Abstract
Phosphatidylinositol 4,5-bisphosphate (PIP2) acts as a signaling lipid, mediating membrane trafficking and recruitment of proteins to membranes. A key example is the PIP2-dependent regulation of the adhesion of L-selectin to the cytoskeleton adaptors of the N-terminal subdomain of ezrin-radixin-moesin (FERM). The molecular details of the mediating behavior of multivalent anionic PIP2 lipids in this process, however, remain unclear. Here, we use coarse-grained molecular dynamics simulation to explore the mechanistic details of PIP2 in the transformation, translocation, and association of the FERM/L-selectin complex. We compare membranes of different compositions and find that anionic phospholipids are necessary for both FERM and the cytoplasmic domain of L-selectin to absorb on the membrane surface. The subsequent formation of the FERM/L-selectin complex is strongly favored by the presence of PIP2, which clusters around both proteins and triggers a conformational transition in the cytoplasmic domain of L-selectin. We are able to quantify the effect of PIP2 on the association free energy of the complex by means of a potential of mean force. We conclude that PIP2 behaves as an adhesive agent to enhance the stability of the FERM/L-selectin complex and identify key residues involved. The molecular information revealed in this study highlights the specific role of membrane lipids such as PIP2 in protein translocation and potential signaling.
Introduction
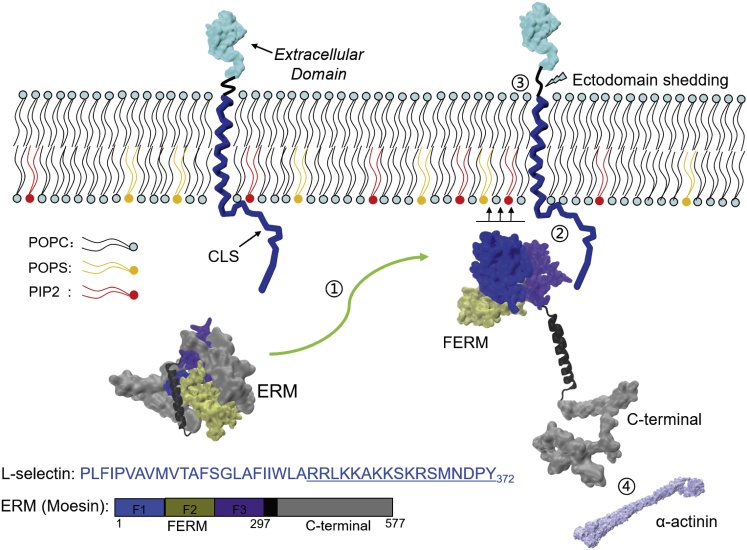
L-selectin is a primary adhesion molecule that mediates the tethering and rolling of leukocytes on endothelial cells (1, 2). L-selectin belongs to the type I transmembrane (TM) glycoproteins and is composed of an extracellular domain, a single TM-spanning region, and a short cytoplasmic domain (3). Although the cytoplasmic domain of L-selectin (CLS) only consists of 17 residues, the short cytoplasmic region is crucial for linking the cytoskeleton adaptor proteins of the ezrin-radixin-moesin (ERM) family (4). Evidence shows that CLS interacts with the N-terminal subdomain of ERM, named FERM, when it dissociates from the helix-connected C-terminal region (Fig. 1). Formation of the L-selectin/FERM complex then allows the binding of the released C-terminal region to the α-actinin cytoskeleton, an important step in the spatial and temporal organization of cells (5). Association of FERM and L-selectin also allows the cleavage of the extracellular L-selectin domain by the membrane-associated metalloproteinases, so-called “ectodomain shedding,” which is critical for leukocyte adhesion and signal transduction (6, 7). Because the association of L-selectin and FERM occurs in proximity to the intracellular membrane leaflet, the lipid composition of the cytoplasmic leaflet plays a crucial role in protein recruitment and orientation. In fact, recent studies have revealed that an increase of negative surface charge mediates the membrane attachment of CLS and the association between L-selectin and FERM (8, 9), thus underlining the importance of anionic lipids in regulating the protein translocation, interaction, and signaling (10, 11).
Figure 1.
A schematic diagram illustrating the conformational transformation and protein association of FERM and L-selectin at the proximity of the inner plasma membrane. (1) The FERM domain approaches the membrane surface when it is released from the helix-connected C-terminal domain of ERM. (2) FERM interacts with the CLS domain at the proximity of the anionic inner membrane, (3) which is favorable for the “ectodomain shedding” of the extracellular membrane-proximal domain. (4) Meanwhile, the C-terminal domain of ERM is able to connect with the α-actinin cytoskeleton protein. The sequence of L-selectin studied here consists of the TM domain and the CLS domain (underlined). The ERM (moesin) is composed of the FERM domain, the C-terminal domain (gray), and the helical linker (black). FERM is represented by the three subdomains F1 (blue, residue: 1–90), F2 (tan, 91–200), and F3 (violet, 201–297). To see this figure in color, go online.
An important anionic lipid of the plasma membrane is phosphatidylinositol 4,5-bisphosphate (PIP2). PIP2 is a type of signaling lipid that exclusively distributes in the inner bilayer leaflet. It can carry up to five negative charges, although a charge from −3 to −4 appears to be more realistic under physiological conditions (12). Previous studies show that PIP2 possesses a remarkable affinity to the juxtamembrane domain of TM proteins and regulates the oligomerization and conformation of the proteins (13, 14). Many biological functions have been described for PIP2, including the production of secondary messengers (15), membrane trafficking (16), mediating structure transformation, and protein association (17, 18). PIP2 is also found to be involved in the recruitment of cytoskeleton proteins (19, 20). Notably, PIP2 is enriched in membrane microdomains like the lamellipodia of migrating cells, where the cytoskeleton adaptors are active in membrane-cytoskeleton cross-linking (21, 22). Furthermore, there is evidence that the generation of the active form of FERM is controlled by PIP2 binding (23, 24, 25), which provides a direct coupling mechanism between the presence of PIP2 on the one hand and the tethering of the cytoskeleton to the cell membrane on the other hand. The molecular details underlying this mechanism remain, however, poorly understood.
A powerful tool to complement experimental studies on protein-lipid interplay is computational microscopy (26, 27, 28), a technique relying on molecular dynamics (MD) simulations. To achieve the time- and length scales that are necessary to observe protein-membrane binding events, the use of coarse-grain (CG) models, in which small groups of atoms are united into effective interaction centers, has proven very efficient (29, 30, 31, 32). In particular, the CG Martini force field (33, 34, 35, 36) has been successfully applied to unravel specific protein-lipid interactions for a wide range of systems (37, 38, 39), including the effect of phosphorylated phosphatidylinositols on protein binding and oligomerization (11, 40, 41, 42, 43, 44, 45, 46).
Here, we use CG simulations based on the Martini model to explore the membrane binding and protein association between the cytoskeleton adaptor FERM domain and the adhesion protein L-selectin dependent on lipid compositions, especially the presence of PIP2. Our results highlight a remarkable contribution of PIP2 to controlling this process. The interface of the FERM/L-selectin protein heterocomplex that forms during the simulation is consistent with previous results (47, 48). The regions of FERM involved with PIP2 carry the well-known lysine-enriched motif (49). The molecular information obtained in this study contributes to our ongoing understanding of the regulation mechanism of PIP2 in protein translocation and association.
Methods
Model setup
In our study, we used the Martini force field (33, 34, 35, 36) to model the interactions between all system components. The Martini force field acts on a CG level of resolution in which, in principle, four heavy atoms and their associated hydrogens are integrated into one bead. Thanks to the reduction in the particle number and the smoothing of the energy surface, an effective speedup of about three orders of magnitude with respect to fully atomistic models is obtained. An extensive discussion of the pros and cons of the Martini model can be found in (33). To test the effect of bilayer composition on the binding of L-selectin and FERM, three asymmetric plasma membrane models were created. In each case, the outer leaflet is composed of 100% palmitoyl-oleoyl-phosphatidylcholine (POPC). The inner leaflet consists of varying percentages of POPC, palmitoyl-oleoyl-phosphatidylserine (POPS), and PIP2, namely 100%POPC, 80%POPC/20%POPS, and 95%POPC/5%PIP2. The charge on each PIP2 lipid was set to −4, which is consistent with the possible charge neutralization by protons or ions (12, 50). Hence, the charge density of the membranes containing phosphatidylserine and PIP2 are the same. Based on these compositions, simulation boxes in two different sizes (a small system containing 225 lipids in either leaflet, 12 × 12 × 14 nm3, and a bigger system containing 361 lipids per leaflet, 15 × 15 × 14 nm3) were constructed using the script insane.py (51). Standard CG water was used to solvate the membranes. For the proteins, the software Pymol (Schrödinger, Cambridge, MA) (52) was used to construct the L-selectin structures (the wild-type (WT) and the K362IK363I mutant), which were then transformed into the CG model by the tool martinize.py (http://www.cgmartini.nl). The L-selectin studied here consisted of the TM domain and the juxtamembrane domain of 17 residues (cf. Fig. 1). The TM domain was defined as α-helix, and the CLS was modeled as random-coil in accordance to the experimental data (9). The CG model of the FERM domain of moesin was built based on the crystal structure (Protein Data Bank: 1SGH) (53). An elastic network was applied on FERM to maintain the conformational stability during the simulations (54). L-selectin was inserted into the bilayer parallel with membrane normal, exposing the CLS to the solvent. For simulations involving the interaction between L-selectin and FERM, the latter was initially placed in the proximity of the membrane with FERM 1.0 nm apart from the membrane surface. The distance between CLS and FERM was set to 5 or 10 nm (for the smaller and bigger membrane patches, respectively) along the diagonal direction of the simulation box to provide the least possible bias to their interaction. Counterions of sodium were added to neutralize all systems.
Simulation details
All simulations were conducted with the software package Gromacs-4.6.3 (55). The systems first underwent an energy minimization for 5000 steps using the steepest-descent method, followed by an NVT equilibration of 200 ns using the Berendsen thermostat (56) with a reference temperature of 323 K and a time constant of 1.0 ps. Subsequently, for each system, an NPT equilibration of 500 ns was conducted with a semiisotropic coupling pressure of 1.0 bar, a compressibility constant of 4.5 × 10−5 bar−1, and a time constant of 5.0 ps. During equilibration, a harmonic potential with a force constant of 1000 kJ ∙ mol−1 ∙ nm−2 was applied in all three dimensions to the protein backbone beads to constrain their positions. After equilibration, production simulations ran for 3 μs without any position restriction of the proteins. For each system, three independent runs (“replicas”) were conducted for analysis. In all cases, a shift function was used for the nonbonded electrostatic and Lennard-Jones interactions. The former decreased to zero from the distance of 0–1.2 nm, and the latter were shifted to zero from 0.9 to 1.2 nm. Periodic boundary conditions were used, and the time step was set as 20 fs, with the atom-pair list updating every 10 steps. These settings closely correspond to the “common” parameter set of the Martini force field (57). The association free energy of L-selectin/FERM (58) in anionic membranes both with and without PIP2 was estimated by the umbrella sampling method. During the pulling process, the backbone of FERM was restricted in position by a harmonic potential of 1000 kJ ∙ mol−1 ∙ nm−2 in all three dimensions. 25–27 separated windows within a pulling distance of 6.0 nm were created to generate well-overlapping configuration distributions. The association energy was computed with the Weighted Histogram Analysis Method (59). Statistical errors were estimated with bootstrap analysis (60). The complete histograms were considered as the independent data points to which random weights were assigned for bootstrap. More details can be seen in our previous study (61). All the simulation snapshots in this study were made with the VMD software package.
Results
Conformation of L-selectin is regulated by PIP2 lipids
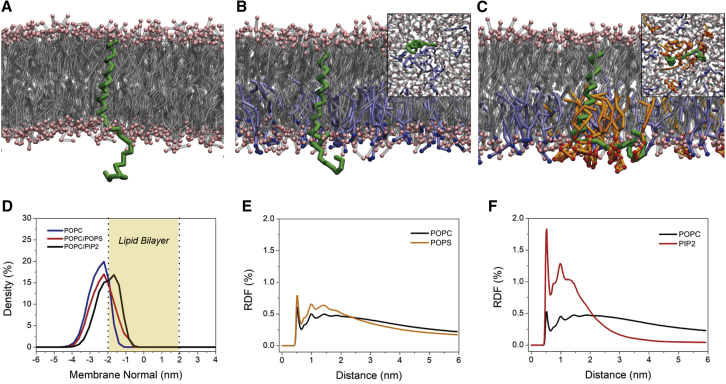
Conformational changes play a crucial role in determining the signaling pathway of TM proteins. Based on a previous study that shows the importance of an anionic membrane surface in regulating the conformation and activity of CLS (8, 9), we simulated three membrane models with different anionic lipid ratios in the presence of L-selectin, thereby using the small simulation box setup (see Methods for details). Starting from a random initial lateral distribution of lipids and a solvent-exposed conformation of CLS as found in experiments (9), the simulations ran for 3.0 μs. Representative snapshots of the simulation and the averaged density distribution profiles of CLS are shown in Fig. 2, A–D. In the membranes solely composed of the neutral POPC lipids, CLS always separates away from the bilayer surface. When 20%POPC lipids are substituted by the anionic POPS in the lower leaflet, CLS inclines to approach to the membrane surface, which is consistent with experimental results (9). Intriguingly, by replacing the POPS lipids by 5%PIP2, a further translocation of CLS, which becomes buried between the lipid headgroups, toward the membrane is observed.
Figure 2.
Conformational changes of L-selectin in the presence of anionic lipids. The lower leaflet is composed of (A) 100%POPC, (B) 80%POPC/20%POPS, and (C) 80%POPC/5%PIP2. L-selectin is depicted in green, with only the backbone beads shown. Lipid tails of POPC, POPS, and PIP2 are shown in silver, ice blue, and orange, respectively. The headgroups of POPC and PIP2 are shown by pink, blue, and red spheres, respectively. Visualizations in a bottom view are supplemented for the membranes containing POPS or PIP2. Solvent molecules are omitted for clarity. (D) The density distributions of CLS in different membrane environments along the membrane normal are given. (E and F) The radial distribution functions of the three lipid species relative to CLS are given. For POPC, only molecules in the lower leaflet are considered. To see this figure in color, go online.
The PIP2 lipids are found to cluster in the vicinity of L-selectin, which can be inferred from the snapshot (Fig. 2 C) and the two significant peaks in the radial distribution function of PIP2 around the L-selectin TM domain (Fig. 2 F). In contrast, the enrichment of anionic POPS lipids is, though present in a higher number than that of POPC, much weaker compared to PIP2 (Fig. 2 E). These results are consistent with previous studies revealing the propensity of PIP2 to cluster around juxtamembrane domains of TM proteins (45, 46) and underline the direct role of PIP2 in regulating both the binding and the conformational state of L-selectin.
FERM domain requires anionic lipids for stable membrane binding
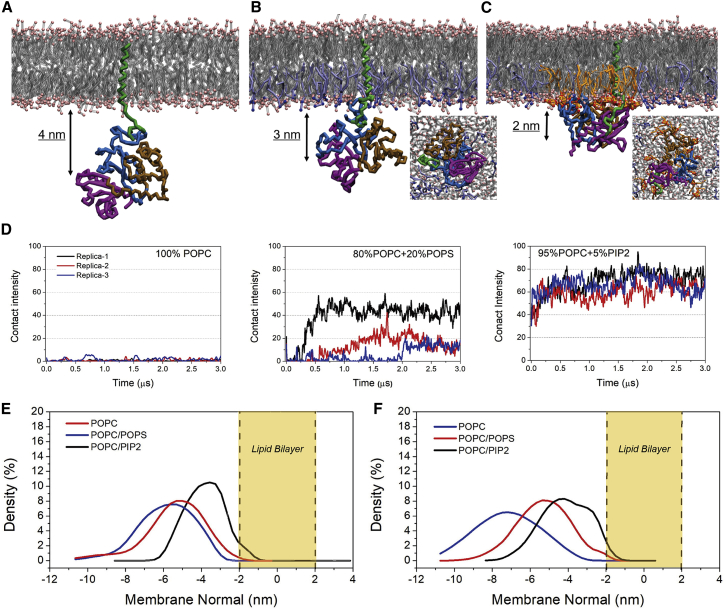
Subsequently, the membrane-affinity of FERM dependent on lipid composition was explored. FERM was initially put below the cytoplasmic membrane leaflet at a distance of 1.0 nm. Meanwhile, the distance between FERM and L-selectin was set to 5.0 nm separation, which is the maximal distance that can be achieved for the small system studied here. Snapshots of the final configurations of the systems, obtained after 3.0 μs, are shown in Fig. 3, A–C. For each of the three membrane compositions studied, we observed spontaneous binding of FERM to L-selectin. The same was observed in all three replicas performed for each case (data not shown). However, the position and orientation of the FERM domain appears to be strongly dependent on membrane composition. In the case of the pure POPC membrane, FERM stays away from the membrane surface (Fig. 3, A and E). The addition of 20%POPS enables FERM to absorb onto the membrane surface (Fig. 3, B and E), in line with experimental data that an anionic membrane surface is necessary for FERM to attach upon (8). The FERM-membrane interaction is most pronounced in the case of the bilayer containing 5%PIP2 (Fig. 3, C and E). The presence of PIP2 lipids allows FERM to embed itself deeply in the bilayer together with the CLS domain. Again, the formation of PIP2 clusters around the FERM-L-selectin complex is observed (Fig. 3 C), similarly to the case of sole L-selectin.
Figure 3.
Tethering of FERM to the membrane with different lipid environments. (A–C) Snapshots of the spatial position of FERM relative to the membrane in different compositions, consistent with the order of Fig. 2, A–C, are shown. Coloring of the FERM domain is consistent with that of Fig. 1. (D) The bilayer contact intensity of FERM with different membrane surfaces is given. The contact intensity was calculated by counting the number of beads of FERM that were at a distance closer than 0.6 nm from any phosphate beads of the lipids. (E) The density distributions of FERM along the membrane normal in the three membrane systems, including L-selectin and (F) excluding L-selectin, are shown. To see this figure in color, go online.
The time-averaged number of contacts between the FERM domain and the membrane lipids further illustrates the strengthened binding when PIP2 is present, whereas for the pure POPC membrane, protein-lipid contacts only happen stochastically (Fig. 3 D). To exclude the influence of L-selectin, the membrane-association of FERM was also investigated in the case of pure membranes. In the absence of the anionic lipids, the FERM domain has no membrane affinity and is found at a larger distance (Fig. 3 F). When anionic lipids are present, however, FERM remains in close proximity to the membrane, showing that anionic lipids by themselves provide the driving force for FERM in membrane adhesion. This result matches to previous experiments showing that the membrane binding of FERM gets strengthened by the presence of anionic lipids in the inner leaflet (20). Taken together, our results provide clear evidence for the importance of anionic lipids, especially PIP2, in stably tethering FERM to the membrane.
Formation of L-selectin/FERM heterocomplex is accelerated by PIP2 lipids
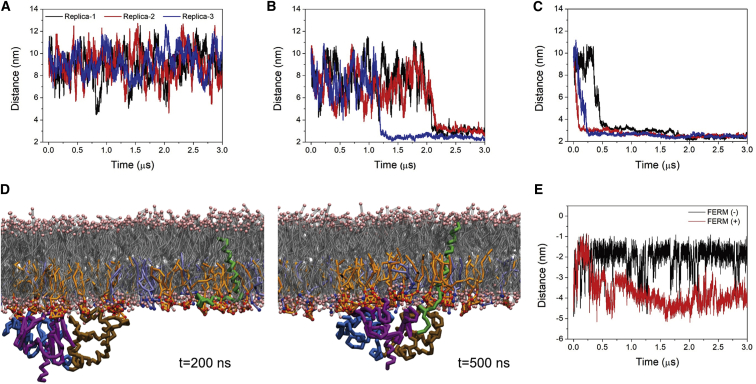
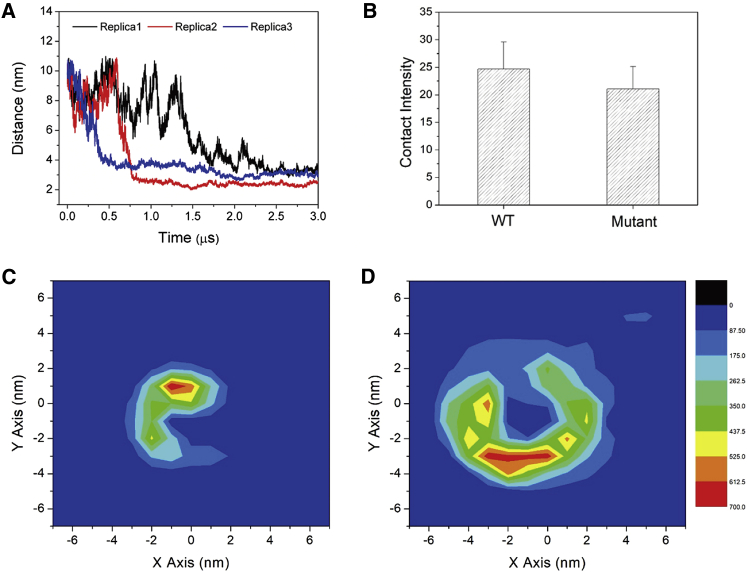
To probe the association kinetics of L-selectin/FERM in more detail, we performed additional simulations using a larger simulation box, in which the initial distance between the two proteins was elongated to 10 nm. It allows investigating the spatiotemporal details for FERM complexation to L-selectin regulated by the membrane environment. Three simulation runs of 3 μs each were performed for each of the three membrane compositions studied. We monitored the distance evolution between L-selectin and FERM as shown in Fig. 4, A–C. We consider the proteins bound if the distance between their centers-of-mass is less than 3.0 nm. In the pure POPC membrane, as expected, no stable binding is observed (Fig. 4 A) (8). In this case, the FERM domain has difficulties attaching to the membrane and gaining access to L-selectin, despite the CLS being pendulous outside the membrane. In the membrane with 20%POPS in the lower leaflet, FERM is able to adhere to the membrane (as shown in Fig. 3 E), and the protein association is observed on a timescale between 1 and 3 μs (Fig. 4 B). Remarkably faster complex formation is observed when PIP2 is present. In each simulation run, binding occurs within 0.5 μs (Fig. 4 C). Once bound, the complex remains intact throughout the rest of simulation (3 μs). Apparently, PIP2 can efficiently bring the two proteins together, presumably by merging the PIP2 clusters that form around each of the individual proteins (Fig. 4 D). Our results thus suggest a clear contribution of PIP2 to the formation of the L-selectin/FERM heterocomplex. The phenomenon is also observed in a recent study on the complex of CD44, another adhesion protein, and FERM (ezrin), showing the association to be highly dependent on the addition of PIP2 (62).
Figure 4.
Formation of the L-selectin/FERM heterocomplex in different membrane environments. (A–C) The distance between the centers-of-mass of L-selectin and FERM as a function of simulation time in the membranes composed of (A) 100%POPC, (B) 80%POPC/20%POPS, and (C) 95%POPC/5%PIP2 is shown. (D) Snapshots of the CLS position relative to the membrane surface in the FERM-unbound and FERM-bound states in the PIP2-contained membrane are shown. (E) The distance evolution of the C-terminal bead of CLS from the center layer of the PIP2-contained membrane with (black line) and without (red line) FERM is shown. To see this figure in color, go online.
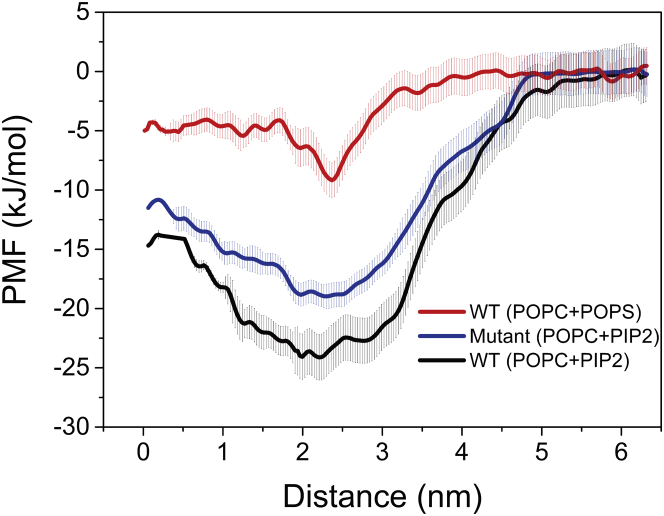
To quantify the contribution of PIP2 to the protein association, the association free energy of the FERM/L-selectin complex in different membranes was computed using the umbrella sampling technique (see Methods for details). The association free energy as a function of the separation distance between the centers-of-mass of the proteins (calculated by the potential of mean force (PMF)) is shown in Fig. 5, in the case of the phosphatidylserine- or PIP2-contained membranes. The energy cost for completely dissociating the protein complex is 9 (± 1.5) kJ/mol in the membrane without PIP2. In contrast, the energy cost increases to 24 (± 2) kJ/mol in the presence of 5% PIP2, indicating the stronger association of the protein complex mediated by PIP2. The result therefore confirms that PIP2 acts as an effective glue for association of FERM and L-selectin.
Figure 5.
Energetics of the FERM/L-selectin complex. PMF profiles are shown as a function of distance between the dissociated FERM and L-selectin. Three independent PMFs were calculated for the L-selectin-WT in systems containing 20%POPS and 5%PIP2 and the L-selectin K362IK363I mutant in 5%PIP2. To see this figure in color, go online.
Conformation of L-selectin changes to maximize binding efficiency to FERM
Upon binding of the FERM domain to L-selectin, we observe a conformational change of L-selectin relative to the membrane surface. The change is clearly visible in the snapshots of the simulations (Fig. 4 D), as well as in the time-dependent distance evolution of the C-terminal of CLS with respect to the center of the membrane (Fig. 4 E). Before interacting with FERM, CLS in the presence of PIP2 is buried at the membrane surface as discussed above (Fig. 2 C). Along with FERM approaching to L-selectin, CLS can detach from the membrane surface, ending up at the surface of FERM in an extended orientation (Fig. 4 E). Note that the membrane-detachment of CLS occurs briefly (0–250 ns) before the complete formation of the L-selectin/FERM complex. A visual inspection shows that the PIP2 lipids, which were previously stabilizing CLS in the membrane-embedded conformation, detach from CLS and reorganize around the heterocomplex during the protein association (Video S1). The resulting solvent-exposed conformation of CLS appears to be favorable for binding to FERM. In a previous study based on the CD44/ezrin heterocomplex, a “sandwich”-like structure of the proteins and PIP2 formed in solution, with PIP2 lipids residing in the interlayer (62). The results of our simulations question this model and suggest a direct interaction between FERM and L-selectin taking place in a more realistic membrane environment, with PIP2 lipids in a stabilizing role by accumulating around the heterocomplex (Fig. 6 C).
Figure 6.
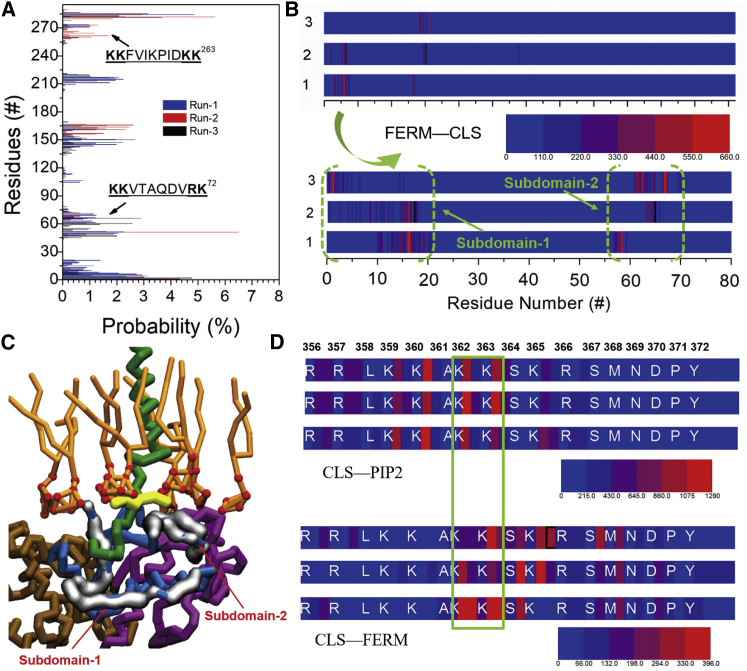
Residue analysis revealing contacts between PIP2 and proteins. (A) The distribution of the residues of FERM in contacting PIP2 is given. The two subdomains, consistent with a previous study (49), are denoted by black arrows, and the sequences are shown. (B) Contact maps of the residues of FERM that interact with CLS are shown. The residue number 0–80 is highlighted in the lower figure. The two subdomains are surrounded by the green dashed brackets. (C) A snapshot of the ternary complex of PIP2/FERM/L-selectin is shown. KK363 residues are shown as yellow branches. The two subdomains revealed in (B) are shown extra in silver. (D) Contact maps of CLS residues in binding to PIP2 (upper panel) and FERM (lower panel) are shown. The residues concurrently contacting PIP2 and FERM are marked by the green rectangle. To see this figure in color, go online.
To provide more insights into the binding mode of the L-selectin/FERM complex, the details of the protein-protein interface and PIP2 binding regions were analyzed. The residues on FERM involved in PIP2 contacts distribute around several conserved regions, including the two well-known regions enriched with positively charged lysine residues (49, 63, 64) (Fig. 6 A). This result thus corroborates the important role of electrostatic interactions in stabilizing FERM on the anionic membrane surface. Consistently, PIP2 mainly interacts with the basic residues on CLS, especially in KKxKK363, which is found at the membrane-proximal region (upper panel in Fig. 6 D). When tethered by FERM, the PIP2-contact intensity decreases dramatically from the position of K365 to the L-selectin C-terminal because of the membrane detachment of CLS. All simulation replicas exhibit two subdomains (residues of 0–20 and 55–70) at the N-terminal of FERM in contact with CLS (Fig. 6 B). It is noteworthy that the second subdomain overlaps with the PIP2-contacted involved region of KKVTAQDVRK72 on FERM, suggesting the guiding role of PIP2 in association of FERM on the L-selectin tail. Meanwhile, FERM-contact residues of CLS distribute at the membrane-distal region, especially of the KKSK362–365 motif (lower panel in Fig. 6 D). These findings match to the known structural results that K362 and K365 act as the key sites in stabilizing the FERM/L-selectin complex (47, 48). Moreover, a previous study shows that FERM and calmodulin (CaM) compete to bind CLS (65), whereas the phosphorylation of S364 can reduce association of CaM on CLS (66). It thus reflects the importance of S364 in the interaction between FERM and CLS, providing the rationale for the competition between FERM and CaM in binding to L-selectin.
Mutations of L-selectin linker region weaken complex formation
It should be noted that the residues of K362K363 on CLS are concurrently involved in interacting with FERM and PIP2 (Fig. 6, C and D). To verify the importance of these residues, we performed additional simulations of an L-selectin mutant in which the K362K363 residues were replaced by isoleucines. Consequently, the association of CLS and FERM is significantly slower as compared to the WT (Fig. 7 A). The lipid/protein complex also became weaker, as demonstrated by the reduced contact intensity of the protein counterparts and the reduced enrichment of PIP2 around CLS (Fig. 7 B). Analysis of the spatial distribution of FERM with respect to L-selectin (Fig. 7, C and D) furthermore shows that FERM displays a wide range of binding geometries in the case of the mutant L-selectin, whereas the heterocomplex involving the WT is relatively well defined. The weakened binding mode of the mutation is also reflected in the association free energy, which is more favorable by ∼5 kJ/mol for the WT (Fig. 5). Taken together, our data show that the K362K363 residues play an important role in binding PIP2 to L-selectin and modulating the stability of the CLS-FERM complex.
Figure 7.
Comparing WT and mutant L-selectin to assess the importance of lysine residues. (A) The time evolution of the distance between L-selectin-K362IK363I and FERM is shown. (B) The contact intensities of L-selectin-WT and -K362IK363I in associating with FERM are shown. (C) The two-dimensional spatial distribution of FERM (center of mass) on the x-y plane relative to the L-selectin-TM domain (centered in the middle of this figure) of the WT and (D) K362IK363I is shown. To see this figure in color, go online.
Discussion
In this study, we investigated the role of anionic lipids in general, and PIP2 in particular, in membrane binding and association of L-selectin and FERM. Our simulations reveal that anionic lipids cluster around CLS and facilitate the attachment of the FERM complex to the membrane, which is a precondition for FERM to associate to CLS. Importantly, we show that PIP2 lipids act as a remarkable accelerator for the association of L-selectin and FERM.
The membrane attachment of FERM observed in this study confirms the importance of the lysine-enriched subdomains, which has been reported in previous experiments (49). In all replicas, FERM adapted a similar orientation on the membrane (Fig. 6), and thus the structural orientation of FERM to the membrane appears determined by the electrostatic interactions between the positively charged residues and the anionic lipids. Moreover, the second lysine-enriched subdomain of FERM binds both PIP2 and CLS (Fig. 6, B and D), which suggests the importance of PIP2 in forming and stabilizing the final protein complex. Based on MD simulations, Herzog et al. also report changes of FERM orientation relative to the membrane when PIP2 is present, proposing that the conformational rolling of FERM is involved in expanding the lifetime of the kinase domain in a FERM-dissociated state (67). Moreover, they highlight the importance of the basic patch residues (220–225) in contacting PIP2, which was also revealed in this work (Fig. 6 A). Together, these data indicate the importance of the anionic lipids in determining how the FERM domain binds and orients at the membrane.
Compared to PIP2, a clustering of POPS around L-selectin and FERM is not evident from our simulations. It is well-known that POPS widely distribute in the inner leaflet of plasma membranes, whereas PIP2 lipids are reported to preferentially locate in specific microdomains of membranes, including the cellular lamellipodia related to cell migrating and adhering (22). The microvillar positioning of L-selectin is necessary for leukocyte tethering (68). A previous study revealed the reduced association of FERM to the K262A mutant of L-selectin and a preclusion of this L-selectin mutant from microvilli (47). Our simulation reflects that K262 is highly involved in binding of CLS to PIP2 (Fig. 6 D), suggesting the pivotal function of PIP2 in regulating the association of L-selectin and FERM by controlling the former distribution in the membranes.
Our simulations reveal that the complexes of L-selectin/PIP2 and FERM/PIP2 are responsible for the rapid association of these proteins. The self-clustering propensity of PIP2 lipids helps in merging the embedded proteins together. An addition of PIP2 efficiently reduces the time required for the docking of CLS to FERM, which is in line with the PIP2-dependent assembling of TM and peripheral proteins (62). Clustering of PIP2, induced by TM and cytosolic proteins, is a known behavior. Notably, PIP2 is found to be necessary for clustering of cellular adhesion protein CD44 on the one hand, and on the other hand it regulates ezrin to transform into the extended structure, which together enables the formation of a heterotetramer complex (62). In addition, coaccumulation of PIP2 and sequestering of proteins in membrane microdomains is reported (11), showing the extensive biological relevance of PIP2 in associating and separating TM proteins. Considering the enrichment of basic residues on the juxtamembrane domain of L-selectin, which is conserved with the known TM proteins in clustering, oligomer/cluster of L-selectins can be sequestered in the PIP2-included membranes. The PIP2-cluster-dependent regulation is believed to be driven by the charge compensation of the anionic PIP2 by positively charged protein residues. Note that the POPS lipids cannot provide a comparable contribution to protein translocation and association like PIP2. Given the similar lipid tails of the two lipid species, the mechanism for accelerating protein association can be addressed to the special structure of PIP2 headgroups, in particular the multivalent negative charge of the PIP2 lipids. Furthermore, efficient stacking of the inositol rings may facilitate PIP2 clustering and drive the PIP2-mediated protein association (Fig. 2, E and F). In addition, a previous study reveals the importance of the hydrogen bond network between the inositol rings, which is found to regulate PIP2 localization and protein recognition (69).
Another interesting aspect of our simulations is the observation that the CLS region becomes linear to the TM domain of L-selectin when bound to FERM, whereas the CLS remains membrane-adsorbed in the presence of PIP2 without FERM. Thus, an autoregulation of L-selectin structure is required for CLS docking to FERM because PIP2 hides CLS in the membrane surface (Fig. 2 C). Based on our data, we propose that FERM competes to attract the PIP2 lipids in the vicinity of L-selectin, which leads to the detachment of CLS from the membrane. We show that the removal of the basic residues on the juxtamembrane domain of L-selectin reduces the protein associating efficiency and loosens the L-selectin/FERM complex. The basic residues therefore act as the key factors in regulating the PIP2 binding and protein-interacting ability. It has been speculated that the conformational alteration of L-selectin from the “hook” shape (induced by PIP2) is related to the membrane cross-regulation for “ectodomain shedding” of L-selectin because the linear shape of L-selectin makes it easier to expose the ectodomain cleavage sites. However, it should be noted that the regulation on the interaction between L-selectin and the cytosolic protein counterparts is multifaceted. Protein modifications such as phosphorylation and ionic mediation also play important roles in steering protein association (70, 71).
In summary, we studied the association of L-selectin and FERM in the vicinity of anionic membranes by CG molecular dynamic simulations. Based on the results, two spatiotemporal aspects explain why PIP2 accelerates the formation of the FERM/L-selectin complex: a strong affinity of PIP2 to the proteins, and the favorable merging of the PIP2 lipid clusters. The molecular details provided in this study contribute to our understanding of the important role of PIP2 lipids in protein association and signaling.
Author Contributions
F.S., S.-Z.L., and S.J.M. designed the research. F.S., L.X., and H.Y. performed the simulations; S.J.M. and C.F.E.S. contributed the analytic methods. F.S., S.-Z.L., and C.F.E.S. analyzed the simulation results. F.S., C.F.E.S., and S.J.M. wrote the article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21372026 and 21672019), the Fundamental Research Funds for the Central Universities (Grant No. XK1701), and the Special Program for Applied Research on Super Computation of the NSFC-Guangdong Joint Fund (The second phase). C.F.E.S. further acknowledges the funding by the European Molecular Biology Organization (ALTF 1340-2016).
Editor: David Cafiso.
Footnotes
One video is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30294-7.
Contributor Information
Siewert J. Marrink, Email: s.j.marrink@rug.nl.
Shi-Zhong Luo, Email: luosz@mail.buct.edu.cn.
Supporting Material
PIP2 lipids, which were previously stabilizing CLS in the membrane-embedded conformation, can be seen detaching from CLS and reorganizing around the heterocomplex during protein association.
References
- 1.Arbonés M.L., Ord D.C., Tedder T.F. Lymphocyte homing and leukocyte rolling and migration are impaired in L-selectin-deficient mice. Immunity. 1994;1:247–260. doi: 10.1016/1074-7613(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 2.Walcheck B., Kahn J., Kishimoto T.K. Neutrophil rolling altered by inhibition of L-selectin shedding in vitro. Nature. 1996;380:720–723. doi: 10.1038/380720a0. [DOI] [PubMed] [Google Scholar]
- 3.González-Amaro R., Sánchez-Madrid F. Cell adhesion molecules: selectins and integrins. Cri. Rev. Immunol. 1999;19:389–429. [PubMed] [Google Scholar]
- 4.Ivetic A., Deka J., Ager A. The cytoplasmic tail of L-selectin interacts with members of the Ezrin-Radixin-Moesin (ERM) family of proteins: cell activation-dependent binding of Moesin but not Ezrin. J. Biol. Chem. 2002;277:2321–2329. doi: 10.1074/jbc.M109460200. [DOI] [PubMed] [Google Scholar]
- 5.Pavalko F.M., Walker D.M., Kansas G.S. The cytoplasmic domain of L-selectin interacts with cytoskeletal proteins via alpha-actinin: receptor positioning in microvilli does not require interaction with alpha-actinin. J. Cell Biol. 1995;129:1155–1164. doi: 10.1083/jcb.129.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peschon J.J., Slack J.L., Black R.A. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 7.Palecanda A., Walcheck B., Jutila M.A. Rapid activation-independent shedding of leukocyte L-selectin induced by cross-linking of the surface antigen. Eur. J. Immunol. 1992;22:1279–1286. doi: 10.1002/eji.1830220524. [DOI] [PubMed] [Google Scholar]
- 8.Deng W., Cho S., Li R. FERM domain of moesin desorbs the basic-rich cytoplasmic domain of l-selectin from the anionic membrane surface. J. Mol. Biol. 2013;425:3549–3562. doi: 10.1016/j.jmb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W., Srinivasan S., Li R. Interaction of calmodulin with L-selectin at the membrane interface: implication on the regulation of L-selectin shedding. J. Mol. Biol. 2011;411:220–233. doi: 10.1016/j.jmb.2011.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 11.van den Bogaart G., Meyenberg K., Jahn R. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLaughlin S., Wang J., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin S., Smith S.O., Murray D. An electrostatic engine model for autoinhibition and activation of the epidermal growth factor receptor (EGFR/ErbB) family. J. Gen. Physiol. 2005;126:41–53. doi: 10.1085/jgp.200509274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michailidis I.E., Rusinova R., Baki L. Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 2011;461:387–397. doi: 10.1007/s00424-010-0904-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Majerus P.W., Connolly T.M., Wilson D.B. The metabolism of phosphoinositide-derived messenger molecules. Science. 1986;234:1519–1526. doi: 10.1126/science.3024320. [DOI] [PubMed] [Google Scholar]
- 16.Martin T.F. PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 17.Legg J.W., Lewis C.A., Isacke C.M. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat. Cell Biol. 2002;4:399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann M., Parra L.M., Herrlich P. Inside-out regulation of ectodomain cleavage of cluster-of-differentiation-44 (CD44) and of neuregulin-1 requires substrate dimerization. J. Biol. Chem. 2015;290:17041–17054. doi: 10.1074/jbc.M114.610204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janmey P.A., Lindberg U. Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 2004;5:658–666. doi: 10.1038/nrm1434. [DOI] [PubMed] [Google Scholar]
- 20.Moore D.T., Nygren P., DeGrado W.F. Affinity of talin-1 for the β3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. USA. 2012;109:793–798. doi: 10.1073/pnas.1117220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erlandsen S.L., Hasslen S.R., Nelson R.D. Detection and spatial distribution of the beta 2 integrin (Mac-1) and L-selectin (LECAM-1) adherence receptors on human neutrophils by high-resolution field emission SEM. J. Histochem. Cytochem. 1993;41:327–333. doi: 10.1177/41.3.7679125. [DOI] [PubMed] [Google Scholar]
- 22.Hanono A., Garbett D., Bretscher A. EPI64 regulates microvillar subdomains and structure. J. Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura F., Huang L., Furthmayr H. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol. Biol. Cell. 1999;10:2669–2685. doi: 10.1091/mbc.10.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayasundar J.J., Ju J.H., Bu Z. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J. Biol. Chem. 2012;287:37119–37133. doi: 10.1074/jbc.M112.380972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braunger J.A., Brückner B.R., Steinem C. Phosphatidylinositol 4,5-bisphosphate alters the number of attachment sites between ezrin and actin filaments: a colloidal probe study. J. Biol. Chem. 2014;289:9833–9843. doi: 10.1074/jbc.M113.530659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingólfsson H.I., Arnarez C., Marrink S.J. Computational ‘microscopy’ of cellular membranes. J. Cell Sci. 2016;129:257–268. doi: 10.1242/jcs.176040. [DOI] [PubMed] [Google Scholar]
- 27.Eggeling C., Honigmann A. Closing the gap: the approach of optical and computational microscopy to uncover biomembrane organization. Biochim. Biophys. Acta. 2016;1858:2558–2568. doi: 10.1016/j.bbamem.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Pöyry S., Vattulainen I. Role of charged lipids in membrane structures - insight given by simulations. Biochim. Biophys. Acta. 2016;1858:2322–2333. doi: 10.1016/j.bbamem.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 29.Ingólfsson H.I., Lopez C.A., Marrink S.J. The power of coarse graining in biomolecular simulations. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2014;4:225–248. doi: 10.1002/wcms.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chavent M., Duncan A.L., Sansom M.S. Molecular dynamics simulations of membrane proteins and their interactions: from nanoscale to mesoscale. Curr. Opin. Struct. Biol. 2016;40:8–16. doi: 10.1016/j.sbi.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pluhackova K., Böckmann R.A. Biomembranes in atomistic and coarse-grained simulations. J. Phys. Condens. Matter. 2015;27:323103. doi: 10.1088/0953-8984/27/32/323103. [DOI] [PubMed] [Google Scholar]
- 32.Hedger G., Sansom M.S.P. Lipid interaction sites on channels, transporters and receptors: recent insights from molecular dynamics simulations. Biochim. Biophys. Acta. 2016;1858:2390–2400. doi: 10.1016/j.bbamem.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marrink S.J., Tieleman D.P. Perspective on the Martini model. Chem. Soc. Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 34.de Jong D.H., Singh G., Marrink S.J. Improved parameters for the martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 35.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 36.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 37.Arnarez C., Marrink S., Periole X. Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. (Camb.) 2016;7:4435–4443. doi: 10.1039/c5sc04664e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta D., Chattopadhyay A. Molecular dynamics simulations of GPCR-cholesterol interaction: an emerging paradigm. Biochim. Biophys. Acta. 2015;1848:1775–1782. doi: 10.1016/j.bbamem.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Hedger G., Rouse S.L., Sansom M.S. Lipid-loving ANTs: molecular simulations of cardiolipin interactions and the organization of the adenine nucleotide translocase in model mitochondrial membranes. Biochemistry. 2016;55:6238–6249. doi: 10.1021/acs.biochem.6b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karandur D., Nawrotek A., Cherfils J. Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane. Proc. Natl. Acad. Sci. USA. 2017;114:11416–11421. doi: 10.1073/pnas.1707970114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gronnier J., Crowet J.M., Mongrand S. Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. eLife. 2017;6:e26404. doi: 10.7554/eLife.26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalli A.C., Rog T., Sansom M.S.P. The integrin receptor in biologically relevant bilayers: insights from molecular dynamics simulations. J. Membr. Biol. 2017;250:337–351. doi: 10.1007/s00232-016-9908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buyan A., Kalli A.C., Sansom M.S. Multiscale simulations suggest a mechanism for the association of the Dok7 PH domain with PIP-containing membranes. PLoS Comput. Biol. 2016;12:e1005028. doi: 10.1371/journal.pcbi.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naughton F.B., Kalli A.C., Sansom M.S. Association of peripheral membrane proteins with membranes: free energy of binding of GRP1 PH domain with phosphatidylinositol phosphate-containing model bilayers. J. Phys. Chem. Lett. 2016;7:1219–1224. doi: 10.1021/acs.jpclett.6b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hedger G., Sansom M.S., Koldsø H. The juxtamembrane regions of human receptor tyrosine kinases exhibit conserved interaction sites with anionic lipids. Sci. Rep. 2015;5:9198. doi: 10.1038/srep09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halim K.B.A., Koldsø H., Sansom M.S. Interactions of the EGFR juxtamembrane domain with PIP 2-containing lipid bilayers: insights from multiscale molecular dynamics simulations. BBA-Gen. Subjects. 2015;1850:1017–1025. doi: 10.1016/j.bbagen.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ivetič A., Florey O., Ridley A.J. Mutagenesis of the ezrin-radixin-moesin binding domain of L-selectin tail affects shedding, microvillar positioning, and leukocyte tethering. J. Biol. Chem. 2004;279:33263–33272. doi: 10.1074/jbc.M312212200. [DOI] [PubMed] [Google Scholar]
- 48.Killock D.J., Parsons M., Ivetic A. In vitro and in vivo characterization of molecular interactions between calmodulin, ezrin/radixin/moesin, and L-selectin. J. Biol. Chem. 2009;284:8833–8845. doi: 10.1074/jbc.M806983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barret C., Roy C., Niggli V. Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP(2)) binding site in the NH(2)-terminal domain of ezrin correlates with its altered cellular distribution. J. Cell Biol. 2000;151:1067–1080. doi: 10.1083/jcb.151.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James D.J., Khodthong C., Martin T.F. Phosphatidylinositol 4,5-bisphosphate regulates SNARE-dependent membrane fusion. J. Cell Biol. 2008;182:355–366. doi: 10.1083/jcb.200801056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wassenaar T.A., Ingólfsson H.I., Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 52.DeLano W.L. DeLano Scientific LLC; San Carlos, CA: 2002. The PyMOL molecular graphics system. [Google Scholar]
- 53.Finnerty C.M., Chambers D., Bretscher A. The EBP50-moesin interaction involves a binding site regulated by direct masking on the FERM domain. J. Cell Sci. 2004;117:1547–1552. doi: 10.1242/jcs.01038. [DOI] [PubMed] [Google Scholar]
- 54.Atilgan A.R., Durell S.R., Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Der Spoel D., Lindahl E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 56.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 57.De Jong D.H., Baoukina S., Marrink S.J. Martini straight: boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 2016;199:1–7. [Google Scholar]
- 58.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 59.Kumar S., Rosenberg J.M., Kollman P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 60.Hub J.S., De Groot B.L., Van Der Spoel D. g_wham: a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010;6:3713–3720. [Google Scholar]
- 61.Sun F., Ding X., Luo S.-Z. A molecular dynamics study of the short-helical-cytolytic peptide assembling and bioactive on membrane interface. J. Phys. Chem. C. 2017;121:17263–17275. [Google Scholar]
- 62.Chen X., Khajeh J.A., Bu Z. Phosphatidylinositol 4,5-bisphosphate clusters the cell adhesion molecule CD44 and assembles a specific CD44-Ezrin heterocomplex, as revealed by small angle neutron scattering. J. Biol. Chem. 2015;290:6639–6652. doi: 10.1074/jbc.M114.589523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niggli V., Andréoli C., Mangeat P. Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 1995;376:172–176. doi: 10.1016/0014-5793(95)01270-1. [DOI] [PubMed] [Google Scholar]
- 64.Fievet B.T., Gautreau A., Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 2004;164:653–659. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kahn J., Walcheck B., Kishimoto T.K. Calmodulin regulates L-selectin adhesion molecule expression and function through a protease-dependent mechanism. Cell. 1998;92:809–818. doi: 10.1016/s0092-8674(00)81408-7. [DOI] [PubMed] [Google Scholar]
- 66.Rzeniewicz K., Newe A., Ivetic A. L-selectin shedding is activated specifically within transmigrating pseudopods of monocytes to regulate cell polarity in vitro. Proc. Natl. Acad. Sci. USA. 2015;112:E1461–E1470. doi: 10.1073/pnas.1417100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herzog F.A., Braun L., Vogel V. Structural insights how PIP2 imposes preferred binding orientations of FAK at lipid membranes. J. Phys. Chem. B. 2017;121:3523–3535. doi: 10.1021/acs.jpcb.6b09349. [DOI] [PubMed] [Google Scholar]
- 68.von Andrian U.H., Hasslen S.R., Butcher E.C. A central role for microvillous receptor presentation in leukocyte adhesion under flow. Cell. 1995;82:989–999. doi: 10.1016/0092-8674(95)90278-3. [DOI] [PubMed] [Google Scholar]
- 69.Wu E.L., Qi Y., Im W. Preferred orientations of phosphoinositides in bilayers and their implications in protein recognition mechanisms. J. Phys. Chem. B. 2014;118:4315–4325. doi: 10.1021/jp500610t. [DOI] [PubMed] [Google Scholar]
- 70.Deng W., Putkey J.A., Li R. Calmodulin adopts an extended conformation when interacting with L-selectin in membranes. PLoS One. 2013;8:e62861. doi: 10.1371/journal.pone.0062861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gifford J.L., Ishida H., Vogel H.J. Structural insights into calmodulin-regulated L-selectin ectodomain shedding. J. Biol. Chem. 2012;287:26513–26527. doi: 10.1074/jbc.M112.373373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PIP2 lipids, which were previously stabilizing CLS in the membrane-embedded conformation, can be seen detaching from CLS and reorganizing around the heterocomplex during protein association.