Abstract
Transforming growth factor-β (TGF-β) induces the expression of Disabled-2 (Dab2), an endocytic adaptor and tumour suppressor, concomitant with the induction of an epithelial–mesenchymal transition (EMT) in mammary epithelial cells. Here we show that following TGF-β-mediated EMT, sustained TGF-β treatment leads to proteolytic degradation of Dab2 by cathepsin B (CTSB), loss of the mesenchymal phenotype and induction of autophagy. CTSB inhibition or expression of a CTSB-resistant Dab2 mutant maintains Dab2 expression and shifts long-term TGF-β-treated cells from autophagy to apoptosis. We further show that Dab2 interacts with Beclin-1 to promote casein-kinase-2-mediated phosphorylation of Beclin-1, preventing Beclin-1–Vps34 interaction and subsequent autophagosome assembly. Thus, CTSB-mediated degradation of Dab2 allows Beclin-1–Vps34 induction of autophagy, whereas sustained Dab2 expression prevents autophagy and promotes apoptosis by stabilizing the pro-apoptotic Bim protein. In vivo studies suggest that Dab2-mediated regulation of autophagy modulates chemotherapeutic resistance and tumour metastasis.
Transforming growth factor-β (TGF-β) is a potent inducer of epithelial–mesenchymal transitions (EMTs), in which cells undergo a switch from a polarized, epithelial phenotype to a highly motile fibroblastic or mesenchymal phenotype1. EMT is a fundamental process during normal embryonic development and in adult tissue homeostasis, but can also become aberrantly activated during metastatic progression and chemoresistance2–4. TGF-β has also been reported to regulate autophagy5; a cellular process involving the lysosomal-mediated degradation of cellular components characterized by the formation of autophagosomes that engulf portions of the cytosol, damaged organelles and protein aggregates that ensures survival during starvation and/or periods of stress. In cancer, autophagy plays both tumour promoter and suppressor roles6. During tumour initiation, autophagy prevents the accumulation of oncogenic protein substrates, toxic unfolded proteins and damaged organelles7; in established tumours, autophagy promotes growth by prolonging tumour cell survival under stress condition, such as starvation and chemotherapy8.
Here we find that the endocytic adaptor and tumour suppressor Disabled-2 (Dab2)9, whose expression is initially induced during TGF-β-mediated EMT10, is attenuated following prolonged exposure of cells to TGF-β (>3 d). We demonstrate that chronic exposure of cells to TGF-β leads to CTSB-mediated proteolysis of Dab2 accompanied by loss of the mesenchymal and gain of the autophagic phenotype. CTSB knockdown (KD) or overexpression of a CTSB-resistant Dab2 mutant prevents TGF-β-induced autophagy and instead stabilizes Bim expression to promote apoptosis11. Further, CTSB-mediated Dab2 degradation attenuates drug-induced apoptosis by promoting autophagy and cell survival, and CTSB KD or overexpression of CTSB-resistant Dab2 enhances drug-induced apoptosis by abrogating autophagy. Thus, we identify Dab2 as an inhibitor of autophagy and a promoter of apoptosis, suggesting that targeting of this molecular mechanism may provide therapeutic benefit.
RESULTS
Chronic TGF-β treatment results in loss of EMT and Dab2 cleavage
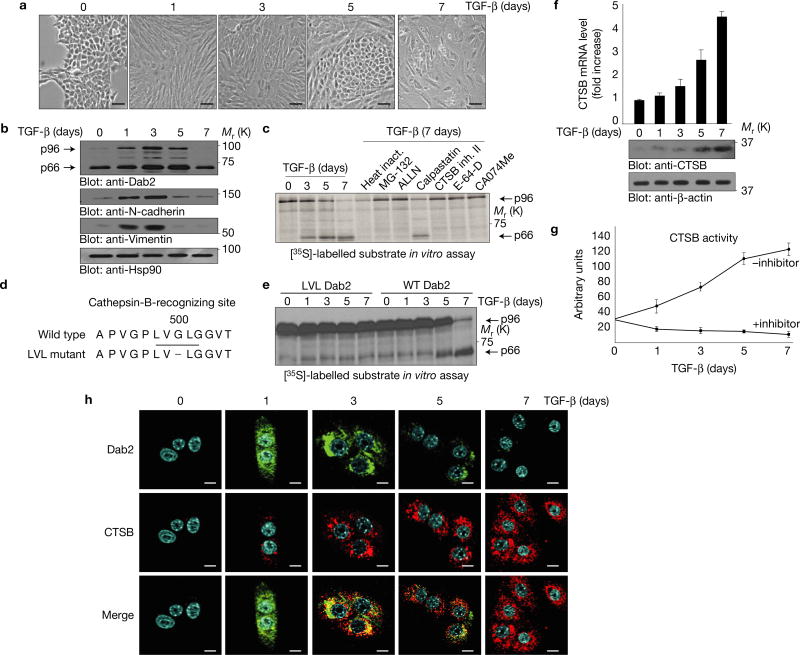
The EMT phenotype (Fig. 1a) in NMuMG cells is induced and persists for 3 days following TGF-β treatment. Thereafter (7 d), cells appeared to transit to a state morphologically suggestive of either autophagy or apoptosis12. Expression levels of the mesenchymal markers N-cadherin and vimentin, initially upregulated during EMT, were attenuated following long-term exposure to TGF-β (Fig. 1b) as was the expression of the p96 isoform of Dab2 (Fig. 1b; top panel). Total messenger RNA levels for the p96 isoform of Dab2 showed no variation with TGF-β treatment confirming our previous results that Dab2 induction by TGF-β is translationally regulated10 (Supplementary Fig. 1a; top panel). A lower-molecular-mass Dab2 band was detected and increased initially during long-term treatment with TGF-β and decreased after 7 days of treatment (Fig. 1b). Similar results were also observed in two other cell lines, mouse Eph4 Ras and human mammary gland epithelial cells (HMLE) (Supplementary Fig. 1b; top panel). Alternative splicing of Dab2 mRNA generating a p96 isoform and a p67 isoform has previously been reported13 but since we could not detect the p67 mRNA isoform in NMuMG cells (Supplementary Fig. 1a; lower panel), we postulated that this lower band (referred to as p66 Dab2) might represent a cleavage product of the p96 isoform of Dab2.
Figure 1.
Chronic TGF-β treatment results in reversal of EMT and Dab2 cleavage by CTSB. (a) Morphological analysis of NMuMG cells treated with TGF-β for the times indicated. Images were captured using a digital camera mounted on an inverted microscope. Scale bars, 100 µm. (b) Whole-cell lysates from cells treated with TGF-β for the times indicated were subjected to immunoblot analysis. Hsp90 expression served as a loading control. (c) In vitro protease inhibitor screen for p96 Dab2 cleavage. In vitro synthesized and [35S]-methionine-labelled p96 Dab2 was incubated for 1 h at 37 °C with extracts from cells treated with TGF-β for the times indicated. Each inhibitor (20 µM) was added to cells 6 h before the preparation of whole-cell lysates and autoradiographic analysis. (d) Amino acid sequence of the wild-type Dab2 and mutant LVL Dab2 cleavage sites. (e) In vitro cleavage analysis of [35S]-methionine-labelled wild-type Dab2 and LVL Dab2 mutant. (f) Real-time PCR analysis of CTSB mRNA and immunoblot analysis of CTSB protein levels in NMuMG cells treated with TGF-β for the times indicated; data are shown as mean ± s.d., n = 5 independent experiments. (g) CTSB activity assay in vitro using extracts from NMuMG cells treated with TGF-β for the times indicated; data are shown as mean ± s.d., n = 3 independent experiments. (h) Immunofluorescence analysis of Dab2 and CTSB in NMuMG cells treated with TGF-β for the times indicated. Scale bars, 10 µm. All experiments were repeated at least three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
CTSB is responsible for Dab2 cleavage
A protease inhibitor screen was performed to determine whether the p66 Dab2 band is generated through proteolytic cleavage of p96 Dab2 (Supplementary Fig. 1c). The specific CTSB inhibitor II and CA074Me inhibited p96 cleavage, suggesting that the p66 band of Dab2 is generated through CTSB-specific cleavage of p96 Dab2. In vitro proteolytic cleavage of Dab2, using [35S]-labelled p96 Dab2 as the substrate and incubating with extracts from cells treated with TGF-β in the presence or absence of protease inhibitors, demonstrated that inhibition of CTSB prevented accumulation of the p66 form of Dab2 (Fig. 1c). A series of Flag-tagged Dab2 constructs representing either the full-length p96 form or various Dab2 truncation mutants (Supplementary Fig. 1d) determined that the CTSB cleavage site is flanked by amino acids Val499 and Gly500 (Supplementary Fig. 1e,f), thereby defining the CTSB recognition sequence as Leu-Val-Gly-Leu (Fig. 1d). To confirm this as the CTSB recognition site, we mutated the Leu-Val-Gly-Leu sequence to Leu-Val-Leu and show that this Dab2 mutant is not cleaved by CTSB (Fig. 1e).
Sustained TGF-β treatment transcriptionally induces CTSB expression and promotes its co-localization with Dab2
TGF-β treatment significantly induced CTSB mRNA (Fig. 1f), protein expression (Fig. 1f), and CTSB activity (Fig. 1g). Immunofluorescence confirmed the induction of Dab2 and CTSB (Fig. 1h) and also their co-localization, with maximal co-localization observed following 3 days of TGF-β treatment. Long-term TGF-β treatment also induced CTSB protein expression in Eph4 Ras and HMLE cells (Supplementary Fig. 1b; middle panel). Silencing of both Smad2 and Smad3, through short hairpin RNA (shRNA)-mediated KD (Supplementary Fig. 1h) significantly attenuated TGF-β-mediated EMT, as monitored by vimentin and N-cadherin regulation; however, TGF-β-induced CTSB expression levels (Supplementary Fig. 1i) and activity (Supplementary Fig. 1k) were unaffected by Smad2/3 silencing, suggesting that CTSB regulation is independent of canonical TGF-β signalling. Smad2/3 phosphorylation also suggested that non-canonical TGF-β signalling is involved in this chronic treatment (Supplementary Fig. 1g). Cellular fractionation analyses (Supplementary Fig. 1j) demonstrated that while most of the TGF-β-induced CTSB is located in the cytosolic fraction some is associated with cell membranes.
Sustained TGF-β treatment induces autophagy
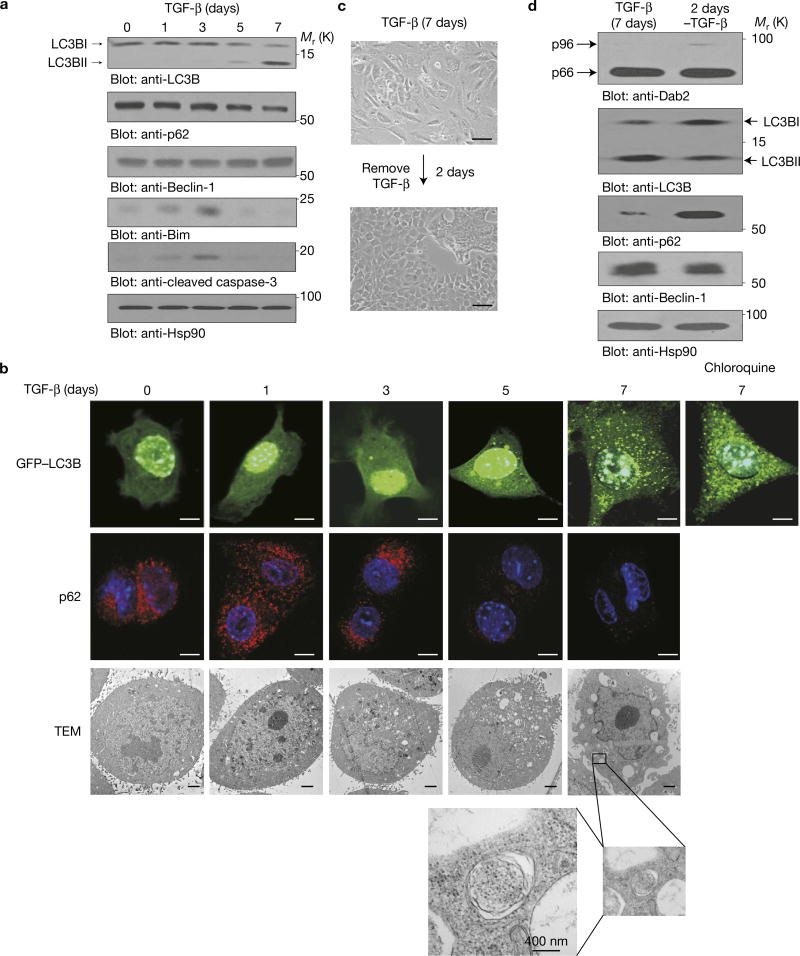
Direct analysis of autophagic (LC3B, p62 and Beclin-1) markers demonstrated that LC3BII was significantly induced by TGF-β treatment whereas p62 was downregulated (Fig. 2a). The autophagic induction of LC3BII by TGF-β was confirmed through the use of chloroquine, which inhibits autolysosome formation and the turnover of LC3BII (that is, autophagic flux)14, and resulted in an increase in TGF-β-induced LC3BII levels (Supplementary Fig. 2a, left panel). Similar results were observed in Eph4 Ras and HMLE cells (Supplementary Fig. 1b bottom panel). The apoptotic markers, Bim and cleaved caspase-3, were initially upregulated, but their expression was attenuated following a 7-day TGF-β exposure. Immunofluorescence confirmed the autophagic effects of chronic TGF-β with increased LC3B puncta in GFP–LC3B-transfected NMuMG cells and decreased endogenous p62 expression (Fig. 2b and Supplementary Fig. 2b). Increased autophagosome structures were also observed by transmission electron microscopy following chronic TGF-β treatment (Fig. 2b). Additionally, the autophagic effects were reversible in that removal of TGF-β reversed autophagy (Fig. 2c,d) within 2 days. These data demonstrate that chronic TGF-β treatment induces autophagy concomitant with the loss of the mesenchymal phenotype.
Figure 2.
Sustained TGF-β treatment induces autophagy. (a) Immunoblot analysis of NMuMG cells treated with TGF-β for the times indicated. (b) Immunofluorescence analysis of NMuMG cells stably expressing GFP–LC3B treated with TGF-β for the times indicated. Scale bars, 10 µm (upper two rows). Transmission electron microscopy (TEM) analysis (lower panels) was performed to detect autophagosomal and autolysosomal structures in cells treated with TGF-β. The framed section highlights an autophagosome. Scale bars, 2 µm. (c) Morphologic analysis of NMuMG cells treated with TGF-β for 7 days before TGF-β removal and growth for an additional 2 days. Scale bars, 100 µm. (d) Immunoblot analysis was performed in NMuMG cells treated with TGF-β for 7 days before TGF-β removal and growth for an additional 2 days. All experiments were repeated at least three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
CTSB-mediated Dab2 degradation promotes TGF-β-induced autophagy
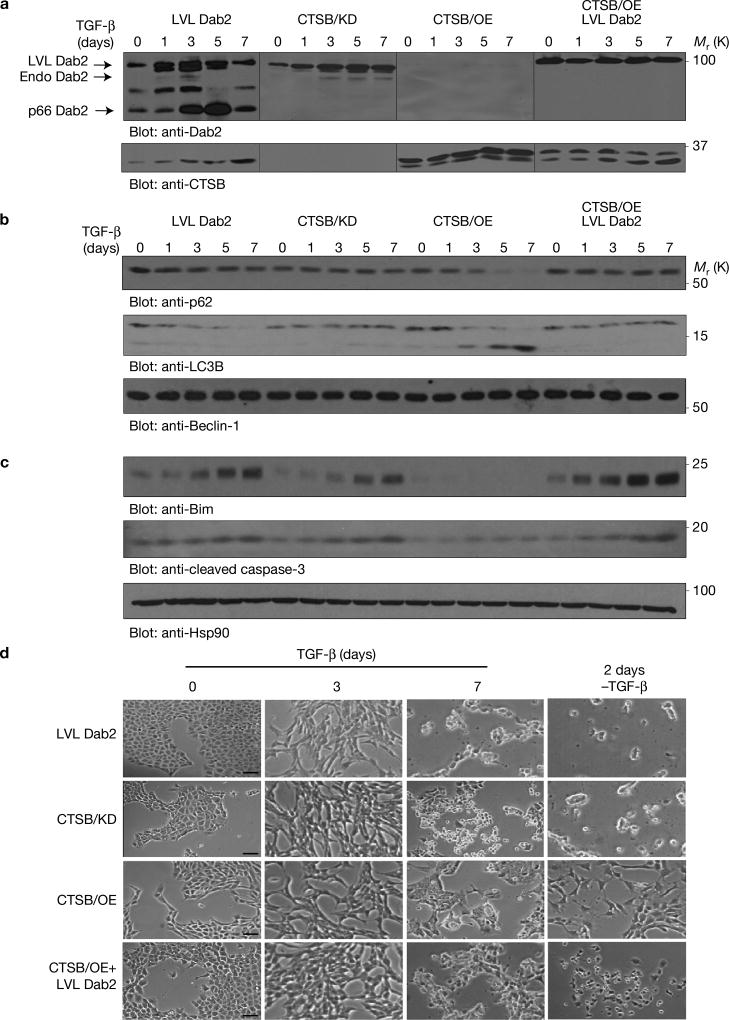
To modulate Dab2 and CTSB expression levels, we expressed a CTSB cleavage mutant construct of Dab2 containing only the Leu-Val-Leu residues of the identified Leu-Val-Gly-Leu CTSB site (Fig. 1d), and designated these cells ‘LVL Dab2’. We silenced CTSB using an shRNA-mediated approach (CTSB/KD cells), overexpressed CTSB expression using a myc-tagged vector (CTSB/OE cells), and also expressed LVL Dab2 in CTSB/OE cells (CTSB/OE+LVL Dab2 cells). Appropriate modulation ofDab2 and CTSB was achieved (Fig. 3a) and TGF-β treatment led to a time-dependent degradation of endogenous Dab2 but not of the LVL Dab2 (Fig. 3a). Endogenous Dab2 levels were stabilized in CTSB/KD cells following TGF-β treatment and essentially non-existent in CTSB/OE cells. In CTSB/OE+LVL Dab2 cells, endogenous Dab2 was absent, suggesting that Dab2 is not stable in the presence of excessive CTSB, but exogenous mutant LVL Dab2 levels were constant throughout the TGF-β treatment (Fig. 3a). In the presence of Dab2 (LVL Dab2, CTSB/KD and CTSB/OE+LVL Dab2 cells), autophagic markers were not affected by TGF-β (Fig. 3b). Only in CTSB/OE cells, lacking Dab2, did TGF-β cause a time-dependent decrease in p62 levels and an increase in LC3BII levels (Fig. 3b and Supplementary Fig. 2a; right panel). In Dab2-containing cells, and not in CTSB/OE cells, TGF-β caused a time-dependent increase in the pro-apoptotic markers (Fig. 3c and Supplementary Fig. 2c). TGF-β induced significant morphological changes in all cells following a 7-day treatment (Fig. 3d). Two days following TGF-β removal, only CTSB/OE cells reversed to a more epithelial phenotype; cells containing Dab2 did not reverse but instead underwent increased cell death (Supplementary Fig. 2d). Analyses of the EMT markers N-cadherin and vimentin and the migration and invasion markers MMP3 and MMP9 demonstrated that EMT was induced within the first 3 days of TGF-β treatment and subsequently downregulated following more chronic TGF-β exposure (Supplementary Fig. 2e), suggesting that Dab2 is modulated by CTSB and serves as a switch to regulate TGF-β-induced autophagy and apoptosis.
Figure 3.
CTSB-mediated Dab2 degradation promotes TGF-β-induced autophagy. (a) NMuMG cells and their derivatives were treated with TGF-β for the times indicated before the preparation of whole-cell lysates and immunoblot analyses using anti-Dab2 and anti-CTSB antibodies. The resultant images are the product of time average data. The lines on the western blots demarcate individual blots that were run in parallel. (b) The same as in a, but with anti-p62, anti-LC3B and anti-Beclin-1 antibodies. (c) The same as in a, but with anti-Bim, anti-cleaved caspase-3 and anti-Hsp90 antibodies. Hsp90 expression served as a loading control. (d) Morphological analysis of modified cell lines after 3 days and 7 days of TGF-β treatment and 2 days after the removal of TGF-β from cells treated for 7 days with TGF-β. Scale bars, 100 µm. The experiments were repeated three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
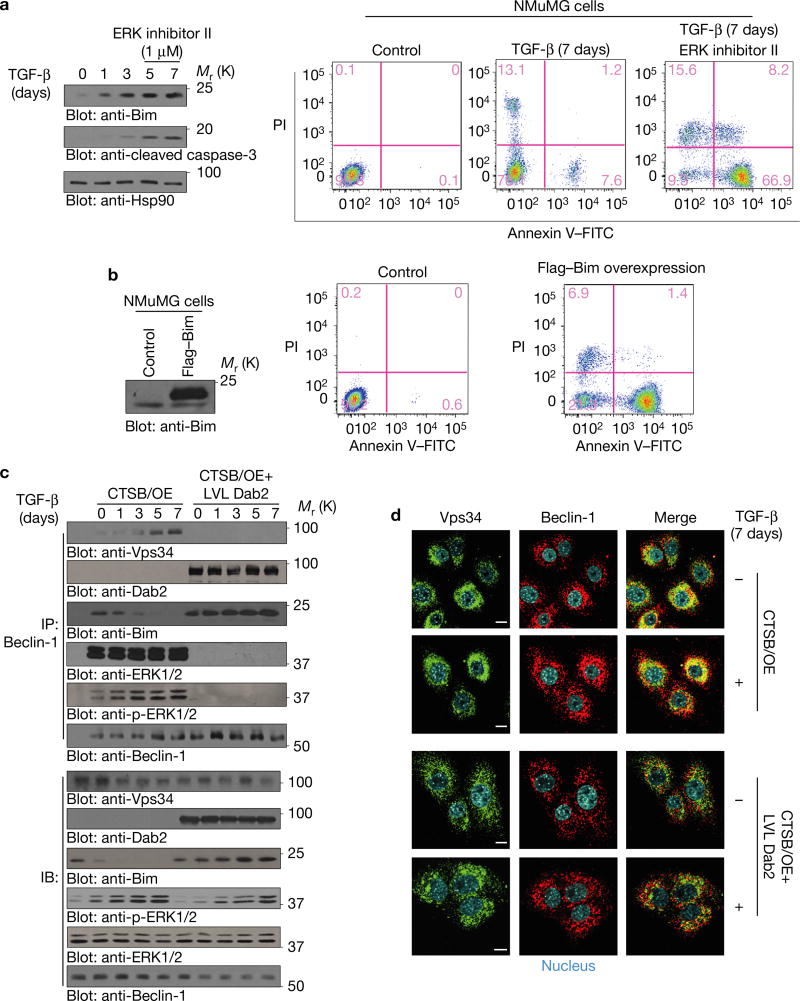
Dab2 inhibits autophagy by blocking Vps34–Beclin-1 interactions and stabilizes Bim by attenuating ERK–Bim interactions
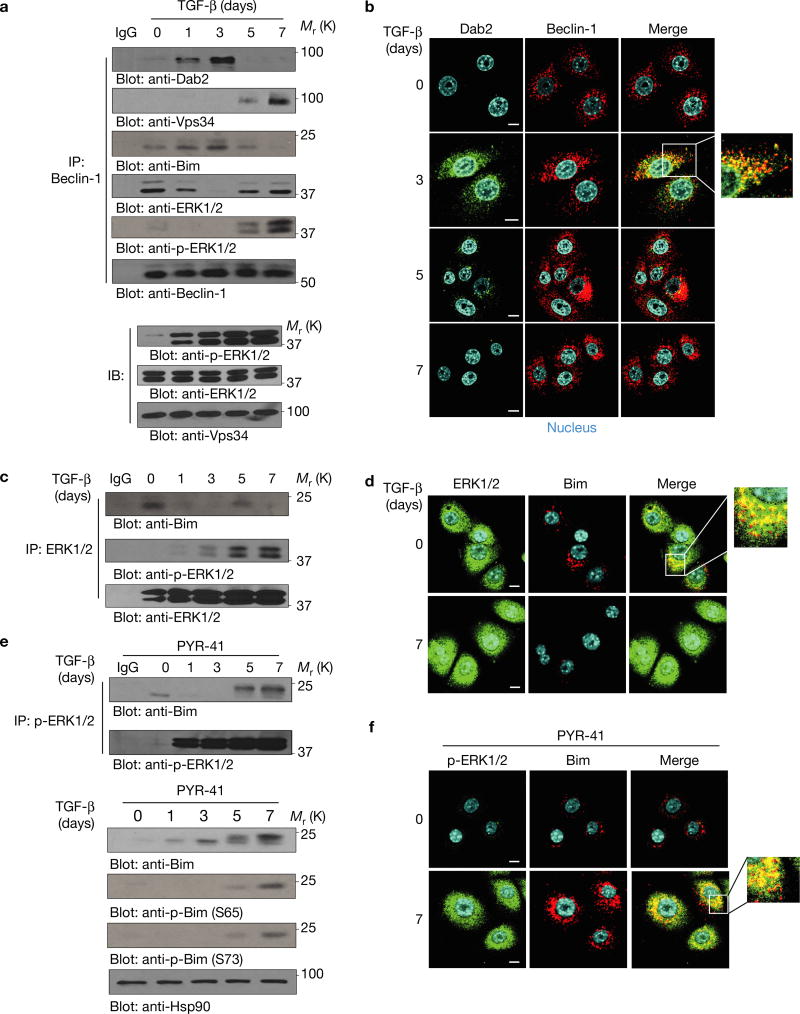
Autophagosome assembly was monitored by examining Beclin-1–Vps34 complex formation. As shown (Fig. 4a–d), TGF-β-induced Dab2 interacted with Beclin-1 and prevented Beclin-1–Vps34, Beclin-1–ERK and ERK–Bim interactions. In the presence of PYR-41 (an inhibitor of ubiquitin-activating enzyme E1 that targets p-Bim ubiquitylation and degradation), p-ERK–Bim interactions (Fig. 4e, top panel) and p-Bim levels (Fig. 4e, bottom panel) were increased following Dab2 degradation. Since ERK-mediated phosphorylation of Bim promotes its proteasomal degradation15, our results suggest that Beclin-1–Bim–Dab2 interactions prevent ERK-mediated Bim Ser65 phosphorylation and stabilize Bim to promote apoptosis. Following chronic TGF-β treatment and Dab2 degradation, Beclin-1–Vps34 interactions were re-established and Bim was phosphorylated and degraded; however, in the presence of the ERK inhibitor, Bim was stabilized resulting in apoptosis (Fig. 5a). Lentivirus-mediated Bim expression also promoted apoptosis (Fig. 5b). In CTSB/OE cells, TGF-β induced Beclin-1–Vps34 interaction while Bim levels were significantly downregulated (Fig. 5c). In CTSB/OE+LVL Dab2 cells, LVL Dab2 prevented Beclin-1–Vps34, Beclin–ERK, ERK–Bim and p-ERK–Bim interactions, resulting in Bim induction and stabilization (Fig. 5c,d and Supplementary Fig. 3a,b). Similarly, by immunofluorescence (Fig. 5d and Supplementary Fig. 3c,d), Beclin-1–Vps34, ERK–Bim and p-ERK–Bim co-localizations were observed following a 7-day TGF-β treatment of CTSB/OE, but these failed to co-localize in CTSB/OE+LVL Dab2 mutant cells. The co-immunoprecipitation data (Supplementary Fig. 4a) further demonstrated that the evolutionarily conserved domain of Beclin-1 contributes to its interaction with Dab2. The data suggest that Dab2–Beclin-1 interactions prevent Beclin-1–Vps34 associations and subsequent autophagosome formation, whereas Dab2–Beclin-1–Bim complex formation prevents ERK–Bim interactions and stabilizes Bim.
Figure 4.
Dab2 inhibits chronic TGF-β-induced autophagy by blocking Vps34–Beclin-1 interaction and promotes apoptosis by attenuating ERK–Bim interaction. (a) Beclin-1 immunoprecipitation using whole-cell lysates from cells treated with TGF-β for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of Dab2, Vps34, Bim, ERK and p-ERK. (b) Immunofluorescence analysis of Dab2 and Beclin-1 in NMuMG cells treated with TGF-β for the times indicated. Photos were taken by confocal microscope. Scale bars, 10 µm. (c) Co-immunoprecipitation analysis using anti-ERK1/2 antibody from whole-cell lysates of NMuMG cells treated with TGF-β for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of p-ERK1/2 and Bim. (d) Immunofluorescence analysis was performed to detect the co-localization between ERK and Bim before and after NMuMG cells were treated for 7 days. Scale bars, 10 µm. (e) Top: p-ERK1/2 immunoprecipitation using whole-cell lysates of NMuMG cells treated with TGF-β for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of Bim. As a control, p-Bim ubiquitylation and degradation is inhibited by PYR-41, a cell-permeable inhibitor of the ubiquitin-activating enzyme E1 (20 µM, 4 h before cell lysates were collected). Bottom: immunoblotting analysis was employed to detect the Bim and p-Bim expression from whole-cell lysates of NMuMG cells treated with TGF-β for the times indicated in the presence of 20 µM PYR-41. (f) Immunofluorescence analysis was performed to detect the co-localization between p-ERK and Bim before and after NMuMG cells were treated for 7 days in the presence of PYR-41. Scale bars, 10 µm. The experiments were repeated three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
Figure 5.
Dab2 blocks ERK–Bim interaction and attenuates ERK-mediated Bim phosphorylation in NMuMG cells. (a) Immunoblotting analysis of apoptosis markers Bim and cleaved caspase-3 from whole-cell lysates of NMuMG cells treated with TGF-β for the times indicated. ERK inhibitor II was applied to cell media 3 h before collecting cell lysates of 5-day-and 7-day-treated cells (left). Flow cytometric analyses of apoptosis using Annexin V staining were evaluated in NMuMG cells treated with TGF-β for 7 days in the presence or absence of ERK inhibitor II (right). (b) Flow cytometric analyses of apoptosis using Annexin V staining were evaluated in NMuMG cells with lentivirus-mediated Bim expression. (c) Co-immunoprecipitation analysis using anti-Beclin-1 antibody of whole-cell lysates from CTSB/OE and CTSB/OE+LVL Dab2 NMuMG cells treated with TGF-β for the times indicated. Immunoprecipitated complexes were analysed to detect the expression of Vps34, Dab2, ERK, p-ERK, Bim and Beclin-1. Immunoblots (lower panels) depict expression levels of Vps34, Dab2, ERK, p-ERK, Bim and Beclin-1 in whole-cell lysates. (d) Immunofluorescence analysis to detect the expression and localization of Vps34 and Beclin-1 in CTSB/OE and CTSB/OE + LVL Dab2 cells treated ± TGF-β for 7 days. Photos were taken by confocal microscope. Scale bars, 10 µm. The experiments were repeated three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
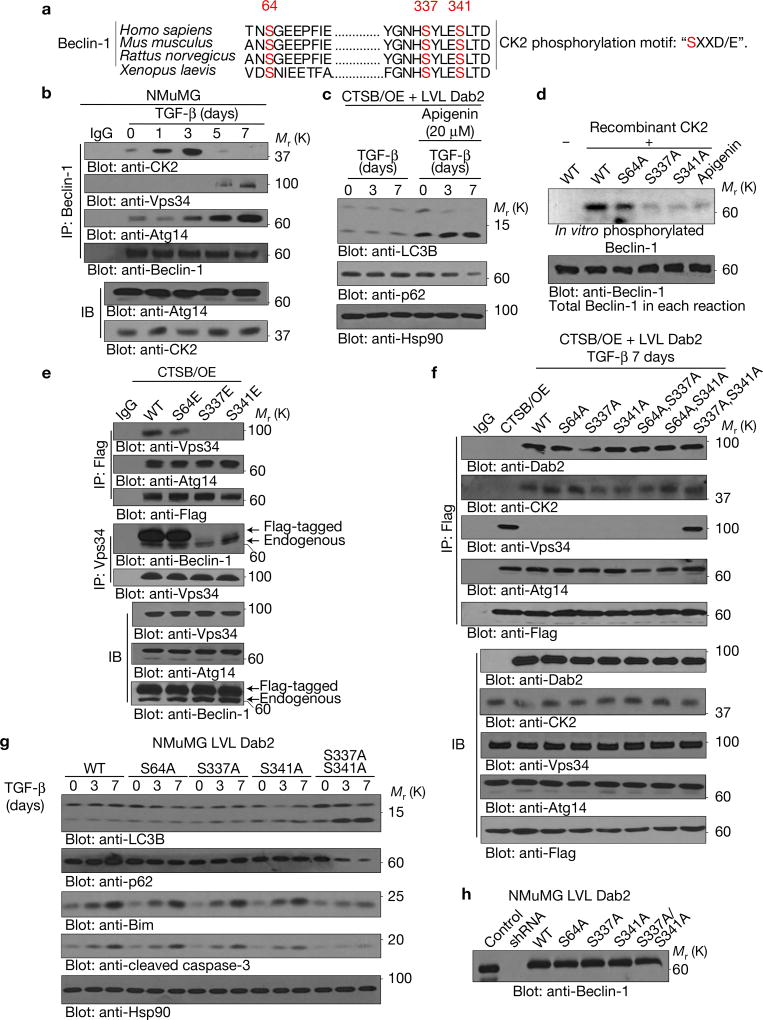
Dab2 promotes casein kinase 2 (CK2)-mediated Beclin-1 phosphorylation resulting in Beclin-1–Vps34 dissociation
Previously we showed that Dab2 interacts with casein kinase 2 (CK2)16, and since Beclin-1 has been shown to contain three CK2 consensus phosphorylation motifs ‘SXXD/E’17 (Fig. 6a), we reasoned that the Dab2–CK2 complex may mediate phosphorylation of Beclin-1 to modulate Beclin-1–Vps34 interactions. As shown (Figs 4a and 6b), Beclin-1 interacted with both Dab2 and CK2 in a TGF-β-dependent manner. Following chronic TGF-β treatment (5 and 7 days), when Dab2 is cleaved by CTSB (Figs 1b and 6b), Beclin-1–CK2 interactions were lost and Beclin-1–Vps34 interactions were re-established (Figs 4a and 6b). Immunofluorescence (Supplementary Fig. 4b) demonstrated that CK2 and Beclin-1 co-localized in CTSB/OE+LVL Dab2 cells, and not in CTSB/OE cells, suggesting that Dab2 mediates CK2–Beclin-1 interactions. The addition of the CK2 inhibitor apigenin to CTSB/OE + LVL Dab2 cells rendered these cells sensitive to TGF-β-induced autophagy (Fig. 6c). Further, Dab2-mediated inhibition of the Beclin-1–Vps34 interactions was abolished in the presence of the CK2 inhibitor apigenin (Supplementary Fig. 4c). To confirm that Beclin-1 is a substrate of CK2, we performed an in vitro kinase assay using recombinant CK2 kinase and [γ-32P]ATP to phosphorylate wild-type (WT) and mutant forms of recombinant Beclin-1. As shown (Fig. 6d), WT Beclin-1 was phosphorylated by CK2, and this phosphorylation was blocked by apigenin. Mutation of Ser64 had no significant effect on Beclin-1 phosphorylation by CK2, whereas mutation of the Ser337 or Ser341 site dramatically inhibited CK2-mediated phosphorylation of Beclin-1, indicating that these two sites are the major CK2 phosphorylation sites.
Figure 6.
Dab2 promotes CK2-mediated Beclin-1 phosphorylation resulting in Beclin-1–Vps34 dissociation. (a) Schematic representation of the three CK2 consensus phosphorylation sites in Beclin-1 (Ser64, Ser337 and Ser341). (b) Beclin-1 immunoprecipitation analysis using whole-cell lysates from cells treated with TGF-β for the times indicated. Immunoprecipitated complexes were immunoblotted to detect the expression of Dab2, CK2, Beclin-1, Atg14 and Vps34. Straight immunoblot analysis depicts the expression level of CK2 and Atg14 in whole-cell lysates. (c) Immunoblot analysis of whole-cell lysates from CTSB/OE + LVL Dab2 cells treated with TGF-β for the times indicated. The CK2 inhibitor apigenin (20 µM) was added 6 h before the preparation of whole-cell lysates. (d) In vitro kinase assay using recombinant CK2 and in vitro-synthesized WT Beclin-1 and Beclin-1 mutants S64A, S337A and S341A as substrates. Reactions were subjected to SDS–PAGE and autoradiography. Apigenin was used as a specific inhibitor of CK2. (e) Co-immunoprecipitation analysis using either anti-Flag or anti-Vps34 antibody of whole-cell lysates from NMuMG CTSB/OE cells transfected with Flag-tagged wild-type Beclin-1 and three phosphomimetic Beclin-1 mutants. Immunoprecipitated complexes were immunoblotted with anti-Vps34, anti-Atg14 and anti-Flag antibodies to determine the interaction between Vps34, Atg14 and Beclin-1. (f) Flag immunoprecipitation using whole-cell lysates from 7-day-TGF-β-treated CTSB/OE+LVL Dab2 cells transfected with Flag-tagged wild-type Beclin-1 and its mutants. Immunoprecipitated complexes were immunoblotted to determine the interaction between Dab2, CK2, Vps34, Atg14 and Flag-tagged-Beclin-1. (g) Immunoblot analysis of whole-cell lysates prepared from TGF-β-treated NMuMG LVL Dab2 cells silenced for endogenous Beclin-1 and re-expressing either Flag-tagged wild-type or the indicated Beclin-1 phosphomutants. Blots were probed with the autophagic markers (LC3B and p62) and apoptosis markers (Bim and cleaved caspase-3). Hsp90 was used as a loading control. (h) Immunoblot analysis of whole-cell lysates from NMuMG LVL Dab2 and Beclin-1 KD cells stably re-expressing Flag-tagged exogenous WT or the indicated Beclin-1 mutants. All experiments were repeated at least three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
Site-directed mutagenesis was employed to generate phosphomimetic mutations in the context of a Flag-tagged Beclin-1 construct. Only the S337E and S341E mutants, not the S64E mutant, abolished Beclin-1–Vps34 interactions (Fig. 6e). We also generated phosphomutants of Beclin-1 including S64A, S337A, S341A, S64A/S337A, S64A/S341A and S337A/S341A (designated as muBeclin-1). Only muBeclin-1 interacted with Dab2, CK2 and Vps34 following 7-day TGF-β treatment in CTSB/OE+LVL Dab2 cells, indicating that CK2-mediated Beclin-1 phosphorylation at Ser337 and/or Ser341 was required for inhibition of Beclin-1–Vps34 interactions (Fig. 6f). Since Atg14 is a Beclin-1-associated autophagy-related key regulator that targets the Beclin-1–Vps34 complex to the probable site of autophagosome formation18,19, we also observed that TGF-β promotes Beclin-1–Atg14 interactions (Fig. 6b). Moreover, Dab2 expression and Beclin-1 mutants showed no effect on Beclin-1–Atg14 interactions (Fig. 6e,f). To determine whether these Beclin-1 mutants could rescue the inhibitory effects of Dab2 on TGF-β-induced autophagy, we silenced endogenous Beclin-1 in NMuMG LVL Dab2 cells, and then stably re-expressed exogenous WT Beclin-1 or various Beclin-1 mutants (Fig. 6h). Only muBeclin-1 (S337A/S341A) attenuated the inhibitory effect of Dab2 on chronic TGF-β-induced autophagy, an observation further confirmed by LC3BII turnover (Supplementary Fig. 4d). The remaining Beclin-1 mutants failed to attenuate autophagy, rather apoptotic effects were observed (Fig. 6g). Since synthesis of phosphatidylinositol-3-phosphate (PtdIns3P) byVps34 is critical for the initial steps of autophagosome biogenesis20,21, we monitored PtdIns3P levels and found that only muBeclin-1(S337A/S341A) could neutralize the inhibitory effect of Dab2 on PtdIns3P induction by TGF-β (Supplementary Fig. 4e). These data indicate that Dab2 inhibits TGF-β-induced autophagy by promoting CK2-mediated Beclin-1 phosphorylation at Ser337 and/or Ser341, thereby preventing Beclin-1–Vps34 interactions and autophagosome formation.
Dab2 promotes chemotherapy-drug-induced cell death by attenuating drug-induced autophagy
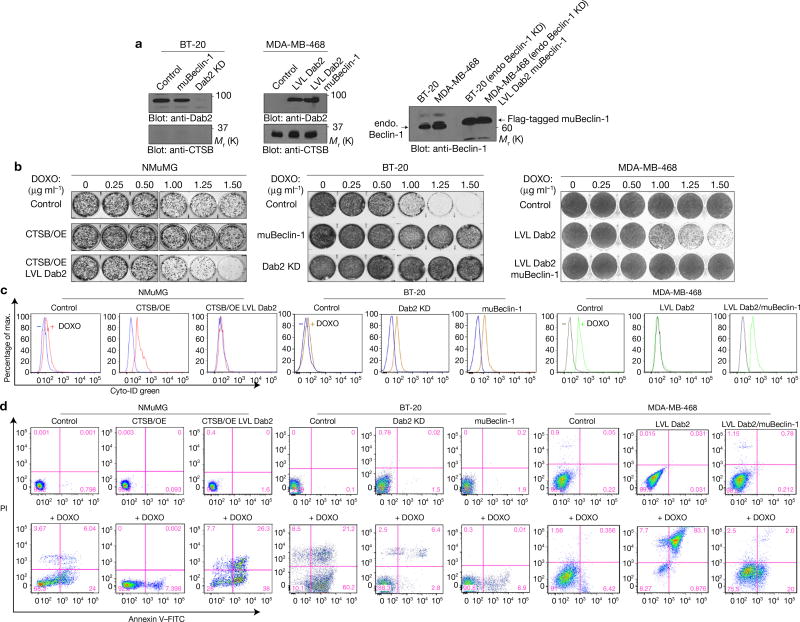
We silenced Dab2 expression in BT-20 cells (high Dab2 and low CTSB) and overexpressed LVL Dab2 in MDA-MB-468 cells (low Dab2 and high CTSB) to examine whether the inhibitory role of Dab2 on autophagy has any effect on chemotherapeutic cytotoxicity. In addition, we also silenced Beclin-1 expression in both cell lines and stably re-expressed muBeclin-1 (Fig. 7a). As shown (Fig. 7b and Supplementary Fig. 5a), NMuMG CTSB/OE cells showed increased survival in the presence of doxorubicin (DOXO) when compared with control NMuMG cells, whereas NMuMG/ CTSB/OE+LVL Dab2 cells were more sensitive to the cytotoxic effects of DOXO. In BT-20 cells, both overexpression of muBeclin-1 and Dab2 KD rendered these cells more chemoresistant. Overexpression of LVL Dab2 in MDA-MBA-468 cells rendered them more chemosensitive, which was amenable to rescue by re-expression of muBeclin-1. As observed, DOXO also induced organized cytoplasmic structures typical of autophagosomes and autolysosomes in cells with low Dab2 or muBeclin-1 whereas in cells with high Dab2 levels it induced dense nuclear masses, indicative of chromatin condensation and severely damaged cytoplasmic structures suggestive of apoptosis (Supplementary Fig. 5b–d). Additionally, LC3BII (autophagic flux) was induced only in low-Dab2-expression cells, while Bim was induced only in high-Dab2-expression cells (Supplementary Fig. 5e,f). Dab2 also inhibited DOXO-induced Beclin-1–Vps34 interactions (Supplementary Fig. 6a).
Figure 7.
Dab2 promotes chemotherapy-drug-induced cell death by attenuating drug-induced autophagy. (a) Immunoblot analysis of whole-cell lysates prepared from BT-20 and MDA-MB-468 cells, and their modified derivatives, depicting the expression levels of Dab2, CTSB, endogenous Beclin-1 and Flag-tagged muBeclin-1. (b) Clonogenic assays depicting the dose-dependent effects of DOXO on cell growth in NMuMG, BT-20 and MDA-MB-468 cells, and their modified derivatives. (c) Flow cytometric analysis of autophagic flux in NMuMG, BT-20 and MDA-MB-468 cells, and their modified derivatives, incubated in the absence (left peak) or presence (right peak) of DOXO (2.0 µg ml−1) for 24 h. FITC-conjugated Cyto-ID was employed to measure autophagic vacuoles with representative histograms. (d) Flow cytometric analysis of apoptosis using Annexin V staining evaluated in NMuMG, BT-20 and MDA-MB-468 cells, and their modified derivatives, incubated in the absence (top panels) or presence (bottom panels) of DOXO (2.0 µg ml−1). The flow cytometry profile represents Annexin V–FITC staining on the x axis and PI on the y axis. The pink numbers represent the percentages of non-apoptotic cells (lower left quadrant), early apoptotic cells (lower right quadrant), later apoptotic cells (upper right quadrant) and dead cells (upper left quadrant). All experiments were repeated at least three times and similar results were observed. Unprocessed original scans of blots are available in Supplementary Fig. 9.
Monitoring autophagy (Fig. 7c) and apoptosis (Fig. 7d) by flow cytometry analysis we also observed that in NMuMG cells (Fig. 7c,d; left panels), a 24 h DOXO treatment resulted in little autophagy and ~30% induction in apoptotic cells. In CTSB/OE cells, DOXO treatment significantly increased autophagic vacuoles (Fig. 7c), while the apoptotic population decreased to ~7% (Fig. 7d). In CTSB/OE+LVL Dab2 cells, little autophagy was observed and apoptotic cells increased to ~64%. In BT-20 cells (Fig. 7c,d: middle panels), Dab2 KD significantly increased DOXO-induced autophagy, while decreasing the apoptotic population from ~81.4% to ~9.2%. In BT-20/muBeclin-1 cells, the autophagic population significantly increased when compared with control BT-20 cells, while the apoptotic cell population decreased from ~81.4% to ~8.9%. Finally, in MDA-MB-468/LVL Dab2 cells (Fig. 7c,d: right panels), DOXO decreased autophagy while increasing apoptosis from ~6.8% to ~84% when compared with MDA-MB-468 cells. In MDA-MB-468/LVL Dab2 cells expressing the muBeclin-1 construct, autophagic induction was increased while apoptosis decreased from ~84% to ~22%. These data suggest that in the absence of Dab2, DOXO induces autophagy and that in its presence apoptosis predominates.
Dab2 enhances DOXO-mediated cell death and attenuates tumour cell metastasis in vivo
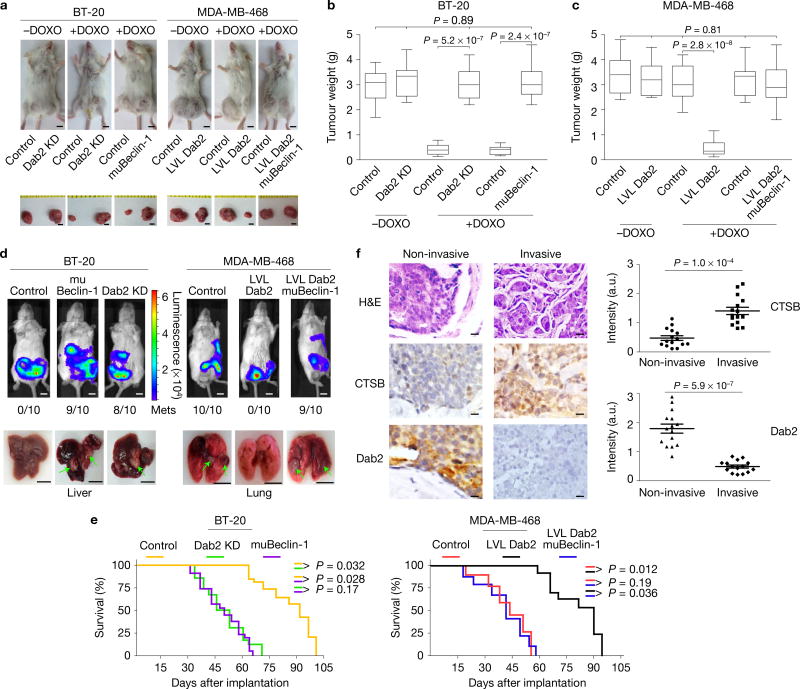
We performed xenograft tumour studies in SCID mice using BT-20, MDA-MB-468, and their modified Dab2 and Beclin-1 derivatives to confirm our results. BT-20 cells formed tumours whose size (Fig. 8a) and weight (Fig. 8b) were not significantly affected by Dab2 KD or expression of muBeclin-1. However, DOXO treatment reduced the size and weight of BT-20 tumours but had little effect on the size and weight of BT-20 Dab2 KD or BT-20 muBeclin-1 tumours. MDA-MB-468 cells elicited the growth of tumours of similar size (Fig. 8a) and weight (Fig. 8c) in the presence or absence of DOXO. MDA-MB-468/LVL Dab2 cells formed tumours of similar size and weight as their parental counterparts, but the presence of Dab2 rendered them more chemosensitive. Expression of the muBeclin-1 in MDA-MB-468/LVL Dab2 cells rendered their tumours highly chemoresistant (Fig. 8a,c). These results confirm the findings of the in vitro analyses of Fig. 7 and suggest that through its ability to inhibit autophagy, Dab2 renders cells more sensitive to DOXO whereas in its absence autophagy induction leads to increased chemoresistance. Whole-animal bioluminescence analyses using cell lines stably expressing luciferase (Fig. 8d) showed that Dab2-containing cells (BT-20 andMDA-MB-468/LVL Dab2) failed to metastasize to the liver or lung. Implantation of cells with low Dab2 (BT-20 Dab2 KD and MDA-MB-468) or with muBeclin-1 resulted in significant metastatic spread to the lungs and liver. BT-20/muBeclin-1 and BT-20/Dab2 KD cells preferentially metastasized to the liver, whereas MDA-MB-468 and LVL Dab2/muBeclin-1 cells metastasized to the lung (Fig. 8d). Mice implanted (Fig. 8e; left and right panels) with Dab2-containing cells (BT-20 and MDA-MB-468/LVL Dab2) showed increased survival rates compared with mice implanted with cells containing little Dab2 (BT-20/muBeclin-1, BT-20/Dab2 KD, MDA-MB-468 and MDA-MB-468/LVL Dab2/muBeclin-1 cells). Tumours developed fromhigh-Dab2 cells (BT-20,MDA-MB-468/LVL Dab2) had low LC3BII and high Bim levels whereas those developed from cells containing little Dab2 (BT-20 Dab2 KD and MDA-MB-468 control) or containingmuBeclin-1 had high LC3BII and low Bim levels (Supplementary Fig. 6b,c).
Figure 8.
Dab2 enhances DOXO-mediated cell death and attenuates tumour cell metastasis in vivo. (a) Parental (control) BT-20 and MDA-MB-468 breast cancer cells (2 × 105 cells) were injected into the right fourth mammary fat pad, and their Dab2 and Beclin-1 mutant derivatives (2 × 105 cells) were injected into the left fourth mammary fat pad. Tumours were excised and imaged. Scale bars, 5 mm. (b,c) Box-and-whisker plots depicting the weight of the respective tumours. Centre values are shown as horizontal lines in the boxes. The vertical lines extending from the top and bottom of the box indicate the maximum and minimum values, respectively. The top horizontal line indicates the score at the 95th percentile, and the lowest horizontal line represents the score at the 5th percentile. n = 10 mice pooled from three independent experiments. One-way ANOVA; P values are indicated. (d) Representative bioluminescence images of metastatic tumours and non-metastatic tumours in mice after the cells in a with stable luciferase expression were implanted into mice mammary gland fat pads. Post implantation (7–8 weeks), mice were subjected to bioluminescence imaging using the IVIS200 image system. Liver and lung metastasis are indicated by green arrows. Scale bars, 5 mm. (e) Kaplan–Meier survival plots showing survival rates for mice implanted with the aforementioned breast cancer cells in mice mammary gland fat pads. (f) Immunohistochemical analysis was performed on 15 invasive and 15 non-invasive human breast cancer tissues using anti-CTSB and anti-Dab2 antibodies. Slides were scanned with a ScanScope CS using bright-field imaging at ×20 magnification. Specimen areas were selected and individual images were saved in a 24-bit RGB TIFF file format with a resolution of 1 µm per pixel using the ImageScope software. Automated analysis of the TIFF image files was performed using the programming language IDL 6.3. Error bars are mean ± s.e.m. n = 15 breast cancer tissues pooled from three independent experiments. Student two-tailed paired test; P <0.05 indicates a significant difference. Scale bars, 20 µm. The experiments were repeated three times and similar results were observed.
DOXO treatment led to decreased proliferation (lower Ki-67 staining) in the BT-20-generated tumours, but not in Dab2-KD- or muBeclin-1-generated tumours (Supplementary Fig. 7a). Apoptosis (TUNEL staining) was increased in response to DOXO in BT-20- but decreased in Dab2-KD- or muBeclin-1-developed tumours, whereas in Dab2-KD- and muBeclin-1-generated tumours DOXO led to a decrease in p62 staining (Supplementary Fig. 7a). In MDA-MB-468-generated tumours, LVL Dab2 expression significantly enhanced DOXO-mediated apoptosis (strong TUNEL staining), low Ki-67 staining, and high levels of p62 indicative of lower level of autophagy (Supplementary Fig. 7b). In liver and lung metastases generated by BT-20 and MD-MB-468 cells high Ki-67 staining indicated active proliferation with little evidence of apoptosis (no TUNEL staining; Supplementary Fig. 8a). Interestingly, p62 levels were relatively lower compared with their corresponding primary tumours (Supplementary Fig. 7), suggesting that autophagy may play a role in tumour metastasis (Supplementary Fig. 8a). Finally, immunohistochemical analysis of 15 invasive and 15 non-invasive breast cancer tissues revealed significantly higher CTSB and lower Dab2 expression levels in invasive versus non-invasive breast cancer tissues (Fig. 8f), confirming that CTSB-mediated Dab2 degradation promotes tumour metastasis.
DISCUSSION
Dab2 has been proposed to act as a tumour suppressor and its expression is often lost in highly metastatic cell lines9,22. We observed that following TGF-β-mediated EMT, prolonged TGF-β treatment leads to a loss of Dab2 expression and of the mesenchymal phenotype. Notably, the loss of Dab2 and the mesenchymal phenotype in the continued presence of TGF-β induces cells into autophagy promoting chemoresistance and metastasis. Dab2 loss is mediated through its proteolytic cleavage by CTSB. We identified a specific CTSB cleavage site in Dab2 and demonstrated that expression of a non-cleavable LVL Dab2 mutant prevents cells from entering autophagy and maintains their chemosensitivity. Thus, CTSB-modulated Dab2 expression appears to regulate metastasis and chemotherapeutic drug-mediated chemosensitivity. Mechanistically (model Supplementary Fig. 8b), Dab2 prevents autophagy by inhibiting Beclin-1–Vps34 interactions and stabilizes Bim by attenuating ERK-mediated Bim phosphorylation to promote apoptosis.
CTSB has been shown to be associated with the plasma membrane of metastatic cells resulting in focal dissolution of the extracellular matrix and leading to increased invasion and metastasis when it functions together with other factors23. While originally thought to function primarily within the endolysosomal compartment, many studies suggest that CTSB can function both extracellularly and intracellularly within locales other than the lysosomes24,25, and even at a neutral pH environment, CTSB still shows significant activity26–28. Herein, we identified a role for CTSB in the proteolytic cleavage of Dab2 and in regulating autophagy and tumorigenesis. Chronic TGF-β treatment leads to the transcriptional induction of CTSB, degradation and loss of Dab2, and activation of the autophagy cascade. The site within the cell where CTSB mediates proteolysis of Dab2 is unknown. As an endosomal vesicle component, it is possible that Dab2 encounters CTSB under conditions of cellular stress when intracellular vesicular fusion events are activated. The Dab2 endosomal vesicle could also serve as an intracellular platform for the fusion and assembly of vesicles containing regulatory molecules and effectors that confer signalling specificity for autophagy and apoptosis. Our data demonstrate that Dab2 modulates Beclin-1–Vps34 interaction and autophagosome formation supporting such an ‘endosomal signalling platform’model. We provide evidence that Dab2–Beclin-1 interactions promote the CK2-dependent phosphorylation of Beclin-1 and that phosphorylation at Ser337 and/or Ser341 attenuates Beclin-1–Vps34 interaction. While CK2 fails to directly interact with Beclin-1 in the absence of Dab2, Beclin-1 serves as a substrate for CK2. Thus, CTSB-mediated proteolysis of Dab2 during chronic TGF-β treatment promotes autophagosome formation and autophagy, whereas in the presence of non-cleavable Dab2, chronic TGF-β treatment leads to apoptosis. It is tempting to speculate that the presence or absence of Dab2 in endocytic vesicles specifies their fusion with vesicles containing signalling effectors mediating either autophagy or apoptosis. Future studies will be aimed at determining the mechanism of this specificity.
METHODS
Cell culture, antibodies and reagents
Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Cat. No. SH30081.01, GE Healthcare Life Sciences) with 10% FBS and a 1% antibiotic and antimycotic combination at 37 °C and 5% CO2 in a cell culture incubator. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. All cell lines were obtained from American Type Culture Collection (ATCC) and authenticated by short tandem repeat profiling and further tested to show no mycoplasma contamination. A Myc-tagged CTSB-expressing construct was purchased from Sino Biological. HA-tagged LVL Dab2 was generated from wild-type Dab2 in our laboratory. Cathepsin B shRNA and Dab2 shRNA constructs were purchased from OriGene Technologies. Flag-tagged Beclin-1 constructs were provided by B. Levine. Beclin-1 KD cells were generated with a psiRNA-Beclin-1 construct from InvivoGen. Dab2 KD cells were generated with a protocol supplied by OriGene Technologies. The Dab2 antibody (Cat. No. 610464, 1:2,000 dilution) and N-cadherin antibody (Cat. No. 610920, 1:10,000 dilution) were purchased from BD Biosciences. The mouse CTSB antibody (Cat. No. SAB1405676, 1:1,000 dilution) was obtained from Sigma and human CTSB antibody (Cat. No. ab125067, 1:2,000 dilution) was purchased from Abcam. The vimentin antibody (Cat. No. 5741, 1:10,000 dilution), LC3B antibody (Cat. No. 2775, 1:1,000 dilution), Beclin-1 antibody (Cat. No. 3738, 1:1,000 dilution), p62 antibody (Cat. No. 8025(D5E2), 1:2,000 dilution), Bim antibody (Cat. No. 2819, 1:3,000 dilution), p-Bim antibody (Cat. No. 4581 and Cat. No. 12433 (D4H12), 1:1,000 dilution), ERK antibody (Cat. No. 9102, 1:5,000 dilution) and p-ERK antibody (Cat. No. 4370, 1:5,000 dilution), cleaved caspase-3 antibody (Cat. No. 9661, 1:1,000 dilution), Atg14 antibody (Cat. No. 5504, 1:1,000 dilution), Smad2 antibody (Cat. No. 5339, 1:3,000 dilution), p-Smad2 (Cat. No. 3108 (138D4), 1:5,000 dilution), p-Smad3 antibody (Cat. No. 9520, 1:2,000 dilution), tubulin antibody (Cat. No. 2146, 1:5,000 dilution), Vps34 antibody (Cat. No. 3811, 1:3,000 dilution) and Histone H3 antibody (Cat. No. 4499, 1:10,000 dilution) were obtained from Cell Signaling Technology. Recombinant CK2 kinase was purchased from New England Biolabs. The CK2α antibody (Cat. No. sc-6479, 1:2,000 dilution), Smad3 antibody (Cat. No. sc-101154, 1:5,000 dilution), and Smad3 shRNA construct (Cat. No. sc-38377-V) were purchased from Santa Cruz Biotechnology. The sodium potassium ATPase alpha 1 antibody (Cat. No. NB300-146, 1:1,000 dilution) was purchased from Novus. The anti-Alexa488 and anti-Alexa546 secondary antibodies (Cat. No. A11001 and A11036, 1:2,000 dilution) were obtained from Life Technologies. MG-132, E64, CTSB inhibitor II, and lactacystin were obtained from EMD Millipore. CA074Me, velcade and ALLN were obtained from Santa Cruz Biotechnology. Puromycin was purchased from Sigma-Aldrich. Neomycin, hygromycin and zeocin were purchased from InvivoGen. Apigenin was obtained from Cayman Chemical. The Smad2 shRNA construct (Cat. No. i049389) was purchased from Applied Biological Materials. Site-directed mutagenesis was performed using a site-directed mutagenesis kit from Life Technologies as described by the manufacturer.
PCR analysis
For PCR analysis with reverse transcription, RNA was isolated using the RNeasy Mini kit from Qiagen (Cat. No./ID: 74104, Valencia). mRNA from each sample (4 µg) was reversely transcribed using the Invitrogen SuperScript III first-strand synthesis system kit (Cat. No. 18080-051, Invitrogen). Reverse transcription products (cDNAs) were subjected to PCR for 20 cycles. The PCR products were stained with ethidium bromide and analysed on 2% agarose gels. For real-time PCR analysis, cDNAs were used as a template for quantitative real-time PCR using SYBR Green Master Mix (Cat. No. 04913922001, Roche) and gene-specific primers. PCR analysis was performed using a MyiQ5 ICycler (BioRad) according to the manufacturer’s protocol. Gene expression was calculated relative to β-actin. The primers used in PCR analysis are listed as follows:
Dab2 p96-forward: 5′-AAGCAGGACTTGGAAAGTTCTGT-3′, reverse: 5′-AA GCAGGACTTGGAAAGTTCTGT-3′;
Dab2 p67-forward: 5′-ACATGTCTACACCTCCTGACCTA-3′, reverse: 5′-CA TTGCCTTTGAAGAGATCCAGAA-3′;
CTSB-forward: 5′-GGCCCAGTGGAGGGTGCCTT-3′, reverse: 5′-TGCGTGG GATTCCAGCCACAA-3′;
β-actin-forward: 5′-TCATGAAGTGTGACGTTGACATCCGT-3′, reverse: 5′-CCTAGAAGCATTTGCGGTGCACGATG-3′.
Subcellular fractionation
The same amount of cells from each 15 cm plate treated with TGF-β was lysed using 500 µl of subcellular fractionation buffer (250 mM sucrose, 20 mM HEPES (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol and protease inhibitor cocktail (1:1,000 dilution)). Plates were scraped and the contents placed in 1.5 ml Eppendorf tubes, and then the lysate was passed through a 25 G needle 10 times using a 1 ml syringe, and left on ice for 20 min. The nuclear pellets were centrifuged out at 720g for 5 min, and then washed once by adding 500 µl of fractionation buffer again. The pellets were then dispersed with pipettes and passed through a 25 G needle 10 times, and centrifuged again at 720g for 10 min. Next, the wash buffer was removed, and the nuclear pellets were resuspended in nuclear buffer (standard lysis buffer with 10% glycerol and 0.1% SDS added), before being sonicated briefly (3 s) on ice at a power setting of 2-continuous. The supernatant was then removed and placed in a fresh labelled Eppendorf tube, centrifuged again at 8,000 r.p.m. (10,000g), and then the supernatant was removed again. This supernatant was the cytosolic and membrane fraction. For a membrane fraction: the supernatant was centrifuged in an ultracentrifuge at 100,000g for 1 h. The pellets were washed by adding 400 µl of the fractionation buffer to them, and were then resuspended by pipetting using a 25 G needle as above, before being re-centrifuged for another 45 min. The membrane pellets were resuspended in the same buffer as used for the nuclei. All fractions were diluted in the same volume of buffer and subjected to immunoblotting with the same volume. The experiment was replicated three times.
Cell invasion assays
Cell invasion assays were performed with a kit purchased from Cell Biolabs according to the manufacturer’s instructions. Briefly, under sterile conditions, invasion chamber plates were warmed to room temperature for 10 min before rehydrating the basement membrane of the cell culture insert by adding 300 µl of serum-free medium to the inner compartment. Cells were seeded at a density of 2.5 × 104 per chamber in DMEM medium. DMEM medium with 10% FBS was used as a chemo attractant. Mitomycin C (5 µg ml−1) was added to media for inhibiting cell proliferation. Post-seeding (12 h), invading cells were fixed and stained with a HAME-3 stain set (Fisher Scientifics). Invading cells were also collected and subjected to OD measurement and quantification. Results are shown as means ± s.d. of three independent experiments performed in triplicate.
Transmission electron microscopy assay
Cells were fixed with 2.5% glutaraldehyde buffered in 0.1 M sodium cacodylate (pH 7.4) at 5 °C for about 4 h on ice, then rinsed in sodium cacodylate buffer and post-fixed in 1% aqueous osmium tetroxide (buffered in 0.1 M sodium cacodylate) at 5 °C for about 2 h, and then rinsed and stored in the buffer at 4 °C. Cells were later dehydrated in an acetone/ethanol series and transferred to propyleneoxide, and then subsequently embedded in Glycidether 100 (formerly Epon). After polymerization, semi-thin sections (1 µm) and ultrathin sections (60–90 nm) were cut and stained with toluidine blue. Digital micrographs were obtained with a JEOL JEM1010 electron microscope.
Measurement of PtdIns3P level
Levels of PtdIns3P were measured in lipid extracts from cells by using a competitive enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s protocol (Echelon Biosciences). Briefly, cell culture medium was gently removed and ice-cold 5% trichloroacetic acid was added to extract lipids. Lyophilized lipids were suspended in PBS–Tween 20 containing 3% protein stabilizer. The ELISA was performed in triplicate. The quantities of PtdIns3P in the samples were determined by comparison with the values of the PtdIns3P standard curve. Concentrations were corrected for cell number. Means ± s.d. of three experimental replicates are shown.
MTT assay and clonogenic assay
For the MTT assay, 4,000 cells per well were seeded in a 96-well dish with 200 µl DMEM containing 10% FBS and incubated in a 37 °C and 5% CO2 incubator. Post-plating (24 h), various concentrations of doxorubicin or paclitaxel or vehicle were added. Following a 48 h treatment, cell viability was assessed by the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide; Sigma) assay according to the manufacturer’s protocol. Viable fraction is expressed as the percentage of vehicle-treated control cells.
For clonogenic assays, cells were seeded in a 24-well plate (1 × 105 cells per well). After 24 h, cells were treated with doxorubicin or paclitaxel or vehicle for an additional 24 h. After culturing in a drug-free culture medium for another 48 h, an aliquot of the harvested cell population was seeded onto a 6-well dish. Crystal violet staining was used to count the colonies after a 10-day culture in the medium. Results are shown as means ± s.d. of three independent experiments performed in triplicate.
Tumorigenesis assay
Tumorigenesis was performed using mammary fat pad injection of BT-20, BT-20/Dab2 KD, BT-20/muBeclin-1, MDA-MB-468, MDA-MB-468/LVL Dab2 and MDA-MB-468/LVL Dab2/muBeclin-1 cells (all cells with luciferase-overexpression (2 × 105 per 100 µl PBS)) into the fourth left and right mammary fat pads of 8-week-old female SCID mice. For chemotherapeutic drug resistance assays, tumours were grown to ~5–6 mm in diameter and treated ± doxorubicin or paclitaxel (i.v. 8–10 mg kg−1 per week) for 3–4 weeks, and then tumours were excised and analysed for weight measurement and imaging. Ten animals were used for each cell type. For the metastasis assay, 7–8 weeks post implantation, all mice were subjected to bioluminescence imaging with luciferin substrate (Caliper LifeSciences) by an IVIS200 image system before the mice were euthanized. All experiments were performed according to approved protocols of the Institutional Animal Care and Use Committee (IACUC), Medical University of South Carolina.
Transfections
All cell transfections were carried out using 5 µg DNA (or shRNA) per 10 ml of medium with cells at 70% confluence cultured in 100 mm plates. The transfection reagent Lipofectamine plus was used according to the protocol provided by the manufacturer (Cat. No. 15338100, ThermoFisher Scientific).
Immunoblotting and immunoprecipitation assays
For immunoprecipitation and western blot analysis, cells were lysed in buffer D, and immunoprecipitation was carried out as previously described10.
Immunofluorescence and imaging
Cells were fixed for 20 min in PBS buffer containing 4% (w/v) paraformaldehyde, followed by permeabilization with PBS containing 0.2% (w/v) Triton X-100 and 2% BSA for 20 min. Then cells were incubated with various primary antibodies for 1 h in permeabilization buffer followed by three washes with PBS; thereafter, cells were incubated in secondary antibodies conjugated with Alex Fluor 488 (green) or Alex Fluor 568 (red) at room temperature for 1 h followed by three washes with PBS before being observed with confocal laser scanning on a Leica TCS SP2 confocal microscope (Leica Microsystems).
Cathepsin B activity assay
Cells treated with a 7-day TGF-β time course. Fresh DMEM and TGF-β ligands were changed every day. Then cells were collected by centrifugation and lysed in 100 µl of chilled CTSB cell lysis buffer from BioVision followed by incubating on ice for 10 min. After incubation, cell lysates were centrifuged at top speed in a microcentrifuge for 5 min and the supernatant was transferred to a new tube. Fifty microlitres of cell lysate and 50 µl of CTSB reaction buffer were added to a 96-well plate, and then 2 µl of 10 mM CTSB substrate Ac-RR-AFC was added to above 100 µl mixture (for negative control, 2 µl of CTSB inhibitor was added). The reaction mixture was incubated at 37 °C for 1 h, followed by reading samples in a fluorometer equipped with a 400-nm excitation filter and a 505-nm emission filter. Fold increase in CTSB activity was determined by comparing the relative fluorescence units with the level of the non-TGF-β-induced control. Results are shown as means ± s.d. of three independent experiments.
In vitro kinase assay
Wild-type Beclin-1 and its various mutant proteins were generated using the T7-Quick coupled TNT system (Promega) and employed as substrates for in vitro phosphorylation assays using purified recombinant CK2 kinase in kinase assay reaction buffer (10 nM Tris, pH 7.4, 20% glycerol, 10 mM MgCl2, 150 mM NaCl, 10 mM dithiothreitol). Apigenin (20 µM) was added directly to the kinase reaction mixture. All substrates were in vitro phosphorylated in the presence of [γ-32P]ATP (PerkinElmer). Kinase reactions were performed for 1 h at 30 °C followed by SDS–PAGE and autoradiography analysis.
Flow cytometry analysis
Cells were harvested after trypsin digestion and washed with PBS containing 2% FBS and 2% BSA. An autophagic vacuole staining solution Cyto-ID-green detection reagent (Enzo Life Sciences), or antibody against Annexin V (FITC-conjugated, Pharmingen) and PI (BD Pharmingen) were added according to the manufacturers’ instructions and incubated on ice for 30 min. Flow cytometric analysis was performed using a FACS LSRII system (BD Biosciences), and the data were analysed using FlowJo software.
H&E staining and immunohistochemistry
For H&E staining, paraffin-embedded sample slides were de-paraffinized, hydrated and then stained with haematoxylin (Sigma-Aldrich) for 1 min. After rinsing, the slides were stained with eosin (Sigma-Aldrich) for 1 min, rinsed and sealed with cover slips using Permount. Immunohistochemistry was performed on paraffin-embedded sections as described. Briefly, tissue sections were incubated with primary antibodies anti-Ki-67, anti-p62, anti-Dab2, anti-Flag, anti-CTSB or anti-Dab2 for 2 h at 25 °C. The sections were washed with PBS before being incubated with biotinylated secondary antibody for 30 min at 25 °C. The stain was developed with diaminobenzidine tetrahydrochloride (DAB) chromogen. All slides were observed and photos were taken under a Leica DM 2000 microscope. The use of human breast tumour tissues and database were approved by the Institutional Review Board for Human Research of the Medical University of South Carolina. All tumour tissue samples were obtained in agreement with informed consent policy.
Statistics and reproducibility
Statistical analyses of enzyme activity assay, cell OD value, immunoblotting quantification and immunohistochemical quantifications were performed using ANOVA or Fisher’s exact test with GraphPad Prism software. For the mouse survival analyses, Kaplan–Meier plots were performed and statistical differences were evaluated using the log rank Mantel–Cox test. All data are shown as mean ± standard deviation (s.d.). n refers to biological replicates. A P value < 0.05 was considered statistically significant. No samples or animals were excluded from the analysis. No statistical method was used to predetermine sample size and experiments were not randomized, and we were not blinded to allocation during experiments and outcome assessment.
Data availability
Source data for Figs 1f,g and 8b,c,f and Supplementary Figs 1j, 2c,d, 4d, 5a and 6c have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We thank G. Hussey, B. Howley, T. Smith and other members of our laboratory for helpful comments and critical insights. We are very grateful to B. Levine (Center for Autophagy Research, University of Texas Southwestern Medical Center, USA) for providing us with Beclin-1 constructs. The work was supported in part by the Flow Cytometry & Cell Sorting, Molecular Imaging, Small Animal Imaging, and the Gene Targeting and Knockout Shared Resources, Hollings Cancer Center, Medical University of South Carolina (P30CA138313). Our study used the services of the MUSC Center for Oral Health Research (COHR), which is partially supported by the National Institute of General Medicine grant P30GM103331. This work was also supported by grants CA555536 and CA154664 from the National Cancer Institute to P.H.H.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
Y.J. and P.H.H. conceived of the project and designed the experiments. Y.J., A.N.W., N.S. and S.N. performed experiments. Y.J. and P.H.H. analysed data and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Massague J. TGFβ in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer KR, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng X, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-β (TGF-β) signaling. Autophagy. 2010;6:645–647. doi: 10.4161/auto.6.5.12046. [DOI] [PubMed] [Google Scholar]
- 6.Avalos Y, et al. Tumor suppression and promotion by autophagy. BioMed Res. Int. 2014;2014:603980. doi: 10.1155/2014/603980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- 8.Degenhardt K, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannigan A, et al. Epigenetic downregulation of human disabled homolog 2 switches TGF-β from a tumor suppressor to a tumor promoter. J. Clin. Invest. 2010;120:2842–2857. doi: 10.1172/JCI36125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhury A, et al. TGF-β-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 2010;12:286–293. doi: 10.1038/ncb2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor L, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Xu XX, Yang W, Jackowski S, Rock CO. Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J. Biol. Chem. 1995;270:14184–14191. doi: 10.1074/jbc.270.23.14184. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luciano F, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Y, He X, Howe PH. Disabled-2 (Dab2) inhibits Wnt/β-catenin signalling by binding LRP6 and promoting its internalization through clathrin. EMBO J. 2012;31:2336–2349. doi: 10.1038/emboj.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinna LA. Protein kinase CK2: a challenge to canons. J. Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 18.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 20.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dote H, et al. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin. Cancer Res. 2004;10:2082–2089. doi: 10.1158/1078-0432.ccr-03-0236. [DOI] [PubMed] [Google Scholar]
- 23.Podgorski I, Sloane BF. Cathepsin B and its role(s) in cancer progression. Biochem. Soc. Symp. 2003;70:263–276. doi: 10.1042/bss0700263. [DOI] [PubMed] [Google Scholar]
- 24.Sinha AA, Jamuar MP, Wilson MJ, Rozhin J, Sloane BF. Plasma membrane association of cathepsin B in human prostate cancer: biochemical and immunogold electron microscopic analysis. Prostate. 2001;49:172–184. doi: 10.1002/pros.1132. [DOI] [PubMed] [Google Scholar]
- 25.Sloane BF. Cathepsin B and cystatins: evidence for a role in cancer progression. Semin. Cancer Biol. 1990;1:137–152. [PubMed] [Google Scholar]
- 26.Werle B, Jülke B, Lah T, Spiess E, Ebert W. Cathepsin B fraction active at physiological pH of 7.5 is of prognostic significance in squamous cell carcinoma of human lung. Br. J. Cancer. 1997;75:1137–1143. doi: 10.1038/bjc.1997.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linebaugh BE, Sameni M, Day NA, Sloane BF, Keppler D. Exocytosis of active cathepsin B enzyme activity at pH 7.0, inhibition and molecular mass. Eur. J. Biochem. 1999;264:100–109. doi: 10.1046/j.1432-1327.1999.00582.x. [DOI] [PubMed] [Google Scholar]
- 28.Mort JS, Recklies AD, Poole AR. Extracellular presence of the lysosomal proteinase cathepsin B in rheumatoid synovium and its activity at neutral pH. Arthritis Rheum. 1984;27:509–515. doi: 10.1002/art.1780270505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figs 1f,g and 8b,c,f and Supplementary Figs 1j, 2c,d, 4d, 5a and 6c have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.