Abstract
The most common malignant neoplasm of the head and neck region is laryngeal cancer which presents a significant international health problem. The present study aims to screen potential proteins related to laryngeal cancer by network analysis to further understanding disease pathogenesis and biomarker discovery. Differentially expressed proteins were extracted from literatures of laryngeal cancer that compare proteome profiling of patient›s tissue with healthy controls. The PPI network analyzed for up and down regulated proteins with Cytoscape Version 3.4. After PPI construction, topological properties of the two networks have been analyzed. Besides, by using MCODE. the Gene Ontology (GO) analysis, the related modules and pathways were examined. Our study screened 275 differentially changed proteins, including 136 up- and 139 down-regulated proteins. For each network, it has been considered 20 key proteins as hub and 20 as bottleneck. A number of 26 hub-bottleneck nodes is introduced for the two networks. A total of 11 modules including 6 downregulated and 5 upregulated network modules were obtained. The most significant GO function in the significant upregulated module was the RNA processing, and the most significant one in the downregulated module with highest score was the respiratory electron transport chain. Among 275 investigated proteins, 12 crucial proteins are determined that 4 of them can be introduce as a possible biomarker panel including YWHAZ, PPP2R1A, HSP90AA1, and CALM3 for human laryngeal cancer.
Key Words: Human laryngeal cancer, PPI network analysis, Biomarker panel, Cytoscape
Introduction
The most common malignant neoplasm of the head and neck regions is laryngeal cancer which presents a significant international health problem. This type of cancer has high rate of mortality because of the poor diagnosis in early stage of the disease. Despite favorable treatment in early-stage laryngeal cancers, survival rates for advanced-stage disease are less than 50%. Surgery and chemotherapy are two suitable treatment options that are used for laryngeal cancer. However, their combination is also is used. Recently the number of patients treated with radiotherapy and chemotherapy is increased (1). However, survival is decreased (2). Laryngeal cancer has been considered as a multifactorial disease associated with the interaction between environmental factors and genetic background (3). Environmental factors of laryngeal cancer are introduced as a lower consumption of vegetables and fruits, and higher consumption of milk, eggs, meat, tea, alcohol, and smoking (4). Recently, various studies have established the changes in molecular level which are associated with the development of laryngeal cancer. For example, several studies have investigated associations between CYP1A polymorphisms and laryngeal cancer risk (5). Alcohol consumption or smoking beside the uridine diphosphate glucuronosyl transferase enzyme (UGTs) rs4148323 act synergistically to increase the risk of laryngeal cancer (6). It has also reported the relationship between this type of cancer and nucleotide excision repair pathway genes such as ERCCs and XPA (7). The proteomics studies on laryngeal cancer show that the changed expression proteins regulate cellular proliferation, differentiation, and apoptosis that may directly related to the pathogenesis of cancer (8). Another one reported that some significantly changed expression proteins were the products of oncogenes and others were related to signal transduction and immune defense (9). Deeb A and colleagues showed that related DNA repair pathways are curtail in larynx cancer patients (10). For better understanding of molecular mechanisms of laryngeal cancer pathogenesis, protein-protein interaction (PPI) network analysis can provide an informative concept and detail schema (11-20). Therefore, we used a systems biology approach (based on the available proteomics literature data) as a rational strategy to reveal novel specific markers and probably therapeutic targets for laryngeal cancer.
Experimental
Data collection
In this study, the inclusion criteria were the studies on the human species using cell line and laryngeal squamous tissue samples involved in the comparison between the tumor and normal tissues. Exclusion criteria were the studies on non-human tissue and studies on samples of biological fluids, including plasma, serum, saliva, and urine. Studies only involved in comparison between the tumor tissue and tumor metastasis one. There was no limitation in methods in proteomic studies. We manually evaluated the publications in line with the above conditions; a total of 275 significantly changed expression proteinsextracted of which 136 proteins belong to up regulated protein group and 139 proteins were as down regulated proteins (See Tables 1 and 2).
Table 1.
The list of up-regulated genes in tissue of human laryngeal cancer
| NO. | Gene name | NO. | Gene name | NO. | Gene name | NO. | Gene name |
|---|---|---|---|---|---|---|---|
| 1 | ACAA1 | 35 | EEF1D | 69 | HSPD1 | 103 | PSMD2 |
| 2 | ACTR2 | 36 | EEF1G | 70 | IDH1 | 104 | RAB2A |
| 3 | AKR1C2 | 37 | EEF2 | 71 | IMPDH2 | 105 | RAP1B |
| 4 | ALB | 38 | EIF2S1 | 72 | ISOC2 | 106 | RPL14 |
| 5 | ALDH3A1 | 39 | EIF3F | 73 | KPNB1 | 107 | RPL6 |
| 6 | ANXA11 | 40 | EIF3H | 74 | LAP3 | 108 | RPS15A |
| 7 | ARHGAP1 | 41 | EIF3I | 75 | LCP1 | 109 | S100A16 |
| 8 | ARHGDIA | 42 | EIF4A1 | 76 | LDHB | 110 | S100A8 |
| 9 | ARHGDIB | 43 | EIF5A | 77 | LGALS7 | 111 | S100A9 |
| 10 | ARL1 | 44 | ENO1 | 78 | LTA4H | 112 | SERPINB3 |
| 11 | ARPC4 | 45 | EPPK1 | 79 | MAPRE1 | 113 | SF3A3 |
| 12 | ATIC | 46 | EPS8L1 | 80 | METAP1 | 114 | SFPQ |
| 13 | ATP6V1A | 47 | ERO1L | 81 | MPO | 115 | SND1 |
| 14 | BLVRB | 48 | FABP5 | 82 | MYL6 | 116 | STAT1 |
| 15 | C1QBP | 49 | FBP1 | 83 | NAP1L1 | 117 | TACSTD2 |
| 16 | CA2 | 50 | FLOT1 | 84 | NCL | 118 | TAGLN2 |
| 17 | CAND1 | 51 | FN1 | 85 | NDRG1 | 119 | TALDO1 |
| 18 | CAP1 | 52 | FSCN1 | 86 | NDUFA8 | 120 | TAPBP |
| 19 | CAPN2 | 53 | FTL | 87 | NP | 121 | TF |
| 20 | CAPNS1 | 54 | FUS | 88 | PABPC1 | 122 | TFRC |
| 21 | CCT6A | 55 | G3BP2 | 89 | PDIA4 | 123 | TKT |
| 22 | CCT7 | 56 | G6PD | 90 | PDXK | 124 | TLN1 |
| 23 | CDC37 | 57 | GAPDH | 91 | PFN1 | 125 | TPI1 |
| 24 | CES1 | 58 | GCN1L1 | 92 | PGAM1 | 126 | TPT1 |
| 25 | CFL1 | 59 | GFAP | 93 | PGK1 | 127 | TRAP1 |
| 26 | CLIC1 | 60 | GNAI2 | 94 | PGM1 | 128 | TXNDC5 |
| 27 | CMPK1 | 61 | GSTP1 | 95 | PLEC1 | 129 | TYMP |
| 28 | COL12A1 | 62 | HADHA | 96 | PLS3 | 130 | USP14 |
| 29 | CPSF6 | 63 | HIST1H1B | 97 | PPA1 | 131 | VASP |
| 30 | CTSB | 64 | HMGA1 | 98 | PPP2R1A | 132 | VCL |
| 31 | CTSC | 65 | HNRNPA1 | 99 | PRKRA | 133 | WARS |
| 32 | CYCS | 66 | HNRNPD | 100 | PRTN3 | 134 | WDR1 |
| 33 | DHX9 | 67 | HNRPDL | 101 | PSMD11 | 135 | XRCC5 |
| 34 | ECH1 | 68 | HSP90B1 | 102 | PSMD13 | 136 | YWHAZ |
Table 2.
The list of down-regulated genes in tissue of human laryngeal cancer
| NO. | Gene name | NO. | Gene name | NO. | Gene name | NO. | Gene name | NO. | Gene name |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A1BG | 29 | CORO1A | 57 | HIST1H1C | 85 | MYH11 | 113 | RPS11 |
| 2 | A2M | 30 | CORO1C | 58 | HNRNPL | 86 | MYH7 | 114 | RPS15 |
| 3 | ABHD14B | 31 | CRYAB | 59 | HP | 87 | MYL2 | 115 | RPS9 |
| 4 | ACADVL | 32 | CSTB | 60 | HSDL2 | 88 | MYLPF | 116 | RRBP1 |
| 5 | ACAT1 | 33 | CTNND1 | 61 | HSP90 | 89 | NDRG2 | 117 | SDHA |
| 6 | ACTG1 | 34 | CYB5R3 | 62 | HSPB1 | 90 | NDUFA10 | 118 | SERPINA1 |
| 7 | AGR2 | 35 | DCN | 63 | HSPG2 | 91 | NDUFA12 | 119 | SFN |
| 8 | AK3 | 36 | DDOST | 64 | IARS2 | 92 | NDUFS2 | 120 | SLC4A1 |
| 9 | ALDH2 | 37 | DLD | 65 | IGHA1 | 93 | OGDH | 121 | SOD1 |
| 10 | ANXA2 | 38 | DYNLL1 | 66 | IGHG1 | 94 | OGN | 122 | SOD3 |
| 11 | APOA1 | 39 | ECHS1 | 67 | IGKC | 95 | ORM1 | 123 | SP140 |
| 12 | APOA2 | 40 | EIF3A | 68 | IMMT | 96 | ORM2 | 124 | SPTAN1 |
| 13 | ASPN | 41 | EPHX1 | 69 | ITIH2 | 97 | PA2G4 | 125 | SPTBN1 |
| 14 | ATP5B | 42 | ERP29 | 70 | JUP | 98 | PCYOX1 | 126 | SSR4 |
| 15 | ATP5D | 43 | EVPL | 71 | KRT19 | 99 | PHB | 127 | TGFBI |
| 16 | ATP5F1 | 44 | F13A1 | 72 | LAMC1 | 100 | PHB2 | 128 | TMED10 |
| 17 | ATP5O | 45 | FAU | 73 | LGALS3 | 101 | PRDX3 | 129 | TNNT3 |
| 18 | BGN | 46 | FGB | 74 | LGALS3BP | 102 | PRELP | 130 | TPM1 |
| 19 | C1QC | 47 | FGG | 75 | LMAN1 | 103 | PSMB1 | 131 | TRIM29 |
| 20 | C3 | 48 | FKBP4 | 76 | LMAN2 | 104 | PSME2 | 132 | TROVE2 |
| 21 | CALM1 | 49 | GGT5 | 77 | LMNA | 105 | PYCR1 | 133 | U2AF1 |
| 22 | CALML3 | 50 | GLUD1 | 78 | LMNB1 | 106 | PYGB | 134 | UNC84B |
| 23 | CANX | 51 | GOT2 | 79 | LRP1 | 107 | RAN | 135 | UQCRB |
| 24 | CFH | 52 | GPD2 | 80 | LTF | 108 | RPL10 | 136 | UQCRC1 |
| 25 | CFL1 | 53 | GRP94 | 81 | LUM | 109 | RPL19 | 137 | UQCRC2 |
| 26 | CKM | 54 | GSN | 82 | LYZ | 110 | RPL23A | 138 | VDAC1 |
| 27 | CKMT1A | 55 | GSTP1 | 83 | MARCKS | 111 | RPL9 | 139 | VDAC2 |
| 28 | COL15A1 | 56 | H2AFY | 84 | MTPN | 112 | RPN1 |
PPI network analysis
PPI network analyzed by Cytoscape Version 3.4 and Betweenness centrality (BC) and node degree the two major centrality parameters were analyzed by using a Cytoscape plug-in called ‘Network Analyzer’ (21). Degree indicates the number of connectivity belongs to a node and nodes having high degree were introduced as hub proteins. BC value the other centrality index reflects the shortest paths that pass through a node (22).
Screening of network modules and functional analysis
The modules of the two constructed networks (including up and down regulated networks) were provided by MCODE analysis and parameters including Node Score Cutoff: 0.2, K-Core: 2, Degree Cutoff: 2 and, Max depth = 100 were used as the cut-off criteria for network module screening. MCODE score > 3 and node > 6 were considered for functional enrichment analysis of the modules. Kappa statistic ≥ 0.4 and Bonferroni step down method for probability value correction were used for annotation analysis of the selected modules.
Results
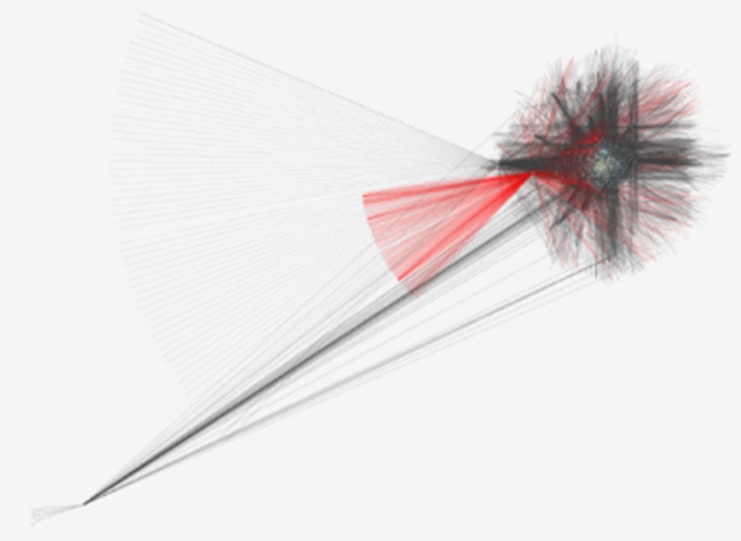
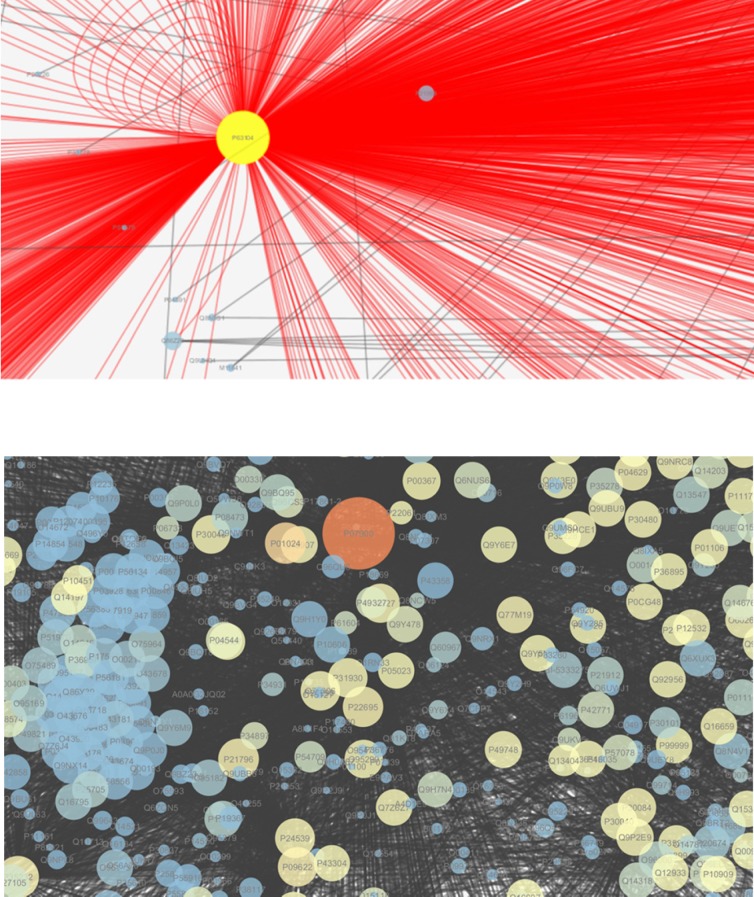
After the submission of up-regulated and down-regulated proteins into Cytoscape, a total of 7312 and 6707 nodes related to the up-regulated and down-regulated proteins are included in the networks, respectively. In the final networks (Figures 1 and 2), the node›s degree was organized based on size; the nodes with high degree have bigger size and the blue to brown color represented low to high BC values for each node. \ The nodes with high degree were considered as key proteins. Then, the top 20 proteins with highest connectivity were identified as the hub proteins for each of the networks and similarly, the top 20 proteins based on betweenness centrality value were selected as bottleneck proteins (See Tables 3 and 4).
Figure 1.
Protein-protein interaction network for up-regulated differentially expressed proteins in tissue of human laryngeal cancer include of 7312 nodes and 33757 edges
Figure 2.
Up: Centrality analysis of protein-protein interaction network for down-regulated differentially expressed proteins in tissue of human laryngeal cancer consist of 6707 nodes and 27422 edges. Down: The dense and central part of upper network is shown in more details
Table 3.
Presentation of the hub proteins in the up-regulated and down-regulated protein–protein interaction networks of laryngeal cancer (top 20 in each PPI network). The hub nodes that play as bottleneck node are asterisked (for more details see Table 4 and discussion
| ID | Degree | ID | Degree | ID | Degree | ID | Degree | |
|---|---|---|---|---|---|---|---|---|
| Up regulated | YWHAZ* | 1634 | CAND1* | 827 | PSMD2* | 636 | ALB* | 524 |
| FN1* | 1538 | PABPC1 | 725 | FUS* | 631 | NCL | 508 | |
| PPP2R1A* | 1208 | MAPRE1* | 716 | KPNB1* | 618 | STAT1* | 503 | |
| CDC37* | 1158 | HNRNPD* | 703 | DHX9 | 554 | ACTR2* | 492 | |
| HNRNPA1* |
1054 |
XRCC5* |
661 |
EEF1G |
538 |
CCT7 |
471 |
|
| Down regulated | HSP90AA1* | 2019 | ACTG1* | 681 | RPL23A | 449 | LMNA* | 407 |
| CALM3* | 1276 | P31947 | 569 | CANX* | 427 | Q13813 | 390 | |
| HSPB1* | 1038 | RPL9P9 | 484 | P20618 | 424 | PHB2 | 364 | |
| RPL10* | 992 | RAN* | 479 | EIF3A | 412 | HNRNPL* | 351 | |
| DYNLL1* | 792 | RPS9 | 450 | IGHG1* | 411 | U2AF1 | 348 |
Table 4.
The list of top 20 up-regulated and down-regulated genes ranked based on BC from largest to smallest values
| ID | BC | ID | BC | ID | BC | ID | BC | |
|---|---|---|---|---|---|---|---|---|
| Up regulated | PDXK | 1.0 | HNRNPA1 | 0.07400 | FUS | 0.04727 | PSMD2 | 0.03599 |
| KHC | 1.0 | CDC37 | 0.06998 | ENO1 | 0.04595 | ALB | 0.03178 | |
| YWHAZ | 0.13462 | GNAI2 | 0.06835 | HNRNPD | 0.03861 | HSPD1 | 0.03167 | |
| FN1 | 0.13420 | PPP2R1A | 0.06310 | ACTR2 | 0.03749 | XRCC5 | 0.03007 | |
| CAND1 |
0.07829 |
MAPRE1 |
0.04832 |
KPNB1 |
0.03667 |
STAT1 |
0.02821 |
|
| Down regulated | HSP90AA1 | 0.20507 | DYNLL1 | 0.06243 | LGALS3 | 0.04051 | APOA1 | 0.02707 |
| CALM3 | 0.13699 | C3 | 0.06131 | A2M | 0.03737 | IGHG1 | 0.02688 | |
| HSPB1 | 0.07676 | CANX | 0.05931 | RAN | 0.03283 | SOD1 | 0.02663 | |
| ACTG1 | 0.07472 | SFN | 0.04720 | FN1 | 0.03122 | HNRNPL | 0.02442 | |
| RPL10 | 0.06626 | LMNA | 0.04078 | PSMB1 | 0.02739 | LGALS3BP | 0.02210 |
Module analysis
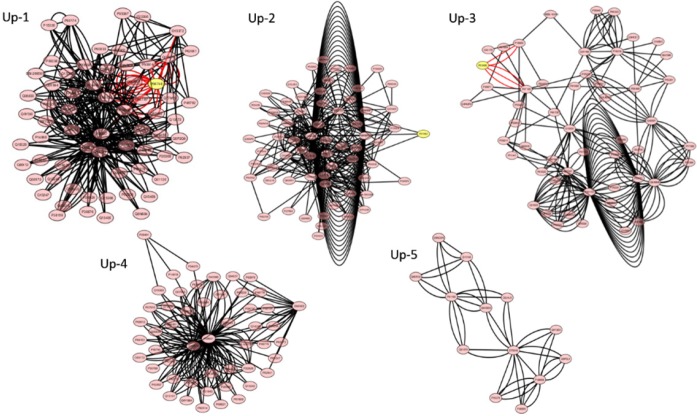
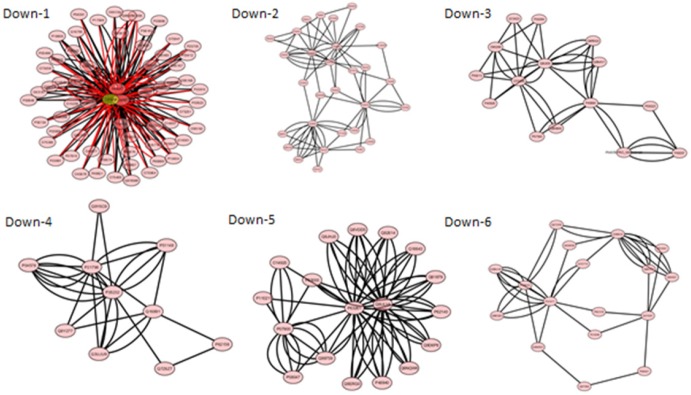
A total of 11 modules including 5 up-regulated and 6 down-regulated network modules were obtained using default criteria. It was selected modules with MCODE score > 3 and node > 6. Five up-regulated modules (Up, 1-5) (Figure 3), and six down-regulated modules (Down, 1-6) (Figure 4) were selected for enrichment analysis.
Figure 3.
Modules of the protein-protein interaction network for up-regulated differentially expressed proteins (MCODE score > 3 and node > 6). The yellow cycles indicate seed proteins and the pink cycles reagent proteins in modules. There are no seed in Up-4 and Up-5 modules
Figure 4.
Modules of the protein-protein interaction network for down-regulated differentially expressed proteins (MCODE score > 3 and node > 6). The yellow cycles indicate seed proteins and the pink cycles reagent proteins in modules. Only Down -1 module has seed and the other ones have no seed
There were some key proteins (hubs) in total of 5 up-regulated modules and 3 up-regulated network modules among them have 3 seed proteins (see Table 5). While, in down-regulated network modules, only Down-1 module has seed. The hubs in this network are distributed as tabulated data in Table 5.
Table 5.
The modules of up regulated and down regulated PPI networks of human tissue of laryngeal cancer. The asterisked proteins are hub-bottleneck nodes
| Category | MCODE score, nodes and edges | Seed | Hub | |
|---|---|---|---|---|
| Up regulated | Up-1 | 7.6, 65 and 358 | NPM1 | HNRNPD*, DHX9, FUS*, NCL and YWHAZ* |
| Up-2 | 5.8, 65 and 320 | HSPA9 | KPNB1*, XRCC5* and CAND1* | |
| Up-3 | 4.0, 52 and 219 | NS | PPP2R1A* | |
| Up-4 | 3.8, 49 and 115 | ---- | HNRNPA1* | |
| Up-5 |
3.3, 13 and 44 |
---- |
ACTR2 |
|
| Down regulated | Down-1 | 5.87, 65 and 219 | UQCRC1 | ---- |
| Down-2 | 4.06 , 30 and 80 | ---- | RPL9P9 ,DYNLL1* | |
| Down-3 | 4.0 , 15 and 43 | ---- | ---- | |
| Down-4 | 4.0 , 10 and 30 | ---- | CALM3* | |
| Down-5 | 4.0 , 18 and 80 | ---- | ACTG1* , HSP90AA1* | |
| Down-6 | 3.25, 17 and 42 | ---- | PHB2, U2AF1 |
Functional enrichment analysis for modules
Four up-regulated modules (Up, 1-4) and three down-regulated modules (Down, 1-3) were enriched based on functional annotation. The top three GO terms for each module are shown in Table 6.
Table 6.
GO functional enrichment analysis of up- regulated and down-regulated PPI network modules. Top three terms of each module are tabulated
| Category | Term | Description | |
|---|---|---|---|
| Up regulated | Up-1 | GO:0006396 | RNA processing |
| GO:0000380 | Alternative mRNA splicing | ||
| GO:0071826 | Ribonucleoprotein complex subunit organization | ||
| Up-2 | GO:0000082 | G1/S transition of mitotic cell cycle | |
| GO:0042769 | DNA damage response | ||
| GO:1901992 | Positive regulation of mitotic cell cycle phase transition | ||
| Up-3 | GO:0031398 | Positive regulation of ubiquitination | |
| GO:0046364 | Monosaccharide biosynthetic process | ||
| GO:0006098 | Pentose-phosphate shut | ||
| Up-4 | GO:0008380 | RNA splicing | |
| GO:0022613 | Ribonucleoprotein complex biogenesis | ||
|
|
GO:0031123 |
RNA 3 -end
processing |
|
| Down regulated | Down-1 | GO:0022904 | Respiratory electron transport chain |
| GO:0046034 | ATP metabolic process | ||
| GO:1902600 | Hydrogen transmembrane transport | ||
| Down-2 | GO:1900739 | Regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway | |
| GO:0031110 | Regulation of microtubule (de) polymerization | ||
| GO:0016259 | Selenocystein metabolic process | ||
| Down-3 | GO:0010257 | NADH dehydrogenase complex assembly | |
| GO:0006099 | Tricarboxylic acid cycle |
Discussion
Protein-protein interaction (PPI) network analysis has a significant growth in cancer studies to facilitate introducing early stage biomarkers (23). In our study, the laryngeal cancer related proteins were analyzed via PPI network construction, hub gene identification, module analysis, and functional enrichment analysis of most significant modules. These stages were carried out for up-regulated proteins and down-regulated ones in human laryngeal cancer tissue, separately. As it is shown in Tables 1 and 2, there are 275 changed expression proteins (including up and down regulated proteins) related to the human tissue of laryngeal cancer. Data management and analysis is a difficult process due to huge numbers of the collected proteins. Since PPI network analysis is a powerful method in categorization and ranking of the candidate and related proteins for a certain disease, here the up and down regulated networks are constructed separately (Figures 1 and 2). Topological analysis of the networks lead to rank of the nodes based on networks properties (18). By using two centrality indices including degree and betweenness, totally 80 nodes are selected among 275 initial proteins as important proteins (see Tables 3 and 4). However, the number of 80 nodes can not be considered as a suitable biomarker panel related to laryngeal cancer and more screening is required. The hub-bottleneck nodes for the up and down regulated networks are shown in Table 3. As it is shown in this Table there are 15 and 11 hub-bottlenecks for up and down regulated networks respectively. Module is a part of a network including closed related proteins havig specific biological function (20). Determined modules of network can provide informative perspective about different roles of the nodes (24). As it is shown in Figures 3 and 4 and Table 5 there are 5 and 6 modules for the up and down regulated networks respectively. Functional enrichment analysis for top score modules indicated that RNA processing and splicing, mitotic cell cycle regulation and sugar biosynthesis are affected by up-regulated modules while metabolic pathways and mitochondria are the main affected subjects by down regulated modules (see Table 6). The most significant pathways in four modules Up, 1-4 were RNA processing, G1/S transition mitotic cell cycle, protein ubiquitination and RNA splicing. It has been revealed overlapping between important pathways involved in the conversion of pre-mRNA to mature mRNA. In previous studies, it shows that polymorphisms of mRNA processing genes can be considered as risk factors for development of laryngeal cancer (25). The most significant pathways in down regulated modules (Down, 1-3) were respiratory electron transport chain, regulation of protein insertion in to mitochondrial membrane involved in apoptotic signaling pathway, and NADH dehydrogenase complex assembly. Proliferating cancer cells, such as laryngeal cancer, preferentially use anaerobic glycolysis rather than oxidative phosphorylation for energy production (26). In one system biology study, the glycolysis/gluconeogenesis pathway has been introduced as the most important pathway in laryngeal cancer (27). Then, the production of energy from mitochondrial respiratory may shift to glycolysis in laryngeal cancer. To prove this hypothesis and determine the energy supply sources of laryngeal cancer cells, more studies are needed. Regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway is the other important pathway in down regulated modules. One of the mechanisms impaired cancer cells is apoptosis. Apoptosis can be activated through several different signaling pathways, but a part of this mechanism is controlled in mitochondrial membrane through insertion apoptotic proteins (28). According to these results, in laryngeal cancer, apoptotic mechanism may disturb through the impairment of transporter proteins which transform apoptotic proteins into mitochondria. According the results of Table 5, the scattering of hubs in up-modules was more than down ones. Interestingly, the finding indicate that the seeds and hubs in up-modules have the similar functions with each other that are associated with regulation of cell cycle (29, 30). Among 26 hub-bottleneck nodes 12 proteins (8 up-regulated and 4 down-regulated proteins) are distributed in 8 modules (see Table 5). These proteins are tabulated in supplementary Table S1 and are ranked based on amounts of degree value. Here two suggestions are feasible: first investigation about expression changes of these 12 genes in the field and the second idea is selection of the top up and down regulated genes for more examinations. We choose cutoff 1200 for degree and therefore YWHAZ and PPP2R1A as the top two up-regulated genes and also HSP90AA1 and CALM3 as the top two down-regulated genes are introduced as human laryngeal cancer. YWHAZ gene with the highest degree and BC scores encodes 14-3-3 protein zeta/delta that has an essential role in tumor cell proliferation (31) through the regulation of multiple cellular processes, such as cell cycle control, anti-apoptosis, signal transduction, inflammation, and cell adhesion/motility (32). YWHAZ has been introduced as candidate proto-oncogene in head and neck squamous cell carcinoma whose reduced expression causes lower level of DNA synthesis rates (33). 14-3-3 proteins could be a key regulatory components in many processes that are crucial for development of cancers (34) such as laryngeal cancer (8). PPP2R1A gene encodes one subunit of protein phosphatase 2. This protein phosphatase is involved in control of cell growth and cell division processes. The role of this subunit in integrity of enzyme is highlighted. Therefore, it is expected that PPP2R1A plays a crucial regulatory role in cell proliferation in cancer cell line(35). HSP90AA1 and CALM3 were found as two top ranked genes in the down-regulated PPI network. These proteins belong to family of proteins which involved in the regulation of specific target proteins in cell cycle control and programmed cell death (36, 37). On the other hand, CALMs in addition to cell cycle, related to centrosome cycle and deregulation of this protein can be the origin of chromosomal instability in cancer (38). Interestingly, all determined possible biomarkers are related to the cell cycle process.
Conclusion
In this study, it has been represented a model of important proteins and pathways that provide a new level of information for laryngeal cancer that increases our knowledge about diagnostic and therapeutic aspects of this disease. Finally, a possible biomarker panel including YWHAZ and PPP2R1A as the two up-regulated genes and HSP90AA1 and CALM3 as the two down-regulated genes for human laryngeal cancer is introduced.
Supplementary Materials
References
- 1.Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, Glisson B, Trotti A, Ridge JA, Chao C, Peters G, Lee DJ, Leaf A, Ensley J, Cooper J. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N. Engl. J. Med. 2003;349:2091–8. doi: 10.1056/NEJMoa031317. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman HT, Porter K, Karnell LH, Cooper JS, Weber RS, Langer CJ, Ang KK, Gay G, Stewart A, Robinson RA. Laryngeal cancer in the United States: Changes in demographics, patterns of care, and survival. Laryngoscope. 2006;116:1–13. doi: 10.1097/01.mlg.0000236095.97947.26. [DOI] [PubMed] [Google Scholar]
- 3.Hakenewerth AM, Millikan RC, Rusyn I, Herring AH, North KE, Barnholtz-Sloan JS, Funkhouser WF, Weissler MC, Olshan1 AF. Joint effects of alcohol consumption and polymorphisms in alcohol and oxidative stress metabolism genes on risk of head and neck cancer. Cancer Epidemiol. Biomarkers Prev. 2011;20:2438–49. doi: 10.1158/1055-9965.EPI-11-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapil U, Singh P, Bahadur S, Dwivedi SN, Singh R, Shukla N. Assessment of risk factors in laryngeal cancer in India: A case-control study. Asian Pac. J. Cancer Prev. 2005;6:202–7. [PubMed] [Google Scholar]
- 5.Zeng W, Li Y, Lu E, Ma M. CYP1A1 rs1048943 and rs4646903 polymorphisms associated with laryngeal cancer susceptibility among Asian populations: A meta-analysis. J. Cell. Mol. Med. 2016;20:287–93. doi: 10.1111/jcmm.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huangfu H, Pan H, Wang B, Wen S, Han R, Li L. Association between UGT1A1 polymorphism and risk of laryngeal squamous cell carcinoma. Int. J. Environ. Res. Public Health. 2016;13:112. doi: 10.3390/ijerph13010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu B, Li J, Gao Q, Yu W, Yang Q, Li X. Laryngeal cancer risk and common single nucleotide polymorphisms in nucleotide excision repair pathway genes ERCC1, ERCC2, ERCC3, ERCC4, ERCC5 and XPA. Gene. 2014;542:64–8. doi: 10.1016/j.gene.2014.02.043. [DOI] [PubMed] [Google Scholar]
- 8.Sewell DA, Yuan C-X, Robertson E. Proteomic signatures in laryngeal squamous cell carcinoma. ORL J. Otorhinolaryngol. Relat. Spec. 2006;69:77–84. doi: 10.1159/000097406. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J-r, Fu Z-x, Li J, Wei L-z, Li J-c. Identification of tumor-associated proteins in well differentiated laryngeal squamous cell carcinoma by proteomics. Clin. Proteomics. 2008;3:42. [PubMed] [Google Scholar]
- 10.Deeb A, Beck J, Schuetz E, Nwizu T, Romick-Rosendale L, Lucas F, Butler R, Wise-Draper T, Mirezwa M, Morris JC, Casper K, Adelstein DJ, Grandis J, Bahassi EM, Urnovitz H, Hashemi Sadraei N. Genomic instability in Larynx cancer. Int. J. Radiat. Oncol. Biol. Phys. 2016;94:938. [Google Scholar]
- 11.Von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 12.Safaei A, Tavirani MR, Oskouei AA, Azodi MZ, Mohebbi SR, Nikzamir AR. Protein-protein interaction network analysis of cirrhosis liver disease. Gastroenterol. Hepatol. Bed Bench. 2016;9:114–23. [PMC free article] [PubMed] [Google Scholar]
- 13.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Tavirani MR, Seyyedi SS. Protein-Protein interaction network could reveal the relationship between the breast and colon cancer. Gastroenterol. Hepatol. Bed Bench. 2015;8:215. [PMC free article] [PubMed] [Google Scholar]
- 14.Jafari M, Sadeghi M, Mirzaie M, Marashi SA, Rezaei-Tavirani M. Evolutionarily conserved motifs and modules in mitochondrial protein–protein interaction networks. Mitochondrion. 2013;13:668–75. doi: 10.1016/j.mito.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Rezaei-Tavirani M, Zamanian-Azodi M, Rajabi S, Masoudi-Nejad A, Rostami-Nejad M, Rahmatirad S. Protein clustering and interactome analysis in Parkinson and Alzheimer’s diseases. Arch. Iran. Med. 2016;19:101–9. [PubMed] [Google Scholar]
- 16.Zali H, Tavirani MR. Meningioma protein-protein interaction network. Arch. Iran. Med. 2014;17:262. [PubMed] [Google Scholar]
- 17.Zamanian Azodi M, Rezaei Tavirani M, Arefi Oskouiea A, Hamdieh M, Derakhshan MK, Ahmadzadeh A, Zayeri F, Nejadia N, Rezaei Tavirani M, Mansourie V, Rostami-Nejad M, Vafaee R. Fluoxetine Regulates Ig Kappa Chain C region expression levels in the serum of obsessive-compulsive disorder patients: A proteomic approach. Iran. J. Pharm. Res. 2017;16:1264–71. [PMC free article] [PubMed] [Google Scholar]
- 18.Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Rezaei-Tavirani M. Ethanol and cancer induce similar changes on protein expression pattern of human fibroblast cell. Iran. J. Pharm. Res. 2016;15:175–84. [PMC free article] [PubMed] [Google Scholar]
- 19.Hajimahdi Z. Small molecules as protein-protein interaction inhibitors. Iran. J. Pharm. Res. 2016;15:1–2. [PMC free article] [PubMed] [Google Scholar]
- 20.Zali H, Zamanian-Azodi M, Rezaei-Tavirani M, Baghban AA. Protein drug targets of lavandula angustifolia on treatment of rat Alzheimer’s diseases. Iran. J. Pharm. Res. 2015;14:291. [PMC free article] [PubMed] [Google Scholar]
- 21.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–4. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 22.Proulx SR, Promislow DE, Phillips PC. Network thinking in ecology and evolution. Trends Ecol. Evol. 2005;20:345–53. doi: 10.1016/j.tree.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov AA, Khuri FR, Fu H. Targeting protein–protein interactions as an anticancer strategy. Trends Pharmacol. Sci. 2013;34:393400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C-Y, Lee T-L, Chiu Y-Y, Lin Y-W, Lo Y-S, Lin C-T, Yang J-M. Module organization and variance in protein-protein interaction networks. Sci. Rep. 2015;5:9386. doi: 10.1038/srep09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osuch-Wojcikiewicz E, Bruzgielewicz A, Niemczyk K, Sieniawska-Buccella O, Nowak A, Walczak A, Majsterek I. Association of polymorphic variants of miRNA processing genes with Larynx cancer risk in a Polish population. Biomed Res. Int. 2015;2015:298378. doi: 10.1155/2015/298378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J, Zhang S, Li Y, Tang Z, Kong W. Hexokinase 2 overexpression promotes the proliferation and survival of laryngeal squamous cell carcinoma. Tumour Biol. 2014;35:3743–53. doi: 10.1007/s13277-013-1496-2. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Du J, Liu J, Wen B. Identification of key target genes and pathways in laryngeal carcinoma. Oncol. Lett. 2016;12:1279–86. doi: 10.3892/ol.2016.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–90. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qing Y, Yingmao G, Lujun B. Role of Npm1 in proliferation, apoptosis and differentiation of neural stem cells. J. Neurol. Sci. 2008;266:131–7. doi: 10.1016/j.jns.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Peng C, Yang P, Cui Y, He M, Liang L, Di Y. HSPA9 overexpression inhibits apoptin-induced apoptosis in the HepG2 cell line. Oncol. Rep. 2013;29:2431–7. doi: 10.3892/or.2013.2399. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H and Otsuji E. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br. J. Cancer. 2013;108:1324–31. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilker E, Yaffe MB. 14-3-3 proteins—a focus on cancer and human disease. J. Mol. Cell. Cardiol. 2004;37:633–42. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 33.Lin M, Morrison CD, Jones S, Mohamed N, Bacher J, Plass C. Copy number gain and oncogenic activity of YWHAZ/14-3-3ζ in head and neck squamous cell carcinoma. Int. J. Cancer. 2009;125:603–11. doi: 10.1002/ijc.24346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman AK, Morrison DK. 14-3-3 Proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–7. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong AL, Han S, Lee S, Park JS, Lu Y, Yu S, Li J, Chun KH, Mills GB, Yang Y. Patient derived mutation W257G of PPP2R1A enhances cancer cell migration through SRC-JNK-c-Jun pathway. Sci. Rep. 2016;6:27391. doi: 10.1038/srep27391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuehlke AD, Beebe K, Neckers L, Prince T. Regulation and function of the human HSP90AA1 gene. Gene. 2015;570:8–16. doi: 10.1016/j.gene.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta. 2014;1843:398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 38.Lingle WL, Lukasiewicz K, Salisbury JL. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Adv. Exp. Med. Biol. 2005;570:393–421. doi: 10.1007/1-4020-3764-3_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.