Abstract
Recent technical and methodological advances have greatly enhanced genome-wide association studies (GWAS). The advent of low-cost, whole-genome sequencing facilitates high-resolution variant identification, and the development of linear mixed models (LMM) allows improved identification of putatively causal variants. While essential for correcting false positive associations due to sample relatedness and population stratification, LMMs have commonly been restricted to quantitative variables. However, phenotypic traits in association studies are often categorical, coded as binary case-control or ordered variables describing disease stages. To address these issues, we have devised a method for genomic association studies that implements a generalized LMM (GLMM) in a Bayesian framework, called Bayes-GLMM. Bayes-GLMM has four major features: (1) support of categorical, binary, and quantitative variables; (2) cohesive integration of previous GWAS results for related traits; (3) correction for sample relatedness by mixed modeling; and (4) model estimation by both Markov chain Monte Carlo sampling and maximal likelihood estimation. We applied Bayes-GLMM to the whole-genome sequencing cohort of the Alzheimer’s Disease Sequencing Project. This study contains 570 individuals from 111 families, each with Alzheimer’s disease diagnosed at one of four confidence levels. Using Bayes-GLMM we identified four variants in three loci significantly associated with Alzheimer’s disease. Two variants, rs140233081 and rs149372995, lie between PRKAR1B and PDGFA. The coded proteins are localized to the glial-vascular unit, and PDGFA transcript levels are associated with Alzheimer’s disease-related neuropathology. In summary, this work provides implementation of a flexible, generalized mixed-model approach in a Bayesian framework for association studies.
Keywords: genome-wide association, whole-genome sequencing, Alzheimer’s disease
LINKING genomic variants to traits is central to discovering the mechanisms of genetic diseases. To date, the National Human Genome Research Institute (NHGRI) has curated >1750 publications of genome-wide association studies (GWAS) that considered at least 100,000 single nucleotide polymorphisms (SNP) (Manolio 2010; Welter et al. 2014). The adoption of high-throughput sequencing technology has facilitated the rapid identification of potentially causal variants. The 1000 Genomes Project has characterized ∼88 million variants by whole-genome sequencing (WGS) of 2504 individuals from 26 populations (Auton et al. 2015). Such sequencing approaches to genomic association will soon enable discovery at a base-pair resolution. Meanwhile, statistical methods for GWAS have evolved from odds ratio tests, to generalized linear regression models (LMs), to more sophisticated multivariate linear mixed models (LMMs). LMM approaches have the capacity to correct population structures and sample relatedness (Henderson 1953), thereby minimizing false positives due to allelic cosegregation. Consequently, the number of LMM-compatible computational tools for genetic studies is rapidly increasing, e.g., ASReml, TASSEL, EMMA, QTLRel, FaST-LMM, DOQTL, GEMMA, and GMMAT (Gilmour et al. 1995; Kang et al. 2008; Zhang et al. 2010; Cheng et al. 2011; Lippert et al. 2011; Gatti et al. 2014; Zhou and Stephens 2014; Chen et al. 2016).
While LMMs are efficient in correcting sample relatedness, response variables are restricted as numerical. Meanwhile, phenotypic traits in GWAS are often categorical, such as binary variables in case-control studies or multi-level ordered categorical variables which correspond to disease stages. To model discrete response variables in the context of mixed models for population relatedness correction, generalized LMMs (GLMMs) are required. Chen et al. (2016) published a method that handles a binary response variable in the context of a mixed model. However, multiple-level categorical variables are not supported. Current approaches commonly transform categorical variables into continuous variables to fit LMMs, following the assumption that the trait has constant residual variance. However, the constant residual variance assumption is often violated by a categorical trait, which can bias effect estimates.
The proliferation of multiple GWAS for a single disease has also generated a need for methods to systematically combine results from multiple studies. Such efforts, often pursued as meta-analyses, can dramatically boost statistical power through an increase in sample size (Kavvoura and Ioannidis 2008). However, association strengths of a given variant or a genetic locus typically fluctuate across studies, which may be due to different population compositions, environmental exposures, clinical reporting standards, and experimental platforms. As a result, it is often difficult or impossible to merge raw data from different studies into a single association model. Furthermore, a more general integration of prior information is often desirable, such as coexpression or other correlations between genes. Integration approaches with more flexibility are needed to address these issues.
To address these challenges, we created the Bayes-GLMM method that exploits the flexibility of a Bayesian modeling framework and the computing efficiency of the recently developed statistical programming language Stan (http://mc-stan.org; Carpenter et al. 2017). As a Bayesian strategy, model parameters are assumed to be stochastic rather than fixed as in the case of frequentist approaches (Gelman et al. 2013). The stochastic nature of Bayesian modeling provides a coherent solution to combine published results of a related GWAS by configuring the prior distributions of the statistics of interest and computing posterior probabilities given new data (Verzilli et al. 2008; Newcombe et al. 2009; Stephens and Balding 2009). Bayes-GLMM priors are determined from reported effect sizes and corresponding P-values, thereby allowing integration of published studies based on summary statistics. Bayes-GLMM is available as an R package for public use.
We applied Bayes-GLMM to the analysis of WGS association studies using resources made available by the Alzheimer’s Disease Sequencing Project (ADSP). Alzheimer’s disease (AD) is the most common form of dementia, predicted to affect 50 million people worldwide by 2020. Unfortunately, there is no known cure. AD is commonly divided into early-onset (EOAD) and late-onset (LOAD) disease. The known genetic causes of EOAD are relatively simple with mutations in amyloid precursor protein (APP) and APP-processing enzymes such as the presenilins (e.g., PSEN1, PSEN2). However, the genetics of LOAD are poorly understood. Variations in apolipoprotein E (APOE) are the greatest genetic risk factor, with the ε4 allele conferring a 30–50% increased risk for AD (Bertram and Tanzi 2008). Recently, rare variants in triggering receptor expressed on myeloid cells 2 (TREM2) were identified that increase risk for AD (Guerreiro et al. 2013; Jonsson et al. 2013). However, few other specific causative variants have been confirmed for AD, although numerous loci have associated by GWAS (Harold et al. 2009; Lambert et al. 2009, 2013; Jones et al. 2010; Jun et al. 2010; Hollingworth et al. 2011; Naj et al. 2011). The lack of causative variants severely hampers diagnosis, animal model creation, and the development of new therapies for LOAD. Here, we report four novel noncoding variants, identified through applying Bayes-GLMM to the ADSP WGS data set. Highlighting the potential of Bayes-GLMM, these putative causative variants provide new avenues for testing the role of novel genes/pathways in LOAD.
Materials and Methods
Overview of the statistical models
Bayes-GLMM implemented GLMMs in a Bayesian framework. Bayesian models are defined by two parts: (1) a likelihood function that describes the data-generating process, and (2) the prior distributions of model parameters. Bayes-GLMM took LM, logistic regression model (logit-LM), and ordered logit-LM (ordered-logit-LM) as likelihoods functions of numerical, binary, and categorical traits, respectively.
LMMs:
In linear modeling, the numerical response variable Y was modeled in the LMM scheme:
In the above equations, X was an n-by-m covariate matrix with sample size n and the number of conditional variables m. β was the corresponding parameter vector in length m. g was the numerical genotype of a variant coded as 0, 1, or 2; representing homozygous reference allele type, heterozygous, and homozygous alternative allele type, respectively. β0 was the variant’s effect size. A standard normal, N(0, 1), was used for β0 of variants with no known effects. Further, β followed N(0, 1) in prior, and σg and σe followed inverse gamma distribution in priors. While a uniformly distributed effect prior may also be used, we found that a normally distributed prior reduced effect estimates by an average of 6% (Supplemental Material, Figure S1 in File S1), which we viewed as a favorable shrinkage to reduce false positives in genome-wide association.
To model the sample relatedness, u was included as a random term that followed a multivariate normal distribution, with prior distribution with expected mean vector 0 and covariance matrix K. was the variance component and K was the kinship matrix of the samples. was parameterized by the Cholesky factoring of K and n independent standard normal distributions:
GLMMs for binary variables:
In logit-LM, the 0/1 response variable Yi followed a binomial distribution with a scalar parameter π representing the probability that Yi equaled 1. π was further transformed by the logit function and modeled in the linear model scheme:
GLMMs for ordered categorical variables:
In ordered-logit-LM, the ordered categorical response variable Yi with J levels followed a multinomial distribution with a vector of parameters π, where πij represents the probability that the ith observation falls in response category j. Cumulative distribution of π was logit-transformed and modeled in the linear model scheme:
The cut-point parameters () in ordered categorical models comprise a vector of monotonically increasing real numbers. In our method, the increasing cut-point vector was specified by the cumulative sum (cumsum) of a primitive parameter which itself is a random sample of Dirichlet distribution, taking advantage of the fact that Dirichlet distribution samples are a vector of positive real numbers that always sum to 1.
Modeling the prior information of variant effects:
To integrate prior information of variant effects, Bayes-GLMM implemented an approach that allowed priors to only modulate information of the data under study. In this method, the prior distribution of variant effect was modeled by a hierarchical model, in which t represented prior information of the given variant and represented the SD of the Gaussian model. t was further modeled by a normal distribution with an expected mean of the standardized effect size prior and the unit deviation. The variable prior was defined by the variant’s prior effect size divided by its SE, which was often reported in published GWAS summary statistics. A standard normal, N(0, 1), was used for β0 of variants with no known effects:
We found this method of using priors appealing in three aspects: (1) it standardized the different interpretations of effect size from different statistical models, (2) it used information on both effect size and its SE, and (3) it softened the strong weight of priors from studies with unbalanced sample sizes.
Model estimations
Our models were built under Stan, which provides a flexible and efficient programming environment for statistical modeling. Inherited from Stan, Bayes-GLMM supported two methods for parameter estimation: limited-memory Broyden–Fletcher–Goldfarb–Shanno (L-BFGS) maximal likelihood estimation (MLE), and Hamilton Markov chain Monte Carlo (HMC) sampling. L-BFGS is in the family of quasi-Newton methods that approximates the original BFGS algorithm using a limited amount of computer memory (Nocedal and Wright 2006). The MLE method made a point estimation for each parameter that maximized the joint posterior of model parameters, whereas the Markov chain Monte Carlo (MCMC) sampling method captured a full posterior distribution for each parameter by iterative sampling. Significance of the estimated effect size β0 can be accessed by combing β0 and its SE, SE(β0). SEs of MLE were computed as the inverse of the square root of the diagonal elements of the observed Fisher information matrix (Pawitan 2001). A standardized z value was computed as β0/SE(β0), which led to a P-value that quantified the probability of obtaining the β0 by chance:
was the MLE of model parameters, I(θ) was the Fisher information matrix, and p was the number of parameters.
In MCMC sampling, we drew 400 samples (200 as burn-in, 200 as effective) for each of three randomly-initiated Markov chains, which resulted in 600 effective samples in total. We used the Gelman–Rubin diagnostic ( in Stan) to assess convergence of multiple chains (Gelman and Rubin 1992). The P-value of variant effect using MCMC sampling results was reported as the tail probability () of the variant effect’s posterior distribution:
Following the normality assumption, Pt was computed by the same procedure as used to compute P-values of MLE estimations, while SE(β0) was taken as the SD of the variant effect’s posterior distribution. We found that tail probabilities computed this way are consistent with the frequentist P-values under a generalized linear model (GLM) scheme (Figure S2 in File S1).
Kinship matrix
We used u as a random term to account for the sample relatedness. u follows the normal distribution where K was the kinship matrix of the samples. For each K entry, genotype-based relatedness for the sample pair, or the identical-by-state coefficient, was computed using the full spectrum of genomic variants in the ADSP samples. PLINK was used for fast kinship estimation on the massive genotype data.
LMMs in the frequentist scheme
To compare the performances of our method to that of an LMM in the frequentist scheme in analyzing the ADSP data set, we built an LMM as follows:
was the numerical mapping of the AD categories: no = 0, possible = 0.25, probable = 0.5, and definite = 1. was the covariate matrix including age and sex, u was the random term, and was the model residual. The LMM was estimated with QTLRel in R (Cheng et al. 2011).
Mouse strains, tissue harvesting, and sectioning
All experiments involving mice were conducted in accordance with policies and procedures described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and were approved by the Institutional Animal Care and Use Committee (IACUC) at The Jackson Laboratory. All mice were bred and housed in a 12-/12-hr light/dark cycle. Male C57BL/6J mice (6 months old) were injected intraperitoneally with a lethal quantity of ketamine/xylazine according to IACUC-approved procedures. Mice were perfused with 1× PBS and whole brains were removed and fixed in 4% paraformaldehyde for 2 hr at 4°. Following fixation, the tissue was rinsed in 1× PBS, incubated in 10% sucrose for 8 hr at 4°, and then incubated in 30% sucrose overnight at 4°. Brains were then frozen in optimal cutting temperature compound and stored at −80° until sectioning. Frozen brains were sectioned at 25 µm and mounted on glass slides, which were stored at −80° until required for immunofluorescence staining.
Immunofluorescence
Brain sections were incubated overnight at 4° in the following primary antibodies: rabbit polyclonal anti-PDGFA (1:50; Bioss Antibodies), sheep polyclonal anti-PRKAR1B (1:50; R&D Systems), goat anti-COL-IV (1:50; EMD Millipore), and goat anti-CD31 (1:50; R&D Systems). Sections were immersed in deionized water for 3 min at 37° and then treated with 0.5 mg/ml pepsin in 0.2N HCl for 15 min at 37°. Slides were then washed twice in 1× PBS for 10 min at room temperature. With the exception of anti-COL-IV, antibodies were diluted in 0.5% PBTB (1× PBS, 0.0.5% Triton X-100, and 0.5% BSA) containing 10% normal donkey serum. Anti-COL-IV was diluted in 0.5% PBS/Tween 20 (PBT). Sections were washed three times in 0.5% PBT and then incubated for 2 hr at room temperature with their respective secondary antibodies (donkey anti-rabbit Alexa Fluor 594, donkey anti-goat Alexa Fluor 488, and donkey anti-sheep Alexa Fluor 594 in a 1:1000 dilution; Life Technologies). All sections were then counterstained with DAPI (1:1000 in 1× PBS) and washed with 1× PBS prior to mounting with Aqua PolyMount. Images were taken using a Leica SP5 confocal microscope located within the imaging facility at The Jackson Laboratory.
Data availability
All ADSP genotype and phenotype data are available via dbGaP under study accession phs000572.v7.p4. C57BL/6J mice are available for purchase from The Jackson Laboratory (strain #000664) at https://www.jax.org/strain/000664. The code used for analysis is available as Bayes-GLMM in a GitHub repository for public use at https://github.com/xulong82/bayes.glmm. Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP) data are available as cited (Lim et al. 2017). Additional information on associated variants can be found in Table S1 and Table S2.
Results
ADSP
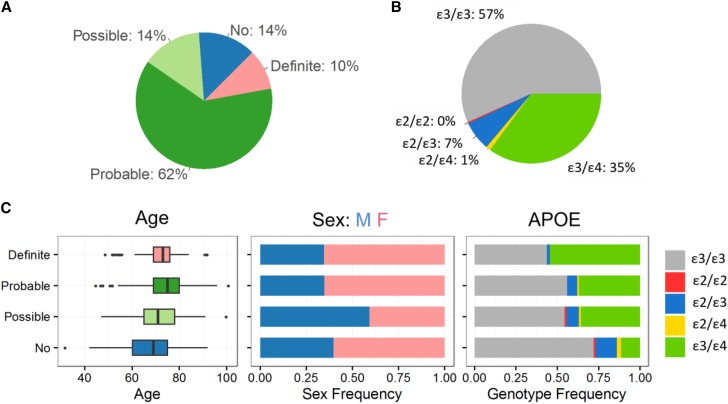
The development of Bayes-GLMM was motivated by the advent of the WGS association studies, such as the ADSP (www.niagads.org/adsp; Materials and Methods). ADSP was initiated to discover novel genomic variants for LOAD. The WGS cohorts of ADSP contained 570 participants from 111 families. This family-based design generated profound sample relatedness that warranted a mixed-model approach. Furthermore, phenotypic traits were categorized into four levels of Alzheimer’s diagnoses: no (N = 78), possible (N = 81), probable (N = 356), and definite (N = 55), which necessitated a generalized categorical model. Family pedigree, race, ethnicity, age, sex, and APOE ε2/ε3/ε4 genotype were also reported for each participant. The population was 61% female. The interquartile range of sample ages was 67–80 years. In APOE genotypes, homozygous APOEε3 comprised 56.7% (N = 323) of the population, followed by 35.1% (N = 200) of APOEε3/APOEε4, 6.84% (N = 39) of APOEε2/APOEε3, 1.05% (N = 6) of APOEε2/APOEε4, and 0.351% (N = 2) of APOEε2/APOEε2 (Figure 1). Individuals homozygous for APOEε4 were excluded from the study.
Figure 1.
Summary statistics of the ADSP WGS cohort. (A) AD diagnosis for 570 individuals across 111 families. (B) APOE allele-type composition. (C) Age distributions of individuals in each AD diagnostic category (left), sex composition in each category (middle), and APOE allele-type composition in each category (right).
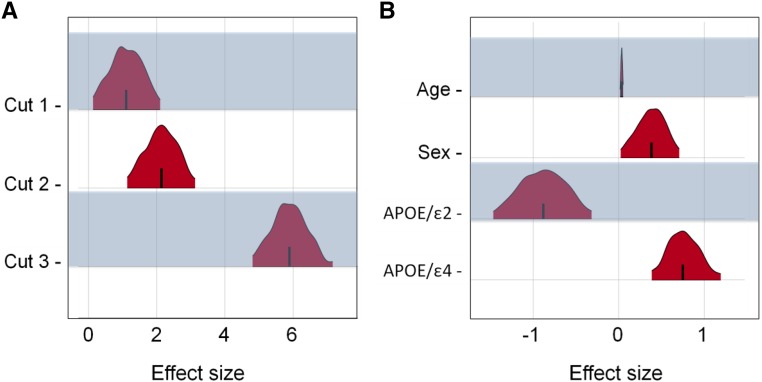
The additive effects of age, sex (female), and APOE allele types (ε2, ε3, and ε4) were tested with Bayes-GLMM together with the cut-points parameters of the ordered categorical model (Figure 2). To account for sample relatedness, kinship structure was computed from autosomal variants and included as the variance–covariance matrix of a random effect that followed a multivariate normal distribution (Materials and Methods). Model parameters were estimated by MCMC sampling. As expected, we observed that the APOEε4 allele significantly increased risk of AD (P = 0.00014), while the APOEε2 allele reduced risk (P = 0.0033) relative to the baseline APOEε3 allele. Sex was also a significant factor, with females at a higher risk (P = 0.032). Increasing age corresponded to a small but significant risk increase (P = 0.00036). The small effect size of age was a result of multiple factors: (1) the relatively large values for age as a model predictor (67–80 years), (2) a narrow age range, and (3) the possible longevity of nonaffected individuals. All covariate pairs were tested with fixed-effect interaction terms, but no significant interactions were observed (Figure S3 in File S1).
Figure 2.
Bayes-GLMM estimation of model parameters by MCMC sampling of GLMM. Areas shown are 95% highest posterior density. (A) Posterior distributions of the ordered categorical model’s cut points. Convergence diagnostics of cut 1, cut 2, and cut 3 were 1.01, 1.00, and 1.03, respectively, which implies strong convergence. (B) Posterior distributions of the model covariate’s effect sizes: age, sex, APOEε2, and APOEε4. of age, sex, APOE/ε2, and APOE/ε4 were 1.00, 1.00, 1.01, and 1.00, respectively.
GWAS of ADSP WGS cohort by Bayes-GLMM
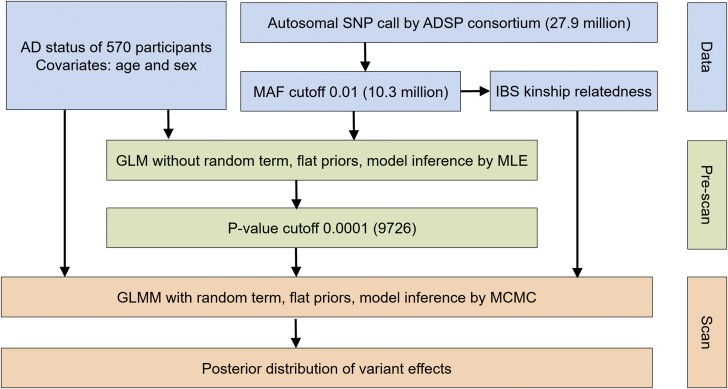
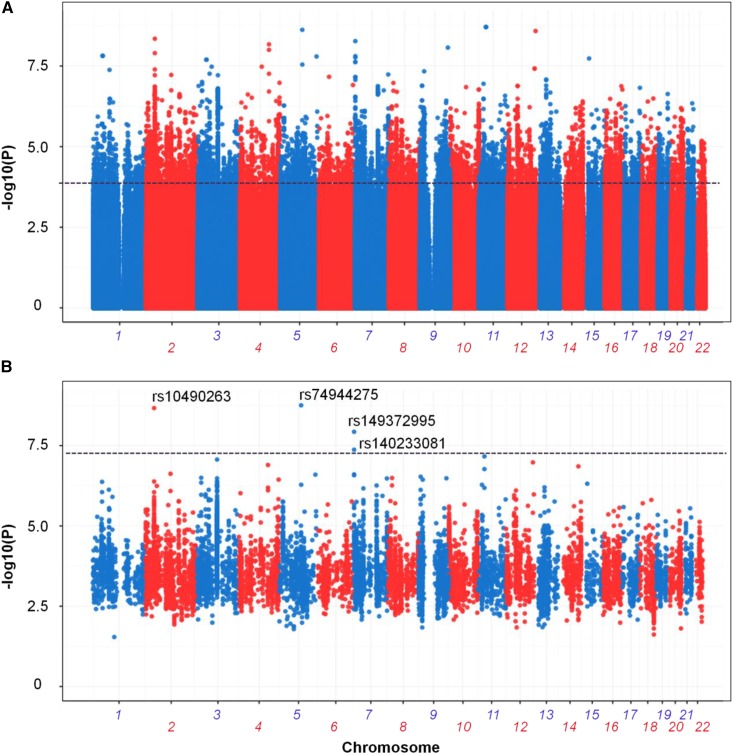
The ADSP consortium identified a total of 27.9 million SNPs from the WGS cohort, of which 10.3 million passed their quality check and had a minor allele frequency >0.01 (Figure S4 in File S1). Associations of the 10.3 million SNPs to AD status were tested by Bayes-GLMM in two steps (Figure 3). In the first step, a GLM (ordered categorical model) was applied to each of the 10.3 million variants without the random term. The purpose of this step was to perform a preliminary screen for potential candidate variants. Model parameters were estimated by the MLE method for computational efficiency. Variants with P < 0.0001 were identified as potential candidate variants (N = 9726; Figure 4A). In the second step, candidate variants from the first step were tested with the full GLMM, including the random term to address sample relatedness. Model parameters were estimated by MCMC sampling to avoid the instability in estimating GLMM by MLE. Final P-values for every variant were obtained from their empirical posterior distributions (Figure 4B).
Figure 3.
Analysis overview of two-step GWAS analysis using Bayes-GLMM. Initial data (blue) were filtered and prescanned with a fixed linear model (green). Results were filtered by significance and scanned using the full GLMM (orange).
Figure 4.
Association results for ADSP WGS cohort by Bayes-GLMM. (A) Results for 10.3 million genomic variants by Bayes-GLMM without kinship correction. Model parameters were estimated by MLE. Variants with P < 0.0001, above the dashed line, were chosen for the full scan (9726 variants). (B) GWAS on filtered variants by GLMM with kinship correction. Model parameters were estimated by MCMC sampling. Dashed line was the cutoff of genome-wide significance (P < 5 × 10−8).
Top LOAD-associated variants from ADSP WGS
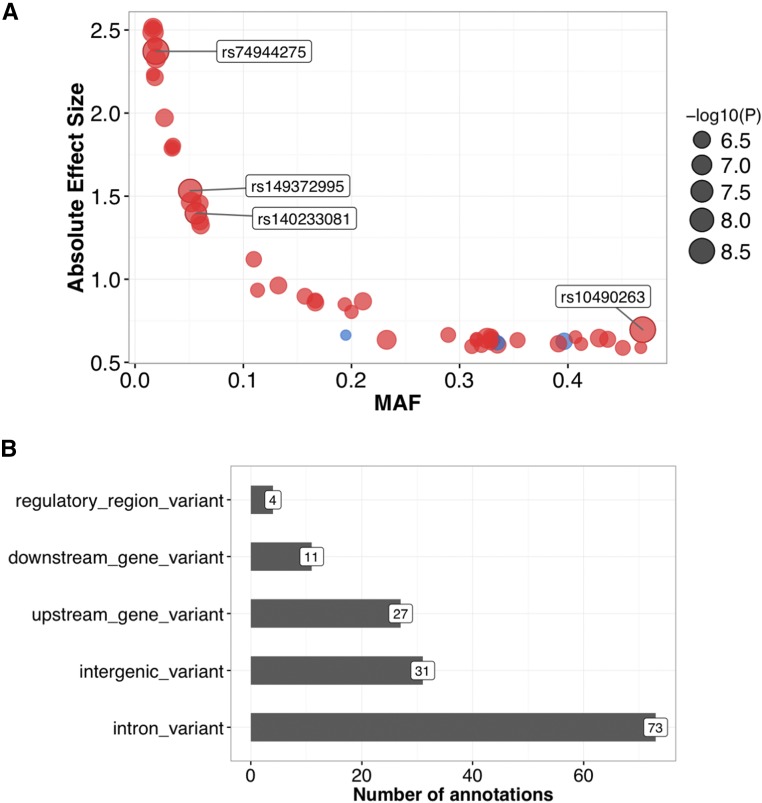
We identified four variants in three independent loci with P < 5 × 10−8, and 55 variants in 28 loci with P < 1 × 10−6 (Table 1). The top two variants meet a stricter significance threshold of P < 5 × 10−9 that would assume ∼10 million independent SNPs. Of the top 55 variants, 52 were associated with an increased LOAD risk. Furthermore, variants with strong effects tended to occur at a lower allele frequency, suggesting that these variants might be under negative selection (Figure 5). The top 55 variants were mapped to 146 genomic annotations using Ensembl Variant Effect Predictor (variants commonly mapped to multiple annotations): 73 were in introns, 31 were in intergenic regions, 27 were upstream of genes (within 5 kb upstream from the 5′ end), 11 were downstream of genes (within 5 kb downstream from the 3′ end), and 4 were regulatory regions (Table S1). The 73 intronic annotations mapped to 19 variants and 18 unique genes. Of the 18 genes, 12 appeared in the NHGRI GWAS catalog as being associated with disease (Welter et al. 2014) (Table S2). Associated traits of the 12 genes included obesity-related traits (PTPRD, SORCS2, and SLC24A4), AD (SLC24A4 and GABRG3), acute lymphoblastic leukemia (ERC2 and ST6GALNAC3), adiponectin levels (CMIP and HIVEP2), bipolar disorder and schizophrenia (ERC2), and type 2 diabetes (PTPRD).
Table 1. Top 55 variants with P < 1 × 10−6.
| RSID | Chromosome | Position | Reference | Alternate | MAF | Effect size | SD | 95% C.I. | P-value |
|---|---|---|---|---|---|---|---|---|---|
| rs74944275 | 5 | 102,726,073 | C | T | 0.019 | 2.371 | 0.394 | [1.633, 3.136] | 1.76 × 10−9 |
| rs10490263 | 2 | 40,973,289 | C | T | 0.469 | 0.697 | 0.116 | [0.483, 0.932] | 2.15 × 10−9 |
| rs149372995 | 7 | 580,540 | A | G | 0.051 | 1.532 | 0.269 | [0.992, 2.063] | 1.18 × 10−8 |
| rs140233081 | 7 | 580,735 | C | A | 0.056 | 1.396 | 0.255 | [0.926, 1.898] | 4.26 × 10−8 |
| rs139258867 | 11 | 33,422,464 | C | T | 0.017 | 2.487 | 0.461 | [1.593, 3.418] | 6.89 × 10−8 |
| rs11709639 | 3 | 94,975,203 | T | A | 0.325 | 0.645 | 0.120 | [0.415, 0.875] | 8.61 × 10−8 |
| rs75841969 | 12 | 127,335,883 | G | A | 0.052 | 1.464 | 0.275 | [0.921, 2.007] | 1.05 × 10−7 |
| rs72720587 | 4 | 137,323,780 | C | T | 0.019 | 2.328 | 0.441 | [1.515, 3.221] | 1.27 × 10−7 |
| rs2018116 | 14 | 92,831,272 | T | C | 0.768 | 0.636 | 0.121 | [0.383, 0.855] | 1.41 × 10−7 |
| rs141404567 | 11 | 33,393,524 | G | A | 0.017 | 2.514 | 0.481 | [1.634, 3.425] | 1.70 × 10−7 |
| rs2010568 | 2 | 118,395,972 | G | C | 0.429 | 0.646 | 0.125 | [0.385, 0.882] | 2.38 × 10−7 |
| rs144152209 | 7 | 585,369 | C | A | 0.060 | 1.349 | 0.262 | [0.879, 1.872] | 2.49 × 10−7 |
| rs74917009 | 5 | 169,915,787 | G | A | 0.027 | 1.971 | 0.382 | [1.233, 2.661] | 2.54 × 10−7 |
| rs144990130 | 7 | 582,328 | G | A | 0.061 | 1.324 | 0.257 | [0.849, 1.823] | 2.65 × 10−7 |
| rs12685122 | 9 | 9,206,006 | T | G | 0.211 | 0.867 | 0.169 | [0.534, 1.196] | 2.95 × 10−7 |
| rs73046027 | 3 | 19,950,385 | C | T | 0.132 | 0.963 | 0.188 | [0.624, 1.349] | 3.18 × 10−7 |
| rs7463321 | 8 | 20,523,821 | T | C | 0.167 | 0.860 | 0.168 | [0.523, 1.198] | 3.23 × 10−7 |
| rs148758667 | 9 | 130,665,077 | G | T | 0.060 | 1.456 | 0.285 | [0.949, 2.026] | 3.30 × 10−7 |
| rs72618491 | 3 | 94,938,828 | G | A | 0.335 | 0.606 | 0.119 | [0.363, 0.85] | 3.31 × 10−7 |
| rs117662279 | 7 | 155,362,626 | G | A | 0.018 | 2.214 | 0.434 | [1.328, 3.048] | 3.34 × 10−7 |
| rs1280103 | 4 | 187,526,002 | C | A | 0.396 | −0.627 | 0.123 | [−0.878, −0.388] | 3.63 × 10−7 |
| rs7856285 | 9 | 18,973,653 | G | A | 0.609 | 0.612 | 0.120 | [0.366, 0.84] | 3.65 × 10−7 |
| rs17383917 | 3 | 94,984,650 | T | C | 0.330 | 0.640 | 0.126 | [0.401, 0.879] | 4.08 × 10−7 |
| rs11124760 | 2 | 41,001,812 | C | T | 0.437 | 0.639 | 0.126 | [0.394, 0.884] | 4.15 × 10−7 |
| rs61768273 | 1 | 44,509,818 | A | T | 0.034 | 1.790 | 0.354 | [1.074, 2.443] | 4.25 × 10−7 |
| rs17383687 | 3 | 94,963,466 | C | T | 0.329 | 0.653 | 0.129 | [0.412, 0.924] | 4.33 × 10−7 |
| rs12497549 | 3 | 20,072,654 | C | T | 0.157 | 0.898 | 0.178 | [0.528, 1.251] | 4.52 × 10−7 |
| rs36147593 | 15 | 27,587,764 | A | G | 0.110 | 1.119 | 0.222 | [0.684, 1.539] | 4.86 × 10−7 |
| rs10933941 | 3 | 94,965,589 | G | A | 0.329 | 0.635 | 0.126 | [0.392, 0.888] | 4.98 × 10−7 |
| rs116407196 | 5 | 102,973,337 | A | G | 0.035 | 1.804 | 0.360 | [1.129, 2.524] | 5.25 × 10−7 |
| rs7122488 | 11 | 21,874,253 | T | C | 0.646 | 0.633 | 0.126 | [0.402, 0.868] | 5.26 × 10−7 |
| rs12639003 | 3 | 94,966,599 | A | G | 0.329 | 0.637 | 0.127 | [0.39, 0.878] | 5.35 × 10−7 |
| rs12549162 | 8 | 20,547,331 | C | G | 0.167 | 0.872 | 0.174 | [0.538, 1.216] | 5.50 × 10−7 |
| rs62483581 | 7 | 106,726,214 | G | A | 0.451 | 0.587 | 0.117 | [0.367, 0.837] | 5.52 × 10−7 |
| rs12485639 | 3 | 94,940,998 | C | A | 0.320 | 0.604 | 0.121 | [0.359, 0.841] | 5.55 × 10−7 |
| rs67822265 | 2 | 53,715,939 | C | T | 0.289 | 0.665 | 0.133 | [0.401, 0.933] | 5.70 × 10−7 |
| rs17383861 | 3 | 94,983,399 | G | A | 0.331 | 0.635 | 0.127 | [0.396, 0.876] | 5.91 × 10−7 |
| rs9826288 | 3 | 95,044,652 | C | T | 0.665 | −0.618 | 0.124 | [−0.855, −0.369] | 6.27 × 10−7 |
| rs2478319 | 13 | 48,111,575 | A | G | 0.689 | 0.596 | 0.120 | [0.358, 0.821] | 6.33 × 10−7 |
| rs72720573 | 4 | 137,257,730 | T | C | 0.018 | 2.416 | 0.485 | [1.488, 3.359] | 6.46 × 10−7 |
| rs140419591 | 11 | 33,327,476 | A | G | 0.017 | 2.516 | 0.506 | [1.553, 3.502] | 6.54 × 10−7 |
| rs78491489 | 7 | 44,335,828 | C | T | 0.113 | 0.934 | 0.188 | [0.61, 1.321] | 6.80 × 10−7 |
| rs6689933 | 1 | 76,837,471 | C | T | 0.684 | 0.633 | 0.128 | [0.401, 0.876] | 7.47 × 10−7 |
| rs17263248 | 3 | 55,574,820 | A | G | 0.200 | 0.804 | 0.163 | [0.5, 1.13] | 7.54 × 10−7 |
| rs10435819 | 9 | 9,197,298 | G | A | 0.194 | 0.849 | 0.172 | [0.49, 1.165] | 7.58 × 10−7 |
| rs61446477 | 3 | 94,964,689 | A | G | 0.329 | 0.639 | 0.129 | [0.38, 0.887] | 7.73 × 10−7 |
| rs72720589 | 4 | 137,333,269 | T | A | 0.017 | 2.233 | 0.452 | [1.326, 3.105] | 7.80 × 10−7 |
| rs7978950 | 12 | 47,361,547 | C | T | 0.412 | 0.611 | 0.124 | [0.382, 0.858] | 7.92 × 10−7 |
| rs2176276 | 3 | 94,989,440 | C | A | 0.329 | 0.613 | 0.124 | [0.389, 0.856] | 8.13 × 10−7 |
| rs1359665 | 13 | 48,097,289 | G | A | 0.684 | 0.642 | 0.130 | [0.384, 0.887] | 8.16 × 10−7 |
| rs4849593 | 2 | 118,369,787 | G | A | 0.407 | 0.652 | 0.132 | [0.404, 0.917] | 8.20 × 10−7 |
| rs72618501 | 3 | 94,974,822 | T | C | 0.330 | 0.633 | 0.129 | [0.398, 0.881] | 8.66 × 10−7 |
| rs61768270 | 1 | 44,498,974 | C | T | 0.034 | 1.788 | 0.364 | [1.134, 2.481] | 8.88 × 10−7 |
| rs1861305 | 2 | 40,950,582 | A | G | 0.468 | 0.589 | 0.120 | [0.356, 0.819] | 8.96 × 10−7 |
| rs4367173 | 4 | 7,383,470 | C | G | 0.195 | −0.664 | 0.136 | [−0.92, −0.4] | 9.58 × 10−7 |
Variants in italics met a standard genome-wide significance of P < 5 × 10−8. RSID, reference SNP cluster identifier; MAF, minor allele frequency.
Figure 5.
Effect sizes and consequences of top variants. (A) Allele frequencies and effect sizes for all variants with Bayes-GLMM-derived P < 1 × 10−6. Positive-effect (i.e., risk-increasing) loci are in red and negative-effect loci (i.e., protective) are in blue. (B) Functional consequences of the top variants. MAF, minor allele frequency.
The four genome-wide significant variants (P < 5 × 10−8) were all intergenic: rs10490263, rs74944275, rs149372995, and rs140233081. These SNPs are located as follows: rs10490263 is 233,714 bp upstream of SLC8A1 and 337 bp upstream of long intergenic noncoding RNA (lincRNA) AC007317.1; rs74944275 is 111,711 bp downstream of C5orf30 and 18,568 bp downstream of lincRNA CTD-2154H6.1; rs140233081 and rs149372995 are in linkage disequilibrium (LD) and are located between PRKAR1B and PDGFA. Additionally, these final two SNPs are 8097 and 8292 bp downstream of PRKAR1B, and 21,254 and 21,059 bp upstream of PDGFA, respectively. To assess the functional relevance of the four variants, we queried the Roadmap Epigenomics (Bernstein et al. 2010) and ENCODE (ENCODE Project Consortium 2012) resources using HaploReg (Ward and Kellis 2012) for chromatin state and protein binding annotations. We found that rs10490263 lies in promoter-associated histone marks in the hippocampus and circulating T cells, and that rs74944275 lies in both promoter- and enhancer-associated histone marks in multiple brain regions. Furthermore, rs149372995 resides in a candidate-binding site of CTCF; rs74944275 resides in a candidate-binding site of CCNT2, Evi-1, GATA, and HDAC2; and rs140233081 and rs149372995 lie in candidate binding sites of NERF1a, SMC3, and TCF12.
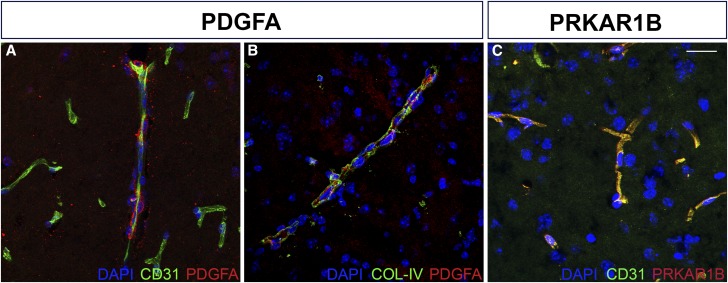
Given the role of CTCF in genome organization and possible gene regulation, we further examined the flanking genes PRKAR1B and PDGFA. We localized the expression of the protein products of these two genes using immunofluorescence. Both PRKAR1B and PDGFA have widespread expression in the mouse brain, but are particularly localized to glia–vascular structures (Figure 6). This could be significant given the recent data suggesting glia–vascular alterations may predispose individuals to, or occur very early in, LOAD (Bell 2012; Zhao et al. 2015; Montagne et al. 2016). Furthermore, we evaluated RNA sequence data from the dorsolateral prefrontal cortex of participants in the ROS and Rush MAP studies, which are two longitudinal cohort studies of aging with prospective brain autopsy (Bennett et al. 2012a,b; De Jager et al. 2014). In these human data, we found that higher PDGFA transcript level is moderately correlated with a greater neuritic plaque burden (P = 0.005, transcriptome-wide false discovery rate = 0.03; β > 0) (Lim et al. 2017), suggesting that the PDGFA association with AD may relate to a role in the accumulation of one of the two key pathologic features of AD.
Figure 6.
PDGFA and PRKAR1B localize to vascular structures in the mouse brain. PDGFA (red) shows close localization to (A) endothelial cells (CD31) and (B) basement membrane (COL-IV), components of the vascular substructure. (C) PRKAR1B (red) shows punctate expression in the region of blood vessels (CD31, green). See Materials and Methods for antibody details. Bar, 20 μm.
Integrating prior knowledge
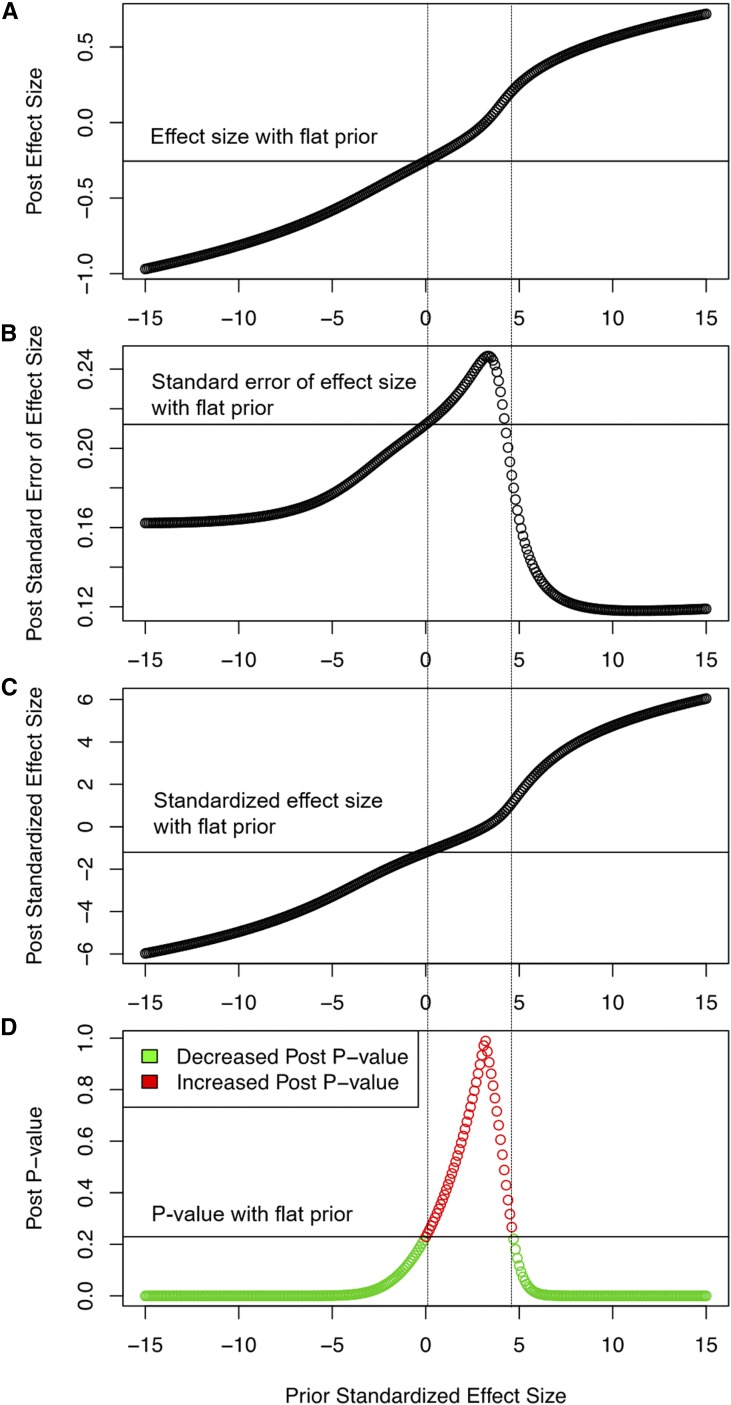
Prior knowledge integration is a prominent feature of Bayesian modeling. In GWAS, prior information of a variant can be implemented with multiple strategies, each allowing posterior estimations to carry different weights of the priors. In brief, we considered the following strategies: (1) summary mean and SE estimated from a previous study, (2) normally distributed mean and inverse-gamma SE distributions based on prior estimates, (3) standardized mean (t-statistic) and inverse-gamma SE distributions based on prior estimates, and (4) normally distributed standardized mean (t-statistic) distribution and inverse-gamma SE distributions based on prior estimates (Table S3 in File S1). In Bayes-GLMM, we implemented the fourth strategy to respect the unique challenges of GWAS, such as the different meanings of effect sizes from studies with different statistical models, variable allele frequencies in multiple study populations, and the particularly small P-values from large-scale studies. We considered priors from the International Genomics of Alzheimer’s Project (IGAP) (Lambert et al. 2013). While none of the top 1000 IGAP variants were genome-wide significant in the ADSP data set, many showed suggestive significance and consistent effect directionality (Figure S5 in File S1). However, drawing mean and SE priors directly from IGAP overwhelmed evidence in our study population and yielded significance estimates strongly correlated with IGAP results (Figure S6 in File S1). Our method took the reported standardized effect sizes as the prior information and integrated them into the hierarchical model of each variant effect (Materials and Methods). To demonstrate the performance of this method, we generated a binary phenotypic trait (coded as 0 or 1) and genotypic trait of a variant (coded as 0, 1, or 2) by Monte Carlo, and used a logit-LM to test their associations. To illustrate the ability of Bayes-GLMM to integrate this information, we assessed the effect of prior information on the estimated variant effect by testing a range of prior standardized effect sizes. This method of prior configuration effectively modulates the information from the data (Figure 7), regardless of the differences between the prior information and the data in hand.
Figure 7.
Effects of priors on (A) posterior effect size, (B) posterior standardized error of the effect size, (C) posterior standardized effect size, and (D) posterior P-values. The x-axis denotes prior standardized effect size. The gray horizontal line in each graph is the respective posterior estimation when the prior standardized effect size is equal to 0. The two vertical dashed lines define a range of prior standardized effect sizes that increased the posterior P-value compared to a flat prior. Post, posterior.
Discussion
We created a new GWAS method, Bayes-GLMM, and applied it to ADSP’s WGS cohort. This method efficiently addresses three major challenges in GWAS: categorical phenotypes, population structure and sample relatedness, and prior knowledge integration. Furthermore, our generalized approach has the flexibility to operate on binary and quantitative traits in addition to ordered categorical phenotypes. These features enabled our identification of four new candidate variants in three loci that significantly increased the risk of AD.
Out of the four new genome-wide significant candidate variants, rs140233081 and rs149372995 are in LD and are located between PRKAR1B and PDGFA, which are potentially relevant to vascular dysfunction. Recent evidence suggests that vascular dysfunction is a critical component of AD pathology (Bell 2012; Zhao et al. 2015; Montagne et al. 2016) and potentially a necessary predisposing feature (Iturria-Medina et al. 2016). Further, vascular dysfunction has been shown to be necessary for the development of AD-like phenotypes in a mouse model of amyloid pathology (Soto et al. 2016). We have localized PDGFA and PRKAR1B to specific components of vascular anatomy. Our immunofluorescence shows PDGFA expression between the collagen-rich tunica external and the endothelium of the tunica intima, supporting the presence of PDGFA in vascular smooth muscle cells (VSMCs). Previous studies have shown PDGF to affect VSMC proliferation by inducing a phenotypic switch from a contractile state to a proliferative one (Owens et al. 2004). Insufficient PDGFA expression, then, may impair vascular regeneration following plaque-related insults, thereby exacerbating AD. This potential mechanism paired with increased PDGFA under amyloid burden expression suggests the two candidate variants could reduce necessary PDGFA expression when plaques are present, thereby attenuating the increase in PDGFA we observed with amyloid burden. PRKAR1B was seen in a punctate fashion, suggesting the presence of cytoplasmic clusters of the protein, and we hypothesize that the PRKAR1B puncta represent accumulation of protein kinase A (PKA) at either the endoplasmic reticulum or the insulin receptor. Calcium release from the endoplasmic reticulum is typically suppressed by phospholamban (PLN); however, such suppression is lifted following PLN phosphorylation by PKA. Changes in the regulation of calcium release due to altered PRKAR1B expression may very well have important consequences for AD, including but not limited to changes in vascular smooth muscle contraction that limit circulation to plaque-burdened brain regions. In addition to its calcium-related role, PKA is essential for signal transduction following activation of the insulin receptor, a process that has been shown to be the mechanism by which PDGF induces phenotypic switching in VSMCs (Zhao et al. 2011). In this way, changes in PRKAR1B may yield corresponding changes in circulation through suppressed arterial muscle contractility or through a direct influence on vascular growth and maintenance.
We consider our method, Bayes-GLMM, to be an important addition to the existing GWAS toolkit. The flexibility of Bayesian modeling allows the convenient configuration of sophisticated models, such as our GLMM. In Bayes-GLMM, logistic and ordered logistic regression likelihoods were used to model binary and ordered categorical variables, respectively. Conditional factors were included as model covariates and, although our study was underpowered for epistasis analysis, interaction terms can be straightforwardly included. Sample relatedness was modeled by a random term that followed a multivariate normal distribution. Model parameters can be estimated by either L-BFGS MLE or HMC sampling, as implemented in Stan.
Although the MLE implementation in Bayes-GLMM was efficient and reliable in estimating GLMs, it was unreliable in estimating GLMMs. We found that the MLE of the random term was skewed toward initial values, suggesting the optimizer was trapped into local optima and limiting reliability in estimating the GLMM. On the other hand, the MCMC sampler allows an improved assessment of the robustness and stability of model inferences by reporting the full posterior distributions of model parameters and the convergence of multiple sampling chains. This information allows one to dissect how multiple factors contribute to model estimation, including poorly defined prior distributions, collinearity of predictors, and inappropriate initial sampling values.
The Bayes-GLMM method was optimized in multiple ways to minimize the computational expense. It was optimized to (1) support parallel computing, (2) conjugate prior distributions, (3) vectorize model statements to exploit efficient matrix operations in Stan, and (4) parameterize multivariate normal distribution for the random effect by Cholesky factoring. Nevertheless, efficiency was still the primary drawback of MCMC sampling. When testing on a 2.3 GHz Intel processor, MLE took ∼0.12 sec to estimate the GLM per variant of the ADSP data set (Materials and Methods; Figure 3). In comparison, the MCMC sampler took ∼30 sec to generate 1000 samples for the same GLM, and 15 min to process 1000 samples for the GLMM model. Our prescan with MLE followed by more precise estimation by MCMC proved a practical approach to overcome these processing limitations when applying Bayes-GLMM in GWAS.
To reduce the computational burden in fitting GLMMs, we suggest that categorical diagnoses could be collapsed into binary variables. For the ADSP data, the “no” and “possible” diagnoses become “control,” while the “probable” and “definite” diagnoses are “case.” Logistic mixed models or binary mixed models were implemented in Bayes-GLMM to accommodate binary variables. The MCMC sampler implemented in Stan took ∼10 min to collect 1000 samples for parameters of such a binary mixed model, as opposed to 15 min for the four-level categorical mixed model. Alternatively, the recently released GMMAT (generalized linear mixed-model association test) method that used a penalized quasi-likelihood method to fit a binary mixed model was significantly faster than the MCMC sampling approach (Chen et al. 2016). However, this practice of collapsing the categorical variable reduced precision due to the information loss in simplifying multiple categories. We tested this practice in the ADSP data and found the association results by binary GLMM and categorical GLMM showed substantial disagreement (Figure S7 in File S1).
Another strategy to reduce computational requirements is to transform categorical variables into continuous variables to accommodate efficient LMM methods (Kang et al. 2010; Chen et al. 2016). However, this practice is prone to yield an incorrect type-I error rate because categorical studies do not satisfy LMM’s constant residual variance assumption; that is, linear models assume residual variances are constant with respect to different values of model predictors. This practice also yields incorrect effect estimates due to the unbalanced sampling in different phenotypic categories, which is prominent in the ADSP study in which the probable diagnosis accounted for 62% of the total and the other three categories accounted for only 10–14%. We also found the inference results of LMM by QTLRel were sensitive to different quantitative coding of categorical variables (Figure S8 in File S1; Materials and Methods). Taking rs34827707 as an example, the likelihood-ratio-test value for rs34827707 dropped from 29 to 15 when changing the coding from no/possible/probably/definite as 0/0.25/0.5/1 to 0/0.33/0.66/1. In contrast, the GLMM robustly estimated three cut points to separate the four categories.
Bayesian modeling naturally allows the integration of prior information by specifying the model parameter’s prior distribution. However, how to best specify a variant’s prior information is an open question when the prior study does not precisely match the experiment design in hand. Association results of each variant in a GWAS are commonly reported by effect size and P-value. While critical in describing the association strength, exact values of effect sizes are often specific to the given study because of dependencies on the statistical model, genotype coding strategies, and covariates. Therefore, it can be misleading to use the reported effect sizes to configure the priors. As opposed to effect sizes, P-values that quantify deviation from a null hypothesis can be less specific to the given study. However, P-values are strongly influenced by the sample size, and P-values from a large-scale study as priors would dominate the posterior estimation of a variant’s association, thereby masking the information of the current study. To tackle this problem, we proposed a strategy that models the variant effect by a hierarchical model, in which variant effect was first modeled by a normal distribution with expected mean represented as the multiplication of the standardized expected mean and the SD. The standardized expected mean was further modeled by a standard normal with expected mean specified as the prior standardized effect. Simulation results showed our method of configuring the priors to be effective in allowing only priors modulating information of the data under study (Figure 7).
While powerful, Bayes-GLMM has several drawbacks. First, the quantitative meaning of parameter values is not readily interpretable in terms of fractional effects. Second, heritability estimation is elusive due to a difficulty in estimating residual variance. Third, as implemented, only one variance component is supported. Although Bayesian modeling can readily encompass multiple variance components, this becomes impractical for GWAS due to computational limitations for most researchers. Fourth, sampling-based estimations remain computationally intensive and may not be suitable for larger data sets (e.g., the full set of ADSP variants). We expect that advances in model estimation techniques, improved algorithms, and broad application of cloud-based computational resources will alleviate these problems in the near future.
To summarize, here we have proposed a method for GWAS with three major features: (1) a generalized model to support multiple types of phenotypic data, (2) a Bayesian strategy to effectively integrate previous GWAS results for the same trait, and (3) a mixed-model implementation to correct population structure. With genome-wide association transitioning to whole-genome and whole-exome platforms, statistical methods for large-scale association studies are essential for uncovering the genetic basis of complex disease. The ability to integrate existing GWAS as prior information can further power these studies to prioritize specific variants at known loci.
Supplementary Material
Supplemental material available at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300673/-/DC1.
Acknowledgments
We thank B. Carpenter and A. Gelman of the Stan Development Team for assistance, G. Churchill for helpful discussions, and M. Miller and the Alzheimer’s Disease Sequencing Project Consortium for assisting with data resources. This work was funded by National Institute on Aging AG-054345 (G.W.C. and G.R.H.); National Institute on Aging P30 AG-10161, R01 AG-15819, R01 AG-179917, R01 AG-36836, and U01 AG-46152 (D.A.B.); The Pyewacket Foundation (G.W.C.); and The Jackson Laboratory Director’s Innovation Fund (G.W.C. and G.R.H.). Acknowledgement for the Alzheimer’s Disease Sequencing Project: Biological samples and Associated Phenotypic Data used in primary data analysis were stored at the ADSP Principal Investigators’ institutions, and at the National Cell Repository for Alzheimer’s Disease (NCRAD) at Indiana University funded by NIA. Associated Phenotypic Data used in primary and secondary data analysis were provided by Principal Investigators, the NIA funded Alzheimer’s Disease Centers (ADCs), and the National Alzheimer’s Coordinating Center (NACC) and stored at the Principal Investigators’ institutions, NCRAD, and the National Institute on Aging Alzheimer’s Disease Data Storage Site (NIAGADS) at the University of Pennsylvania, funded by NIA. Genomic data were quality control checked at the Genome Center for Alzheimer’s Disease (GCAD) at the University of Pennsylvania. ADSP data were obtained from dbGaP, Study Accession phs000572.v7.p4.
Author contributions: G.W.C., G.R.H., X.W., and M.S. designed the study. X.W., V.M.P., G.A., A.M., P.J.M., G.W.C., and K.R.M.K. imported and analyzed data. S.R.C., C.A., and G.R.H. performed mouse experiments. C.C.W, D.A.B., and P.L.D.J. provided and analyzed ROS/MAP data. X.W., G.W.C., and G.R.H. primarily wrote the manuscript, with additional contributions from all other authors.
Footnotes
Communicating editor: N. Wray
Literature Cited
- Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell R. D., 2012. The imbalance of vascular molecules in Alzheimer’s disease. J. Alzheimers Dis. 32: 699–709. [DOI] [PubMed] [Google Scholar]
- Bennett D. A., Schneider J. A., Arvanitakis Z., Wilson R. S., 2012a Overview and findings from the religious orders study. Curr. Alzheimer Res. 9: 628–645. 10.2174/156720512801322573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D. A., Schneider J. A., Buchman A. S., Barnes L. L., Boyle P. A., et al. , 2012b Overview and findings from the rush memory and aging project. Curr. Alzheimer Res. 9: 646–663. 10.2174/156720512801322663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E., Stamatoyannopoulos J. A., Costello J. F., Ren B., Milosavljevic A., et al. , 2010. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 28: 1045–1048. 10.1038/nbt1010-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L., Tanzi R. E., 2008. Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 9: 768–778. 10.1038/nrn2494 [DOI] [PubMed] [Google Scholar]
- Carpenter B., Gelman A., Hoffman M. D., Lee D., Goodrich B., et al. , 2017. Stan: a probabilistic programming language. J. Stat. Softw. 76: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Wang C., Conomos M. P., Stilp A. M., Li Z., et al. , 2016. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am. J. Hum. Genet. 98: 653–666. 10.1016/j.ajhg.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R., Abney M., Palmer A. A., Skol A. D., 2011. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12: 66 10.1186/1471-2156-12-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P. L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L. C., et al. , 2014. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 17: 1156–1163. 10.1038/nn.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 4: 1623–1633. 10.1534/g3.114.013748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Rubin D. B., 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7: 457–472. 10.1214/ss/1177011136 [DOI] [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Dunson D. B., Vehtari A., et al. , 2013. Bayesian Data Analysis. Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
- Gilmour A. R., Thompson R., Cullis B. R., 1995. Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51: 1440–1450. 10.2307/2533274 [DOI] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., et al. , 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368: 117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D., Abraham R., Hollingworth P., Sims R., Gerrish A., et al. , 2009. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41: 1088–1093 [erratum: Nat. Genet. 41: 1156; corrigenda: Nat. Genet. 45: 712 (2013)] 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. R., 1953. Estimation of variance and covariance components. Biometrics 9: 226–252. 10.2307/3001853 [DOI] [Google Scholar]
- Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J. C., et al. , 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat. Genet. 43: 429–435. 10.1038/ng.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y., Sotero R. C., Toussaint P. J., Mateos-Pérez J. M., Evans A. C., et al. , 2016. Early role of vascular dysregulation on late-onset Alzheimer’s disease based on multifactorial data-driven analysis. Nat. Commun. 7: 11934 10.1038/ncomms11934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L., Holmans P. A., Hamshere M. L., Harold D., Moskvina V., et al. , 2010. Genetic evidence implicates the immune system and cholesterol metabolism in the aetiology of Alzheimer’s disease. PLoS One 5: e13950 [corrigenda: PLoS One 6 (2011)] 10.1371/journal.pone.0013950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., et al. , 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368: 107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G., Naj A. C., Beecham G. W., Wang L. S., Buros J., et al. , 2010. Meta-analysis confirms CR1, CLU, and PICALM as alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch. Neurol. 67: 1473–1484. 10.1001/archneurol.2010.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. 10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavvoura F. K., Ioannidis J. P., 2008. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum. Genet. 123: 1–14. 10.1007/s00439-007-0445-9 [DOI] [PubMed] [Google Scholar]
- Lambert J. C., Heath S., Even G., Campion D., Sleegers K., et al. , 2009. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41: 1094–1099. 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- Lambert J. C., Ibrahim-Verbaas C. A., Harold D., Naj A. C., Sims R., et al. , 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45: 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. S., Klein H. U., Yu L., Chibnik L. B., Ali S., et al. , 2017. Diurnal and seasonal molecular rhythms in human neocortex and their relation to Alzheimer’s disease. Nat. Commun. 8: 14931 10.1038/ncomms14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C. M., Davidson R. I., et al. , 2011. FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. 10.1038/nmeth.1681 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., 2010. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363: 166–176. 10.1056/NEJMra0905980 [DOI] [PubMed] [Google Scholar]
- Montagne A., Nation D. A., Pa J., Sweeney M. D., Toga A. W., et al. , 2016. Brain imaging of neurovascular dysfunction in Alzheimer’s disease. Acta Neuropathol. 131: 687–707. 10.1007/s00401-016-1570-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj A. C., Jun G., Beecham G. W., Wang L. S., Vardarajan B. N., et al. , 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat. Genet. 43: 436–441. 10.1038/ng.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe P. J., Verzilli C., Casas J. P., Hingorani A. D., Smeeth L., et al. , 2009. Multilocus Bayesian meta-analysis of gene-disease associations. Am. J. Hum. Genet. 84: 567–580. 10.1016/j.ajhg.2009.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocedal J., Wright S. J., 2006. Numerical Optimization. Springer-Verlag, New York. [Google Scholar]
- Owens G. K., Kumar M. S., Wamhoff B. R., 2004. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 84: 767–801. 10.1152/physrev.00041.2003 [DOI] [PubMed] [Google Scholar]
- Pawitan Y., 2001. In All Likelihood: Statistical Modeling and Inference Using Likelihood. Oxford University Press, Oxford. [Google Scholar]
- Soto I., Grabowska W. A., Onos K. D., Graham L. C., Jackson H. M., et al. , 2016. Meox2 haploinsufficiency increases neuronal cell loss in a mouse model of Alzheimer’s disease. Neurobiol. Aging 42: 50–60. 10.1016/j.neurobiolaging.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Balding D. J., 2009. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 10: 681–690. 10.1038/nrg2615 [DOI] [PubMed] [Google Scholar]
- Verzilli C., Shah T., Casas J. P., Chapman J., Sandhu M., et al. , 2008. Bayesian meta-analysis of genetic association studies with different sets of markers. Am. J. Hum. Genet. 82: 859–872. 10.1016/j.ajhg.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward L. D., Kellis M., 2012. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40: D930–D934. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D., MacArthur J., Morales J., Burdett T., Hall P., et al. , 2014. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42: D1001–D1006. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ersoz E., Lai C. Q., Todhunter R. J., Tiwari H. K., et al. , 2010. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42: 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Biswas S. K., McNulty P. H., Kozak M., Jun J. Y., et al. , 2011. PDGF-induced vascular smooth muscle cell proliferation is associated with dysregulation of insulin receptor substrates. Am. J. Physiol. Cell Physiol. 300: C1375–C1385. 10.1152/ajpcell.00670.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Nelson A. R., Betsholtz C., Zlokovic B. V., 2015. Establishment and dysfunction of the blood-brain barrier. Cell 163: 1064–1078. 10.1016/j.cell.2015.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Stephens M., 2014. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 11: 407–409. 10.1038/nmeth.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ADSP genotype and phenotype data are available via dbGaP under study accession phs000572.v7.p4. C57BL/6J mice are available for purchase from The Jackson Laboratory (strain #000664) at https://www.jax.org/strain/000664. The code used for analysis is available as Bayes-GLMM in a GitHub repository for public use at https://github.com/xulong82/bayes.glmm. Religious Orders Study (ROS) and Rush Memory and Aging Project (MAP) data are available as cited (Lim et al. 2017). Additional information on associated variants can be found in Table S1 and Table S2.