Abstract
Structural RNA domains are widely involved in the regulation of biological functions, such as gene expression, gene modification, and gene repair. Activity of these dynamic regions depends sensitively on the global fold of the RNA, in particular, on the binding affinity of individual conformations to effector molecules in solution. Consequently, both the 1) structure and 2) conformational dynamics of noncoding RNAs prove to be essential in understanding the coupling that results in biological function. Toward this end, we recently reported observation of three conformational states in the metal-induced folding pathway of the tRNA-like structure domain of Brome Mosaic Virus, via single-molecule fluorescence resonance energy transfer studies. We report herein selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE)-directed structure predictions as a function of metal ion concentrations ([Mn+]) to confirm the three-state folding model, as well as test 2° structure models from the literature. Specifically, SHAPE reactivity data mapped onto literature models agrees well with the secondary structures observed at 0–10 mM [Mg2+], with only minor discrepancies in the E hairpin domain at low [Mg2+]. SHAPE probing and SHAPE-directed structure predictions further confirm the stepwise unfolding pathway previously observed in our single-molecule studies. Of special relevance, this means that reduction in the metal-ion concentration unfolds the 3′ pseudoknot interaction before unfolding the long-range stem interaction. This work highlights the synergistic power of combining 1) single-molecule Förster resonance energy transfer and 2) SHAPE-directed structure-probing studies for detailed analysis of multiple RNA conformational states. In particular, single-molecule guided deconvolution of the SHAPE reactivities permits 2° structure predictions of isolated RNA conformations, thereby substantially improving on traditional limitations associated with current structure prediction algorithms.
Introduction
The structure and dynamics of catalytically active, noncoding RNAs represent a highly active field that promises new insights into the regulation of fundamental biological processes with applications in nanotechnology and nanomedicine (1, 2, 3, 4). Biological functions that are regulated through noncoding RNAs are observed across many forms of life, including bacteria (5), viruses (6, 7, 8), plants (9), and humans (1, 10). Processes that involve RNA-mediated regulation cover essential cellular mechanisms such as gene expression (5), gene modification (11, 12), and gene repair (13, 14). As a result of this functional diversity, noncoding RNAs are observed in great variety, e.g., from individual small interfering RNA to structure-forming domains on long mRNA transcripts that act as riboswitches (10, 15) and regulatory elements on viral genomes.
Despite this variability in function, these elements all have one common denominator: their biological activity depends on the formation of recognizable three-dimensional structures commensurate with timescales for the subsequent biological processes in living systems (16). Essential in the formation of active structures are tertiary contacts (17) that stabilize the global fold of the RNA (18). Intramolecular interactions require that the molecule overcome the electrostatic repulsion from the negatively charged phosphate backbone (19). This interplay of attractive tertiary interactions, electrostatic repulsion from the RNA backbone, and charge neutralization from the ionic environment, provides an intricate landscape for RNA folding (20). As a result, RNA molecules can sample multiple conformations, leading to heterogeneity and conformational distributions in the ensemble equilibrium (21, 22).
Under physiological conditions, such structural dynamics often provide an important mechanism for active sampling of environmental factors, e.g., metabolites in riboswitch regulation. Valuable insights into the function of active domains can thus be gleaned from studying RNA structure and folding dynamics. To this end, a particularly useful probe is the process of metal-ion [Mn+]-induced RNA folding, whereby increase in the counterion concentration reduces electrostatic repulsion through site-specific coordination and nonspecific electrostatic screening (19, 23, 24). [Mn+]-induced folding then provides a means to control the thermodynamic equilibrium and inspect the structure interconversion mechanisms. Of special relevance, RNA structure and dynamics can be accessed in combination with 1) selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) structure probing and 2) single-molecule Förster resonance energy transfer studies (smFRET) (25, 26, 27, 28), which this study demonstrates to be a particularly powerful, synergistic combination.
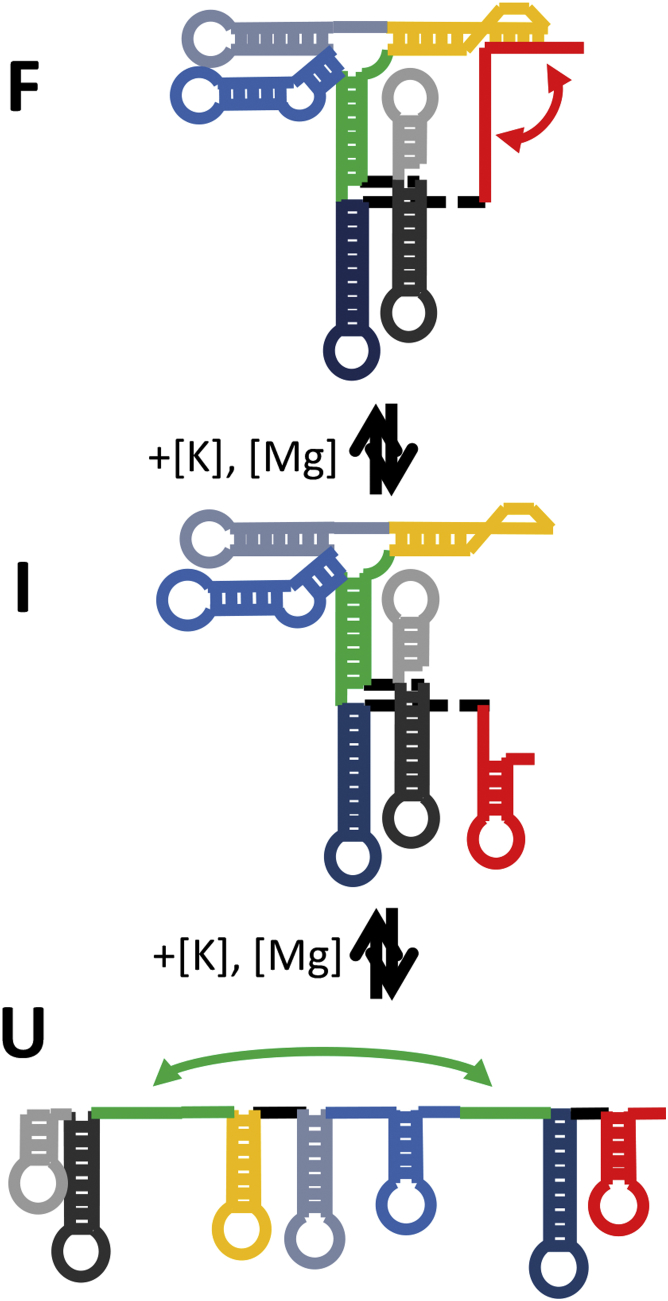
By way of example, we have recently performed smFRET folding studies of the tRNA-like structure (TLS) domain in Brome Mosaic Virus (BMV) (8). This TLS domain has previously been implicated as an essential structural feature in the regulation of the BMV replication cycle (29, 30). Population analysis has revealed an intricate folding scheme of at least three distinct conformations, depending on mono- and divalent ion concentration levels in solution (see Fig. 1). Under saturating [Mn+] conditions, the TLS presents itself in a compact (F) conformation stabilized by two intramolecular 1) stem and 2) pseudoknot (PSK) interactions. Reduction in the metal-ion concentration unfolds the 3′ PSK interaction before unfolding the long-range stem interaction. This leads to a three-state, sequential folding pathway (F ↔ I ↔ U) that therefore predicts significant concentrations of interconverting F and I under physiologically relevant salt conditions. In this article, we perform SHAPE probing studies on the BMV TLS domain to demonstrate the complementarity of [Mn+]-dependent structure probing and single-molecule folding in correctly deconstructing the multiple RNA conformations that can occur under physiologically relevant and structurally heterogeneous solution environments.
Figure 1.
Stepwise folding dynamics in the TLS of BMV. The F conformation at high salt is stabilized by two intermolecular interactions—a stem (green) and a PSK (red). Decrease in [Mn+] first breaks the 3′ PSK interaction before breaking the long-range stem. To see this figure in color, go online.
In the following, we present results from SHAPE structure and smFRET investigations of the TLS domain for BMV RNA3 over a systematic series of monovalent [Na+] and divalent [Mg2+] cation conditions. Structure probing and single-molecule folding studies are performed at identical conditions and compared to currently accepted 2° structure models for F and U in the literature. SHAPE-directed secondary structure predictions supplement single-molecule folding studies, and thereby demonstrate strong confirmation of our previously proposed three-step folding model. We further show that deconvolution of SHAPE probing data using smFRET fractional populations provides a powerful means to compensate for 1) conformational heterogeneity and 2) overinterpretation of basepairing probabilities at low [Mn+] conditions. This work highlights the powerful combination of smFRET methods with SHAPE-directed 2° structure predictions, in particular, for detailed elucidation of biologically active conformational states as a function of environmental conditions.
Materials and Methods
RNA preparation for single-molecule and structure probing experiments
SHAPE constructs are prepared following Wilkinson et al. (31) by extension of the desired gene product, the 169-nucleotide (nt) minimum TLS element of BMV RNA3, by 14-nt 5′ and 43-nt 3′ structure cassettes. The resulting 251-nt DNA template (including a 7-nt spacer and 18-nt T7 promoter) is built up from eight custom primers purchased from Integrated DNA Technologies (Coralville, IA) using the Phusion High Fidelity PCR Kit (New England Biolabs, Ipswitch, MA). The quality of the PCR reaction is assessed in denaturing polyacrylamide gel electrophoresis and DNA sequencing. PCR products are transcribed in vitro using the T7 High Yield RNA Synthesis Kit (New England Biolabs). Transcribed RNA is purified on a native polyacrylamide gel, passively eluted into TE buffer (10 mM TrisHCl, 1 mM EDTA), and concentrated using Amicon Ultra 15 Centrifugal Filter Units (Millipore, Billerica, MA).
RNA constructs for single-molecule FRET are prepared as described previously. In brief, the RNA construct is designed around a FRET-pair consisting of a 3′ Cy3 and a 5′ Cy5 fluorophore for observation of conformational dynamics. For utilization in smFRET microscopy, the 169-nt TLS sequence is 5′ extended by 20 nt and the resulting 214-nt DNA template (including a 7-nt spacer and 18-nt T7 promoter) transcribed, as described above. The FRET pair is incorporated posttranscriptionally via 3′ hydrazide end-labeling and annealing of the 5′ extension to a complementary Cy5- and biotin-labeled oligomer (5′-UGU GU/iCy5/GUG UGU GUG UGC GGG CCC/BioTEG/-3′, Integrated DNA Technologies) (32). The doubly labeled FRET-competent Cy3/Cy5 RNA constructs are purified using ion-pair reverse-phase chromatography.
SHAPE assay
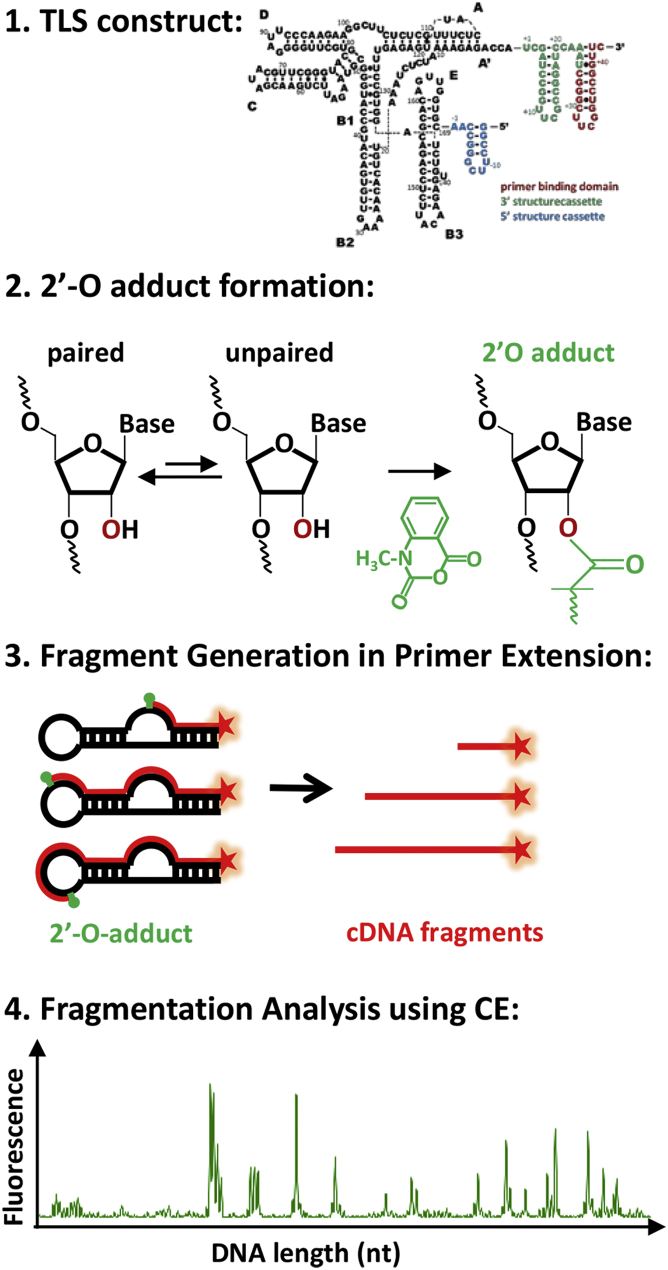
SHAPE assays are performed according to McGinnis et al. (33) as outlined in Fig. 2. Briefly, 2 pmol RNA is denatured at 95°C for 5 min and refolded at the desired probing conditions. Solutions are equilibrated at RT for 15 min and split in two separate reaction tubes for 1) SHAPE probing (+) and 2) control (−) experiments. N-methylisatoic anhydride, freshly dissolved in dimethyl sulfoxide, is added to the probing (+) reaction at 5 mM, whereas neat dimethyl sulfoxide is added to the control (−) reaction. Both samples are incubated for 4 h at RT to allow for 2′O-adduct formation, then ethanol-precipitated and resuspended in 10 μL HPLC water. 2′O-adduct sites are converted to DNA fragments using primer extension with SuperScript III Reverse Transcriptase (Life Technologies, Carlsbad, CA). Specifically, resuspended RNA is denatured at 65°C for 5 min and annealed to fluorescent dye-labeled cDNA primers (VIC; Applied Biosystems, Foster City, CA) at 37°C for 1 min. TLS-cDNA complexes are then primer-extended to yield fluorescently (VIC) labeled ssDNA fragments of lengths corresponding to the 2′O-adduct position at the modified RNA. Sequencing reactions are performed on untreated RNA using the same primer extension procedure but with the addition of 5 mM dideoxynucleotides (Trilink Biotechnologies, San Diego, CA) and NED-labeled cDNA primers. Samples from sequencing reaction and primer extensions are finally combined, ethanol-precipitated, washed twice with 70% cold ethanol, and resuspended in 10 μL HiDi Formamide. Samples are then shipped to GENEWIZ (South Plainfield, NJ) overnight and fragments analyzed using capillary electrophoresis on an ABI 3730 data analyzer instrument.
Figure 2.
SHAPE assay. Outline of the four main steps in the SHAPE assay: (1) incorporation of the SHAPE structure cassettes, (2) SHAPE chemistry in 2′O-adduct formation, (3) fragmentation and cDNA formation in primer extension, and (4) analysis of fragmentation patterns via capillary electrophoresis. To see this figure in color, go online.
Analysis of SHAPE reactivities has been performed using QuShape software algorithms (34). SHAPE reactivities are determined from the difference in fluorescence intensities of the probing and control experiments. To improve the statistical analysis, the SHAPE reactions are performed at least threefold for each positive and negative control experiment. For each condition, at least six combinations of + and – N-methylisatoic anhydride runs are performed, with averages and standard deviations calculated to determine the statistical significance of the SHAPE signal for each nucleotide as a function of probing conditions. RNA degradation, premature enzyme termination, and other experimental factors can affect the reproducibility of the SHAPE signal at specific nucleotides within the sequence. Such nucleotides were identified as outliers (>3σ) and removed from secondary structure predictions (see Supporting Material). SHAPE reactivities are normalized using a model free boxplot analysis and converted into pseudo free energies, which in turn are used to guide secondary structure predictions via RNAstructure (35, 36) and SHAPEKnots (37) software analysis.
Freely diffusing single-molecule burst spectroscopy
Single-molecule experiments are performed on an inverted scanning confocal microscope (IX70; Olympus, Center Valley, PA) with alternating laser excitation and single photon detection, as described previously (38, 39). Briefly, two synchronized picosecond pulsed lasers (532 nm, LYNX; Time-Bandwidth Products, Zurich, Switzerland and 635 nm, PDL800; PicoQuant, Berlin, Germany) at a repetition rate of 20.6 MHz are spatially overlapped and temporally delayed to obtain a train of equally spaced pulses of alternating excitation wavelength. The laser beams are expanded to overfill the back aperture of the objective and focused to the diffraction limit by a 1.2 NA water immersion objective (UPlanApo 60XW; Olympus), which delivers 0.5–100 μW at the focal plane. Fluorescence is then collected by the same objective, spatially filtered on a 50-μm confocal pinhole, split into donor (Cy3) and acceptor (Cy5) channels (645DCXR; Chroma Technology, Rockingham, VT), bandpass-filtered (HQ585/70M and HQ700/75M; Chroma Technology), and detected with individual single-photon counting avalanche photodiodes (SPCM-AQR-14; Perkin-Elmer Optoelectronics, Fremont, CA). Photon arrival events are recorded on a four-channel time-correlated single-photon counting module (SPC-134; Becker & Hickl, Berlin, Germany) and stored time-tagged and time-resolved with respect to both 1) the excitation pulse and 2) start of the experiment, with <100 ps and 50 ns uncertainties, respectively.
Single-molecule burst studies are performed on 125 pM RNA constructs in 50 mM HEPES buffer (pH 7.5; H9897; Sigma-Aldrich, St. Louis, MO) containing the triplet quencher Trolox (2 mM, 9-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid, 238813; Sigma-Aldrich) and the enzymatic oxygen scavenging system PCA/PCD (10 mM, 3,4-dihydroxybenzoic acid, 37580; 0.1 mg/mL, protocatechuate 3,4-dioxygenase, P2879; Sigma-Aldrich) to increase the photostability of the Cy3/Cy5 FRET pair (40, 41). Salt-dependent investigations of the folding-unfolding dynamics are performed by adding desired concentrations of NaCl and MgCl2. Samples are studied in a microliter flowcell assembled from a microscope slide and a No. 1.5 cover glass (22 × 22 mm; Corning, Corning, NY) secured by two pieces of double-sided tape separated by 1–2 mm to form a narrow channel with a ∼5 μL sample volume. This channel is flushed with 50 μL of sample solution just before the experiment.
Analysis of freely diffusing molecules has been described elsewhere in detail (38, 42). Briefly, fluorescence time traces for both excitation of donor and acceptor molecules are obtained by sorting single-photon arrival events into 1 ms bins. For fluorescence bursts that exceed a threshold count (typically 25 kHz) in both traces (25), FRET efficiencies (EFRET) are calculated from cross-talk- and background-corrected donor (IcD) and acceptor (IcA) intensities according to EFRET = IcA/(IcA + IcD) (38). Energy transfer efficiencies are then histogrammed for ∼103–104 individual burst events, with the resulting distributions analyzed by nonlinear least-squares fits to a sum of Gaussians that characterize the statistical probability of finding an RNA molecule at a given FRET value and conformation.
Results
Single-molecule studies of Mn+-induced TLS folding
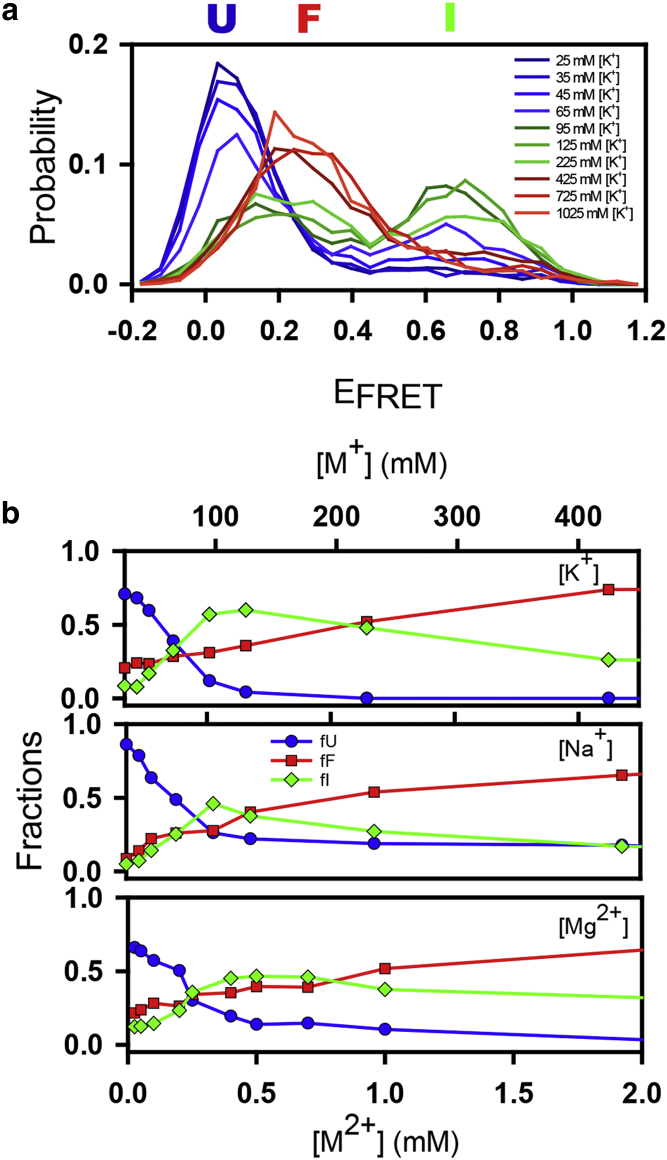
Our previous Na+-and Mg2+-induced single-molecule folding studies revealed three distinct conformations for the BMV3 TLS domain. To show that these conformations are relevant at physiological salt conditions, we here extend our single-molecule experiments to include K+-induced folding. [K+]-dependence in TLS folding is then observed by systematically increasing the [K+] from 0 to 1000 mM under standard HEPES buffer conditions (50 mM HEPES, pH 7.5). Accordingly, the placement of Cy3 donor and Cy5 acceptor labels near the 3′ and 5′ termini (Fig. 3) permits one to quantitatively characterize the TLS conformational folding dynamics by statistical analysis of single-molecule burst FRET efficiencies. For consistency, hemi-sodium HEPES buffer was used in all burst experiments, which adds a background [Na+] concentration of 25 mM. This is accounted for in Fig. 3 by plotting both [Na+] and [K+] burst population series versus the total [M+].
Figure 3.
Single-molecule burst fluorescence. (a) Single-molecule probability distributions are shown of FRET efficiencies at 0–1000 mM [K+]. (b) Fractional distributions are shown of single-molecule burst studies on [Mn+]-induced TLS folding experiments. TLS constructs in U conformation are shown in blue, I conformation in green, and F conformation in red. To see this figure in color, go online.
Three distinct populations are observed that evolve from low through high EFRET levels and then back to medium EFRET with increasing [K+] (see Fig. 3 for details). These three values correspond to the 1) unfolded (U) (EFRET ≈ 0.040(2)) and 2) folded (F) (EFRET ≈ 0.685(3)) states known from the literature, as well as 3) a heretofore unknown intermediate conformational state (I) with medium FRET efficiency (EFRET ≈ 0.211(2)). To minimize parameter correlation, the full set of K+-dependent EFRET histograms is simultaneously fit to a sum of three Gaussians (i = 1–3) with common but adjustable peak centers (EFRETi) and widths (FWHMi). Integrating areas under the Gaussian peaks permits accurate determination of the three subpopulations over the entire [K+] range, with the resulting fits reconfirming the presence of three distinct TLS conformational populations.
The results clearly indicate that the U conformation predominates for [K+] = 0 mM, whereas the F conformation becomes the major component for [K+] > 250 mM. For intermediate [K+] values, a more complex distribution of all three conformations is naturally observed, with the fractional population of the corresponding I maximized at ∼70–200 mM [K+]. Comparison with our previous [Na+] and [Mg2+]-induced folding studies (Fig. 3 b) shows that this three-step folding mechanism is retained across all three metal-ion titration series. These results suggest an electrostatics-driven exchange of all three TLS conformations, with both I and F dominating the equilibrium under physiological salt conditions. Interestingly, comparing Na+- and K+-induced folding (Fig. 3 b, top two panels) indicates that K+ drives TLS folding more efficiently than Na+. In particular, Na+-induced folding leads to a plateau with 20% nonfolded molecules, whereas in the presence of K+, this residual 20% drops to zero and predominantly converts into the I conformation between 70 and 250 mM [M+]. Consequently, in the 0.5–1 mM [Mg2+] range, the I conformation is expected to prevail over the F conformation even more so in the presence of K+ than Na+.
SHAPE probing of Mn+-induced TLS folding
To further correlate these single-molecule conformations with RNA 2° structure, we perform both burst fluorescence and SHAPE probing at identical salt and buffer conditions. Conditions are specifically chosen to include previous structure probing studies that led to the currently accepted U and F structure models (50 mM [Na+] and 50 mM [Na+] + 10 mM [Mg2+], respectively). Specific Mn+ concentrations probed are: 1) 25, 50, 100, and 330 mM [Na+] and 2) 0, 0.1, 0.25, 0.5, 1, and 10 mM [Mg2+] at 50 mM [Na+].
SHAPE reactivity data, in particular when comparing conformations probed at varying [Mn+] conditions, can be somewhat counterintuitive to interpret. We have thus added to the Supporting Material a more detailed explanation of the SHAPE signal analysis and how the changes therein can be correlated with structural rearrangements. In brief, hybridized and constrained nucleotides exhibit negligible SHAPE reactivity (R = 0), whereas flexible nucleotides do provide partial access to the SHAPE reagents (R > 0). Hence, it is the lower end of this reactivity scale that we will be most interested in. The reactivities are normalized onto a 0 < R < 2 scale, with only a few hyperreactive nucleotides well outside this scale (R > 2). Most importantly, structural changes in hybridization, i.e., nucleotides shifting from single-stranded to double-stranded or vice versa, are observed on the lower end of this reactivity scale (R = 0–1). Reactivity changes above R > 1, although often much larger in magnitude, are more reflective of changes in the microconformation of a flexible (single-stranded) nucleotide and thus not indicative of structural changes in the RNA construct.
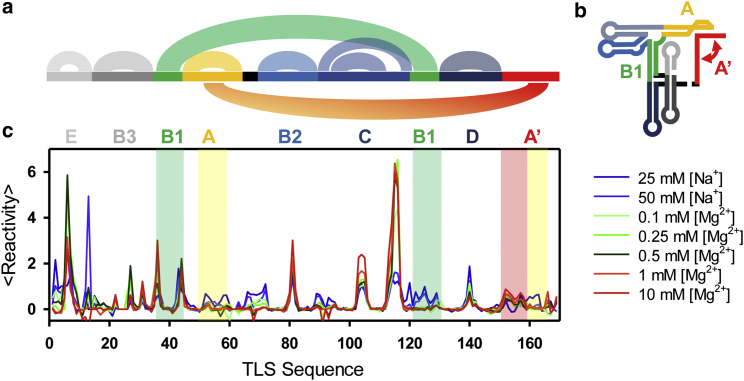
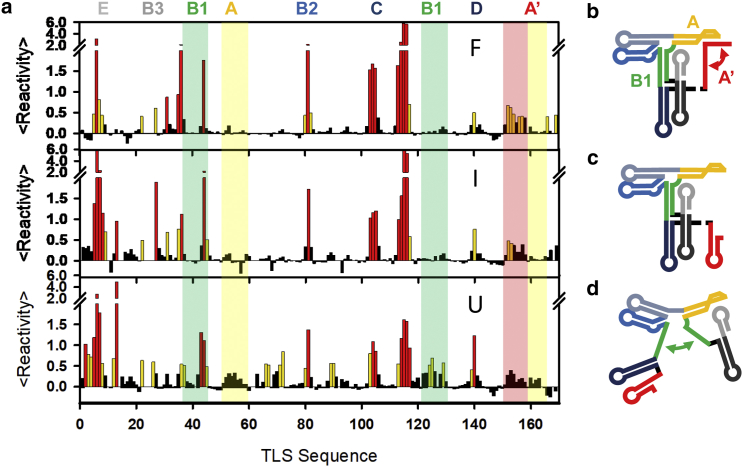
To help facilitate comparison of SHAPE probing results with RNA 2° structure, the literature model for F is illustrated with color-coding of the basepairing regions in Fig. 4. A regional map of structural elements (Fig. 4 a) is provided as arched lines across the nucleotide sequence representing nucleotide basepairing (e.g., E, B2, B3, D). Second layer arched curves indicate long-range interactions (e.g., B1-B1 and A-A′). Furthermore, regions indicated in structural reorganization are highlighted in the SHAPE reactivity plots. To illustrate changes in SHAPE reactivities upon structural reorganization, SHAPE traces for [Mg2+] values of 10, 0.5, and 0 mM corresponding to F, I, and U conformations in single-molecule studies, respectively, are discussed in more detail below.
Figure 4.
[Mn+]-dependent SHAPE probing of the TLS domain. (a) Color-coded diagram is given of 2° and 3° interactions in the TLS construct in F conformation, (b) color-coded regional map is given of the TLS construct in F conformation, and (c) changes are shown in normalized SHAPE reactivities with increasing [Mn+]. To see this figure in color, go online.
SHAPE analysis at high [Mg2+] confirms the literature 2° structure model
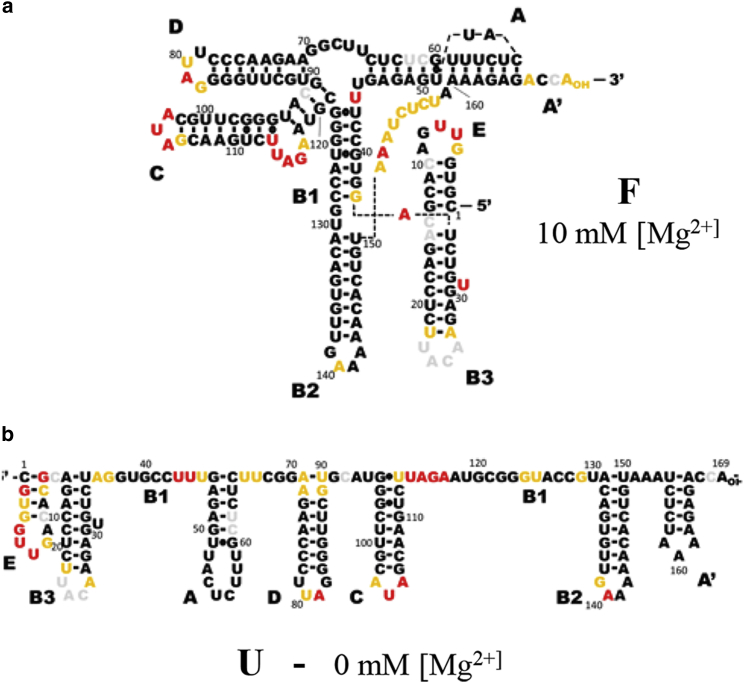
As a first step, we compare SHAPE probing experiments at 10 mM [Mg2+] (in 50 mM [Na+] and 50 mM HEPES buffer) to the 2° structure model of the F conformation from the literature. SHAPE reactivities are represented in Fig. 5 a as a heat map indicating nonreactive nucleotides in black (R < 0.4), moderately reactive nucleotides in orange (0.4 < R < 0.85), and highly reactive nucleotides in red (R > 0.85) (36, 37, 43). Finally, the same data is color-coded onto the 2° structure model to provide a heat map of SHAPE reactivities in Fig. 6.
Figure 5.
SHAPE analysis at the U, I, and F conditions. (a) Normalized SHAPE reactivities are shown at 10 mM [Mg2+] + 50 mM [Na+] (top), 0.5 mM [Mg2+] + 50 mM [Na+] (middle), and 25 mM [Na+] (bottom). (b–d) Color-coded regional map is given of the (b) F, (c) I, and (d) U conformations. To see this figure in color, go online.
Figure 6.
SHAPE reactivity mapped onto literature structure models. (a) SHAPE reactivities obtained at 10 mM [Mg2+] + 50 mM [Na+] are mapped onto the literature F model. (b) SHAPE reactivities are obtained at 50 mM [Na+] mapped onto the literature U model. Colors represent high reactivity (red), moderate reactivity (orange), and low reactivity (black). To see this figure in color, go online.
The mapping of SHAPE reactivities onto the 2° structure at single-nucleotide resolution shows overall good agreement with the literature model (Fig. 5 a (top) and Fig. 6 a). Stem regions composed of hybridized nucleotides show up as predominantly black, indicating low reactivity and thus low flexibility. Few exceptions are observed (e.g., G37), in general for terminal bases that display partial flexibility and appear as moderately reactive (orange). No hybridized region with more than one highly reactive nucleotide in a row is observed, indicating that the stem domains at high [Mg2+] are well assigned in the literature model (44). Similarly, most nucleotides in loop and bulge regions show up as red (highly reactive) and orange (moderately reactive), indicative of their flexible nature and high accessibility to the SHAPE chemical reagent. In total, only eight bases within loop regions remain nonreactive, i.e., G8, A9, U52, A53, U79, G139, A141, and A142, which may indicate that these bases exhibit low flexibility.
However, despite these few individual nucleotides, the overall secondary structure is remarkably well captured in the literature model. In particular, the reactivity at high [Mg2+] in structurally important regions at the 3′ PSK of region A (52–59) and A′ (160–165), the linker between A′ and B2 (151–159), the C bulge (113–117), and the long-range stem interaction B1 (122–128 and 37–43) are well matched with secondary structure predictions and expectations from the literature model. The high fraction of nonreactive sites further support a highly structured domain for ∼3/4 of the nucleotides. This is also in good agreement with recent observations from H2O2 probing and short-angle x-ray scattering experiments, which suggest a highly structured and compact core for BMV TLS at high [Mg2+] (45, 46).
SHAPE analysis at low [Mg2+] shows slight discrepancies from the literature model
Next, we compare SHAPE probing experiments at 0 mM [Mg2+] (in 50 mM [Na+] and 50 mM HEPES buffer) to the 2° structure model of the U conformation from the literature. SHAPE structure probing data for the U conformation, depicted in Fig. 5 a (bottom), shows a significant increase in SHAPE reactivity, which signals an increased flexibility and conformational dynamics in the unfolded U versus folded F conformations. The majority of differences are observed in a few key regions indicative of a specific structural reorganization. More quantitatively, the regions that undergo the most relevant signal changes (regions with SHAPE changes in the 0–1 range) are 1) the hairpin E, 2) the loop of stem A (52–59), 3) the bulge connecting A with D (66–72), 4) the 5′ region of the extended C stem (89–92), 5) the 3′ side of the long-range stem B1 (122–129), and 6) the 3′ terminal A′ domain (151–158).
The majority of these changes correspond to an increase in reactivity. For example, nucleotides 122–129 of stem B1 are nonreactive at high salt (F) conditions, but achieve significant reactivity for low salt (U) conditions, indicating that a decrease in ionic strength results in loss of basepairing for this long-range stem interaction. By way of contrast, nucleotides 151–158 in A′ exhibit a significant decrease in SHAPE reactivity, indicating that a previously single-stranded region has been converted to a basepaired region. Both observations, as well as the increased reactivity in stem loops A and C, confirm a loss upon unfolding (F → U) of the two long-range intramolecular interactions previously reported in the literature for the F TLS conformation.
On closer inspection, however, inconsistencies with the literature model for the U conformation are also clearly apparent (see Fig. 6 b); e.g., the 5′ side of stem B1 (37–43) shows little increase in reactivity, whereas the A-D bulge domain (66–72) does. SHAPE reactivities in the stem domains of A, A′, B2, B3, C, and D are consistently small, indicating that, with exceptions for a few terminal nucleotides (e.g., U113), these regions remain hybridized down to low [Mg2+]. Consequently, it would appear that all stem loops remain intact over the full [Mg2+] range probed, with the possible exception of A′ and E. Indeed, the stem of loop E shows significant increase in reactivity, indicating perhaps that the 5′ terminal domain becomes more dynamic at low salt conditions. In addition, a significant number of nucleotides in the loop and single-stranded regions remain nonreactive at low [Mg2+], i.e., in contrast with the proposed secondary structure model in the literature for the U conformation.
SHAPE analysis at 0.5 mM [Mg2+] shows high similarity to the F conformation
As discussed above, a completely new conformation (I) for BMV RNA3 TLS domain has been recently noted in smFRET experiments at intermediate Mg2+ values. To directly observe the 2° structure elements responsible for the TLS folding dynamics, SHAPE probing experiments are displayed in Fig. 5 a (middle) at moderate [Mg2+] conditions (0.5 mM [Mg2+]), where the I conformation reaches its highest fractional population. Interestingly, SHAPE studies for the I conformation reveal features nearly indistinguishable to those observed above for the F conformation. Consequently, a reduction of [Mg2+] from 10 to 0.5 mM results in only minor structural rearrangements. Of the domains involved in the long-range interactions, only the terminal A′ domain (nucleotides 151–158) exhibits significant changes. For this domain, decreasing [Mg2+] leads to a reduction in reactivity from moderate to low, signaling a shift in nucleotide hybridization in the 3′ PSK region.
Complete removal of [Mg2+], on the other hand, leads to considerable changes in the nucleotide reactivities, indicating additional rearrangements in the secondary structure of the TLS domain (Fig. 5 b). Although the C loop (103–105) and C-bulge (113–117) as well as a few individual nucleotides (35, 36, and 81) exhibit further decreases in reactivity, the majority of changes observed between I and U probing conditions reflect increases in reactivity. The most dominant regions that exhibit such effects are the E stem (2, 3, 4), B1 stem (38–43 and 128–122), the A loop (52–59) and the bulge connecting stem loops A and D (66–72). While loss of B1 hybridization is expected from the literature models, changes in E stem, A loop, and the A-D bulge indicate deviations from these predictions. To extract the most valuable structural information from such data, the full set of SHAPE reactivities are leveraged as restriction parameters for refining 2° structure for each of the U, I, and F conformations, as described in the following sections.
Discussion
SHAPE refinement predicts F dominates at physiological salt conditions
An extremely powerful use of the SHAPE probing data is for refinement of 2° structure predictions, with the SHAPE reactivities implemented as experimental constraints (36, 37). Indeed, de novo prediction of RNA secondary structures often achieves only quite limited accuracies of 50–70%, due to neglect of base stacking, noncanonical interactions or PSKs that can significantly modify the free energy landscape for RNA folding. By way of contrast, the use of data input from SHAPE probing studies as restriction parameters typically improves the accuracy of RNA structure predictions to 90–95%. Furthermore, such SHAPE probing studies can be performed in a range of salt and buffer conditions as well as in the presence of proteins, ligands and even inside cells (47, 48).
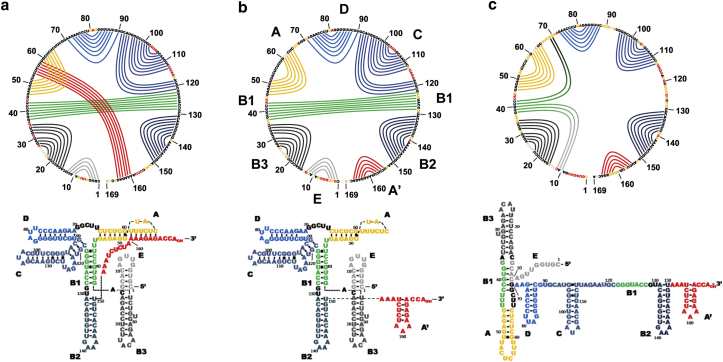
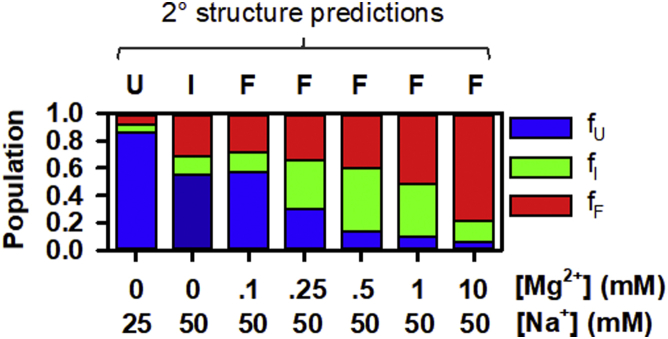
To make use of such capabilities for improved TLS secondary structure predictions, we applied SHAPE reactivity data taken over a range of 1) 25–300 mM [Na+] and 2) 50 mM [Na+] + 0–10 mM [Mg2+] as refinement parameters for RNAStructure and SHAPEKnots algorithms (35, 36, 37). The lowest free energy SHAPE-directed structure predictions are represented in Fig. 8 as 2° structure models (bottom) as well as circle plots (top). In good agreement with our previous smFRET folding experiments, three distinct conformations are again observed. While the structural features of the lowest free energy predictions are discussed in detail below, a comparison of structure prediction and single-molecule distributions is presented in Fig. 7 for all salt conditions tested. Although 2° structure predictions are observed for F, I, and U conformations, the F conformation is clearly predicted to predominate throughout the range of cation probe conditions. For instance, a contribution of as little as 28% of the solution ensemble at 0.1 mM [Mg2+] + 50 mM [Na+] still predicts the F conformation to predominate. The I conformation is predicted as the lowest free energy structure at 50 mM [Na+] with the U conformation suggested to be the lowest structure at 25 mM [Na+] or lower.
Figure 8.
SHAPE-directed structure prediction. Circle diagrams (top) and 2° structure representations of all three TLS conformations are shown: (a) predicted F, (b) predicted I, (c) and predicted U conformations. To see this figure in color, go online.
Figure 7.
Comparison of single-molecule folding and SHAPE-guided 2° structure predictions. Fractional populations from single-molecule folding are displayed in a bar graph for all [Mn+] conditions explored. Results for the lowest free energy conformer from SHAPE-guided structure predictions are presented above the bar graph. Comparison of both techniques indicates that predictions overestimate the stability of the F conformation at low cation concentrations. To see this figure in color, go online.
SHAPE-directed structure predictions largely confirm the literature models
Having access to SHAPE-directed structure predictions yields a quantitative metric with which to refute, refine, or confirm the literature TLS models at high (10 mM [Mg2+]) and low (50 mM [Na+]) salt conditions. In conjunction with commonly used 2° structure models, we therefore use circle plots to highlight intramolecular interactions and facilitate visualizing changes thereof in response to altering salt conditions. To this end, the TLS sequence is displayed clockwise from 5′ to 3′ (on a circle starting with stem loop E). Relevant intramolecular interactions are represented by lines connecting hybridized nucleotides and color-coded across both representations. The use of circle plots allows rapid differentiation between domains involved in 2°, i.e., stem loops, and 3° structure interactions, i.e., long-range interactions and PSKs. For example, stem loops are easily recognized as parallel curves centered around a locus of nonhybridized (loop) nucleotides, e.g., B2 (dark blue) or A (yellow). Long-range interactions can be seen as lines crossing the circle. Finally, PSKs show up as lines that intercept stem loops and/or long-range interactions. Individual nucleotides are heat-map color-coded with respect to SHAPE reactivity to demonstrate the impact of experimental data on SHAPE-guided structure prediction.
At high [Mg2+], these SHAPE reactivity-refined structural predictions (Fig. 8 a) show a number of stem loops, B2, C, D (blue), A (yellow), B3, E (gray), as well as a 1-nt bulge in stem B3 and a 4-nt bulge in stem C. In addition, a long-range interaction is observed in B1 (green) as well as a 6-nt PSK interaction connecting the loop of stem A with the 3′ terminal nucleotides 160–165. As indicated more clearly in the 2° structure representation, these interactions at high [Mg2+] are in excellent agreement with the F structure reported in the literature.
Complete removal of [Mg2+] (i.e., Fig. 8 b) maintains all secondary interactions, except for the loop in stem A and the 3′ terminus. The 3′ terminal 20 nucleotides refold into an alternate stem loop A′, at the loss of the long-range PSK interaction. Further reduction of the [Na+] results in several additional changes (Fig. 8 c). First of all, the stem loop C loses its bulge due to lack of stabilization of its peripheral 4-nt stem interaction between nucleotides 91–94 and 122–119. In addition, the long-range stem interaction B1 unfolds and contributes partially to the formation of interactions between nucleotides 41, 42 and 70, 69 (A-D bulge) as well as 38–40 with 12–10 (E domain), which have not been reported in the current U literature model.
SHAPE probing confirms that three-state TLS folding is dynamic
A key finding of this study is that both experimental approaches, 1) Mn+-induced single-molecule folding (Fig. 3) and 2) Mn+-dependent SHAPE structure probing (Figs. 4, 5, and 6), successfully reveal the presence of three different conformational states for the TLS domain in solution. Specifically, both methods clearly provide evidence for a stepwise folding mechanism in which the PSK in F, as a function of decreasing [Mn+], unfolds independently to form a new intermediate I, before breaking the B1 stem interaction to unfold completely into the U species.
In smFRET folding studies (Fig. 3), the U, I, and F conformations predominate the solution ensemble at 0, 0.5, and 10 mM [Mg2+] (all at 50 mM [Na+]), respectively. SHAPE reactivities observed under identical conditions (Fig. 5) clearly indicate the release of the 3′ PSK upon conversion from F to I and loss of the long-range stem interaction B1 in conversion from I to U species. This conformational dynamic is further substantiated by SHAPE-refined 2° structures predicted in Fig. 8 for high, intermediate, and low Mn+ conditions. However, relatively minor but nonvanishing differences are observed in the Mn+ concentrations required to stabilize TLS conformations in single-molecule folding studies versus SHAPE-refined structure predictions, as discussed below.
Specifically, at 10 mM [Mg2+] the F conformation is observed in both experimental techniques. Upon lowering the [Mn+] concentration, SHAPE analysis systematically predicts TLS conformations to predominate at lower salt conditions than smFRET, e.g., 0.5 vs. 0 mM [Mg2+] for I and 50 vs. 25 mM [Na+] for U species. However, this trend is consistent with the anticipated overestimation of the nearest-neighbor basepairing parameters that were obtained under higher salt conditions (1 M [NaCl]). Indeed, because positively charged cations screen Coulombic repulsion from the negatively charged RNA backbone, the Gibbs free energy for basepair formation becomes more favorable with higher [Mn+], and thus weakens base interactions at lower ionic strength. RNAstructure prediction with SHAPE restraints uses nearest-neighbor thermodynamic parameters that were determined at 1 M [Na+]. Although salt corrections of thermodynamic basepairing parameters have been extensively studied and are available for routine structure predictions, at this point, RNAstructure does not support salt corrections in concert with using SHAPE restraints (35, 36, 37). This is likely due to SHAPE restraints being used to calculate pseudo free-energy terms (ΔGSHAPE) as additive contributions for the calculation of nearest-neighbor basepairing probabilities. Thus, ΔGSHAPE terms are designed to drive the structure prediction toward the most likely geometry. Yet, constructs probed under low salt conditions exhibit an overestimate of unreactive nucleotides as 1) their thermodynamic parameters are overestimated due to the mismatch in salt conditions and 2) SHAPE free-energy terms add a negative ΔGSHAPE contribution to nucleotides with R < 0.4. This effect leads to overestimation of basepairing probabilities and thus prediction of the mostly basepaired, F conformation at low [Mn+] conditions. The lower [Mn+] required to stabilize U and I conformations compared with single-molecule studies is thus consistent with the underestimated stabilization arising from the much higher [Mn+] conditions assumed in the SHAPE probing and SHAPE-directed structure predictions (49, 50).
Combining SHAPE with smFRET allows deconvolution of RNA conformations from ensemble SHAPE traces
This work further highlights the complementarity of single-molecule folding and SHAPE probing studies in the evaluation of RNA structures and their [Mn+]-dependent conformational dynamics. SHAPE-directed structure prediction has been shown to significantly improve the predictive accuracy of RNA secondary structures (36, 37). Nonetheless, SHAPE probing is inherently an ensemble technique that probes all molecules in solution simultaneously. SHAPE reactivities thus represent an average of the fractional contributions for all conformations in the ensemble equilibrium at each respective [Mn+]. This limited dynamic range typically requires structure determinations to be conducted at elevated cation concentrations to achieve homogeneity in the 2° and 3° RNA structures (19, 20, 51). As a result, the structural information obtained does not necessarily represent the biologically active state of the RNA construct. Simply stated, accurate structure-function predictions benefit considerably from studies under environmental conditions for which the biologically relevant RNA folding dynamics are indeed taking place.
SmFRET is ideally suited for this purpose, as it provides access to kinetics and thermodynamics of intramolecular interactions over a large range of environmental conditions (25, 42). Single-molecule detection further allows for clean rejection of signals from contaminants and nonactive molecules, thus permitting detailed analysis of thermodynamic folding parameters. In particular, the fractional populations for each state can be determined over a wide range of [Mn+] in the presence and absence of small molecule ligands, proteins, and even inside living cells.
Combination of both smFRET and SHAPE probing data at identical salt conditions provides us with the unique opportunity to apply single-molecule fractional populations to deconvolve SHAPE probing patterns into the individual contributions from each TLS conformation. To obtain 2° structure information of pure TLS conformations we solve a system of linear equations for each nucleotide according to
| (1) |
where aij are the fractional populations of the U, I, and F conformations (i = 1–3) at j salt condition, xi are the SHAPE reactivities of pure TLS conformations, and bj are the experimentally observed SHAPE reactivities.
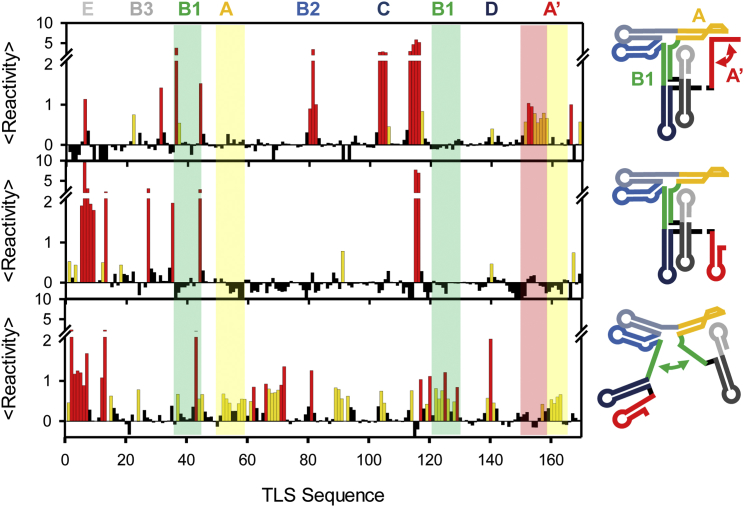
Implementing this analysis for the TLS data in Figs. 3 and 4, we obtain the SHAPE probing profiles in Fig. 9 for isolated U, I, and F conformations. In comparison to the 0, 0.5, and 10 mM [Mg2+] profiles in Fig. 5, a significant increase in dynamic range is readily apparent. Most importantly, small fractions of highly reactive nucleotides in F do not significantly contribute to I and U deconvoluted SHAPE traces. This is observed, for instance, in the B2 loop (80–82), the C loop (103–106), the C bulge (113–116), and the A′ domain (150–160). Similarly, highly reactive nucleotides in U and I do not affect F, e.g., the stem and loop in E. Furthermore, some nucleotides in I lack the reactivity expected based on the TLS folding model in Fig. 1, e.g., the A (52–59) and A′ loops (160–162). Such an absence of SHAPE reactivity might indicate that these nucleotides do not sample SHAPE reactive conformations or contribute to additional alternative conformations.
Figure 9.
Single-molecule folding guided deconvolution of SHAPE probing. Clean SHAPE profiles after deconvolution with fractional population from single-molecule folding experiments are displayed for F (top), I (middle), and U (bottom). Regions that undergo structural reorganizations are highlighted and color-coded with respect to the 2° structure models on the right. To see this figure in color, go online.
The benefits of extracting isolated SHAPE profiles for individual conformations in a complex dynamic equilibrium ensemble extends also to 2° structure predictions. Decreasing the impact of highly reactive nucleotides in the conformational ensemble improves the predictive accuracy for the F, I, and U conformations, as presented in Fig. 8. Indeed, increasing the dynamic range of salt concentrations probed from smFRET deconvolution of SHAPE traces reduces the overestimation of nearest-neighbor thermodynamic parameters. Thus, the combination of single-molecule folding and SHAPE structure probing provides valuable 2° structure information for individual conformations in a complex mixture under heterogeneous biologically relevant conditions. Although not implicitly studied herein, the above principles apply equally well to RNA interactions with proteins and small-molecule metabolites.
In summary, SHAPE reactivity studies function superbly in refinement of 2° structures and thus predictions of RNA conformations at specific mono- and divalent cation concentrations, thereby providing nucleobase-resolved structural information under biologically relevant environmental conditions. Conversely, interconversion kinetics among these RNA conformations, their fractional populations, and equilibrium constants are more accurately extracted from single-molecule folding studies. The synergistic combination of SHAPE reactivity probing with smFRET solution dynamics provides a considerably more powerful, conformation-resolved picture of the biologically relevant folding energy landscape.
Biological implications
This combination of SHAPE probing and single-molecule RNA folding suggests an intriguing mechanistic view for BMV TLS structure-function relations. This mechanism is based on an RNA switch between individual virus replication steps that is driven by selective stabilization of one of the TLS conformations. A number of viral enzymes as well as host factors have been reported to interact with a specific structural feature in the BMV TLS domains. Of particular interest are the 3′ PSK and 3′ terminal stem loops that are observed in the F and I conformation, respectively. Specific interaction with one of these structural features likely leads to selective stabilization of that conformation. As the 3′ PSK (F conformation) has been associated with virus replication and the 3′ hairpin (I conformation) with virion assembly, this mechanism could effectively result in a switch of the virus replication stage. A crucial component of this RNA switch is that both 3′ terminal features are accessible at environmental conditions, thus sampling the solution ensemble for replication-stage-specific host factors.
The SHAPE-directed structure-probing studies in this work have confirmed the presence of a stepwise folding mechanism first reported in smFRET studies from our laboratory (8). In this three-conformation kinetic model, the TLS domain maintains a dynamic equilibrium among U, I, and F conformations, whereby the relative abundances of each conformation depend on [Mg2+] and [Na+]. At zero Mg2+, the U conformation predominates, which in turn can fold spontaneously into the I and fully F conformations with increasing divalent ion concentration. Under physiologically relevant cation conditions (150 mM [Na+], 0.5–1 mM [Mg2+]), however, the long-range stem interaction is already formed. As a result, only I and F species contribute significantly to the TLS conformational equilibria, with very little U present. Most importantly, the intermediate I conformation is structurally quite similar to F, with the only changes being 1) loss of PSK interaction between six nucleotides in the stem loop A (54–59) and 3′ terminal domain (160–165), and 2) refolding of the 3′ terminus into an alternate 5-nt stem loop A′ (154–166).
Precisely due to the modest extent of such structural changes, switching between I/F conformations offers a feasible switching mechanism for regulation of the RNA replication process (22, 27, 51). For example, the F conformation and in particular, the 3′ terminal PSK, has been implicated as a critical structural feature for aminoacylation, which is an essential property in all tRNAs and commonly observed in TLS domains. Aminoacylation requires interaction of aminoacyl tRNA synthetase with the TLS domain and therefore recognition of its TLS features, which requires accessibility of the 3′– terminal CCAOH that is available only in the PSK interaction. Conversely, the lack of a 3′ terminal PSK in I would inhibit recognition and thus potentially render the aminoacylation process inactive. Interestingly, aminoacylation assays have shown that tyrosylation of the TLS 3′ termini can be achieved at stoichiometric ratios, suggesting that TLS-enzyme binding shifts the equilibrium toward the F conformer (52). This clearly would be consistent with the presence of a dynamic equilibrium between I and F conformers under physiological salt conditions.
Taking this idea one step further, one could envision a mechanism for which interaction with a host factor or viral protein might shift this dynamic equilibrium from F back toward the I conformer. Because the 3′ terminal CCAOH group in the F conformer is required for recognition of viral and host factors, such an equilibrium shift would be equivalent to an on-off switch for virus replication. In fact, such a mechanism has been observed for members of the related Ilarvirus genus of the Bromoviridae that share similarities with BMV, such as Alfalfa Mosaic Virus (30, 53). The members of this genus require interaction of coat protein (CP) with a series of stem loops near the 3′-end for switching between viral replication steps. This 3′-terminal domain is capable of folding into an alternative tRNA-like conformer that has some resemblance to the TLS of BMV. As for BMV, the tRNA-like conformer is recognized by the viral replicase and thus is required for minus-strand synthesis. Binding of CP to the 3′-terminal hairpin could result in unfolding of the PSK and thus inhibition of minus-strand synthesis. Mg2+, on the other hand, was found to interfere with CP binding due to stabilization of the PSK interaction (30). In agreement with such a mechanism, BMV has developed a complex scheme resulting in delayed translation of its CP (54). Consequently, the [CP] increases at later steps during the viral replication cycle, thus presenting a plausible factor for timed regulation of replication. In a similar manner, riboswitches turn off gene expression by binding to a downstream product, thus regulating expression as a function of the RNA-ligand binding equilibrium.
In summary, this study of solution dynamics for BMV TLS reveals a dynamic equilibrium between I and F conformations at biologically relevant salt conditions. The F conformation provides access to the 3′ PSK for aminoacylation, which is essential in virus replication. The I conformation, on the other hand, provides access to the 3′ stem loop for CP binding in virus assembly. Sampling of both conformations offers an elegant concentration-dependent solution for switching between replication and assembly in viruses of the Bromoviridae family.
Conclusions
We have performed detailed SHAPE probing studies of the TLS domain of BMV, yielding nucleotide-specific SHAPE reactivity data as a systematic function of divalent [Mg2+] and monovalent [Na+] concentrations. These reactivities, in turn, have been used to provide nucleotide-specific pseudo free energies, which permit refinement of thermodynamic parameters for folding and thus remarkably detailed predictions on the stability of various TLS conformations as a function of the cation environment. Specifically, mapping of the SHAPE reactivities onto secondary structure models provides strong experimental validation for current structural models of the TLS domain at both vanishing (0 mM) and saturating (10 mM) levels of [Mg2+], corresponding to fully U and F conformations. Furthermore, the SHAPE probing studies also predict the presence of a new I TLS conformation at 0.25–0.50 mM Mg2+ concentrations, corresponding to the unfolding of the 3′ terminal PSK, but without subsequent breakage of the long-range B1 stem interaction to form the unfolded U conformation. This is in excellent agreement with and confirms the presence of a novel three-step folding pathway (U → I → F) first proposed and motivated by previous smFRET experiments (8).
Furthermore, the SHAPE-directed structure predictions support a stepwise unfolding mechanism of these two tertiary interactions as a function of Mg2+. With decreasing [Mg2+], the 3′ terminal PSK in the fully F TLS conformation unfolds first, stabilized by formation of an alternate 3′ terminal hairpin (I), which is then followed by unfolding of a long-range B1 stem interaction to form U. The SHAPE studies clearly reveal that under physiological divalent and monovalent cation conditions, TLS has essentially no population in the U state, but still exists in a dynamic equilibrium between I and F conformations. This is particularly important, as recognition of the 3′ terminal hairpin on the F conformer is known to be a trigger for viral replication, and yet this moiety disappears in the I conformation with unfolding of the PSK. Thus, environmentally controlled shifts in the equilibrium between F and I conformation could act as an effective switch for the viral replication pathway. This dynamic equilibrium offers an intriguing mechanism for differential recognition by F and I conformations of host factors during the virus life cycle and thus potential regulation of viral replication.
We further observe that SHAPE probing at low [Mn+] conditions can lead to overestimation of basepairing probabilities in a 2° structure prediction. This is in part due to the structural heterogeneity in the conformational equilibrium that leads to overinterpretation of highly reactive nucleotides. In addition, nearest-neighbor thermodynamic parameters incorporated in RNAstructure and ShapeKnots software correspond to basepairing probabilities under quite high 1 M [Na+] concentrations. Deconvolution of SHAPE probing data using smFRET fractional populations provides a powerful means to overcome both effects by providing clean SHAPE traces of conformational states as restriction parameters for 2° structure prediction. These combined efforts highlight the strong potential for combining 1) single-molecule folding studies with 2) SHAPE-directed structure-probe analysis for detailed elucidation of biologically active conformational states as a function of environmental cation conditions.
As a final comment, although the combination of smFRET and SHAPE methodologies are clearly quite powerful, such capabilities do currently rely on Gibbs free energies for basepair formation obtained for fixed and often quite nonphysiological cation conditions, despite the fact that these free energies clearly depend on the identity and concentration of the cation environment. We note that extension of such combined smFRET and SHAPE efforts could benefit enormously from further systematic study of monovalent- and divalent cation [Mn+]-dependent thermodynamics for basepair formation, and which are extensively used as input into current RNA structural prediction algorithms.
Author Contributions
M.V. performed research. M.V. analyzed data. M.V. and D.J.N. contributed analytic tools. M.V. and D.J.N. designed research. M.V and D.J.N. wrote the article.
Acknowledgments
The authors thank Robert T. Batey, Jeremy Trausch, and Jake Polaski for help with the RNA synthesis and SHAPE assays.
Funding for this work was provided by the National Science Foundation (CHE 1266416 and PHY 1734006) with support for initial equipment from the National Institute for Standards and Technology and the W.M. Keck Foundation initiative in RNA Sciences at the University of Colorado, Boulder.
Editor: Karin Musier-Forsyth.
Footnotes
Supporting Materials and Methods, four figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30253-4.
Supporting Material
References
- 1.Walter N.G., Woodson S.A., Batey R.T., editors. Non-Protein Coding RNAs. Springer; Berlin, Germany: 2009. [Google Scholar]
- 2.Cech T.R. Crawling out of the RNA world. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Cech T.R. The RNA worlds in context. Cold Spring Harb. Perspect. Biol. 2012;4:a006742. doi: 10.1101/cshperspect.a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cech T.R. How a chemist looks at RNA. Angew. Chem. Int. Ed. Engl. 2013;52:75–78. doi: 10.1002/anie.201205427. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A., Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreher T.W. Viral tRNAs and tRNA-like structures. Wiley Interdiscip. Rev. RNA. 2010;1:402–414. doi: 10.1002/wrna.42. [DOI] [PubMed] [Google Scholar]
- 7.Dreher T.W. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- 8.Vieweger M., Holmstrom E.D., Nesbitt D.J. Single-molecule FRET reveals three conformations for the TLS domain of brome mosaic virus genome. Biophys. J. 2015;109:2625–2636. doi: 10.1016/j.bpj.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Axtell M.J. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 10.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs J., Glanz S., Kück U. RNA trans-splicing: identification of components of a putative chloroplast spliceosome. Eur. J. Cell Biol. 2010;89:932–939. doi: 10.1016/j.ejcb.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Zong X., Tripathi V., Prasanth K.V. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins K. Forms and functions of telomerase RNA. Springer Ser. Biophys. 2009;13:285–301. [Google Scholar]
- 14.Wan G., Liu Y., Lu X. Noncoding RNAs in DNA repair and genome integrity. Antioxid. Redox Signal. 2014;20:655–677. doi: 10.1089/ars.2013.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grigg J.C., Ke A. One platform, five brands: how nature cuts the cost on riboswitches. J. Mol. Biol. 2013;425:1593–1595. doi: 10.1016/j.jmb.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA. 2nd Ed. Cold Spring Harbor Laboratory; Harbor, NY: 1999. [Google Scholar]
- 17.Batey R.T., Rambo R.P., Doudna J.A. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Tinoco I., Jr., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 19.Draper D.E. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Draper D.E., Grilley D., Soto A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 21.Treiber D.K., Williamson J.R. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 22.Treiber D.K., Williamson J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 23.Anderson C.F., Record M.T., Jr. Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 24.Draper D.E., Misra V.K. RNA shows its metal. Nat. Struct. Biol. 1998;5:927–930. doi: 10.1038/2901. [DOI] [PubMed] [Google Scholar]
- 25.Deniz A.A., Dahan M., Schultz P.G. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grunwell J.R., Glass J.L., Schultz P.G. Monitoring the conformational fluctuations of DNA hairpins using single-pair fluorescence resonance energy transfer. J. Am. Chem. Soc. 2001;123:4295–4303. doi: 10.1021/ja0027620. [DOI] [PubMed] [Google Scholar]
- 27.Bokinsky G., Zhuang X. Single-molecule RNA folding. Acc. Chem. Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 28.Rothwell P.J., Berger S., Seidel C.A.M. Multiparameter single-molecule fluorescence spectroscopy reveals heterogeneity of HIV-1 reverse transcriptase:primer/template complexes. Proc. Natl. Acad. Sci. USA. 2003;100:1655–1660. doi: 10.1073/pnas.0434003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman M.R., Kao C.C. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J. Mol. Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 30.Olsthoorn R.C., Mertens S., Bol J.F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson K.A., Merino E.J., Weeks K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 32.Hermanson G.T., editor. Bioconjugate Techniques. Elsevier; Amsterdam, Netherlands: 1995. [Google Scholar]
- 33.McGinnis J.L., Duncan C.D., Weeks K.M. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karabiber F., McGinnis J.L., Weeks K.M. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013;19:63–73. doi: 10.1261/rna.036327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deigan K.E., Li T.W., Weeks K.M. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hajdin C.E., Bellaousov S., Weeks K.M. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc. Natl. Acad. Sci. USA. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiore J.L., Hodak J.H., Nesbitt D.J. Monovalent and divalent promoted GAAA tetraloop-receptor tertiary interactions from freely diffusing single-molecule studies. Biophys. J. 2008;95:3892–3905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiegland L.R., Garst A.D., Nesbitt D.J. Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry. 2012;51:9223–9233. doi: 10.1021/bi3007753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiore J.L., Holmstrom E.D., Nesbitt D.J. Entropic origin of Mg2+-facilitated RNA folding. Proc. Natl. Acad. Sci. USA. 2012;109:2902–2907. doi: 10.1073/pnas.1114859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deniz A.A., Laurence T.A., Weiss S. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 43.McGinnis J.L., Dunkle J.A., Weeks K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rietveld K., Pleij C.W., Bosch L. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond J.A., Rambo R.P., Kieft J.S. Comparison and functional implications of the 3D architectures of viral tRNA-like structures. RNA. 2009;15:294–307. doi: 10.1261/rna.1360709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felden B., Florentz C., Westhof E. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- 47.Kladwang W., VanLang C.C., Das R. Understanding the errors of SHAPE-directed RNA structure modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonard C.W., Hajdin C.E., Weeks K.M. Principles for understanding the accuracy of SHAPE-directed RNA structure modeling. Biochemistry. 2013;52:588–595. doi: 10.1021/bi300755u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner D.H., Mathews D.H. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owczarzy R., Moreira B.G., Walder J.A. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 51.Draper D.E. RNA folding: thermodynamic and molecular descriptions of the roles of ions. Biophys. J. 2008;95:5489–5495. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dreher T.W., Hall T.C. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3′-adenylation. J. Mol. Biol. 1988;201:41–55. doi: 10.1016/0022-2836(88)90437-8. [DOI] [PubMed] [Google Scholar]
- 53.Chen S.-C., Olsthoorn R.C.L. In vitro and in vivo studies of the RNA conformational switch in Alfalfa mosaic virus. J. Virol. 2010;84:1423–1429. doi: 10.1128/JVI.01443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kao C.C., Ni P., Dragnea B. The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein. Mol. Plant Pathol. 2011;12:403–412. doi: 10.1111/j.1364-3703.2010.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.