Abstract
The 14-3-3 family comprises a group of small proteins that are essential, ubiquitous, and highly conserved across eukaryotes. Overexpression of the 14-3-3 proteins σ, ϵ, ζ, and η correlates with high metastatic potential in multiple cancer types. In Dictyostelium, 14-3-3 promotes myosin II turnover in the cell cortex and modulates cortical tension, cell shape, and cytokinesis. In light of the important roles of 14-3-3 proteins across a broad range of eukaryotic species, we sought to determine how 14-3-3 proteins interact with myosin II. Here, conducting in vitro and in vivo studies of both Dictyostelium (one 14-3-3 and one myosin II) and human proteins (seven 14-3-3s and three nonmuscle myosin IIs), we investigated the mechanism by which 14-3-3 proteins regulate myosin II assembly. Using in vitro assembly assays with purified myosin II tail fragments and 14-3-3, we demonstrate that this interaction is direct and phosphorylation-independent. All seven human 14-3-3 proteins also altered assembly of at least one paralog of myosin II. Our findings indicate a mechanism of myosin II assembly regulation that is mechanistically conserved across a billion years of evolution from amebas to humans. We predict that altered 14-3-3 expression in humans inhibits the tumor suppressor myosin II, contributing to the changes in cell mechanics observed in many metastatic cancers.
Keywords: 14-3-3 protein, cytoskeleton, Dictyostelium, fluorescence correlation spectroscopy (FCS), human, myosin, surface plasmon resonance (SPR), bipolar filament assembly
Introduction
14-3-3 proteins are small acidic regulatory proteins that form obligatory dimers and are involved in numerous cellular processes, including cell cycle control, DNA damage repair, apoptosis, and many signaling pathways (1–4). 14-3-3s act to scaffold, sequester, or change the conformation of their binding partners, interactions that are often mediated by phosphorylation of the target protein (5). However, many phosphorylation-independent interactions have also been reported, including to glycoprotein Ib, p75NTR-associated cell death executor (NADE), inositol polyphosphate 5-phosphatase, and the bacterial toxin exoenzyme S (exoS) (6–8). 14-3-3 proteins are essential in all eukaryotes and are highly conserved both structurally and by sequence. Multiple paralogs of 14-3-3 are often expressed in a single cell and can form homodimers or heterodimers with each other (9, 10). Although their structural similarity confers a large degree of functional overlap to these paralogs, differences in the paralog-specific effects or interactors are also common (5). Therefore, the expression patterns of 14-3-3s, and their relative ability to heterodimerize, can affect a host of regulatory pathways. In fact, altered expression or dysregulation of 14-3-3s is associated with numerous disease states, including cancers and neurological diseases such as Parkinson's disease and Alzheimer's disease (11–14).
Human 14-3-3 σ, which is widely expressed in epithelial tissues, has been particularly well studied in the cancer literature and is often down-regulated in breast, colorectal, liver, and bladder cancers (15–18). 14-3-3 σ is both induced by and an activator of the p53 transcription factor in response to DNA damage, where it is involved in the maintenance of the G2/M checkpoint. Because bypassing this checkpoint is a major promotor of genetic instability, it is no surprise that loss of 14-3-3 σ expression is common in tumors. However, a more inexplicable finding is that 14-3-3 σ is often overexpressed in pancreatic, lung, and gastric cancers, where it correlates with increased metastasis and poorer clinical outcomes (19–21). How this regulatory protein can function as a tumor suppressor or promoter under different conditions remains largely unexplored.
Nonmuscle myosin II is a hexameric motor protein comprised of two heavy chains, two essential light chains, and two regulatory light chains, which associate to form a “functional monomer.” These monomers are capable of regulated assembly via interaction of the tail domains to form bipolar thick filaments (BTFs),2 the force-generating units of myosin II that bind to actin filaments and hydrolyze ATP. Myosin IIs are major players in cell mechanics, acting to guide and drive cellular processes, including cytokinesis, cell migration, and tissue invasion, and responses to external mechanical cues (22–24). Previous work in our laboratory identified 14-3-3 as a genetic suppressor of nocodazole and an interactor of myosin II in Dictyostelium discoideum, a social ameba (25). Through this interaction, 14-3-3 modulates myosin II activity, cortical tension, cell shape control, and cytokinesis (26). Here we demonstrate that 14-3-3 binds directly to the tail of myosin II and inhibits its assembly into BTFs, which increases the soluble fraction of myosin in the cell and promotes myosin turnover.
In Dictyostelium, assembly of BTFs is heavily regulated by myosin heavy chain kinase (MHCK) phosphorylation of the myosin tail at three key threonines (1823, 1833, and 2029). Phosphorylated myosin remains largely unassembled, whereas unphosphorylated myosin assembles readily (27). As expected, mutation of these three threonines to aspartic acids (3×Asp, a phosphomimetic myosin tail) leads to severe defects in myosin assembly, and mutation of the threonines to alanine (3×Ala, a nonphosphorylatable tail) leads to myosin II overassembly in vivo (28, 29). 14-3-3 provides further myosin assembly modulation by tuning the balance of assembly and disassembly as well as maintaining a dynamic pool of available myosin subunits (25).
Given the importance of 14-3-3 proteins across eukaryotic species and the interaction between 14-3-3 and myosin II in Dictyostelium, we used a suite of in vitro and in vivo approaches to determine how 14-3-3 regulates myosin II assembly. We then explored the breadth of these interactions in human proteins using all seven 14-3-3 paralogs and the three nonmuscle myosin IIs. Thus, we present the quantitative landscape of 14-3-3–nonmuscle myosin II interactions that can modulate myosin II filament assembly.
Results
14-3-3 binds to the minimal assembly domain of Dictyostelium myosin II
Our previous work demonstrated that 14-3-3 interacted with myosin and reduced assembly, but the mechanism of this inhibition was unclear. 14-3-3 proteins are well known to interact with phosphorylated proteins, but because the preparation of myosin II from Dictyostelium involves sequential rounds of assembly and disassembly, the final product is expected to be largely unphosphorylated, as heavy chain phosphorylation is inhibitory to filament formation. Therefore, any in vitro interaction between purified myosin II and 14-3-3 (such as in our previous experiments (25)) is likely to be phosphorylation-independent.
To eliminate the possibility of residual phosphorylation within the myosin preparation, we examined 14-3-3's effect on bacterially expressed Dictyostelium myosin II tail fragments. We generated several mCherry-tagged myosin II tail fragment constructs (Fig. 1A): AD (the minimal myosin assembly domain, residues 1531–1824 (30)), ADCT (which begins at the minimal myosin II assembly domain and ends at the C terminus of the heavy chain, residues 1531–2116), ADCT 3×Ala (ADCT with the three regulatory threonines at 1823, 1833, and 2029 mutated to alanines), and ADCT 3×Asp (ADCT with the same threonines mutated to aspartic acids). The mCherry fusion allowed for the use of fluorescence techniques and provided a globular head to promote the formation of ordered filaments as described by Hostetter et al. (31). Each construct was expressed in Escherichia coli, which lacks the eukaryotic kinases to phosphorylate Dictyostelium myosin II, and was purified using column chromatography.
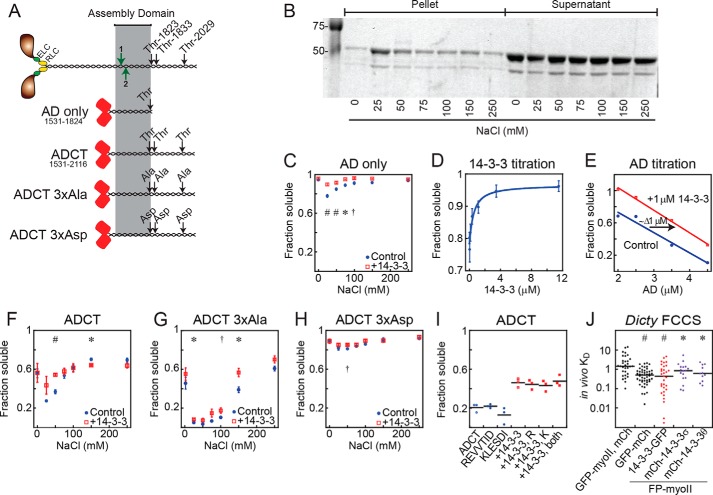
Figure 1.
In Dictyostelium, 14-3-3 solubilizes myosin II by directly binding to the minimal assembly domain in a phosphorylation-independent manner. A, schematic of myosin II and constructs derived from it. Red barrels, mCherry; highlight, minimal assembly domain; black arrows, critical regulatory threonines or substitutions thereof; green arrows, ELM-predicted 14-3-3 interaction sites in the AD region: 1, 1582–1588 and 2, 1593–1598. B, example gel of the AD-only salt-dependent sedimentation assay. Coomassie Blue was used to visualize protein. A lower soluble fraction corresponds to more complete assembly. Sedimentations were performed at 22 °C, 1 μmdimer each protein unless otherwise stated. C, AD is solubilized by 14-3-3. Sample size: n = 11 AD only, n = 9 AD plus 14-3-3. Results of Student's t test; *, p < 0.05; #, p < 0.005; †, p = 0.06. All points represent mean ± S.E. D, titration of 14-3-3 against AD at 25 mm NaCl. KD = 450 ± 250 nmdimer (χ2 = 0.0015). Sample size: n = 6 at each concentration. All points represent mean ± S.E. E, titration of AD at 25 mm NaCl. Increasing amounts of AD lead to increased assembly, and addition of 14-3-3 reduces assembly. F–H, salt-dependent sedimentation of ADCT (F, n = 4, m = 3), ADCT 3×Ala (G, n = 4, m = 5), ADCT 3×Asp (H, n = 4, m = 3–4). Addition of 14-3-3 reduces assembly in all cases. Sample sizes: n, myosin only; m, myosin plus 14-3-3. Results of Student's t test; *, p < 0.05; #, p < 0.005; †, p = 0.06. All points represent mean ± S.E. I, sedimentation of mCherry-ADCT at 100 mm NaCl, alone or in combination with 14-3-3 and/or the 14-3-3 candidate binding site peptides REVVTID (also called peptide R) and KLESDI (also called peptide K). These peptides do not affect baseline ADCT assembly, nor do they competitively inhibit 14-3-3–driven solubilization of ADCT. Lines represent means. J, FCCS in myoII-null Dictyostelium cells expressing fluorescently tagged proteins as listed. GFP-myoII, mCh represents the negative control of independently expressed, noninteracting proteins. GFP-mCh is a positive control fusion protein and was expressed in WT cells. Either GFP-myosin II or mCh-myosin II were co-expressed with a 14-3-3 protein fused to the opposite fluorescent protein (FP) in each case. Dictyostelium 14-3-3-GFP and human mCh-14-3-3 σ and mCh-14-3-3 θ associated with Dictyostelium myosin II in living Dictyostelium cells. Lines represent medians. Results of Wilcoxon–Mann-Whitney test versus negative control; *, p < 0.05; #, p < 0.0005.
We first examined whether the minimal myosin assembly domain alone could be solubilized by 14-3-3. We used sedimentation assays to monitor assembly (Fig. 1B) and confirmed that our mCherry-AD construct assembled in a salt-dependent manner like that of full-length Dictyostelium myosin II, although to a lesser degree. Somewhat unexpectedly, this construct was solubilized by 14-3-3, indicating that at least one binding site for 14-3-3 lies within the myosin II assembly domain itself (Fig. 1C).
To determine the affinity of the interaction between 14-3-3 and the myosin II assembly domain, we performed titration experiments. When 14-3-3 was titrated against 1 μmdimer mCherry-AD, we obtained the expected saturation binding curve, with an apparent KD of 450 ± 250 nmdimer (Fig. 1D). Because the assembly of myosin II itself is also concentration-dependent (assembly will proceed until the concentration of unassembled myosin II is equal to the critical concentration for assembly), we performed the reverse experiment and assessed assembly of the minimal AD at varying myosin concentrations with and without 1 μmdimer 14-3-3. Indeed, the fraction of myosin in the soluble phase decreased with increasing myosin concentration, and the soluble fraction increased at each point upon addition of 14-3-3 (Fig. 1E). These results demonstrate a direct, nonphosphorylation-dependent, assembly-inhibiting interaction between Dictyostelium 14-3-3 and myosin II, which maps within the minimal assembly domain of the myosin tail.
14-3-3 reduces assembly of Dictyostelium myosin II in a phosphorylation-independent manner
Because assembly of our minimal construct was affected by 14-3-3, phosphorylation is not absolutely necessary for 14-3-3 to interact with myosin. However, because of the importance of heavy chain phosphorylation in myosin II assembly and the large number of proteins known to interact with 14-3-3 in a phospho-dependent manner, we reasoned that the tail regulatory region may contain additional binding sites, perhaps allowing the effect to be reversibly tuned. We tested this theory using our ADCT constructs, which included the WT, phosphomimetic, and nonphosphorylatable forms of the regulatory region.
After verifying that the mCherry-tagged ADCT construct assembled appropriately, we observed that this assembly was reduced in the presence of 14-3-3 (Fig. 1F), mirroring the behavior of the AD construct as well as that of full-length myosin II (25). The 3×Ala ADCT construct assembled to a higher degree than WT ADCT, and the 3×Asp ADCT construct assembled poorly, consistent with previous in vitro and in vivo work (28). Addition of purified 14-3-3 also reduced assembly of both of these ADCT constructs (Fig. 1, G and H) as well, implying that the regulatory threonines of the myosin II tail are not also 14-3-3 binding sites.
We then used the Eukaryotic Linear Motif (ELM) resource (http://elm.eu.org/)3 to see whether we could identify any potential 14-3-3 binding sites in the assembly domain (Fig. 1A). Indeed, two potential 14-3-3 interaction sites were predicted within the minimal assembly domain of Dictyostelium myosin II (note that the ELM prediction algorithm changed in 2016 so that it no longer predicts these sites). Because other unphosphorylated peptides bind 14-3-3, tightly inhibiting its interaction with target proteins (32), we synthesized the nonphosphorylated REVVTID and KLESDI peptides (the two predicted binding sites). We performed competition assays using the ADCT construct. Neither of these peptides, individually or in combination, altered myosin assembly, or 14-3-3's ability to solubilize myosin (Fig. 1I). Because of the sensitivity of myosin II's assembly activity to perturbations within this minimal assembly domain and the lack of any well-defined rules for predicting unphosphorylated 14-3-3 binding sequences, we did not map the binding region at a higher resolution.
Both Dictyostelium and human 14-3-3s bind to Dictyostelium myosin II in vivo
The effects of Dictyostelium 14-3-3 on myosin II turnover in vivo have been previously demonstrated by fluorescence recovery after photobleaching (FRAP), and co-immunoprecipitation experiments have confirmed their interaction (25). To examine the in vivo interaction in more detail, we turned to fluorescence correlation spectroscopy (FCS). Here the fluorescence emissions of a ∼1 fL confocal volume are monitored over time, and an autocorrelation analysis is performed on these data. As fluorescently tagged proteins of interest pass through the volume, their average residence time (inversely proportional to the diffusion rate) and the number of fluorescing species in the volume are represented on the autocorrelation curve. Furthermore, by differentially labeling two species of interest, the technique can be extended to fluorescence cross-correlation spectroscopy (FCCS), which provides a measurement of the interaction strength, or “apparent in vivo KD,” of the two labeled species as well as their bound and free diffusion times. This parameter is “apparent” because it is an equilibrium term being used to interpret binding associations in living cells, where competing interactions with unlabeled proteins exist.
To measure 14-3-3–myosin II interaction in vivo, we expressed mCherry-myosin II and 14-3-3-GFP in a myoII-null background and assessed associations using FCCS (Fig. 1J). By using this design, we eliminated unlabeled myosin II. However, because 14-3-3 is an essential protein, its gene cannot be deleted. For a positive control, we used a GFP-mCherry fusion protein, which yielded an apparent KD of 0.62 ± 0.075 μm. For a negative control, we analyzed free mCherry with GFP-myosin II, which returned an apparent KD of 1.43 ± 0.63 μm. We interpreted these numbers to reflect the dynamic range of the technique for GFP-myosin II, which is a large protein with a relatively slow diffusion coefficient. We then measured the interaction between 14-3-3-GFP and mCh-myosin II, obtaining an apparent KD of 0.90 ± 0.21 μm. Thus, we can detect and quantify Dictyostelium 14-3-3 interactions with myosin II in living cells.
Because of the high sequence identity between 14-3-3s across species and the high structural identity between Dictyostelium and human myosin IIs, we speculated that human 14-3-3s may also have an effect on myosin II in Dictyostelium. To explore this possibility, we generated mCherry-tagged constructs of human 14-3-3 σ and θ and used FCCS to explore their interaction with Dictyostelium myosin II in vivo. We found that expressing GFP-myosin II with mCherry-14-3-3 σ or θ returned positive correlations with an apparent KD of 1.2 ± 0.31 μm and 0.89 ± 0.23 μm, respectively. These results motivated us to test the interaction between human 14-3-3s with human nonmuscle myosin IIs.
Mammalian myosin II tail fragments form ordered filaments in vitro
Mammalian nonmuscle myosin II assembly is also regulated by phosphorylation, much like in Dictyostelium, but heavy chain tail phosphorylation is less fully characterized. Because our data indicated that 14-3-3 binding occurs within the myosin II tail in Dictyostelium, we chose to focus our studies on this region within mammalian myosin IIs as well. We generated and purified mCherry-fused tail fragments of each mammalian myosin II (human IIA (MYH9), human IIB (MYH10), and mouse IIC (MYH14)) that comprised the assembly competence domain and the nonhelical tailpiece (Fig. 2A). We later cloned and generated the human IIC fragment as well (MYH14). These truncated constructs allowed us to study assembly-competent myosin tails in an unphosphorylated state. Untagged tail fragments of this region have been described previously (33).
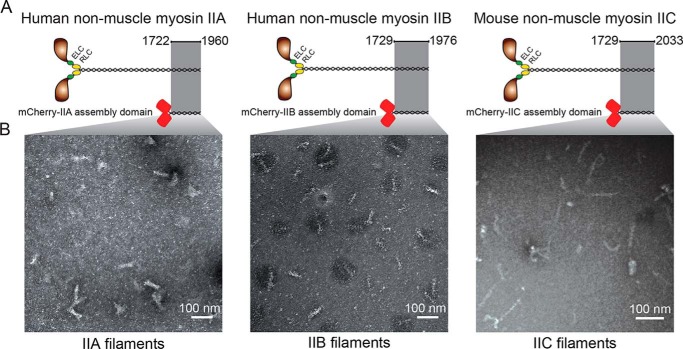
Figure 2.
Mammalian myosin II tail fragments form ordered filaments. A, schematic showing the mammalian myosin IIs and constructs derived from them. Highlight, regions necessary for filament formation (assembly competence domain and nonhelical tailpiece); red barrels, mCherry. B, representative negative-stain EM images of human mCherry-IIA, human mCherry-IIB, and mouse mCherry-IIC filaments. IIA was assembled at 25 mm NaCl, IIB and IIC were assembled at 150 mm NaCl. Images were uniformly adjusted for brightness/contrast and γ to enhance visibility.
All three nonmuscle myosin II constructs formed ordered filaments, as observed by negative-stain EM (Fig. 2B). Filaments were assembled at 150 mm NaCl prior to being placed on the grid for mCherry-IIB and mCherry-IIC, whereas 25 mm NaCl was necessary to promote sufficient filament formation for mCherry-IIA. Assembly of mCherry-IIB at a lower ionic strength resulted in the formation of larger fiber-like structures, suggestive of side-polar assembly of the tail fragments. These large fibers were similar in appearance to EM images obtained using an unlabeled IIB assembly domain construct (33). We measured the filaments formed by each construct and found that mCherry-IIA formed filaments of 69.7 ± 8.6 nm and (mouse) mCherry-IIC of 78 ± 14 nm. The mCherry-IIB construct formed the best-ordered filaments, with a clear banding pattern and average filament length of 68 ± 8.6 nm. We were unable to obtain EM images of unassembled tail fragments. This is not surprising because unassembled tail fragments are on the order of ∼120 kDa, which can be difficult to visualize with conventional uranyl acetate–negative staining (34). We concluded that our tail fragments could recapitulate myosin assembly and were suitable for studying the effects of 14-3-3 on these myosins.
Human 14-3-3 σ reduces the size of assembled myosin II filaments
To test our hypothesis that 14-3-3s affect myosin assembly in the mammalian system, we first used analytical size exclusion chromatography to determine the degree of filament assembly. We began our studies with the σ paralog of 14-3-3 and myosin IIB, which are both well studied in the literature and have great relevance for human disease. We first performed these assays on mCherry-IIB alone, which was preassembled at varying salt concentrations and then loaded on the column for analysis. As we decreased the salt concentration from 500 mm (a level that precludes assembly) to 150 mm (matching the physiological ionic strength in human cells), the partition coefficient (KAV) of the eluted peak decreased, corresponding to the formation of larger assemblies (Fig. 3A). Furthermore, the shape of the peak, which, at high ionic strength, appeared as a single Gaussian corresponding to one species, shifted to a skewed distribution as ionic strength decreased, signifying that multiple larger species were eluting. Mixing 14-3-3 σ with preassembled tail fragments at 150 mm NaCl prior to the run shifted the eluted peak to a smaller size, indicating that 14-3-3 σ disassembled myosin II filaments (Fig. 3B).
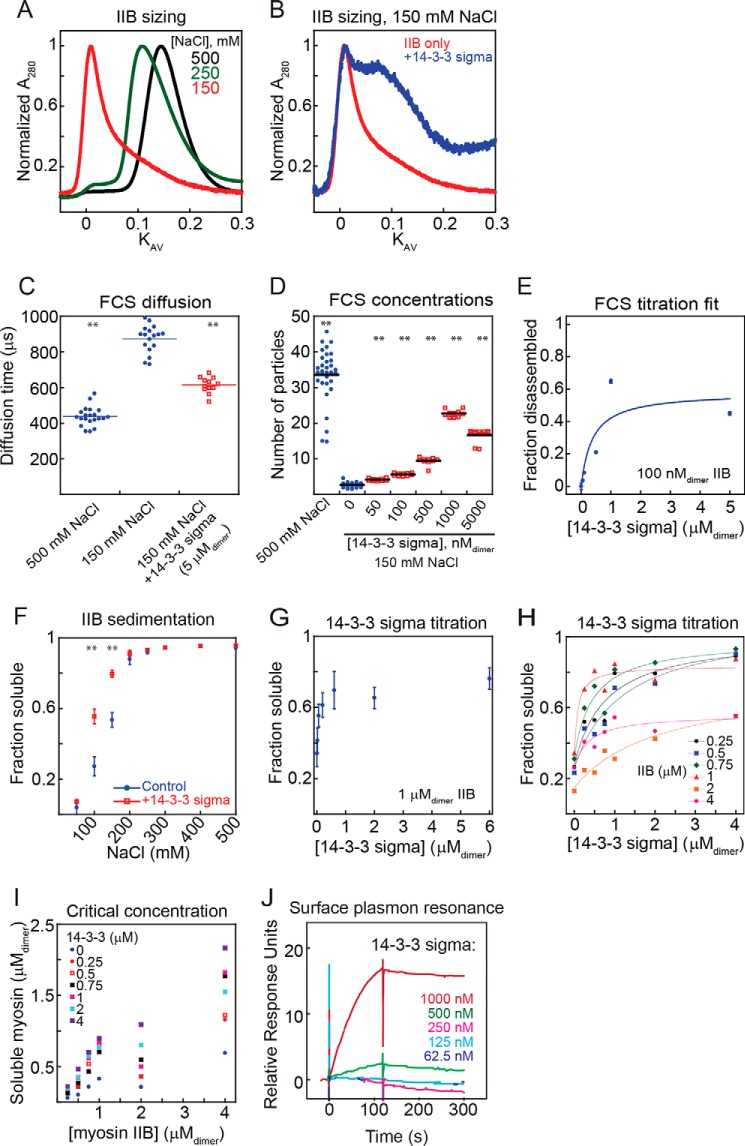
Figure 3.
14-3-3 σ solubilizes myosin IIB filaments. A, analytical sizing on mCherry-IIB preassembled at the listed salt concentrations. A decrease in eluted peak KAV indicates filament assembly. B, analytical sizing on mCherry-IIB (loaded at 5 μmdimer) preassembled at 150 mm NaCl, alone or with 14-3-3 σ (loaded at 12.5 μmdimer). 14-3-3 σ leads to a pronounced shift toward smaller filaments. C, FCS diffusion time for mCherry-IIB filaments assembled at 100 nmdimer, 37 °C, with or without 14-3-3 σ. IIB assembled at 500 mm NaCl is included as an unassembled control. Diffusion time at 150 mm NaCl is markedly larger, indicating filament assembly. Addition of excess 14-3-3 σ reduces the diffusion time, indicating a reduction in filament size. Points are technical replicates. Shown are post-ANOVA results of Fisher's LSD test versus IIB only at 150 mm NaCl; **, p < 0.005. D, FCS independently diffusing particle counts for mCherry-IIB assembled at 100 nmdimer, 37 °C, 150 mm NaCl unless otherwise listed. A lower particle count (at constant protein concentration) corresponds to greater assembly. Addition of 14-3-3 σ leads to disassembly of filaments in a dose-dependent manner. Points are technical replicates. Shown are post-ANOVA results of Fisher's LSD test versus IIB only at 150 mm NaCl; **, p < 0.005. E, FCS particle counts from D, normalized between measurements at 500 mm NaCl (disassembled) and 150 mm NaCl (normal assembly), plotted versus 14-3-3 concentration and fit with a quadratic equation. KD = 380 ± 390 nmdimer (χ2 = 0.077). Points represent mean ± S.E. F, salt-dependent sedimentation of 1 μmdimer mCherry-IIB with or without equimolar 14-3-3 σ. Addition of 14-3-3 σ increases the soluble fraction, indicating disassembly. Sample sizes at all salt concentrations ranged from n = 3–10. All points represent mean ± S.E. Student's t test versus control; **, p < 0.005. G, titration of 14-3-3 σ against 1 μmdimer mCherry-IIB at 150 mm NaCl. Addition of 14-3-3 σ leads to disassembly of filaments in a dose-dependent manner. Points represent mean ± S.E., n = 6. H, titration of various concentrations of 14-3-3 σ against various concentrations of mCherry-IIB at 150 mm NaCl. 14-3-3 leads to saturable disassembly of myosin across the entire concentration regime tested. Trend lines are provided to aid visualization of each titration. I, concentration of myosin in soluble fraction plotted versus total myosin. Increasing 14-3-3 leads to more myosin in the supernatant. J, surface plasmon resonance titration of 14-3-3 σ against immobilized monomeric mCherry-IIB. 14-3-3 σ binds to monomeric tail fragments in a dose-dependent manner.
Human 14-3-3 σ perturbs myosin II assembly in solution
To gain more information about myosin assembly in solution and at physiological temperatures, we turned again to FCS as described above. For these studies, we used purified proteins, which gave us full control over protein concentrations and the ionic strength of the solution. For each condition, we obtained measurements of the average number of independently diffusing fluorescent species (reported as “number of particles”) within the confocal volume as well as their average diffusion time (35). First we performed FCS on mCherry-IIB tail fragments at high salt, where assembly is inhibited. The measured diffusion time was 438 ± 11 μs, which is in good agreement with the theoretical value of 388 μs (at 37 °C) based on the measured Stokes radius from our analytical gel filtration experiments. We then lowered the ionic strength to 150 mm NaCl and repeated our observations, obtaining a diffusion time of 873 ± 19 μs, indicating the presence of larger, more slowly diffusing structures. Finally, we added 14-3-3 σ while maintaining the ionic strength constant, obtaining a diffusion time of 615 ± 13 μs, which is lower than the assembled state but still higher than the fully disassembled state (Fig. 3C). These changes in diffusion time match our analytical sizing results and demonstrate that 14-3-3 σ reduces the size of myosin II filaments. However, diffusion time does not directly indicate the size of the myosin II filaments; even if the addition of 14-3-3 completely disassembled myosin in solution, we would expect an increase in the baseline diffusion time because of the larger size of the myosin–14-3-3 complex.
To estimate the size of the myosin II filaments, we monitored the number of independent particles as the ionic strength was lowered. Because all myosins are fluorescently labeled in these experiments, the number of independently diffusing fluorescent species will drop as myosin II filaments assemble at constant total concentration. Indeed, transitioning from 500 mm to 150 mm NaCl produced a marked decrease in independent diffusors within the measurement volume, as predicted. We then titrated in increasing amounts of 14-3-3 σ while holding the ionic strength constant and observed that the number of independent diffusors increased in a 14-3-3 concentration-dependent manner (Fig. 3D). Fitting a binding model to these data (Fig. 3E) yielded an apparent KD of 380 ± 390 nmdimer, which is in a similar range to the KD obtained for the Dictyostelium 14-3-3–myosin II interaction.
Next we turned to bulk myosin sedimentation assays to examine the effects of 14-3-3 on large, assembled filaments, a size range that is poorly sampled in the previously discussed techniques. We first performed a salt-dependent assembly assay on mCherry-IIB to confirm its proper assembly and noted the expected behavior, with high assembly at low ionic strengths and no assembly at high ionic strengths. These findings agree with previous work by Nakasawa et al. (33) on myosin IIB tail fragments. We then added an equimolar amount of 14-3-3 σ and observed robust mCherry-IIB solubilization, with the greatest effect occurring at 150 mm NaCl (Fig. 3F). We also titrated 14-3-3 σ against a constant 1 μmdimer of mCherry-IIB at 150 mm NaCl and observed saturable solubilization (Fig. 3G). We then repeated this experiment at a range of mCherry-IIB and 14-3-3 σ concentrations, obtaining saturable solubilization in each case (Fig. 3H). We plotted the soluble pool as a function of total mCherry-IIB, expecting this to plateau at a single critical concentration at each 14-3-3 level. However, we found that the free concentration rose with higher amounts of mCherry-IIB without plateauing, possibly reflecting the paucidisperse nature of the system (Fig. 3I). We have also found that, in cells, the myosin IIB and 14-3-3 dimer concentrations are on the order of 100 nm (36) and 250 nm (data not shown), respectively, indicating that the lower end of the titration may be the more physiologically relevant range.
Because our readout for 14-3-3 binding in the previous assays was its effect on myosin filaments, we obtained a direct readout of binding independent of myosin assembly. To do this, we employed surface plasmon resonance (SPR). In this method, a “bait” analyte is immobilized on the surface of a sample chip. Light reflected off the bottom of this chip excites an electromagnetic surface wave, or plasmon, whose propagation properties are highly sensitive to surface conditions. Upon the addition of liquid-phase “prey,” any binding to the bait will alter surface conditions enough to produce a detectable signal. Because SPR requires no labeling of the species being analyzed and is performed with immobilized bait, it presents a unique opportunity to measure the binding of 14-3-3 to fully unassembled myosin monomers under physiological conditions. We immobilized these monomers to polyclonal antibodies in a high-salt buffer to maintain their monomeric state. When immobilized and therefore locked in unassembled conditions, we switched to a physiological ionic strength and tested 14-3-3 σ for binding. We observed that 14-3-3 σ indeed bound to unassembled myosin IIB monomers in a dose-dependent manner (Fig. 3J). This binding event in vivo would cause similar effects to those we observed in Dictyostelium, shifting the system equilibrium to a less assembled state by lowering the effective concentration of assembly-competent monomers.
14-3-3's effect on myosin II assembly is 14-3-3 paralog–specific
In higher eukaryotes, the spectrum of possible 14-3-3–myosin II interactions is very broad, as there are three paralogs of nonmuscle myosin II and seven paralogs of 14-3-3 in the mammalian genome. Having explored 14-3-3 σ's interaction with myosin IIB in detail, we then examined whether any other interactions might occur between human myosin IIs and 14-3-3s. After expressing and purifying His6-tagged versions of the remaining human paralogs of 14-3-3 (β, ϵ, γ, η, θ, and ζ), we expanded our sedimentation studies to all three human myosin II paralogs, both alone and in the presence of each of the seven human 14-3-3 paralogs, capturing the entire landscape of 14-3-3's regulation of myosin II assembly. After obtaining high-purity preparations of each protein (Fig. 4A), we performed sedimentation assays at the physiological ionic strength of 150 mm NaCl and 37 °C. All 14-3-3s had effects on at least one paralog of myosin II, most of them acting as myosin solubilizers (Fig. 4, B–D). 14-3-3 σ solubilized all three paralogs of myosin II, and 14-3-3 ζ had no significant effect on IIB or IIC but promoted assembly of IIA.
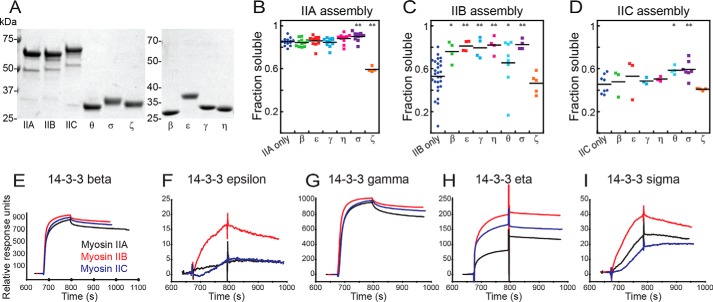
Figure 4.
The landscape of 14-3-3 interaction with human myosin IIs. A, images of Coomassie Brilliant Blue–stained SDS-PAGE gels of purified proteins used in this study. B–D, sedimentation assays of 1 μmdimer mCherry-IIA (B), mCherry-IIB (C), and mCherry-IIC (D), assembled at 150 mm NaCl, 37 °C, alone or in the presence of an equimolar 14-3-3 paralog (β, ϵ, γ, η, θ, σ, or ζ). Shown are post-ANOVA results of Fisher's LSD test versus negative control; *, p < 0.05; **, p < 0.005. E–I, surface plasmon resonance traces for each of the three immobilized, monomeric myosins bound by 14-3-3s: β (E), ϵ (F), γ (G), η (H), and σ (I).
We then returned to SPR to test whether the 14-3-3s could bind to immobilized monomeric myosin II, as was the case with 14-3-3 σ. We tested 14-3-3s β, ϵ, γ, η, and σ, each of which was able to bind to the three myosin paralogs in the SPR framework (Fig. 4, E–I). Overall, these results provide a biochemical framework for the 14-3-3 paralogs to bind the myosin II tails, helping control the degree of myosin II assembly and thereby modulating myosin II activity, with myosin IIB being particularly sensitive to this mode of regulation.
Discussion
We have demonstrated that 14-3-3 acts as a buffering system for myosin II motor proteins, directly interacting with the C-terminal tail domain. All three paralogs of mammalian myosin II are modulated by 14-3-3s, and their ability to inhibit or drive assembly provides a diverse repertoire of outcomes for myosin II assembly. We demonstrated that, in Dictyostelium, 14-3-3 is able to solubilize both phosphomimetic and nonphosphorylatable myosin II tail fragments, and in mammals, most 14-3-3s solubilize unphosphorylated tail fragments. Therefore, 14-3-3 modulation of myosin II assembly does not require phosphorylation of the myosin tail. In vivo, the overall level of myosin II assembly is likely achieved by a combination of canonical phospho-regulation and 14-3-3 binding. Because myosin II phosphorylation and dephosphorylation are enzymatic steps, whereas 14-3-3 binding is nonenzymatic, phosphorylation could serve to govern the timing and gross magnitude of myosin II assembly, whereas 14-3-3 would modulate myosin II assembly continuously to ensure the availability of assembly-competent myosin II monomers around the cell.
This regulatory paradigm becomes particularly interesting in the context of cells that express multiple 14-3-3 paralogs. Because different 14-3-3s have varying effects on myosin assembly, 14-3-3 paralogs likely compete for myosin II within a single cell. The relative expression levels of each paralog would, therefore, alter the average level of myosin II assembly. Furthermore, myosin II must compete with other 14-3-3 interactors for occupancy. Because the majority of known 14-3-3 interactors are phosphorylated, these binding events may be of higher affinity than the myosin–14-3-3 interaction. Therefore, the degree of 14-3-3 contribution to myosin II assembly becomes a function of relative protein levels. Myosin II is highly expressed in most cells, 3.4 μmhexamer in Dictyostelium (37) but in the 100–800 nmhexamer range in mammalian cells, depending on the paralog and cell type (36). 14-3-3s are also abundant in cells, and the single 14-3-3 in Dictyostelium is found at 700 nmdimer (25), whereas 14-3-3 sigma is found at ∼250 nmdimer in HeLa cells. If Dictyostelium myosin II and 14-3-3 were the only interactors, then, at these concentrations and the measured KD of 450 nmdimer, ∼90% of the 14-3-3 would be bound to myosin. However, there are many other 14-3-3 targets in the cell at varying levels of expression, although often only the phosphorylated forms of these targets will bind 14-3-3 and compete with myosin. Thus, changes in available 14-3-3, either through shifts in its own expression or in the availability of binding partners, could alter the levels of myosin II assembly around the cell. This is similar to what has been observed in Dictyostelium, where altering 14-3-3 levels has dramatic effects on myosin II assembly as well as on cortical tension (25). As myosin II force production is a major contributor to cell mechanics in both systems, this would make myosin regulation by 14-3-3 a significant determinant of overall cell mechanics.
This concept is quite compelling in the context of 14-3-3 σ, which has many protective roles for epithelial cells but whose expression patterns change upon tumorigenesis (38). In breast, colon, lung, pancreatic, and kidney cancers, 14-3-3 σ expression levels are altered. The loss of 14-3-3 σ expression in early-stage tumor growth is easy to rationalize, as this paralog plays roles in apoptosis, cell cycle control, and DNA damage repair. However, the field has been unable to explain why this tumor suppressor protein would be overexpressed in metastatic cancers. Our finding that 14-3-3 σ is a solubilizer of nonmuscle myosin II provides one possible explanation. Metastatic cancers are often mechanically softer than regular tissue or nonmetastatic cancer (39, 40). These tissues are also more mechanoresponsive (36, 41). This seeming contradiction can be reconciled by considering the increased myosin II turnover caused by 14-3-3 σ sequestration of myosin II monomers. In fact, S100A4 proteins, which inhibit myosin IIA assembly by a similar sequestration mechanism (42, 43), are also often overexpressed in metastatic cancers, and decreasing myosin II expression overall has also been shown to lead to increased invasiveness and tumor formation in vivo (44). As in Dictyostelium, excess 14-3-3 σ would be expected to promote more dynamic myosin II assemblies with greater turnover, promoting increased mechanosensitivity and, thereby, the ability of myosin II to polarize, driving 3D motility (45). Thus, we have demonstrated a role for 14-3-3 proteins in a cell mechanics pathway that is conserved from amebas to humans and that may play an important role in both healthy and disease states.
Experimental procedures
Reagents and protein purification
Bacterial expression plasmids coding for an N-terminal His6 tag, fused to the mCherry fluorophore, and fused to the assembly domains of Dictyostelium myosin II (residues 1533–1823), human myosin IIA (residues 1722–1960), human myosin IIB (residues 1729–1976), and mouse myosin IIC (residues 1782–2033) were generated in pBiEx1 using standard cloning techniques. Human myosin IIC (residues 1787–2036) was generated by reverse transcriptase PCR and inserted into pBiEx1 using standard cloning techniques. Bacterial expression plasmids for His6-tagged Dictyostelium 14-3-3, human 14-3-3 ϵ, human 14-3-3 σ, and human 14-3-3 ζ were generated in pBiEx1 as well. Bacterial expression plasmids for human 14-3-3s β (Addgene plasmid 39128), γ (Addgene plasmid 39129), η (Addgene plasmid 38814), and a truncated θ construct (Addgene plasmid 38931) were gifts from Nicola Burgess-Brown. To generate full-length θ, the missing residues were added by PCR.
Proteins were expressed in BL-21 StarTM (DE3) (Invitrogen) E. coli in Lysogeny Broth shaking culture overnight at room temperature. Bacteria were harvested by centrifugation and lysed by lysozyme treatment followed by sonication, and the lysate was clarified by centrifugation. Polyethyleneimine (PEI) was added to a final concentration of 0.1% to precipitate nucleic acids, which were then removed by centrifugation. 14-3-3 precipitated in the PEI pellet for Dictyostelium 14-3-3 and human 14-3-3 ϵ. This pellet was resuspended in column running buffer (10 mm HEPES (pH 7.1), 500 mm NaCl, and 10 mm imidazole), dialyzed against the same for a minimum of 4 h, clarified by centrifugation and filtration, and run on a nickel-nitrilotriacetic acid metal affinity column to obtain high-purity 14-3-3. The myosin II constructs and human 14-3-3s β, γ, η, σ, θ, and ζ remained in the PEI supernatant and were precipitated by adding ammonium sulfate to 50% saturation and centrifuging. The pellet was resuspended in column running buffer, dialyzed against the same for a minimum of 4 h, clarified by centrifugation and filtration, and run on a nickel-nitrilotriacetic acid metal affinity column followed by a sizing column for the myosin II constructs. The myosin II constructs were then concentrated and further purified by dialyzing against assembly buffer (10 mm HEPES (pH 7.1) and 50 mm NaCl) until precipitate formed, followed by centrifugation and resuspension of the pellet in storage buffer (10 mm HEPES (pH 7.1) and 500 mm NaCl). Protein purity was verified by SDS-PAGE followed by Coomassie Blue staining, and concentration was quantified by UV absorbance using the calculated extinction coefficient for each protein's amino acid sequence. The REVVTID and KLESDI peptides were synthesized by the Synthesis and Sequencing Facility at the Johns Hopkins University School of Medicine using an Aapptec Focus Synthesizer.
Analytical gel filtration
Analytical gel filtration was performed using a BioLogic DuoFlow FPLC system (Bio-Rad) and a Superdex 200 10/300 GL column (GE Healthcare). After equilibration with running buffer (10 mm HEPES (pH 7.1) and 500 mm NaCl), the void volume and total bed volume were obtained with blue dextran in water. The column was then calibrated using sizing standards of aprotinin, cytochrome c, carbonic anhydrase, BSA, and thyroglobulin. The volume of elution was measured from the beginning of sample injection to the peak of the UV trace, and a KAV for each species was obtained using Equation 1. A calibration plot of known RH versus measured KAV was then generated and fit with an exponential function. Experimental runs were performed in the same fashion, allowing for the calculation of RH for each species. Assembly was monitored by re-equilibrating the column at a lower salt concentration (150 mm NaCl), diluting a sample to the same concentration, and monitoring the shift in the UV peak. All runs were performed at 0.5 ml/min flow rate, 0.5 ml total injection volume, and 4 °C.
| (1) |
Assembly assay
In vitro assembly of myosin II was conducted according to the method of Zhou et al. (25) with modifications. The incubation time and temperature were adjusted to 30 min at the physiological temperature for each myosin species (22 °C for Dictyostelium myosin, 37 °C for human myosins). These temperatures were also used during the centrifugation step. Band intensities post-staining were quantified and background-subtracted using ImageJ (National Institutes of Health). The integrated density of the supernatant, divided by the summed integrated densities of the supernatant and the pellet, yielded the fraction soluble (FS) for each sample. Apparent KD was determined by fitting these data to Equation 2 (where AT is the input concentration of 14-3-3, BT represents input myosin II minus the fraction soluble in the absence of 14-3-3, and FS0 represents the fraction soluble in the absence of 14-3-3) to a titration experiment using a least-squares approach. Fits were performed using KaleidaGraph (Synergy Software), and we report parameter value ± S.E. (46).
| (2) |
Fluorescence correlation spectroscopy
All FCS measurements were performed as described previously (47). Briefly, we used a Zeiss LSM780-FCS confocal microscope with a Zeiss C-Apochromat ×40/1.2 water immersion objective and a stage warmer to maintain the sample at 37 °C. The confocal volume was calibrated using a 50 nm solution of rhodamine-6G in water, with a diffusion coefficient of 400 μm2/s at 22.5 °C (48), which equates to a diffusion coefficient of 572.3 μm2/s at 37 °C (Equation 3).
| (3) |
To measure myosin filament assembly, mCherry-labeled myosin tail fragments were diluted in a solution containing 10 mm HEPES (pH 7.1) at a desired concentration of NaCl, and the temperature was equilibrated to 37 °C for at least 10 min. The final concentration of tail fragments was either 100 nmdimer or 50 nmdimer, and the salt concentrations used were 500 mm NaCl (for completely disassembled filaments), 250 mm NaCl (for partial assembly), or 150 mm NaCl (for assembly at a physiological ionic strength). Other proteins (14-3-3 σ, purified BSA at 0.5 μg/ml) were included as needed. Because the concentration of the fluorescent species is a crucial parameter to normalization, we verified that raw counts remained the same across conditions, indicating that neither nonspecific aggregation nor adherence to the walls of the chamber was a factor. We also observed no effects on raw counts or the independent particle count when BSA was added to the solution as a nonfluorescent blocking protein.
After equilibration, FCS measurements were taken as 10 consecutive 5-s scans. Any scans that demonstrated aggregates, baseline drift, or other aberrant features were discarded, and an autocorrelation curve was generated for the remaining scans and fitted with a one-component, one-triplet state model using ZEN imaging software (Zeiss). Particle counts and diffusion times were extracted from these fits, and a minimum of 10 points per sample were averaged to generate the final measured values.
For in vivo FCCS, Dictyostelium cells were cultured as described previously (25). For studies with FP-myosin II, we used the mhcA null strain HS1 (49), and for the WT background, we used KAx3 cells (50). The plasmids for GFP, mCherry, GFP-myosin-II, mCherry-myosin-II, and 14-3-3-GFP have also been described previously (25). The GFP-mCherry linked construct was generated by adding a five-amino acid linker between GFP and mCherry.
FCCS experiments were performed as described previously at room temperature (∼25 °C) (51). The in vivo KD was calculated by Equation 4, where V is volume, Gx is the cross-correlation, and Ga and Gb refer to the auto-correlation values for GFP and mCherry as extracted from the Zen imaging software (51).
| (4) |
EM
Carbon-coated EM grids were rendered hydrophilic by glow discharge. Freshly prepared grids were placed in sample droplets for 30 s, followed by 2 washes in water and staining using 1% uranyl acetate. Samples were then observed using a Philips BioTwin CM120 transmission electron microscope.
SPR
A rabbit polyclonal antibody against mCherry was covalently attached to an SPR sensor chip. mCherry-labeled myosin tail fragments in a buffer containing 10 mm HEPES and 500 mm NaCl (pH 7.1) were then added to the chip, allowing the antibodies to capture these fragments in the monomeric state. Excess protein was removed by washing, and the remaining immobilized fragments were equilibrated to 150 mm NaCl. Unlabeled 14-3-3 was then added in the same buffer while monitoring the resonance angle, generating a readout of binding. After each experiment, the chip was regenerated to remove bound protein, and fresh myosin fragments were immobilized to the surface. Experiments were carried out at 37 °C.
Author contributions
H. W.-F. and D. N. R. designed the study. P. K. performed the FCCS experiments on Dictyostelium and assisted with data analysis. J. O. performed the experiments on the Dictyostelium ADCT, 3×Asp, and 3×Ala constructs. H. W.-F. performed all other experiments and data analysis and generated figures. H. W.-F., P. K., and D. N. R. wrote and edited the manuscript.
Acknowledgments
We thank the Johns Hopkins University Microscope Facility, particularly Barbara Smith, for providing equipment and technical assistance for the EM and FCS experiments. We thank Vasudha Srivastava for FCS training and experimental suggestions. We thank Robert Bloch and Yinghua Zhang at the University of Maryland Biosensor Core, who performed the SPR studies. We would also like to thank all D. N. R. laboratory members for their help and feedback. The FCS instrument was purchased using National Institutes of Health Shared Instrumentation Grant S10 OD016374.
This work was supported by National Institutes of Health Grant R01 GM66817 and the Thomas Wilson Foundation (to D. N. R). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.
- BTF
- bipolar thick filament
- AD
- assembly domain
- ADCT
- C terminus assembly domain
- ELM
- Eukaryotic Linear Motif
- FCS
- fluorescence correlation spectroscopy
- FCCS
- fluorescence cross-correlation spectroscopy
- SPR
- surface plasmon resonance
- PEI
- polyethyleneimine
- ANOVA
- analysis of variance
- LSD
- least significant difference.
References
- 1. Ford J. C., al-Khodairy F., Fotou E., Sheldrick K. S., Griffiths D. J., and Carr A. M. (1994) 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265, 533–535 10.1126/science.8036497 [DOI] [PubMed] [Google Scholar]
- 2. Hermeking H., Lengauer C., Polyak K., He T. C., Zhang L., Thiagalingam S., Kinzler K. W., and Vogelstein B. (1997) 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3–11 10.1016/S1097-2765(00)80002-7 [DOI] [PubMed] [Google Scholar]
- 3. Peng C.-Y., Graves P. R., Thoma R. S., Wu Z., Shaw A. S., and Piwnica-Worms H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277, 1501–1505 10.1126/science.277.5331.1501 [DOI] [PubMed] [Google Scholar]
- 4. Zhang L., Chen J., and Fu H. (1999) Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 8511–8515 10.1073/pnas.96.15.8511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bridges D., and Moorhead G. B. (2005) 14-3-3 proteins: a number of functions for a numbered protein. Sci. STKE 2005, re10 [DOI] [PubMed] [Google Scholar]
- 6. Masters S. C., Pederson K. J., Zhang L., Barbieri J. T., and Fu H. (1999) Interaction of 14-3-3 with a nonphosphorylated protein ligand, exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38, 5216–5221 10.1021/bi982492m [DOI] [PubMed] [Google Scholar]
- 7. Calverley D. C., Kavanagh T. J., and Roth G. J. (1998) Human signaling protein 14-3-3ζ interacts with platelet glycoprotein Ib subunits Ibα and Ibβ. Blood 91, 1295–1303 [PubMed] [Google Scholar]
- 8. Campbell J. K., Gurung R., Romero S., Speed C. J., Andrews R. K., Berndt M. C., and Mitchell C. A. (1997) Activation of the 43 kDa inositol polyphosphate 5-phosphatase by 14-3-3ζ. Biochemistry 36, 15363–15370 10.1021/bi9708085 [DOI] [PubMed] [Google Scholar]
- 9. Liang X., Butterworth M. B., Peters K. W., Walker W. H., and Frizzell R. A. (2008) An obligatory heterodimer of 14-3-3β and 14-3-3epsilon is required for aldosterone regulation of the epithelial sodium channel. J. Biol. Chem. 283, 27418–27425 10.1074/jbc.M803687200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu K., Lu G., Sehnke P., and Ferl R. J. (1997) The heterologous interactions among plant 14-3-3 proteins and identification of regions that are important for dimerization. Arch. Biochem. Biophys. 339, 2–8 10.1006/abbi.1996.9841 [DOI] [PubMed] [Google Scholar]
- 11. Wilker E., and Yaffe M. B. (2004) 14-3-3 proteins: a focus on cancer and human disease. J. Mol. Cell Cardiol. 37, 633–642 10.1016/j.yjmcc.2004.04.015 [DOI] [PubMed] [Google Scholar]
- 12. Fountoulakis M., Cairns N., and Lubec G. (1999) Increased levels of 14-3-3 γ and ϵ proteins in brain of patients with Alzheimer's disease and Down syndrome. J. Neural Transm. Suppl. 57, 323–335 [DOI] [PubMed] [Google Scholar]
- 13. Slone S. R., Lavalley N., McFerrin M., Wang B., and Yacoubian T. A. (2015) Increased 14-3-3 phosphorylation observed in Parkinson's disease reduces neuroprotective potential of 14-3-3 proteins. Neurobiol. Dis. 79, 1–13 10.1016/j.nbd.2015.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perathoner A., Pirkebner D., Brandacher G., Spizzo G., Stadlmann S., Obrist P., Margreiter R., and Amberger A. (2005) 14-3-3sigma expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Clin. Cancer Res. 11, 3274–3279 10.1158/1078-0432.CCR-04-2207 [DOI] [PubMed] [Google Scholar]
- 15. Ostergaard M., Rasmussen H. H., Nielsen H. V., Vorum H., Orntoft T. F., Wolf H., and Celis J. E. (1997) Proteome profiling of bladder squamous cell carcinomas: identification of markers that define their degree of differentiation. Cancer Res. 57, 4111–4117 [PubMed] [Google Scholar]
- 16. Ferguson A. T., Evron E., Umbricht C. B., Pandita T. K., Chan T. A., Hermeking H., Marks J. R., Lambers A. R., Futreal P. A., Stampfer M. R., and Sukumar S. (2000) High frequency of hypermethylation at the 14-3-3 σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 6049–6054 10.1073/pnas.100566997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata N., Yamamoto H., Sasaki S., Itoh F., Suzuki H., Kikuchi T., Kaneto H., Iku S., Ozeki I., Karino Y., Satoh T., Toyota J., Satoh M., Endo T., and Imai K. (2000) Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 σ gene in human hepatocellular carcinoma. Oncogene 19, 5298–5302 10.1038/sj.onc.1203898 [DOI] [PubMed] [Google Scholar]
- 18. Suzuki H., Itoh F., Toyota M., Kikuchi T., Kakiuchi H., and Imai K. (2000) Inactivation of the 14-3-3 sigma gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 60, 4353–4357 [PubMed] [Google Scholar]
- 19. Li Y. L., Liu L., Xiao Y., Zeng T., and Zeng C. (2015) 14-3-3sigma is an independent prognostic biomarker for gastric cancer and is associated with apoptosis and proliferation in gastric cancer. Oncol. Lett. 9, 290–294 10.3892/ol.2014.2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi W., Liu X., Qiao D., and Martinez J. D. (2005) Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int. J. Cancer 113, 359–363 10.1002/ijc.20492 [DOI] [PubMed] [Google Scholar]
- 21. Rodriguez J. A., Li M., Yao Q., Chen C., and Fisher W. E. (2005) Gene overexpression in pancreatic adenocarcinoma: diagnostic and therapeutic implications. World J. Surg. 29, 297–305 10.1007/s00268-004-7843-0 [DOI] [PubMed] [Google Scholar]
- 22. Zhang W., and Robinson D. N. (2005) Balance of actively generated contractile and resistive forces controls cytokinesis dynamics. Proc. Natl. Acad. Sci. U.S.A. 102, 7186–7191 10.1073/pnas.0502545102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kee Y.-S., and Robinson D. N. (2008) Motor proteins: myosin mechanosensors. Curr. Biol. 18, R860–R862 10.1016/j.cub.2008.07.071 [DOI] [PubMed] [Google Scholar]
- 24. Elliott H., Fischer R. S., Myers K. A., Desai R. A., Gao L., Chen C. S., Adelstein R. S., Waterman C. M., and Danuser G. (2015) Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat. Cell Biol. 17, 137–147 10.1038/ncb3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Q., Kee Y.-S., Poirier C. C., Jelinek C., Osborne J., Divi S., Surcel A., Will M. E., Eggert U. S., Müller-Taubenberger A., Iglesias P. A., Cotter R. J., and Robinson D. N. (2010) 14-3-3 coordinates microtubules, rac, and myosin II to control cell mechanics and cytokinesis. Curr. Biol. 20, 1881–1889 10.1016/j.cub.2010.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson D. N. (2010) 14-3-3, an integrator of cell mechanics and cytokinesis. Small GTPases 1, 165–169 10.4161/sgtp.1.3.14432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bosgraaf L., and van Haastert P. J. (2006) The regulation of myosin II in Dictyostelium. Eur. J. Cell Biol. 85, 969–979 10.1016/j.ejcb.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 28. Egelhoff T. T., Lee R. J., and Spudich J. A. (1993) Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell 75, 363–371 10.1016/0092-8674(93)80077-R [DOI] [PubMed] [Google Scholar]
- 29. Surcel A., Ng W. P., West-Foyle H., Zhu Q., Ren Y., Avery L. B., Krenc A. K., Meyers D. J., Rock R. S., Anders R. A., Freel Meyers C. L., and Robinson D. N. (2015) Pharmacological activation of myosin II paralogs to correct cell mechanics defects. Proc. Natl. Acad. Sci. U.S.A. 112, 1428–1433 10.1073/pnas.1412592112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Halloran T. J., Ravid S., and Spudich J. A. (1990) Expression of Dictyostelium myosin tail segments in Escherichia coli: domains required for assembly and phosphorylation. J. Cell Biol. 110, 63–70 10.1083/jcb.110.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hostetter D., Rice S., Dean S., Altman D., McMahon P. M., Sutton S., Tripathy A., and Spudich J. A. (2004) Dictyostelium myosin bipolar thick filament formation: Importance of charge and specific domains of the myosin rod. PLoS Biol. 2, e356 10.1371/journal.pbio.0020356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang B., Yang H., Liu Y.-C., Jelinek T., Zhang L., Ruoslahti E., and Fu H. (1999) Isolation of high-affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 10.1021/bi991353h [DOI] [PubMed] [Google Scholar]
- 33. Nakasawa T., Takahashi M., Matsuzawa F., Aikawa S., Togashi Y., Saitoh T., Yamagishi A., and Yazawa M. (2005) Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry 44, 174–183 10.1021/bi048807h [DOI] [PubMed] [Google Scholar]
- 34. Ohi M., Li Y., Cheng Y., and Walz T. (2004) Negative staining and image classification: powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 10.1251/bpo70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fitzpatrick J. A., and Lillemeier B. F. (2011) Fluorescence correlation spectroscopy: linking molecular dynamics to biological function in vitro and in situ. Curr. Opin. Struct. Biol. 21, 650–660 10.1016/j.sbi.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 36. Surcel A., Schiffhauer E. S., Thomas D., Zhu Q., DiNapoli K., Herbig M., Otto O., Guck J., Jaffee E., Iglesias P., Anders R., and Robinson D. (2017) Harnessing the adaptive potential of mechanoresponsive proteins to overwhelm pancreatic cancer dissemination and invasion. BioRxiv 10.1101/190553 [DOI] [Google Scholar]
- 37. Robinson D. N., Cavet G., Warrick H. M., and Spudich J. A. (2002) Quantitation of the distribution and flux of myosin-II during cytokinesis. BMC Cell Biol. 3, 4 10.1186/1471-2121-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guweidhi A., Kleeff J., Giese N., El Fitori J., Ketterer K., Giese T., Büchler M. W., Korc M., and Friess H. (2004) Enhanced expression of 14-3-3σ in pancreatic cancer and its role in cell cycle regulation and apoptosis. Carcinogenesis 25, 1575–1585 10.1093/carcin/bgh159 [DOI] [PubMed] [Google Scholar]
- 39. Gal N., and Weihs D. (2012) Intracellular mechanics and activity of breast cancer cells correlate with metastatic potential. Cell Biochem. Biophys. 63, 199–209 10.1007/s12013-012-9356-z [DOI] [PubMed] [Google Scholar]
- 40. Martinez Vazquez R., Nava G., Veglione M., Yang T., Bragheri F., Minzioni P., Bianchi E., Di Tano M., Chiodi I., Osellame R., Mondello C., and Cristiani I. (2015) An optofluidic constriction chip for monitoring metastatic potential and drug response of cancer cells. Integr. Biol. (Camb.) 7, 477–484 10.1039/C5IB00023H [DOI] [PubMed] [Google Scholar]
- 41. Bordeleau F., Alcoser T. A., and Reinhart-King C. A. (2014) Physical biology in cancer: 5: the rocky road of metastasis: the role of cytoskeletal mechanics in cell migratory response to 3D matrix topography. Am. J. Physiol. Cell Physiol. 306, C110–C120 10.1152/ajpcell.00283.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ford H. L., Silver D. L., Kachar B., Sellers J. R., and Zain S. B. (1997) Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry 36, 16321–16327 10.1021/bi971182l [DOI] [PubMed] [Google Scholar]
- 43. Malashkevich V. N., Varney K. M., Garrett S. C., Wilder P. T., Knight D., Charpentier T. H., Ramagopal U. A., Almo S. C., Weber D. J., and Bresnick A. R. (2008) Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry 47, 5111–5126 10.1021/bi702537s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schramek D., Sendoel A., Segal J. P., Beronja S., Heller E., Oristian D., Reva B., and Fuchs E. (2014) Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343, 309–313 10.1126/science.1248627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chi Q., Yin T., Gregersen H., Deng X., Fan Y., Zhao J., Liao D., and Wang G. (2014) Rear actomyosin contractility-driven directional cell migration in three-dimensional matrices: a mechano-chemical coupling mechanism. J. R. Soc. Interface 11, 20131072 10.1098/rsif.2013.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Press W., Teukolsky S., Vetterling W., and Flannery B. (1992) Numerical Recipes in C: The Art of Scientific Computing Second Edition, pp. 663–665, Cambridge University Press, Cambridge, UK [Google Scholar]
- 47. Srivastava V., and Robinson D. N. (2015) Mechanical stress and network structure drive protein dynamics during cytokinesis. Curr. Biol. 25, 663–670 10.1016/j.cub.2015.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gendron P. O., Avaltroni F., and Wilkinson K. J. (2008) Diffusion coefficients of several rhodamine derivatives as determined by pulsed field gradient–nuclear magnetic resonance and fluorescence correlation spectroscopy. J. Fluoresc. 18, 1093–1101 10.1007/s10895-008-0357-7 [DOI] [PubMed] [Google Scholar]
- 49. Ruppel K. M., Uyeda T. Q., and Spudich J. A. (1994) Role of highly conserved lysine 130 of myosin motor domain: in vivo and in vitro characterization of site specifically mutated myosin. J. Biol. Chem. 269, 18773–18780 [PubMed] [Google Scholar]
- 50. Lee S., Shen Z., Robinson D. N., Briggs S., and Firtel R. A. (2010) Involvement of the cytoskeleton in controlling leading-edge function during chemotaxis. Mol. Biol. Cell 21, 1810–1824 10.1091/mbc.E10-01-0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kothari P., Schiffhauer E. S., and Robinson D. N. (2017) Cytokinesis from nanometers to micrometers and microseconds to minutes. Methods Cell Biol. 137, 307–322 10.1016/bs.mcb.2016.03.038 [DOI] [PubMed] [Google Scholar]