Abstract
Inflammatory cell death, or pyroptosis, is triggered by pathogenic infections or events. It is executed by caspase-1 (in the canonical pyroptosis pathway) or caspase-11 (noncanonical pathway), each via production of a cell-lytic domain from the pyroptosis effector protein gasdermin D through specific and limited proteolysis. Pyroptosis is accompanied by the release of inflammatory mediators, including the proteolytically processed forms of interleukin-1β (IL-1β) and IL-18. Given the similar inflammatory outcomes of the canonical and noncanonical pyroptosis pathways, we hypothesized that caspase-1 and -11 should have very similar activities and substrate specificities. To test this hypothesis, we purified recombinant murine caspases and analyzed their primary specificities by massive hybrid combinatorial substrate library (HyCoSuL) screens. We correlated the substrate preferences of each caspase with their activities on the recombinant natural substrates IL-1β, IL-18, and gasdermin D. Although we identified highly selective and robust peptidyl substrates for caspase-1, we were unable to do so for caspase-11, because caspase-1 cleaved even the best caspase-11 substrates equally well. Caspase-1 rapidly processed pro-IL-1β and -18, but caspase-11 processed these two pro-ILs extremely poorly. However, both caspase-1 and -11 efficiently produced the cell-lytic domain from the gasdermin D precursor. We hypothesize that caspase-11 may have evolved a specific exosite to selectively engage pyroptosis without directly activating pro-IL-1β or -18. In summary, comparing the activities of caspase-1 and -11 in HyCoSuL screens and with three endogenous protein substrates, we conclude that caspase-11 has highly restricted substrate specificity, preferring gasdermin D over all other substrates examined.
Keywords: interleukin, caspase, fluorescence, cell death, enzyme kinetics, GSDMD, proptosis
Introduction
The two cellular pathways that transduce recognition of pathogens by innate immune effector cells to induce cell death and accompanying cytokine release are the canonical pathway (which integrates extracellular pathogen recognition) and the non-canonical pathway (which integrates intracellular cytoplasmic pathogen recognition) (1–4). The canonical pathway operates by recognizing pathogen-associated molecular patterns through membrane-associated Toll-like receptors to engage signal transduction with up-regulation of components of inflammasomes. These macromolecular platforms assemble in the cytoplasm and are essential for recruitment and activation of inflammatory caspases (5, 6), and sensitization of cells to a second signal that activates caspase-1 in an inflammasome-dependent manner. The non-canonical pathway is driven by intracellular cytosolic sensing of pathogens by caspase-11, the mouse ortholog of human caspase-4 and -5, and does not require inflammasome components (1, 7, 8). Although the activation mechanism of caspase-1 and -11 is not defined, it most likely mirrors the proximity-induced dimerization model of initiator caspases in apoptosis (9, 10).
Caspase-1 and caspase-11 cleave gasdermin D (GSDMD),3 forming pores in liposomes by aggregation of N-terminal GSDMD fragments. Although not yet recorded in vivo, these pores are thought to also form in the cell membrane (11–14). The subsequent release of pro-inflammatory mediators via cellular membrane pores defines pyroptosis, a form of regulated inflammatory cell death (15). The requirement of two inflammatory caspases for the different inflammatory pathways is not well-understood.
Caspase-1, formerly known as ICE (interleukin-converting enzyme), was originally identified as the protease responsible for cleaving and activating the 33-kDa pro-IL-1β to its mature 17-kDa form in human monocytic cells (16–18), a role mirrored in mice (18, 19). Caspase-1 also processes pro-IL-18 into its mature form (20–22). Although these inflammatory messengers lack a signal export sequence, after maturation they are nevertheless released from the cell (23). Akin to caspase-11, caspase-1 produces the same cell lytic N-terminal domain of GSDMD (11–14, 24, 25). Together these caspase-1-mediated limited proteolytic events provide a rational explanation of how leaderless cytokines are activated and released from bacterially-infected macrophages, no export mechanism is required because the release is from N-GSDMD-permeabilized cells.
Thus there exist three well-recognized substrates for caspase-1, and at least one substrate in common with caspase-11 (GSDMD). Genetic evidence based on induction of non-canonical pyroptosis indicates that caspase-1 and the canonical inflammasome components Nlrp3 and ASC are required for processing of pro-IL-1β and pro-IL-18 with the implication that caspase-11 does not, or cannot, process these cytokines (1). Given the 53% sequence identity in their catalytic domains of caspase-1 and -11 and the regulatory role caspase-11 has been implicated to play, a question that has remained unanswered is whether efficient catalysis of caspase-11 on pro-IL-1β and pro-IL-18, the only two members of the interleukin family that are processed by caspase-1, differs from caspase-1 hydrolysis. The biochemical characterization of caspase-1 and -11 presented here addresses this dichotomy.
Screening of proteolytic enzymes with short peptide substrate libraries is a common approach to define their inherent substrate preferences in the active site, and thus distinguish closely related proteases. Several chemical- and biological-based approaches have been developed to dissect protease preferences (26). The most commonly used technology utilized Positional Scanning Synthetic Combinatorial Libraries (PS-SCL), introduced by Thornberry and co-workers (27, 28) to screen human caspases. This seminal research provided a robust screening platform and a framework defining the major differences in specificity between inflammatory and apoptotic caspases. These tetrapeptides have served for many years as gold standards for developing substrates, inhibitors, and activity-based probes to study caspases (29) and many other proteases (30). A substantial extension of PS-SCL termed hybrid combinatorial substrate library (HyCoSuL) (31) employs a wide set of unnatural amino acids in peptide mixtures, allowing for detailed exploration of protease-active sites, as we have employed HyCoSuL to discriminate between human apoptotic caspases (32). Accordingly, we employed HyCoSuL here to explore specificity differences between mouse caspase-1 and -11, and deployed the complementary technique of natural protein profiling and kinetics to address the individual roles of caspase-1 and -11 in pyroptosis and inflammatory cytokine activation.
Results
Expression and purification of mouse inflammatory caspases
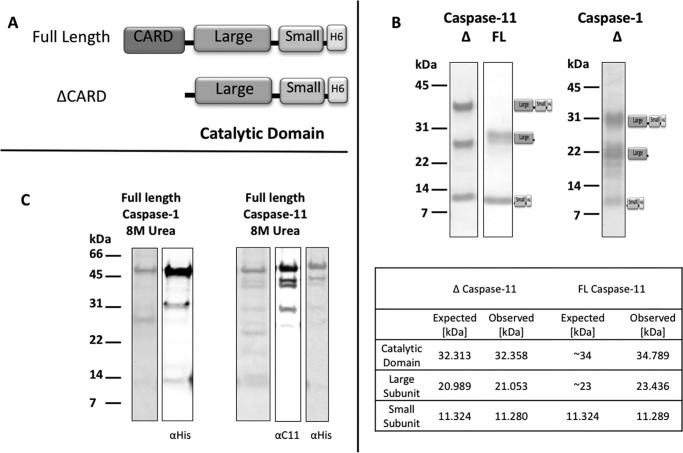
To explore and differentiate the substrate specificity of mouse caspase-1 and caspase-11, we expressed full-length C-terminally His-tagged versions of each enzyme as well as caspase activation and recruitment domain (CARD)-deleted (ΔCARD) versions (Fig. 1). When expressed in Escherichia coli, the purified soluble material did not render a full-length enzyme (Fig. 1B). The purified material from ΔCARD constructs of caspase-1 and -11 revealed two species: an unprocessed catalytic domain comprised of large and small subunits, and processed large and small subunits (Fig. 1B). The identity of these ΔCARD caspase-11 derivatives was confirmed by MS/MALDI (Fig. 1B). MALDI analysis of caspase-11 recovered from full-length construct expression revealed removal of the CARD consistent with cleavage at Phe77/Ser78, which is likely due to cleavage by an endogenous E. coli protease.
Figure 1.
Expression and purification of mouse caspase-1 and -11. A, two constructs, full-length (FL) and CARD-deleted (Δ), were designed for each enzyme, see “Materials and methods” for details of the constructs. B, purified ΔCARD caspase-1 and -11 resulted in a mixture of soluble, unprocessed catalytic subunit with processed large and small subunits. MALDI-MS analysis revealed molecular mass derivatives of ΔCARD caspase-11 that coincides with the expected engineered protein molecular weights. The CARD of FL caspase-11 was removed during expression to generate derivatives whose mass is consistent with processing at Glu97/Ser98. C, cell pellets expressing full-length enzymes were resuspended in 8 m urea, 50 mm Tris-Cl, 100 mm NaCl buffer, pH 8.0. Purified unfolded full-length enzymes were detected on 4–12% BisTris SDS-PAGE with Instant Blue and anti-His or caspase-11 antisera.
CARD-containing proteins belong to the death domain superfamily, which consists of 35 family members, all varying in solubility degree when expressed in E. coli (33). The disappearance of CARDs from the full-length expression constructs raised three possibilities: 1) the overexpression resulted in autoprocessing and removal of the CARDs, 2) CARD is insoluble and was trapped in inclusion bodies; and/or 3) CARD is cleaved by an E. coli protease. To investigate these possibilities we dissolved cell pellets in 8 m urea and purified all His-tagged proteins under denaturing conditions. Immunoblotting revealed full-length versions of both enzymes (Fig. 1C), suggesting that the CARD rendered the full-length caspases insoluble upon expression in E. coli and we are thus only able to purify CARD-removed proteins.
Although CARD-containing proteins, such as human ASC and ICEBERG, have been refolded from denatured preparations (33), our efforts to capture activity following denaturation and renaturation of caspase-1 or -11 were futile after following previously reported protocols (34). Accordingly, because of higher yields, we decided to continue our work with the ΔCARD-purified derivatives of these enzymes.
Caspase-1 and caspase-11 substrate specificity
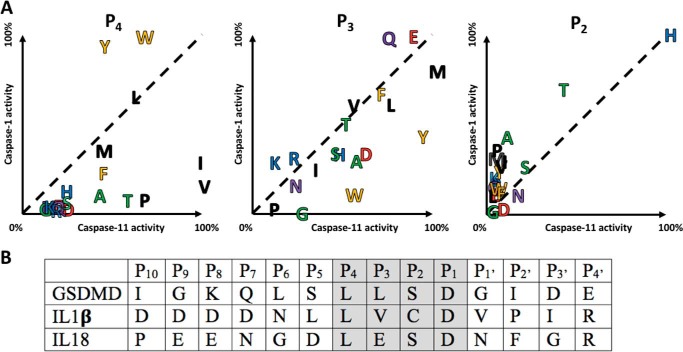
The enzymes were scanned using a HyCoSuL, which employs 19 natural amino acids and 110 unnatural amino acids (32). This tactic allows us to explore in depth the chemical space represented by caspase-active sites. The general formula of the HyCoSuL library is Ac-P4-P3-P2-Asp-ACC where the primary specificity determinant (P1) is fixed as Asp, and the upstream P2–P4 positions are randomized. Caspase-1 and caspase-11 are not synonymous on this library and each displays characteristic differences, as visualized by comparison of preferences for natural amino acids (Fig. 2A). The analysis reveals caspase-1 and -11 both have a strong preference for histidine in the P2 position, and broad tolerance in the P3 position. Caspase-11 accepts a wide variety of residues at P3, but this position is more restrictive for caspase-1. At the P4 position caspase-1 prefers tryptophan, whereas caspase-11 favors valine. The region of endogenous substrates spanning the cleavage sites for caspase-1 and -11 (Fig. 2B) reveals that P4 and P3 somewhat match the optimal substrate sequence motifs, but that P2 does not, with the optimal histidine not utilized in endogenous protein substrates.
Figure 2.
Comparative subsite preferences. A, mouse caspase-1 and caspase-11 were screened on a natural amino acid positional scanning library, see Figs. S1–S3 for complete data. Relative activities reveal distinctive preferences at the P4 and P3 positions, and a common preference at the P2 position. There appeared to be no positions that could be used to discriminate caspase-1 from caspase-11 (except possibly Lys in the P3 position), but caspase-11 showed a preference over caspase-1 for β-branched residues in P4 and large hydrophobic residues in P3. Amino acids close to the dotted diagonal line are equally tolerated by both caspases. Residue coloring reflects common properties of the amino acid side chains. B, alignment of endogenous mouse substrates of the inflammatory caspases from the P10–P4′ region, with the region corresponding to synthetic substrates shaded.
Expanding the screen with unnatural amino acids reveals that in the P2 position caspase-11 can also recognize His(Bzl), whereas caspase-1 has much broader specificity recognizing multiple amino acids, including His(Bzl). Both enzymes present broad specificity in P3 with caspase-11 even tolerating d-amino acids to a small extent, however, several amino acids (Bip, Tyr(Bzl), Bpa) are significantly better recognized by caspase-11 than by caspase-1. The P4 position shows broad specificity for aliphatic and aromatic unnatural amino acids for both caspases with Trp(Me) being the best for caspase-1 and Tle (tert-leucine) being the most active toward caspase-11. Detailed amino acid screening profiles for caspase-1 and -11 are found in Figs. S1–S3.
These characteristics suggested that we may be able to generate individual caspase-selective substrates, and to this end we synthesized the top hits for P2, P3, and P4 using natural amino acids and determined kcat/Km values (Table 1 and Table S1). WEHD and LEHD-based substrates had already been used to measure inflammatory caspase activity (35), and thus served as prior indicators. Indeed, WEHD is the best substrate for caspase-1 with a kcat/Km of 522,000 m−1 s−1. We found the WQPD substrate sequence to be highly selective for caspase-1 over caspase-11 (selectivity factor >5,140), and over a panel of other caspases (Table S3). The best substrates for caspase-11 included PYHD and PMHD tetrapeptides, however, caspase-1 cleaved these substrates even more efficiently with 2–3-fold higher kcat/Km values. Based on these results, caspase-1 is a more active enzyme and outpaces caspase-11 on all substrates containing natural amino acids.
Table 1.
Unique preferred substrates with natural amino acids
Based on screening of a natural amino acid HyCoSuL library, single substrates were synthesized for caspase-1 and caspase-11. Each substrate was assayed with either caspase-1 or caspase-11. All synthesized natural amino acid substrates were preferred by caspase-1, even those that had the highest catalytic numbers for caspase-11. The selectivity factor represents the catalytic parameter kcat/Km for caspase-1 divided by that for caspase-11. Each experiment was repeated at least three times, and the kcat/Km values are presented as an average. Standard deviations were below 15%.
| Substrate sequence: Ac-P4-P3-P2-P1-ACC |
kcat/Km |
Selectivity factor | |
|---|---|---|---|
| Caspase-1 | Caspase-11 | ||
| m−1s−1 | |||
| Reference substrates | |||
| WEHD | 522,000 | 4,100 | 127 |
| LEHD | 179,000 | 3,850 | 47 |
| Caspase-1 preferred | |||
| WQPD | 257,000 | <50 | >5,140 |
| FEAD | 171,000 | <50 | >3,420 |
| WQVD | 328,000 | 244 | 1,340 |
| Caspase-11 preferred | |||
| PYHD | 22,400 | 7,070 | 3.2 |
| PMHD | 16,100 | 7,200 | 2.2 |
Taking advantage of the unique side chains that unnatural amino acids present to explore the active sites of caspase-1 and -11, we synthesized distinctive substrates based on the preferred unnatural amino acid library screening results (Table 2, Table S2). To account for potential subsite cooperativity issues (36, 37) we covered a large amount of sequence combinations, synthesizing 57 individual substrates, 20 with natural amino acids and 37 with natural and unnatural amino acids (Tables S1 and S2). We found several substrates that display significant selectivity for caspase-1 over caspase-11. All these substrates share some structural similarities: cyclic, aliphatic (Cha), or aromatic (Phe and Trp derivatives) amino acids at the P4 position, Glu or Met(O2) at P3, and Tic or His(Bzl) at P2. Despite the large diversity of unnatural amino acids used in HyCoSuL none of these substrates was more selective than the substrate containing the natural amino acids WQPD, and the Cha-Glu-Tic-Asp unnatural substrate was only slightly more active than the reference natural amino acid WEHD sequence.
Table 2.
Unique preferred substrates with unnatural amino acids
Based on screening of an unnatural amino acid HyCoSuL library, optimal single substrates were synthesized for caspase-1 and caspase-11. Each substrate was assayed with either caspase-1 or caspase-11. Most synthesized unnatural amino acid substrates were preferred by caspase-1, with only the best caspase-11 substrates demonstrating about equal catalytic rates with caspase-1. The selectivity factor represents the catalytic parameter kcat/Km for caspase-1 divided by that for caspase-11. Each experiment was repeated at least three times, and the kcat/Km values are presented as an average. Standard deviations were below 15%.
| Substrate sequence: Ac-P4-P3-P2-P1-ACC |
kcat/Km |
Selectivity factor | |
|---|---|---|---|
| Caspase-1 | Caspase-11 | ||
| m−1s−1 | |||
| Caspase-1 preferred | |||
| Cha-Glu-Tic-Asp | 544,000 | 609 | 893 |
| Phe(2Cl)-Glu-Tic-Asp | 525,000 | 1,010 | 519 |
| Trp(Me)-Glu-His(Bzl)-Asp | 524,000 | 1,700 | 106 |
| Trp-Met(O2)-His-Asp | 512,000 | 7,140 | 72 |
| Caspase-11 preferred | |||
| Tle-Bpa-His(Bzl)-Asp | 89,000 | 112,000 | 0.80 |
| Tle-Bip-His-Asp | 80,200 | 109,000 | 0.74 |
| Pro-Bip-His(Bzl)-Asp | 25,900 | 69,000 | 0.38 |
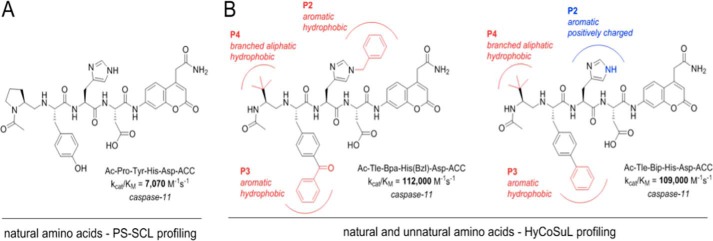
In contrast to caspase-1, more success in obtaining better substrates for caspase-11 was achieved. We found that the use of branched, aliphatic amino acids (Tle) at P4, and large, aromatic amino acids (Bip, Bpa) at P3 generated substantially improved substrates, as the Ac-Tle-Bpa-His(Bzl)-Asp-ACC was hydrolyzed 15.6- and 27-fold faster than the best substrate containing natural amino acids (Ac-PMHD-ACC) and reference (Ac-WEHD-ACC) substrates, respectively (Fig. 3). The extended nature of the cyclic side chains that comprise these unnatural amino acids may indicate that they could reach surface hydrophobic pockets that natural amino acids cannot explore, much as seen in the case of binding of peptides based on unnatural amino acids to the serine protease neutrophil elastase (38). Unfortunately, even the best caspase-11 substrates were also efficiently hydrolyzed by caspase-1 so we could not obtain a substrate reasonably selective for caspase-11. Nevertheless, we demonstrated that the application of unnatural amino acids is a good strategy to enhance catalysis for caspase-11.
Figure 3.
Structures of the best natural and unnatural fluorogenic substrates for caspase-11. A, one of the most active natural substrate contains the Pro-Tyr-His-Asp sequence. B, two of the most active caspase-11 substrates containing unnatural amino acids. Extended cyclic and hydrophobic side chains enhance catalysis compared with substrates with natural amino acids.
In vitro cleavage of pro-IL-1β, pro-IL-18, and GSDMD by mouse inflammatory caspases
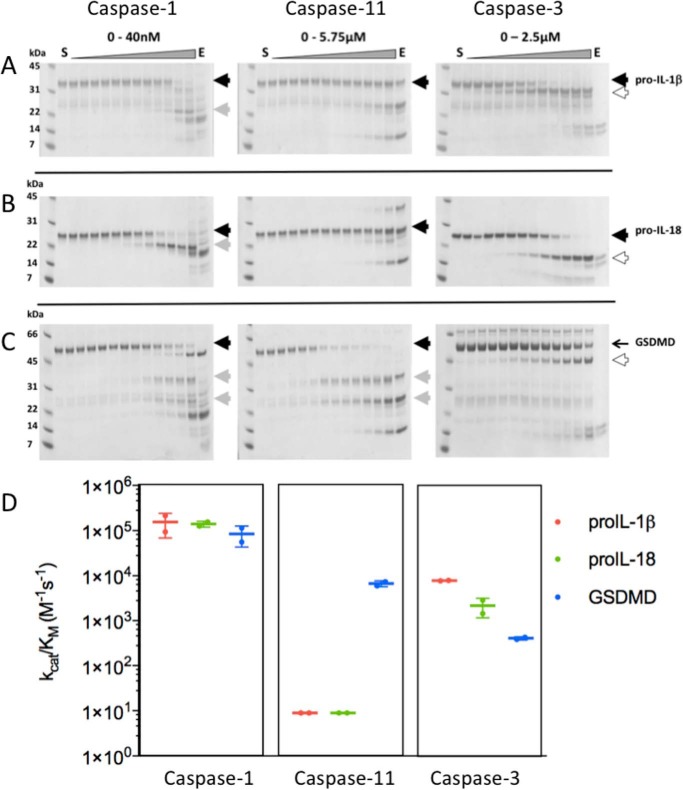
To compare the catalytic efficiency of mouse inflammatory caspases for endogenous substrates, we incubated the active site-titrated enzymes with 4 μm recombinant mouse pro-IL-1β, pro-IL-18, and GSDMD (Fig. 4). To determine the enzyme concentration needed to cleave half of the substrate, and thus derive kcat/Km values according to Equation 2, the incubated samples were run on SDS-PAGE gels, and the stained gels were quantified by densitometry scanning. Because the apoptotic executioner caspase-3 has also been implicated in cleavage of the proteins we included this enzyme in our analyses.
Figure 4.
Cleavage rates of substrate proteins by caspases. To test catalytic efficiency of caspases on A, pro-IL-1β, B, pro-IL-18, and C, GSDMD, the indicated enzymes were active site titrated using Z-VAD-fmk, incubated with 4 μm substrates for 30 min at 37 °C, and run in SDS-PAGE. Gel densitometry of the remaining uncleaved substrate yielded IC50 values from which catalytic rates kcat/Km were determined according to Equation 1 (“Materials and methods”). The precursor proteins are shown by a solid black arrowheads, derivatives of each substrate are indicated by the gray or white arrowheads. Importantly, although the apoptotic caspase-3 cleaved the proteins, each site was distinct from those generated by inflammatory caspase-1 and -11 (white arrowheads). The gels above are representative of two biological replicates employing different caspase preparations. D, comparative kcat/Km values are described by the colored symbols, as indicated in the key. Caspase-3 cleaved all three proteins, but at sites distinct from those generated by caspase-1 and -11, as discussed in the text. Data are averages from two biological replicates, and the error bars indicate standard deviations.
The respective kcat/Km are presented in Fig. 4, from which it is apparent that caspase-1 efficiently processes pro-IL-1β, pro-IL-18, and GSDMD to give the expected derivatives. Caspase-11 cleaves GSDMD with somewhat comparable efficiency, but is unable to cleave the pro-interleukins, and the kcat/Km of 9.0 m−1 s−1 is a maximal estimate, and orders of magnitude slower than caspase-1 cleavage of the pro-interleukins. Caspase-3 efficiently cleaves pro-interleukins, but displays a low kcat/Km on GSDMD (Fig. 4). Importantly, the caspase-3 cleavage sites are different from the inflammatory caspase cleavage sites, and the consequence of this is discussed later.
Discussion
Genetic evidence indicates that caspase-1 processes pro-IL-1β and pro-IL-18 but caspase-11 does not (1, 39). Both of these caspases are nevertheless able to induce pyroptosis by cleaving GSDMD (24, 25). Thus there must be biochemical characteristics that distinguish these two otherwise closely related caspases. Working with recombinantly expressed enzymes, we found these enzymes to be soluble and active after purification. The lack of CARD domain does not inhibit their activity as seen in the library profiles and in vitro cleavages of endogenous substrates. A question of whether CARDs influence specificity cannot yet be determined. However, substrate specificity mapping of the CARD-containing caspase-9 reveals no role in specificity or catalysis for its CARD, and from this perspective we predict that the specificity and activity of caspase-1 and -11 would not be influenced by their CARDs.
Since the discovery of human caspase-1 more than two decades ago, various studies have focused on understanding its kinetic parameters by designing multiple length substrates using different approaches (18, 28, 40–42). The optimal length of peptide substrates is four residues (41). Using this peptide length, initial substrate specificity studies conducted on caspase-11, and its closest human orthologs caspase-4 and -5, reported similar specificity to caspase-1 (28, 43), which we confirm in Tables S3 and S4.
To better understand the distinctions between caspase-1 and -11 we employed a screening strategy utilizing HyCoSuL, followed by synthesis of individual substrates containing unnatural amino acids. Analysis of these substrates allowed us to draw several conclusions. First, the use of unnatural amino acids had no impact on the activity or the selectivity of caspase-1 substrates, as the sequence WEHD was still among the best recognized substrates, and WQPD was the most selective substrate over caspase-11 with a selectivity factor over 5,140. Thus, the WQPD substrate can serve as an excellent distinguishing marker for caspase-1 detection in cells undergoing pyroptosis. On the other hand, unnatural amino acids were very useful in caspase-11 profiling. We found that two substrate containing unnatural amino acids (Tle-Bip-His-Asp and Tle-Bpa-His(Bzl)-Asp) displayed a 15-fold higher kcat/Km toward caspase-11 than the best substrate containing natural amino acids (PMHD). The high increase in enzyme activity was due to Tle at P4 and Bip/Bpa at P3. We reasoned that these amino acids produce new binding interactions with the enzyme that could not be obtained using natural analogs. For instance, Ac-Pro-Bip-His(Bzl)-Asp-ACC displayed almost 3-fold higher kcat/Km for caspase-11 over caspase-1, whereas all alleged caspase-11 selective substrates containing only natural amino acids were hydrolyzed substantially faster by caspase-1. However, the very best selective substrates for caspase-11, suboptimal for caspase-1, are still cleaved by caspase-1 with equal or greater efficiency. Even the very best peptide substrates for caspase-11 have kcat/Km values of less than 104 m−1 s−1, which is generally very poor for caspase peptidyl substrates (32).
At this point we reasoned that the active form of caspase-11 produced in E. coli might lack activity due to folding or conformational issues, and so we tested both caspases on natural substrates obtained from recombinant expression. We were struck by the very poor cleavage of pro-IL-1β and pro-IL-18 by caspase-11. The kcat/Km values are so poor (>10,000-fold lower than caspase-1) that we estimate, through applying Equation 3 and an estimated enzyme concentration of 10 nm, that it would take over 3 months for caspase-11 to cleave 50% of pro-IL-1β and pro-IL-18. In contrast, caspase-1 would take about 5–10 min to accomplish cleavage under the same conditions. However, the cleavage rate of GSDMD by caspase-1 and -11 is more comparable with only a 13-fold difference in kcat/Km values (half-times for cleavage of 10 min to 2 h).
Taking these results into account we conclude that caspase-11 has a very restricted substrate specificity, preferring GSDMD over all other substrates examined. The P4–P1 residues in GSDMD are LLSD and those in pro-IL-18 are LESD, and so the only difference in this region that occupies the active site cleft is at P3, where GSDMD has Leu, and pro-IL-18 has Glu (Fig. 2B). Examination of Fig. 2 reveals that Glu is preferred to Leu at P3, so from this perspective pro-IL-18 should be a better substrate than GSDMD for caspase-11. Clearly this is not the case, so another explanation must be considered to explain the rapid and preferred cleavage of GSDMD by caspase-11.
Mechanistically, this is likely to represent a case where exquisite substrate specificity is created by an exosite on the scaffold of an otherwise poor enzyme. Originally identified in thrombin as a mechanism to enhance cleavage of its key substrate fibrinogen (44), exosites are emerging as a critical substrate recognition function in several distinct protease families including matrix metalloproteases, small ubiquitin-like modifier (SUMO), and ubiquitin-deconjugating enzymes, and cathepsins (45–48). Exosites are generally distant from the enzyme active site and tend to direct proteases toward specific substrates that are normally not well-cleaved. Caspase-11 may represent such a case, cleaving peptide substrates and pro-interleukins poorly, but cleaving GSDMD well, and has thus developed a highly selective substrate specificity via a yet to be identified exosite. Notably, both pro-IL-1β and pro-IL-18 possess negatively charged residues in the P7–P10 region upstream of the cleavage site (Fig. 2B). This may preclude efficient binding by caspase-11 and constitute part of an exosite.
Despite the finding that caspase-11 is an unlikely candidate for activation of pro-interleukins there is nevertheless evidence based on in vivo studies and ablation of inflammasome components in mice that non-canonical stimuli lead to pro-interleukin processing (1). One potential pathway is via direct activation of pro-caspase-1 by active caspase-11. We have not been able to produce pro-caspase-1 by recombinant expression, so have been unable to test this straightforwardly. However, mice and human cells ablated in Nlrp3 and ASC, key inflammasome components, are unable to process the pro-interleukins, leading to the conclusion that canonical activation of caspase-1 is required and activation should not be directly via caspase-11 (1, 49–51) (Fig. 5). The identity of the pathway(s) leading to the indirect activation of caspase-1 by caspase-11 remains elusive.
Figure 5.
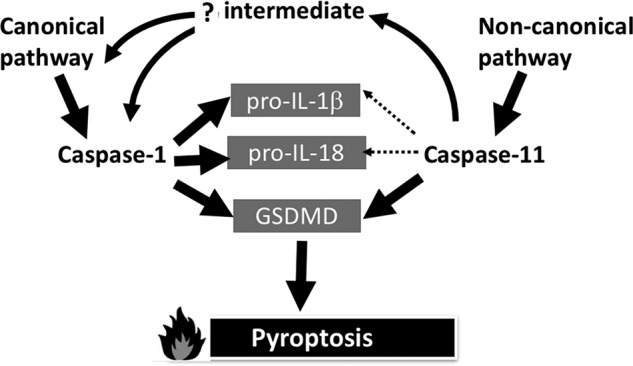
Pathway model of inflammatory caspase functions. Based on the kcat/Km values presented in this paper, caspase-11 is unlikely to be involved in directly processing pro-interleukins, as emphasized by the relative weights of the arrowed lines. However, monocytic cells treated with non-canonical stimuli that activate caspase-11 still retain the ability to process the pro-interleukins. As discussed in the text, there cannot be a direct activation of caspase-1 by caspase-11, so there must be an intermediate factor(s) that respond to active caspase-11 by ultimately activating caspase-1, most likely via the canonical pathway. In contrast, both caspases are able to directly activate GSDMD in a timely manner to generate pyroptosis.
Although we have focused on the pro-inflammatory caspase-1 and -11, it is noteworthy that the apoptotic caspase-3 also cleaves pro-IL-1β, pro-IL-18, and GSDMD, but at different sites. Cleavage of human GSDMD by caspase-3 inactivates lytic potential, preventing caspase-1 and -11 from generating a cell-permeabilizing product (52). Cleavage of human pro-IL-18 by caspase-3 generates biologically inactive products (53). Together these observations give credence to the hypothesis that apoptotic caspases blunt inflammation by inactivating pro-inflammatory cytokines (54) and GSDMD.
Future studies will seek to delineate the pathway that leads from active caspase-11 to active caspase-1. The pathway may be cell autonomous or it may involve sensing of cellular contents released from pyroptotic cells by adjacent cells, leading to canonical caspase-1 activation. Moreover, evidence presented here implies that an exosite exists on caspase-11 to secure rapid cleavage of GSDMD. Given that we have not been able to capture a substrate selective for caspase-11 over caspase-1, despite massive peptide-based screening, it is unlikely that a simple active site-directed strategy will succeed in developing compounds to interfere with the non-canonical pathway over the canonical one. Accordingly, directing compounds against the putative exosite may lead to highly selective compounds targeting caspase-11, but this will first depend on the characterization of the exosite.
Materials and methods
Protein construct design, expression, and purification
Constructs encoding full-length mouse caspase-1 and caspase-11 in pcDNA3(+) Zeocin were the kind gifts of Irma Stowe and Nobuhiko Kayagaki (Genentech, San Francisco, CA). Sequences encoding mouse full-length GSDMD, pro-IL-1β, and pro-IL-18 were purchased from Integrated DNA Technologies (San Diego, CA) and cloned into the pET29b(+) (Novagen) containing a C-terminal His tag. CARD deleted (ΔCARD) modified versions of mouse caspase-1 (minus amino acids 2–129) and caspase-11 (minus amino acids 2–99) were also cloned into the pET29b(+) containing a C-terminal His tag. ΔCARD-modified versions of human caspase-1 (minus amino acids 2–118) and caspase-4 (minus amino acids 2–93) were cloned into pET15b(+) containing an N-terminal His tag. ΔCARD Caspase-5 (minus amino acids 2–132) was cloned into the pET21b(+) containing a C-terminal His tag. All other caspases were expressed as previously described (55). All constructs were transformed into BL21 (DE3)-competent E. coli. Expression was induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside for 4 h at 25 °C shaking at 250 rpm. Cells were collected and pelleted at 4 °C for 10 min at 3,900 × g. The caspase cell pellets were resuspended in 50 mm Tris-Cl, 100 mm NaCl, pH 8.0, and lysed via sonication. The GSDMD, pro-IL-1β, and pro-IL-18 cell pellets were resuspended in 50 mm HEPES, 100 mm NaCl, pH 8.0.
Cell lysates were centrifuged at 4 °C for 30 min at 28,000 × g, supernatants were filtered through a 0.22-μm filter (Millipore) and the soluble fractions were applied to a 1-ml Ni-chelating-Sepharose resin (GE Healthcare Life Sciences) in a chromatography column. The enzyme-bound resin was washed in 50 mm Tris-Cl, 500 mm NaCl, pH 8.0, and then enzymes were eluted in 25 mm Tris-Cl, 100 mm NaCl, pH 8.0, with stepwise increments of imidazole from 12.5 to 100 mm. Human caspase-3 was prepared as previously described (55). Purified protein concentration was determined by absorbance at 280 nm. Active enzyme concentration was calculated by carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone (Z-VAD-fmk, Cayman Chemical, 14467-5) titration, see below. Proteins were analyzed by 4–12% BisTris SDS-PAGE gels (Novex Life Technologies) and stained with Instant Blue (Expedeon), or transferred onto a nitrocellulose membrane and detected with antibodies.
Urea protein denaturation and refolding
Full-length caspase-1 and -11 were expressed and purified as described above, except that cell pellets were resuspended in 8 m urea, 50 mm Tris-Cl, 100 mm NaCl, pH 8.0. The enzyme-bound resin was washed in 8 m urea, 50 mm Tris-Cl, 500 mm NaCl, pH 8.0, and then enzymes were eluted in 8 m urea, 25 mm Tris-Cl, 100 mm NaCl, pH 8.0, with stepwise increments of imidazole from 12.5 to 200 mm. Refolding procedures were followed as described previously (36).
Antibodies
Caspase-11 antibody was from Novusbio (Littleton, CO) (NB120–10454). His-tag polyclonal antibody was from Cell Signaling Technology (Danvers, MA) (number 2365).
MALDI time-of-flight (TOF)-MS
Purified recombinant ΔCARD caspase-11 was analyzed by MALDI TOF/TOF mass spectrometry on a Bruker Daltonics Autoflex II TOF/TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA), in the Sanford Burnham Prebys Proteomics Facility, to accurately determine the molecular weights of the bands observed in SDS-PAGE. Enzyme was desalted and concentrated using C18 ZipTip (EMD Millipore). A ZipTip was equilibrated 5 times with 10 μl of 75% acetonitrile (ACN) and 0.1% trifluoroacetic acid (TFA) in water, and washed 5 times in 10 μl of 0.1% TFA, binding of the protein was accomplished by pipetting up and down 15 times, and the ZipTip was washed once more as above. Proteins were eluted in 4 μl of 75% ACN and 0.1% TFA. Sinapinic acid (20 mg/ml) for MALDI matrix was dissolved in 1 ml of 50% ACN and 0.1% TFA by vortexing for 1 min. Matrix was spun at 10,000 × g for 1 min to pellet undissolved matrix. One to one ratios of matrix supernatant and protein were crystallized on MALDI target plates and allowed to dry for 5 min at room temperature. Mass spectra were processed with Flex Analysis 2.4.
Enzymatic assays
Enzymatic activities of purified recombinant inflammatory caspase-1 and -11 were measured in an opaque 96-well plate (Costar, Corning). The final assay volume was 100 μl including enzyme and substrate. Caspase buffer was 10 mm PIPES, 10% sucrose, 100 mm NaCl, 0.1% CHAPS, 1 mm EDTA, and 10 mm DTT, pH 7.2. To maintain high enzyme activity, the caspase buffer used for individual substrate kinetics was supplemented with 0.75 m sodium citrate. Caspases were incubated in buffer for 10 min at 37 °C before adding substrate, and activity was measured by fluorescence detection using a CLARIOstar plate reader (BMG Labtech) operating in the kinetic mode (ACC fluorophore; excitation 355 nm, emission 460 nm; and AFC fluorophore; excitation 400 nm, emission 505 nm), and analyzed with MARS Data Analysis Software (BMG Labtech) and Prism (GraphPad Software).
Z-VAD-fmk active site titration
To determine the active concentration of inflammatory enzymes so that substrate kcat/Km values could be calculated, a serial dilution series of Z-VAD-fmk was incubated with purified enzyme for 30 min at 37 °C. The fluorogenic substrate Ac-LEHD-AFC was added to a final concentration of 100 μm in 100 μl of assay buffer and enzymatic activity was measured as above. Velocities were plotted against the inhibitor concentration allowing the calculation of the active enzyme concentration (55).
Screening of caspase-1 and -11 substrate specificities
A HyCoSuL substrate scanning library was utilized to define the substrate specificity cleavage preferences of caspase-1 and -11, essentially as reported before for apoptotic caspases (32). In brief, three fluorogenic substrate sublibraries (P4, P3, and P2) were each screened at 100 μm with mouse recombinant caspase-1 and -11 in a 100-μl final volume. Both enzymes were used immediately after purification because caspase-11 tends to become unstable upon storage. The progress of substrate hydrolysis was monitored for 45 min, but for each substrate only the linear portion of the hydrolysis curve (15–30 min) was analyzed to calculate the reaction velocity (relative fluorescent units/s). Screening of each sublibrary was performed three times, and the averages were used to calculate caspase-1 and -11 specificity matrixes. For each sublibrary the amino acid with the highest cleavage rate (relative fluorescent units/s) was set to 100%, and other amino acids were adjusted accordingly.
Synthesis and kinetic analysis of substrates containing natural and unnatural amino acids
Individual ACC-labeled substrates were obtained via solid-phase peptide synthesis in N,N-dimethylformamide and purified as described previously (32). Briefly, ACC-OH was attached to the Rink Amide resin, Fmoc-Asp(tBu)-OH was coupled, and elongated by coupling P2-P4 Fmoc-protected amino acids and acetylation of the N terminus. Side chain protecting groups were removed and the peptide substrate was cleaved from the resin using a TFA procedure. Crude products were precipitated in diethyl ether and purified by HPLC. Purity of each substrate was confirmed via analytical HPLC and the molecular weight was determined by mass spectrometry on a high resolution mass spectrometer WATERS LCT premier XE with electrospray ionization (ESI) and TOF module.
For each fluorogenic substrate we determined the kcat/Km parameters toward caspase-1 and -11. To do this, serial dilutions of substrate were placed in the eight wells of 96-well plates followed by addition of caspase. The reaction was monitored for 30 min and the hydrolysis velocity from each well (V) was plotted against substrate concentration (S) and the V/S slope was calculated. The kcat/Km parameter was then calculated according to the Equation 1.
| (1) |
Each experiment was performed at least three times and the average values with S.D. were calculated.
In vitro cleavage of recombinant protein substrates
Active site-titrated recombinant ΔCARD caspase-1 or -11 was subjected to 2-fold series dilutions and incubated separately for 30 min at 37 °C with 4 μm pro-IL-1β, pro-IL-18, or GSDMD caspase buffer in a final assay volume of 60 μl. Reactions were terminated by heating at 86 °C with 30 μl of 3× SDS loading buffer to give a final concentration of 1× loading buffer. Samples were analyzed on 4–12% BisTris SDS-PAGE gels by Instant Blue staining. The gels were then scanned and imported to Image Studio (LI-COR Biosciences) for protein band intensity quantification. Band intensity values were plotted against enzyme concentration and IC50 values were determined via GraphPad Prism. Those values were then used to measure catalytic efficiencies of mouse capase-1 and -11 for natural substrates according to the relationship described previously (56).
| (2) |
Rearranging this equation and applying a pseudo-first order approximation yields the half-life for cleavage of a substrate.
| (3) |
Here, kcat/Km is the second order rate constant for substrate hydrolysis, E½ is the concentration of protease for which half the substrate is consumed, E is the concentration of enzyme, and t is the incubation time (57).
Author contributions
M. L. G. R. and M. P. data curation; M. L. G. R., M. P., and G. S. S. formal analysis; M. L. G. R. and M. P. investigation; M. L. G. R., M. P., S. J. S., K. G., and M. D. methodology; M. L. G. R., M. P., and G. S. S. writing-original draft; M. L. G. R., M. P., S. J. S., K. G., M. D., and G. S. S. writing-review and editing; M. P., M. D., and G. S. S. conceptualization; M. P. and S. J. S. validation; M. D. and G. S. S. funding acquisition; G. S. S. resources; G. S. S. supervision; G. S. S. project administration.
Supplementary Material
Acknowledgment
We thank Irma Stowe and Nobuhiko Kayagaki for providing mouse caspase cDNAs.
This work was supported by National Institutes of Health Grants R01 GM99040 and P30 CA30199 (to G. S. S.), National Science Center in Poland Grant OPUS, UMO-2011/03/B/ST5/01048 (to M. D.), and the European Union's Horizon 2020 research and innovation program under Marie Skłodowska-Curie Grant 661187 (to M. P.). This work was supported in part by a fellowship from Genentech. G. S. S. consults for Genentech. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Tables S1–S6 and Figs. S1–S3.
- GSDMD
- gasdermin D
- HyCoSuL
- hybrid combinatorial substrate library
- CARD
- caspase activation and recruitment domain
- Z
- carbobenzoxy
- fmk
- fluoromethylketone
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ACN
- acetonitrile
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- Bzl
- benzyl.
References
- 1. Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W. P., Roose-Girma M., and Dixit V. M. (2011) Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- 2. Lamkanfi M., and Dixit V. M. (2014) Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 10.1016/j.cell.2014.04.007 [DOI] [PubMed] [Google Scholar]
- 3. Viganò E., Diamond C. E., Spreafico R., Balachander A., Sobota R. M., and Mortellaro A. (2015) Human caspase-4 and caspase-5 regulate the one-step non-canonical inflammasome activation in monocytes. Nat. Commun. 6, 8761 10.1038/ncomms9761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang J., Zhao Y., and Shao F. (2015) Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol. 32, 78–83 10.1016/j.coi.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 5. Martinon F., and Tschopp J. (2004) Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell 117, 561–574 10.1016/j.cell.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 6. Zamboni D. S., and Lima-Junior D. S. (2015) Inflammasomes in host response to protozoan parasites. Immunol. Rev. 265, 156–171 10.1111/imr.12291 [DOI] [PubMed] [Google Scholar]
- 7. Hagar J. A., Powell D. A., Aachoui Y., Ernst R. K., and Miao E. A. (2013) Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 10.1126/science.1240988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., and Shao F. (2014) Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- 9. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., and Salvesen G. S. (2001) Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. U.S.A. 98, 14250–14255 10.1073/pnas.231465798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boatright K. M., and Salvesen G. S. (2003) Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15, 725–731 10.1016/j.ceb.2003.10.009 [DOI] [PubMed] [Google Scholar]
- 11. Aglietti R. A., Estevez A., Gupta A., Ramirez M. G., Liu P. S., Kayagaki N., Ciferri C., Dixit V. M., and Dueber E. C. (2016) GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. U.S.A. 113, 7858–7863 10.1073/pnas.1607769113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D. C., and Shao F. (2016) Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 10.1038/nature18590 [DOI] [PubMed] [Google Scholar]
- 13. Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V. G., Wu H., and Lieberman J. (2016) Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 10.1038/nature18629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sborgi L., Rühl S., Mulvihill E., Pipercevic J., Heilig R., Stahlberg H., Farady C. J., Müller D. J., Broz P., and Hiller S. (2016) GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 35, 1766–1778 10.15252/embj.201694696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salvesen G. S., Hempel A., and Coll N. S. (2016) Protease signaling in animal and plant-regulated cell death. FEBS J. 283, 2577–2598 10.1111/febs.13616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Black R. A., Kronheim S. R., Merriam J. E., March C. J., and Hopp T. P. (1989) A pre-aspartate-specific protease from human leukocytes that cleaves pro-interleukin-1b. J. Biol. Chem. 264, 5323–5326 [PubMed] [Google Scholar]
- 17. Kostura M. J., Tocci M. J., Limjuco G., Chin J., Cameron P., Hillman A. G., Chartrain N. A., and Schmidt J. A. (1989) Identification of a monocyte specific pre-interleukin 1b convertase activity. Proc. Natl. Acad. Sci. U.S.A. 86, 5227–5231 10.1073/pnas.86.14.5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Howard A. D., Kostura M. J., Thornberry N., Ding G. J., Limjuco G., Weidner J., Salley J. P., Hogquist K. A., Chaplin D. D., Mumford R. A., Schmidt J. A., and Tocci M. J. (1991) IL-1 converting enzyme requires aspartic acid residues for processing of the IL-1b precursor at two distinct sites and does not cleave 31-kDa IL-1a. J. Immunol. 147, 2964–2969 [PubMed] [Google Scholar]
- 19. Brough D., and Rothwell N. J. (2007) Caspase-1-dependent processing of pro-interleukin-1β is cytosolic and precedes cell death. J. Cell Sci. 120, 772–781 10.1242/jcs.03377 [DOI] [PubMed] [Google Scholar]
- 20. Ghayur T., Banerjee S., Hugunin M., Butler D., Herzog L., Carter A., Quintal L., Sekut L., Talanian R., Paskind M., Wong W., Kamen R., Tracey D., and Allen H. (1997) Caspase-1 processes IFN-γ-inducing factor and regulates LPS-induced IFN-γ production. Nature 386, 619–623 10.1038/386619a0 [DOI] [PubMed] [Google Scholar]
- 21. Gu Y., Kuida K., Tsutsui H., Ku G., Hsiao K., Fleming M. A., Hayashi N., Higashino K., Okamura H., Nakanishi K., Kurimoto M., Tanimoto T., Flavell R. A., Sato V., Harding M. W., Livingston D. J., and Su M. S. (1997) Activation of interferon-γ inducing factor mediated by interleukin-1β converting enzyme. Science 275, 206–209 10.1126/science.275.5297.206 [DOI] [PubMed] [Google Scholar]
- 22. Kuida K., Lippke J. A., Ku G., Harding M. W., Livingston D. J., Su M. S., and Flavell R. A. (1995) Altered cytokine export and apoptosis in mice deficient in interleukin-1-beta converting enzyme. Science 267, 2000–2003 10.1126/science.7535475 [DOI] [PubMed] [Google Scholar]
- 23. Auron P. E., Warner S. J., Webb A. C., Cannon J. G., Bernheim H. A., McAdam K. J., Rosenwasser L. J., LoPreste G., Mucci S. F., and Dinarello C. A. (1987) Studies on the molecular nature of human interleukin 1. J. Immunol. 138, 1447–1456 [PubMed] [Google Scholar]
- 24. Kayagaki N., Stowe I. B., Lee B. L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., et al. (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- 25. Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F. (2015) Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- 26. Poreba M., and Drag M. (2010) Current strategies for probing substrate specificity of proteases. Curr. Med. Chem. 17, 3968–3995 [DOI] [PubMed] [Google Scholar]
- 27. Rano T. A., Timkey T., Peterson E. P., Rotonda J., Nicholson D. W., Becker J. W., Chapman K. T., and Thornberry N. A. (1997) A combinatorial approach for determining protease specificities: application to interleukin-1b converting enzyme (ICE). Chem. Biol. 4, 149–155 10.1016/S1074-5521(97)90258-1 [DOI] [PubMed] [Google Scholar]
- 28. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., and Nicholson D. W. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 10.1074/jbc.272.29.17907 [DOI] [PubMed] [Google Scholar]
- 29. Poreba M., Szalek A., Kasperkiewicz P., Rut W., Salvesen G. S., and Drag M. (2015) Small molecule active site directed tools for studying human caspases. Chem. Rev. 115, 12546–12629 10.1021/acs.chemrev.5b00434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harris J. L., Backes B. J., Leonetti F., Mahrus S., Ellman J. A., and Craik C. S. (2000) Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc. Natl. Acad. Sci. U.S.A. 97, 7754–7759 10.1073/pnas.140132697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poreba M., Salvesen G. S., and Drag M. (2017) Synthesis of a HyCoSuL peptide substrate library to dissect protease substrate specificity. Nat. Protoc. 12, 2189–2214 10.1038/nprot.2017.091 [DOI] [PubMed] [Google Scholar]
- 32. Poreba M., Kasperkiewicz P., Snipas S. J., Fasci D., Salvesen G. S., and Drag M. (2014) Unnatural amino acids increase sensitivity and provide for the design of highly selective caspase substrates. Cell Death Differ. 21, 1482–1492 10.1038/cdd.2014.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen W., Yun S., Tam B., Dalal K., and Pio F. F. (2005) Target selection of soluble protein complexes for structural proteomics studies. Proteome Sci. 3, 3 10.1186/1477-5956-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Burgess R. R. (2009) Refolding solubilized inclusion body proteins. Methods Enzymol. 463, 259–282 10.1016/S0076-6879(09)63017-2 [DOI] [PubMed] [Google Scholar]
- 35. Garcia-Calvo M., Peterson E. P., Rasper D. M., Vaillancourt J. P., Zamboni R., Nicholson D. W., and Thornberry N. A. (1999) Purification and catalytic properties of human caspase family members. Cell Death Diff. 6, 362–369 10.1038/sj.cdd.4400497 [DOI] [PubMed] [Google Scholar]
- 36. Ng N. M., Pike R. N., and Boyd S. E. (2009) Subsite cooperativity in protease specificity. Biol. Chem. 390, 401–407 [DOI] [PubMed] [Google Scholar]
- 37. Poreba M., Szalek A., Rut W., Kasperkiewicz P., Rutkowska-Wlodarczyk I., Snipas S. J., Itoh Y., Turk D., Turk B., Overall C. M., Kaczmarek L., Salvesen G. S., and Drag M. (2017) Highly sensitive and adaptable fluorescence-quenched pair discloses the substrate specificity profiles in diverse protease families. Sci. Rep. 7, 43135 10.1038/srep43135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lechtenberg B. C., Kasperkiewicz P., Robinson H., Drag M., and Riedl S. J. (2015) The elastase-PK101 structure: mechanism of an ultrasensitive activity-based probe revealed. ACS Chem. Biol. 10, 945–951 10.1021/cb500909n [DOI] [PubMed] [Google Scholar]
- 39. He Y., Hara H., and Núñez G. (2016) Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 41, 1012–1021 10.1016/j.tibs.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sleath P. R., Hendrickson R. C., Kronheim S. R., March C. J., and Black R. A. (1990) Substrate specificity of the protease that processes human interleukin-1b. J. Biol. Chem. 265, 14526–14528 [PubMed] [Google Scholar]
- 41. Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J., Elliston K. O., Ayala J. M., Casano F. J., Chin J., Ding G. J., et al. (1992) A novel heterodimeric cysteine protease is required for interleukin-1β processing in monocytes. Nature 356, 768–774 10.1038/356768a0 [DOI] [PubMed] [Google Scholar]
- 42. Thornberry N. A., and Molineaux S. M. (1995) Interleukin-1b converting enzyme: A novel cysteine protease required for IL-1b production and implicated in programmed cell death. Protein Sci. 4, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kang S. J., Wang S., Hara H., Peterson E. P., Namura S., Amin-Hanjani S., Huang Z., Srinivasan A., Tomaselli K. J., Thornberry N. A., Moskowitz M. A., and Yuan J. (2000) Dual role of caspase-11 in mediating activation of caspase-1 and caspase-3 under pathological conditions. J. Cell Biol. 149, 613–622 10.1083/jcb.149.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fenton J. W. 2nd, Olson T. A., Zabinski M. P., and Wilner G. D. (1988) Anion-binding exosite of human α-thrombin and fibrin(ogen) recognition. Biochemistry 27, 7106–7112 10.1021/bi00418a066 [DOI] [PubMed] [Google Scholar]
- 45. Overall C. M. (2002) Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 22, 51–86 10.1385/MB:22:1:051 [DOI] [PubMed] [Google Scholar]
- 46. Drag M., and Salvesen G. S. (2008) DeSUMOylating enzymes–SENPs. IUBMB Life 60, 734–742 10.1002/iub.113 [DOI] [PubMed] [Google Scholar]
- 47. Boucher D., Blais V., and Denault J. B. (2012) Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc. Natl. Acad. Sci. U.S.A. 109, 5669–5674 10.1073/pnas.1200934109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma V., Panwar P., O'Donoghue A. J., Cui H., Guido R. V., Craik C. S., and Brömme D. (2015) Structural requirements for the collagenase and elastase activity of cathepsin K and its selective inhibition by an exosite inhibitor. Biochem. J. 465, 163–173 10.1042/BJ20140809 [DOI] [PubMed] [Google Scholar]
- 49. Schmid-Burgk J. L., Gaidt M. M., Schmidt T., Ebert T. S., Bartok E., and Hornung V. (2015) Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 45, 2911–2917 10.1002/eji.201545523 [DOI] [PubMed] [Google Scholar]
- 50. Baker P. J., Boucher D., Bierschenk D., Tebartz C., Whitney P. G., D'Silva D. B., Tanzer M. C., Monteleone M., Robertson A. A., Cooper M. A., Alvarez-Diaz S., Herold M. J., Bedoui S., Schroder K., and Masters S. L. (2015) NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur. J. Immunol. 45, 2918–2926 10.1002/eji.201545655 [DOI] [PubMed] [Google Scholar]
- 51. Rühl S., and Broz P. (2015) Caspase-11 activates a canonical NLRP3 inflammasome by promoting K+ efflux. Eur. J. Immunol. 45, 2927–2936 10.1002/eji.201545772 [DOI] [PubMed] [Google Scholar]
- 52. Taabazuing C. Y., Okondo M. C., and Bachovchin D. A. (2017) Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 24, 507–514 10.1016/j.chembiol.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akita K., Ohtsuki T., Nukada Y., Tanimoto T., Namba M., Okura T., Takakura-Yamamoto R., Torigoe K., Gu Y., Su M. S., Fujii M., Satoh-Itoh M., Yamamoto K., Kohno K., Ikeda M., and Kurimoto M. (1997) Involvement of caspase-1 and caspase-3 in the production and processing of mature human interleukin 18 in monocytic THP.1 cells. J. Biol. Chem. 272, 26595–26603 10.1074/jbc.272.42.26595 [DOI] [PubMed] [Google Scholar]
- 54. Afonina I. S., Müller C., Martin S. J., and Beyaert R. (2015) Proteolytic processing of interleukin-1 family cytokines: variations on a common theme. Immunity 42, 991–1004 10.1016/j.immuni.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 55. Denault J. B., and Salvesen G. S. (2003) Expression, purification, and characterization of caspases. Curr. Protoc. Protein Sci. Chapter 21, Unit 21 13 10.1002/0471140864.ps2113s30 [DOI] [PubMed] [Google Scholar]
- 56. Stennicke H. R., and Salvesen G. S. (2000) Caspase assays. Methods Enzymol. 322, 91–100 10.1016/S0076-6879(00)22010-7 [DOI] [PubMed] [Google Scholar]
- 57. Pop C., Salvesen G. S., and Scott F. L. (2008) Caspase assays: identifying caspase activity and substrates in vitro and in vivo. Methods Enzymol. 446, 351–367 10.1016/S0076-6879(08)01621-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.