Abstract
Canonical K+ channels are tetrameric and highly K+-selective, whereas two-pore–domain K+ (K2P) channels form dimers, but with a similar pore architecture. A two-pore–domain potassium channel TWIK1 (KCNK1 or K2P1) allows permeation of Na+ and other monovalent ions, resulting mainly from the presence of Thr-118 in the P1 domain. However, the mechanistic basis for this reduced selectivity is unclear. Using ion-exchange–induced difference IR spectroscopy, we analyzed WT TWIK1 and T118I (highly K+-selective) and L228F (substitution in the P2 domain) TWIK1 variants and found that in the presence of K+ ions, WT and both variants exhibit an amide-I band at 1680 cm−1. This band corresponds to interactions of the backbone carbonyls in the selectivity filter with K+, a feature very similar to that of the canonical K+ channel KcsA. Computational analysis indicated that the relatively high frequency for the amide-I band is well explained by impairment of hydrogen bond formation with water molecules. Moreover, concentration-dependent spectral changes indicated that the K+ affinity of the WT selectivity filter was much lower than those of the variants. Furthermore, only the variants displayed a higher frequency shift of the 1680-cm−1 band upon changes from K+ to Rb+ or Cs+ conditions. High-speed atomic force microscopy disclosed that TWIK1's surface morphology largely does not change in K+ and Na+ solutions. Our results reveal the local conformational changes of the TWIK1 selectivity filter and suggest that the amide-I bands may be useful “molecular fingerprints” for assessing the properties of other K+ channels.
Keywords: potassium channel, metal ion-protein interaction, infrared spectroscopy (IR spectroscopy), biophysics, Fourier transform IR (FTIR), ion channel, molecular dynamics, quantum chemistry, fluorescence
Introduction
Potassium ion (K+) conductance through K+ channels is very important for cellular processes. Most K+ channels are highly K+–selective and almost impermeable to sodium ions (Na+). How K+ channels accomplish this high degree of K+ selectivity has been extensively investigated. In particular, crystallographic studies of KcsA, a bacterial K+ channel, have revealed the molecular mechanisms underlying K+ selectivity. Crystal structures of KcsA showed that KcsA has a 4-fold rotational symmetry axis along the pore and revealed how the selectivity filter in the pore interacts with permeant cations, such as K+, and impermeant cations, such as Na+ (1–5). According to the crystal structures of KcsA, K+ ions bind to the S1–S4 sites in the selectivity filter (see Fig. 1A), where a K+ ion is coordinated to eight backbone carbonyls of the filter at each binding site (2). By contrast, nonpermeating cations, such as Na+ and Li+, were accommodated by different binding sites (B-sites, indicated in orange in Fig. 1A), where each cation is coordinated to four carbonyls with an in-plane configuration (4–7). Furthermore, in environments of low-K+ concentration or Na+ only, the selectivity filter of KcsA “collapses” to a nonconducting conformation (5, 7). However, Li+ does not cause the collapse (4), and the filter collapse does not occur in other bacterial K+ channels, such as MthK (8). Based on these structural data, the so-called “snug-fit model” (1, 3, 6) was proposed, and dynamic features of the filter structure (6, 9) were also considered to play important roles in the K+ selectivity.
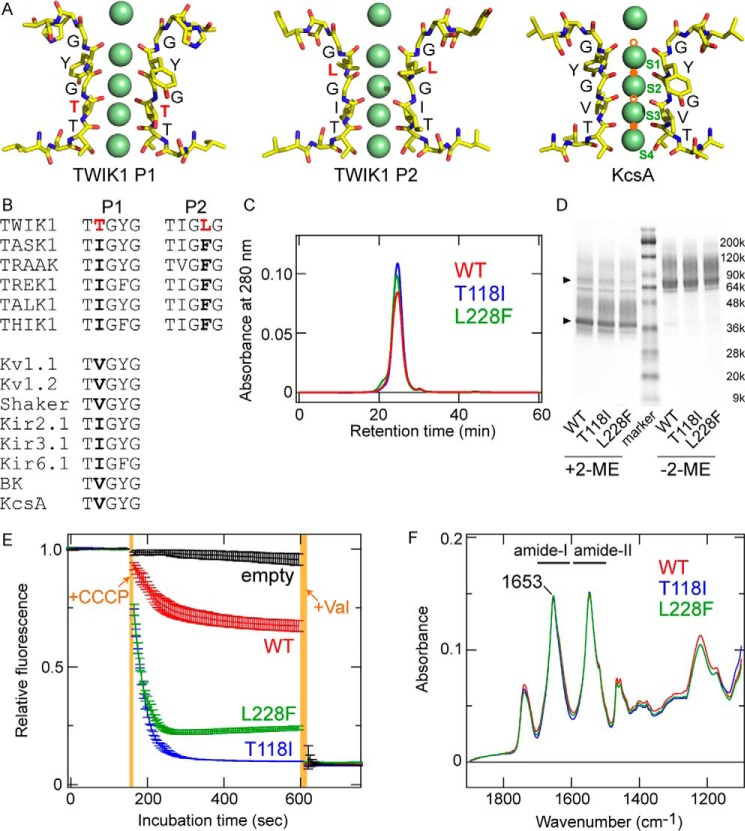
Figure 1.
Molecular characteristics of TWIK1. A, comparison of atomic models of the selectivity filter for TWIK1 (PDB code 3UKM) and KcsA (PDB code 1K4C). Green spheres indicate K+ ions resolved in the crystal structures. The K+-binding sites, S1–S4, are labeled in the KcsA structure. Note that the structural models were constructed from the averaged electron density maps, and all of the K+ ions do not exist simultaneously (2). The pore domains (P1 and P2) in TWIK1 possess unique filter residues, Thr-118 and Leu-228, respectively. In the KcsA structure, Na+/Li+-binding sites (B-sites) are indicated by orange circles. The lower filled orange circle indicates the Li+-binding site (4) as well as a potential Na+-binding site (5, 6), and the upper filled orange circle indicates the Na+-binding site (5). Open orange circles are hypothetical binding sites for Na+/Li+ (6). B, amino acid sequence alignment in the selectivity filter of K+ channels. A and B, unique residues Thr-118 and Leu-228 are highlighted in red. C, size-exclusion chromatographs of purified TWIK1: WT (red), T118I mutant (blue), and L228F mutant (green). D, Coomassie Brilliant Blue-stained SDS-polyacrylamide gel with purified WT TWIK1 and mutants (T118I and L228F) in the presence (+2-ME) or absence (−2-ME) of a reducing reagent (final concentration of 5% 2-mercaptoethanol (2-ME)). The triangles indicate major bands of the purified TWIK1 samples with or without reduction. E, traces of flux assay fluorescence changes using liposomes containing TWIK1: WT (red), T118I mutant (blue), or L228F mutant (green) and “empty” liposomes without channel proteins (black). Error bars indicate S.D. values from three independent measurements. +CCCP indicates the time point of addition for the CCCP protonophore, and +Val indicates the time point of addition for the K+-selective ionophore valinomycin. Orange-shaded areas indicate the dead time for adding and mixing CCCP or valinomycin. F, absolute IR absorption spectra of WT (red), T118I (blue), and L228F (green) after normalization at the amide-I peak (at 1653 cm−1).
The KcsA selectivity filter interactions with cations, such as K+ and Na+, are typically detected in the amide-I region (1700 to 1600 cm−1, mainly composed of C=O stretching vibrations of the protein main chains) using ion-exchange–induced difference attenuated total reflection FTIR (ATR-FTIR)3 spectroscopy (10–12). The observed IR absorption bands were then assigned to specific vibrational modes corresponding to the S2/S3 and B-sites using transmittance IR spectroscopy aided by computer simulations (13). Recently, 2D IR spectroscopy was applied to isotopically labeled (13C and 18O) KcsA for analyzing the dynamics of the carbonyls in the filter (14). ATR-FTIR spectroscopy has also been applied to study various ion channels (15, 16) and transporters (17–19).
The amino acid sequence motif of the selectivity filter (T(V/I)G(Y/F)G) is highly conserved among prokaryotic and eukaryotic K+ channels (Fig. 1B). In fact, crystallographic studies have revealed that the atomic structures of the filter and K+-binding sites (S1–S4) are virtually identical in many K+ channels, such as KcsA, voltage-sensitive (Kv) K+ channels, and inward-rectifying (Kir) K+ channels (20–22). This strongly suggests that the molecular mechanism underlying K+ selectivity is highly conserved.
However, not all K+ channels are always highly K+-selective (22). For example, the mammalian two-pore–domain K+ channel TWIK1 (23) (also named “K2P1” or “KCNK1”) is known to conduct Na+ under certain conditions, such as low extracellular pH and/or low extracellular K+ concentration (24–27). The Na+ current through TWIK1 is reported to regulate the resting potential and to be associated with Na+ influx during hypokalemia (26).
TWIK1, together with other K2P channels, functions as a dimer, and each subunit possesses two-pore loop (P1 and P2) regions. These pore loop regions consist of the selectivity filter and break the 4-fold rotational symmetry axis along the pore (28–31). Moreover, TWIK1 has unconventional amino acid residues in each pore loop region: Thr-118 (TTGYG) in the P1 region and Leu-228 (TIGLG) in the P2 region (Fig. 1, A and B). The T118I mutation in TWIK1 changes the P1 sequence to a conventional sequence (TIGYG) and makes the channel highly K+-selective, like typical K+ channels (24, 26). This result clearly indicates that the unconventional selectivity filter sequence is responsible for the Na+ permeability of TWIK1 and implies that the filter structure of TWIK1 is somewhat different from typical K+ channels. In 2012, a crystal structure of TWIK1 was reported (30). Surprisingly, the arrangement of the carbonyl groups in the selectivity filter accommodating K+ ions was almost identical to those of KcsA (Fig. 1A) (2), another K+-selective two-pore–domain channel TRAAK (31), and other K+ channels. The crystal structure of Na+-bound TWIK1 is not yet available, and the crystal structure with K+ does not explain how TWIK1 achieves its unique ion selectivity.
In this study, we applied ion-exchange–induced difference ATR-FTIR spectroscopy for elucidating the molecular mechanisms underlying the unique ion selectivity of TWIK1 relying on the higher structural sensitivity of this spectroscopic technique. The IR spectral changes of WT TWIK1, the highly K+-selective mutant T118I and the L228F mutant were systematically analyzed upon replacing K+ with Na+ or other monovalent cations. We found that the manner of interaction between WT TWIK1 to K+ and to non-K+ cations were different from those of the mutants and KcsA. High-speed atomic force microscopy (HS-AFM) observation confirmed that the shape of a TWIK1 molecule barely changes under the IR measurement conditions, suggesting that our IR observations detected the local structural changes induced by binding of the different cations. Moreover, the normal vibrational modes of the carbonyl groups in the TWIK1 and KcsA selectivity filters were calculated. The amide-I band at 1680 cm−1, commonly observed in K+ conditions, originated from the selectivity filters and could be used for analyzing the K+ affinity and the local structural perturbations upon changing the cations. Our data showed that these characteristics correlated well with the K+ selectivity of WT TWIK1 and the TWIK1 mutants.
Results
Functional expression of TWIK1 and its reconstitution into liposomes
ATR-FTIR spectroscopic analysis requires several micrograms of purified membrane protein, and FSEC analysis was used to assess the suitability of membrane proteins for expression in cultured mammalian cells (32). Mouse TWIK1 showed a preferable FSEC profile (a single peak with high fluorescence intensity) (Fig. S1); thus, mouse WT TWIK1 as well as mutants T118I and L228F, with a His8 tag on the C terminus, were expressed in COS-1 cells, extracted, and purified on Ni-Sepharose and size-exclusion columns (see “Experimental procedures”). The purified TWIK1 samples showed a monodisperse peak on the size-exclusion chromatograph (Fig. 1C), thus suggesting that each TWIK1 sample had a homogeneous conformation with little aggregation or digestion. The Coomassie Brilliant Blue-stained SDS-polyacrylamide gel of each purified sample showed one major band with minor band smearing (Fig. 1D). The major bands were ∼75 kDa (dimer) and ∼38 kDa (monomer) in size before and after reduction using 2-mercaptoethanol, respectively. This result is consistent with the fact that TWIK1 forms a dimer connected by a disulfide bond between the protomers (30, 33).
To assess the functionality of the purified TWIK1 samples, we reconstituted the purified TWIK1 proteins into liposomes and conducted a K+ flux assay (30, 31). Here, we performed the K+ flux assay using lipid vesicles containing TWIK1 variants T118I or L228F, and the WT, thus confirming the high ion selectivity of the T118I mutant. In the flux assay, the K+ flux out of the vesicles is compensated by proton influx via a protonophore, carbonyl cyanide m-chlorophenylhydrazone (CCCP). Thus, the amount of K+ efflux can be detected by a fluorescence decrease of a proton indicator, 9-amino-6-chloro-2-methoxyacridine (ACMA) (30, 31). Under the same experimental conditions (see “Experimental procedures”), WT TWIK1 and the variants reached different plateau levels for K+ efflux (Fig. 1E). In the case of the WT, the amount of K+ released from the vesicles was relatively small, and the addition of a K+-selective ionophore, valinomycin, caused further K+ release, as shown in a previous crystallization study (30) that compared a WT TWIK1 construct for crystallization, which showed a lesser amount of K+ release compared with the highly K+-selective Kv1.2. By contrast, vesicles containing the TWIK1 T118I variant released much more K+, and valinomycin induced little additional K+ efflux. The L228F variant showed a profile relatively close to that of the T118I variant. Thus, our data suggest that the L228F mutant possesses higher potassium selectivity than the WT. It should be noted that the K+ efflux levels were nearly same for all three types of vesicles after the addition of valinomycin as well as vesicles without channels (Fig. 1E).
In our flux assay, only K+ ions were found in the vesicles ([K+]in = 150 mm) as cations, and Na+ was predominantly outside of the vesicles ([K+]out = ∼4 mm and [Na+]out = ∼146 mm). The amount of outward K+ flux is reflected in the equilibrium potential of the vesicles. The equilibrium potential is determined by the ratios of K+ efflux and Na+ influx; this depends on the relative permeability of reconstituted TWIK1 for K+ and Na+. Thus, the difference in the amount of K+ release, detected by fluorescence decrease from ACMA, would represent the relative permeability of K+ over Na+. Based on our experimental results, the K+ selectivity was estimated to be T118I (≈ valinomycin) > L228F > WT. The higher K+ selectivity of the T118I mutant is consistent with previous electrophysiological studies (24, 26). However, a previous electrophysiological study reported that the L228F mutation has little effect on the K+ selectivity of TWIK1 (24). Our results showed a small but obvious change in K+ selectivity for the L228F mutant (Fig. 1E). The discrepancy of the L228F mutation effect may be due to the different experimental methods employed (electrophysiological method with heterologous expression in Xenopus oocytes) (24) and/or different constructs used for the assays (I293A/I294A/K274E mutations were additionally introduced to the L228F mutant of human TWIK1) (24).
For ATR-FTIR spectroscopy, we reconstituted WT TWIK1 and the variants into liposomes using the same lipid composition as for the flux assay but with a higher protein to lipid molecules ratio (about 100-fold) to increase the IR signals from the proteins (see “Experimental procedures” and Fig. S2). The absolute IR absorption spectra in the 1800 to 1100-cm−1 region were very similar between the WT and the variants (Fig. 1F). Their amide-I peaks were located at 1653 cm−1, and the band shapes were almost identical, indicating that the overall protein secondary structures were largely unchanged by the substitutions.
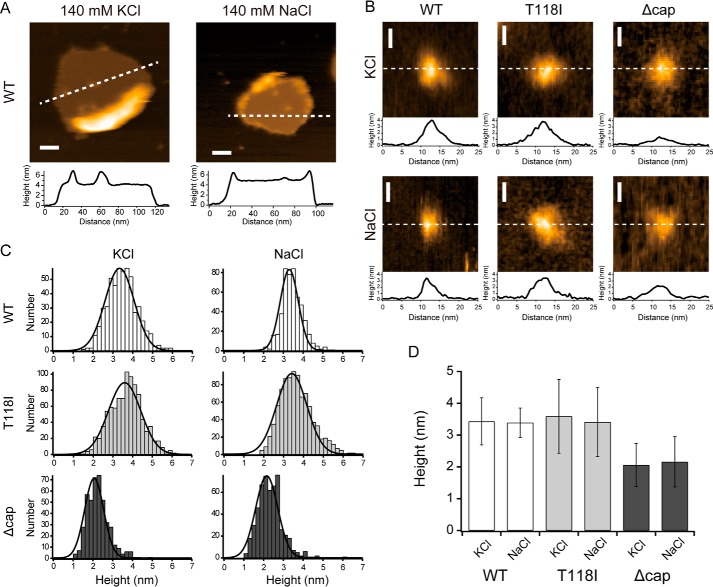
To further confirm the status of the liposome-reconstituted TWIK1 proteins, we assessed their molecular shapes at the single-molecule level using HS-AFM (34, 35). The HS-AFM images of WT TWIK1 in liposomes perfused with a buffer containing K+ showed a homogeneous conical structure (diameter = ∼10 nm) with a height of ∼3.3 nm from the membrane surface (membrane thickness = ∼5 nm) (Fig. 2, A, B, and D). The diameter and height values were consistent with the crystal structure (30), and these values were not significantly changed when the ion was changed to Na+. The TWIK1 T118I mutant also possessed similar structural dimensions to those of the WT (Fig. 2, B–D). The conical structure was presumably attributed to the extracellular “cap” domain in TWIK1. The Δcap mutant (lacking the “cap” domain, i.e. amino acid residues from positions 47 to 101) showed a similar diameter and a height reduced by ∼1 nm (∼2.1 nm from the membrane) (Fig. 2, B–D). Thus, the conical structures of ∼1.2 nm height correspond to the cap domain (see Fig. S3).
Figure 2.
HS-AFM images and height analysis of TWIK1 channels. A, typical HS-AFM images of WT TWIK1 channels reconstituted in a lipid bilayer and measured at 140 mm KCl (left) and 140 mm NaCl (right). The broken lines on the images indicate positions for the corresponding cross-sectional profiles displayed immediately below. Scale bar, 20 nm. B, single-molecule images of WT, T118I, and Δcap-TWIK1 observed at 140 mm KCl (upper row) and 140 mm NaCl (lower row). Broken lines on the images indicate positions for the cross-sectional profiles displayed at the bottom side of each image. C, histograms of height distributions measured for WT, T118I, and Δcap-TWIK1 under different salt conditions (140 mm KCl and 140 mm NaCl). The height of each TWIK1 channel was measured for the difference between the highest part and the lipid surface on the AFM image. Histograms were constructed using 533 channels for WT in KCl, 494 channels for the WT in NaCl, 923 channels for T118I in KCl, 907 channels for T118I in NaCl, 446 channels for Δcap in KCl, and 534 channels for Δcap in NaCl. Each histogram could be well fitted using a Gaussian distribution, which is depicted by a solid line. D, comparison of molecular heights from the lipid surface for three different TWIK1 molecules under the different salt conditions (140 mm KCl and 140 mm NaCl). The heights and error bars correspond to the center values of the fitted Gaussian distributions shown in C and the full widths at half-maximum (FWHMs) of the distributions, respectively. The heights ± FWHMs were 3.34 ± 1.04 nm (WT in KCl), 3.30 ± 0.65 nm (WT in NaCl), 3.59 ± 1.16 nm (T118I in KCl), 3.41 ± 1.08 nm (T118I in NaCl), 2.06 ± 0.68 nm (Δcap in KCl), and 2.66 ± 0.79 nm (Δcap in NaCl).
These results clearly indicate successful purification of functional TWIK1 proteins. Moreover, the absolute amide-I bands and the AFM images were very similar between the WT and the pore-domain mutants. Therefore, we can discuss effects of the T118I and L228F mutations on their IR difference spectra without needing to consider mutation-induced morphology changes or large differences in the secondary structures, such as deformation of α-helices.
Ion-exchange–induced difference IR spectroscopy upon replacement of K+ with Na+
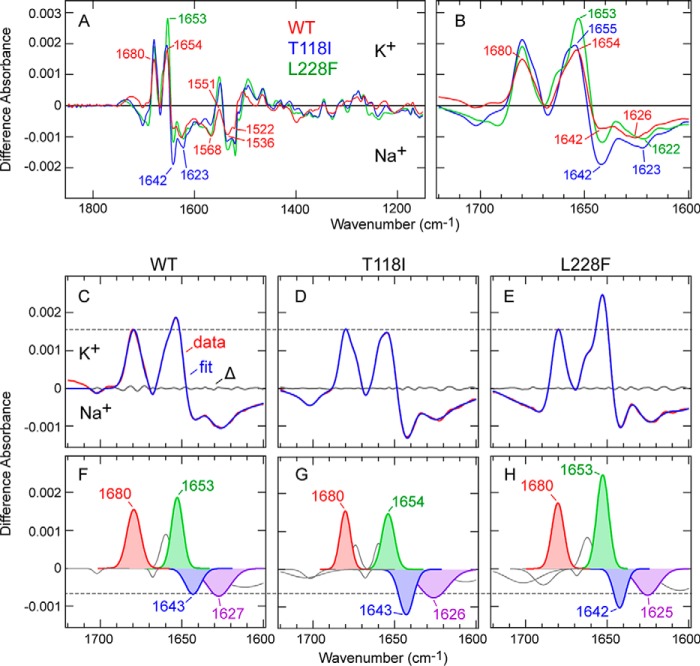
The difference IR spectra of the WT and the variants (T118I and L228F), i.e. the IR spectra obtained before buffer exchange minus those obtained after buffer exchange from 140 mm K+ to 140 mm Na+, are shown in Fig. 3A. Because the difference spectra were calculated through subtraction of the absolute spectra in K+ conditions minus the spectra in Na+ conditions, the positive and negative bands should reflect the molecular structures in the presence of K+ and Na+, respectively. The spectral changes in the amide-I region are highlighted in Fig. 3B. The difference spectrum of the WT shows clear absorption changes in the amide-I region (1700 to 1600 cm−1): positive bands at 1680 and 1654 cm−1 as well as negative bands at 1642 and 1626 cm−1 (Fig. 3B). Changes were also clearly observed in the amide-II region (1600 to 1500 cm−1): a positive band at 1551 cm−1 as well as negative bands at 1568, 1536, and 1522 cm−1 (Fig. 3A). Previous experimental (10, 11, 13) and spectral simulation (13) studies of KcsA concluded that difference absorption bands in the amide-I region upon ion exchange can be assigned to the vibrational modes of the selectivity filter interacting with K+ ions (∼1680 cm−1) or Na+ ions (∼1630 cm−1) and the pore helices (∼1660 cm−1).
Figure 3.
Difference IR spectra of WT TWIK1 and the mutants (T118I and L228F) upon the K+ to Na+ exchange. A, difference spectra of TWIK1: WT (red), T118I (blue), and L228F (green) on exchange from 140 mm K+ to 140 mm Na+. Wavenumber values for specific bands are indicated. B, spectral changes in the amide-I region are highlighted. C–E, spectral changes in the amide-I region (red) of WT TWIK1 (C), T118I (D), and L228F (E) are fitted with seven to eight Gaussian peaks (blue). Residuals are shown in gray. F–H, Gaussian peaks of WT TWIK1 (F), T118I (G), and L228F (H) used in the fittings (C–E). The major peaks discussed in the main text are highlighted.
The previous IR spectroscopic studies on KcsA observed a positive (K+ side) band at ∼1680 cm−1 (10), and computational analysis assigned it to two vibrational modes (S2 mode at 1680 cm−1 and S3 mode at 1674 cm−1), representing interactions of the backbone carbonyls in the selectivity filter with two K+ ions in the S2 and S4 sites (denoted as [S2, S4] here) as well as S1 and S3 ([S1, S3]) sites in the crystal structure of KcsA, respectively (13). In the difference spectrum of WT TWIK1, a positive band at the identical position (1680 cm−1) was observed (Fig. 3B). The arrangement of the carbonyl groups in the selectivity filter and the K+-binding sites are almost identical between the crystal structures of WT TWIK1 (30) and KcsA (Fig. 1A) (7). Therefore, it is reasonable to assign the 1680-cm−1 band to the S2/S3 mode in TWIK1 as well, which is well supported by our quantum chemical computational study, described later. The 1680-cm−1 band was observed in both the L228F and T118I variants as well (Fig. 3B). The difference spectra normalized to the band intensity at 1680 cm−1 are shown in Fig. 3, C–E.
The previous KcsA study also assigned the other major positive peak at ∼1660 cm−1 in the amide-I region to the “helix mode,” representing vibrational changes of α-helices surrounding the selectivity filter (pore α-helices) upon K+ to Na+ exchange (13). In the TWIK1 IR difference spectrum, a positive peak at 1654 cm−1 was observed (Fig. 3B), and the band would correspond to the helix mode. The ∼6-cm−1 difference between KcsA and TWIK1 suggests that slight structural differences exist in the pore α-helices between the two channels.
Stevenson et al. (13) assigned the strong negative (Na+ side) band at 1625 cm−1 to the “B-site mode,” representing four carbonyl groups interacting Na+ at the “B-site” in the KcsA structure (shown in orange in KcsA, Fig. 1A). In the TWIK1 IR difference spectrum (Fig. 3B), the negative band at 1626 cm−1 might correspond to the B-site mode, despite the considerably weaker intensity (10, 12, 13). Additionally, another negative band at 1642 cm−1 was clearly observed in TWIK1. One of negative bands at 1650 and 1639 cm−1 in the difference spectrum of KcsA (10, 12) may correspond to the 1642-cm−1 band in TWIK1. Although the frequencies are different, these results suggest that the manner of interaction with Na+ in TWIK1 is somewhat different from that in KcsA.
Comparison of the K+ minus Na+ difference IR spectra among WT, T118I, and L228F mutants
Upon the K+ to Na+ exchange, TWIK1 mutant proteins T118I and L228F showed similar difference spectra to that of the WT (Fig. 3, A and B). In particular, the positive band at 1680 cm−1 was also observed at the same frequency in the mutants. This suggests that these mutations in the selectivity filter retain their manner of interaction with K+ ions in the [S2, S4] or [S1, S3] configurations. Similarly, the frequencies of the positive band at 1654 cm−1 as well as the negative bands at ∼1642 and ∼1626 cm−1 in the mutants were nearly identical to those in the WT (Fig. 3B). By contrast, the intensities of these bands were greater in the mutants (Fig. 3B). Because the peak intensities have already been normalized against total protein using the peak intensities of the absolute amide-I peaks at 1653 cm−1, the differences can be interpreted by two factors as follows: lower affinity for K+ in WT than in the mutants and differences in the manner of interaction with Na+ and/or K+ of WT and that of the mutants. To compare band intensities more accurately, we conducted band fitting on each difference spectrum after normalization of the spectra with the peak intensities at 1680 cm−1. We successfully fitted the difference spectra of the amide-I region using seven to eight Gaussian peaks (Fig. 3, C–E) and four peaks (at 1680, ∼1653, ∼1643, and ∼1627 cm−1) corresponding to major bands in each spectrum (Fig. 3, F–H). The area of the fitted bands at ∼1643 cm−1 in the mutants was larger than that in the WT (WT, 0.0081; T118I, 0.0133; and L228F, 0.0098); the area of the fitted band at 1654 cm−1 in T118I was smaller than that of the WT, and the positive band at 1653 cm−1 in L228F was larger than that of the WT (WT, 0.0164; T118I, 0.0141; and L228F, 0.0229). However, the bands overlapped with the amide-I peak of the absolute spectra of the TWIK1 samples (Fig. 1F); thus, the effect of spectral correction for swelling effects blurs the spectral changes near 1653 cm−1, which may result in a drift of the baseline for the ∼1643- and ∼1653-cm−1 bands. The sum of the areas of the 1643- and 1653-cm−1 bands of the WT were very similar to that of T118I and smaller than that of L228F (WT, 0.0245; T118I, 0.0274; and L228F, 0.0327). Thus, the differences in Fig. 3, A and B, were attributed to the difference in affinity for K+ and that for Na+, at least for the T118I mutant.
Concentration-dependent experiments for analyzing the relative K+ affinity of TWIK1
Next, we assessed and compared the relative affinity for K+ in WT TWIK1 and the mutant proteins (T118I and L228F) by concentration dependence of spectral changes upon K+ to Na+ exchange (10). Spectral changes in the amide-I region were measured upon buffer exchange from 0 mm K+ (200 mm Na+) to 0.1, 0.3, 1, 3, 10, 20, 50, 100, or 200 mm K+ (199.9, 199.7, 199, 197, 190, 180, 150, 100, or 0 mm Na+, respectively). To minimize the effect of the ionic strength change on the IR spectra, the total concentrations of K+ and Na+ were kept constant (200 mm). The measured difference spectra are shown in Fig. 4, A (WT), B (T118I), and C (L228F).
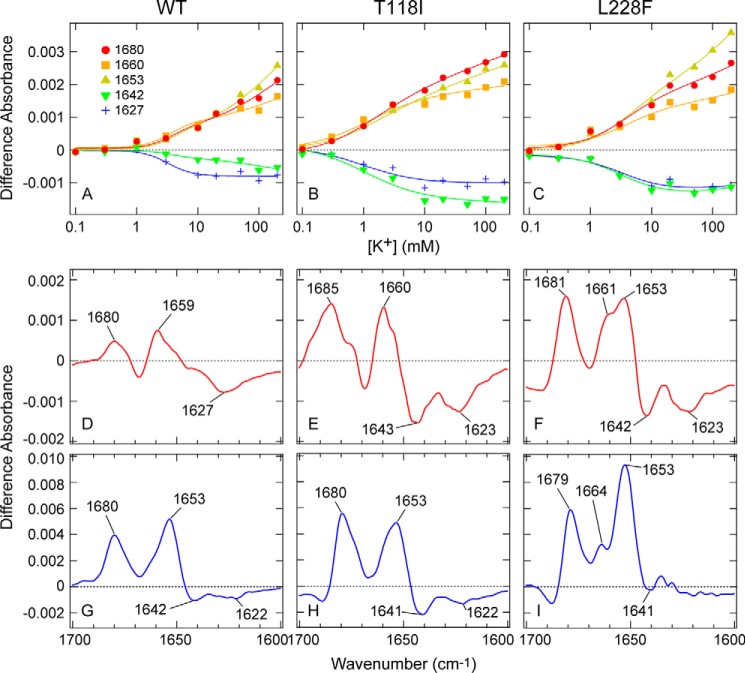
Figure 4.
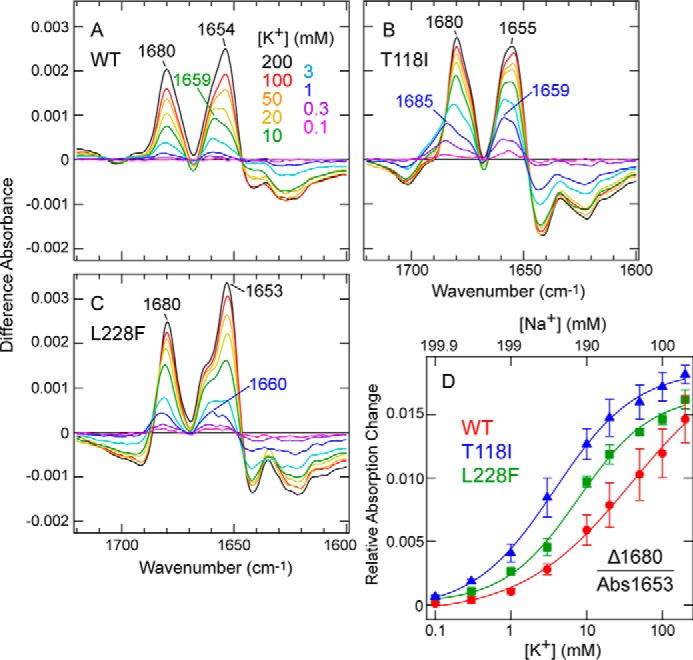
Concentration dependence of spectral changes for WT TWIK1 and the mutants (T118I and L228F) upon exchange from 0 mm K+ (200 mm Na+) to various concentrations of K+ (Na+) up to 200 mm K+ (0 mm Na+). A–C, difference spectra for WT TWIK1 (A), T118I (B), and L228F (C) on exchange from 0 mm K+ (200 mm Na+) to 0.1 (pink), 0.3 (purple), 1 (blue), 3 (pale blue), 10 (green), 20 (yellow), 50 (orange), 100 (red), and 200 (black) mm K+ (199.9, 199.7, 199, 197, 190, 180, 150, 100, and 0 mm Na+, respectively). K+ buffer concentrations are indicated. The peaks at 1680 cm−1 in the spectra of WT, T118I, and L228F are indicated and used for the concentration dependence plot in D. In the presence of low (less than 1 mm) K+ concentrations, the 1680-cm−1 bands are shifted slightly to 1685 and 1683 cm−1 for mutants T118I and L228F, respectively. D, concentration dependence plot of the positive band absorption intensity at 1680 cm−1 relative to the absolute absorbance at 1653 cm−1 (see Fig. 1F) as a function of the K+ concentration. Concentration-dependent data and curves of WT (red), T118I (blue), and L228F (green) are shown. The error bars represent S.D. values of three (T118I and L228F) or four (WT) independently prepared samples. The half-maximal values of WT, T118I, and L228F, based on the Hill equation fitting using Igor Pro software (WaveMetrics, Inc., Lake Oswego, OR), are 39.6 ± 19.8, 3.8 ± 0.4, and 7.7 ± 1.1 mm, respectively.
Because the positive band at 1680 cm−1 has been tentatively assigned to the S2/S3 mode of TWIK1, it is a useful marker band for the selectivity filter interacting with K+ ions. Thus, we plotted the band intensity at 1680 cm−1 as a function of K+ concentration (Fig. 4D). Because the difference spectra were normalized against the absolute absorption of amide-I (at 1653 cm−1) (Fig. 1F), this plot clearly indicated that the spectral changes of the WT at 1680 cm−1 were less sensitive to K+ concentration increases than those of the mutants (T118I and L228F). The half-maximum values of K+ were 39.6, 3.8, and 7.7 mm for the WT, T118I, and L228F, respectively. These values indicated that the selectivity filter in the WT possessed a lower affinity for K+ (this could be explained by an increased affinity for Na+) than that in T118I and L228F.
The previous KcsA study reported that, in the presence of low-K+ concentrations (< ∼10 mm), the 1680-cm−1 band shifted to a higher frequency (1688 cm−1) (10). A similar but smaller shift (∼5 cm−1) was observed in the T118I mutant in the presence of very low-K+ concentrations (< ∼1 mm) (Fig. 4B). In the WT, the up-shift of the 1680-cm−1 band was unclear, but the band at 1653 cm−1 (helix mode) was shifted to ∼1659 cm−1 in the presence of < ∼10 mm K+ (Fig. 4A). These results suggest that the K+/Na+ concentration-dependent spectral changes in the amide-I region can be resolved to at least two phases.
To resolve the K+ concentration-dependent two-phase spectral changes, we constructed a model of K+ binding to TWIK1 proteins as shown in Schemes 1 and 2,
and
where Ch denotes potassium channel. In our two-phase model, K+-binding reactions are divided into “higher affinity” (with lower Kd (Kd1) values) and “lower affinity” (with higher Kd (Kd2) values) binding reactions. Based on this model, we conducted global fitting procedures on K+/Na+ concentration-dependent spectral changes of the WT TWIK1 and the mutants over the entire amide-I region (Fig. 4, A–C). Five fitting curves for each sample are representatively shown in Fig. 5, A–C, which are traces for the major bands observed in the difference spectra (Fig. 3, F–H). We successfully resolved the two-phase binding reactions as two difference spectra (“higher affinity” (ΔAKn+Ch − Ch): red curves in Fig. 5, D–F and “lower affinity” (ΔAKn + m+Ch* − Ch): blue curves in Fig. 5, G–I). The global fitting calculations provided two dissociation constants for the “higher affinity” and “lower affinity” binding reactions (Kd1 and Kd2, respectively) (Table 1), which were associated with the two difference spectra for each sample.
Figure 5.
Global fitting of the difference spectra series of WT TWIK1 and the mutants. A–C, concentration dependence plots of the absorption intensity at 1680 (red-filled circle), 1660 (orange-filled square), 1653 (yellow-filled rectangles), 1642 (green-filled vertical rectangles), and 1627 (blue plus mark) cm−1 of WT (A), T118I (B), and L228F (C) as a function of the K+ concentration. The plots are globally fitted using Equation 1 and shown with solid curves. D–I, ΔAKn+Ch − Ch coefficient spectra (D, WT; E, T118I; and F, L228F) and ΔAKn + m+Ch* − Ch coefficient spectra (G, WT; H, T118I; and I, L228F) obtained from the global fitting of the difference spectra series using Equation 1.
Table 1.
The parameters for Equation 1 obtained by global fitting of the experimental data
| WT | T118I | L228F | |
|---|---|---|---|
| Kd1K+ (mM) | 12.7 ± 3.3 | 1.2 ± 0.2 | 4.1 ± 0.4 |
| n | 2.1 ± 0.2 | 0.89 ± 0.10 | 1.1 ± 0.1 |
| Kd2K+ (mM) | 44.4 ± 18.2 | 3.9 ± 2.4 | 42.3 ± 26.3 |
| m | 0.68 ± 0.19 | 0.25 ± 0.44 | 0.52 ± 0.33 |
In the resolved difference spectra corresponding to the higher affinity binding (Fig. 5, D–F), the helix mode bands are shifted to ∼1660 cm−1 in the WT and the T118I mutant. In the L228F mutant, the helix mode splits into two bands, at 1661 and 1653 cm−1. This shift is consistent with the helix mode shift in the low-K+ concentration case (Fig. 4, A–C). In addition, the amplitude of the ∼1660-cm−1 band was larger (∼1.5-fold) in the mutants than in the WT (Fig. 5, D–F). Furthermore, the negative band at ∼1642 cm−1 was observed only in the mutant (T118I (Fig. 5E) and L228F (Fig. 5F)) and not in the WT (Fig. 5D). Furthermore, in the T118I mutant, the 1680-cm−1 band (corresponding to the S2/S3 mode) shifted to 1685 cm−1 (Fig. 5E), indicating the interaction manner between the selectivity filter and K+ was different from that with the WT. Taken together, the higher affinity interaction with K+ caused more distinct spectral changes of the helix mode and adjacent bands in the variants than in the WT, suggesting that structural changes of region(s), including the pore helix, occur more drastically upon K+ to Na+ exchange in the mutants than they do in the WT, especially during the initial binding process.
In addition to the spectral changes, the calculated Kd1 values were also quite different between the mutants and the WT. The calculated Kd1 value of the WT was ∼10-fold or ∼3-fold larger than the values of the mutants (Table 1). Thus, in TWIK1 (WT), K+ ions initially bind to the selectivity filter with reduced affinity and cause smaller structural changes around the selectivity filter region when compared with the mutants that have higher ion selectivity.
In contrast to the higher affinity interaction, the difference spectra corresponding to the lower affinity binding were relatively similar between the WT and T118I mutant (Fig. 5, G and H), whereas that of the L228F mutant was somewhat different (Fig. 5I). This result suggests that the lower affinity interaction with K+ causes similar structural changes, at least in the WT and the T118I mutant. In the difference spectra, the helix mode bands were observed at ∼1653 cm−1, suggesting that in the lower affinity binding, K+ ions cause structural changes in the pore helix that are different from those in the higher affinity binding. Although the spectral changes were relatively small, the dissociation constant for the lower affinity binding (Kd2) between the WT and T118I was quite (∼10-fold) different (Table 1). Interestingly, the Kd2 value of L228F was very similar to that of the WT, whereas the Kd1 values were ∼3-fold different (Table 1). In other words, Thr-118 affects both the higher affinity and the lower affinity binding, but Leu-228 affects only the higher affinity binding. We speculate that the higher K+ selectivity in the T118I mutant (Fig. 1E) may be related to having higher affinities for K+ in both states of the sequential binding process, and the lower affinity for K+ at the second binding process in L228F may account for the slightly lower K+ selectivity in the L228F mutant (Fig. 1E).
The detailed IR spectroscopic analyses of WT TWIK1 and the mutants revealed two manners of interaction (higher affinity and lower affinity) with K+ ions. Our experimental results and calculated fittings strongly suggest that the unique residues (Thr-118 and Leu-228) in the selectivity filter make the protein structure of TWIK1 insensitive to Na+/K+ exchanges, leading to decreased affinity for K+ in the selectivity filter. Based on the experimental data, we discuss how TWIK1 reduces the ion selectivity under the “Discussion.”
Ion-exchange–induced difference IR spectra of TWIK1 upon exchange from K+ to various cations (alkali-metal cations, NH4+, or NMDG+)
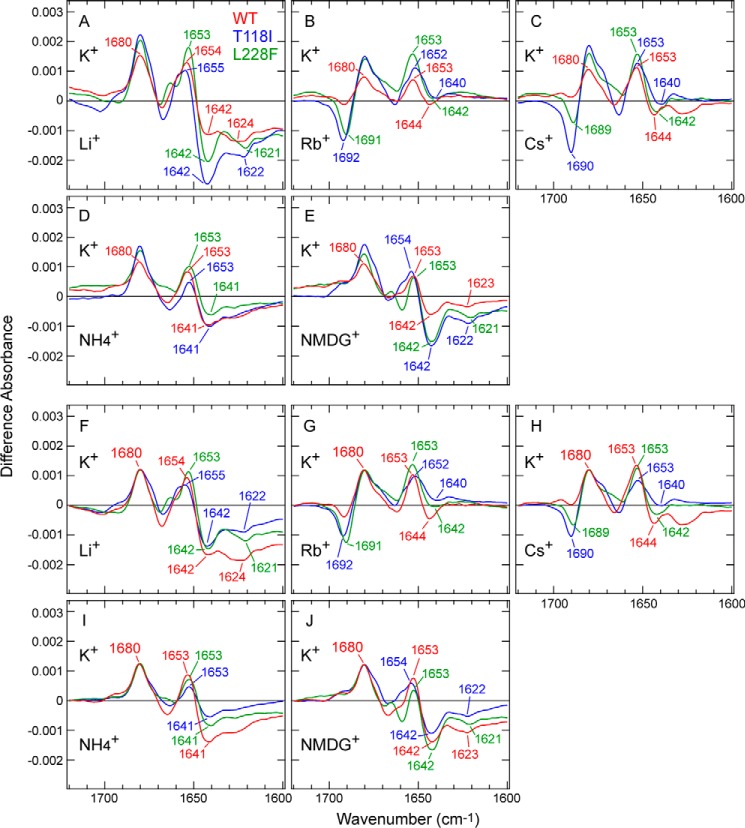
IR spectral changes upon K+ to Na+ exchange revealed how differently the selectivity filter interacts with K+ and Na+ in WT TWIK1 and in the mutants. To explore the ion interactions with TWIK1 further, we next measured the spectral changes caused by replacing K+ with monovalent cations other than Na+. Similar to the experiments for Na+ described above, we measured the difference spectra of WT TWIK1 and the mutant proteins (T118I and L228F) by replacing 140 mm K+ with 140 mm Li+, Rb+, Cs+, NH4+, or N-methyl-d-glucamine (NMDG+) (Fig. 6, A–E). To clarify the spectral differences, the difference spectra were normalized by the 1680-cm−1 band (Fig. 6, F–J). Thus, the differences in protein structural changes upon cation exchange can be discussed without considering the cation affinity differences, as already explained during the analysis of K+ to Na+ exchange (Fig. 3).
Figure 6.
Comparison of the ion-exchange–induced difference spectra in the amide-I region for WT TWIK1 and the mutants upon exchange from K+ to various monovalent cations. A–E, difference spectra for WT TWIK1 (red), T118I (blue), and L228F (green) upon exchanges from 140 mm K+ to 140 mm Li+ (A), Rb+ (B), Cs+ (C), NH4+ (D), and NMDG+ (E). Wavenumber values for specific bands in the amide-I region are indicated. F–J, difference spectra in A–E are normalized against the band intensity at 1680 cm−1, and the absorbance difference at 1720 cm−1 is set to zero for offsetting differences in baselines.
Upon K+ to Li+ exchange, WT TWIK1 and the mutants showed similar spectral changes to those upon K+ to Na+ exchange (Figs. 3B and 6A). The overall spectral amplitude of the WT was less than those of the mutants (Fig. 6A), whereas the spectral shape was relatively similar to those of the mutants after normalization (Fig. 6F). Therefore, the differences (Fig. 6A) can mostly be explained by the different affinities for K+ between the WT and the mutants. These results also suggest that all three TWIK1 samples (the WT and the mutants) interact with Li+ in a similar fashion to Na+.
Upon K+ to Rb+ exchange (Fig. 6B), the TWIK1 samples showed different spectral changes from those of K+ to Na+/Li+ exchange. The overall spectral amplitudes of all three samples were smaller than those obtained upon exchange to Na+ or Li+ (Figs. 3B and 6A). Therefore, it is generally acceptable that the structural changes of TWIK1 upon exchanging K+ to Rb+ were smaller than those involving Na+ and Li+; this was also the case for KcsA (11, 12). In more detail, the negative bands at 1642 and 1626 cm−1, both of which can be observed upon K+ to Na+/Li+ exchange (Fig. 3, B and C as well as Fig. 6, A and F), almost disappear in the WT and the mutants (Fig. 6, B and G). This means that the structural features characterized by those bands in the presence of Na+ or Li+ were not conserved in the presence of Rb+. However, we found a unique feature for WT TWIK1, a negative (Rb+ side) band at 1692 (T118I) or 1691 cm−1 (L228F) was distinctly observed in the mutants, whereas the band intensity was much reduced in the WT, despite the peak intensity normalization at 1680 cm−1 (Fig. 6G). A similar band was observed at 1692 cm−1 in KcsA with Rb+ (11, 12), which means that WT TWIK1 interacts with Rb+ differently. In other words, the carbonyl groups in the selectivity filter of the TWIK1 mutants interact with Rb+ in a similar but distinguishable coordination manner from that with K+, just like with KcsA. However, such specific interactions were largely diminished in the WT, suggesting that the WT interacts with Rb+ in a different manner. K+ to Cs+ exchange (Fig. 6, C and H) caused similar spectral changes to the K+ to Rb+ exchange (Fig. 6, B and G), indicating that the manner of interaction between WT TWIK1 and Cs+ is similar to those with Rb+.
To deepen our IR analysis, we investigated molecular cations, such as NH4+ and NMDG+, which possess a much larger ionic radius compared with the monatomic cations. Notably, both the K+ to NH4+ and K+ to NMDG+ exchanges caused similar spectral changes in the WT TWIK1 and the mutants (Fig. 6, D, E, I, and J). The spectral shapes were very similar to those obtained upon K+ to Na+ and K+ to Li+ exchanges rather than those obtained upon K+ to Rb+ and K+ to Cs+ exchanges.
Previous electrophysiological studies revealed that many different cations, such as K+, Na+, Li+, Rb+, Cs+, and NH4+, can permeate through TWIK1. Therefore, interaction of NH4+ to TWIK1 could be thought to be similar to those of Na+ and Li+. However, a very large cation, such as NMDG+, should be impermeable (26, 27). Thus, we expected the spectral changes caused by the K+ to NMDG+ exchange would provide useful clues for understanding the characteristics of the selectivity filter without bound cations (especially in the negative bands of the difference spectra). Surprisingly, the K+ to NMDG+ exchange induced spectral changes in the WT and the mutants (Fig. 6E), exhibiting similar spectral shapes to those of the K+ to Na+/Li+ exchanges (Figs. 3B and 6A). Notably, negative bands at 1642 and 1623 cm−1 emerged upon the K+ to NMDG+ exchange (Fig. 6E), similar to the case of the K+ to Na+/Li+ exchange (Figs. 3B and 6A). Because NMDG+ is thought to be too large to enter the selectivity filter, the ∼1642- and 1623-cm−1 bands should be assigned to vibrational modes in the selectivity filter without bound cations. Thus, similar structural changes around the selectivity filter would occur from Na+/Li+ binding and/or only by depletion of K+ ions in the filter (such as water–filter interactions).
Computational analysis of the normal modes of the carbonyl groups in the TWIK1 selectivity filter using quantum mechanical and molecular mechanical (QM/MM) treatments
The ion-exchange–induced difference spectroscopy of TWIK1 revealed that the specific vibration of the carbonyl groups interacting with K+ ions at 1680 cm−1 was similar to that of the well-known bacterial potassium channel, KcsA. To assign the 1680-cm−1 band in TWIK1 to a specific vibrational mode, we performed normal mode analysis on the carbonyl groups in the TWIK1 selectivity filter using the QM/MM method, in which the peptide backbones, K+ ions, and water molecules in the selectivity filter were treated by quantum chemistry, and the rest of the system by molecular mechanics. In the X-ray crystal structure (30), there are five K+ ions in the filter in the equilibrium ensemble, although all five K+ ions do not exist in the filter simultaneously. Therefore, we analyzed the normal modes of TWIK1 with two types of K+ configuration: three K+ ions existing at the S0, S2, and S4 sites (denoted as [S0, S2, S4]) and two K+ ions existing at the S1 and S3 sites (denoted as [S1, S3]).
As illustrated in Fig. 7, the normal modes with strong IR intensities were calculated to higher frequencies compared with a control model molecule, N-methylacetamide (NMA), both in [S0, S2, S4] and [S1, S3] configurations. The calculated peak frequency difference, 61 cm−1 (1708 and 1647 cm−1 for TWIK1 and NMA, respectively), was in good agreement with the experimental difference, 54 cm−1 (1680 and 1626 cm−1 for TWIK1 and NMA (36), respectively). Here, to avoid vibrational coupling with the O–H bending modes of H2O molecules (accurate calculation of the O–H bending modes requires a larger QM subsystem and more sampling), we calculated the frequencies and the normal modes using D2O molecules. We also performed the ion-exchange–induced difference spectra in D2O conditions and confirmed that the difference spectra were very similar to those obtained in H2O (Fig. S4). The vectors in Fig. 7, A–F, show the direction and amplitude of the C=O stretching vibrations. The normal modes in Fig. 7, A, C, E, and F, are relatively localized on three nearby carbonyl groups, including a central carbonyl group (Gly-119 in P1 or Ile-226 in P2). The normal modes in Fig. 7, B and D, are mainly localized on a carbonyl group located at the outer mouth of the filter (Tyr-120 in P1 or Leu-228 in P2). The normal modes with five highest intensity IR absorbance are listed in Table 2 for both the [S0, S2, S4] and [S1, S3] configurations. Together, the combined peak occurs at ∼1710 cm−1 (Fig. 7G). Similar results were also obtained for KcsA (Fig. S5). Therefore, it is very reasonable to assign the experimentally observed 1680-cm−1 band to the carbonyl groups in the selectivity filter interacting with K+ ions.
Figure 7.
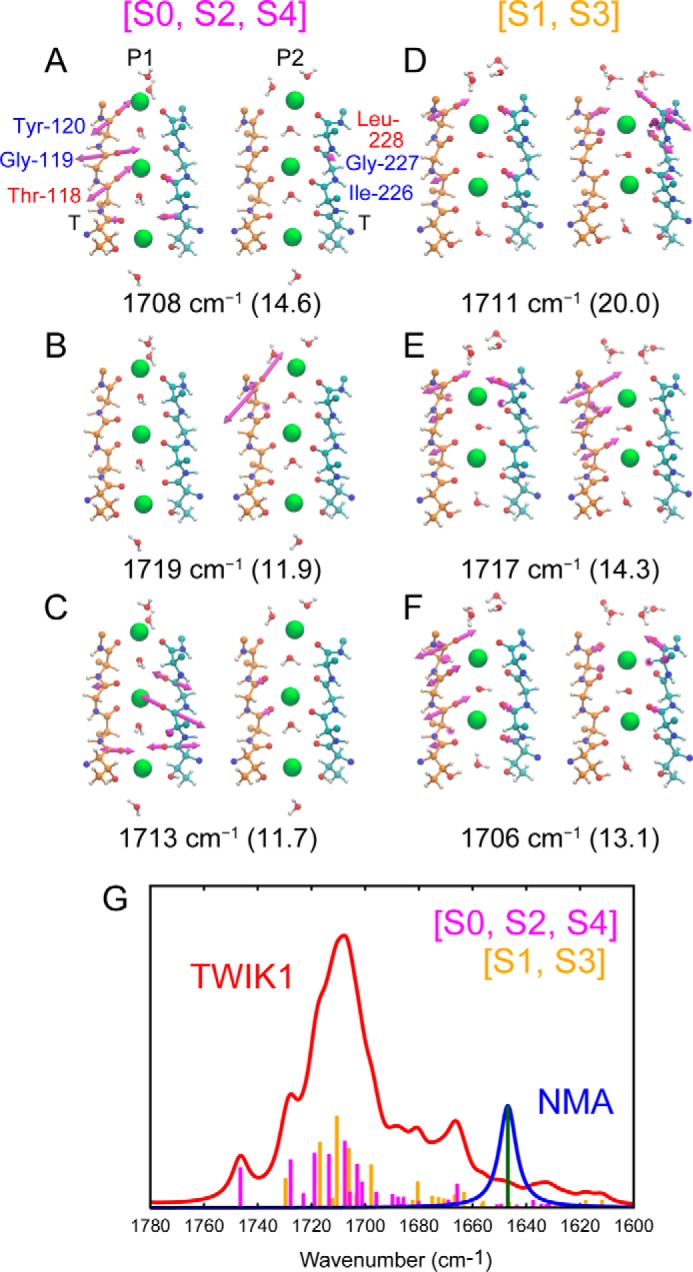
Computational analysis of the normal modes of the TWIK1 selectivity filter. The vibrational vectors for the normal modes of the three highest IR intensities for [S0, S2, and S4] (A–C) and [S1, S3] (D–F). The P1 and P2 domains in the selectivity filter are shown on the left and right sides, respectively. The frequency for each normal mode is shown at the bottom with its intensity in parentheses. G, calculated IR absorption spectra of the selectivity filter: TWIK1 (red) and NMA (blue). The IR intensities of all vibrational modes of the carbonyl groups in the selectivity filter are shown as bar graphs (magenta, TWIK1 [S0, S2, S4]; orange, TWIK1 [S1, S3]; and green, NMA).
Table 2.
Normal modes of the carbonyl groups in the selectivity filter of TWIK1 (five highest IR absorbance intensities)
| TWIK1 [S0, S2, S4] |
TWIK1 [S1, S3] |
||||
|---|---|---|---|---|---|
| Frequency | Intensity of IR absorbance | Contribution to the modea | Frequency | Intensity of IR absorbance | Contribution to the mode |
| cm−1 | % | cm−1 | % | ||
| 1708 | 14.6 | 21 (1.5A), 17 (2.5A), 11 (0.5A) | 1711 | 20.0 | 18 (0.5B′), 6 (0.5A′), 4 (1.5B′) |
| 1719 | 11.9 | 64 (0.5A′) | 1717 | 14.3 | 16 (0.5A′), 9 (0.5B), 6 (0.5A) |
| 1713 | 11.7 | 31 (2.5B), 16 (3.5A), 10 (3.5B) | 1706 | 13.1 | 14 (0.5A), 8 (2.5A), 8 (0.5B′) |
| 1728 | 10.5 | 56 (0.5B), 4 (0.5B′) | 1698 | 9.4 | 23 (2.5A′), 21 (2.5B′), 6 (3.5B′) |
| 1703 | 9.5 | 43 (0.5A), 6 (2.5A′), 4 (1.5A′) | 1730 | 6.5 | 31 (0.5B), 3 (0.5A′), 3 (0.5B′) |
a The number indicates the position of the carbonyl groups according to the K+-binding site number. 1.5 indicates a carbonyl group between S1 and S2 sites. A and B correspond to each monomer of TWIK1, which are colored orange and sky blue, respectively, in Fig. 7. A′ and B′ indicate the P2 domain.
Discussion
In this study, we successfully applied ion-exchange–induced difference ATR-FTIR spectroscopy to a mammalian two-pore–domain K+ channel, TWIK1, and found that the amide-I bands were assignable to the selectivity filter and pore helices affected by the unconventional amino acid residues (Thr-118 and Leu-228). We now discuss how TWIK1 achieves its unique characteristics and compare our experimental results with previously reported findings from electrophysiological studies (24, 26, 27), crystallographic analyses (30), and molecular dynamics simulations (37, 38).
Functional properties of purified TWIK1 proteins
Electrophysiological analyses have revealed that TWIK1 can conduct not only K+ and Rb+ but also Na+, Li+, NH4+, and Cs+ (24, 26, 27). In particular, the Na+ permeability of TWIK1 is unique in K+ channels. This low ion selectivity results from the presence of unconventional amino acid residues in the WT, Thr-118 in the P1 domain (24, 26), and Leu-228 in the P2 domain (see below) (Fig. 1B). Our flux assay showed a significant difference in the K+ flux between WT TWIK1 and T118I as well as L228F mutants to reach the equilibrium potentials determined by their permeability to K+ and Na+ (Fig. 1E), suggesting that the WT TWIK1 protein itself possesses a lower selectivity for K+ against Na+ than do the selectivity filter mutants. These results are consistent with previous electrophysiological studies that showed that the TWIK1 T118I mutant behaves like a “normal” K+ channel with high K+ selectivity (24, 26). Although the previous study showed that the L228F substitution had a small effect on K+ selectivity (24), our flux assay showed a significant increase of K+ efflux by the L228F mutation (Fig. 1E). The K+ ion selectivity of the L228F mutant is thought to be more evidently observable under our experimental conditions than in the previous study (see “Results”). Taken together, our purified TWIK1 samples show functional characteristics relating to the unconventional residues in the selectivity filter. Because leucine and phenylalanine residues have relatively similar properties in terms of hydrophobicity and bulkiness, the L228F mutation may only cause a slight structural change in the selectivity filter. However, this might influence the environment around Thr-118 through secondary effects (Fig. S6).
Interaction of TWIK1 with K+
Because the IR spectral changes of TWIK1 upon the K+ to Na+ exchange (Fig. 3) are similar to those of KcsA (10, 13), we could assign some of the vibrational bands by referring to KcsA reports. We also performed QM/MM calculations to obtain the normal modes of the carbonyl groups in the TWIK1 selectivity filter interacting with K+ ions (Fig. 7). Thus, we successfully assigned the positive (K+ side) band at 1680 cm−1 (Fig. 3B) to the C=O stretching vibration of carbonyl groups in the selectivity filter interacting with K+ ions (see Fig. 1A). This mode likely corresponds to the “S2/S3 mode” in KcsA (13). Because the 1680-cm−1 band was also observed in the T118I and L228F mutants (Fig. 3B), the manner of interaction with K+ is not affected by the presence of Thr-118 and Leu-228. This result is consistent with the crystal structure of TWIK1 (WT) as its filter structure and K+-binding sites (S1–S4) are virtually identical to those of KcsA (see Fig. 1A). By contrast, concentration dependence of the spectral changes upon the K+ to Na+ exchange showed that the intensity of the 1680-cm−1 band in the WT was much less sensitive to K+ concentration increases (half-maximum value = 39.6 mm) than in the mutants T118I (3.8 mm) and L228F (7.7 mm) (Fig. 4D). These results suggest that the affinity of the selectivity filter for K+ is negatively affected by Thr-118 as well as Leu-228 (it may be also possible that the affinity of the filter for Na+ is relatively increased by these residues). High affinity for K+ in the selectivity filter is considered to be important for obtaining high K+ selectivity (4, 39, 40). Thus, the lower affinity of the binding sites for K+ relative to Na+ is consistent with the overall lower K+ selectivity of TWIK1.
The IR spectral studies on KcsA have reported that in the presence of low-K+ concentrations (< ∼10 mm) the 1680-cm−1 band is shifted to a higher frequency (1688 cm−1) (10). The band shift might be correlated to the “collapse” of the selectivity filter in the KcsA crystal structures in a “low-K+” environment ([K+] = 3 mm) (7, 10). However, such a frequency shift was also observed in Rb+ and Cs+ conditions in which the filter does not collapse. Therefore, the upshift may be caused by changes in the ion occupancy at the S2 and S3 sites (12). Such a shift was not observed in WT TWIK1 (Fig. 4A), but a smaller shift was observed in T118I in the presence of very low-K+ concentrations (< ∼1 mm) (Fig. 4B). This result implies that WT TWIK1 releases K+ ions without changing the ion occupancy in the selectivity filter as the K+ concentration drops below the Kd1 value (12.7 ± 3.3 mm). Thus, in WT TWIK1, Thr-118 may retard the filter response to changes in the ion occupancy state in the selectivity filter at low-K+ concentrations (Fig. 5, D and E).
Why do the filter carbonyl groups interacting with K+ ions shift their vibrations to a higher frequency? This could be explained by the depletion of hydrogen bonds from water molecules. In the presence of K+ ions in the filter, isolated water molecules between the K+ ions exist persistently, and influx of water molecules is hampered by the resting K+ ions at each binding site (see Movies S1 and S2, and snapshots of the filter structure in Fig. 7, A and B), which is also true for KcsA (Fig. S5 as well as Movies S3 and S4). Therefore, the carbonyl groups only interact with K+ ions and do not form stable hydrogen bonds with water molecules.
Interaction of TWIK1 with Rb+ and Cs+
In the TWIK1 mutants, T118I and L228F, the IR difference spectra upon the K+ to Rb+/Cs+ exchanges showed a negative (Rb+/Cs+ side) band at ∼1690 cm−1 (Fig. 6, B and C). A very similar band was observed in KcsA (11). These results suggest that interactions of the TWIK1 mutants with these cations are similar to those of KcsA. The previous KcsA study identified the ∼1690-cm−1 band to be the 1680-cm−1 band in the K+-bound form (the S2/S3 mode); the band only upshifted, and the ∼10-cm−1 shift reflects a slight difference in the manner of interaction between K+ and Rb+/Cs+ (11). This difference may be correlated with ion occupancy in the selectivity filter. Actually, in crystal structures of Rb+/Cs+-bound KcsA, the ion-binding sites in the selectivity filter were only occupied by three cations, although four K+ ions can be found in the K+-binding sites (41). In the TWIK1 mutants, Rb+/Cs+ binds similar ion-binding sites to those in KcsA. By contrast, WT TWIK1 possesses a dramatically reduced band intensity at ∼1690 cm−1 upon the K+ to Rb+/Cs+ exchange (Fig. 6, B and C). This can be interpreted to indicate that Rb+/Cs+ unstably interacts with the WT selectivity filter. Therefore, the carbonyl groups in the selectivity filter of WT TWIK1 could be so flexible that the filter accommodates the larger cations (Rb+ or Cs+) without specific static interactions as observed in KcsA. A previous molecular dynamics (MD) study of WT TWIK1 supported this interpretation as Thr-118 and Leu-228 were shown to increase the conformational freedom of the selectivity filter (38).
Interaction of TWIK1 with other monovalent cations
The previous IR study on KcsA by Stevenson et al. (13) assigned a negative (Na+ side) 1630-cm−1 band as the B-site mode, representing interactions of Na+ with four carbonyl groups in the selectivity filter (see Introduction). For TWIK1, a negative band at 1626 cm−1 (Fig. 3B) could correspond to the same B-site mode, although the intensity was relatively smaller than the 1627-cm−1 band observed in KcsA. Importantly, another negative band at ∼1642 cm−1 was prominent in the mutants, T118I and L228F, but was reduced in the WT (Fig. 3). These results may imply that Na+ interacts with WT TWIK1 and the mutants in a somewhat different manner from that observed for KcsA. Moreover, the ∼1642-cm−1 band was also prominent in the absence of monovalent cations in the pore region when K+ was exchanged to NMDG+ (Fig. 6, E and J). Therefore, we do not interpret the ∼1642 cm−1 to a Na+-binding mode. It may be a helix mode in the absence of K+ that is downshifted from ∼1653 cm−1. Because the K+ to Li+, K+ to NH4+, and K+ to NMDG+ exchanges caused similar spectral changes in the amide-I region to that obtained upon replacing K+ with Na+ (Figs. 3 and 6), the negative bands could not provide any specific interactions with these cations, which is further discussed in the next paragraph.
Like other K+ channels, a large cation, such as NMDG+, cannot permeate through TWIK1 (26), suggesting that NMDG+cannot enter the pore (selectivity filter region). Notably, the K+ to NMDG+ exchange caused similar spectral changes in the WT TWIK1 and the mutants to those in the K+ to Na+ exchange (Figs. 3B and 6E). Upon replacement of K+ with NMDG+ in the perfused buffer solution, K+ ions in the selectivity filter were forced out, and water molecules were expected to enter the pore. This may suggest that upon removal of K+ from the pore, water molecules can induce most of the structural changes in the selectivity filter and the pore helices of TWIK1 observed upon the K+ to Na+ exchange. In other words, K+ is required for the formation of the filter structure that is seen in the crystal structure (30).
How do Thr-118 and Leu-228 lower the ion selectivity in TWIK1?
TWIK1 mutants T118I and L228F, with higher K+ selectivity (Fig. 1E), show higher affinity for K+ ions (Figs. 4 and 5). Previous electrophysiological studies reported that TWIK1 conducted Na+ when extracellular K+ concentrations were less than 3 mm ([Na+]out >137 mm) (26). Our concentration-dependent experiment (Fig. 4) suggested that under similar conditions ([K+] = 1 mm and [Na+] = 199 mm), most of the WT TWIK1 proteins released K+, at least from the S2/S3 sites based on the band intensity at 1680 cm−1. The enhanced exclusion of K+ from the pore could encourage Na+ permeation because the presence of a K+/Na+ mixture in the pore is suggested to be energetically unfavorable for K+/Na+ permeation (6).
The mutants also showed vibration bands specific for Rb+ and Cs+ at ∼1690 cm−1. These bands were not observed in the WT (Fig. 6, B, C, G, and H) and suggest that cation binding changes the structure and electrostatic environment of the selectivity filter in the mutants, but these changes are minimal in the WT. As discussed above, the missing bands in the WT could be partly due to inhomogeneous broadening and decoupling induced by the increased flexibility of carbonyls in the selectivity filter. Therefore, each of the carbonyl groups in the filter forms hydrogen bonds with water molecules randomly and is decoupled from the specific C=O stretching vibration we followed at ∼1680 cm−1. The flexibility of the filter could encourage nonselective permeation because one Na+-permeable KcsA E71A mutant possessed a highly flexible selectivity filter that could rotate carbonyls at position 76 (corresponding to position 118 or 226 in TWIK1), resulting in the loss of stable interactions with cations (42). One of the corresponding residues of Glu-71 is Tyr-217 in the P2 domain of TWIK1. The previous MD simulation demonstrated that Tyr-217 disrupted a hydrogen bond with Asp-230, which corresponds to Asp-80 in KcsA, and this led to destabilization of the alignments of the carbonyl groups in the selectivity filter (38). To clarify the difference in dynamics around the selectivity filter of TWIK1 and KcsA, the trajectories of the MD simulations are animated as shown in Movies S5–S8. Although our MD simulation was only performed with the duration of 10 ns for equilibration of the system needed for the QM calculation, the disruption of the hydrogen bond between Tyr-217 and Asp-230 was observed in a trajectory of TWIK1 with [S0, S2, S4] configuration (Movie S6). Switch of hydrogen-bonding interaction was also observed for Thr-118 (38) (Movies S5 and S7), which forms a hydrogen bond with a hydroxyl group of the side chain at Ser-116 or a carbonyl group of the main chain at Ser-222. To reach a reliable conclusion, we need to perform the MD simulations with longer duration and to systematically examine the initial parameters, such as protonation states of key residues pointed out by the previous MD simulation (His-211 and Asp-230) (38).
Taken together, Thr-118 and Leu-228 have two synergistic effects in the pore region: one is reduction of affinity for K+ relative to Na+, and the other is impairment of stable interactions with cations other than K+.
Contribution of the “cap” domain to the ion selectivity of TWIK1
Our IR analyses revealed properties of the selectivity filter and pore helices that were related to the ion selectivity; however, other regions in TWIK1 can contribute to its unique channel properties. The crystal structures of TWIK1 and TRAAK revealed the extracellular cap domain specific to two-pore–domain K+ channels and suggested that the cap regulates ion permeation (30, 31). Our HS-AFM observation visualized the cap domain in a single molecule of WT TWIK1 and the T118I mutant for the first time (Fig. 2). The conical shape and height of the cap did not change under our measurement conditions (Fig. S7 as well as Movies S9–S11). The regulation of ion permeation by this domain may be achieved by small and/or fast structural changes beyond the time and spatial resolutions of the HS-AFM (time resolution of 0.2–0.5 s/frame as well as spatial resolutions of ∼2 nm for lateral direction and ∼0.1 nm for vertical direction). In addition, a previous MD simulation study showed that a hydrophobic barrier in a deep inner pore of TWIK1 was important for K+ conductance (37). Thus, the ion permeation would be regulated cooperatively by the pore region as well as other regions.
Conclusion
As we show here, the vibrational bands in the amide-I region are very useful for understanding the molecular mechanisms of not only KcsA but also a mammalian K+ channel. Thus, ATR-FTIR spectroscopy can be applicable for exploring how various other K+ channels may operate, and the vibrational bands we noticed here can be used as “molecular fingerprints” to assess the functional properties of a diverse group of K+ channels.
Experimental procedures
Expression and purification of TWIK1 channels
A full-length gene of mouse TWIK1 with a His8 tag on the C terminus was inserted into the EcoRI/NotI site of a pMT vector. The mutations, T118I and L228F, were induced by conventional PCR using mutated primers. The His8-tagged WT TWIK1 and the mutants were transiently expressed in 20 plates (100 mm diameter) containing COS-1 cells. The cells were harvested 48 h after transfection, as described previously (43). The transfected cells were collected and solubilized with 1% DDM, 140 mm KCl, 20 mm imidazole, and 20 mm HEPES at pH 7.4, and then centrifuged for 20 min at 100,000 × g and 4 °C. The supernatant was mixed with Ni-Sepharose 6 Fast Flow (GE Healthcare) for 5 h at 4 °C, then transferred into Bio-Spin columns (Bio-Rad), washed with 0.05% DDM, 2 mm ATP, 0.5 m KCl, 3 mm MgCl2, 20 mm imidazole, 20 mm HEPES, pH 7.4, and subsequently washed with 0.05% DDM, 140 mm KCl, 50 mm imidazole, and 20 mm HEPES at pH 7.4. The His8-tagged proteins were eluted with 0.05% DDM, 140 mm KCl, 300 mm imidazole, and 20 mm HEPES, at pH 7.4. For Ni-Sepharose purification of the Δcap mutant lacking the cap domain (amino acid residues from positions 47 to 101 are deleted), we used a DDM/cholesteryl hemisuccinate mixture (5:1 (w/w)) instead of only DDM to stabilize the mutant proteins. The eluted samples were injected onto a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with 0.05% DDM, 140 mm KCl, and 20 mm HEPES at pH 7. The elution rate was set at 0.5 ml/min, and the absorbance at 280 nm was monitored using a SPD-20A detector (Shimadzu). The fraction eluted from 22.5 to 27.5 min was collected and concentrated using an Amicon Ultra (MWCO 10K, Millipore, Temecula, CA) concentrator.
Flux assay
The flux assays for purified TWIK1 samples were conducted according to the procedure from previous studies of two-pore K+ channels (30, 31). The purified TWIK1 samples were reconstituted into liposomes by mixing with a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) (Avanti Polar Lipids, Alabaster, AL) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-1′-rac-glycerol (POPG) (Avanti Polar Lipids) lipid mixture (7.5 and 2.5 mg/ml) solubilized in 8% n-octyl-β-d-maltopyranoside (OM) (Anatrace, Maumee, OH) at the protein/lipid weight ratio of 1:100 (molar ratio of 1:∼5400). The protein/lipid mixture was dialyzed against 150 mm KCl and 10 mm HEPES at pH 7 for 4 days (the buffer was exchanged once each day) to remove the detergents. The “empty” liposome samples (Fig. 1E) were prepared without channel proteins. The protein-containing liposome samples, after dialysis, were flash-frozen in liquid N2 and stored at −80 °C.
On the day of the assay, the frozen liposome samples were thawed and sonicated for 10 s. 7.5 μl of the samples were mixed into 300 μl (40-fold) of 150 mm NaCl, 10 mm HEPES at pH 7 containing 2 μm 9-amino-6-chloro-2-methoxyacridine (ACMA). The mixture was loaded in a cuvette and placed into a FP-6500 fluorimeter (Jasco, Tokyo, Japan). The excitation and emission wavelengths were 410 nm (1 nm slit) and 490 nm (10 nm slit). After 150 s of measurements, protonophore CCCP was added at a final concentration of 1 μm (see Fig. 1E). A K+-selective ionophore, valinomycin (final concentration of 20 nm), was added at 600 s (see Fig. 1E). The measurements were conducted at 25 °C, and the stock solutions of ACMA, CCCP, and valinomycin were prepared using DMSO at concentrations of 2 and 1 mm and 20 μm, respectively.
Reconstitution of the TWIK1 channel into lipid vesicles for IR measurements
The purified TWIK1 samples were reconstituted to liposomes by mixing with POPE/POPG (7.5 and 2.5 mg/ml) solubilized in 8% OM at the protein/lipid molar ratio of 1:50. Detergents were removed by mixing with Bio-beads (Bio-Rad) for 1.5 h. The samples, after Bio-bead treatment, were centrifuged for 25 min at 100,000 × g, 4 °C, and the pellet was rinsed twice with 140 mm KCl and 20 mm HEPES at pH 7. The pellet was suspended with 10 mm KCl and 2 mm potassium phosphate at pH 7, and the final protein concentration in the suspension was ∼1 mg/ml.
ATR-FTIR measurements
A suspension of liposome samples (∼6 μl) containing ∼6 μg of purified TWIK1 protein was placed on the surface of a diamond ATR crystal (Smith Detection, DurasamplIR II, nine internal reflections with 45° of the incident angle). After drying under a gentle stream of N2, the sample was washed with 140 (for K+–monovalent cation exchange, Figs. 2 and 4) or 200 mm KCl (for concentration-dependent measurement, Fig. 4) and 20 mm HEPES at pH 7 and a flow rate of 1 ml/min for ∼60 min. The ATR-FTIR spectra of the samples were recorded at 25 °C with 2 cm−1 spectral resolution using a Vertex 70 spectrometer (Bruker Optics, Ettlingen, Germany) equipped with a liquid nitrogen-cooled MCT detector. For K+ monovalent cation exchange measurements (Figs. 3 and 6), the perfused buffer solutions were changed to 140 mm KCl and 20 mm HEPES at pH 7 as well as 140 mm NaCl, LiCl, RbCl, CsCl, NH4Cl, or NMDG and 20 mm HEPES at pH 7. For K+/Na+ concentration-dependent measurements (Fig. 4), the perfused buffer solutions were changed to 0:200, 100:100, 150:50, 180:20, 190:10, 197:3, 199:1, 199.7:0.3, or 199.9:0.1 mm NaCl/KCl in the presence of 20 mm HEPES at pH 7. Before the spectra were measured, the sample was washed with a buffer for ∼14 min. The IR spectra were recorded by collecting 768 interferograms; this took ∼11 min. Then, the buffer was changed; the sample was washed, and the spectra were measured again. After the absorption spectra, measurements before and after buffer exchanges were completed four times; the difference spectra were calculated by subtraction, correction of membrane swelling changes, and nonspecific distortions associated with ion exchange according to Lórenz-Fonfría et al. (44). The absolute absorption spectra of TWIK1 samples immersed in buffer solution (Fig. 1F) were obtained by subtracting the buffer contribution according to Lórenz-Fonfría et al. (44).
Global fitting of the concentration-dependent data
The equation used for fitting the difference absorbance spectra is presented as Equation 5, which was constructed based on a sequential binding model (see supporting Methods). Global fitting was performed using IgorPro Version 6.37 software.
| (1) |
High-speed AFM observation
HS-AFM experiments were performed with a laboratory-built HS-AFM operated in the tapping mode (35). A cantilever with a nominal resonant frequency of ∼0.8 MHz and a spring constant of ∼0.2 newtons/m was used. The cantilever's free oscillation amplitude was ∼1 nm, and the reference amplitude for feedback control was 70–90% of the free oscillation amplitude.
A proteoliposome composed of TWIK1 channels was prepared using the IR measurement samples procedure (see above). The sample was deposited onto a freshly cleaved mica substrate after the substrate was sonicated for 1 min. After an incubation time of 3 min, the residual lipids and molecules were thoroughly washed using the observation buffer (20 mm HEPES and 140 mm KCl or NaCl at pH 7). Then, the AFM probe was approached to the sample surface, and imaging was carried out.
Computational analysis of the normal modes of the selectivity filter
The X-ray crystal structure of the TWIK1 dimer (PDB code 3UKM) (30) was used for the initial geometry. Missing loops were modeled with the ModLoop program (45), and the cavity was filled with water molecules using the SOLVATE program (https://www.mpibpc.mpg.de/grubmueller/solvate).4 For K+ ions in the filter, two configurations were considered: K+ ions in the S0, S2, and S4 positions were kept in one state ([S0, S2, S4] configuration), whereas those in the S1 and S3 positions were kept in the second state ([S1, S3] configuration). K+ ions in the remaining positions were replaced with water molecules. The TWIK1 models were embedded in a pre-equilibrated POPC lipid bilayer and then solvated with a rectangular water box (104 × 87 × 128 Å) with periodic boundary conditions. Na+ ions were added to neutralize the system, and the total system consisted of ∼120,000 atoms.
We first carried out the MD simulations for 10 ns with a molecular mechanics force field to equilibrate the system. Amber ff99SB-ILDN (47), LIPID14 (48), and TIP3P (49) force fields were used for the proteins, POPC lipids, and water molecules, respectively. The Lennard-Jones parameters developed by Joung and Cheatham (50) were used for the cations. The particle mesh Ewald method was used for long-range electrostatic interactions. Bonds involving hydrogen atoms were constrained using the SHAKE method. The equations of motion were integrated using the leapfrog algorithm with a time step of 2 fs at 300 K. During the MD simulation, a weak harmonic restraint (force constant of 1 kcal/(mol Å2)) was applied to the backbone heavy-atom positions in the proteins. The MD simulations were performed using the AMBER program package (51).
The final snapshots of the MD simulations were used as the initial geometries for the QM/MM calculations. In the QM/MM method, the QM subsystem consisted of the filter part of the protein (four pentapeptide backbones with threonine side chains), K+ ions, and water molecules that hydrogen bond with the carbonyl groups in the filter. The link atom method was used for the QM/MM boundary. Including the link hydrogen atoms, the QM subsystem consisted of 166 and 168 atoms for the [S0, S2, S4] and [S1, S3] configurations, respectively (see Fig. 7). B3LYP density functional with the 6–31G (d,p) basis set was used for the QM subsystem. We adopted a site-site representation of the QM/MM electrostatic interaction energy (52, 53) and the RESP scheme (54) to describe the partial charges of the QM subsystem. The long-range electrostatic interactions in the QM/MM calculation were smoothly truncated at 15 Å using the tapering method implemented in the TINKER program (55). After geometry optimization of the QM subsystem, the vibrational frequencies of the QM subsystem were calculated with a scaling factor of 0.940. In the IR spectrum calculations, we assumed that the probability for the [S0, S2, S4] configuration was identical to that of the [S1, S3] configuration. The QM/MM calculations and our in-house QM/MM routine were performed using the GAMESS program package (56).
For comparison, we also calculated the IR spectra of the KcsA channel in K+ solutions. The X-ray crystal structures of KcsA in high K+ concentration conditions (PDB code 1K4C) (2) were used for the initial geometry. As in the case of TWIK1, two configurations, [S0, S2, S4] and [S1, S3], were considered for KcsA. In addition, we calculated the IR spectrum of aqueous NMA as a reference. The NMA system consisted of 1 NMA and 3091 water molecules, and the QM subsystem consisted of 1 NMA and 2 water molecules that hydrogen-bonded with the carbonyl group of NMA.
Author contributions
H. T. and Y. K. resources; H. T., M. H., H. M., and Y. F. formal analysis; H. T. and M. H. validation; H. T., M. H., H. M., H. W., C. G., K. N., T. U., and Y. F. investigation; H. T., M. H., H. W., C. G., T. U., and Y. F. visualization; H. T., M. H., T. U., and Y. F. methodology; H. T. and Y. F. writing-original draft; H. T., M. H., K. N., Y. K., T. U., and Y. F. writing-review and editing; M. H. and C. G. software; Y. F. conceptualization; Y. F. supervision; Y. F. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. David Farrens (Oregon Health and Science University) for providing us with expression vectors and COS-1 cell lines. We also thank Hiroe Motomura and the Functional Genomics Facility, NIBB Core Research Facilities (Okazaki, Japan) for technical support and Dr. Víctor A. Lórenz-Fonfría for valuable suggestions on global fitting analysis. We thank Enago for the English language review. The computations were performed at the Research Center for Computational Science, Okazaki, Japan.
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI Grants 25840122 and 17K15109 (to H. T.); 16H00778, 16KT0165, and 17K05757 (to M. H.); and 22247024, 22770159, 24650203, 26640047, and 26708002 (to Y. F.); NINS program for cross-disciplinary study (to Y. F. and K. N.); the Cooperative Study Program of National Institute for Physiological Sciences; and JST CREST Grant JPMJCR17N5. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7, supporting Methods, supporting Movies S1–S11, and supporting Refs. 1–2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ATR-FTIR
- attenuated total reflection-Fourier-transform infrared
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- AFM
- atomic force microscopy
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- DDM
- n-dodecyl-β-d-maltoside
- FSEC
- fluorescence-detected size-exclusion chromatography
- K2P
- two-pore–domain potassium
- NMDG
- N-methyl-d-glucamine
- OM
- n-octyl-β-d-maltopyranoside
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
- 1-1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- PDB
- Protein Data Bank
- FWHM
- full width at half-maximum
- QM/MM
- quantum mechanical/molecular mechanical
- NMA
- N-methylacetamide
- MD
- molecular dynamics
- HS-AFM
- high-speed atomic force microscopy.
References
- 1. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., and MacKinnon R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- 2. Zhou Y., Morais-Cabral J. H., Kaufman A., and MacKinnon R. (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414, 43–48 10.1038/35102009 [DOI] [PubMed] [Google Scholar]
- 3. Gouaux E., and Mackinnon R. (2005) Principles of selective ion transport in channels and pumps. Science 310, 1461–1465 10.1126/science.1113666 [DOI] [PubMed] [Google Scholar]
- 4. Thompson A. N., Kim I., Panosian T. D., Iverson T. M., Allen T. W., and Nimigean C. M. (2009) Mechanism of potassium-channel selectivity revealed by Na+ and Li+ binding sites within the KcsA pore. Nat. Struct. Mol. Biol. 16, 1317–1324 10.1038/nsmb.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lockless S. W., Zhou M., and MacKinnon R. (2007) Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5, e121 10.1371/journal.pbio.0050121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nimigean C. M., and Allen T. W. (2011) Origins of ion selectivity in potassium channels from the perspective of channel block. J. Gen. Physiol. 137, 405–413 10.1085/jgp.201010551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou M., Morais-Cabral J. H., Mann S., and MacKinnon R. (2001) Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature 411, 657–661 10.1038/35079500 [DOI] [PubMed] [Google Scholar]
- 8. Ye S., Li Y., and Jiang Y. (2010) Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat. Struct. Mol. Biol. 17, 1019–1023 10.1038/nsmb.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noskov S. Y., Bernèche S., and Roux B. (2004) Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature 431, 830–834 10.1038/nature02943 [DOI] [PubMed] [Google Scholar]
- 10. Furutani Y., Shimizu H., Asai Y., Fukuda T., Oiki S., and Kandori H. (2012) ATR-FTIR spectroscopy revealed the different vibrational modes of the selectivity filter interacting with K+ and Na+ in the open and collapsed conformations of the KcsA potassium channel. J. Phys. Chem. Lett. 3, 3806–3810 10.1021/jz301721f [DOI] [PubMed] [Google Scholar]
- 11. Furutani Y., Shimizu H., Asai Y., Oiki S., and Kandori H. (2015) Specific interactions between alkali metal cations and the KcsA channel studied using ATR-FTIR spectroscopy. Biophys. Physicobiol. 12, 37–45 10.2142/biophysico.12.0_37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furutani Y. (2017) Ion-protein interactions of a potassium ion channel studied by attenuated total reflection Fourier transform infrared spectroscopy. Biophys. Rev. 2017, 10.1007/s12551-017-0337-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stevenson P., Götz C., Baiz C. R., Akerboom J., Tokmakoff A., and Vaziri A. (2015) Visualizing KcsA conformational changes upon ion binding by infrared spectroscopy and atomistic modeling. J. Phys. Chem. B 119, 5824–5831 10.1021/acs.jpcb.5b02223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kratochvil H. T., Carr J. K., Matulef K., Annen A. W., Li H., Maj M., Ostmeyer J., Serrano A. L., Raghuraman H., Moran S. D., Skinner J. L., Perozo E., Roux B., Valiyaveetil F. I., and Zanni M. T. (2016) Instantaneous ion configurations in the K+ ion channel selectivity filter revealed by 2D IR spectroscopy. Science 353, 1040–1044 10.1126/science.aag1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inaguma A., Tsukamoto H., Kato H. E., Kimura T., Ishizuka T., Oishi S., Yawo H., Nureki O., and Furutani Y. (2015) Chimeras of channel rhodopsin-1 and -2 from Chlamydomonas reinhardtii exhibit distinctive light-induced structural changes from channelrhodopsin-2. J. Biol. Chem. 290, 11623–11634 10.1074/jbc.M115.642256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kasuya G., Fujiwara Y., Tsukamoto H., Morinaga S., Ryu S., Touhara K., Ishitani R., Furutani Y., Hattori M., and Nureki O. (2017) Structural insights into the nucleotide base specificity of P2X receptors. Sci. Rep. 7, 45208 10.1038/srep45208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., and Kandori H. (2013) A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 10.1038/ncomms2689 [DOI] [PubMed] [Google Scholar]
- 18. Granell M., León X., Leblanc G., Padrós E., and Lórenz-Fonfría V. A. (2010) Structural insights into the activation mechanism of melibiose permease by sodium binding. Proc. Natl. Acad. Sci. U.S.A. 107, 22078–22083 10.1073/pnas.1008649107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doki S., Kato H. E., Solcan N., Iwaki M., Koyama M., Hattori M., Iwase N., Tsukazaki T., Sugita Y., Kandori H., Newstead S., Ishitani R., and Nureki O. (2013) Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. U.S.A. 110, 11343–11348 10.1073/pnas.1301079110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Long S. B., Tao X., Campbell E. B., and MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- 21. Tao X., Avalos J. L., Chen J., and MacKinnon R. (2009) Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 10.1126/science.1180310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy J. G., and Nimigean C. M. (2012) Structural correlates of selectivity and inactivation in potassium channels. Biochim. Biophys. Acta 1818, 272–285 10.1016/j.bbamem.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lesage F., Guillemare E., Fink M., Duprat F., Lazdunski M., Romey G., and Barhanin J. (1996) TWIK1, a ubiquitous human weakly inward rectifying K+ channel with a novel structure. EMBO J. 15, 1004–1011 [PMC free article] [PubMed] [Google Scholar]
- 24. Chatelain F. C., Bichet D., Douguet D., Feliciangeli S., Bendahhou S., Reichold M., Warth R., Barhanin J., and Lesage F. (2012) TWIK1, a unique background channel with variable ion selectivity. Proc. Natl. Acad. Sci. U.S.A. 109, 5499–5504 10.1073/pnas.1201132109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma L., Xie Y. P., Zhou M., and Chen H. (2012) Silent TWIK1 potassium channels conduct monovalent cation currents. Biophys. J. 102, L34–L36 10.1016/j.bpj.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma L., Zhang X., and Chen H. (2011) TWIK1 two-pore–domain potassium channels change ion selectivity and conduct inward leak sodium currents in hypokalemia. Sci. Signal. 4, ra37 10.1126/scisignal.2001726 [DOI] [PubMed] [Google Scholar]
- 27. Ma L., Zhang X., Zhou M., and Chen H. (2012) Acid-sensitive TWIK and TASK two-pore–domain potassium channels change ion selectivity and become permeable to sodium in extracellular acidification. J. Biol. Chem. 287, 37145–37153 10.1074/jbc.M112.398164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Enyedi P., and Czirják G. (2010) Molecular background of leak K+ currents: two-pore–domain potassium channels. Physiol. Rev. 90, 559–605 10.1152/physrev.00029.2009 [DOI] [PubMed] [Google Scholar]
- 29. Lesage F., and Barhanin J. (2011) Molecular physiology of pH-sensitive background K(2P) channels. Physiology 26, 424–437 10.1152/physiol.00029.2011 [DOI] [PubMed] [Google Scholar]
- 30. Miller A. N., and Long S. B. (2012) Crystal structure of the human two-pore–domain potassium channel K2P1. Science 335, 432–436 10.1126/science.1213274 [DOI] [PubMed] [Google Scholar]
- 31. Brohawn S. G., del Mármol J., and MacKinnon R. (2012) Crystal structure of the human K2P TRAAK, a lipid- and mechano-sensitive K+ ion channel. Science 335, 436–441 10.1126/science.1213808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawate T., and Gouaux E. (2006) Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- 33. Lesage F., Reyes R., Fink M., Duprat F., Guillemare E., and Lazdunski M. (1996) Dimerization of TWIK1 K+ channel subunits via a disulfide bridge. EMBO J. 15, 6400–6407 [PMC free article] [PubMed] [Google Scholar]
- 34. Uchihashi T., Iino R., Ando T., and Noji H. (2011) High-speed atomic force microscopy reveals rotary catalysis of rotorless F(1)-ATPase. Science 333, 755–758 10.1126/science.1205510 [DOI] [PubMed] [Google Scholar]
- 35. Uchihashi T., Kodera N., and Ando T. (2012) Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 7, 1193–1206 10.1038/nprot.2012.047 [DOI] [PubMed] [Google Scholar]
- 36. Chen X. G., Schweitzerstenner R., Asher S. A., Mirkin N. G., and Krimm S. (1995) Vibrational Assignments of trans-N-methylacetamide and some of its deuterated isotopomers from band decomposition of Ir, visible, and resonance Raman Spectra. J. Phys. Chem. 99, 3074–3083 10.1021/j100010a017 [DOI] [Google Scholar]
- 37. Aryal P., Abd-Wahab F., Bucci G., Sansom M. S., and Tucker S. J. (2014) A hydrophobic barrier deep within the inner pore of the TWIK1 K2P potassium channel. Nat. Commun. 5, 4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oakes V., Furini S., Pryde D., and Domene C. (2016) Exploring the dynamics of the TWIK1 channel. Biophys. J. 111, 775–784 10.1016/j.bpj.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neyton J., and Miller C. (1988) Discrete Ba2+ block as a probe of ion occupancy and pore structure in the high-conductance Ca2+-activated K+ channel. J. Gen. Physiol. 92, 569–586 10.1085/jgp.92.5.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neyton J., and Miller C. (1988) Potassium blocks barium permeation through a calcium-activated potassium channel. J. Gen. Physiol. 92, 549–567 10.1085/jgp.92.5.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Y., and MacKinnon R. (2003) The occupancy of ions in the K+ selectivity filter: charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 333, 965–975 10.1016/j.jmb.2003.09.022 [DOI] [PubMed] [Google Scholar]
- 42. Cheng W. W., McCoy J. G., Thompson A. N., Nichols C. G., and Nimigean C. M. (2011) Mechanism for selectivity-inactivation coupling in KcsA potassium channels. Proc. Natl. Acad. Sci. U.S.A. 108, 5272–5277 10.1073/pnas.1014186108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tsukamoto H., and Farrens D. L. (2013) A constitutively activating mutation alters the dynamics and energetics of a key conformational change in a ligand-free G protein-coupled receptor. J. Biol. Chem. 288, 28207–28216 10.1074/jbc.M113.472464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lórenz-Fonfría V. A., León X., and Padrós E. (2012) Studying substrate binding to reconstituted secondary transporters by attenuated total reflection infrared difference spectroscopy. Methods Mol. Biol. 914, 107–126 [DOI] [PubMed] [Google Scholar]
- 45. Fiser A., and Sali A. (2003) ModLoop: automated modeling of loops in protein structures. Bioinformatics 19, 2500–2501 10.1093/bioinformatics/btg362 [DOI] [PubMed] [Google Scholar]
- 46. Deleted in proof.
- 47. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., and Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dickson C. J., Madej B. D., Skjevik A. A., Betz R. M., Teigen K., Gould I. R., and Walker R. C. (2014) Lipid14: the amber lipid force field. J. Chem. Theory Comput. 10, 865–879 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., and Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 10.1063/1.445869 [DOI] [Google Scholar]
- 50. Joung I. S., and Cheatham T. E. 3rd. (2008) Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 112, 9020–9041 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Case D. A., Cerutti D. S., Cheatham I. T., Darden T. A., Duke R. E., Giese T. J., Gohlke H., Goetz A. W., Greene D., Homeyer N., Izadi S., Kovalenko A., Lee T. S., LeGrand S., Li P., et al. (2017) AMBER17. University of California, San Francisco [Google Scholar]
- 52. Hayashi S., and Ohmine I. (2000) Proton transfer in bacteriorhodopsin: structure, excitation, IR spectra, and potential energy surface analyses by an ab initio QM/MM method. J. Phys. Chem. B 104, 10678–10691 10.1021/jp001508r [DOI] [Google Scholar]
- 53. Higashi M., and Truhlar D. G. (2008) Electrostatically embedded multiconfiguration molecular mechanics based on the combined density functional and molecular mechanical method. J. Chem. Theory Comput. 4, 790–803 10.1021/ct800004y [DOI] [PubMed] [Google Scholar]
- 54. Bayly C. I., Cieplak P., Cornell W. D., and Kollman P. A. (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges–the Resp model. J. Phys. Chem. 97, 10269–10280 10.1021/j100142a004 [DOI] [Google Scholar]
- 55. Ponder J. W. (1997) TINKER. 3.5 Ed., Washington University, St. Louis, MO [Google Scholar]
- 56. Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S. J., Windus T. L., Dupuis M., and Montgomery J. A. (1993) General atomic and molecular electronic-structure system. J. Comp. Chem. 14, 1347–1363 10.1002/jcc.540141112 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.