Abstract
Polyglutamine (polyQ) diseases, including Huntington’s disease, result from the aggregation of an abnormally expanded polyQ repeat in the affected protein. The length of the polyQ repeat is essential for the disease’s onset; however, the molecular mechanism of polyQ aggregation is still poorly understood. Controlled conditions and initiation of the aggregation process are prerequisites for the detection of transient intermediate states. We present an attenuated total reflection Fourier-transform infrared spectroscopic approach combined with protein immobilization to study polyQ aggregation dependent on the polyQ length. PolyQ proteins were engineered mimicking the mammalian N-terminus fragment of the Huntingtin protein and containing a polyQ sequence with the number of glutamines below (Q11), close to (Q38), and above (Q56) the disease threshold. A monolayer of the polyQ construct was chemically immobilized on the internal reflection element of the attenuated total reflection cell, and the aggregation was initiated via enzymatic cleavage. Structural changes of the polyQ sequence were monitored by time-resolved infrared difference spectroscopy. We observed faster aggregation kinetics for the longer sequences, and furthermore, we could distinguish β-structured intermediates for the different constructs, allowing us to propose aggregation mechanisms dependent on the repeat length. Q11 forms a β-structured aggregate by intermolecular interaction of stretched monomers, whereas Q38 and Q56 undergo conformational changes to various β-structured intermediates, including intramolecular β-sheets.
Introduction
Over the last decades, many research studies focused on understanding the development of the genetically inherited neurodegenerative disorders called “polyQ diseases.” PolyQ diseases include Huntington’s, Kennedy’s disease (spinal and bulbar muscular atrophy), several forms of spinocerebellar ataxia, and dentatorubral-pallidoluysian atrophy (1, 2, 3). These diseases are accompanied by the progressive death of neurons. The neurotoxicity correlates with the length of poly-CAG repeats in the gene encoding a stretch of consecutive glutamine (Q) residues in the corresponding proteins. The critical polyglutamine (polyQ) length (with spinocerebellar ataxia 6 as an exception) lies in the mid-thirty of glutamine repeats (4), and the expanded polyQ tracts lead to the formation of insoluble fibrous inclusions in the affected cells (5, 6). Although the correlation between the abnormal polyQ expansion and protein aggregation is obvious, the molecular mechanisms of polyQ misfolding and aggregation remain poorly understood. Studying aggregation intermediates and identifying potential toxic forms is highly relevant for polyQ pathologies. Aggregation mechanisms and misfolding pathways of polyQ repeats have been proposed by the use of peptides. The nucleation-elongation model assumes that an unfolded polyQ monomer is in rapid equilibrium with a thermodynamically unfavorable β-sheet nucleus that serves as a template for the addition of further unfolded monomers, resulting in an elongated β-structured fibril (7). Another model, called association-conformational conversion, proposes that hydrophobic interaction between the disordered monomers induce the association of soluble oligomers, and further conformational rearrangements result in insoluble fibrillar aggregates (8). Both suggested mechanisms assume a number of intermediate states governed by the polyQ repeats. However, the models are based on peptide studies, which, because of the lack of glutamine solubility, are limited to polyQ sequences below the disease threshold. Thus, conformational analysis of polyQ repeat lengths close to and above the disease threshold is highly desirable because those are the critical and disease-relevant ones. A suitable approach that allows the controlled initiation of the aggregation process as well as a molecular characterization and differentiation of structural intermediates has to be applied. Huntington’s chorea or Huntington’s disease (HD) is the best-studied polyQ-related disorder so far. The abnormally elongated polyQ stretch in the affected Huntingtin protein (Htt) promotes aggregation that consequently causes HD. The onset and severity of disease are strongly correlated with the length of the polyQ repeats. A polyQ length of fewer than 35 glutamines does not cause the disease, 35–39 polyQ repeats may or may not result in HD, 40–60 polyQ repeats lead to the adult onset, and more than 60 repeats cause juvenile forms of HD (9). It has also been shown that the Huntingtin-encoded polyglutamine expansions form insoluble amyloid-like protein aggregates both in vitro and in vivo (10).
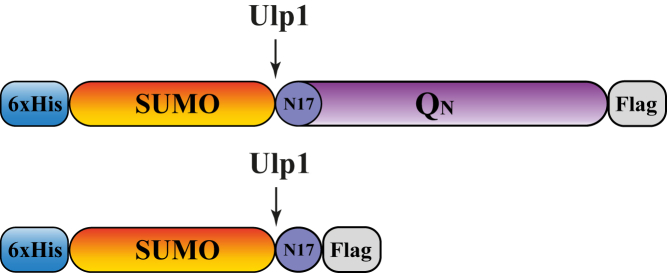
In this study, we developed an attenuated total reflection Fourier-transform infrared (ATR-FTIR) spectroscopic approach to investigate aggregation pathways of engineered polyQ constructs. Infrared spectroscopy (IR) is very well suited to analyze secondary structures in proteins and conformational dynamics and moreover to characterize aggregates of amyloid proteins (11, 12). IR has been used before to characterize short polyglutamine-containing peptides (13, 14, 15, 16, 17, 18, 19), but not for polyQ repeat lengths close to or above the disease threshold. Here, we combined ATR-FTIR difference spectroscopy with an immobilization approach, facilitating the controlled initiation of the aggregation reaction by enzymatic cleavage. All studied polyQ constructs had the same design (20) and varied only in the number of glutamines (Fig. 1). The constructs with polyglutamine lengths below (N = 11), close to (N = 38), and above (N = 56) the disease threshold mimic the normal and abnormally expanded polyglutamine sequence. The N-terminal 17-amino-acid sequence (N17) of the Htt is adjacent to the polyQ sequence and assumed to play an essential role for the aggregation process (13, 21, 22). Thus, N17 was integrated into the constructs as well. The SUMO (small ubiquitin-like modifier) protein keeps the fusion protein soluble and facilitates Ulp1 (ubiquitin-like-specific protease 1) cleavage. Ulp1 belongs to the cysteine protease family and identifies a Gly-Gly motif in the C-terminal exposed β-strand of the tertiary structure of SUMO. The catalytic mechanism occurs with help of the catalytic triad, cysteine, histidine, and aspartic acid in the active center of the Ulp1 protease (23, 24). A construct without polyglutamine sequence is used as a control to identify conformational changes caused by other constituting construct parts.
Figure 1.
Composition of the polyQ construct including the polyglutamine region (QN) (violet), the N-terminal fragment of the mammalian Htt protein (dark blue), the SUMO domain (orange), and a His- and a Flag-tag (light blue and gray). The polyglutamine sequences vary from below (polyQ11), close to (polyQ38), and above (polyQ56) the HD disease threshold. The His-tag is used for affinity purification and enables protein immobilization. The SUMO part enhances the solubility of the construct. Enzymatic cleavage with Ulp1 triggers the aggregation of the polyglutamine sequence. The Flag-tag can be used for immunodetection via Western blot. The construct without a polyglutamine sequence was used for control measurements. To see this figure in color, go online.
Materials and Methods
Materials
The phospholipids, 1-palmitoyl-2-oleoyl-sn-glycero-3 phosphocholine (POPC) and 1,2-dioleoyl-sn-glycero-3-((N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl) (DOGS-NTA), in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL). The isopropylthio β-D-l-galactopyranoside (IPTG), GnHCl, TRIS-HCl, and HEPES were purchased from Sigma-Aldrich (Taufkirchen, Germany) and used as received. All buffers were prepared using ultrapure water. Protino matrix (Ni-NTA silica matrix) was purchased from Macherey-Nagel (Düren, Germany).
Expression and purification of polyQ constructs
The polyQ constructs consist of the polyglutamine repeat, the N-terminal fragment N17 of the mammalian Htt (MATLEKLMKAFESLKSF), the SUMO domain, a His-tag (HHHHHH), and a Flag-tag (DYKDDDDK). The expression and purification of the polyQ constructs were described previously (20). Briefly, the respective pLANA-Qn plasmids were transformed into the Escherichia coli expression-competent strain BL21(DE3). The culture of the bacterial strains with plasmids was inoculated overnight. The protein expression was induced by addition of 1 mM IPTG when the OD600 of the culture solution reached 0.6. After 5–6 h expression at 37°C, the culture was harvested by centrifugation. Afterwards, the cells were lysed by a French press in denaturing lysis buffer containing 6 M GnHCl. After a further centrifugation step, the supernatant was incubated with 1.5 g Protino matrix for 30 min at 4°C while rotating. Because polyQ proteins have a tendency to form insoluble inclusions, the purification was performed under denaturing conditions, using 6 M GnHCl at the beginning and slowly decreasing the concentration of the denaturant to 1 M GnHCl. After several washing steps, the Ni-bound constructs were eluted with elution buffer containing 250 mM imidazole-HCl. The purest fraction was dialyzed against 5 L low-salt buffer at 4°C overnight. To avoid uncontrolled aggregation, the purified constructs were frozen in liquid nitrogen and stored at −80°C.
ATR measurements
The measurements were performed with a vertical ATR cell (Specac, Fort Washington, PA) mounted into a Vertex 80 V spectrometer (Bruker Optik, Bremen, Germany). The internal reflection element (IRE) is a germanium (Ge) crystal of trapezoidal shape (52 × 20 × 2 mm). Both sides of the IRE were used, resulting in 25 reflections. Measurements were performed at room temperature with a spectral resolution of 4 cm−1 and a scanner velocity of 80 kHz. Scans were performed in a double-sided forward-backward mode. The resulting interferograms were apodized with the Blackman-Harris three-term function, and a zero-filling factor of 4 was used. For instrument control and data acquisition, the OPUS software version 7.2 (Bruker Optik) was applied. The experiments were carried out using the ATR cell as a flow-through system connected to a peristaltic pump. The sample was circulated in a closed cycle with a pumping velocity of 1.4 mL/min.
Preparation of the ATR crystal
Before the experiment, the surface of the Ge crystal was activated. Both sides of the crystal were cleaned with Millipore water and a mixture of chloroform/methanol = 1:1. The crystal was polished with 0.1 μm diamond polishing paste (Sommer, Polimant-Color; Demotec, Nidderau, Germany) and optical soft paper tissue. Afterwards, the surface of the IRE was washed again with Millipore water and dried under nitrogen flow. To generate a hydrophilic crystal surface, the IRE was incubated with a concentrated sulfuric acid (H2SO4) solution for 10 min, followed by gently rinsing with Millipore water and drying under a nitrogen flow. The activation procedure was performed three times.
Lipid bilayer formation and NTA-Ni2+ complexation
27.4 μL of 25 mg/mL POPC stock solution, 4.06 μL of 25 mg/mL NTA-DOGS stock solution, and 50 μL of chloroform were combined in a 1.5 mL tube and vortexed for a short time to prepare a POPC/NTA-DOGS lipid mixture with a molar ratio of 9:1. This molar ratio was chosen to avoid further uncontrolled interactions between adjacent immobilized polyQ constructs. Chloroform was removed first under a mild nitrogen flow until there was no solvent visible anymore. Vacuum (generated by a water suction pump) was applied for 2 h to ensure that chloroform was removed completely (25). The 1.5 mL of the lipid buffer (10 mM TRIS/HCl (pH = 7.4), 5 mM MgCl2) was added to the dry lipid film, and the mixture was shaken at 2000 min−1 for 1 h at room temperature. This procedure led to the formation of multilamellar vesicles (26). To convert them into unilamellar vesicles, the solution was extruded by a mini handheld extruder (Mini-Extruder; Avanti Polar Lipids) using a polycarbonate membrane with 50 nm pore size. 1.5 mL of the unilamellar vesicle solution containing 0.9 mM POPC and 0.1 mM NTA-DOGS was directly brought into contact with the clean hydrophilic surface of the Ge crystal to induce vesicle spreading and lipid bilayer (LB) formation. The LB was incubated with NiCl2 in lipid buffer for 1 h. Unbound Ni2+ ions were removed by washing first with lipid buffer and afterwards with polyQ buffer (30 mM HEPES (pH = 7.4), 5 mM NaCl, 5 mM MgCl2, 5% Glycerol (v/v), 1 mM β-mercaptoethanol). Spectral acquisition was started immediately after bilayer preparation.
Protein immobilization
The polyQ immobilization was achieved by chemical conjugation between the His-tag at the N-terminus of the polyQ construct with the nickel-chelating nitriletriacetic acid (Ni2+-NTA) complex of the LB head groups. 500 μL of polyQ construct in H2O/protein buffer (c = 0.1 mg/mL) was injected into the ATR flow-through cell by means of a peristaltic pump, which helps to distribute the proteins homogeneously inside the ATR cell and furthermore to maintain the same injection rate for different constructs. Spectra were recorded for 1 h to monitor the immobilization process. Afterwards, the system was washed with 3 mL of protein buffer to remove unbound constructs.
Curve fitting of the amide I region
A secondary structure analysis was performed by applying a curve-fitting procedure to the amide I band (1600–1700 cm−1) using OPUS software (Bruker Optik). The number of components and their frequency positions were determined by the negative minima in the second derivatives of the ATR spectra. This method allows us to enhance the separation of overlapping bands (27). All original protein spectra were baseline-corrected between 2800 and 1500 cm−1 for further analysis. Spectra were fitted with a linear combination of (10–20%) Lorentzian and (80–90%) Gaussian band profiles. Band (frequency) positions were fixed in accordance to the second-derivative spectra. Bandwidths and intensities were fitted without restriction by using the Levenberg-Marquardt algorithm. The root mean-square error evaluating the difference between the original and reconstructed spectra was ∼10−4. The reproducibility of the fits was confirmed by repeated measurements of independent samples.
Results
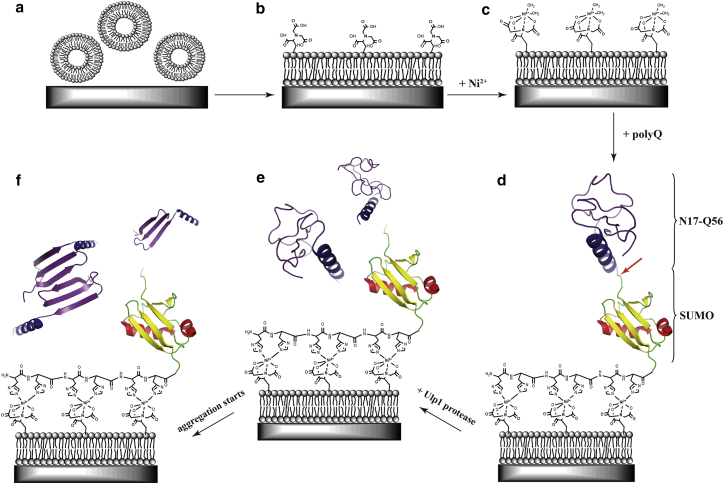
PolyQ constructs of the design His-SUMO-N17-QN-Flag were studied with various glutamine lengths (N = 11, 38, 56). From here on they are termed as polyQ11, polyQ38, and polyQ56, respectively. The construct His-SUMO-N17-Flag without glutamine domain was used as negative control. The experimental procedure includes several steps, as illustrated schematically in Fig. 2. First, an LB with vacant NTA groups was formed on the ATR crystal surface by lipid vesicle spreading (Fig. 2, a and b). The addition of Ni2+ results in Ni2+-NTA complexation, providing specific binding sites for the polyQ constructs (Fig. 2 c). Protein immobilization was achieved via His-tag binding to the Ni2+-NTA complex (as illustrated for polyQ56 as an example in Fig. 2 d). Ulp1 protease was added and cleaved the construct between SUMO and N17 (red arrow in Fig. 2 d), resulting in the release of the intact N17-QN-Flag (Fig. 2 e). Thus, the aggregation reaction started. Structural transitions of the released fragments (containing only the polyQ sequence, N17, and Flag-tag) were monitored (in the aqueous phase) by time-dependent IR measurements (Fig. 2 f). The LB, immobilized SUMO, and Ulp1 were taken as background spectrum after Ulp1 addition to cancel out their absorbance in the IR spectra, assuming that their vibrational modes change only marginally if at all.
Figure 2.
Schematic representation of the ATR-FTIR experiment shown for polyQ56 as an example. (a) This panel shows the vesicle deposition onto an activated Ge crystal. (b) The formation of the LB containing POPC/DOGS with vacant NTA groups. (c) The complexation of NTA groups by the addition of Ni2+ in the lipid buffer is. (d) PolyQ construct immobilization. The construct binds to the Ni2+-NTA via the His-tag. The SUMO (yellow β-sheet, red α-helices) and N17-Q56 (dark blue N17, violet Q56 region). (e) The addition of Ulp1 protease. The enzyme cleavage site is located between SUMO and N17 and is shown by the red arrow in (d). The polyQ-containing fragments are released. (f) The formation of the polyQ aggregates. The polyQ-containing fragments undergo conformational changes and form extended β-sheet aggregates, which are monitored spectroscopically in a time-resolved manner. To see this figure in color, go online.
LB formation
The LB serves as support material for the polyQ construct immobilization. IR difference spectra monitoring the formation of the LB and the Ni2+-NTA complexation are presented in Figs. S1 and S2.
Secondary structure analysis of immobilized constructs
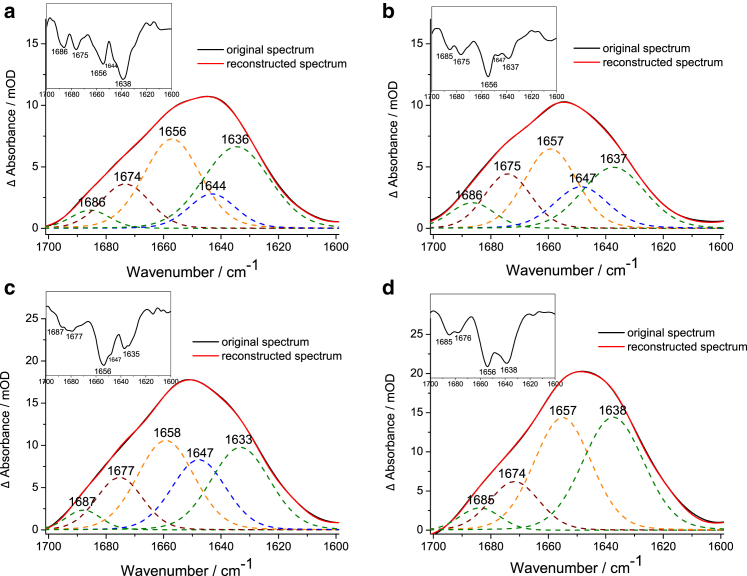
The amide I band (mainly C=O stretching vibration of the polypeptide backbone) is commonly used for secondary structure analysis in IR spectra. The assignment of amide I frequencies to secondary structure elements are summarized in various literature reports, e.g., in (28, 29, 30, 31, 32). The amide I band of α-helical structure is located between 1650 and 1660 cm−1, the disordered structure absorbs between 1640 and 1650 cm−1, and β-sheet absorbs between 1630 and 1640 cm−1. Aggregated proteins often show a band below 1630 cm−1, which is characteristic for extended β-sheets (11, 12, 33, 34). If the β-strands within the β-sheet are antiparallel to each other, there is an additional weak band between 1670 and 1695 cm−1 (35). Turn and loop structures absorb between 1660 and 1680 cm−1. Because the amide I band of the polyQ constructs consists of several overlying band components, we applied curve fitting to the entire amide I contour of the immobilized construct to determine the contribution from different structural elements (Fig. 3). The fit was initialized using the number of band components and their wavenumber positions obtained from the second-derivative spectra, which are presented as insets for all spectra. All protein constructs include SUMO, N17, and Flag domains. Therefore, the amide I spectra include structural information from these segments. The SUMO structure contains β-sheet and α-helix structures (36), and N17 possesses α-helical conformation (13, 21, 37). Because all constructs have the same design, spectral differences will only arise from the glutamine sequence, which is lacking in the control construct. The fitting procedure revealed five relevant band components for the polyQ constructs (Fig. 3, a–c) and four for the control construct without a glutamine sequence (Fig. 3 d). The bands between 1633–1638 cm−1 and 1685–1687 cm−1 were assigned to the native intramolecular antiparallel β-sheets from SUMO. The band between 1656 and 1658 cm−1 arises from the α-helical conformations of SUMO and N17, and, in the cases of polyQ11, polyQ38, and polyQ56, contains a (weak) contribution of the glutamine C=O side-chain-stretching vibrations (13). The bands at 1674–1677 cm−1 indicate turn and loop structures. A band shoulder at 1644–1647 cm−1 can be observed in the second-derivative spectra for polyQ11, polyQ38, and polyQ56, but not for the control construct. Thus, this band component was assigned to the disordered structure of the polyglutamine sequence within the immobilized polyQ construct. To confirm the band assignments above, the SUMO protein alone was immobilized and measured under the same conditions as the other constructs (Fig. S3). Comparison of the amide I band shapes reveals high similarity between immobilized SUMO and the control construct without glutamines (Fig. 3 d), as expected, with a stronger component at 1657 cm−1 for the control construct because of the contribution from the α-helix of N17.
Figure 3.
Amide I spectra of the immobilized constructs (a) polyQ11, (b) polyQ38, (c) polyQ56, and (d) control (without glutamines). The constructs were bound to the Ni2+-NTA on the LB via the His-tag. The bands at 1633–1638 cm−1 and 1685–1687 cm−1 (green) indicate the β-sheets from SUMO. The band at 1674–1677 cm−1 arises from turn and loops. The band at 1656–1658 cm−1 (yellow) refers to the contributions from α-helices of SUMO and N17, overlapped by glutamine side-chain vibrations. The band at 1644–1647 cm−1 (blue) occurs from the disordered glutamine backbone and is not present in the control. Second-derivative spectra (insets) reveal the frequency positions of the amide I components for curve fitting and reconstruction of the band components. To see this figure in color, go online.
Conformational changes of polyQ upon aggregation initiation
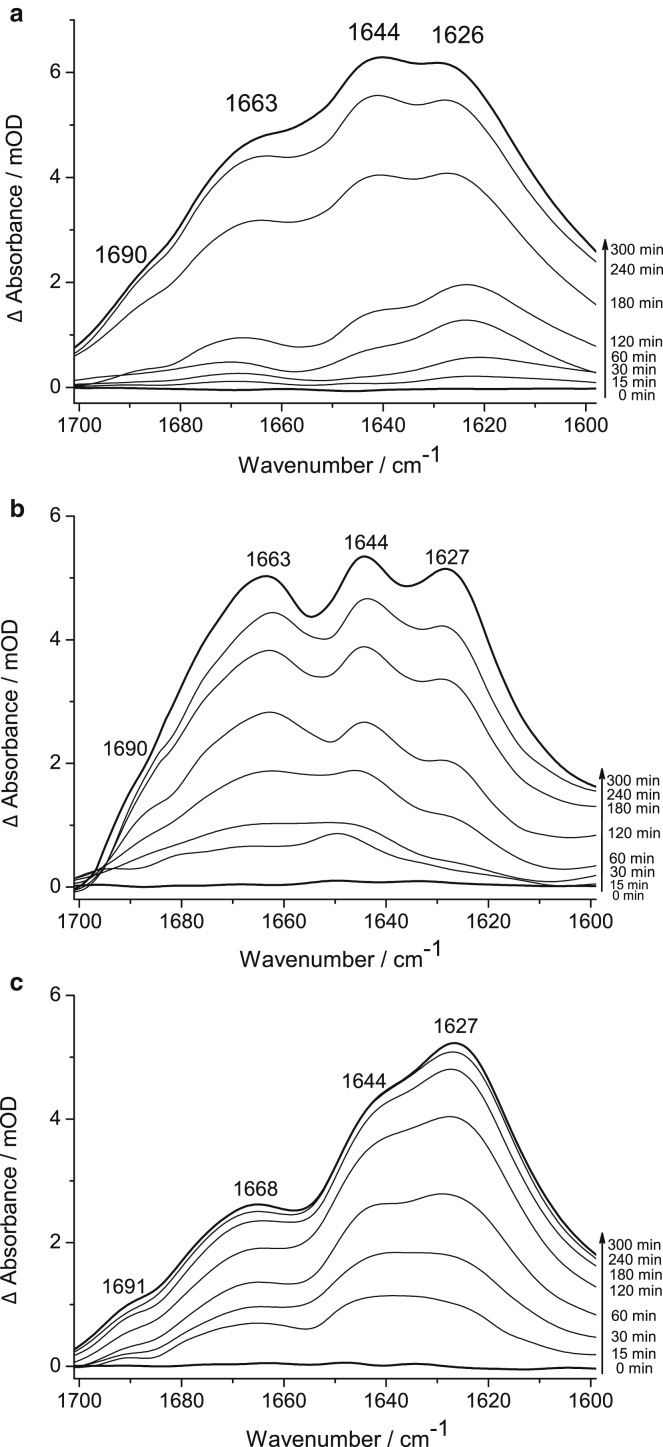
After immobilization of the constructs and amide I analysis (Fig. 3), the aggregation reaction was initiated by enzymatic cleavage with Ulp1. The released fragments consist only of N17, the polyQ sequence, and the Flag-tag, and are called N17-Q11-Flag, N17-Q38-Flag, and N17-Q56-Flag, in contrast to the entire constructs (including SUMO and His-tag), which were termed polyQ11, polyQ38, and polyQ56. Spectra were recorded in a time-dependent manner, as described in Materials and Methods, and show the conformational changes of the polyglutamine-containing fragments upon aggregation (Fig. 4, a–c). Therefore, a new background was measured immediately after addition of the protease to cancel out the (nonchanging) vibrations of the added protease Ulp1 and of the immobilized construct parts left over after cleavage (SUMO and His-tag). It is worth noting that the difference spectra reveal only positive bands. They are caused by new secondary structures that are formed during the aggregation process as well as by the progressive sedimentation of the aggregates, which also results in a band-intensity increase when the aggregates come closer to the ATR crystal surface. Sedimentation effects were reduced by a vertical ATR cell geometry in combination with the flow-through operating. Negative bands were not observed. There are no spectral indications that the aggregation process induces any significant structural changes in SUMO or in Ulp1, nor that adsorbed aggregates disrupt the LB. We could prove that the cleavage process successfully resulted in the initiation of the aggregation reaction because the rise of a new band was monitored at 1626–1627 cm−1. This band indicates the formation of aggregated β-structure and is not present in the spectra of the uncleaved constructs in Fig. 3. Applying this experimental procedure, we exclusively monitor the conformational changes of the glutamine sequence upon aggregation. We assume that the α-helical structure of N17 and also the eight amino acids of the Flag-tag stay in their conformation during the aggregation process.
Figure 4.
Conformational changes of released N17-QN-Flags upon Ulp1 cleavage and aggregation initiation, (a) N17-Q11-Flag, (b) N17-Q38-Flag, and (c) N17-Q56-Flag. The band components at 1626/1627 and 1690/1691 cm−1 are assigned to antiparallel β-structured aggregates, 1644 cm−1 to disordered, and 1662/1668 cm−1 to loop and turn structures within the polyglutamine sequences. The nonchanging contributions of the LB, the immobilized part of the construct (His-tag and SUMO), Ulp1, and H2O/buffer were subtracted as background. N17 and Flag-tag are monitored as well but are assumed to perform only minor conformational changes upon polyQ aggregation if at all.
In agreement with other studies showing that longer polyglutamine chains are prone to induce faster structural rearrangements into aggregates (13, 38, 39), we observed the formation of β-structured aggregates at 1627 cm−1 with the highest relative intensity of all amide I components for N17-Q56-Flag (Fig. 4 c). The β-sheets are in an antiparallel organization, as the occurrence of the additional band at 1691 cm−1 indicates. The other amide I components indicate that disordered (1644 cm−1) as well as loop/turn structures (1668 cm−1) are also present. N17-Q11-Flag and N17-Q38-Flag, the fragments with the polyglutamine repeats below and close to the disease threshold, also rearrange into aggregates, but slower (Fig. 4, a and b). To analyze intermediates and differences in the aggregation mechanisms dependent on the polyQ repeat length, we performed a band deconvolution and curve fitting of the amide I regions at different times (10, 20, 40, 60, and 180 min) for all fragments, as reported in detail in the next paragraph.
Determination of intermediate structures upon polyQ aggregation
Molecular characterization of intermediate structures was performed for all polyglutamine-containing fragments (N17-Q11-Flag, N17-Q38-Flag, and N17-Q56-Flag) by analysis of the amide I region 10, 20, 40, 60, and 180 min after aggregation initiation. We used two representations of the spectra, absorption (Figs. S4–S6) and second-derivative (Figs. 5, 6, and 7). The intensities of individual amide I components were compared in the absorption spectra, whereas second derivatives of the (same) spectra were used to resolve the small spectral changes in band shapes that occur when new amide I components arise. A particular focus was placed on the spectral region ∼1615–1630 cm−1, where frequency shifts reveal conformational changes within β-structured aggregates.
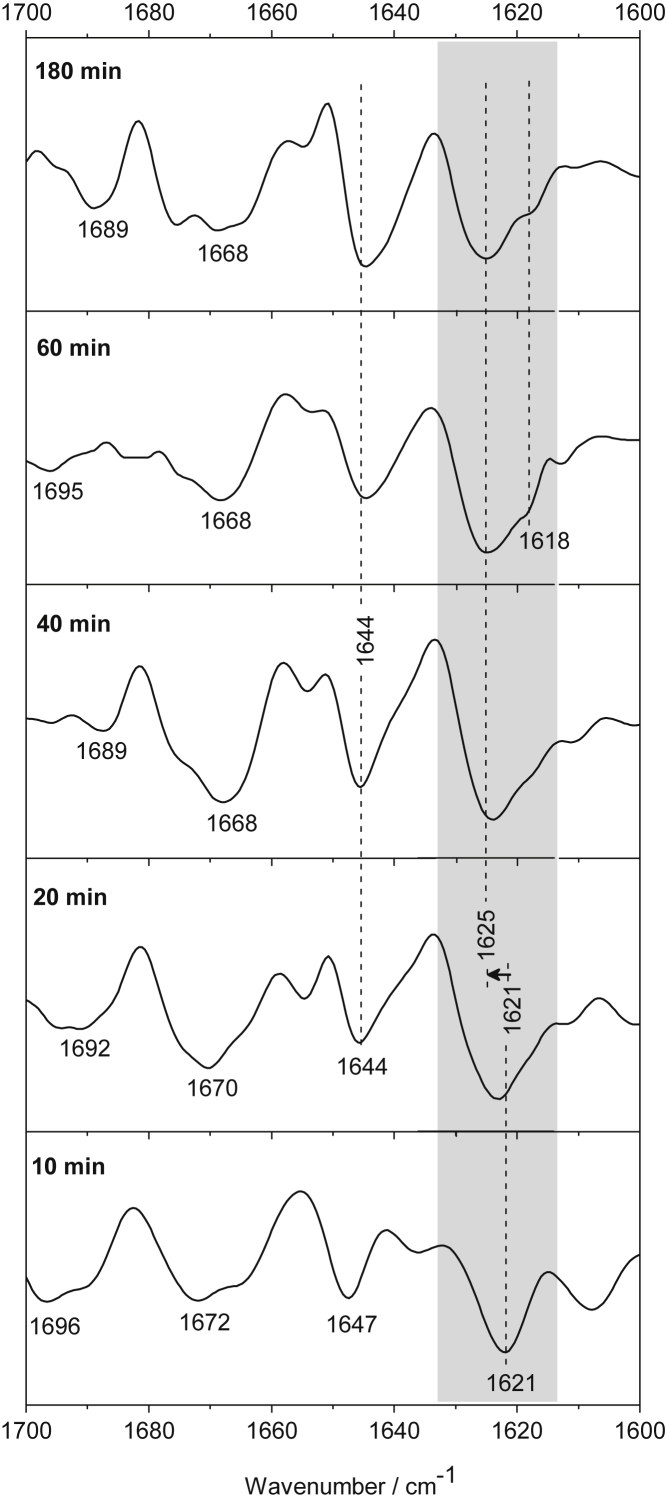
Figure 5.
Second-derivative spectra reveal structural transitions of N17-Q11-Flag, 10, 20, 40, 60, and 180 min after aggregation initiation. New band components arise in a frequency region (gray) that is characteristic for β-structured aggregates. The frequency positions indicate the formation of intermolecular β-sheets rearranging over time. The corresponding absorption spectra are presented in Fig. S4.
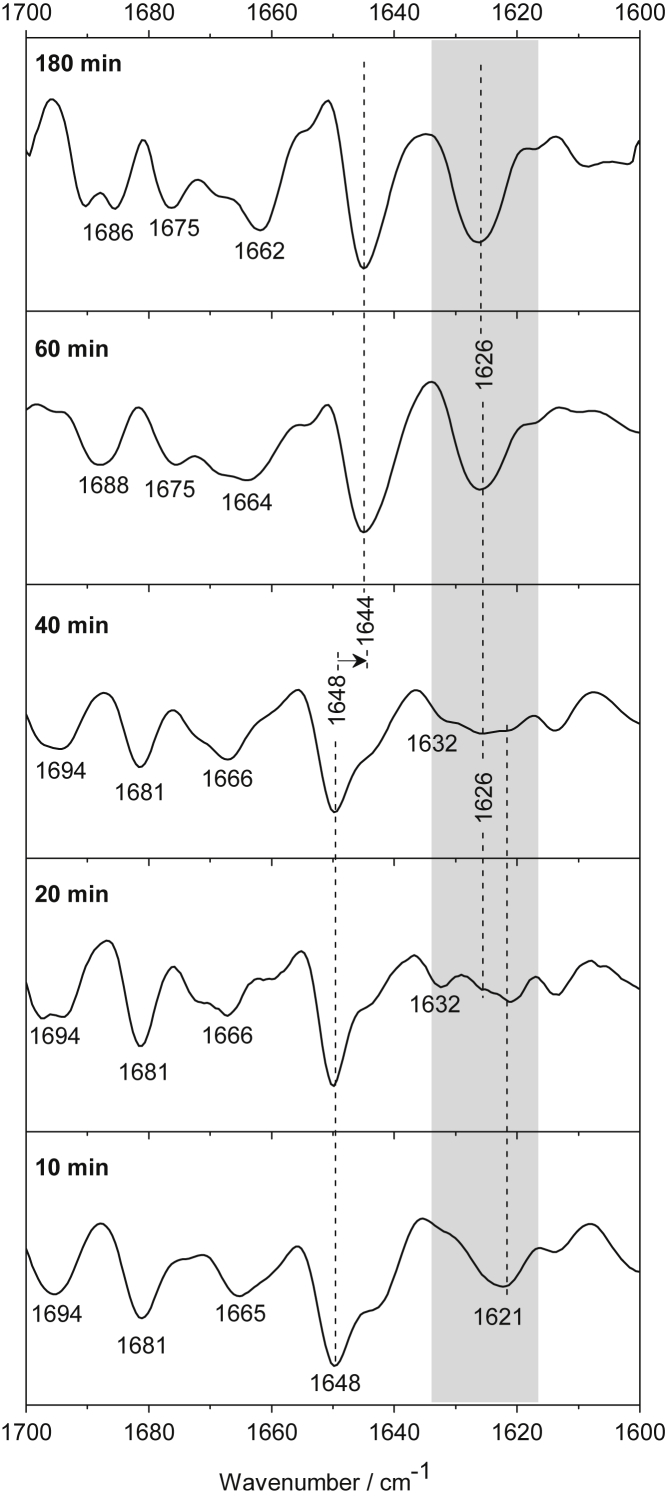
Figure 6.
Second-derivative spectra characterize structural transitions of the N17-Q38-Flag, 10, 20, 40, 60, and 180 min after aggregation initiation. The spectra indicate that a predominantly disordered structure (1648 cm−1) with minor contributions of intermolecular β-sheets (1621 cm−1) undergoes conformational changes to intramolecular β-sheets (1632 cm−1) and expanded β-structured aggregates (1626 cm−1). Corresponding absorption spectra are presented in Fig. S5.
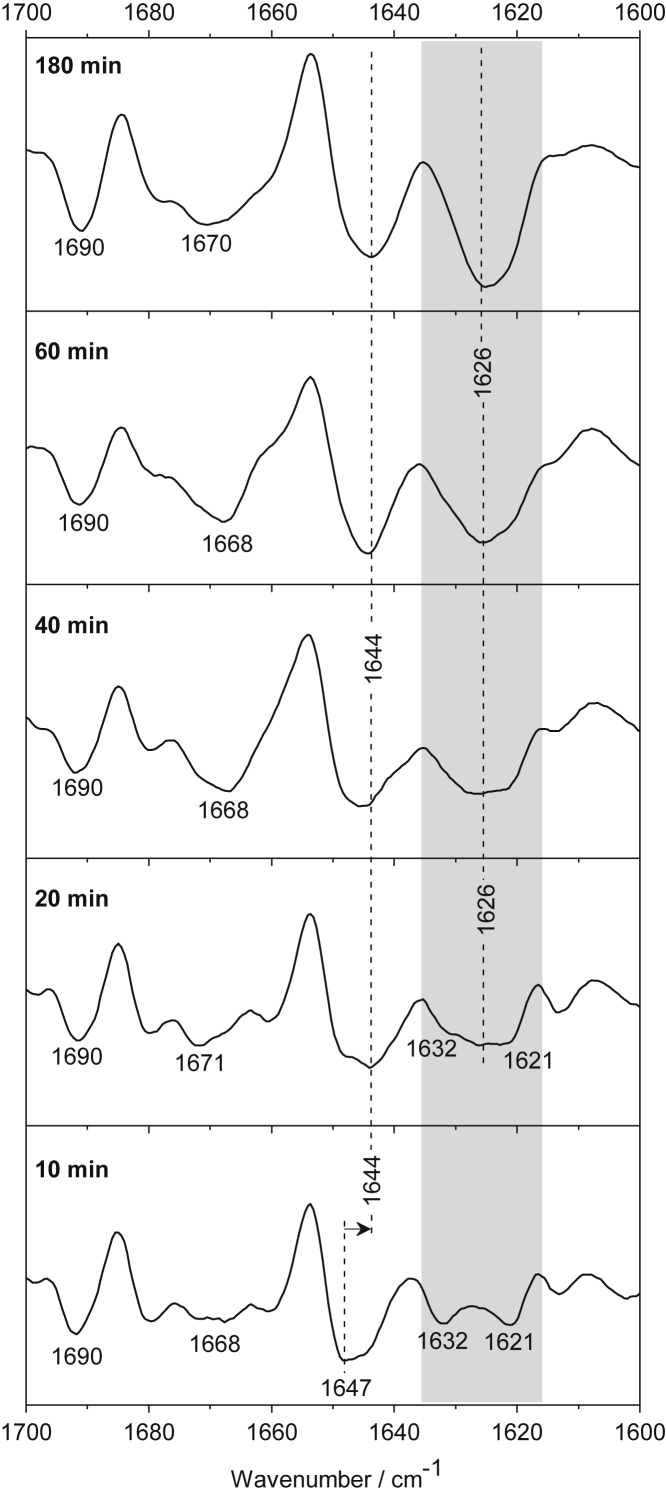
Figure 7.
Second-derivative spectra of the N17-Q56-Flag characterize aggregate formation after 10, 20, 40, 60, and 180 min. The minima at 1621 and 1632 cm−1 reveal that inter- and intramolecular β-sheets are already formed at early stages and further rearrange into expanded β-structured aggregates (1626 cm−1). The corresponding absorption spectra are presented in Fig. S6.
N17-Q11-Flag
Despite the short polyglutamine length of 11 residues, we observed that N17-Q11-Flag has the ability to form β-structures. The rise of the band at 1621 cm−1 within 10 min reveals the beginning of the aggregation process (Fig. 5). The frequency position indicates intermolecular interactions that are likely formed between glutamine stretches. After ∼40 min, the band shifts up to 1625 cm−1, and an additional band component appears at 1618 cm−1. These frequency shifts reveal structural rearrangements and/or the growing of the β-structured aggregate. The occurrence of several components below 1630 cm−1 hints at the heterogeneity of β-structured aggregates that develop over time. The disordered structure (1644 cm−1), which is minor at the beginning, becomes quite intense after 60 min (clearly obvious in Fig. S4), indicating that disordered polyQ regions also contribute to the structure of the aggregates beside the β-sheet. The weak bands at 1689/1697 cm−1 show that the β-strands are oriented in an antiparallel manner, and the bands within the 1672–1668 cm−1 region reveal some contribution of loop/turn configurations.
N17-Q38-Flag
At the beginning of the aggregation process (after 10–20 min), the main structural component of the N17-Q38-Flag fragments is disordered, as indicated by the most intense amide I component at 1648 cm−1 in the absorption spectra (Fig. S5) and the minimum in the second derivative, respectively (Fig. 6). Intermolecular β-sheets are also present in the early stage of the aggregation process (1621 cm−1), but only to a minor degree. It is noticeable that, after 20 min, a minimum at 1632 cm−1 appears and hints at the formation of an intramolecular β-sheet, demonstrating that N17-Q38-Flag fragments are long enough to form intramolecular β-sheets. The bands at 1694/1681 cm−1 indicate that the β-structures are in antiparallel organization. Interestingly, between 20 and 40 min, a new peak at 1626 cm−1 appears in the spectra concurrently with still-existing minima at 1621 and 1632 cm−1, showing the coexistence of different β-structures. With further measuring time (60–180 min), the contour of the band at 1626 cm−1 becomes sharper, revealing the formation of an expanded β-sheet structure. Presumably, this band characterizes the formation of a β-structured fibril.
N17-Q56-Flag
The longest polyglutamine-containing fragment, N17-Q56-Flag, reveals significant structural transitions immediately after aggregation initiation (Fig. 7). When the aggregation process starts (within 10 min), N17-Q56-Flag is in disordered structure (1647 cm−1), but already with significant contributions from intramolecular and intermolecular β-sheets, as observed by the distinct minima at 1632 and 1621 cm−1, respectively. As in N17-Q38-Flag, a new peak at 1626 cm−1 arises, but earlier (after 20 min). The band at 1626 cm−1 continuously sharpens and increases in the second-derivative and absorption spectra (Fig. S6) and dominates in the spectra after 40 min, indicating the expansion of β-structured aggregates, presumably fibrillar structures. The band at 1690 cm−1 hints that the β-structures are in antiparallel arrangement. Besides β-structure, the aggregates possess significant fractions of disordered (1647/1644 cm−1) and loop/turn structures (1668–1670 cm−1).
Control experiments
The construct without any polyglutamine sequences was measured as a control (Fig. 1). Release of the N17-Flag from the immobilized construct by incubation with Ulp1 protease was performed under the same experimental conditions as for the polyglutamine-containing constructs. After enzymatic cleavage, no band arises that would indicate the formation of any β-sheet structures (Fig. S7). A weak band at 1657 cm−1 is observable, which we assign to small changes in the α-helical structure of N17 when it is released into the solution, but obviously neither aggregation nor unfolding occurs. As an additional control experiment and to validate any contribution from the enzyme, we measured the amide I absorption of Ulp1 (Fig. S8). The secondary structure of Ulp1 consists of α-helices and β-sheets (24). Curve fitting of the amide I spectrum revealed a band component at 1654 cm−1, indicating the α-helical fraction of Ulp1, as well as band components at 1633 and 1682 cm−1. These frequency positions are characteristic for antiparallel β-sheets within proteins. No band components were detected below 1630 cm−1, and thus there are no indications for Ulp1 aggregation.
Discussion
We established an IR spectroscopic approach to analyze polyQ aggregation pathways dependent on the polyQ repeat length. To characterize transient conformations and to identify differences in aggregation pathways, the analytical method needs to fulfill several prerequisites. First, the expanded polyQ sequences have to be kept soluble until the aggregation reaction is initiated. Therefore, we used polyQ constructs comprising a SUMO protein at the N-terminus, which keeps the polyQ sequence soluble until it is cleaved. In addition, all constructs had the same constituents and varied only in the length of the polyQ stretch from below, close to, and above the HD threshold. Second, the time point of aggregation initiation has to be controlled if kinetics and transient states are to be investigated. Enzymatic cleavage was used to trigger the aggregation reaction and to monitor aggregation-induced conformational changes of the released polyQ sequence. The released part of the construct included the Htt’s N-terminus N17, which is assumed to play a crucial role in the aggregation process, beside the polyQ stretch. Thus, the design of the constructs facilitated them to mimic on one hand the biologically relevant sequence lengths and on the other hand the Htt proteinogenic neighboring residues. Third, the experimental approach should be suitable for low concentrations because aggregation processes are concentration dependent. Using ATR mode, the strong absorbance of water (overlapping the amide I region) is reduced in comparison to the transmission mode. Thus, we could work with absolute concentrations less than 50 μg. This was the amount of polyQ constructs used for the experiments; however, only a fraction is immobilized and measured in the experiments. The immobilization approach allows us to vary the density of the immobilization sites and thus to avoid intermolecular aggregation of the constructs attached to the IRE before the aggregation process is initiated. Using a vertical ATR cell geometry in combination with a flow-through system minimizes sedimentation effects. Fourth, β-structured aggregates are identifiable by IR spectroscopy. Aggregates absorb at wavenumbers (typically below 1630 cm−1) that are lower than those of native β-sheets in proteins and thus can be distinguished. The formation of transient structures can be observed and characterized by frequency variations dependent on time.
A major aim of our study was to investigate aggregation processes of polyQ repeat lengths below, close to, and above the disease threshold. IR absorption spectra (Figs. S4–S6) and corresponding second derivatives (Figs. 5, 6, and 7) were analyzed dependent on time to characterize different intermediate stages of the polyQ fragments N17-Q11-Flag, N17-Q38-Flag, and N17-Q56-Flag. Second-derivative spectra resolve small spectral changes better, and thus the rise of new structures; however, they do not report on band intensities. Absorption spectra allow the comparison of band intensities and hence provide information about the contribution of individual structural elements. To determine amide I components, we performed a secondary structure analysis by band deconvolution and curve fitting. A well-reconstructed amide I absorption spectrum can be achieved in several ways, e.g., by varying the number of band components or by the choice of the band profiles, and the interpretation of deconvoluted amide I components certainly has to be done very carefully. To perform comparative amide I band deconvolutions as objectively as possible, we used the same analysis criteria for all polyQ constructs. Frequency positions of amide I components were derived from the second-derivative spectra, and band components of similar frequencies were assigned to the same structural elements, i.e., disordered (1644–1648 cm−1), loop/turn (1665–1681 cm−1), intramolecular β-sheet (∼1632 cm−1), intermolecular β-sheet (1626 cm−1 and below), and antiparallel organization of the strands (1685–1696 cm−1). Analysis of aggregation pathways is complex because the polyQ fragments can possess several structural arrangements simultaneously, and common as well as individual properties were found for the aggregation processes of the various polyQ repeat lengths.
All fragments show the formation of β-structured aggregates in time, but there are fundamental differences between the short fragment N17-Q11-Flag (Figs. 5 and S4), which is far below the disease threshold, and the fragments close to and above it. For N17-Q11-Flag, there is no amide I component observable at 1632 cm−1, although it is already detectable in the early stages of both N17-Q32-Flag (Figs. 6 and S5) and N17-Q56-Flag (Figs. 7 and S6). The band at 1632 cm−1 is indicative of the transition of the polyQ sequence to an intramolecular β-sheet, probably with very flexible turn/loop regions. In contrast, the N17-Q11-Flag exhibits only bands at frequencies below 1625 cm−1, which are indicative for expanded β-structure. We conclude that the glutamine sequence of the N17-Q11-Flags is too short to form hairpins, but the fragments can arrange in an expanded intermolecular β-sheet that consists of stretched monomers in an antiparallel orientation, as indicated by the band components >1688 cm−1. In the early stages of the aggregation process of N17-Q11-Flag, the growth of the β-structure dominates. However, the fraction of disordered structure gains significantly after 60 min, which is clearly observable in the absorption spectra (Fig. S4). This suggests that disordered fragments still remain after seeding the aggregate growth and were probably not incorporated into the expanded β-structure. For N17-Q38-Flag and N17-Q56-Flag fragments, we observe the formation of inter- and intramolecular β-structures (at 1621 and 1632 cm−1, respectively) in the early stages of the aggregation process. Because of the length and high flexibility of the polyQ sequences, we assume that inter- and intramolecular β-structures are formed simultaneously in the early stages of the aggregation process. The intramolecular sheets might serve as a template to which other disordered monomers bind, assisting the elongation of the aggregate to an expanded β-structure. The growth of an expanded β-aggregate is supported by a band component at ∼1626 cm−1 evolving and strongly increasing at later stages (Figs. S5 and S6). In comparison to N17-Q38-Flag, conformational changes of β-structures occur significantly faster in N17-Q56-Flag and result in a β-structured aggregate (1626 cm−1) that dominates over all other structural components.
We observed that disordered polyQ regions contribute significantly to the aggregates for all investigated constructs. The importance of disordered polyQ regions has also been found by other studies, which have shown that aggregates in the cerebral area consist of amyloid conformation and so-called β-sheet/unordered structure (40, 41). It is assumed that such a combination of structures is more toxic for cells than amyloid inclusions formed only by β-sheets, presumably due to the intense dynamics of the partially disordered state within these conformational ensembles. Thus, they are capable of deleterious interactions with various cellular elements. It is noticeable that the disordered structure for all polyglutamine-containing fragments exhibits the same band frequency at 1644 cm−1. In addition, all polyQ fragments contain a high amount of loop/turn structures (the region 1660–1680 cm−1). The fragments with longer polyglutamine sequences, N17-Q38-Flag and N17-Q56-Flag, showed significant conformational transitions to loops/turns already at the beginning of the aggregation process. We assume that the just-released fragments are mainly in a disordered structure and form loose β-turns as an initial structure for the following conversion into β-sheets. This suggestion is supported by x-ray studies showing that a Huntingtin-exon 1 fragment with Q36 exhibits a conformational flexibility and possesses a sequential transition from a loop to a β-hairpin conformation (37). In contrast to N17-Q38-Flag and N17-Q56-Flag, the short N17-Q11-Flag showed a negligible fraction of loop/turn structures in the early spectra.
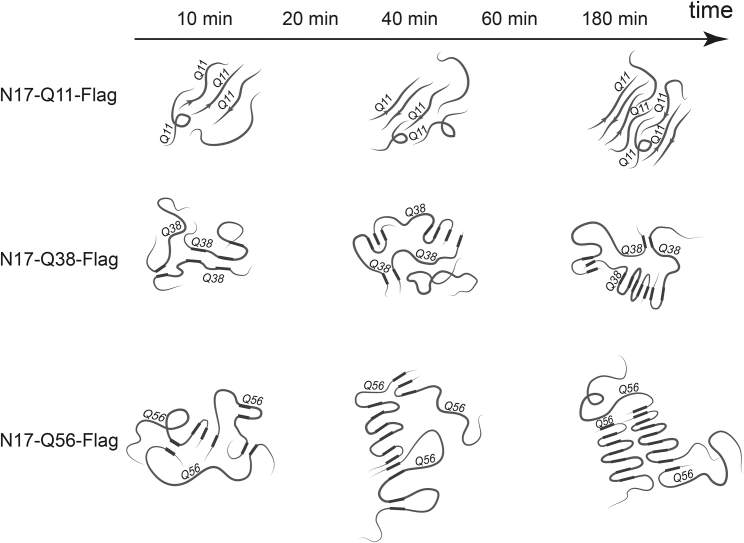
Based on these analyses, we tentatively propose aggregation pathways for N17-QN-Flag dependent on the glutamine repeat length (N = 11, 38, 56) as illustrated in Fig. 8. Aggregation starts from disordered monomers. For N17-Q11-Flag, the short polyQ repeats assemble as fairly stretched and opposite-directed monomers to an intermolecular antiparallel β-structured aggregate that still contains disordered regions. The growth of such assemblies can lead to the formation of large aggregates, as an atomic force microscopy study has shown for a short synthetic polyglutamine-containing peptide (K-Q7-K) (39). For the longer fragments, N17-Q38-Flag and N17-Q56-Flag, the monomers undergo conformational changes, including loops and loose turns, and thus can form intramolecular interactions, even if weak. Sequence parts assemble more and more to expanded β-structured aggregates with intermolecular interactions formed between strands. Although the disordered structure is initially predominant, we also observe a minor fraction of intramolecular β-sheets which are only present for the longer glutamine sequences. With the following rearrangements, new β-structures form, likely because of stronger hydrogen bonding or/and tighter packing of the β-strands within the aggregates. The further growing process might occur by addition of other disordered monomers. Presumably, disordered monomers attach to the already ordered β-sheets and then undergo structural rearrangements. For N17-Q56-Flag, this process occurs faster because longer glutamine chains are more likely to build amyloid-like aggregates (38).
Figure 8.
Proposed aggregation pathways of N17-QN-Flag fragments dependent on the glutamine repeat length. N17-Q11-Flag fragments tend to form extended β-sheets with strands that consist of stretched monomers. 11 glutamines are too short to form turns. The β-sheets are stabilized by intermolecular interactions. In contrast, the longer N17-Q38-Flag and N17-Q56-Flag sequences are highly flexible to change conformation and to form turns and loops. Sequence parts assemble to intra- and intermolecular β-structured aggregates, which still have a significant number of disordered regions. The aggregation kinetics of N17-Q56-Flag is faster than that of N17-Q38-Flag. It is likely that the Q56 conformers accumulate further into higher-ordered aggregate morphologies.
Conclusions
An IR spectroscopic approach was established to study polyQ aggregate formation on a molecular level using polyQ constructs with glutamine lengths below, close to, and above the HD threshold. By a combination of ATR-FTIR difference spectroscopy and protein immobilization, we were able to investigate small concentrations, thereby reducing the probability of spontaneous aggregation. Controlled initiation of the aggregation process facilitated the structural characterization of intermediates in dependence of time. We proposed aggregation pathways for the various polyQ repeat lengths.
Acknowledgments
We thank R. Schlömer for excellent technical assistance and gratefully acknowledge financial support by the Deutsche Forschungsgemeinschaft (SFB 969, A1, and A2).
Editor: Arne Gericke.
Footnotes
Eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30298-4.
Supporting Citations
Reference (42) appears in Supporting Material.
Supporting Material
References
- 1.Margolis R.L., Ross C.A. Expansion explosion: new clues to the pathogenesis of repeat expansion neurodegenerative diseases. Trends Mol. Med. 2001;7:479–482. doi: 10.1016/s1471-4914(01)02179-7. [DOI] [PubMed] [Google Scholar]
- 2.Hands S.L., Wyttenbach A. Neurotoxic protein oligomerisation associated with polyglutamine diseases. Acta Neuropathol. 2010;120:419–437. doi: 10.1007/s00401-010-0703-0. [DOI] [PubMed] [Google Scholar]
- 3.Gusella J.F., MacDonald M.E. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat. Rev. Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 4.Bates G., Benn C. Huntington’s Disease. Oxford University Press; 2002. The polyglutamine diseases; pp. 429–472. [Google Scholar]
- 5.Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 6.Milewski M., Gawliński P., Bal J. Complex interplay between the length and composition of the huntingtin-derived peptides modulates the intracellular behavior of the N-terminal fragments of mutant huntingtin. Eur. J. Cell Biol. 2015;94:179–189. doi: 10.1016/j.ejcb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Chen S., Ferrone F.A., Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc. Natl. Acad. Sci. USA. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walters R.H., Murphy R.M. Aggregation kinetics of interrupted polyglutamine peptides. J. Mol. Biol. 2011;412:505–519. doi: 10.1016/j.jmb.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates G.P., Dorsey R., Tabrizi S.J. Huntington disease. Nat. Rev. Dis. Primers. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 10.Scherzinger E., Lurz R., Wanker E.E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 11.Goormaghtigh E., Raussens V., Ruysschaert J.M. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta. 1999;1422:105–185. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 12.Sarroukh R., Goormaghtigh E., Raussens V. ATR-FTIR: a “rejuvenated” tool to investigate amyloid proteins. Biochim. Biophys. Acta. 2013;1828:2328–2338. doi: 10.1016/j.bbamem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman M., Kodali R., Wetzel R. Slow amyloid nucleation via α-helix-rich oligomeric intermediates in short polyglutamine-containing huntingtin fragments. J. Mol. Biol. 2012;415:881–899. doi: 10.1016/j.jmb.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heck B.S., Doll F., Hauser K. Length-dependent conformational transitions of polyglutamine repeats as molecular origin of fibril initiation. Biophys. Chem. 2014;185:47–57. doi: 10.1016/j.bpc.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Natalello A., Frana A.M., Regonesi M.E. A major role for side-chain polyglutamine hydrogen bonding in irreversible ataxin-3 aggregation. PLoS One. 2011;6:e18789. doi: 10.1371/journal.pone.0018789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen S., Dash D., Brahmachari S.K. Role of histidine interruption in mitigating the pathological effects of long polyglutamine stretches in SCA1: a molecular approach. Protein Sci. 2003;12:953–962. doi: 10.1110/ps.0224403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma D., Sharma S., Brahmachari S.K. Peptide models for inherited neurodegenerative disorders: conformation and aggregation properties of long polyglutamine peptides with and without interruptions. FEBS Lett. 1999;456:181–185. doi: 10.1016/s0014-5793(99)00933-3. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M., Morishima I., Nukina N. Intra- and intermolecular β-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J. Biol. Chem. 2001;276:45470–45475. doi: 10.1074/jbc.M107502200. [DOI] [PubMed] [Google Scholar]
- 19.Thakur A.K., Jayaraman M., Wetzel R. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scior A., Preissler S., Deuerling E. Directed PCR-free engineering of highly repetitive DNA sequences. BMC Biotechnol. 2011;11:87. doi: 10.1186/1472-6750-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivanandam V.N., Jayaraman M., van der Wel P.C. The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 2011;133:4558–4566. doi: 10.1021/ja110715f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetzel R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J. Mol. Biol. 2012;421:466–490. doi: 10.1016/j.jmb.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malakhov M.P., Mattern M.R., Butt T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 24.Mossessova E., Lima C.D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 25.Richter R., Mukhopadhyay A., Brisson A. Pathways of lipid vesicle deposition on solid surfaces: a combined QCM-D and AFM study. Biophys. J. 2003;85:3035–3047. doi: 10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Güldenhaupt J., Adigüzel Y., Gerwert K. Secondary structure of lipidated Ras bound to a lipid bilayer. FEBS J. 2008;275:5910–5918. doi: 10.1111/j.1742-4658.2008.06720.x. [DOI] [PubMed] [Google Scholar]
- 27.Saakov V.S. Derivative Spectrophotometry and Electron Spin Resonance (ESR) Spectroscopy for Ecological and Biological Questions. Springer; 2013. Basis of derivative spectrophotometry; pp. 5–70. [Google Scholar]
- 28.Venyaminov SYu, Kalnin N.N. Quantitative IR spectrophotometry of peptide compounds in water (H2O) solutions. I. Spectral parameters of amino acid residue absorption bands. Biopolymers. 1990;30:1243–1257. doi: 10.1002/bip.360301309. [DOI] [PubMed] [Google Scholar]
- 29.Jackson M., Mantsch H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995;30:95–120. doi: 10.3109/10409239509085140. [DOI] [PubMed] [Google Scholar]
- 30.Fabian H., Mäntele W. Handbook of Vibrational Spectroscopy. Biochemical Applications John Wiley & Sons Ltd.; 2002. Infrared spectroscopy of proteins; pp. 1–27. [Google Scholar]
- 31.Arrondo J.L.R., Muga A., Goñi F.M. Quantitative studies of the structure of proteins in solution by Fourier-transform infrared spectroscopy. Prog. Biophys. Mol. Biol. 1993;59:23–56. doi: 10.1016/0079-6107(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 32.Dong A., Matsuura J., Carpenter J.F. Infrared and circular dichroism spectroscopic characterization of structural differences between β-lactoglobulin A and B. Biochemistry. 1996;35:1450–1457. doi: 10.1021/bi9518104. [DOI] [PubMed] [Google Scholar]
- 33.Barth A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta. 2007;1767:1073–1101. doi: 10.1016/j.bbabio.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Jackson M., Haris P.I., Chapman D. Fourier transform infrared spectroscopic studies of Ca(2+)-binding proteins. Biochemistry. 1991;30:9681–9686. doi: 10.1021/bi00104a016. [DOI] [PubMed] [Google Scholar]
- 35.Miyazawa T., Blout E.R. The infrared spectra of polypeptides in various conformations: amide I and II bands. J. Am. Chem. Soc. 1961;83:712–719. [Google Scholar]
- 36.Ding H., Xu Y., Shi Y. Solution structure of human SUMO-3 C47S and its binding surface for Ubc9. Biochemistry. 2005;44:2790–2799. doi: 10.1021/bi0477586. [DOI] [PubMed] [Google Scholar]
- 37.Kim M. Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues. Prion. 2013;7:221–228. doi: 10.4161/pri.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morley J.F., Brignull H.R., Morimoto R.I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Legleiter J., Mitchell E., Muchowski P.J. Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J. Biol. Chem. 2010;285:14777–14790. doi: 10.1074/jbc.M109.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.André W., Sandt C., Hoffner G. Structure of inclusions of Huntington’s disease brain revealed by synchrotron infrared microspectroscopy: polymorphism and relevance to cytotoxicity. Anal. Chem. 2013;85:3765–3773. doi: 10.1021/ac400038b. [DOI] [PubMed] [Google Scholar]
- 41.Nekooki-Machida Y., Kurosawa M., Tanaka M. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl. Acad. Sci. USA. 2009;106:9679–9684. doi: 10.1073/pnas.0812083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rigler P., Ulrich W.P., Vogel H. Reversible immobilization of peptides: surface modification and in situ detection by attenuated total reflection FTIR spectroscopy. ChemPhysChem. 2003;4:268–275. doi: 10.1002/cphc.200390043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.