C, Maisonnasse A, Crauser D, Tchamitchian S, Bonnet M, Cousin M, Kretzschmar A, Brunet J-L, Le Conte Y. 2016. Combined neonicotinoid pesticide and parasite stress alter honeybee queens' physiology and survival. Sci. Rep. 6, 31430 ( 10.1038/srep31430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morimoto T, Kojima Y, Toki T, Komeda Y, Yoshiyama M, Kimura K, Nirasawa K, Kadowaki T. 2011. The habitat disruption induces immune-suppression and oxidative stress in honey bees. Ecol. Evol. 1, 201–217. ( 10.1002/ece3.21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoban S, et al. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am. Nat. 188, 379–397. ( 10.1086/688018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feder JL, Egan SP, Nosil P. 2012. The genomics of speciation-with-gene-flow. Trends Genet. 28, 342–350. ( 10.1016/j.tig.2012.03.009) [DOI] [PubMed] [Google Scholar]
- 69.Theodorou P, Radzevičiūtė R, Kahnt B, Soro A, Grosse I, Paxton RJ. 2018. Data from: Genome-wide single nucleotide polymorphism scan suggests adaptation to urbanization in an important pollinator, the red-tailed bumblebee (Bombus lapidarius L.). Dryad Digital Repository. ( 10.5061/dryad.1hn0951) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Theodorou P, Radzevičiūtė R, Kahnt B, Soro A, Grosse I, Paxton RJ. 2018. Data from: Genome-wide single nucleotide polymorphism scan suggests adaptation to urbanization in an important pollinator, the red-tailed bumblebee (Bombus lapidarius L.). Dryad Digital Repository. ( 10.5061/dryad.1hn0951) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Demultiplexed, raw RAD-Seq reads are available under accession no. SRP099050 at the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/Traces/sra). The variant calling file (VCF) is available on the Dryad Digital Repository at https://doi.org/10.5061/dryad.1hn0951 [69].