Abstract
Rheumatoid factors (RFs) are autoantibodies against the fragment-crystallizable (Fc) region of IgG. In individuals with hematological diseases such as cryoglobulinemia and certain B cell lymphoma forms, the RFs derived from specific heavy- and light-chain germline pairs, so-called “stereotypic RFs,” are frequently produced in copious amounts and form immune complexes with IgG in serum. Of note, many structural details of the antigen recognition mechanisms in RFs are unclear. Here we report the crystal structure of the RF YES8c derived from the IGHV1-69/IGKV3-20 germline pair, the most common of the stereotypic RFs, in complex with human IgG1-Fc at 2.8 Å resolution. We observed that YES8c binds to the CH2–CH3 elbow in the canonical antigen-binding manner involving a large antigen–antibody interface. On the basis of this observation, combined with mutational analyses, we propose a recognition mechanism common to IGHV1-69/IGKV3-20 RFs: (1) the interaction of the Leu432–His435 region of Fc enables the highly variable complementarity-determining region (CDR)-H3 to participate in the binding, (2) the hydrophobic tip in the CDR-H2 typical of IGHV1-69 antibodies recognizes the hydrophobic patch on Fc, and (3) the interaction of the highly conserved RF light chain with Fc is important for RF activity. These features may determine the putative epitope common to the IGHV1-69/IGKV3-20 RFs. We also showed that some mutations in the binding site of RF increase the affinity to Fc, which may aggravate hematological diseases. Our findings unravel the structural basis for germline-encoded antibody autoreactivity.
Keywords: autoimmunity, complex, crystal structure, IgG, structural biology
Introduction
Rheumatoid factors (RFs)4 are autoantibodies that specifically recognize the fragment-crystallizable (Fc) region of IgG and have long been used as an important diagnostic factor of rheumatoid arthritis (RA). Immunochemical RF research has been developed historically from studies using paraprotein-IgM RFs from B cell malignancies, such as mixed cryoglobulinemia (MC) and Waldenstrom's macroglobulinemia (WM) (1). There are differences in genetic diversity and binding pattern to the Fc region between RFs from B cell malignancies and those from RA. The genetic origin of RFs from RA is more diverse than that of RFs from B cell malignancy (2–7). The binding pattern of RFs to the different IgG subclasses (IgG1, IgG2, IgG3, or IgG4) is diverse in RFs from RA (8, 9), and it is suggested that the epitopes are distributed widely in the Fc region (10). In contrast, in B cell malignancies such as MC and WM, RFs with specific combinations of idiotypes are frequently observed (11). In particular, combinations of the heavy chain with the Wa or Ga cross-reactive idiotype and the light chain with the 17.109 cross-reactive idiotype are the most common and are related to the IGHV1-69 and IGKV3-20 germline genes, respectively (1).
In serum from healthy human donors immunized with mismatched red blood cells (12) as well as patients with MC caused by hepatitis C virus (HCV) infection (13), mucosa-associated lymphoid tissue–type lymphoma (14), HCV-associated lymphoma (15), and B cell chronic lymphocytic leukemia (16), high frequencies of RFs with combinations of heavy chains and light chains that are from specific germline genes, called “stereotypic RFs”, have been reported (17). Of the stereotypic RFs, those from IGHV1-69/IGKV3-20 are the most prominent. The heavy chain variable germline IGHV1-69 is frequently used as the heavy chain of the neutralizing antibodies for HCV (18, 19), HIV (20), influenzae virus (21), Middle East respiratory syndrome coronavirus (22), and the Staphylococcus aureus virulence factor NEAT (23). The κ light chain variable germline IGKV3-20 is commonly used as the partner of IGHV1-69. The RFs derived from IGHV1-69/IGKV3-20 are highly related to HCV-associated lymphoma (24). Of note, more than 90% of patients with MC are HCV carriers (25). In patients with HCV-associated MC, IgM antibodies from IGHV1-69/IGKV3-20 have RF activity with or without somatic hypermutations (26). These findings indicate that IGHV1-69/IGKV3-20-derived antibodies have some characteristics reflective of RF activity.
Despite physiological importance, the structural details of the antigen recognition mechanism have not been clarified for the IGHV1-69/IGKV3-20 RFs. Research has shown that IGHV1-69/IGKV3-20 RFs from healthy immunized donors, RA patients, and MC patients have long third complementarity-determining regions of the heavy chain (CDR-H3s) with a highly diverse amino acid composition, despite being restricted in length (12–15 amino acids) (14, 26, 27). On the other hand, it has been shown that the epitope of this type of RF is mostly located at the CH2–CH3 elbow in the Fc region (9). These findings lead to the following questions. Do the IGHV1-69/IGKV3-20 RFs recognize the same epitope with the same binding mechanism? If so, how does the highly diverse CDR-H3 participate in RF binding? Why does the CH2–CH3 cleft region of Fc tend to be the epitope of these RFs? And last, why is this germline combination prone to having RF activity? So far, the crystal structures of two RFs have been determined in complex with Fc. One of these is a RA patient–derived IgM rheumatoid factor, RF-AN, that recognizes the CH2–CH3 cleft via a site distal to the conventional antigen-binding site (28). The other is a high-affinity IgM rheumatoid factor, RF61, that recognizes the CH3 domain close to the C terminus (29). However, these RFs are not stereotypic RFs and therefore cannot provide answers to the aforementioned questions.
Here we report the 2.8 Å resolution crystal structure of an IGHV1-69/IGKV3-20 stereotypic monoclonal human RF, YES8c, in complex with the Fc fragment of human IgG1 (IgG1-Fc). YES8c is a monoreactive IgM RF obtained from bone marrow B cells from an RA patient with macroglobulinemia (30). Crystal structure and mutagenesis studies revealed a common mechanism used by a wide range of IGHV1-69/IGKV3-20 stereotypic RFs to interact with the same position in the Fc fragment.
Results
Sequence and binding characteristics of YES8c-Fab
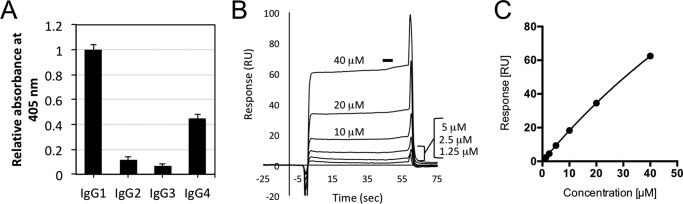
Amino acid sequence analysis of YES8c showed that the germline of the V, D, and J regions in the heavy chain are IGHV1-69, IGHD2-2, and IGHJ4, respectively (Fig. S1A). The germline of the V and J regions in the light chain are IGKV3-20 and IGKJ2, respectively (Fig. S1B). Therefore, YES8c is an IGHV1-69/IGKV3-20 stereotypic RF. According to previous research, YES8c is a monoreactive IgM RF that does not show cross-reactivity to self-antigens other than IgG-Fc; however, the precise binding characteristics are unknown (30). There are two large groups of the IgG subclass-specific RFs, Ga (binds to IgG1, 2 and 4) and Pan (binds to all subclasses) (10). We investigated the subclass specificity of YES8c. Although YES8c showed a preference for IgG1, it interacted with IgG1 and IgG4 based on ELISA results (Fig. 1A). This specificity was also confirmed by surface plasmon resonance (SPR) (Fig. S2). Based on these results, YES8c was categorized as another subclass specificity group. We measured the affinity of YES8c Fab for IgG1 by SPR (Fig. 1, B and C). The dissociation constant (Kd) was 160 ± 30 μm.
Figure 1.
Binding specificity of YES8c to IgG subclasses. A, binding of the purified YES8c-Fab to the immobilized four human IgG subclasses (IgG1, IgG2, IgG3, and IgG4) was measured by ELISA. Data show mean ± S.D. (error bars) from triplicates from one representative experiment of two independent experiments with similar results. B, representative sensorgrams derived from injection of different concentrations of YES8c-Fab over immobilized human IgG1 by surface plasmon resonance analysis. The black horizontal bar indicates the region of the sensorgrams from which equilibrium binding responses are derived. RU, resonance unit. C, plot of the equilibrium binding response against the concentration of YES8c-Fab for the sensorgrams shown in B. The line represents a nonlinear fit of the 1:1 binding model. The Kd is 160 ± 30 μm (mean ± S.D.) from three independent experiments.
YES8c–IgG1-Fc complex structure and implications for low-affinity interactions
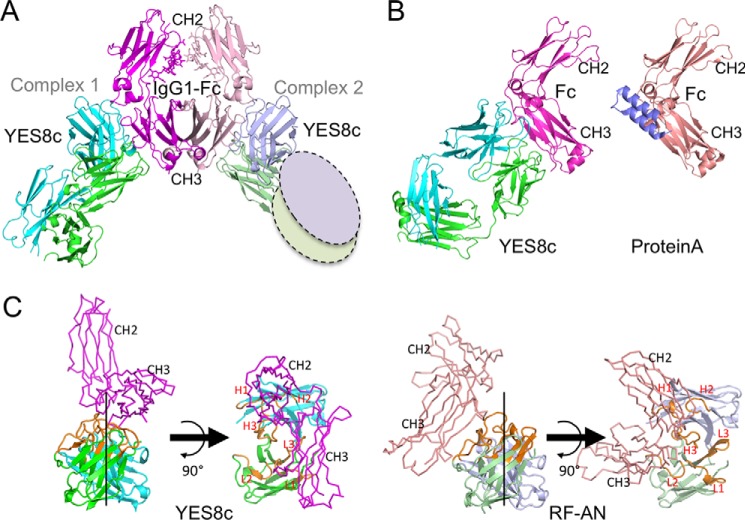
The structure of YES8c-Fab in complex with human IgG1-Fc was determined at 2.8 Å resolution (Table 1). Two YES8c–IgG1-Fc complexes were observed in the asymmetric crystal unit (Fig. 2A). Each YES8c–Fc complex was designated complex 1 or complex 2. In complex 2, the electron density of the constant region of YES8c was too weak to build atomic models, which might be due to crystal packing (Fig. S3). The heavy chain of YES8c recognizes the CH2–CH3 cleft, and the light chain mainly recognizes the CH3 domain. The binding site of the heavy chain overlaps with that of S. aureus protein A, which competitively inhibits the binding of most RFs to Fc regions (Fig. 2B) (31, 32).
Table 1.
Crystallographic data collection and refinement statistics
| YES8c-Fab–IgG1-Fc | |
|---|---|
| Data collection | |
| Space group | P65 |
| Number of crystals | 1 |
| Cell dimensions | |
| a, b, c (Å) | 189.9, 189.9, 79.6 |
| α, β, γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 20.0–2.80 (2.85–2.80)a |
| Rmerge | 0.074 (0.355)a |
| CC½ of the highest-resolution shell | 0.917 |
| I/σI | 19.1 (2.6)a |
| Completeness (%) | 98.7 (95.1)a |
| Redundancy | 4.6 (3.8)a |
| Refinement | |
| Resolution (Å) | 20.0–2.80 |
| No. of reflections | 40,124 |
| Rwork | 0.221 |
| Rfree | 0.270 |
| No. of atoms | |
| Protein | 8,175 |
| Water | 64 |
| Average B-factor | 52.0 |
| RMSDs | |
| Bond lengths (Å) | 0.003 |
| Angles (°) | 0.632 |
| Ramachandran plot (%) | |
| Favored | 95.12 |
| Allowed | 4.04 |
| Disallowed | 0.85 |
a The highest shell statistics are in parentheses.
Figure 2.
Crystal structure of YES8c in complex with IgG1-Fc. A, overall structure of the YES8c–Fab–Fc complex in an asymmetric unit. In complex 1, the Fc and heavy and light chains are shown in magenta, cyan, and green, respectively, and in light pink, light blue, and light green in complex 2. The ovals show the missing constant region (CL and CH1). B, the YES8c–Fc complex and protein A–Fc complex (PDB code 1L6X) from the same viewpoint. C, comparison between the YES8c and RF-AN binding mode. In YES8c, the Fc and heavy and light chains are shown in magenta, cyan, and green, respectively, and in light pink, light blue, and light green in RF-AN. CDRs are shown in orange in the both structure.
In the case of the rheumatoid factor RF-AN, the Fc region was bound to the edge of the conventional antigen-binding site of Fab, deviating from the pseudo-axis between VH and VL (Fig. 2C) (28). In contrast, in the case of YES8c, the Fc binds near the center of the axis (Fig. 2C). Consequently, 22 residues of YES8c participate in binding to Fc. This number is larger than those of two structurally determined RFs, RF-AN (nine residues) and RF61 (14 residues), whose affinities are higher than YES8c (Table 2). YES8c has a large buried surface area (BSA) within the complex, 2101 Å2 for complex 1 and 1996 Å2 for complex 2, compared with RF-AN (1458 Å2) and RF61 (1689 Å2), where the BSA represents the interfacial area of the YES8c–Fc complex. The YES8c BSA value ranks in the top 30% of the published protein–antibody complex structures (33). In the YES8c complex, seven to eight hydrogen bonds and no salt bridges are observed between the RFs and the Fc (Table 2 and Table S1). The number of van der Waals interactions is similar to that of RF-AN and RF61.
Table 2.
The number of contact residues, buried surface area, number of non-covalent interactions, and shape complementarity of the RF–Fc complexes
| YES8c (Complex 1) | YES8c (Complex 2) | RF-AN | RF61 | |
|---|---|---|---|---|
| Contact residues | ||||
| L chain | 10 | 11 | 2 | 5 |
| H chain | 14 | 10 | 7 | 9 |
| Buried surface area (Å) | ||||
| Total | 2,101 | 1,996 | 1,458 | 1,689 |
| L chain | 450 | 427 | 196 | 255 |
| H chain | 593 | 577 | 552 | 578 |
| Fc | 1,058 | 992 | 710 | 856 |
| Salt bridgesa | 0 | 0 | 1 | 2 |
| Hydrogen bondsa | 7 | 8 | 5 | 12 |
| van der Waals contactsa | 71 | 81 | 66 | 74 |
| Shape complementarity (Sc score) | ||||
| LH-Fc | 0.65 | 0.67 | 0.69 | 0.69 |
| L-Fc | 0.57 | 0.58 | 0.44 | 0.65 |
| H-Fc | 0.73 | 0.72 | 0.75 | 0.69 |
a The criteria of the atomic contacts are as follows: C-C, 4.1 Å; C-N, 3.8 Å; C-O, 3.7 Å; O-O, 3.3 Å; O-N, 3.4 Å; N-N, 3.4 Å; C-S, 4.1 Å; O-S, 3.7 Å; N-S, 3.8 Å.
Fc recognition by the heavy chain of YES8c
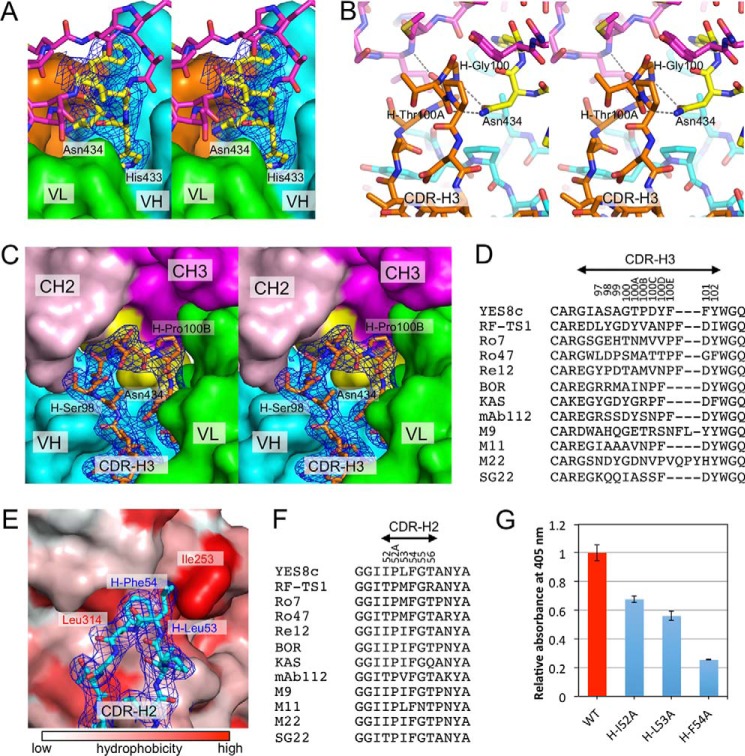
The residues in the heavy chain were highly conserved among the 12 IGHV1-69/IGKV3-20 RFs, except for a part of CDR-H1 and the entirety of CDR-H3 (Fig. S4A). The interfacial area (∼590 Å2) and the shape complementarity coefficient (34) (0.72 and 0.73) between the YES8c heavy chain and Fc are larger than those of the light chain (Table 2). A structural feature of the YES8c–Fc complex is that the protrusion at Leu432–His435 in the Fc region sticks into the pocket formed between VL and VH and binds to the residues in CDR-H2 and CDR-H3 (Fig. 3A). The interfacial area of the Leu432–His435 region (∼320 Å2) occupies about one-third of the interfacial area on the Fc surface (Fig. S5A). In particular, the side chain of Asn434 forms hydrogen bonds with the main chain atoms (N of H-Gly100 and O of H-Thr100A) of CDR-H3 from inside the binding site (Fig. 3B). Consequently, the residues in CDR-H3 (especially from H-Ala99 to H-Pro100B) are located in the space between CH2 and CH3 (Fig. 3C). As pointed out in previous reports, the amino acid composition of CDR-H3 in IGHV1-69/IGKV3-20 RFs is diverse, although the length is restricted to 12–15 residues (Fig. 3D) (14, 17, 27).
Figure 3.
Fc recognition by the heavy chain of YES8c. A, stereo view of the protruding region in Leu432–His435 of Fc bound to YES8c. The heavy chain and the light chain of YES8c are shown as cyan and green surfaces, respectively, and the Fc is shown as magenta sticks. The CDR-H3 is shown in orange. The Leu432–His435 region is shown in yellow. A 2m|Fo| − D|Fc| map for the Leu432–His435 region is contoured at 1 σ (blue mesh). B, stereo view of the interaction between CDR-H3 (orange) and the Asn434 (yellow). Hydrogen bonds are shown as dashed lines. C, stereo view of the interaction between CDR-H3 and the CH2–CH3 cleft in Fc. The surfaces of CH2 and CH3 are shown in light pink and magenta, respectively. The Leu432–His435 region is shown in yellow. VH (excluding CDR-H3) and VL are shown as cyan and green surfaces, respectively. CDR-H3 is shown as orange sticks. A 2m|Fo| − D|Fc| map for the CDR-H3 is contoured at 1 σ (blue mesh). D, alignment of the amino acid residues in CDR-H3 of YES8c and the other IGHV1-69/IGKV3-20 RFs. The residues are numbered based on the Kabat numbering scheme. E, interactions between CDR-H2 of YES8c and Fc. The surface of Fc is shown with the hydrophobicity scale (48). A 2m|Fo| − D|Fc| map for the CDR-H2 is contoured at 1 σ (blue mesh). F, alignment of the amino acid residues in CDR-H2 of YES8c and the IGHV1-69/IGKV3-20 derived rheumatoid factors. G, binding of the mutants in the CDR-H2 of YES8c to the immobilized IgG1 by ELISA. Data show mean ± S.D. (error bars) from triplicates from one representative experiment of two independent experiments with similar results.
The hydrophobicity of the CDR-H2 tip is a characteristic of the heavy chain derived from the IGHV1-69 germline. In YES8c, the hydrophobic residues (H-Leu53 and H-Phe54) at the CDR-H2 tip interact with the hydrophobic moiety in the CH2–CH3 cleft comprised of Ile253 and Leu314 (Fig. 3E). The residues in CDR-H2 form a large interaction interface (Fig. S5B). Comparing the sequence of the IGHV1-69/IGKV3-20 RFs, the hydrophobicity in CDR-H2 is highly conserved (Fig. 3F). To investigate the role of the residues in CDR-H2, the binding of the Ala mutants of YES8c was measured by ELISA (Fig. 3G). The bindings were lower in H-I52A and H-L53A mutants compared with the WT. The binding was significantly lower in the H-F54A mutant. SPR analysis confirmed the large decrease in binding of the H-F54A mutant (Fig. S6).
Fc recognition by the light chain of YES8c
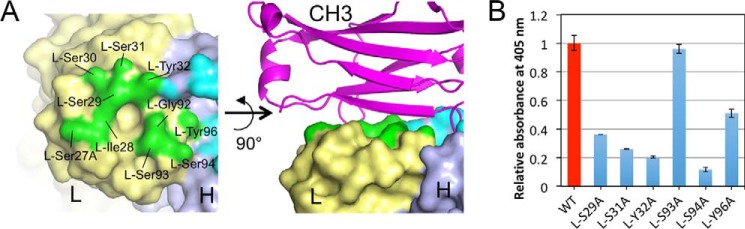
The residues in the light chain were highly conserved among the IGHV1-69/IGKV3-20 RFs throughout the chain (Fig. S4B). For the light chain of YES8c, the number of residues participating in Fc binding (10–11 residues), and the binding interface area (430–450 Å2) is much larger than that of RF-AN (two residues and 196 Å2) and RF61 (five residues and 255 Å2) (Table 2). CDR-L1 and CDR-L3 form a flat binding face and recognize the flat β-sheet region of the CH3 domain (Fig. 4A). The shape complementarity score between the light chain and the Fc region was low (0.57 and 0.58) (Table 2). A slight difference in the binding orientation between the light chain and Fc was observed when comparing complex 1 and complex 2 (Fig. S7). This caused differences in the interfacial area at Asn384 and Lys439 in Fc, and L-Ser30 in YES8c (Fig. S5, A and B). To investigate the role of the residues on the light chain involved in Fc recognition, the binding of the Ala mutants was assessed (Fig. 4B). In the ELISA, the binding of the L-S29A, L-S31A, L-Y32A, and L-S94A mutant was significantly lower than that of the WT, and the binding decreased by one-half for L-Y96A, whereas the L-S93A mutant had no effect on the interaction. SPR analysis confirmed the large decreases in binding of L-S94A and L-Y96A (Fig. S8).
Figure 4.
Binding by the light chain of YES8c. A, interaction footprints in the light chain of YES8c to Fc (left panel) and the 90° rotated view (right panel). The contacting area of the light chain to Fc is shown in green. The other surface of the light chain is shown in pale yellow. Fc is shown as a magenta schematic model. B, binding of mutants in the light chain of YES8c to the immobilized IgG1 by ELISA. Data show mean ± S.D. (error bars) from triplicates from one representative experiment of two independent experiments with similar results.
The effects of somatic hypermutations on YES8c
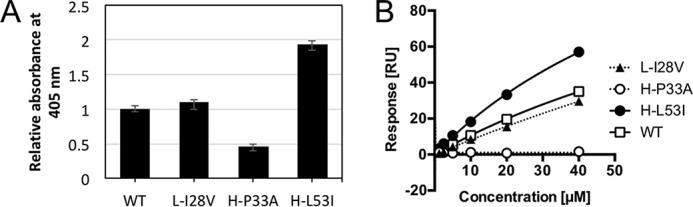
According to previous reports, some antibodies acquire RF activity as a result of somatic hypermutations (SHMs), and others possess RF activity without SHMs (26, 35, 36). Although the structures of RF-AN and RF61 indicated the importance of SHMs for RF activity, binding analyses using mutants have not been performed. In this study, we investigated the effects of SHMs on YES8c by mutation analysis. In the light chain, there are three amino acid substitutions (L-V28I, L-K39R, and L-S53T) (Fig. S1B). In the heavy chain, there are seven amino acid substitutions in the V region (H-A33P, H-M48V, H-I53L, H-K62R, H-S76R, H-L82V, and H-S82BV) and one substitution in the J region (H-D100EF) (Fig. S1A). Among them, the residues near the Fc binding site in YES8c (Leu28, His33, and His53) were substituted with the germline residues (L-I28V, H-P33A, and H-L53I), and the bindings were analyzed by ELISA (Fig. 5A). In the L-I28V mutant, the binding was similar to that of the WT. The binding was lower in the H-P33A mutant than in the WT. In contrast, binding increased in the H-L53I mutant compared with WT. Binding of the mutants was also analyzed by SPR (Fig. 5B and Fig. S9). A binding response similar to that of the WT was observed for L-I28V. A response was not observed for H-P33A, even at the highest concentration. The affinity of H-L53I (Kd = 84 μm) was 2-fold higher than that of WT.
Figure 5.
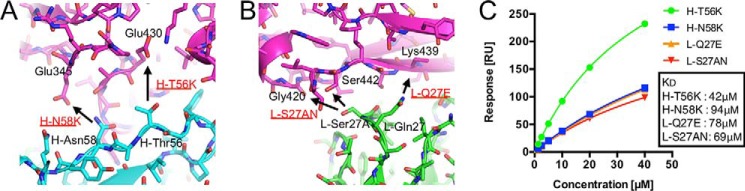
Binding of the germline-reverted mutant RFs to IgG1-Fc. A, binding of the germline-reverted mutants to IgG1-Fc was measured by ELISA. Data show mean ± S.D. (error bars) from triplicates from one representative experiment of two independent experiments with similar results. B, plot of equilibrium binding responses against concentration of YES8c-Fab mutants for the sensorgrams shown in Fig. S10. The solid line represents the nonlinear fit of the 1:1 binding model for the WT and the H-L53I mutant that gave Kd values of 130 μm and 84 μm, respectively. The responses for L-I28V and H-P33A could not be fit with the 1:1 binding model. RU, resonance unit.
Binding affinity increments of YES8c for IgG1-Fc by amino acid substitutions
Abnormal increments in the binding affinity between RFs and IgGs could be associated with disease progression. As noted above, the binding affinity of the H-L53I mutant was increased. Here, to confirm the possibility of an increase in binding affinity through another amino acid substitution, mutants were designed based on the crystal structure. The H-T56K, H-N58K, and L-Q27E mutants were predicted to form salt bridges with Glu430, Glu345, and Lys439 of the Fc, respectively (Fig. 6, A and B). The L-S27AN mutant was predicted to form hydrogen bonds with Gly420 and Ser442 (Fig. 6B). The YES8c mutants were prepared, and the binding affinity of each mutant for IgG1-Fc was measured by ELISA. The YES8c mutants (H-T56K, H-N58K, L-Q27E, and L-S27AN) displayed increased binding to IgG1-Fc (Fig. S10A). The affinity increases were confirmed by SPR analysis. The Kd values of these mutants (42, 94, 78, and 69 μm) were lower than that of WT (160 μm) (Fig. 6C and Fig. S10, B–E). It is noteworthy that these mutations can occur by single base substitution, such as H-Leu53 (CTC) to Ile (ATC), H-Thr56 (ACA) to Lys (AAA), H-Asn58 (AAC) to Lys (AAA), L-Gln27 (CAG) to Glu (GAG), and L-Ser27A (AGT) to Asn (AAT).
Figure 6.
The YES8c sites relative to the increase in binding affinity for IgG1-Fc. A, the residues (H-Thr56 and H-Asn58) in CDR-H2 located near the acidic residues on Fc (Glu345 and Glu430). B, the residues (L-Gln27 and L-Ser27A) in CDR-L1. C, plot of equilibrium binding responses against concentration of YES8c-Fab mutants for the sensorgrams shown in Fig. S11, B–E. The solid line represents a nonlinear fit of the 1:1 binding model for H-T56K, H-N58K, L-Q27E, and L-S27AN. The Kd values are indicated. RU, resonance unit.
Discussion
The RFs derived from the IGHV1-69/IGKV3-20 germline combination have long been studied as paraprotein-IgM RFs observed in MC and WM. IGHV1-69/IGKV3-20 RFs are detected at a high level as a stereotypic RF in patients with B cell lymphoma caused by HCV infection (13–15). The germline combinations for RF-AN and RF61 are IGHV3-9/IGLV3-21 and IGHV4-39/IGLV1-47, respectively, and the crystal structures of both RFs could not explain the binding properties of stereotypic RFs (28, 29). Here we determined the crystal structure of the IGHV1-69/IGKV3-20 RF, designated YES8c, in complex with IgG1-Fc.
Binding analysis showed that YES8c interacted with IgG1 and IgG4 despite some preference for IgG1. The epitope residues were different for IgG3 and the other subclasses at the 384, 422, 435, and 436 positions (Table S2). In particular, position 435, which is considered the key residue for the Ga specificity (9), is histidine in IgG1, 2, and 4 but arginine in IgG3. The structure of the complex suggested that the side chain at this position collides with H-Phe54 containing an H435R substitution (Fig. S11). However, the reason for the decreased binding to IgG2 is unclear. The low affinity of YES8c Fab is consistent with that found in a previous study for RF-AN (28). Despite the low affinity, RFs exert an enhanced avidity in the IgM form, which has ten Fab “arms.”
The YES8c–Fc complex had a larger interfacial area, and more residues participated in the binding than the other structurally determined RFs (RF-AN and RF61). The structure also explains that the low affinity of YES8c for IgG1-Fc was due to the lack of energetically strong interactions, such as salt bridges and hydrogen bonds. This “loose” binding mode may be an important feature for IGHV1-69/IGKV3-20 RFs to commonly recognize the similar epitope of Fc despite variations in CDR-H3 and SHMs. Although both RF-AN and YES8c recognize the CH2–CH3 elbow region of Fc, the binding modes are different. Fc binds to RF-AN outside of the antibody axis and is located on the edge of the conventional antigen-binding region (28). In contrast, Fc bound to YES8c near the center of the antibody axis. The binding mode of CDR-H3, which is located at the center of the antigen-binding site, is grossly different between RF-AN and YES8c. In RF-AN, the interfacial area provided by CDR-H3 occupies 44% of the total antigen-binding region. The side chains of key CDR-H3 residues (H-Arg96, H-Tyr98, and H-Val99) form salt bridges, hydrogen bonds, and hydrophobic interactions with the Fc region and greatly contribute to antigen recognition. This implies that CDR-H3 mainly mediates the binding of RF-AN to IgG–Fc. In contrast, CDR-H3 in YES8c occupies only 21% of the antigen-binding area, and weaker interactions are formed. This indicates that other regions of YES8c outside of CDR-H3 more predominantly contribute to Fc binding.
With a larger interfacial area and complementarity score than the light chain, the heavy chain likely plays a central role in the YES8c–Fc interaction. We propose a model that allows for the diversity of the CDR-H3 loop of IGHV1-69/IGKV3-20 RFs in the RF-Fc binding site. In the YES8c–IgG1-Fc structure, the protrusion at Leu432–His435 in Fc sticks into the VL-VH pocket in YES8c, and Asn434 interacts with the main chain of CDR-H3 from inside. This may prevent the CDR-H3 loop from inserting into the binding interface. Consequently, a large variation in CDR-H3 could be accommodated in the space between CH2 and CH3 with only a slight change in the loop structure. However, it has been suggested that stereotypic RFs are restricted with regards to the CDR-H3 length (14, 27). Most of the IGHV1-69/IGKV3-20 RFs have a CDR-H3 composed of 12–15 amino acids. Considering the YES8c–Fc structure, it is likely that the interaction between CDR-H3 and Fc is not possible when CDR-H3 is shorter than 12 amino acids. In contrast, if CDR-H3 is longer than 15 amino acids, then the loop may not be accepted in the space between CH2 and CH3, or it may interrupt the binding of the protrusion at Leu432–His435 by covering the VL–VH pocket. To validate this model, structure determination of the other IGHV1-69/IGKV3-20 RFs with different CDR-H3 will be required.
Hydrophobic residues in CDR-H2 are considered to participate in the pattern recognition–like interaction of naïve antibodies derived from the IGHV1-69 germline. According to the crystal structures of the anti-influenza hemagglutinin antibody CR6261 (37), anti-HCV E2 envelope antibody AR3C (18), and anti-HIV-1 gp41 antibody D5 (20), the CDR-H2 of the IGHV1-69–derived antibody recognizes a hydrophobic patch in the antigen. Therefore, the naïve antibodies derived from IGHV1-69 are considered pattern recognition antibodies with specificity for CDR-H2. Our mutational analysis showed that the binding of H-F54A mutants to IgG1-Fc largely decreased, indicating that the hydrophobic tip of CDR-H2 (H-Phe54) contributes to the RF activity of YES8c.
The YES8c–Fc structure showed that the interface between the light chain and Fc was flat and the binding was loose. However, the number of YES8c light chain residues interacting with IgG1-Fc and the interfacial area between the light chain and Fc are much larger than found for RF-AN and RF61. The residues involved in the YES8c-Fc interaction were highly conserved among the IGHV1-69/IGKV3-20 RFs (Fig. S4B). Additionally, a large decrease in binding was observed for substitution of L-Ser29, L-Ser31, L-Tyr32, and L-Ser94 to Ala. These findings suggest that the recognition of the highly conserved hydrophilic patch by the light chain is important for the recognition of IGHV1-69/IGKV3-20 RFs despite its loose and flat interface.
In this study, the binding of the germline-reverted mutants with IgG1-Fc was explored. Binding of the H-P33A mutant was largely decreased, indicating that the SHM at this position is important for conferring RF activity to YES8c. In contrast, the affinity of the H-L53I mutant to IgG1-Fc doubled, with no changes observed for the L-I28V mutant. It remains unclear whether a combined germline-reverted mutant has RF activity. In the case of RF-TS1, an IGHV1-69/IGKV3-20 RF, the naïve antibody without SHMs had a higher affinity for Fc than RF-TS1 (36). On the other hand, in mixed cryoglobulinemia related to HCV, the IGHV1-69/IGKV3-20 IgM acquired RF activity with SHMs (26). Furthermore, we showed that the affinity of WT YES8c for IgG1-Fc was further increased by a single-base substitution in the antigen-binding site (H-L53I, H-T56K, H-N58K, L-Q27E, and L-S27AN), indicating that further SHMs could make YES8c an aggravated pathogenic factor.
YES8c binds to the CH2–CH3 elbow in the canonical antigen-binding manner. Combined with mutational analyses, we propose a recognition mechanism common to the IGHV1-69/IGKV3-20 stereotypic RFs that mediates RF activity. Additionally, amino acid substitutions increased the affinity of YES8c to Fc, showing the potential of this RF for aggravating the virulence factor. Our findings may help to rationally design therapeutic antibodies with reduced binding to RFs (38, 39). The other types of stereotypic RFs, RFs derived from IGHV3-7 and IGHV4–59 germlines, are targets for future structural studies to explore the Fc recognition mechanisms of RFs that are frequently observed in hematological diseases.
Experimental procedures
Preparation of YES8c-Fab expressed in Escherichia coli
YES8c-Fab was prepared for crystallization as described in our previous report (40). In brief, the heavy chain composed of the variable domain of the heavy chain (VH) from YES8c IgM and CH1 from IgG1 and the light chain composed of variable domain of the light chain (VL) from YES8c IgM and the κ light chain (CLκ) were separately expressed in E. coli as inclusion bodies. The inclusion bodies were solubilized in a buffer containing 6 m guanidine hydrochloride, and then the heavy chain and light chain were mixed and refolded using a stepwise dialysis method. The refolded Fab was purified by Resource S cation exchange chromatography (GE Healthcare).
Preparation of YES8c-Fab mutants in the silkworm baculovirus expression system
The precise methods are described in the Supplemental Experimental Procedures. In brief, the recombinant bacmid DNAs of the heavy and light chains were generated separately using the BmNPV/T3 bacmid system (41). The baculoviruses for the heavy and light chains were generated separately by transfection of bacmid DNA into Bombyx mori–derived cells. The recombinant virus mixture of the heavy and light chains was injected into silkworm larvae. After infection, the serum from silkworm larvae was collected. The Fab fragment was purified by immobilized metal affinity chromatography, followed by streptactin affinity chromatography. The two-dimensional structures of the Fab mutants were confirmed by CD spectroscopy (Jasco, Tokyo, Japan).
Preparation of the IgG1-Fc fragment
The humanized IgG1 mAb rituximab (Chugai Pharma, Tokyo, Japan) was treated with 1/10 (w/w) of papain activated with 10 mm l-cysteine for 3 h at 37 °C. The papain-digested IgG1 was applied to COSMOGEL® Ig-Accept protein A resin (Nakalai, Kyoto, Japan). The resin was washed with 5 column volumes of wash buffer (50 mm Tris-HCl and 100 mm NaCl (pH 7.0)). Protein was eluted with elution buffer (0.1 m Gly-HCl (pH 3.0)), and the eluent was mixed with 1 m Tris-HCl (pH 8.5). The Fc fragment was further purified by gel filtration using a Superdex200 10/300 column with running buffer (20 mm Tris-HCl and 150 mm NaCl (pH 7.5)).
ELISA
Human IgG2 (ab90284), IgG3 (ab118426), and IgG4 (ab90286) were purchased from Abcam. IgG was immobilized on a 96-well Maxisorb immunoplate (Thermo Fisher Scientific) by incubation with 50 μl/well of 20 μg/ml IgG in immobilization buffer (50 mm Na2CO3-NaHCO3 and 10% NaN3 (pH 9.6)) for 1 h at 25 °C. The plate was washed three times with PBS-T (PBS with 0.05% Tween 20) and then blocked with 5% skim milk for 1 h at 25 °C. After washing with PBS-T, YES8c-Fab diluted with PBS-T was applied and incubated for 1 h at 25 °C. After washing with PBS-T, the wells were treated with Strep-Tactin® alkaline phosphatase conjugate (Iba Lifesciences) diluted with PBS-T and incubated for 1 h at 37 °C. After washing with PBS-T, 50 μl/well of 1 mg/ml p-nitrophenyl phosphate was added, and then absorbance at 405 nm was measured using a Scientific Multiscan plate reader (Thermo Fisher Scientific).
Surface plasmon resonance
The affinity of YES8c-Fab for IgG1-Fc was measured by SPR using a Biacore X100 or Biacore 3000 instrument (GE Healthcare). HBS-EP buffer (10 mm HEPES (pH 7.4), 150 mm NaCl, 3 mm EDTA, and 0.005% (v/v) surfactant P20) was used as the running buffer. YES8c-Fab was dialyzed overnight into HBS-EP buffer. Approximately 1000 resonance units of human IgG were immobilized on a CM5 sensor chip by amine coupling. The reference flow cell was prepared by blocking with ethanolamine after activation with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and N-hydroxysuccinimide. Sequentially diluted YES8c-Fab solutions were injected serially over the flow cells for 60 s at a flow rate of 30 μl/min. The maximum concentration of YES8c-Fab was limited to 40 μm because of its tendency for nonspecific binding at high concentration. The reference response was subtracted from the sensorgram to obtain the actual binding response. The equilibrium dissociation constant (Kd) was obtained by fitting a plot of the responses at equilibrium against the concentration with one site-specific binding model using Prism 6.0 software.
Crystallization, data collection, and structure determination
YES8c-Fab expressed in E. coli and IgG1-Fc were mixed at a molar ratio of 2:1 and concentrated to 10 mg/ml. The crystals were obtained by sitting drop vapor diffusion in a reservoir solution (0.1 m Tris-HCl (pH 8.5), 0.2 m sodium acetate, and 18% (w/v) PEG4000) and incubated for 1 week at 20 °C. The crystals were soaked in soaking buffer (reservoir solution with 20% glycerol) and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the beamline AR-NE3A at the Photon Factory (Tsukuba, Japan). The diffraction data were processed using HKL2000 (HKL Research Inc.). The structure was determined by molecular replacement using Phaser-MR (42) (PHENIX software package (43)) with the Fab structure (PDB code 1VGE) and human Fc structure (PDB code 3DO3) as search models. Model rebuilding and refinement were performed using Coot (44) and phenix.refine (45), respectively. The final Rwork/Rfree factors were 0.221/0.270. The crystallography statistics are shown in Table S1. The intermolecular atomic contacts were determined using the program CONTACT in the CCP4 suite (46). The buried surface area was calculated with a default probe radius of 1.4 Å using the program AREAIMOL (47) in the CCP4 suite. The shape complementarity was calculated using the program SC in CCP4 with a default probe radius of 1.7 Å.
Author contributions
M. S. and T. U. designed and supervised the research. M. S., Y. I., K. S., and J. M. L. prepared proteins. M. S. and K. S. performed crystallization. M. S. performed diffraction data collection and structure determination. Y. I. and K. S. performed the binding study. Data were analyzed by M. S., Y. I., J. M. L., T. K., and T. U. M. S., J. M. L., T. K., and T. U. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences for technical assistance. We thank Prof. Kouhei Tsumoto at the University of Tokyo and Dr. Jose Caaveiro at Kyushu University for beneficial discussions. We thank Dr. Koki Makabe at Yamagata University for assistance with data collection at the Photon Factory. We thank Dr. Imanishi (National Institute of Agrobiological Sciences, Japan) for providing the NIAS-Bm-oyanagi2 (BmO2) cell line.
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI grants 23655160 and 15K15184 (to T. U.) and the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from AMED. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S11, Tables S1 and S2, and Experimental procedures.
The atomic coordinates and structure factors (code 5XMH) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- RF
- rheumatoid factor
- Fc
- fragment crystallizable
- RA
- rheumatoid arthritis
- MC
- mixed cryoglobulinemia
- WM
- Waldenstrom's macroglobulinemia
- HCV
- hepatitis C virus
- BSA
- buried surface area
- SPR
- surface plasmon resonance
- SHM
- somatic hypermutation
- CDR
- complementarity-determining region
- VH
- variable domain of the heavy chain
- VL
- variable domain of the light chain
- CH
- constant domain of the heavy chain.
References
- 1. Randen I., Thompson K. M., Pascual V., Victor K., Beale D., Coadwell J., Førre O., Capra J. D., and Natvig J. B. (1992) Rheumatoid factor V genes from patients with rheumatoid arthritis are diverse and show evidence of an antigen-driven response. Immunol. Rev. 128, 49–71 10.1111/j.1600-065X.1992.tb00832.x [DOI] [PubMed] [Google Scholar]
- 2. Pascual V., Randen I., Thompson K., Sioud M., Forre O., Natvig J., and Capra J. D. (1990) The complete nucleotide sequences of the heavy chain variable regions of six monospecific rheumatoid factors derived from Epstein-Barr virus-transformed B cells isolated from the synovial tissue of patients with rheumatoid arthritis: further evidence that some autoantibodies are unmutated copies of germ line genes. J. Clin. Invest. 86, 1320–1328 10.1172/JCI114841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Victor K. D., Randen I., Thompson K., Forre O., Natvig J. B., Fu S. M., and Capra J. D. (1991) Rheumatoid factors isolated from patients with autoimmune disorders are derived from germline genes distinct from those encoding the Wa, Po, and Bla cross-reactive idiotypes. J. Clin. Invest. 87, 1603–1613 10.1172/JCI115174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pascual V., Victor K., Randen I., Thompson K., Steinitz M., Førre O., Fu S. M., Natvig J. B., and Capra J. D. (1992) Nucleotide sequence analysis of rheumatoid factors and polyreactive antibodies derived from patients with rheumatoid arthritis reveals diverse use of VH and VL gene segments and extensive variability in CDR-3. Scand. J. Immunol. 36, 349–362 10.1111/j.1365-3083.1992.tb03108.x [DOI] [PubMed] [Google Scholar]
- 5. Ermel R. W., Kenny T. P., Chen P. P., and Robbins D. L. (1993) Molecular analysis of rheumatoid factors derived from rheumatoid synovium suggests an antigen-driven response in inflamed joints. Arthritis Rheum. 36, 380–388 10.1002/art.1780360314 [DOI] [PubMed] [Google Scholar]
- 6. Mantovani L., Wilder R. L., and Casali P. (1993) Human rheumatoid B-1a (CD5+ B) cells make somatically hypermutated high affinity IgM rheumatoid factors. J. Immunol. 151, 473–488 [PMC free article] [PubMed] [Google Scholar]
- 7. Youngblood K., Fruchter L., Ding G., Lopez J., Bonagura V., and Davidson A. (1994) Rheumatoid factors from the peripheral blood of two patients with rheumatoid arthritis are genetically heterogeneous and somatically mutated. J. Clin. Invest. 93, 852–861 10.1172/JCI117040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Artandi S. E., Canfield S. M., Tao M. H., Calame K. L., Morrison S. L., and Bonagura V. R. (1991) Molecular analysis of IgM rheumatoid factor binding to chimeric IgG. J. Immunol. 146, 603–610 [PubMed] [Google Scholar]
- 9. Artandi S. E., Calame K. L., Morrison S. L., and Bonagura V. R. (1992) Monoclonal IgM rheumatoid factors bind IgG at a discontinuous epitope comprised of amino acid loops from heavy-chain constant-region domains 2 and 3. Proc. Natl. Acad. Sci. U.S.A. 89, 94–98 10.1073/pnas.89.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonagura V. R., Artandi S. E., Davidson A., Randen I., Agostino N., Thompson K., Natvig J. B., and Morrison S. L. (1993) Mapping studies reveal unique epitopes on IgG recognized by rheumatoid arthritis-derived monoclonal rheumatoid factors. J. Immunol. 151, 3840–3852 [PubMed] [Google Scholar]
- 11. Silverman G. J., Goldfien R. D., Chen P., Mageed R. A., Jefferis R., Goñi F., Frangione B., Fong S., and Carson D. A. (1988) Idiotypic and subgroup analysis of human monoclonal rheumatoid factors: implications for structural and genetic basis of autoantibodies in humans. J. Clin. Invest. 82, 469–475 10.1172/JCI113620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thompson K. M., Randen I., Børretzen M., Førre O., and Natvig J. B. (1994) Variable region gene usage of human monoclonal rheumatoid factors derived from healthy donors following immunization. Eur. J. Immunol. 24, 1771–1778 10.1002/eji.1830240808 [DOI] [PubMed] [Google Scholar]
- 13. Charles E. D., Green R. M., Marukian S., Talal A. H., Lake-Bakaar G. V., Jacobson I. M., Rice C. M., and Dustin L. B. (2008) Clonal expansion of immunoglobulin M+CD27+ B cells in HCV-associated mixed cryoglobulinemia. Blood 111, 1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bende R. J., Aarts W. M., Riedl R. G., de Jong D., Pals S. T., and van Noesel C. J. (2005) Among B cell non-Hodgkin's lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J. Exp. Med. 201, 1229–1241 10.1084/jem.20050068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Re V., De Vita S., Gasparotto D., Marzotto A., Carbone A., Ferraccioli G., and Boiocchi M. (2002) Salivary gland B cell lymphoproliferative disorders in Sjogren's syndrome present a restricted use of antigen receptor gene segments similar to those used by hepatitis C virus-associated non-Hodgkins's lymphomas. Eur. J. Immunol. 32, 903–910 10.1002/1521-4141(200203)32:3%3C903::AID-IMMU903%3E3.0.CO%3B2-D [DOI] [PubMed] [Google Scholar]
- 16. Stamatopoulos K., Belessi C., Moreno C., Boudjograh M., Guida G., Smilevska T., Belhoul L., Stella S., Stavroyianni N., Crespo M., Hadzidimitriou A., Sutton L., Bosch F., Laoutaris N., Anagnostopoulos A., et al. (2007) Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood 109, 259–270 10.1182/blood-2006-03-012948 [DOI] [PubMed] [Google Scholar]
- 17. Bende R. J., Slot L. M., Hoogeboom R., Wormhoudt T. A., Adeoye A. O., Guikema J. E., and van Noesel C. J. (2015) Stereotypic rheumatoid factors that are frequently expressed in mucosa-associated lymphoid tissue-type lymphomas are rare in the labial salivary glands of patients with Sjogren's syndrome. Arthritis Rheumatol. 67, 1074–1083 10.1002/art.39002 [DOI] [PubMed] [Google Scholar]
- 18. Kong L., Giang E., Nieusma T., Kadam R. U., Cogburn K. E., Hua Y., Dai X., Stanfield R. L., Burton D. R., Ward A. B., Wilson I. A., and Law M. (2013) Hepatitis C virus E2 envelope glycoprotein core structure. Science 342, 1090–1094 10.1126/science.1243876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan C. H., Hadlock K. G., Foung S. K., and Levy S. (2001) V(H)1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood 97, 1023–1026 10.1182/blood.V97.4.1023 [DOI] [PubMed] [Google Scholar]
- 20. Luftig M. A., Mattu M., Di Giovine P., Geleziunas R., Hrin R., Barbato G., Bianchi E., Miller M. D., Pessi A., and Carfí A. (2006) Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nat. Struct. Mol. Biol. 13, 740–747 10.1038/nsmb1127 [DOI] [PubMed] [Google Scholar]
- 21. Ekiert D. C., Bhabha G., Elsliger M. A., Friesen R. H., Jongeneelen M., Throsby M., Goudsmit J., and Wilson I. A. (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251 10.1126/science.1171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ying T., Prabakaran P., Du L., Shi W., Feng Y., Wang Y., Wang L., Li W., Jiang S., Dimitrov D. S., and Zhou T. (2015) Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat. Commun. 6, 8223 10.1038/ncomms9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yeung Y. A., Foletti D., Deng X., Abdiche Y., Strop P., Glanville J., Pitts S., Lindquist K., Sundar P. D., Sirota M., Hasa-Moreno A., Pham A., Melton Witt J., Ni I., Pons J., et al. (2016) Germline-encoded neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire. Nat. Commun. 7, 13376 10.1038/ncomms13376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanovski M., Silvestri F., Pozzato G., Anand S., Mazzaro C., Burrone O. R., and Efremov D. G. (1998) Somatic hypermutation, clonal diversity, and preferential expression of the VH 51p1/VL kv325 immunoglobulin gene combination in hepatitis C virus-associated immunocytomas. Blood 91, 2433–2442 [PubMed] [Google Scholar]
- 25. Agnello V., Chung R. T., and Kaplan L. M. (1992) A role for hepatitis C virus infection in type II cryoglobulinemia. N. Engl. J. Med. 327, 1490–1495 10.1056/NEJM199211193272104 [DOI] [PubMed] [Google Scholar]
- 26. Charles E. D., Orloff M. I., Nishiuchi E., Marukian S., Rice C. M., and Dustin L. B. (2013) Somatic hypermutations confer rheumatoid factor activity in hepatitis C virus-associated mixed cryoglobulinemia. Arthritis Rheum. 65, 2430–2440 10.1002/art.38041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Børretzen M., Randen I., Natvig J. B., and Thompson K. M. (1995) Structural restriction in the heavy chain CDR3 of human rheumatoid factors. J. Immunol. 155, 3630–3637 [PubMed] [Google Scholar]
- 28. Corper A. L., Sohi M. K., Bonagura V. R., Steinitz M., Jefferis R., Feinstein A., Beale D., Taussig M. J., and Sutton B. J. (1997) Structure of human IgM rheumatoid factor Fab bound to its autoantigen IgG Fc reveals a novel topology of antibody-antigen interaction. Nat. Struct. Biol. 4, 374–381 10.1038/nsb0597-374 [DOI] [PubMed] [Google Scholar]
- 29. Duquerroy S., Stura E. A., Bressanelli S., Fabiane S. M., Vaney M. C., Beale D., Hamon M., Casali P., Rey F. A., Sutton B. J., and Taussig M. J. (2007) Crystal structure of a human autoimmune complex between IgM rheumatoid factor RF61 and IgG1 Fc reveals a novel epitope and evidence for affinity maturation. J. Mol. Biol. 368, 1321–1331 10.1016/j.jmb.2007.02.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ezaki I., Kanda H., Sakai K., Fukui N., Shingu M., Nobunaga M., and Watanabe T. (1991) Restricted diversity of the variable region nucleotide sequences of the heavy and light chains of a human rheumatoid factor. Arthritis Rheum. 34, 343–350 10.1002/art.1780340312 [DOI] [PubMed] [Google Scholar]
- 31. Deisenhofer J. (1981) Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 20, 2361–2370 10.1021/bi00512a001 [DOI] [PubMed] [Google Scholar]
- 32. Sasso E. H., Barber C. V., Nardella F. A., Yount W. J., and Mannik M. (1988) Antigenic specificities of human monoclonal and polyclonal IgM rheumatoid factors: the C γ 2-C γ 3 interface region contains the major determinants. J. Immunol. 140, 3098–3107 [PubMed] [Google Scholar]
- 33. Raghunathan G., Smart J., Williams J., and Almagro J. C. (2012) Antigen-binding site anatomy and somatic mutations in antibodies that recognize different types of antigens. J. Mol. Recognit. 25, 103–113 10.1002/jmr.2158 [DOI] [PubMed] [Google Scholar]
- 34. Lawrence M. C., and Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 10.1006/jmbi.1993.1648 [DOI] [PubMed] [Google Scholar]
- 35. Randen I., Brown D., Thompson K. M., Hughes-Jones N., Pascual V., Victor K., Capra J. D., Førre O., and Natvig J. B. (1992) Clonally related IgM rheumatoid factors undergo affinity maturation in the rheumatoid synovial tissue. J. Immunol. 148, 3296–3301 [PubMed] [Google Scholar]
- 36. Carayannopoulos M. O., Potter K. N., Li Y., Natvig J. B., and Capra J. D. (2000) Evidence that human immunoglobulin M rheumatoid factors can be derived from the natural autoantibody pool and undergo an antigen driven immune response in which somatically mutated rheumatoid factors have lower affinities for immunoglobulin G Fc than their germline counterparts. Scand. J. Immunol. 51, 327–336 10.1046/j.1365-3083.2000.00731.x [DOI] [PubMed] [Google Scholar]
- 37. Lingwood D., McTamney P. M., Yassine H. M., Whittle J. R., Guo X., Boyington J. C., Wei C. J., and Nabel G. J. (2012) Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570 10.1038/nature11371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sène D., Ghillani-Dalbin P., Amoura Z., Musset L., and Cacoub P. (2009) Rituximab may form a complex with IgMκ mixed cryoglobulin and induce severe systemic reactions in patients with hepatitis C virus-induced vasculitis. Arthritis Rheum. 60, 3848–3855 10.1002/art.25000 [DOI] [PubMed] [Google Scholar]
- 39. Jones J. D., Shyu I., Newkirk M. M., and Rigby W. F. (2013) A rheumatoid factor paradox: inhibition of rituximab effector function. Arthritis Res. Ther. 15, R20 10.1186/ar4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujii T., Ohkuri T., Onodera R., and Ueda T. (2007) Stable supply of large amounts of human Fab from the inclusion bodies in E. coli. J. Biochem. 141, 699–707 10.1093/jb/mvm075 [DOI] [PubMed] [Google Scholar]
- 41. Ono C., Nakatsukasa T., Nishijima Y., Asano S., Sahara K., and Bando H. (2007) Construction of the BmNPV T3 bacmid system and its application to the functional analysis of BmNPV he65. J. Insect Biotechnol. Sericol. 76, 161–167 [Google Scholar]
- 42. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saff E. B., and Kuijlaars A. B. J. (1997) Distributing many points on a sphere. Math. Intell. 19, 5–11 [Google Scholar]
- 48. Eisenberg D., Schwarz E., Komaromy M., and Wall R. (1984) Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142 10.1016/0022-2836(84)90309-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.