Abstract
Bayesian multiple-regression methods incorporating different mixture priors for marker effects are used widely in genomic prediction. Improvement in prediction accuracies from using those methods, such as BayesB, BayesC, and BayesCπ, have been shown in single-trait analyses with both simulated and real data. These methods have been extended to multi-trait analyses, but only under the restrictive assumption that a locus simultaneously affects all the traits or none of them. This assumption is not biologically meaningful, especially in multi-trait analyses involving many traits. In this paper, we develop and implement a more general multi-trait BayesC and BayesB methods allowing a broader range of mixture priors. Our methods allow a locus to affect any combination of traits, e.g., in a 5-trait analysis, the “restrictive” model only allows two situations, whereas ours allow all 32 situations. Further, we compare our methods to single-trait methods and the “restrictive” multi-trait formulation using real and simulated data. In the real data analysis, higher prediction accuracies were observed from both our new broad-based multi-trait methods and the “restrictive” formulation. The broad-based and restrictive multi-trait methods showed similar prediction accuracies. In the simulated data analysis, higher prediction accuracies to the “restrictive” method were observed from our general multi-trait methods for intermediate training population size. The software tool JWAS offers open-source routines to perform these analyses.
Keywords: multi-trait, mixture priors, genomic prediction, Bayesian regression, pleiotropy, GenPred, Shared data resources, Genomic Selection
GENOMIC prediction was proposed by Meuwissen et al. (2001) to incorporate marker effects from whole-genome data into genetic evaluation. In genomic prediction, all the marker or haplotype effects are estimated simultaneously, and these estimates can then be used to predict breeding values of individuals not in the training population used to estimate the effects.
Bayesian multiple-regression methods incorporating mixture priors for marker effects are used widely in genomic prediction, including various extensions to the BayesB method of Meuwissen et al. (2001). BayesB accommodates models where the prior for each marker effect follows a mixture distribution with a point mass at zero with probability π and a univariate-t distribution with probability (Meuwissen et al. 2001; Gianola et al. 2009; Cheng et al. 2015b). Another model, BayesC, assumes a mixture with a point mass at zero with probability π and a univariate normal distribution with probability for all marker effects, and its extension known as BayesCπ further treats π as an unknown parameter with a uniform prior distribution (Habier et al. 2011).
Bayesian multiple-regression methods were first proposed for single-trait analyses but have been extended to some particular forms of multi-trait analyses (Calus and Veerkamp 2011; Jia and Jannink 2012). Those extensions have pertained to a particular, somewhat restrictive mixture model. The “restrictive” multi-trait BayesC presented by Jia and Jannink (2012) assumes any particular locus affects none of the traits or simultaneously affects all traits. This assumption of genetic architecture in that multi-trait BayesC model is violated if some loci have no effect on at least one of the traits while having an effect on the remaining traits.
In this paper, we propose a more general class of multi-trait BayesC and BayesB methods, where each locus can have an effect on any combination of traits. For example, in a 5-trait analysis, the restricted model only allows two situations, whereas ours allows all 32 situations. The previous restrictive multi-trait models are special cases of this general class of models. Further, our model allows the use of a single-site Gibbs sampler that requires less computing effort than some alternative Markov chain Monte Carlo approaches, especially for analyses involving many traits. Methodologies for the new models are compared to single-trait methods and the previous multi-trait methods using real and simulated data.
Materials and Methods
Multi-trait marker effects model
For simplicity of our description, but without loss of generality, we will assume individuals have all traits measured with a general mean as the only fixed effect, and write the multi-trait model for individual i from n genotyped individuals as
where is a vector of phenotypes of t traits for individual i, is a vector of overall means for t traits, is the genotype covariate at locus j for individual i (coded as 0,1, and 2), p is the number of genotyped loci, is a vector of allele substitution effects or marker effects of t traits for locus j, and is a vector of random residuals of t traits for individual i. The fixed effects, or general mean in this case, are assigned flat priors. The residuals, are a priori assumed to be independently and identically distributed multivariate normal vectors with null mean and covariance matrix which, in turn, is assumed to have an inverse Wishart prior distribution,
We will show that, employing the concept of data augmentation, the vector of marker effects at a particular locus can be written as where is a diagonal matrix whose kth diagonal entry is an indicator variable indicating whether the marker effect of locus j for trait k is zero or nonzero, and follows a multivariate normal distribution in multi-trait BayesC or a multivariate t distribution in multi-trait BayesB.
Multi-trait BayesC model
Priors for marker effects:
The prior for the allele substitution or marker effect of trait k for locus j, is a mixture with a point mass at zero and a univariate normal distribution conditional on
and the covariance between effects for traits k and at the same locus, i.e., and is
The vector of marker effects at a particular locus is written as where is a diagonal matrix with elements where is an indicator variable indicating whether the marker effect of locus j for trait k is zero or nonzero, and the vector follows a multivariate normal distribution with null mean and covariance matrix The covariance matrix is a priori assumed to follow an inverse Wishart distribution, Thus, the multi-trait BayesC model with data augmentation is written as
| (1) |
In the most general case, any marker effect might be zero for any possible combination of t traits resulting in possible combinations of For example, in a t=2 trait model, there are combinations for In the restrictive special case of this model described by Jia and Jannink (2012), only two combinations, i.e., and have nonzero probability. Suppose, in general, we use numerical labels “1,” “2,” “l” for the possible outcomes for then the prior for is a categorical distribution
where with being the prior probability that the vector corresponds to the vector labeled A Dirichlet distribution with all parameters equal to one, i.e., a uniform distribution, can be used for a prior for
As shown below, we consider two Gibbs samplers to draw samples for all the parameters in this model. Gibbs sampler I is a single-site sampler, where only one of the t indicator labels is sampled at a time. Thus, in a 2-trait model, for example, this sampler cannot move from to in a single step without stepping through or for Therefore, Gibbs sampler I cannot be used for the restrictive model which excludes and from the state space for Gibbs sampler II, however, samples all elements of jointly, and can move from to in a single step. However, Gibbs sampler II is computationally more intensive because it requires drawing samples from a multivariate normal distribution of order t, the number of traits.
Gibbs sampler I for multi-trait BayesC:
Suppose the prior for is a categorical distribution for which the support is the set of outcomes of For convenience, from now on let “1” denote trait k and “2” the other traits. In our sampling scheme, and are sampled from their joint full conditional distributions, which can be written as the product of the full conditional distribution of given and the marginal full conditional distribution of Let denote all other parameters except and then our sampling scheme can be written as
The full conditional distributions of and for Gibbs sampler I, whose derivations are in the Appendix, are given below. The full conditional distributions of is
with
where and are the partitions of corresponding to trait k and covariances between trait k and other traits, respectively. and are the partitions of corresponding to trait k and covariances between trait k and other traits, respectively.
The marginal full conditional probability that is
where
The full conditional distribution for can be written as
where is the number of loci or markers for which
The full conditional distributions for the covariance matrix for residuals, is an inverse Wishart distribution, where is the matrix for residuals whose ith row is The full conditional distribution for the covariance matrix for is an inverse Wishart distribution, where is the matrix whose ith row is
Gibbs sampler II for multi-trait BayesC:
The Gibbs sampler above, where only one of the t indicator labels is sampled at a time, cannot be used for the restrictive model assuming any particular locus affects all traits or none of them. Further, if some particular are near zero, the chain might exhibit mixing problems. Another, more general, but computationally intensive, Gibbs sampler that samples all elements of jointly and may exhibit improved mixing is proposed below.
The full conditional distributions of and for Gibbs sampler II, whose derivations are in the Appendix, are given below.
Let denote all other parameters except and then our sampling scheme can be written as
The full conditional distribution of is
where and
The marginal full conditional probability of is
where
This Gibbs sampler can accommodate the “restrictive” multi-trait BayesC that was proposed by Jia and Jannink (2012), which only allows to be a vector of all ones or a vector of all zeros.
Multi-trait BayesB model
The multi-trait BayesC model proposed above can be modified to accommodate the general multi-trait BayesB model. Model Equation (1) can also be used for the multi-trait BayesB method. The differences in multi-trait BayesB method is that the prior for is a multivariate t distribution, rather than a multivariate normal distribution. This is equivalent to assuming has a multivariate normal distribution with null mean and locus-specific covariance matrix which is assigned an inverse Wishart prior,
The derivations of the full conditional distributions of parameters of interest for Gibbs samplers are shown in the Appendix. In the multi-trait BayesB model, the full conditional distributions for all parameters except are similar to the multi-trait BayesC model. The full conditional distribution for the covariance matrix for is an inverse Wishart distribution,
Data analyses
Real data:
Published genotypic and deregressed breeding values based on phenotypic data for Loblolly Pine (Pinus taeda L.) were used (Resende et al. 2012; Daetwyler et al. 2013). Two disease traits, namely presence or absence of rust (Rust_bin) and gall volume (Rust_gall_vol) were analyzed. These are the two traits used in Jia and Jannink (2012). The reported heritabilities were 0.21 for Rust_bin and 0.12 for Rust_gall_vol. Loci with missing genotypes were imputed as the mean of the observed genotype covariates at that locus but loci with a missing rate 50% were excluded. After these quality control edits, 4828 SNPs on 807 individuals with deregressed phenotypes and genotypes on both traits remained.
Prediction accuracy was calculated as the correlation between the vector of deregressed phenotypes and the vector of estimated breeding values. Cross-validation using 10 folds formed the basis for comparing these methods. Paired t-tests were used for tests of significance of difference in prediction accuracies between two methods, where prediction accuracies for two different methods from each validation fold were considered as paired samples. The general multi-trait BayesC model (MT-BayesC-G) was compared to a similar model where the prior for is a multivariate normal rather than a mixture of multivariate normals (MT-BayesC0), the more restricted multi-trait BayesC proposed by Jia and Jannink (2012) (MT-BayesC-R) and the usual single trait formulations of the mixture models (ST-BayesC0, ST-BayesCπ). Since BayesC0 is equivalent to random regression best linear unbiased prediction (RR-BLUP), ST-BayesC0 and MT-BayesC0 are denoted as ST-RR-BLUP and MT-RR-BLUP below. The prior for the residual covariance matrix in all multi-trait methods was an inverse Wishart distribution, for which the mean of is the SD of diagonal elements are and the SD of off-diagonal elements are 0. This same prior was used for the marker effects covariance matrix The priors for the residual variance and marker effects variance in single-trait analyses were scaled inverted chi-squared distributions with scale parameter and degrees of freedom for which the mean of the prior was also 0.001. In the data analyses, multi-trait BayesB methods provided similar results as multi-trait BayesC methods. Thus, only results from BayesC analyses were presented below to demonstrate the superiority of our multi-trait methods.
Simulated data:
Simulated data described below were used to quantify the superiority of the general multi-trait Bayesian methods. Two scenarios were simulated. In scenario 1, as a known ideal condition, the simulated genome consisted of 100 loci on each of two chromosomes that were in Hardy-Weinberg and linkage equilibria. All these loci were considered as QTL or causative variants and used in the analyses. The QTL on the first chromosome had effects only on trait 1 and those on the second chromosome only on trait 2. The effects of these QTL were simulated from a standard normal distribution and then were equally scaled to provide unit genetic variance for each trait in the simulated population of 8000 unrelated individuals. The phenotypes for these traits were obtained by adding independent residuals to the genetic values. Two situations were simulated: (1) heritabilities for both traits were 0.5; (2) heritability for trait 1 was 0.2 and for trait 2 was 0.8. The XSim package was used in the simulation (Supplemental Material, File S1) (Cheng et al. 2015a).
In scenario 2, both markers and QTL were simulated. The simulated genome consisted of 100 evenly spaced loci on each of three chromosomes of length 10 cM. Ten loci were randomly selected on each chromosome as QTL. Allele states were sampled from a Bernoulli distribution with frequency 0.5 in the base population. Starting from a base population of 500 males and 500 females, random mating was simulated for 500 generations to generate linkage disequilibrium. Random mating was continued for five more generations to increase the population size to 4000 males and 4000 females, which were used in the analyses. The effects of QTL on the first two chromosomes were simulated following the same strategy in scenario 1, i.e., the QTL on the first chromosome had effects only on trait 1, and those on the second chromosome only on trait 2. All QTL on the third chromosome had effects on both traits. The effects of these QTL on the third chromosome were simulated from a standard bivariate normal distribution with correlation 0.5. The phenotypes for these traits were obtained by adding independent residuals to the genetic values. In total, 8000 individuals were simulated with heritability 0.2 for trait 1 and 0.8 for trait 2.
The same validation approaches were used for these two simulation scenarios. A total of 500 individuals were used for testing, and for each training population of size N, 100 replicates of the training population were sampled from the remaining individuals. The values considered for N were 50, 100, 200, 400, 1000, 2000, 4000, or 7000. The true genetic and residual variances were used to compute the scale parameters for the priors of the variance components. The general multi-trait BayesC model (MT-BayesC-G) was compared to the more restricted multi-trait BayesC (MT-BayesC-R) using this dataset.
All analyses were performed using JWAS (Cheng et al. 2018), an open-source, publicly available package for single-trait and multi-trait whole-genome analyses written in the freely available Julia language.
Data availability
The genotypic and phenotypic data used in the real data analysis are publicly available (Resende et al. 2012). The scripts used to generate the simulated data are provided as supplementary information. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Real data
The prediction accuracies from all methods for Rust_bin and Rust_gall_vol are in Figure 1. The prediction accuracies from all single-trait analyses using JWAS are similar to those in Resende et al. (2012).
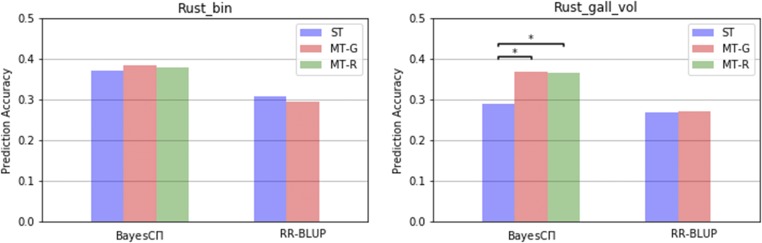
Figure 1.
Comparison of single-trait and multi-trait methods for Rust_bin and Rust_gall_vol traits. ST, MT-G and MT-R indicate single-trait, our general multi-trait and restricted multi-trait analyses, respectively. * indicates a statistically significant (P < 0.01) difference between methods.
The predictions of Rust_bin exhibited no significant difference in accuracy between multi-trait and single-trait analyses within each method (ST-RR-BLUP vs. MT-RR-BLUP; ST-BayesCπ vs. MT-BayesC-R; ST-BayesCπ vs. MT-BayesC-G).
In contrast, prediction accuracies for the lower heritability Rust_gall_vol with MT-BayesC-G were significantly higher than those from ST-BayesCπ. MT-BayesC-G and MT-BayesC-R showed similar prediction accuracies. The posterior means of for both methods are in Table 1. When RR-BLUP was used for the analysis, however, the advantage of the multiple-trait analysis (MT-RR-BLUP) over the single-trait analysis (ST-RR-BLUP) for Rust_gall_vol was not observed.
Table 1. Estimation of π for alternative multi-trait BayesC methods.
Posterior mean of were given for different categories of δ. Different categories of δ are denoted as where if a marker has a null effect on Rust_bin, otherwise and similarly for representing sampled effects for Rust_gall_vol.
Combinations that do not exist in the restricted model.
Simulated data
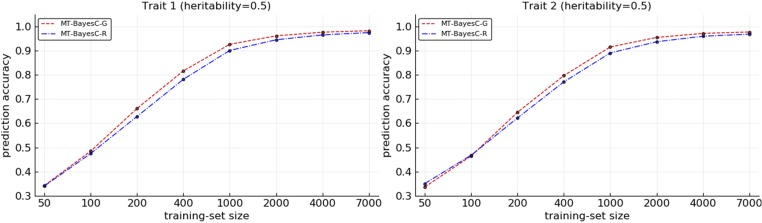
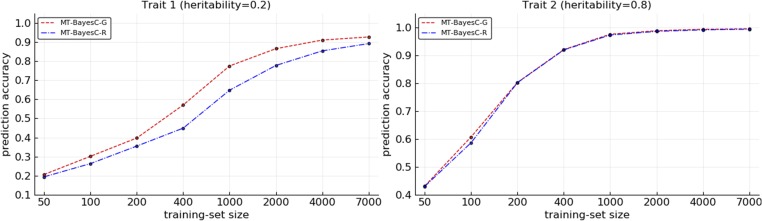
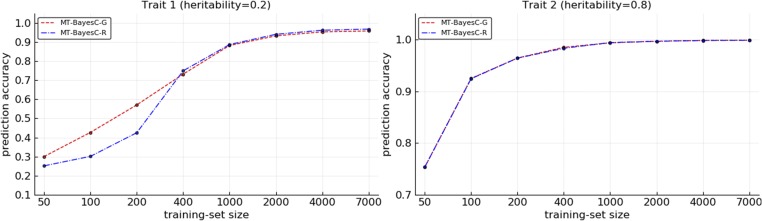
The prediction accuracies from MT-BayesC-G and MT-BayesC-R methods were compared for varying size (N) of training populations under two simulation scenarios. In simulation scenario 1, Figure 2 shows the prediction accuracies where heritabilities for both traits were 0.5. Figure 3 shows the prediction accuracies where heritabilities for trait 1 and trait 2 were 0.2 and 0.8, respectively. When both methods had similar prediction accuracy. For both traits, as N increased, initially, MT-BayesC-G became superior to MT-BayesC-R, but, as expected, the accuracies of these methods asymptotically converged (Karaman et al. 2016). In most cases, the differences in accuracies for both traits were small. However, in Figure 3, the differences in accuracies for trait 1, for which the heritability was 0.2, were substantial for intermediate values of N. Figure 4 shows the prediction accuracies for simulation scenario 2. The pattern observed is similar to Figure 3 under simulation scenario 1. MT-BayesC-G was superior to MT-BayesC-R for intermediate training population, but as N increased, the accuracies of these methods asymptotically converged (Karaman et al. 2016).
Figure 2.
Comparison of multi-trait BayesC methods for situation 1 under simulation scenario 1.
Figure 3.
Comparison of multi-trait BayesC methods for situation 2 under simulation scenario 1.
Figure 4.
Comparison of multi-trait BayesC methods under simulation scenario 2.
Discussion
Real data
Significant differences between multi-trait and single-trait analyses were only observed for Rust_gall_vol within BayesCπ methods (MT-BayesC-G vs. ST-BayesCπ; MT-BayesC-R vs. ST-BayesCπ). MT-BayesC-G and MT-BayesC-R outperformed ST-BayesCπ for Rust_gall_vol, and the accuracy gain was (from 0.287 to 0.364). The lower-heritability trait Rust_gall_vol benefited from information on the other correlated trait Rust_bin. Thus higher prediction accuracy from MT- BayesC-G were observed in trait Rust_gall_vol but not for the high heritability Rust_bin. Results in Jia and Jannink (2012) showed no difference between MT-BayesC and ST-BayesCπ because a reduced marker panel (500 markers) was used.
The fact that RR-BLUP showed no improvement in multi-trait analyses suggested that benefits from MT-BayesC-G may be due to the estimation of the hyper-parameter In the MT-BayesC-G, the mean of the posterior probability that a marker has a null effect on Rust_gall_vol was ∼0.97, calculated as the summation of posterior mean of for categories and The posterior mean of π, the probability that a marker has a null effect, in ST-BayesCπ for Rust_gall_vol was 0.74, different from the equivalent value, 0.97, in MT-BayesC-G shown above. Thus a ST-BayesC analysis with was undertaken. Prediction accuracy from this ST-BayesCπ analysis with was 0.361, which was similar to the accuracy from MT-BayesC-G. This shows that including an additional correlated trait, especially one with high heritability, will bring in more data into the analysis, helping variable selection in a low-heritability trait to become more effective and result in improved prediction accuracy.
The difference between MT-BayesC-G and MT-BayesC-R is that MT-BayesC-R assumes a locus has an effect on all traits or none of them. This assumption regarding genetic architecture is likely to be seldom true. MT-BayesC-G and MT-BayesC-R, however, showed similar prediction accuracies. This can be explained by the estimation of in MT-BayesC-G and MT-BayesC-R in Table 1. The posterior probability means for and were almost zero in MT-BayesC-G and for and are similar in MT-BayesC-G and MT-BayesC-R, suggesting that the assumption of genetic architecture whereby the same loci affect both traits as explicit in MT-BayesC-R may be valid for these two disease traits. Note that the lack of difference between the methods may also result from the limited size of the training population.
Simulated data
In scenario 1, we simulated bivariate data where each QTL had an effect on only one or the other of the traits. In MT-BayesC-R, if a locus has an effect on one of the traits, that locus is included in the model for all traits. So, in the simulated data, MT-BayesC-R would need to include all loci in the model for both traits. Thus for the trait that had heritability 0.2, the contribution of noise to the prediction from loci on chromosome 2, which had no effect on this trait, is large relative to the real signal from QTL on chromosome 1. In contrast, the general variable selection in MT-BayesC-G allows loci on chromosome 2, which have no effect on trait 1, to be excluded from the model for trait 1. Thus when sufficient data were available for variable selection to exclude loci on chromosome 2 for trait 1, MT-BayesC-G showed a substantial advantage over MT-BayesC-R. On the other hand, for the trait with heritability 0.8, the contribution of noise to the prediction from the loci on chromosome 1, which had no effect on this trait, is small relative to the signal from loci on chromosome 2. Thus MT-BayesC-G and MT-BayesC-R had similar accuracies. As the training population size increased, the contribution of noise to the prediction of a trait from loci which had no effect on this trait, vanished even when the heritability was low. This was observed for both traits as apparent in Figure 2 and Figure 3. Since only bivariate data with different heritabilities showed substantial differences in prediction accuracies, traits with different heritabilities were simulated in scenario 2. In scenario 2, both markers and QTL were simulated. As expected, MT-BayesC-G showed higher prediction accuracy to MT-BayesC-R for intermediate training population, but as N increased, the accuracies of these methods asymptotically converged (Karaman et al. 2016).
Further, in both real and simulated analyses, MT-BayesC-G gave equal or higher prediction accuracy than MT-BayesC-R. In addition, MT-BayesC-R requires drawing samples from a multivariate normal distribution of order t, whereas Gibbs sampler I, which can be used for MT-BayesC-G, requires sampling from a univariate normal. Thus, in addition to MT-BayesC-G giving equal or better performance than MT-BayesC-R, MT-BayesC-G can also be computationally more efficient.
Priors
In practice, genetic variances from previous conventional analyses are usually used to construct priors for marker effect variances. For single trait analyses, under some assumptions, it can be shown that the marker effect variance can be obtained as
| (2) |
where is the genetic variance, is the allele frequency for locus j, and π is the probability that a marker has a null effect (Habier et al. 2007; Gianola et al. 2009; Fernando and Garrick 2013). Following a similar strategy, the marker effect covariance matrix in a two-trait analysis can be obtained as
| (3) |
where is the genetic covariance matrix and and are the probabilities a marker has null effects on the first trait but not the second trait, on the second trait but not the first trait, or on neither trait. Thus the probability that a marker has an effect on the first trait can be obtained as which is the denominator of the upper left element in (3). This strategy relating marker effect covariance matrix to genetic covariance matrix can be readily extended to >2 traits. Note that positive definite matrix may result in negative definite matrix using (3), especially when the prior for the probability a marker has null effects is far from the real value. In that case, the diagonal elements of which are the marker effect variances for different traits, can be obtained using (2), where π may be estimated from previous single-trait analyses, and the off-diagonal elements of may be set to zero to guarantee positive definiteness of
Multi-trait variable selection:
In regard to a single trait, a locus either has an effect, or it does not. Hence, the scalar parameter π (and its complement ) completely defines this circumstance. In a multi-trait setting, it is conceivable that loci that influence one trait, may or may not influence other traits. In that circumstance, a vector is required to define the genetic architecture. The number of parameters that constitute the vector is which grows rapidly with the number of traits. In most cases, the researcher will have little or no knowledge of the likely extent of pleiotropy of loci that influence two traits, other than knowing or having an estimate of the genetic covariance. There are two simple ways to reduce this complexity in priors.
First, one can assume, as did Jia and Jannink (2012), that, in the context of variable selection, a locus should be selected for all of the traits or selected for none of the traits, reducing the required probabilities to being analogous to the single trait π and This approach has the advantage of simplicity, but the disadvantage that many effects might need to be estimated for loci that have no effect on a trait, and this may erode the accuracy of prediction. This should not be a problem for asymptotically large datasets, as in that case the fitted locus effects should converge to zero for those traits not influenced by that locus.
A second simple way to accommodate the multiple trait circumstance is to assume the parameters can be derived from t trait-specific parameters. However, when the probability that a single trait locus has an effect is small for each of two or more traits, the pair-wise probability that a locus affects all the traits will be the product of those small probabilities, making it very difficult for loci to enter the model for all traits simultaneously.
The better way to solve this problem is to use a hyper-parameter that completely defines the alternative models that are required to capture all the alternative forms of genetic architecture. We have shown here how this can be done, with two alternative Gibbs sampling strategies. One involves single-site sampling for one locus and trait at a time. The other samples all the alternative combinations of effects for one locus considering all traits simultaneously. We have shown that both are practical with real data and can result in improved accuracies of prediction in certain circumstances in terms of genetic architecture and size of dataset.
Conclusions
Many researchers are interested in genome-wide association studies and finding causal genes and variants. For those researchers, pleiotropy is of considerable interest, and they would want to know which loci affect which traits, from a purely biological perspective. Practitioners are often interested in “breaking” the genetic correlation, by selecting parents to give a favorable selection response in respect to multiple trait consequences. In either of these circumstances, with intermediate- rather than asymptotically large datasets, we believe the methods described here and available in the open-source, freely-available JWAS package offer real promise.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.118.300650/-/DC1.
Acknowledgments
We thank bioRxiv for making this manuscript available early online as preprints. This work was supported by the United States Department of Agriculture, Agriculture and Food Research Initiative National Institute of Food and Agriculture Competitive grant no. 2015-67015-22947.
Appendix
Gibbs Sampler Algorithm for Multi-Trait BayesC-G
Single-site Gibbs sampler for multi-trait BayesC-G
The full conditional distribution of can be written as
where Further, by dropping factors that do not involve
where and
Note that when
When
Thus when the full conditional distribution of is
When the full conditional distribution of becomes
The marginal full conditional distribution of can be written as
The factor can be written as
Note that are same when or 1. Thus the ratio becomes
Thus the conditional probability of is
where and
The full conditional distribution for can be written as
where is the number of markers with
Joint Gibbs sampler for multi-trait BayesC-G
Let denote all other parameters except and then our sampling scheme can be written as
The marginal full conditional distribution of can be written as
Denote then
where and
Note that is same for different Thus the marginal full conditional distribution of can be written as
where
The full conditional distribution of is
Gibbs Sampler Algorithm for Multi-Trait BayesB
Single-site Gibbs sampler for multi-trait BayesB
For convenience, from now on let “1” denote trait k and “2” the other traits. Thus, can be denoted as and can be denoted as The Gibbs sampler for and is derived as below. In our sampling scheme, and are sampled from their joint full conditional distributions, which can be written as the product of the full conditional distribution of given and the marginal full conditional distribution of Let denote all other parameters except and then our sampling scheme can be written as
The full conditional distribution of can be written as
where Further, by dropping factors that do not involve
where and
Note that, when
When
Thus, when the full conditional distribution of is
When the full conditional distribution of becomes
The marginal full conditional distribution of can be written as
The factor can be written as
Note that are same when or 1. Thus the ratio becomes
Thus, the conditional probability of is
where and
Joint Gibbs sampler for multi-trait BayesB
Let denote all other parameters except and then our sampling scheme can be written as
The marginal full conditional distribution of can be written as
Denote then
where and
Note that is same for different Thus the marginal full conditional distribution of can be written as
where
The full conditional distribution of is
Footnotes
Communicating editor: M. Calus
Literature Cited
- Calus M. P., Veerkamp R. F., 2011. Accuracy of multi-trait genomic selection using different methods. Genet. Sel. Evol. 43: 26 10.1186/1297-9686-43-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Garrick D., Fernando R., 2015a XSim: simulation of descendants from ancestors with sequence data. G3 5: 1415–1417. 10.1534/g3.115.016683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Qu L., Garrick D. J., Fernando R. L., 2015b A fast and efficient Gibbs sampler for BayesB in whole-genome analyses. Genet. Sel. Evol. 47: 80 10.1186/s12711-015-0157-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Fernando R. L., Garrick D. J., 2018. JWAS: Julia implementation of whole-genome analysis software. Proceedings of the World Congress on Genetics Applied to Livestock Production, 11.859. Auckland, New Zealand. [Google Scholar]
- Daetwyler H. D., Calus M. P. L., Pong-Wong R., de los Campos G., Hickey J. M., 2013. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193: 347–365. 10.1534/genetics.112.147983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando R. L., Garrick D., 2013. Bayesian methods applied to GWAS, pp. 237–274 in Genome-Wide Association Studies and Genomic Prediction. Humana Press, Totowa, NJ: 10.1007/978-1-62703-447-0_10 [DOI] [PubMed] [Google Scholar]
- Gianola D., de los Campos G., Hill W. G., Manfredi E., Fernando R., 2009. Additive genetic variability and the Bayesian alphabet. Genetics 183: 347–363. 10.1534/genetics.109.103952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Dekkers J. C. M., 2007. The impact of genetic relationship information on genome-assisted breeding values. Genetics 177: 2389–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., Garrick D. J., 2011. Extension of the Bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Jannink J.-L., 2012. Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192: 1513–1522. 10.1534/genetics.112.144246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman E., Cheng H., Firat M. Z., Garrick D. J., Fernando R. L., 2016. An upper bound for accuracy of prediction using GBLUP. PLoS One 11: e0161054 10.1371/journal.pone.0161054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende M. F. R., Muñoz P., Resende M. D. V., Garrick D. J., Fernando R. L., et al. , 2012. Accuracy of genomic selection methods in a standard data set of Loblolly Pine (Pinus taeda L.). Genetics 190: 1503–1510. 10.1534/genetics.111.137026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genotypic and phenotypic data used in the real data analysis are publicly available (Resende et al. 2012). The scripts used to generate the simulated data are provided as supplementary information. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.