Abstract
Metabolomics, in which small-molecule metabolites (the metabolome) are identified and quantified, is broadly acknowledged to be the omics discipline that is closest to the phenotype1–3. Although appreciated for its role in biomarker discovery programs, metabolomics can also be used to identify metabolites that could alter a cell’s or an organism’s phenotype. Metabolomics activity screening (MAS) as described here integrates metabolomics data with metabolic pathways and systems biology information, including proteomics and transcriptomics data, to produce a set of endogenous metabolites that can be tested for functionality in altering phenotypes. A growing literature reports the use of metabolites to modulate diverse processes, such as stem cell differentiation, oligodendrocyte maturation, insulin signaling, T-cell survival and macrophage immune responses. This opens up the possibility of identifying and applying metabolites to affect phenotypes. Unlike genes or proteins, metabolites are often readily available, which means that MAS is broadly amenable to high-throughput screening of virtually any biological system.
Historically, metabolites have been either supplemented or eliminated from growth media and diets to modulate cellular activity and affect phenotype. For example, in the phenylalanine hydroxylase deficiency disease phenylketonuria, deficient metabolism of phenylalanine results in severe and adverse symptoms that can only be ameliorated by strict adherence to a low-phenylalanine diet from birth4. A prominent example of a frequently supplemented metabolite is niacin (vitamin B3), which has an important role in energy transfer and maintenance of metabolic activity5. Metabolites can also function as metabolic coenzymes (e.g., coenzyme Q10 (CoQ10) and thiamine) and modulation of coenzymes can alter phenotypes by altering regulation of enzyme reactions. For example, statins, a class of cholesterol-lowering drugs, have the side effect of inhibiting the endogenous synthesis of CoQ10 (ref. 6). CoQ10 (ubiquinone) is a commonly prescribed supplement for patients receiving statins to regain mitochondrial energy homeostasis.
Metabolomics is used to identify the set of metabolites that are associated with physiological conditions or aberrant processes. To date, the main focus of the field has been on using this information to identify biomarkers and active or dysregulated pathways. In this Perspective, we discuss how to screen metabolomics data for metabolites that can be used to either induce or suppress biological functions. Unlike proteins, or genes, endogenous metabolites are readily amenable to biological testing and clinical applications.
Metabolomics activity screening
Untargeted (global) metabolomics uses liquid chromatography high-resolution mass spectrometry (LC-MS) to carry out comprehensive comparative analysis of metabolites. LC-MS is well-suited to metabolomic analyses, because it has high sensitivity, specificity, and reproducibility. It enables a broad statistical assessment of the metabolites extracted from a sample, and can be used to reveal unanticipated metabolic perturbations. There are numerous commercial and freely available data-processing packages, such as XCMS Online7, Mzmine8, and MetaboAnalyst9, that can be applied to analyze LC-MS data. These suites of algorithms can identify chromatographic peaks, align them, and then statistically assess the comparative data, based on calculated probability, fold change, and intensity. Metabolites that are differentially regulated can be identified using databases (e.g., METLIN (https://metlin.scripps.edu), the human metabolome database (HMDB; http://www.hmdb.ca), and LIPID MAPS; http://www.lipidmaps.org/)10–12, whose features and limitations have been reviewed13. The main advantage of untargeted LC-MS metabolomics is that it is an unbiased way to identify metabolites associated with a particular condition, whether it is stem-cell differentiation14,15, immune-cell activation16–19, remyelination in multiple sclerosis20, chronic pain21, or type 2 diabetes22,23, to name but a few of the hundreds of examples that have been reported.
Endogenous metabolites identified in metabolomics data sets can be screened to identify metabolites that modulate phenotype. Unlike genes and proteins, metabolites are readily available, typically inexpensive, and have relatively simple structural features making them very amenable to screening.
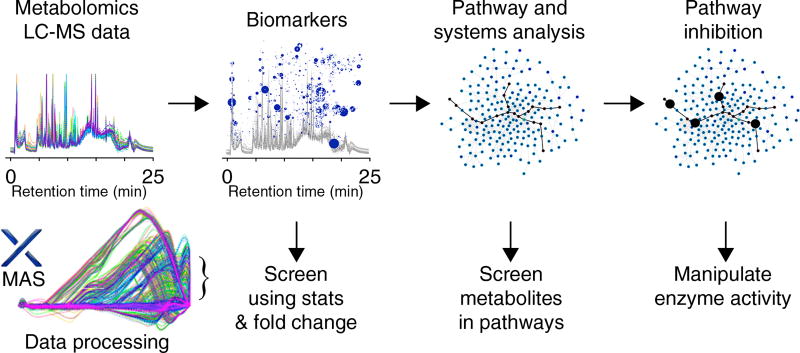
Various MAS workflows can be designed to identify metabolites from metabolomics experiments for activity testing (Fig. 1). The most straightforward approach selects metabolites based on statistical significance and fold change, which is also the standard method for screening metabolites in global metabolomics experiments. For example, in a comparative analysis using a cell model, any metabolites that have statistical significance represented by a P-value lower than 0.001, and fold changes greater than two, would qualify for further testing, although these values are user-defined and can vary. A secondary level of candidate selection would be to test metabolites from pathways identified as being active, a feature that has been recently automated in XCMS Online24. This ‘biologically driven’ selection method would include metabolites identified as dysregulated and metabolites involved in pathways of interest. Metabolites can be plotted onto pathway maps and ranked on the basis of the number of pathways involving each metabolite, leveraging pathway specificity. A third level of candidate selection can be mediated by manipulating the activity of enzymes in pathways using inhibitors or molecular biology approaches.
Figure 1.
MAS for the identification of endogenous metabolites that modulate phenotype. Metabolomics data analysis and identification of candidates for screening are carried out by XCMS Online or other data-processing approaches. Initial candidates are generated using statistical and fold-change cut-offs and can then be further investigated using high-throughput screening to identify biologically active metabolites. Pathway analysis can provide additional metabolite candidates, while a third level of screening would identify candidates following perturbations with known pathway inhibitors.
An important part of metabolite selection, beyond evaluating statistical significance, fold change, and pathways, is metabolite identification. For this purpose, multiple databases have been created that allow metabolites to be putatively identified using accurate mass and tandem mass spectrometry data10. Metabolite identification is validated by comparison with an authentic standard with tandem MS data generation as well as chromatography retention time (when available). Further validation in which experimental samples are analyzed using a targeted approach with triple quadrupole MS to compare against the original quantification (performed in the untargeted experiments) can also be used. These multiple levels of authentication help minimize misidentifications, which commonly occurred in the past when only precursor m/z values were used.
It is worth noting that while databases for initial identification information are not complete, their growth has been tremendous in the last decade. Currently, users examine multiple databases when performing searches because the databases are not completely overlapping25.
Phenotype-modulating metabolites identified using MAS
Metabolomics has been applied to provide insights into immunomodulation16–19, cardiovascular disease26–28, and diabetes22,23, with specific examples from our group, including stem cell differentiation (G.S. and colleagues)14, the role of microbiome metabolism (G.S. and colleagues)29, molecular origins of chronic pain (G.S. and colleagues)21, and, most recently, remyelination for neuron repair (J.R.M.-B., G.S, and colleagues)20. Comparatively, though, little effort has been dedicated to examining the activity of these biomarkers. In the following paragraphs, we briefly outline five examples of biologically active metabolites as unraveled by MAS.
Modulating stem cell differentiation
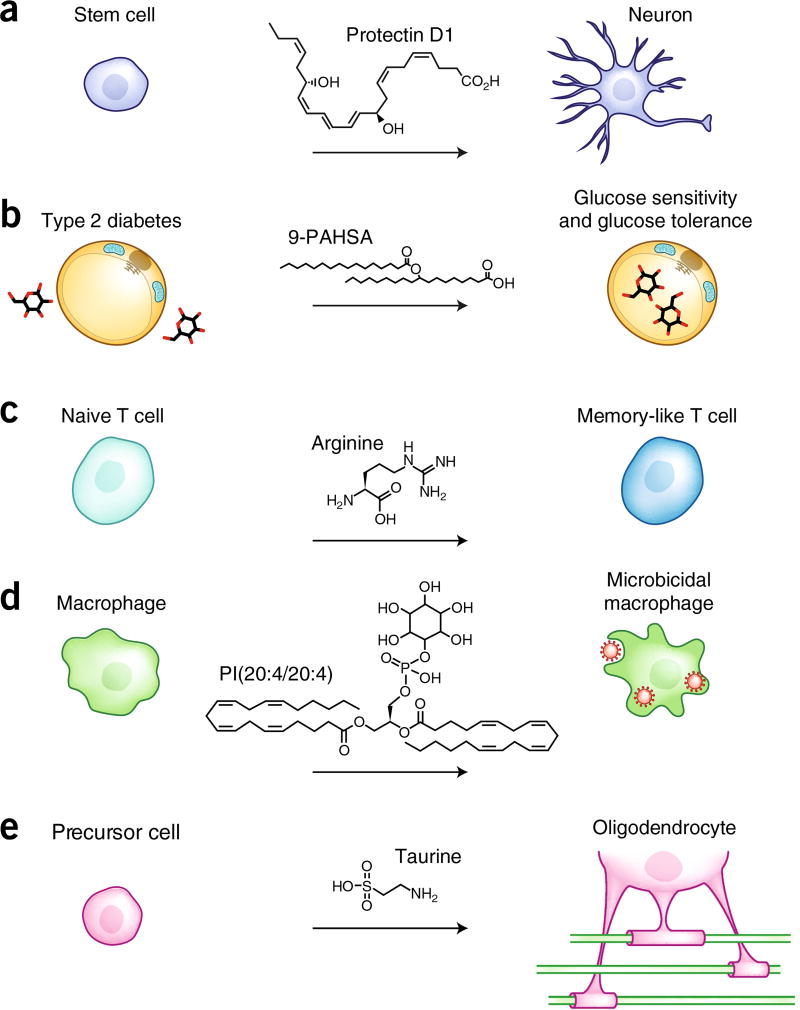
One of our (G.S. and colleagues)14,30 earliest efforts in stem cell analysis was designed to identify metabolites associated with cell differentiation. In these experiments, the metabolome of pluripotent stem cells, differentiated neurons and cardiomyocytes were quantitatively compared. Globally, the differentially regulated metabolites indicated that oxidation was a primary driver for cell differentiation. For example, arachidonic acid, a polyunsaturated fatty acid and the metabolic precursor to >100 functionally diverse metabolites, is highly upregulated in stem cells. Arachidonic acid in stem cells is important for maintaining ‘chemical plasticity’ and in mediating differentiation by regulation of redox status and activation of oxidative pathways. A crucial downstream molecule in these experiments, protectin D1 (derived from docosahexaenoic acid, also a polyunsaturated fatty acid) was used to promote differentiation and neurogenesis at concentrations as low as 50 nM (Fig. 2a).
Figure 2.
MAS demonstrated in stem-cell differentiation, a mouse model of type 2 diabetes, T-cell function and activity, macrophage response to a fungal stimulus, and a remyelination model for multiple sclerosis. (a) Experiments with embryonic stem cells identified the metabolites involved in their differentiation. Among them, protectin D1, a lipid, was found to enhance differentiation to neurons by a factor of 15 (ref. 14). (b) 9-PAHSA was discovered in adipose tissue and plasma of glucose-tolerant mice. This metabolite was identified as a key molecule that maintains correct glucose homeostasis in a model of type 2 diabetes induced by a high-fat diet22. (c) l-arginine levels decreased in activated naive T-cells. When l-arginine levels were raised externally, this amino acid actively increased survival and anti-tumor activity of T cells by modulating the activity of several transcriptional factors19. (d) Minor phospholipid species PI(20:4/20:4) is actively synthesized by activated macrophages. When exogenously added, this lipid amplified microbicidal capacity of macrophages in response to the fungal stimulus zymosan16. (e) Taurine, that was observed to be highly upregulated during OPC differentiation, enhanced the effect of a novel drug treatment (miconazole) to induce OPC differentiation into mature oligodendrocytes, a promising cell target for multiple sclerosis treatment20.
Overall, the results from these experiments suggested that the activation of oxidation is a metabolic requirement of stem-cell differentiation. Specifically, endogenous metabolites that promote pluripotency induce stem cells to a more reduced state whereas those that promote differentiation induce a more oxidized state. Moreover, it is well known that hypoxia maintains the pluripotent and undifferentiated phenotype of stem or precursor cells both, in vitro and in vivo31. Interestingly, these results also showed that endogenous metabolites are not merely substrates and products of metabolic reactions, but rather are involved in modulating stem cell differentiation and can be used to enhance their regenerative potential.
Modulating type 2 diabetes
Branched fatty acid esters of hydroxy fatty acids (FAHFAs) were discovered as dysregulated metabolites in mice protected against diabetes and further used to modulate the type 2 diabetes phenotype22. A class of uncharacterized endogenous metabolites were found to be highly upregulated in the adipose tissue and plasma of mice overexpressing the glucose transporter Glut4 compared to their wild-type littermates, in an untargeted metabolomics study. Even though the m/z of these compounds did not correspond to any known metabolite in METLIN and LIPID MAPS, its structure was characterized as FAHFA using fragmentation spectra in negative-ion mode. Glut4-overexpressing transgenic mice have an elevated lipogenesis and glucose tolerance, despite being obese, with elevated levels of circulating fatty acids. Hence, it was hypothesized that FAHFAs could affect glucose and insulin homeostasis. Once chemically characterized and synthesized, palmitic acid 9-hydroxystearic acid (9-PAHSA), one of the most abundant FAHFAs, was tested in an in vivo model of type 2 diabetes. Diabetic mice fed a high-fat diet, that were orally administered 9-PAHSA, showed an overall higher glucose tolerance and insulin sensitivity compared with controls (Fig. 2b). Moreover, in adipocytes, the improvement in glucose metabolism resulted from 9-PAHSA-triggered binding and activation of the GPR120 receptor, a well-known anti-inflammatory and insulin-sensitizing mediator in response to omega-3 fatty acids.
Because type 2 diabetes is accompanied by a low-grade inflammation in adipose tissue that may contribute to the insulin-resistant state, 9-PAHSA was further tested as a possible immunomodulator of the adipose-tissue-associated inflammatory response. Mice orally supplemented with 9-PAHSA showed an effective reduction in the in vivo inflammatory response of adipose tissue macrophages to a high-fat diet. In summary, 9-PAHSA was discovered and tested as a possible phenotype modulator. When exogenously administered, 9-PAHSA increased insulin sensitivity and glucose tolerance in a mouse model of type 2 diabetes22.
Modulating T-cell survival and anti-tumor activity
Metabolic modulation through l-arginine prompted a central memory-like T-cell phenotype with enhanced survival capacity and anti-tumor activity both in vitro (human) and in vivo (mouse model)19. In that study, untargeted flow injection metabolomics analyses32 were performed to determine the dynamic changes in arginine metabolism during a time-course experiment. Results were validated by monitoring cell uptake of isotopically labeled l-arginine to determine its fate/flux as well as enzyme inhibitors and clones.
These observations were then further explored to demonstrate that higher l-arginine levels induced structural alterations in three transcriptional regulators (BAZ1B, PSIP1, and TSN) and modulated T-cell metabolic ‘fitness’ and survival (Fig. 2c).
Modulating innate immune response
Correct regulation of the innate immune response is a key factor in the maintenance of whole-body homeostasis. Dysregulation of the immune response may underpin several illnesses related to an excessive or chronic activation or immunosuppression. Relevant to this, the uncommon phosphatidylinositol species 1,2-diarachidonyl-glycero-3-phosphoinositol (PI(20:4/20:4)) was found to be upregulated in mouse-resident peritoneal macrophages stimulated with the yeast cell wall preparation zymosan, a classic stimulus of the innate immune response16. This lipid species, previously characterized using the LIPID MAPS database, is rapidly formed and degraded upon stimulation, suggesting a role in regulating cell signaling events, such as generation of reactive oxygen species and secretion of lysozyme, two pivotal molecules produced by macrophages for pathogen killing. When added exogenously, macrophages incorporate this molecule into their phospholipid pool and show a higher superoxide anion production and lysozyme secretion than control cells and macrophages enriched with a scrambled phosphatidylinositol species (Fig. 2d), suggesting this molecule plays a key role in the coordination of the macrophage response to zymosan.
Modulating oligodendrocyte maturation
We (J.R.M.-B., G.S., and collaborators)20 have also used MAS to analyze oligodendrocyte precursor cell (OPC) differentiation in multiple sclerosis, an autoimmune disease characterized by demyelination of axons and neuronal dysfunction. Disease remission in multiple sclerosis is dependent on remyelination, which involves the differentiation of OPCs and leads to the formation of mature oligodendrocytes33. Premyelinating oligodendrocytes are present in chronic lesions of patients and inhibition of OPC differentiation is associated with multiple sclerosis disease progression. Therefore, a promising complementary treatment of multiple sclerosis involves the identification of pharmacological agents that stimulate remyelination by enhancing OPC differentiation. Multiple drug candidates have been identified using high-throughput screening34, which induce OPC differentiation in vitro and enhance remyelination in vivo.
We used MS-based metabolomics to investigate how endogenous metabolites play a role in the process of OPC differentiation20. Among other related metabolites, taurine, an amino sulfonic acid, was found to be significantly elevated (~20-fold) over the course of in vitro oligodendrocyte differentiation (Fig. 2e). When added exogenously at physiologically relevant concentrations, taurine not only enhanced drug-induced OPC differentiation but also facilitated the in vitro myelination of co-cultured axons. Unlike in l-arginine T-cell modulation, and overturning the common assumption that upregulated metabolites are end-point biomarkers, the addition of upregulated taurine had a positive effect, further stimulating remyelination during OPC differentiation. Mechanistically, taurine-induced activities that enhance OPC differentiation and myelination appear to be driven by taurine’s ability to increase serine levels, which is an initial building block required for the synthesis of the glycosphingolipid components of myelin.
Outlook
In the past, several metabolites have been discovered as effective phenotype modulators, using approaches other than MAS, including cellular fractioning, ligand-binding assays, and enzymatic assays. Examples include sphingosine-1-phosphate (immunomodulation)35, docosahexaenoic acid (cognitive function)36, carnitine (fertility)37, anandamide (neurological disorders)38, and melatonin (sleep)39, to name a few. However, the use of metabolomics data to characterize dysregulated metabolites of interest is gaining more attention because this approach is able to detect a wide range of small molecules at low concentrations, increasing throughput. Thus, metabolomics has been successful in identifying active metabolites for phenotype modulation (Table 1). It is clear that metabolomics can enable identification of molecules with interesting and potentially beneficial functions.
Table 1.
Metabolomics activity screening examples
| Metabolite | System | Original observation | Induced phenotype | Reference |
|---|---|---|---|---|
| Protectin D1 | Stem cell differentiation | Polyunsaturated fatty acid precursors decrease during differentiation | Promotes differentiation into neurons | 14 |
| cis-9,10-octadecenoamide (oleamide) | Sleep induction | Accumulated in cerebrospinal fluid of sleep-deprived felines | Induces sleep | 41 |
| Trimethylamine N-oxide | Cardiovascular disease | Augmented in plasma of subjects with cardiovascular risk | Increases scavenger receptors expression, foam cell formation and atherosclerotic lesions | 27,28 |
| N,N-dimethylsphingosine | Chronic pain | Increased in a rat model of chronic neuropathic pain | Elicits neuropathic pain behavior and cytokine release | 21 |
| PI(20:4/20:4) | Pathogen killing | Upregulated in zymosan-stimulated macrophages | Increases superoxide anion production and lysozyme release | 16 |
| 3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid | Gestational and type 2 diabetes | Elevated in plasma in human and mice models of gestational diabetes and type 2 diabetes | Impairs glucose tolerance and β-cell function | 23 |
| 9-PAHSA | Type 2 diabetes | Increased in plasma and adipose tissue of diabetes-protected mice. Decreased in diabetic humans | Improves glucose metabolism and insulin sensitivity | 22 |
| S-adenosyl methionine | Stem cell differentiation | Downregulated in naive embryonic stem cells | Induces differentiation to primed stem cells | 15 |
| cis-7-hexadecenoic acid (16:1n-9) | Cardiovascular disease | Elevated in atherosclerosis-initiating foamy monocytes and macrophages | Decreases inflammatory response to bacterial lipopolysaccharide in monocytes and macrophages | 17 |
| Dioxolane A3 | Acute inflammation | Increased in thrombin-activated platelets | Promotes neutrophil recruitment and activation | 18 |
| Proline, isoleucine, and phenylalanine | Synthetic mutualism | Secreted by Zymomonas mobilis | Results in rescue and growth of Escherichia coli auxotrophs in co-culture with Z. mobilis | 42 |
| l-arginine | Adaptive immune response | Decreased in activated naive T cells | Induces differentiation into memory-like cell, increases survival and anti-tumor activity | 19 |
| Taurine | Multiple sclerosis | Upregulated during oligodendrocyte precursor cell differentiation | Enhances oligodendrocyte differentiation and myelination | 20 |
Notwithstanding MAS’s clear utility, challenges exist that could impede the broad its implementation. It is unclear how to accurately identify either the best candidate molecules for further testing or which molecule among the numerous other dysregulated metabolites is likely to be the most effective phenotype modulator. Statistical analyses, metabolite classification schemes based on prior metabolite activity knowledge, and pathway redundancy have all been used to prioritize the best candidates and reduce the need for large-scale biological validation experiments. Follow-up experiments including the use of pathway metabolites, pathway inhibitors and stable isotope labeling as well as flux analysis are valid strategies to further reduce the list of candidate metabolites. Another challenge for untargeted metabolomics studies is the identification of ‘unknown’ molecules. This is attributed to the chemical diversity and heterogeneity of the metabolome, and substantial effort has been dedicated to the development of advanced computational tools for tandem MS prediction and metabolite characterization. Although thousands of metabolites are commercially available, a limiting aspect of MAS can be the lack of commercially available reference materials for activity validation, particularly for lesser known or characterized metabolites. In those cases, the potential solutions available at present involve synthetic or isolation strategies, and for the most part, the former is the more common approach because large amounts of sample are rarely available to undertake isolation attempts40.
Discovery metabolomics has largely been used to identify biomarkers and characterize mechanisms of biological action. Going forward, the use of MAS to identify biologically active endogenous metabolites that can be used to intentionally alter phenotype might prove to be a far more effective application of metabolomics. Metabolites identified using MAS can be used to induce a phenotypic response alone, or in conjunction with a drug. MAS might conceivably permit dose or side effect reduction while maintaining or even improving therapeutic outcomes.
Applications of MAS could be expanded to disease modulation, biofilm initiation or suppression, drug–exposome interactions, plant biology and immunotherapy. Perhaps what is most intriguing is that rather than identifying metabolites to understand pathways, we can apply metabolites to modulate physiology, thereby turning the tables on conventional thinking.
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health (Grants R01 GM114368-03, P30 MH062261-10, P01 DA026146-02), and support was also received from Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research under Contract DE-AC02-05CH11231.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
References
- 1.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patti GJ, Yanes O, Siuzdak G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 4.Woolf LI, Griffiths R, Moncrieff A. Treatment of phenylketonuria with a diet low in phenylalanine. BMJ. 1955;1:57–64. doi: 10.1136/bmj.1.4905.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamanna VS, Kashyap ML. Mechanism of action of niacin. Am. J. Cardiol. 2008;101(8A):20B–26B. doi: 10.1016/j.amjcard.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 6.Banach M, et al. Statin therapy and plasma coenzyme Q10 concentrations--A systematic review and meta-analysis of placebo-controlled trials. Pharmacol. Res. 2015;99:329–336. doi: 10.1016/j.phrs.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautenhahn R, et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart DS, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahy E, et al. A comprehensive classification system for lipids. J. Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Vinaixa M, et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. Trends Analyt. Chem. 2016;78:23–35. [Google Scholar]
- 14.Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sperber H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gil-de-Gómez L, et al. A phosphatidylinositol species acutely generated by activated macrophages regulates innate immune responses. J. Immunol. 2013;190:5169–5177. doi: 10.4049/jimmunol.1203494. [DOI] [PubMed] [Google Scholar]
- 17.Guijas C, Meana C, Astudillo AM, Balboa MA, Balsinde J. Foamy monocytes are enriched in cis-7-hexadecenoic fatty acid (16:1n-9), a possible biomarker for early detection of cardiovascular disease. Cell Chem. Biol. 2016;23:689–699. doi: 10.1016/j.chembiol.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Hinz C, et al. Human platelets utilize cycloxygenase-1 to generate dioxolane A3, a neutrophil-activating eicosanoid. J. Biol. Chem. 2016;291:13448–13464. doi: 10.1074/jbc.M115.700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger R, et al. L-arginine modulates t cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–842.e13. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beyer BA, et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 2018;14:22–28. doi: 10.1038/nchembio.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patti GJ, et al. Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nat. Chem. Biol. 2012;8:232–234. doi: 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yore MM, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prentice KJ, et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 2014;19:653–666. doi: 10.1016/j.cmet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Huan T, et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods. 2017;14:461–462. doi: 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlgemuth G, et al. SPLASH, a hashed identifier for mass spectra. Nat. Biotechnol. 2016;34:1099–1101. doi: 10.1038/nbt.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koeth RA, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panopoulos AD, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA. 2005;102:4783–4788. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuhrer T, Heer D, Begemann B, Zamboni N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 2011;83:7074–7080. doi: 10.1021/ac201267k. [DOI] [PubMed] [Google Scholar]
- 33.Franklin RJM, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- 34.Deshmukh VA, et al. A regenerative approach to the treatment of multiple sclerosis. Nature. 2013;502:327–332. doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernández-Pisonero I, et al. Synergy between sphingosine 1-phosphate and lipopolysaccharide signaling promotes an inflammatory, angiogenic and osteogenic response in human aortic valve interstitial cells. PLoS One. 2014;9:e109081. doi: 10.1371/journal.pone.0109081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cardoso C, Afonso C, Bandarra NM. Dietary DHA and health: cognitive function ageing. Nutr. Res. Rev. 2016;29:281–294. doi: 10.1017/S0954422416000184. [DOI] [PubMed] [Google Scholar]
- 37.Ng CM, Blackman MR, Wang C, Swerdloff RS. The role of carnitine in the male reproductive system. Ann. NY Acad. Sci. 2004;1033:177–188. doi: 10.1196/annals.1320.017. [DOI] [PubMed] [Google Scholar]
- 38.Wise LE, Shelton CC, Cravatt BF, Martin BR, Lichtman AH. Assessment of anandamide’s pharmacological effects in mice deficient of both fatty acid amide hydrolase and cannabinoid CB1 receptors. Eur. J. Pharmacol. 2007;557:44–48. doi: 10.1016/j.ejphar.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Hardeland R, et al. Melatonin—a pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 2011;93:350–384. doi: 10.1016/j.pneurobio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 40.Cohen LJ, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cravatt BF, et al. Chemical characterization of a family of brain lipids that induce sleep. Science. 1995;268:1506–1509. doi: 10.1126/science.7770779. [DOI] [PubMed] [Google Scholar]
- 42.Kosina SM, et al. Exometabolomics assisted design and validation of synthetic obligate mutualism. ACS Synth. Biol. 2016;5:569–576. doi: 10.1021/acssynbio.5b00236. [DOI] [PubMed] [Google Scholar]