Abstract
Two lytic bacteriophages, WCHABP1 and WCHABP12, were recovered from hospital sewage and were able to infect 9 and 12 out of 18 carbapenem-resistant Acinetobacter baumannii clinical strains, which belonged to different clones. Electron microscopy scan showed that both bacteriophages had the similar morphology as those of the Myoviridae family. Whole genomic sequencing revealed 45.4- or 45.8-kb genome with a 37.6% GC content for WCHABP1 and WCHABP12, both of which showed significant DNA sequence similarity with bacteriophages of the Ap22virus genus within the Myoviridae family. Taxonomic analysis was therefore performed following the proposal approved by the International Committee on Taxonomy of Viruses, which confirmed that WCHABP1 and WCHABP12 represented two new species of the Ap22virus genus. No tRNAs but 88 and 89 open reading frames (ORFs) were predicted for the two bacteriophages, among which 22 and 21 had known function and encoded proteins for morphogenesis, packaging, lysis, and nucleiotide metabolism. The C-terminal amino acids of the large unit of fiber tail proteins varied between the bacteriophages, which may explain their different host ranges. For most lytic bacteriophages, a set of holin and endolysin are required for lysis. However, no known holin-encoding genes were identified in WCHABP1 and WCHABP12, suggesting that they may use alternative, yet-to-be-identified, novel holins for host cell membrane lysis. To test the efficacy of the bacteriophages in protecting against A. baumannii infection, a Galleria mellonella larva model was used. Only <20% G. mellonella larvae survived at 96 h after being infected by carbapenem-resistant A. baumannii strains, from which the two bacteriophages were recovered. With the administration of WCHABP1 and WCHABP12, the survival of larvae increased to 75%, while the treatment of polymyxin B only slightly increased the survival rate to 25%. The isolation of two new lytic bacteriophages in this study could expand our sight on Acinetobacter bacteriophages and may offer new potential therapeutic alternatives against A. baumannii.
Keywords: bacteriophage, phage therapy, carbapenem resistance, Acinetobacter baumannii, Galleria mellonella
Introduction
On 27 February 2017, World Health Organization (WHO) released a list of 12 types of antimicrobial-resistant “priority pathogens” that pose the greatest threat to human health (World Health Organization, 2017). In the list, Acinetobacter along with Pseudomonas and various species of the Enterobacteriaceae (e.g., Klebsiella, Escherichia coli, Serratia, and Proteus) are labeled as “Critical,” for which new antimicrobial agents are urgently needed. Acinetobacter baumannii is a non-fermentative, non-motile, catalase-positive, gram-negative bacterium. It is widely dispersed in soil and water as well as the healthcare environment with the ability to cause various infections such as bacteremia and pneumonia (Dijkshoorn et al., 2007; Peleg et al., 2008). In addition to its capability of survival on dry and moist surfaces (Berlau et al., 1999), a variety of intrinsic and acquired mechanisms for antimicrobial resistance and virulence (Di et al., 2011; Perez et al., 2011) have made A. baumannii a successful pathogen worldwide (Perez et al., 2007).
One century ago, since the first day of their discovery, the compelling feature of bacteriophages as a natural enemy of bacteria has drawn attention of early researchers to exploit them as a mean to treat bacterial infections (d'herelle, 1917; Bruynoghe and Maisin, 1921; Helvoort, 2001). However, the successive discovery of penicillin and other antibiotics soon put bacteriophages on the shelf, with exception in the Soviet Union and Poland (Kutter et al., 2015). The emergence of multi-drug resistant (MDR) bacteria calls for alternatives of antibiotics and bacteriophages therefore gain interests for extensive studies again (Smith and Huggins, 1983; Soothill, 1992).
As a potential alternative to antibiotics and a possible solution for impending infections caused by MDR bacteria, there are still constraints for bacteriophages to be widely applied in real clinical practice despite the presence of reported successful cases (Abubakar et al., 2016). The main challenges for such a bacteriophage therapy include that the relatively narrow lytic range against bacterial strains and the proneness of bacteria to develop resistance. To tackle the challenges, multiple lytic bacteriophages have been used in combination as cocktails (Merabishvili et al., 2009). The success of cocktail bacteriophage therapy against predominantly MDR bacteria such as A. baumannii relies on isolation of novel lytic bacteriophages. Here we report two newly isolated lytic bacteriophages named WCHABP1 and WCHABP12 of the Ap22virus genus within the Myoviridae family against carbapenem-resistant A. baumannii clinical isolates.
Methods
Bacterial strains
This study included a total of 18 nonduplicate carbapenem-resistant (MICs of imipenem ≥ 8 mg/L) but polymyxin B-susceptible (MICs <2 mg/L) A. baumannii clinical strains recovered from blood, sputum, urine, ascites, bronchoalveolar lavage fluid (BALF), and drainages of different patients at West China Hospital, Sichuan University, China, from April to June 2016 (Table 1) as hosts for bacteriophage isolation. The species identification of them was established by partially sequencing the recA gene as described previously (Krawczyk et al., 2002). These isolates belonged to 18 different clones as determined using a pulsed field gel electrophoresis (PFGE) analysis (data not shown). Clones were defined for isolates with an 80% or above pattern similarity. Antimicrobial susceptibility testing was performed using the Vitek II automated microbiological system (bioMérieux, Marcy-l'Étoile, France) or broth microdilution method of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2017) (for colistin, imipenem, polymycin B, and tigecycline). Breakpoints defined by U. S. Food and Drug Administration were used for tigecycline; otherwise, those defined by CLSI were applied. The results are shown in Table S1.
Table 1.
Host ranges of WCHABP1 and WCHABP12.
| Strain | Source | MICs (mg/L) | EOP | |||
|---|---|---|---|---|---|---|
| Imipenem | Colistin | Tigecycline | WCHABP1 | WCHABP12 | ||
| Ab1138 | Blood | 64 | 1 | 1 | 0.2 | |
| Ab1186 | Ascites | 64 | 1 | 0.5 | 1+ | 0.8 |
| Ab1262 | Sputum | 64 | 1 | 0.5 | 1+ | |
| Ab1334 | BALF | 16 | 0.5 | 0.5 | ||
| Ab1337 | Sputum | 64 | 1 | 0.5 | 0.6 | |
| Ab1369 | Sputum | 64 | 1 | 0.5 | ||
| Ab1391 | Sputum | 64 | 1 | 0.5 | 0.8 | |
| Ab1397 | Sputum | 64 | 1 | 0.5 | 0.8 | |
| Ab1412 | Ascites | 64 | 1 | 0.5 | 0.8 | |
| Ab1415 | Sputum | 64 | 1 | 0.5 | 0.1 | 0.3 |
| Ab1454 | Urine | 64 | 1 | 0.5 | ||
| Ab1478 | Blood | 64 | 0.5 | 0.5 | 0.1 | |
| Ab1497 | Sputum | 64 | 1 | 0.5 | 0.7 | 0.6 |
| Ab1531 | Sputum | 32 | 1 | 0.5 | 0.8 | 0.6 |
| Ab1585 | Sputum | 16 | 1 | 1 | 0.5 | 0.8 |
| Ab1588 | Ascites | 16 | 0.5 | 1 | ||
| Ab1623 | Blood | 64 | 0.5 | 1 | 0.8 | 0.2 |
| Ab1673 | Blood | 64 | 0.5 | 1 | 0.4 | 0.3 |
The original host strain, from which the bacteriophage was recovered.
Bacteriophage isolation, titering, and concentration
Bacteriophages were isolated from sewage samples collected at the influx of the wastewater treatment plant at West China Hospital in September 2016. The isolation and propagation of bacteriophages were performed as described previously (Merabishvili et al., 2009) and titering was carried out using the agar overlay method (Kropinski et al., 2009). PEG 8000/NaCl precipitation was used to concentrate bacteriophages for further use as described previously (Peng et al., 2014).
Determination of the host range
The bacteriophage stock was diluted 1:106 with Luria-Bertani (LB) broth (Hopebio, Qingdao, China) and was adjusted to a final titer of 1 × 104 pfu/ml. Aliquots of 10 μl diluted bacteriophages were mixed with tested bacterial host strains or the original strains from which the bacteriophages were isolated. The mixtures were subjected to agar overlay method. The efficiency of plating (EOP) was calculated by dividing the bacteriophage titer of tested strain by that of original strain. A particular bacteriophage-bacterial strain combination was considered as high, medium and low production efficiency if the average EOP value was ≥ 0.5, ≥ 0.1 to < 0.5, and ≥ 0.001 to < 0.1, respectively; while a < 0.001 average EOP value suggests inefficient production (Khan and Nilsson, 2015).
Electron microscopy
The concentrated bacteriophage stocks were negatively stained with phosphotungstic acid (PTA) and were then imaged using a H-600 II transmission electron microscope at 75 kV (Hitachi, Tokyo, Japan) to acquire morphological information of single bacteriophage particles.
Multiplicity of infection (MOI) assay
Bacteriophage stocks were diluted by LB broth (Hopebio, Qingdao, China) into 10-fold series. Aliquots of each dilution were mixed with their corresponding host cultures at 108 cfu/ml to result in different ratio (from 10−4 to 10 pfu/cfu) and were incubated at 37°C for 4 h. Bacteriophage progenies of each mixture ratio were titered to determine the highest production as the optimal MOI (Peng et al., 2014).
Adsorption rate assay
Aliquots of bacteriophages were incubated with host strain cultures at the optimal MOI under 37°C. Samples (100 μl) were collected at 0, 3, 6, 9, 12, and 15 min, respectively, and were centrifuged to remove the bacterial cells. The supernatants were titered for determination of unabsorbed bacteriophages at each time interval (Peng et al., 2014).
One-step growth curve
Bacteriophages (10 μl at a titer of 1 × 109 pfu/ml) were mixed with 5 ml of their host strain cultures at about 2 × 107 cfu/ml to reach a MOI of 0.1. The mixture was incubated at 37°C for 10 min to allow the complete adsorption and was then centrifuged at 15,000 g for 1 min to remove the unabsorbed bacteriophage particles by discarding the supernatants. The pellets were resuspended in LB broth and were incubated at 37°C. Aliquots (100 μl) were taken every 10 min from the beginning to 90 min and were titered for free bacteriophages after being centrifuged at 15,000 g for 1 min to remove bacterial cells. An additional 100 μl aliquot was taken at the beginning (0 min). A half of the aliquot (50 μl) was centrifuged and the remaining half was not centrifuged, both of which were titered for bacteriophages. Infected cells were calculated by subtracting the number of free bacteriophages of the centrifuged part from the number of bacteriophages of the un-centrifuged part, then the burst size could be calculated as dividing the maximal progeny counts by the number of infected cells (Peng et al., 2014).
Bacteriophage genome sequencing and bioinformatics analysis
Bacteriophage DNA was prepared using the standard phenol-chloroform extraction (Mandell and Hershey, 1960) after concentration. Whole genome sequencing was performed using the HiSeq 2500 Sequencer (Illumina, San Diego, CA, USA) with 150-bp paired-end (the final coverage was ~200x). De novo assembly was performed using SPAdes v3.10.1 (Bankevich et al., 2012) with auto-cutoff and careful mode. Annotation of the genomic sequence was carried out using Prokka v1.11 (Seemann, 2014) followed by manually confirmation via running BLASTp and PSI-BLAST (http://blast.ncbi.nlm.nih.gov/) (Altschul et al., 1990, 1997) against the non-redundant protein database with a significant E-value of <10−3. For annotated proteins, the conserved protein domains/motif and additional function inference were detected using InterProScan (http://www.ebi.ac.uk/interpro/search/sequence-search) (Quevillon et al., 2005), HMMER (https://www.ebi.ac.uk/Tools/hmmer), and Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd) (Marchlerbauer et al., 2011). The ExPASy server (http://us.expasy.org/tools/protparam.html) was used to predict molecular weight and isoelectric point. Prediction of transmembrane helices was performed using TMHMM 2.0 (Krogh et al., 2001) and signal peptides were screened using SignalP (Petersen et al., 2011). Potential tRNA genes were identified using tRNAscan-SE (Lowe, 1997) and ARAGON (Laslett and Canback, 2004).
The comparative genomic analysis between our bacteriophages and those deposited in GenBank was performed using BLASTn and BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011). The newest proposal of International Committee on Taxonomy of Viruses (ICTV) (https://talk.ictvonline.org/files/ictv_official_taxonomy_updates_since_the_8th_report/m/prokaryote-official/5901) (Adams et al., 2017) was taken as the reference to determine the taxonomy of the bacteriophages. Mauve (http://asap.ahabs.wisc.edu/mauve/) and CoreGenes (Zafar et al., 2002; Mahadevan et al., 2009a,b) were used to compare the genomic organization and core genes for a given group of bacteriophages. Phylogenetic tree for taxonomic analysis was constructed via phylogeny.fr with “One Click mode” using MUSCLE for multiple alignments, PhyML for phylogeny, and Gblocks for eliminating poorly aligned positions and divergent regions (Anisimova and Gascuel, 2006).
Nucleotide accession no. Genome sequences of bacteriophages WCHABP1 and WCHABP12 in the present study have been deposited into GenBank under accession no. KY829116 and KY670595.
Analysis of the genome ends
Genome ends were determined as described previously (Casjens and Gilcrease, 2009). Briefly, 1 μg of each bacteriophage genome DNA was digested with the restriction enzyme HindIII (TaKaRa, Dalian, China). The mixture was heated at 80°C for 15 min to inactivate the digestion and was divided into two equal aliquots. One was rapidly chilled in ice water bath and the other was left to slowly cool to the room temperature. The resulting DNA bands were separated in a 1% agarose gel by electrophoresis. In parallel, 1 μg of Lambda DNA standard digested with the restriction enzyme EcoRV (TaKaRa) was used as control.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis
Bacteriophage particles were boiled in loading buffer (50 mM tris-HCl, 2% Sodium dodecyl sulfate, 0.1% bromophenol blue, 10% glycerol, and 1% β-mercaptoethanol) for 5 min. Denatured proteins were then separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) with a 5% concentration gel and a 12% separation gel and were stained with Coomassie Blue G-250 as described previously (Boulanger, 2009).
Galleria mellonella bacteriophage therapy assay
G. mellonella infected by A. baumannii was used to study in vivo antibacterial efficacy of our bacteriophages (Peleg and Jara, 2009). Larvae of G. mellonella of 250–350 mg in weight (Huiyude, Tianjin, China) were stored at 4°C and were used within 1 week after delivery. Briefly, 16 randomly chosen larvae were used for each group. Host bacterial cultures of strain Ab1186 and Ab1262, from which the two bacteriophages were recovered, were washed by phosphate-buffered saline (PBS; Beyotime, Shanghai, China) and were then diluted 1:10 to an appropriate cell density (107 cfu/ml) as determined using a McFarland turbidimetry and direct plate colony counting. Aliquots of 10 μl PBS-washed bacterial cultures (105 cfu) were injected into the hemocoel of each larva through the last left proleg using a microsyringe (Gaoge, Shanghai, China), which led to a pre-determined LD80 dose for each inoculum (105 cfu/larva). Bacteriophage stocks were re-concentrated as described above and were re-suspended and diluted with PBS to the optimal MOI titer (for host cell at 105 cfu/ml, the bacteriophage dosage was 104 pfu/ml to reach a final MOI of 0.1 pfu/cfu). Aliquots of 10 μl bacteriophage suspensions (104 pfu/larva) were then injected into the larva via a different proleg within 30 min from the bacterial inoculation. In parallel, larvae injected with bacterial cells at the same concentration were treated with polymyxin B (Meilun, Dalian, China) at 2.5 mg/kg (Hornsey and Wareham, 2011) to compare the therapeutic efficacy between antibiotics and bacteriophages. For each bacteriophage, four control groups were set up including larvae infected with bacterial cells and treated with PBS solution to observe the bacterial virulence (group 1), those injected with either the bacteriophage suspension or polymyxin B to assess their potential toxicity (group 2 and 3); those injected with PBS only to observe the potential physical trauma from injection (group 4). Larvae were then incubated in plastic containers at 37°C and the number of dead, determining as no movement under touch, was counted at 24 h intervals up to 96 h after the incubation. When more than two larvae died in any negative control (group 2, 3, and 4) at the end of observation, the assay was considered invalid and was repeated. Assays were performed in triplicate using different batches of larvae.
The statistical analysis was performed by GraphPad Prism v.7.0 (Software Inc., La Jolla, CA, USA) to plot the survival curves with the Kaplan-Meier method following a log-rank test to calculate the differences in survival. A P-value of < 0.05 was considered to be statistically significant.
Results
Two bacteriophages of the Myoviridae family were recovered
Two bacteriophages, designated WCHABP1 and WCHABP12, were obtained from hospital sewage with the ability to infect 9 and 12 out of the 18 host strains, exhibiting that both bacteriophages can infect multiple strains of A. baumannii with varied efficiency (Table 1). Upon infecting their original host strain, clear plaques of 8–9 and 5–7 mm in diameter with all surrounded by a 1–3 mm halo were formed by bacteriophage WCHABP1 and WCHABP12, respectively.
Both bacteriophages produced the highest progeny at MOI of 0.1 (1.55 × 1010 pfu/ml for WCHABP1 and 2.2 × 1010 for WCHABP12), which was therefore used in subsequent experiments. The adsorption rate of them was comparable where more than 99% of bacteriophage particles could adsorb onto the host cell within 10 min (Figure 1). After the adsorption, for both WCHABP1 and WCHABP12, the eclipse period was between 10 and 20 min and the plateau was reached at 60 min with a slightly different final titer (1.5 × 109 pfu/ml for WCHABP1 and 2.1 × 109 pfu/ml for WCHABP1; Figure 1). Considering that the initial number of infected cells were 1.1 × 107 cfu/ml for WCHABP1 and 1.2 × 107 cfu/ml for WCHABP12, the burst size of the two bacteriophages was 136 and 175 pfu per infected cell, respectively.
Figure 1.
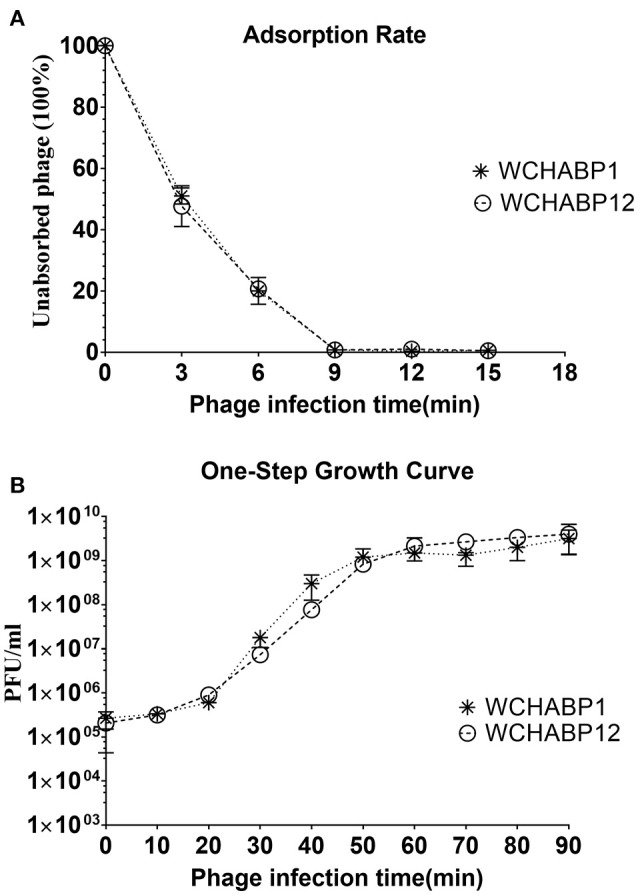
Growth and lytic characteristics. (A) adsorption rate; (B) One-step growth curve. WCHABP1 and WCHABP12 are labeled by asterisk and circle, respectively.
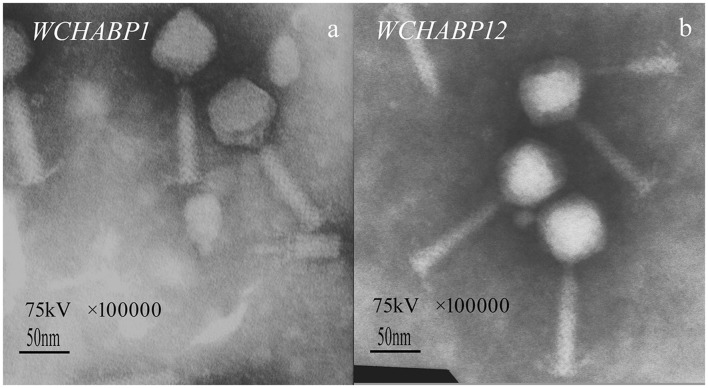
The electron microscopy revealed that both WCHABP1 and WCHABP12 had a ~75 nm icosahedral head and a ~105 nm tail, on which crossed striation could be seen (Figure 2). The morphologic features suggested that both WCHABP1 and WCHABP12 belong to the Myoviridae family.
Figure 2.
Electron microscopy of bacteriophage morphology. (a) WCHABP1; (b) WCHABP12. The black scale bar represents 50 nm.
The two bacteriophages represent two new species of the Ap22virus genus within the Myoviridae family
Genomes of the WCHABP1 and WCHABP12 were 45,888 and 45,415 bp in length with the same GC content of 37.6 mol%. The number of ORFs predicted was 88 for WCHABP1 and 89 for WCHABP12, respectively. Genomes of WCHABP1 and WCHABP12 were highly similar with each other (88.0% overall DNA sequence identity) and had 51.0 to 56.0% overall DNA sequence identity to a set of bacteriophages (Table 2) belonging to the Ap22virus genus of the Myoviridae family.
Table 2.
Genomic properties of WCHABP1 and WCHABP12 compared with other bacteriophages of the Ap22virus genus.
| Bacteriophage | GenBank accession no. | Genome length (kb) | GC content (%) | Overall DNA sequence identity (%)* | Homologous proteins (%)* | No. of CDS | No. of tRNAs |
|---|---|---|---|---|---|---|---|
| AP22 | HE806280 | 46.3 | 37.7 | – | – | 89 | 0 |
| IME-AB2 | JX976549 | 46.6 | 37.5 | 49.0 | 64.0 | 82 | 0 |
| AB1 | HM368260 | 45.1 | 37.7 | 56.0 | 61.8 | 85 | 0 |
| YMC-13-01-C62 | KJ817802 | 44.8 | 37.6 | 53.0 | 65.4 | 84 | 0 |
| WCHABP1 | KY829116 | 45.8 | 37.6 | 58.0 | 74.2 | 89 | 0 |
| WCHABP12 | KY670595 | 45.4 | 37.6 | 62.0 | 75.3 | 88 | 0 |
Bacteriophage AP22 was used as the reference for determining overall DNA sequence identity and homologous proteins
To investigate the taxonomy of WCHABP1 and WCHABP12, we followed the approach of the recently approved taxonomic proposal for the Ap22virus genus (Adams et al., 2017). Bacteriophages of this genus possess genome of ~45 kb (around 37.6 mol% G+C content) and encode ORFs close to 85 in number with no tRNAs. Members of the Ap22virus genus had an average of 53% overall DNA sequence identity and 64% homologous proteins compared with the reference species bacteriophage AP22. WCHABP1 and WCHABP12 exhibited a higher overall DNA sequence identity (58 and 62%) and homologous proteins (74 and 75%) with AP22, carrying almost the same number of ORFs as AP22 (88 and 89 vs. 89). The multiple genome alignment of WCHABP1, WCHABP12, and bacteriophages of the Ap22virus genus showed that all of these genomes were in a resembling gene cluster and order (Figure S1). Phylogenetic trees constructed with bacteriophage major capsid and baseplate J-like protein revealed that both WCHABP1 and WCHABP12 were clustered together within the Ap22virus genus (Figure S2). All above findings suggest that WCHABP1 and WCHABP12 belonged to the Ap22virus genus. In addition, WCHABP1 and WCHABP12 had <95.0% identity between each other and with any other bacteriophages of the Ap22virus genus. Therefore, WCHABP1 and WCHABP12 present two new species of the Ap22virus genus.
Genomes of the two bacteriophages encode proteins for morphogenesis, genome packaging, nucleotide metabolism, and host lysis
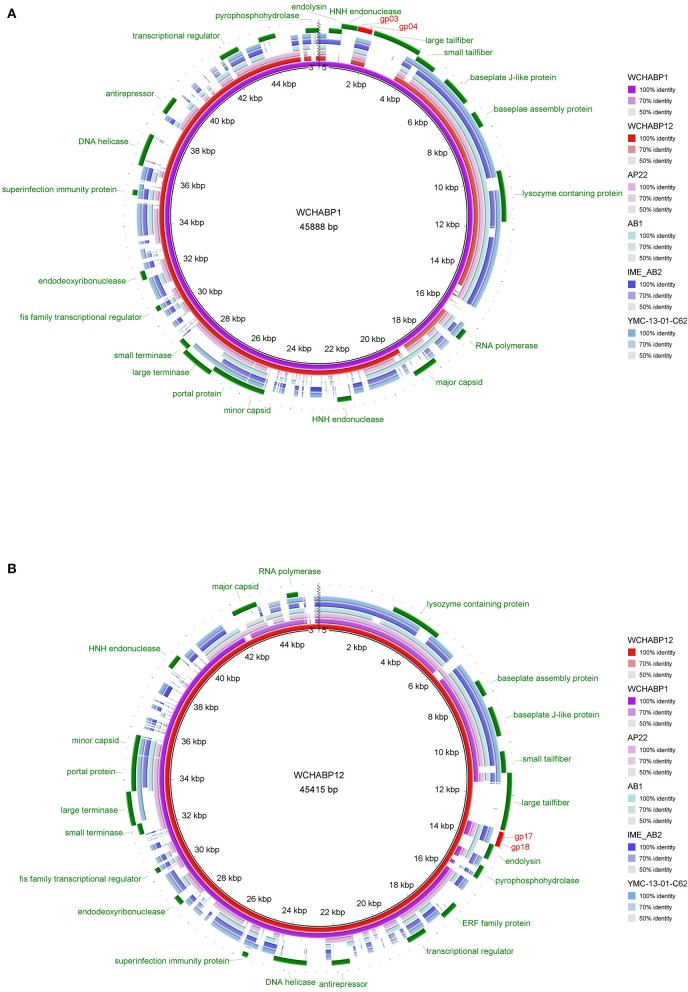
Among ORFs of WCHABP1 and WCHABP1, 22 (WCHABP1) and 21 (WCHABP12) could be assigned to known function including morphogenesis, genome packaging, nucleotide metabolism, and host lysis. Neither tRNAs nor genes involving in the lysogenic process such as integrase and repressor were identified in both bacteriophages (Table 2, Figure 3).
Figure 3.
Multiple genome alignments of WCHABP1, WCHABP12, and bacteriophages of the Ap22virus genus. (A) the genome of WCHABP1 is used as the reference. (B) the genome of WCHABP12 is used as the reference. The alignment is a pairwise BLASTn performed using BRIG. Accession numbers of genomes of bacteriophages of the Ap22virus genus are listed in Table 2.
Morphology module
The SDS-PAGE analysis showed similar protein patterns between WCHABP1 and WCHABP12 (Figure S3). The protein bands mainly converged at 15, 35, 40–55, and 70 kDa, corresponding to the respective capsid, portal and tail fiber proteins of the two bacteriophages in light of the predicted molecular weight.
Bioinformatics analysis predicted seven ORFs encoding structural proteins for the two bacteriophages. These structural proteins included two capsid proteins (gp27 and gp43 of WCHABP1; gp66 and gp82 of WCHABP12), one portal protein (gp44 of WCHABP1; gp65 of WCHABP12), two baseplate proteins (gp08 and gp10 of WCHABP1; gp11 and gp13 of WCHABP12), and two tail fiber proteins (gp05 and gp06 of WCHABP1; gp15 and gp16 of WCHABP12). The structural proteins encoded by each of the two bacteriophage genomes are highly similar (>60% overall amino acids identity, a detailed list is shown in Table 3) with the exception of the large tail fiber subunits (gp05 of WCHABP1 and gp16 of WCHABP12). The N-terminal 149 amino acids of the large tail fiber subunit of WCHABP1 and WCHABP12 were of 100% coverage and 76% identity, while the remaining C-terminal amino acids had no significant similarity.
Table 3.
Genes with known function in bacteriophage WCHABP1 and WCHABP12.
| WCHABP1 | Predicated function | WCHABP12 | Coverage, identity (%, amino acid sequence) | ||||
|---|---|---|---|---|---|---|---|
| ORFs | Location | Length (bp) | ORFs | Location | Length (bp) | ||
| gp05 | 2148–4052 | 1905 | Large tail fiber subunit | gp16 | 11145–13343 | 2199 | 41, 76 |
| gp06 | 4054–4899 | 846 | Small tail fiber subunit | gp15 | 10298–11143 | 846 | 100, 95 |
| gp08 | 5503–6687 | 1185 | Baseplate J-like protein | gp13 | 8510–9694 | 1185 | 100, 99 |
| gp10 | 7183–7827 | 645 | Baseplate assembly protein | gp11 | 7367–8014 | 648 | 100, 94 |
| gp14 | 9691–11721 | 2031 | Lysozyme domain containing protein | gp06 | 3007–5037 | 2031 | 100, 94 |
| gp23 | 16332–16769 | 438 | RNA polymerase | gp86 | 44161–44598 | 438 | 100, 93 |
| gp27 | 17988–18980 | 993 | Major capsid | gp82 | 41925–42932 | 1008 | 98, 60 |
| gp33 | 21655–22212 | 558 | HNH endonuclease | gp76 | 38683–39240 | 558 | 100, 100 |
| gp43 | 25134–25904 | 771 | Minor capsid | gp66 | 34991–35761 | 771 | 100, 99 |
| gp44 | 25907–27337 | 1431 | Portal protein | gp65 | 33558–34988 | 1431 | 100, 100 |
| gp45 | 27340–28641 | 1302 | Large terminase subunit | gp64 | 32254–33555 | 1302 | 100, 100 |
| gp46 | 28622–29053 | 432 | Small terminase subunit | gp63 | 31842–32273 | 432 | 100, 100 |
| gp52 | 30486–30686 | 201 | Fis family transcriptional regulator | gp57 | 30209–30409 | 201 | 100, 100 |
| gp56 | 31829–32191 | 363 | Endodeoxyribonuclease | gp53 | 28704–29066 | 363 | 100, 100 |
| gp64 | 35219–35431 | 213 | Superinfection immunity protein | gp45 | 25464–25676 | 213 | 100, 100 |
| gp69 | 36413–37714 | 1302 | DNA helicase | gp40 | 23181–24482 | 1302 | 100, 100 |
| gp72 | 38691–39404 | 714 | Antirepressor protein | gp37 | 21491–22204 | 714 | 100, 100 |
| gp80 | 41808–42599 | 792 | Transcriptional regulator | gp29 | 18296–19087 | 792 | 100, 100 |
| gp84 | 43473–44153 | 681 | ERF family protein | gp25 | 16742–17422 | 681 | 100, 100 |
| gp89 | 45367–45888 | 522 | Nucleoside triphosphate pyrophosphohydrolase | gp21 | 14909–15442 | 534 | 97, 63 |
| gp01 | 396–911 | 501 | Endolysin | gp19 | 13985–14593 | 609 | – |
| gp02 | 873–1484 | 612 | HNH endonuclease | – | |||
In addition, both WCHABP1 and WCHABP12 had a gene encoding a lysozyme-containing protein, which is also possessed by other bacteriophages of the Ap22virus genus. The lysozyme is a glycoside hydrolase that catalyzes the peptidoglycan of the bacterial cell wall (Strynadka and James, 1996). The lysozyme-containing protein lies close to the baseplate protein and tail fiber proteins, suggesting that it might be a part of tail. Lysozyme could be released from the inner tail tube to create a small hole in the cell wall peptidoglycan layer that allows the double-stranded DNA (dsDNA) genome to enter the cell (Duda, 2008). Nonetheless, the exact function of the lysozyme-containing protein in the bacteriophages warrants further studies.
Packaging module and genome termini
Two ORFs encoding terminase, which is the key component of the packaging machine, were identified in both bacteriophage genomes (gp45 and gp46 of WCHABP1; gp63 and gp64 of WCHABP12). Terminase for bacteriophages usually comprises two subunits, where the large subunit translocates bacteriophage DNA into empty capsids and cuts it at a unique and precise dsDNA sequence to accomplish the packaging process, while the small subunit contributes to binding of the packaging initiation site and regulates the ATPase activity for the large subunit (Feiss and Rao, 2012). Amino acid sequences of terminase subunits between WCHABP1 and WCHABP12 were identical (Table 3). The restriction map of genomes of WCHABP1 and WCHABP12 showed no change in band patterns after endonuclease restriction with rapid or slow cooling, suggesting the absence of cohesive ends in their genomes (Figure S4).
Nucleotide metabolism module
Both WCHABP1 and WCHABP12 had 10 genes involving in genome replication, transcription, establishment of infection, and other processes, including a RNA polymerase, a DNA helicase, a transcriptional regulator, a fis family transcriptional regulator, an ERF (essential recombination function) family protein, a nucleoside triphosphate pyrophosphohydrolase, a HNH endonuclease, an endodeoxyribonuclease, a superinfection immunity protein and a bacteriophage antirepressor protein (Table 2). Products of these genes were highly similar (93–100% overall amino acids identity) between the two bacteriophages except for the nucleotide triphosphate pyrophosphohydrolase (gp89 of WCHABP1 and gp21 of WCHABP12) with a 97% coverage but only 63% amino acid identity. Both gp89 of WCHABP1 and gp21 of WCHABP12 contained an NTP-PPase_u3 domain (also known as MazG domain, CCD accession cd11540), suggesting a similar function. Previous study suggests that the MazG domain-containing proteins might contribute to extending the logarithmic phase of bacterial growth, facilitating the production of bacteriophage progeny via the reactivation of metabolic pathways that are usually suppressed under nutrient starvation (Bryan et al., 2008).
Besides the identical HNH endonuclease seen in both WCHABP1 (gp33) and WCHABP12 (gp76), WCHABP1 had another HNH endonuclease (gp02), which is absent from WCHABP12 and any other members of the Ap22virus genus and has a relatively low similarity (31% coverage and 31% identity) to the aforementioned HNH endonuclease of WCHABP1 and WCHABP12. The gp02 product of WCHABP1 consisted of a HNH endonuclease domain (Pfam accession PF13392.5) and an AP2 domain (Pfam accession PF00847.19), while the gp33 product of WCHABP1 and gp76 product of WCHABP12 contained a HNH endonuclease domain (Pfam accession PF13392.5) and a NUMOD4 motif (Pfam accession PF07463.10). The HNH endonuclease domain is involved in the process of homing, while both AP2 domain and NUMOD4 motif play a role in DNA binding (Ohmetakagi and Shinshi, 1995; Sitbon and Pietrokovski, 2003) but without significant amino acid similarity.
Lysis control
WCHABP1 and WCHABP12 had one gene encoding the endolysin. However, endolysins of the two bacteriophages had no significant similarity in amino acid sequences. The endolysin of WCHABP1 (gp01) had 171 amino acids and contains a glycosyl hydrolase 108 family domain (Pfam accession PF05838.11) with catalytic activity as a N-acetylmuramidase (Stojković and Rothman-Denes, 2007) and a peptidoglycan domain for substrate binding (Pfam accession PF09374.9). By contrast, the endolysin of WCHABP12 (gp19) had 202 amino acids with a single domain belonging to the glycoside hydrolase family 19 (Pfam accession PF00182.18), which is able to hydrolyze the β-1,4-N-acetyl-D-glucosamine linkages in chitin polymers and leads to the breakage of chitin-containing cell walls (Eijsink et al., 2008). The different types of endolysins between WCHABP1 and WCHABP12 suggest that even closely-related bacteriophages may target different substrates of the host cell wall.
Most dsDNA bacteriophages with a genome size larger than 10 kb employ a complete set of a holin-endolysin system to release the phage progeny (Young and White, 2008). The holin first permeabilizes the host cell membrane and endolysin can therefore reach the periplasm to break down the peptidoglycan layer (Young et al., 2000). However, for both WCHABP1 and WCHABP12 no known holin-encoding genes were identified using both a BLASTp search and domain/motif prediction.
In vivo therapeutic efficacy
For larvae infected by carbapenem-resistant A. baumannii strains, the treatment of WCHABP1 and WCHABP12 significantly improved the survival rates (Figure 4 and Table S2). For those infected by strain Ab1186, survival of larvae at 96 h was 75.00 and 18.75% (p < 0.01) with and without the treatment of WCHABP1, respectively. Similarly, for those infected by strain Ab1262, survival was 75 and 12.5% (p < 0.01) with and without the treatment of WCHABP12, respectively. By contrast, the treatment of polymyxin B only slightly increased the survival, which is not statistically significant. With and without the treatment of polymyxin B, the survival rate of those infected by strain Ab1186 was 25 and 18.75% (p = 0.46), respectively, and that of those infected by Ab1262 was 25 and 12.5% (p = 0.47), respectively. The treatment of the combination of bacteriophage and polymyxin B did not further enhance the survival of larvae infected by strain Ab1186 or Ab1262 compared with the treatment of bacteriophage alone (68.75 vs. 75%, p = 0.687).
Figure 4.
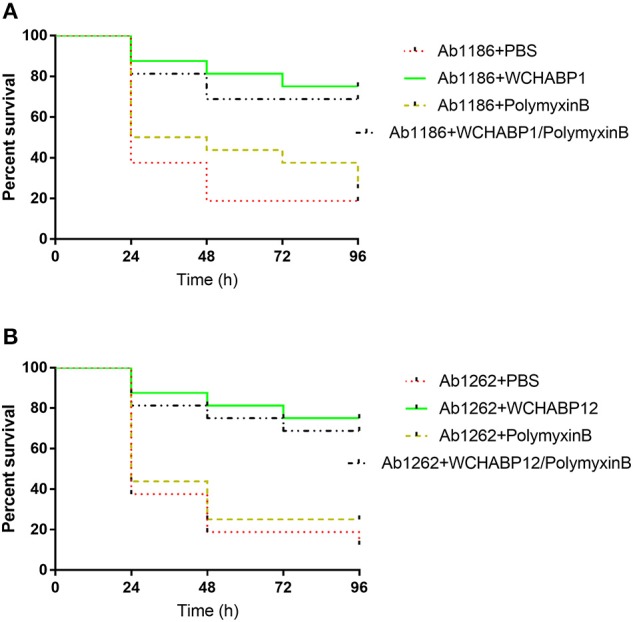
Survival of G. mellonella larvae after infection by carbapenem-resistant A. baumannii clinical strains. (A) WCHABP1; (B) WCHABP12. Ab1186 and Ab1262 are two carbapenem-resistant A. baumannii clinical strains, from which bacteriophages WCHABP1, WCHABP12 were recovered. The detailed survival rates are listed in Table S2.
Discussion
The recent success in treating the infection caused by MDR A. baumannii by topical and systematic administration of varied bacteriophages together with antibiotics once again highlights the potential of bacteriophages as promising alternatives against bacterial infections that are difficult to be treated otherwise (https://www.sciencedaily.com/releases/2017/04/170425124826.htm). Nonetheless, constant works on isolation and characterization of new bacteriophages as well as their in vitro and in vivo test for both safety and therapeutic efficacy are still imperative. In this study, we reported two bacteriophages, WCHABP1 and WCHABP12, belonging to the Ap22virus genus of the Myoviridae family with a large burst size and short lytic life cycle against the carbapenem-resistant A. baumannii host strains. The bacteriophages identified in the present study together with others discovered in previous studies (Lee et al., 2011; Jin et al., 2012; Popova et al., 2012; Ghajavand et al., 2017) may expand our arsenal against A. baumannii.
The injection of bacteriophages WCHABP1 and WCHABP12 did not cause any death of larvae, suggesting that they were not toxic for the larvae. The administration of WCHABP1 and WCHABP12 was able to rescue most larvae infected by carbapenem-resistant A. baumannii susceptible to the lysis of the bacteriophages, suggesting both bacteriophages new potent weapons against A. baumannii. By contrast, the treatment of polymyxin B alone was only able to rescue few larvae infected by these strains that were susceptible to the antibiotic, which is consistent with the findings of another study (Hornsey et al., 2013). A few factors may contribute to the failure of polymyxin B treatment such as emerging tolerance and resistance to the antibiotic after exposure (Li et al., 2006; Harding et al., 2018).
For products encoded by the genomes of WCHABP1, WCHABP12, and other members of the Ap22virus genus, there are several noteworthy features. First, the C-terminal amino acid sequence of the large tail fiber subunit had no significant similarity between WCHABP1 and WCHABP12. Such highly variable C-terminal sequences of large tail fiber subunits are also present among other members of the Ap22virus genus. Previous studies have revealed that the large tail fiber subunit of various bacteriophages including those belonging to the Myoviridae family has a conserved N-terminus and a variable C-terminus, which determines the host specificity by binding the receptor on the surface of host cell (Rakhuba et al., 2010; Kurtböke, 2012). The variable C-terminal sequence of large tail fiber subunits may therefore account for the different host ranges of bacteriophages of the Ap22virus genus. Second, like those of other bacteriophages of the Ap22virus genus, the large terminase subunit of WCHABP1 and WCHABP12 had a terminase_3 family domain (Pfam accession pfam04466) and were distantly homologous (26% identity with a E-value of 2e-22) with that of bacteriophage SPP1 (GenBank accession NC_004166), which is a well-studied model system for headful DNA packaging mechanism (also known as pac-mechanism; Oliveira et al., 2013). This suggests that both WCHABP1 and WCHABP12 also employ a pac-mechanism for genome packaging and their genomes are circularly permuted and terminally redundant (Casjens and Gilcrease, 2009; Oliveira et al., 2013). Third, surprisingly, unlike the typical holin-endolysin set seen in most bacteriophages, no known holin-encoding genes were identified for bacteriophages of the Ap22virus genus. Two ORFs (gp3 and gp4 of WCHABP1; gp17 and gp18 of WCHABP12) were present between genes encoding endolysin and the large tail fiber subunit and were highly similar among members of the Ap22virus genus. The products encoded by the two ORFs had two and three transmembrane domains as identified using TMHMM. As most holins contain transmembrane domains (Young et al., 2000) and their encoding genes usually locate proximately to endolysin-encoding genes, it is reasonable to hypothesize that the two ORFs may encode a new, yet-to-be-characterized holin. Alternatively, as shown in a previous study there are means other than holin for bacteriophages to permeabilize the host cell membrane via inhibiting the specific host enzymes and impairing peptidoglycan biosynthesis (Bernhardt et al., 2001). Therefore, the exact lysis mechanism employed by bacteriophages of the Ap22virus genus remains unclear and warrants further studies.
Although genomes of bacteriophages WCHABP1 and WCHABP12 were highly similar, there were several differences, which have been demonstrated above and are summarized here. Endolysins of WCHABP1 and WCHABP12 had no significant similarity in amino acid sequences. WCHABP1 had one more HNH endonuclease than WCHABP12. The C-terminal amino acids of the large unit of tail fiber protein between the two bacteriophages had no significant similarity. In addition, WCHABP1 had three genes without known function that were absent from WCHABP12, while two genes with unknown function of WCHABP12 were not present in WCHABP1.
In conclusion, two new bacteriophages, exhibiting activity to infect and kill multiple carbapenem-resistant A. baumannii clinical strains, were recovered. The two bacteriophages represent two new species of the Ap22virus genus within the Myoviridae family. Administration of the two bacteriophages was effective to treat carbapenem-resistant A. baumannii infections in the G. mellonella larvae model. The findings could expand our sight on Acinetobacter bacteriophages and offer new potential therapeutic alternatives.
Author contributions
ZZ designed the study. WZ performed the experiments and prepared figures. WZ, YF, and ZZ analyzed and interpreted the data. WZ and ZZ wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (project no. 81222025, 81572030, and 81661130159) and the Newton Advanced Fellowship, Royal Society, UK (NA150363).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00850/full#supplementary-material
References
- Abubakar S., Suleiman B. H., Abbagana B. A., Mustafa I. A., Musa I. A. (2016). Novel uses of bacteriophages in the treatment of human infections and antibiotic resistance. Am. J. Biosci. 4, 34–40. 10.11648/j.ajbio.20160403.13 [DOI] [Google Scholar]
- Adams M. J., Lefkowitz E. J., King A. M. Q., Harrach B., Harrison R. L., Knowles N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 162, 1–34. 10.1007/s00705-017-3358-5 [DOI] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlau J., Aucken H. M., Houang E., Pitt T. L. (1999). Isolation of Acinetobacter spp. including A. baumannii from vegetables: implications for hospital-acquired infections. J. Hosp. Infect. 42, 201–204. 10.1053/jhin.1999.0602 [DOI] [PubMed] [Google Scholar]
- Bernhardt T. G., Wang I. N., Struck D. K., Young R. (2001). A protein antibiotic in the phage Qβ virion: diversity in lysis targets. Science 292, 2326–2329. 10.1126/science.1058289 [DOI] [PubMed] [Google Scholar]
- Boulanger P. (2009). Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. Methods Mol. Biol. 502, 227. 10.1007/978-1-60327-565-1_13 [DOI] [PubMed] [Google Scholar]
- Bruynoghe R., Maisin J. (1921). Essais de thérapeutique au moyen du bacteriophage. CR Soc. Biol. 85, 1120–1121. [Google Scholar]
- Bryan M. J., Burroughs N. J., Spence E. M., Clokie M. R. J., Mann N. H., Bryan S. J. (2008). Evidence for the intense exchange of mazG in marine cyanophages by horizontal gene transfer. PLoS ONE 3:e2048. 10.1371/journal.pone.0002048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. R., Gilcrease E. B. (2009). Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502, 91–111. 10.1007/978-1-60327-565-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S27. (Wayne, PA: Clinical and Laboratory Standards Institute; ). [Google Scholar]
- d'herelle F. (1917). Sur un microbe invisible antagoniste des bacilles dysentériques. CR Acad. Sci. Paris 165, 373–375. [Google Scholar]
- Di P. A., Giannouli M., Triassi M., Brisse S., Zarrilli R. (2011). Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin. Microb. Infect. 17, 197–201. 10.1111/j.1469-0691.2010.03254.x [DOI] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5, 939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Duda R. L. (2008). Icosahedral tailed dsDNA bacterial viruses. Encycl. Virol. 30–37. 10.1016/B978-012374410-4.00754-8 [DOI] [Google Scholar]
- Eijsink V. G. H., Vaaje-Kolstad G., Vårum K. M., Horn S. J. (2008). Towards new enzymes for biofuels: lessons from chitinase research. Trends Biotechnol. 26, 228–235. 10.1016/j.tibtech.2008.02.004 [DOI] [PubMed] [Google Scholar]
- Feiss M., Rao V. B. (2012). The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 726, 489–509. 10.1007/978-1-4614-0980-9_22 [DOI] [PubMed] [Google Scholar]
- Ghajavand H., Esfahani B. N., Havaei A., Fazeli H., Jafari R., Moghim S. (2017). Isolation of bacteriophages against multidrug resistant Acinetobacter baumannii. Res. Pharm. Sci. 12, 373–380. 10.4103/1735-5362.213982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C. M., Hennon S. W., Feldman M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. 10.1038/nrmicro.2017.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helvoort T. V. (2001). Felix d'Herelle and the origins of molecular biology. J. Hist. Med. Allied Sci. 56, 305–307. 10.1093/jhmas/56.3.305 [DOI] [Google Scholar]
- Hornsey M., Wareham D. W. (2011). In vivo efficacy of glycopeptide-colistin combination therapies in a Galleria mellonella model of Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 55, 3534–3537. 10.1128/AAC.00230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsey M., Phee L., Longshaw C., Wareham D. W. (2013). In vivo efficacy of telavancin/colistin combination therapy in a Galleria mellonella model of Acinetobacter baumannii infection. Int. J. Antimicrob. Agents 41, 285–287. 10.1016/j.ijantimicag.2012.11.013 [DOI] [PubMed] [Google Scholar]
- Jin J., Li Z. J., Wang S. W., Wang S. M., Huang D. H., Li Y. H., et al. (2012). Isolation and characterization of ZZ1, a novel lytic phage that infects Acinetobacter baumannii clinical isolates. BMC Microbiol. 12:156. 10.1186/1471-2180-12-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. M., Nilsson A. S. (2015). Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 10:e0118557 10.1371/journal.pone.0118557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk B., Lewandowski K., Kur J. (2002). Comparative studies of the Acinetobacter genus and the species identification method based on the recA sequences. Mol. Cell. Probes 16, 1–11. 10.1006/mcpr.2001.0388 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kropinski A. M., Mazzocco A., Waddell T. E., Lingohr E., Johnson R. P. (2009). Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 501, 69–76. 10.1007/978-1-60327-164-6_7 [DOI] [PubMed] [Google Scholar]
- Kurtböke I. (2012). Bacteriophages. Rijeka: InTech; 10.5772/1065 [DOI] [Google Scholar]
- Kutter E. M., Kuhl S. J., Abedon S. T. (2015). Re-establishing a place for phage therapy in western medicine. Future Microbiol. 10, 685–688. 10.2217/fmb.15.28 [DOI] [PubMed] [Google Scholar]
- Laslett D., Canback B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16. 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. N., Tseng T. T., Lin J. W., Fu Y. C., Weng S. F., Tseng Y. H. (2011). Lytic myophage Abp53 encodes several proteins similar to those encoded by host Acinetobacter baumannii and phage phiKO2. Appl. Environ. Microbiol. 77, 6755–6762. 10.1128/AEM.05116-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rayner C. R., Nation R. L., Owen R. J., Spelman D., Tan K. E., et al. (2006). Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 50, 2946–2950. 10.1128/AAC.00103-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. (1997). tRNAscan-SE: a program for improved transfer RNA detection in genomic sequence (release: 1.23). Nucleic Acids Res. 25, 955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan P., King J. F., Seto D. (2009a). CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res. Notes 2:168. 10.1186/1756-0500-2-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan P., King J. F., Seto D. (2009b). Data mining pathogen genomes using geneorder and coregenes and CGUG: gene order, synteny and in silico proteomes. Int. J. Comput. Biol. Drug Des. 2, 100–114. 10.1504/IJCBDD.2009.027586 [DOI] [PubMed] [Google Scholar]
- Mandell J. D., Hershey A. D. (1960). A fractionating column for analysis of nucleic acids. Anal. Biochem. 1, 66–77. 10.1016/0003-2697(60)90020-8 [DOI] [PubMed] [Google Scholar]
- Marchlerbauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., Deweesescott C., et al. (2011). CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabishvili M., Pirnay J.-P., Verbeken G., Chanishvili N., Tediashvili M., Lashkhi N., et al. (2009). Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 4:e4944. 10.1371/journal.pone.0004944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmetakagi M., Shinshi H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. 10.1105/tpc.7.2.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L., Tavares P., Alonso J. C. (2013). Headful DNA packaging: bacteriophage SPP1 as a model system. Virus Res. 173, 247–259. 10.1016/j.virusres.2013.01.021 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Jara S. D. (2009). Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob. Agents Chemother. 53, 2605–2609. 10.1128/AAC.01533-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F., Mi Z., Huang Y., Yuan X., Niu W., Wang Y., et al. (2014). Characterization, sequencing and comparative genomic analysis of vB_AbaM-IME-AB2, a novel lytic bacteriophage that infects multidrug-resistant Acinetobacter baumannii clinical isolates. BMC Microbiol. 14:181. 10.1186/1471-2180-14-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Hujer A. M., Hujer K. M., Decker B. K., Rather P. N., Bonomo R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51, 3471–3484. 10.1128/AAC.01464-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F., Ponceterashima R., Adams M. D., Bonomo R. A. (2011). Are we closing in on an “elusive enemy”? The current status of our battle with Acinetobacter baumannii. Virulence 2, 86–90. 10.4161/viru.2.2.15748 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., Von H. G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Popova A. V., Zhilenkov E. L., Myakinina V. P., Krasilnikova V. M., Volozhantsev N. V. (2012). Isolation and characterization of wide host range lytic bacteriophage AP22 infecting Acinetobacter baumannii. FEMS Microbiol. Lett. 332, 40–46. 10.1111/j.1574-6968.2012.02573.x [DOI] [PubMed] [Google Scholar]
- Quevillon E., Silventoinen V., Pillai S., Harte N., Mulder N., Apweiler R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhuba D. V., Kolomiets E. I., Dey E. S., Novik G. I. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59, 145–155. 10.1016/j.micres.2015.01.008.1.94 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinform 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sitbon E., Pietrokovski S. (2003). New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem. Sci. 28, 473–477. 10.1016/S0968-0004(03)00170-1 [DOI] [PubMed] [Google Scholar]
- Smith H. W., Huggins M. B. (1983). Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J. Gen. Microbiol. 129:2659. 10.1099/00221287-129-8-2659 [DOI] [PubMed] [Google Scholar]
- Soothill J. S. (1992). Treatment of experimental infections of mice with bacteriophages. J. Med. Microbiol. 37, 258–261. 10.1099/00222615-37-4-258 [DOI] [PubMed] [Google Scholar]
- Stojković E. A., Rothman-Denes L. B. (2007). Coliphage N4 N-acetylmuramidase defines a new family of murein hydrolases. J. Mol. Biol. 366, 406–419. 10.1016/j.jmb.2006.11.028 [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. (1996). Lysozyme: a model enzyme in protein crystallography. EXS 75, 185–222. 10.1007/978-3-0348-9225-4_11 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2017). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization. [Google Scholar]
- Young I., Wang I., Roof W. D. (2000). Phages will out: strategies of host cell lysis. Trends Microbiol. 8, 120–128. 10.1016/S0966-842X(00)01705-4 [DOI] [PubMed] [Google Scholar]
- Young R. F., III., White R. L. (2008). Lysis of the host by bacteriophage. Encycl. Virol. 248–258. 10.1016/B978-012374410-4.00752-4 [DOI] [Google Scholar]
- Zafar N., Mazumder R., Seto D. (2002). Coregenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinform. 3:12. 10.1186/1471-2105-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.