Abstract
RNA polymerase (RNAP) is the primary machine responsible for transcription. Its ability to distinguish between correct (cognate) and incorrect (noncognate) nucleoside triphosphates (NTPs) is important for fidelity control in transcription. In this work, we investigated the substrate selection mechanism of T7 RNAP from the perspective of energetics. The dissociation free energies were determined for matched and unmatched base pairs in the preinsertion complex using the umbrella sampling method. A clear hydrogen-bond-rupture peak is observed in the potential of mean force curve for a matched base pair, whereas no such peaks are present in the position of mean force profiles for unmatched ones. The free-energy barrier could prevent correct substrates from being separated from the active site. Therefore, when NTPs diffuse into the active site, correct ones will stay for chemistry once they establish effective base pairing contacts with the template nucleotide, whereas incorrect ones will be withdrawn from the active site and rejected back to solution. This result provides an important energy evidence for the substrate selection mechanism of RNAP. Then we elucidated energetics and molecular details for correct NTP binding to the active site of the insertion complex. Our observations reveal that strong interactions act on the triphosphate of NTP to constrain its movement, whereas relatively weak interactions serve to position the base in the correct conformation. Triple interactions, hydrophobic contacts from residues M635 and Y639, base stacking from the 3′ RNA terminal nucleotide, and base pairing from the template nucleotide act together to position the NTP base in a catalytically competent conformation. At last, we observed that incorrect NTPs cannot be as well-stabilized as the correct one in the active site when they are misincorporated in the insertion site. It is expected that our work can be helpful for comprehensively understanding details of this basic step in genetic transcription.
Introduction
Transcription that transfers genetic information coded in template DNA to messenger RNA is the first step of gene expression (1). RNA polymerase (RNAP) is the primary molecular machine to conduct transcription, in which RNA transcript is synthesized through repeated nucleotide addition cycles. Structural studies of bacteriophage T7 RNAP indicate each nucleotide addition cycle can be divided into at least four steps: preinsertion, insertion, product, and posttranslocation (2). At the initial stage, the fingers domain (a five-α-helix subdomain including O-helix) is in semiopen conformation. A nucleoside triphosphate (NTP) diffuses through a substrate entry channel to attain the active site. The correct one will stay in the active site once it establishes effective Watson-Crick base-pairing contact with the template nucleotide (TN), whereas the incorrect one will be withdrawn from the active site and rejected back to solution. In the insertion phase, the fingers domain closes to create a steady environment for chemistry. The fitting of experimental data shows that RNAP works efficiently with an elongation rate of ∼130 nt/s, and the probability of error is as low as ∼1/10,000 without proofreading (3). Therefore, the ability of RNAP to distinguish between correct and incorrect nucleotides is essential for its high efficiency and fidelity control of transcription. Previous works proposed that nucleotide selection occurs in the preinsertion phase. A critical amino-acid residue, Tyr639 of T7 RNAP, may participate in nucleotide selection. It occupies the insertion site, allowing the insertion of matched base pairs and blocking these unmatched ones (4). Furthermore, an Mg2+-ion-mediated interaction between the hydroxyl of Tyr639 and the 2′OH of the ribose in a ribonucleotide (or NTP) contributes to the discrimination of the ribo- versus deoxyribonucleotides (5, 6, 7). Yu et al. found that some water molecules assist Tyr639 in ribose recognition (8). It is generally assumed that the correct substrate binds to the active site with a higher affinity compared with incorrect ones. Therefore, it is of particular interest to ask whether RNAP can distinguish correct and incorrect substrates from energy. Yu et al. determined binding free energies for a noncognate nucleotide (GTP) and a cognate one (ATP) in the preinsertion site using an alchemical simulation and found a small binding free-energy difference (∼1.8 kcal · mol−1) existing between the two nucleotides (9). However, the energetics is still far from being clearly described. In addition, Huang et al. proposed a three-component mechanism to elucidate how multisubunit RNAP positions an inserted NTP in a catalytically competent conformation (10). Is this mechanism equally suitable for single-subunit RNAP? In addition, when incorrect NTPs are misincorporated into the active site, can they be as well-stabilized as the correct one? These questions are also very important and remain unanswered.
Molecular dynamics (MD) simulation, by modeling atomistic interactions, is a powerful tool that can provide dynamic information and help address many important issues (10, 11, 12, 13). To address the above questions, we launched a series of all-atom MD simulations to investigate different NTPs binding to the active site of T7 RNAP. Umbrella sampling (US) technique was used to measure dissociation free energies for matched and unmatched base pairs; molecular mechanics generalized Born/surface area (MM/GBSA) analysis was employed to estimate the binding energy (ΔGbinding) for NTP binding to the active site of T7 RNAP. A small energy barrier is observed in the dissociation free-energy profile for a matched base pair, which may prevent the correct NTP from dissociating from the active site; for incorrect NTPs, no such energy barrier is observed. Hence, they can be easily withdrawn from the active site and rejected back to solution. Thereafter, our simulations confirm that the three-component mechanism is suitable for single-subunit T7 RNAP. Triple interactions, hydrophobic contacts from residues M635 and Y639, base stacking from the 3′ RNA terminal nucleotide (R3), and base pairing from the TN act together to position the NTP base in a catalytically competent conformation. In addition, incorrect NTPs cannot be as well-stabilized as the correct one in the active site when they are misincorporated into the insertion site. It is expected that our work will shed more light on the molecular mechanism of RNA synthesis.
Materials and Methods
Model construction
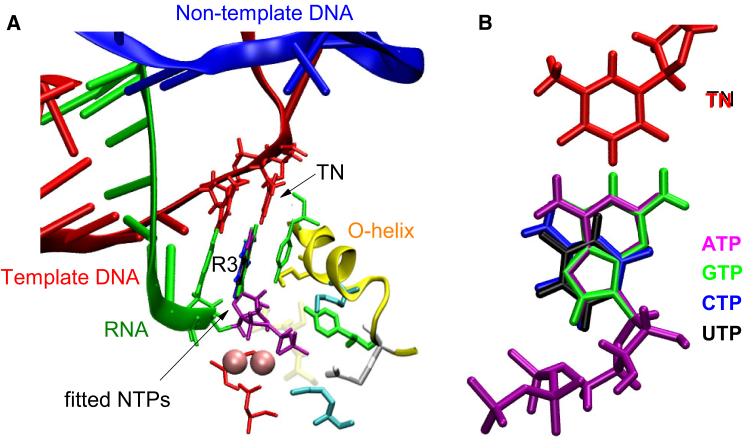
Our model systems were constructed on the basis of existing crystal structures of preinsertion (Protein Data Bank: 1S0V) and insertion (Protein Data Bank: 1S76) complexes. The active sites in the two crystal structures are in semiopen (or open for clarity) and closed conformations, respectively. They both contain a bound nonhydrolyzable ATP analog (α,β methylene ATP), and both of the TNs are thymines. Thus, ATP is the correct or cognate substrate. To construct our models, the ATP analog was modified into an ATP molecule just by replacing the carbon atom linking the Pα and Pβ atoms with an oxygen atom. Other NTP-bound T7 RNAP complexes were prepared by replacing the ATP base with the bases of CTP, GTP, and UTP, respectively, although the sugar and triphosphate moieties remained unchanged. These bases were installed to fit the position of the original ATP base, which is parallel to R3 in vertical direction (see Fig. 1 A) and base pairing with the TN in lateral direction (see Fig. 1 B). As a result, these newly generated NTPs maintain almost the same conformation as the original ATP.
Figure 1.
Schematic diagram of model systems construction. (A) This panel shows the structure of the T7 RNAP active site, where the bases of GTP (colored in green), CTP (blue), and UTP (black) were fitted to the original position of the ATP base (purple), and their sugar and triphosphate parts (colored in purple) were transplanted from the ATP. (B) This panel shows the front view of the four NTPs, the bases of which are ready for pairing with the TN. To see this figure in color, go online.
Simulation details
Most simulations were performed in the GROMACS 4.6.5 software package (14, 15, 16), except those for binding energy calculations, which were carried out in the AMBER 12 package (AMBER Software, San Francisco, CA) (17). The ff99SB force field with ParmBSC0 nucleic-acid parameters (18) was used to describe protein, DNA, RNA, and ions. Other AMBER parameters for NTPs were developed by Meagher et al. (19) The initial T7 RNAP complex was immersed in a rectangular box of 106.13 Å × 101.58 Å × 100.00 Å filled with ∼31,000 three-point transferable intermolecular potential (TIP3P) water molecules (20). Then 145 Na+ and 98 Cl− ions were added to neutralize the system at an ionic concentration of 0.15 mol · L−1. The final system reached ∼110,000 atoms. The cutoff distance for van der Waals interactions was set to 1.2 nm, with a switching distance of 1.0 nm. Electrostatic interaction was treated using the particle mesh Ewald method (21, 22) with a 1.2 nm cutoff distance. Motion equations were numerically solved at a time step of 2 fs. Each solvated system was first subjected to an energy minimization using steepest-descent minimization and then a 1000 ps MD simulation with position restraint on heavy atoms. Equilibrium simulations were performed at a constant temperature of 300 K and a constant pressure of 105 Pa. A velocity rescaling thermostat (23) and Berendsen barostat (24) were applied to control temperature and pressure, respectively. Trajectories were saved at 2 ps intervals. Conformational characterizations were carried out with the built-in tools of the GROMACS package. The potential of mean force (PMF) calculations were performed using the US program in GROMACS package. Binding energy calculations were conducted using the MM/GBSA module (25) in AmberTools15 (AMBER Software). All snapshots for the T7 RNAP complex were produced using VMD software (26).
US simulation
The US method was used to estimate dissociation free energies for the four base pairs in the active site in the preinsertion complex. The distance between two centers-of-mass of NTP and TN bases was chosen as the reactive coordinate for free-energy calculation, referred to as ζ. The steered molecular dynamics (SMD) technique was used to produce a set of seed frames for US simulations. Firstly, each solvated T7 RNAP complex was simply equilibrated for 2 ns in an unbiased MD simulation. Then we performed a 10 ns SMD simulation to pull each NTP base away from the TN base at a speed of 0.4 Å/ns. Because ATP and GTP have similar base structures, their SMD distance ranges were set as 0.6∼1.0 nm; similarly, the SMD distances for CTP and UTP were set to be 0.9∼1.3 nm. We extracted a total of 40 frames with a spacing of 0.01 nm from each SMD trajectory and used them as seed configurations for the subsequent 40 US simulations. Each US window was simulated for 10 ns, with the earlier 2 ns trajectory discarded as preequilibrium and the later 8 ns sampled at a frequency of every 2 ps. PMF profiles were generated with the GROMACS tool g_wham. Error was estimated through 200 rounds of bootstrapping analysis (27).
MM/GBSA method
MM/GBSA analysis was employed to estimate binding energy (ΔGbinding) with the following equation:
where Gcomplex, Greceptor, and Gligand are free energies of complex, receptor, and ligand, respectively. Free energy is estimated as a sum of five terms:
where Eint, Evdw, and Eele denote the internal energy, van der Waals energy, and electrostatic energy, respectively; Gpsolv and Gnpsolv account for the polar and nonpolar contributions to solvation free energy; and T and S denote the absolute temperature and molecule entropy. In this work, entropy is not considered, for we are more concerned about the relative changes in the binding affinity. Each simulation for binding energy calculation was run for 100 ns. The earlier 20 ns of trajectory was discarded as preequilibrium, and the later 80 ns of trajectory was sampled at a frequency of 20 ps intervals for binding energy calculation.
Results and Discussion
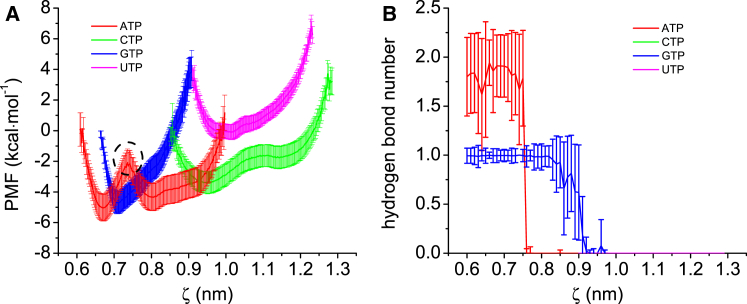
Substrate selection occurs in the preinsertion phase. The correct NTP stays in the active site once it establishes effective Watson-Crick base-pairing contact with the TN, whereas incorrect ones will be withdrawn from the active site and rejected back to solution. To explain this phenomenon, we determined dissociation free energies for both matched and unmatched base pairs in the preinsertion complex using the US method. As mentioned above, ATP is the only correct NTP for the current TN, and they form a matched base pair ATP:TN; CTP, GTP, and UTP form unmatched base pairs with the TN. We launched a total of 1600 ns US simulations to estimate the dissociation PMF profiles for the four base pairs, as shown in Fig. 2 A. One can see that the profile (red line) for the ATP:TN base pair is different from the other three profiles. It displays a significant peak at the position of ζ = ∼0.75 nm (cycled by dashed line), which separates the profile into two free-energy wells that represent two intermediate states during ATP:TN dissociation. To explain the cause of this peak, we observed the change of the number of hydrogen bonds in the process of base-pair dissociation, as shown in Fig. 2 B. We can find that in the section of ζ < ∼0.75 nm, the ATP:TN base pair has about two hydrogen bonds, whereas in the section of ζ > ∼0.75 nm, the hydrogen bond number rapidly decreases to zero, which indicates that the two hydrogen bonds are completely broken. Therefore, the left well in the PMF profile denotes the conformation in which ATP maintains Watson-Crick base-pairing contact with the TN, whereas the right one denotes the conformation of an open ATP:TN base pair; furthermore, the peak represents the energy barrier for the rupture of the hydrogen bonds in the ATP:TN base pair. It is evident that the energy barrier may hinder the dissociation of the matched base pair. On the other hand, in the PMF profiles for the other three NTPs, no significant peaks are observed. It implies these incorrect NTPs cannot establish effective base-pairing contact with the current TN. Note that GTP can form one hydrogen bond with the TN, although it is not a correct substrate (see Fig. 2 B). However, the association is too weak to generate an energy barrier in the PMF profile. In fact, nascent base pair is very fragile and easily damaged by thermal perturbation in the preinsertion phase, but it is very robust in the insertion phase (28). Therefore, the fingers domain should be closed so as to protect the nascent base pair. Fortunately, previous works demonstrated that the fingers domain closing is a spontaneous process with a cognate NTP (29, 30). Therefore, the picture of substrate selection mechanism becomes clear. If an incorrect NTP diffuses into the active site, because it cannot form effective base-pairing contact with the TN, it will be withdrawn from the active site without any energy-barrier blocking. On the contrary, if the incoming NTP is a correct one that can establish a matched base pair with the TN, the energy barrier contributed by the hydrogen bonds will hinder its dissociation from the active site. Then the matched base pair slides into the insertion site upon pushing the originally occupied Y639 aside, which further triggers the closure of the fingers domain and creates a steady environment for the next chemical step.
Figure 2.
(A) PMF profiles for the four base pairs’ dissociations. The peak in dashed cycle represents the energy barrier of hydrogen-bond rupture in the ATP:TN base pair. (B) This panel shows the hydrogen-bond-number evolution during base-pair dissociation for the four NTPs. To see this figure in color, go online.
For multisubunit RNAP II, Huang et al. proposed a three-component (hydrophobic contact, base stacking, and base pairing) mechanism for correctly positioning incoming NTP in a catalytically competent conformation in the active site (10). Here, to verify whether this mechanism is suitable for single-subunit RNAP, we examined the energetic and molecular details of ATP binding to the active site of T7 RNAP, the structure of which is displayed in Fig. 3 A. It can be seen that the ATP is surrounded by many amino acid residues of protein, nucleotides of template DNA strand and nascent RNA transcript, two magnesium ions, etc. Here, we determined binding energies using the MM/GBSA method. The results show the ATP binds to the active site with a negative total binding energy (ΔGbinding) of −182.10 ± 15.35 kcal · mol−1, suggesting that the correct substrate forms favorable contacts with surrounding groups in the active site. We decomposed the binding energy into five components (see Fig. 3 B): internal energy (ΔEint), van der Waals energy (ΔEvdw), electrostatic energy (ΔEele), polar solvation free energy (ΔGpsolv), and nonpolar solvation energy (ΔGnpsolv). Among them, internal energy is ignored here (ΔEint ≈ 0 kcal · mol−1); electrostatic energy and polar solvation energy approximately cancel each other out, yielding a much smaller total electrostatic energy contribution (ΔGele = ΔEele + ΔGpolar). Because NTPs are charged molecules, the ΔGele plays a dominant role in NTP binding. To obtain more details, we further decomposed each energy component into per-residue terms, with the top 12 contributors displayed in Fig. S1. Two magnesium ions form the strongest electrostatic interactions with the ATP triphosphate (ΔEele, MA = −345.55 ± 32.82 kcal · mol−1, and ΔEele, MB = −385.33 ± 23.74 kcal · mol−1). The two ions are located in a triangular region of three negatively charged groups (D812, D537, and the triphosphate of ATP (see Fig. 3 A)), serving as an adhesive to draw the three negatively charged groups together. On the other hand, three positively charged residues (R627, K631, and K472) hold the triphosphate in other directions (see Fig. 3 A; Fig. S1 B). As a result, the triphosphate’s motion is constrained by these charged groups, resulting in a fixed distance between NTP and TN for base pairing. Because most of the NTP’s charges are distributed in its triphosphate, it is not surprising that most strong interactions act on the triphosphate moiety. On the other hand, one can see the triphosphate bound with two ions cross-links the O-helix to the two aspartic acid residues, creating a closed environment in the active site for chemistry.
Figure 3.
(A) Structural representation for ATP binding to the active site of the insertion complex. (B) This panel shows the binding free energy (ΔGbinding) and its components. To see this figure in color, go online.
Furthermore, some relatively weak interactions are observed to play important roles in stabilizing and positioning the ATP base in a correct conformation. As shown in Fig. S1 A, two nucleic groups, TN and R3, form significant contacts with the ATP base, ΔGbinding, TN = −4.13 ± 0.39 kcal · mol−1 and ΔGbinding, R3 = −5.38 ± 0.88 kcal · mol−1, which are mainly contributed by the total electrostatic energy contribution (ΔGele, TN = −3.81 ± 0.38 kcal · mol−1) and van der Waals energy (ΔEvdW, R3 = −4.99 ± 0.64 kcal · mol−1), respectively. The results show that the ATP has established favorable base-pairing contact with the TN and base-stacking interaction with the R3. Additionally, two hydrophobic residues, M635 and Y639, are observed to make favorable van der Waals contacts (ΔEvdW, M635 = −1.61 ± 0.28 kcal · mol−1 and ΔEvdW, Y639 = −1.65 ± 0.29 kcal · mol−1) with the ATP base. Under the triple interactions (base pairing from TN, base stacking from R3, and hydrophobic contacts from M635 and Y639), the vertical and lateral motions of the ATP base are both constrained, generating a catalytically competent conformation for chemistry. The above analysis shows that the three-component mechanism is suitable for single-subunit T7 RNAP as well. In a different way, the hydrophobic contact of L1081 in RNAP II is undertaken by two residues, M635 and Y639, together in T7 RNAP. It should be noted that a positively charged residue H784, located below the middle of the two bases of R3 and ATP, has significant association with the ATP base (ΔGbinding, H784 = −2.64 ± 0.34 kcal · mol−1, Fig. S1 A). Experimental work proposed that alanine substitution of H784 increases both misincorporation and mismatch extension (31). In addition, one can conclude that strong interactions mainly act on the triphosphate to give a fixed distance for base pairing, whereas weak interactions help to position the base in a correct conformation.
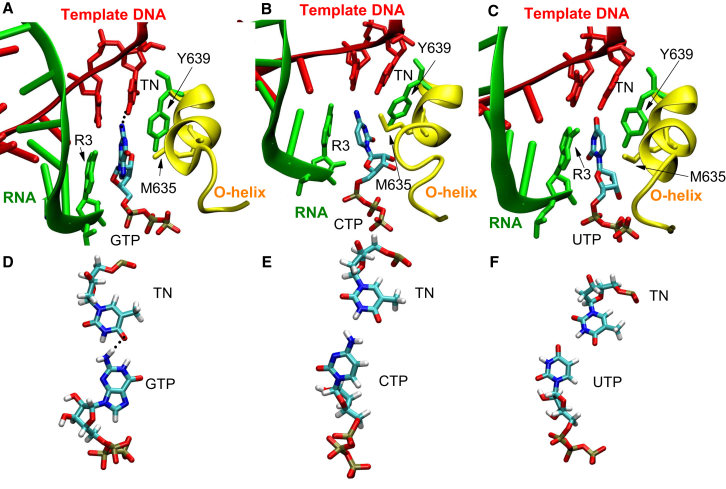
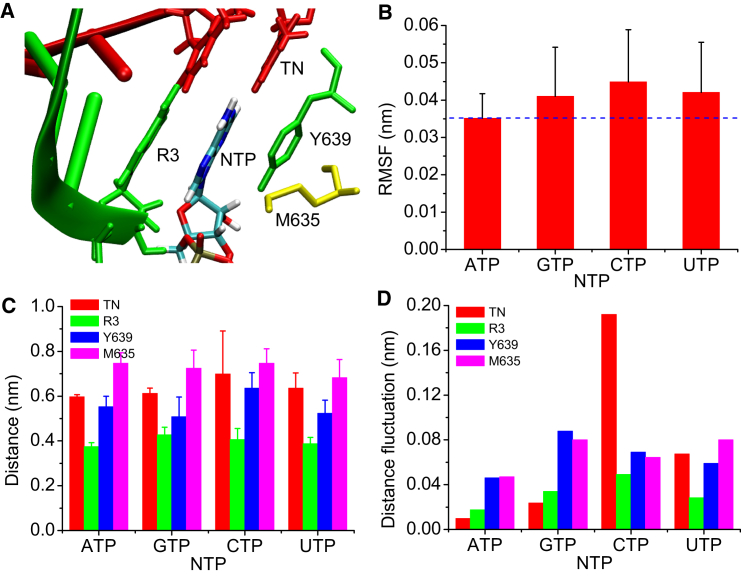
At last, it is of particular interest to investigate whether incorrect NTPs could be well-stabilized when they were misincorporated into the insertion site. We performed eight 100 ns unbiased MD simulations for each NTP-bound T7 RNAP insertion complex. Fig. 4 presents representative snapshots for three incorrect NTPs binding to the active site of the insertion complex. One can see that GTP, structurally similar to ATP, is able to form one hydrogen bond with the TN and favorable base-stacking contact with the R3; CTP and UTP are unable to base pair with the TN, causing their bases not to be properly positioned. We determined root-mean-square-deviation contributions from triphosphate, sugar, and base for each NTP (see Fig. S2). The result shows the three components with root-mean-square-deviation contributions from low to high are as follows: triphosphate < sugar < base. It is evident that the movement of triphosphate in each NTP is constrained by electrostatic interaction network, whereas the base part still has a certain flexibility. Because the four NTPs have the same triphosphate and sugar structures, we only examined their bases to compare their stabilities in the active site. As mentioned above, triple interactions from four residues give rise to a catalytically competent conformation of the correct substrate in the insertion site. Fig. 5 A displays the relative positions for these groups. Here, we determined the root mean-square fluctuation (RMSF) for the bases of the four NTPs to evaluate their stabilities. Fig. 5 B shows the RMSF values from low to high as follows: ATP < GTP < UTP < CTP. One can see the correct substrate has a slightly smaller RMSF value than the incorrect NTPs. Furthermore, to evaluate the associations between each NTP and four key residues (TN, R3, Y639, and M635), we determined their distances as shown in Fig. 5 C, with the corresponding distance fluctuations shown in Fig. 5 D. We can find that ATP has relatively smaller distance fluctuations as compared with the other three NTPs. The observations suggest that correct NTP has established preferable contacts with key residues, and thus can be better stabilized in the active site. Therefore, it is deduced that even if incorrect NTPs are misincorporated into the insertion site, their instability may affect the closure of the finger domain, eventually leading to their dissociations from the active site. However, more theoretical or experimental works are needed to support this deduction.
Figure 4.
Conformations for these misincorporated NTPs in the active site. (A) GTP, (B) CTP, and (C) UTP complexes are shown, respectively. (D–F) These panels display the relative positions of each NTP and the TN in the three unmatched base pairs, respectively. To see this figure in color, go online.
Figure 5.
Characterization of the contacts between the four NTPs and some key residues. (A) An enlarged drawing of the active site, showing the relative positions for an inserted NTP base and four key residues, is presented. (B) This panel gives the RMSFs for the four NTP bases. (C) This panel shows the distance between the two centers-of-mass of each NTP base and the side chain (or base) of each key residue. (D) The corresponding distance fluctuations are shown. To see this figure in color, go online.
Conclusions
In this study, through launching a series of all-atom MD simulations, we investigated the substrate selection mechanism of RNAP from the perspective of energetics. A single-subunit RNAP bacteriophage T7 RNAP was employed to construct our model systems. Because substrate selection occurs in the preinsertion phase, we determined the dissociation free energies for matched and unmatched base pairs in the preinsertion complex. The resulting PMF profiles display a clear hydrogen-bond-rupture peak for the dissociation of matched base pairs, whereas no obvious peaks are observed for unmatched base pairs. The observations support the mechanism that when a NTP diffuses into the active site, the correct one can establish base-pairing contact with the TN and stay in the active site for chemistry because the energy barrier may prevent it from dissociating from the active site; incorrect NTPs, because they cannot base pair effectively with the TN, will be withdrawn from the active site and rejected back to solution.
Then we examined the interaction mechanism of correct NTP binding to the active site of T7 RNAP. One can find that strong interactions are mainly acting on the NTP triphosphate so as to constrain its movement and give a fixed distance for base pairing; relatively weak interactions serve to position the NTP base in the correct conformation. The triphosphate, which binds two magnesium ions, cross-links the O-helix to two aspartic acid residues, creating a closed environment in the active site for chemistry in the insertion phase. Four residues (TN, R3, Y639, and M635) play an important role in maintaining the correct conformation of the base.
Furthermore, we explored whether incorrect substrates can be as well-stabilized as the correct one when they are misincorporated into the insertion site. Our observations show that the correct substrate forms preferable contacts with key residues and can be well-stabilized, whereas incorrect substrates exhibit a certain flexibility in the insertion site, suggesting that their associations with these key residues are not as robust as those for the correct one. Therefore, we deduce that the fingers domain might be hard to close with misincorporated NTPs, which would be withdrawn from the active site eventually. We hope that our work can be helpful for comprehensively understanding the details of this basic step in genetic transcription.
Author Contributions
S.W. and Q.L. conceived and designed the research. S.W., X.P., and L.L. performed the research. S.W., X.P., and Q.L. analyzed the data. S.W. and J.W. wrote the article.
Acknowledgments
We thank Prof. Hualin Shi at the Institute of Theoretical Physics, Chinese Academy of Sciences, Prof. Jin Yu at the Beijing Computational Science Research Center, and Prof. Yi Wang at the Chinese University of Hong Kong for helpful advice.
This work was supported by the National Natural Science Foundation of China (No. 11405113).
Editor: Karin Musier-Forsyth.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30293-5.
Contributor Information
Shaogui Wu, Email: wsgchem@foxmail.com.
Quan Li, Email: liquan6688@163.com.
Supporting Material
References
- 1.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Oster G. A small post-translocation energy bias aids nucleotide selection in T7 RNA polymerase transcription. Biophys. J. 2012;102:532–541. doi: 10.1016/j.bpj.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Temiakov D., Patlan V., Vassylyev D.G. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 5.Brieba L.G., Sousa R. Roles of histidine 784 and tyrosine 639 in ribose discrimination by T7 RNA polymerase. Biochemistry. 2000;39:919–923. doi: 10.1021/bi992324+. [DOI] [PubMed] [Google Scholar]
- 6.Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Eckstein F., Sousa R. Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry. 1997;36:8231–8242. doi: 10.1021/bi962674l. [DOI] [PubMed] [Google Scholar]
- 8.Duan B., Wu S., Yu J. A critical residue selectively recruits nucleotides for t7 RNA polymerase transcription fidelity control. Biophys. J. 2014;107:2130–2140. doi: 10.1016/j.bpj.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E C., Duan B., Yu J. Nucleotide selectivity at a preinsertion checkpoint of T7 RNA polymerase transcription elongation. J. Phys. Chem. B. 2017;121:3777–3786. doi: 10.1021/acs.jpcb.6b11668. [DOI] [PubMed] [Google Scholar]
- 10.Huang X., Wang D., Levitt M. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc. Natl. Acad. Sci. USA. 2010;107:15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva D.A., Weiss D.R., Huang X. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. USA. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Da L.T., Pardo-Avila F., Huang X. Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat. Commun. 2016;7:11244. doi: 10.1038/ncomms11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Xiao X., Pu X. Probing immobilization mechanism of alpha-chymotrypsin onto carbon nanotube in organic media by molecular dynamics simulation. Sci Rep. 2015;5:9297. doi: 10.1038/srep09297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kutzner C., van der Spoel D., Grubmüller H. Speeding up parallel GROMACS on high-latency networks. J. Comput. Chem. 2007;28:2075–2084. doi: 10.1002/jcc.20703. [DOI] [PubMed] [Google Scholar]
- 15.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 17.Pearlman D.A., Case D.A., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 18.Pérez A., Marchán I., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meagher K.L., Redman L.T., Carlson H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 20.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 21.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 22.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 23.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 24.Berendsen H.J., Postma J.P.M., Haak J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 25.Miller B.R., III, McGee T.D., Jr., Roitberg A.E. MMPBSA.py: an efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 27.Hub J.S., De Groot B.L., Van Der Spoel D. g_wham—a free weighted histogram analysis implementation including robust error and autocorrelation estimates. J. Chem. Theory Comput. 2010;6:3713–3720. [Google Scholar]
- 28.Wu S., Li L., Li Q. Mechanism of NTP binding to the active site of T7 RNA polymerase revealed by free-energy simulation. Biophys. J. 2017;112:2253–2260. doi: 10.1016/j.bpj.2017.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang B., Predeus A.V., Feig M. Energetic and structural details of the trigger-loop closing transition in RNA polymerase II. Biophys. J. 2013;105:767–775. doi: 10.1016/j.bpj.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Shao Q., Zhu W. Exploring transition pathway and free-energy profile of large-scale protein conformational change by combining normal mode analysis and umbrella sampling molecular dynamics. J. Phys. Chem. B. 2014;118:134–143. doi: 10.1021/jp4105129. [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Brieba L.G., Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.