Abstract
Genetic variation in mitochondrial DNA (mtDNA) provides adaptive potential although the underlying genetic architecture of fitness components within mtDNAs is not known. To dissect functional variation within mtDNAs, we first identified naturally occurring mtDNAs that conferred high or low fitness in Saccharomyces cerevisiae by comparing growth in strains containing identical nuclear genotypes but different mtDNAs. During respiratory growth under temperature and oxidative stress conditions, mitotype effects were largely independent of nuclear genotypes even in the presence of mito–nuclear interactions. Recombinant mtDNAs were generated to determine fitness components within high- and low-fitness mtDNAs. Based on phenotypic distributions of isogenic strains containing recombinant mtDNAs, we found that multiple loci contributed to mitotype fitness differences. These mitochondrial loci interacted in epistatic, nonadditive ways in certain environmental conditions. Mito–mito epistasis (i.e., nonadditive interactions between mitochondrial loci) influenced fitness in progeny from four different crosses, suggesting that mito–mito epistasis is a widespread phenomenon in yeast and other systems with recombining mtDNAs. Furthermore, we found that interruption of coadapted mito–mito interactions produced recombinant mtDNAs with lower fitness. Our results demonstrate that mito–mito epistasis results in functional variation through mitochondrial recombination in fungi, providing modes for adaptive evolution and the generation of mito–mito incompatibilities.
Keywords: mtDNA, mitochondrial recombination, mito–mito epistasis, genetic interactions, Saccharomyces yeast
MITOCHONDRIAL DNA (mtDNA) haplotypes are frequently associated with environmental temperature gradients across eukarya, suggesting that mitochondrial performance plays an important role in adaptation (Mishmar et al. 2003; Lucassen et al. 2006; Chatelain et al. 2011; Scott et al. 2011; Lagisz et al. 2013; Dingley et al. 2014; Melo-Ferreira et al. 2014; Silva et al. 2014; Consuegra et al. 2015; Li et al. 2016). To understand the adaptive potential of mtDNAs, it is necessary to dissect the genetic and environmental factors that influence the functional variation in mtDNAs. This is particularly challenging in systems where mtDNA inheritance is uniparental because the lack of recombination makes it difficult to differentiate between mitochondrial alleles that contribute to functional variation and those that are neutral.
Homologous recombination between mtDNAs should promote the reorganization of mitochondrial genes and increase the efficacy of selection on adaptive loci. Mitochondrial recombination is common in systems with biparental mtDNA inheritance, such as fungi and many plants (Barr et al. 2005; Gualberto and Newton 2017). In Saccharomyces yeasts, the diverse and highly reticulated mtDNAs show signatures of recombination and horizontal gene transfer within and between species (Peris et al. 2014, 2017; Wolters et al. 2015; Wu et al. 2015; Leducq et al. 2017). In predominantly asexual fungi, mitochondrial recombination occurs more frequently than expected (Brankovics et al. 2017). The machinery for homologous recombination is found in the mitochondria of mammals (Dahal et al. 2017) and there is some evidence of mitochondrial recombination in mammals (Piganeau et al. 2004), other vertebrates (Ciborowski et al. 2007; Ujvari et al. 2007; Sammler et al. 2011; Wang et al. 2015; Park et al. 2016), and invertebrates (Ladoukakis and Zouros 2001; Passamonti et al. 2003). Mitochondrial recombination occurs with enough frequency that it should play an important role in the evolution of mtDNAs, especially in fungi.
The effects of mitochondrial recombination on selection and adaptive potential are not understood. In Saccharomyces yeasts, mitochondrial recombination can occur in zygotes containing different mtDNAs but, because heteroplasmic mtDNA states are not maintained, a single mtDNA haplotype (either parental or recombinant) becomes fixed after ∼20 generations (Berger and Yaffe 2000). Hybrids of Saccharomyces cerevisiae and S. uvarum contained different species-specific mtDNA genetic markers, depending on whether the hybrids were created in a laboratory (Verspohl et al. 2018) or isolated from industrial settings (Masneuf et al. 1998; Rainieri et al. 2008), suggesting that environmental conditions influence the selection for mitochondrial alleles or entire mitotypes. Supporting this, mitochondrial allele inheritance during hybridization of S. cerevisiae and S. paradoxus was altered by changing laboratory conditions during matings (Hsu and Chou 2017).
Mitochondrial alleles that participate in mito–nuclear interactions will also influence the adaptive success of recombinant mtDNAs. Mito–nuclear incompatibilities occur between (Sulo et al. 2003; Chou et al. 2010; Spirek et al. 2014) and within (Paliwal et al. 2014; Hou et al. 2015) Saccharomyces species and species-specific compatible mito–nuclear genetic combinations were universally maintained in rare, viable meiotic progeny from S. cerevisiae/S. bayanus hybrids (Lee et al. 2008). The extent of recombination in these hybrid studies is not known due to the limited number of mitochondrial markers followed. Laboratory-derived isogenic S. paradoxus hybrids containing different recombinant mtDNAs were phenotypically variable, consistent with the presence of functionally distinct mitochondrial or mito–nuclear alleles (Leducq et al. 2017). Selection on such functional units could explain the existence of recombinant mtDNAs found in natural S. paradoxus hybrids (Leducq et al. 2017; Peris et al. 2017).
Mitochondrial genes encode for physically interacting subunits of respiratory complexes and so there is potential that mitochondrial genes coevolve through compensatory mutations or selection for adaptive mito–mito allele combinations in recombinant mtDNAs. In support of this, comparative analysis of mtDNA and in silico analysis of protein structures revealed coevolving amino acids within mitochondrial genes within primates (Azevedo et al. 2009). In S. cerevisiae, mitochondrial suppressor mutations that counteracted respiratory-deficient mitochondrial mutations were characterized (Fox and Staempfli 1982; di Rago et al. 1995). These examples of functional interactions suggest that epistasis between mitochondrial alleles (mito–mito epistasis) could contribute to functional variation in mtDNAs.
In this work, we identified mitotypes that conferred strong adaptive potential by examining fitness in S. cerevisiae strains containing different combinations of mtDNA and nuclear genomes. Certain mitotypes provided growth advantages during respiratory growth under temperature and oxidative stress conditions, irrespective of nuclear backgrounds, indicating direct effects on fitness. To determine whether these growth advantages were due to one or more interacting mitochondrial loci, we generated and phenotyped strains containing recombinant mtDNAs. The effects of these mitochondrial alleles were nonadditive and revealed that mito–mito epistasis contributes to phenotypic variation. Furthermore, interruption of coadapted mito–mito interactions produced recombinant mtDNAs with lower fitness in media mimicking natural yeast environments, indicating that mito–mito epistasis may play an important role in hybridization and mtDNA evolution.
Materials and Methods
Strains and media
Strain names and genotypes are provided (Supplemental Material, Table S1 and Table S2). The creation of strains containing synthetic combinations of nuclear and mitochondrial genotypes was previously described (Paliwal et al. 2014). Basically, mtDNAs from mtDNA donor haploid strains were serially passaged through a strain containing a KAR1-1 mutation to mtDNA recipient haploid strains. The KAR1-1 mutation inhibits nuclear fusion and allows the formation of heterokaryotic zygotes containing mixed cytoplasmic components including mitochondria (Rose and Fink 1987). Haploid progeny containing mtDNA from the donor strain and the nuclear genotype of the recipient strain were identified by scoring auxotrophic markers.
Five different media were used (Table S3). These included rich, undefined media containing fermentable (YPD) or nonfermentable (YPEG) carbon sources, minimal defined media containing fermentable (CSM) or nonfermentable (CSMEG) carbon sources, and a minimal media emulating oak tree exudate (SOE). Nonfermentable media require mitochondrial respiration for growth. The oxidative stress agent menadione was added to 20 µM when indicated (YPEGM). Agar (2%) was added to solid medium. Undefined media types contained yeast extract for which the exact nutrient composition is not known, while defined media contained precisely specified nutrient compositions.
Colony arrays
Cells from the haploid mito–nuclear (nine nuclear × nine mito) and recombinant mtDNA (YJM975 × Y12) diploid strain collections were printed onto OmniTrays (Nunc) in randomized 1536 colony arrays using a BM3-BC colony processing robot (S&P Robotics). Arrays were first printed to YPD and then to test media. Strains were acclimated to test media (YPD, CSM, SOE, YPEG with and without menadione, and CSMEG, Table S3) for 3 days at 30, 35, and 37°, and then reprinted to the same media. Each strain in the mito–nuclear collection was printed in 15 replicates. Strains in the recombinant mtDNA collection, including two biological replicates for each parental mtDNA, were printed in 25 replicates. Photographs were taken at times 0 and 48 hr. Colony sizes were extracted from images using ImageJ 1.440 software (Diss et al. 2013). Size differences were determined using custom R scripts available at https://github.com/JFWolters/Wolters-Genetics-2018/.
A block design was used for additional phenotyping of diploid strains containing mitochondrial recombinants from all crosses. The phenotyping strategy was completed as before with the following modifications. Phenotyping was conducted on YPD, YPEG, and SOE media at 30 and 37°. The block design was similar to that used in Strope et al. (2015), modified such that each strain existed on the plate as a four-row by eight-column colony block. Colonies on the edges of blocks were excluded from analysis to remove neighbor effects. Images were taken over 7 days. Image analysis was completed using Gitter (Wagih and Parts 2014). A custom R script pipeline using logistic regressions to fit growth curves and estimate growth rate is available at https://github.com/JFWolters/Robot-Image-Analysis-master.
Competition assay
Haploid strains (MATa) containing the YJM975 nuclear genotype and Y12 or YJM975 mitotypes (strains SP15a1ρ26 and NCYC3954, respectively, Table S1) were grown overnight in liquid YPD media at 30°, mixed in equal volumes, spread as lawns onto solid YPEG media, and grown for 2 days at 30 and 37°. The pooled cells were then retrieved from the solid plates in sterile water, diluted, plated for single colonies on YPEG media, and incubated at 37° for 4 days. Cells harboring the Y12 mitotype produced large colonies as compared to cells with the YJM975 mitotype. Numbers of large and small colonies were used to determine the proportion of each genotype. Significance was assessed using a test of equal proportions and P-values were assessed vs. a stringent α after Bonferroni correction (three tests, α = 0.017).
Mitochondrial recombination
To produce mitochondrial recombinants, matings between haploid strains containing congenic nuclear backgrounds (YJM975) and different mitotypes (YJM975 × Y12, YJM975 × YPS606, YJM975 × Y55, and YPS606 × Y55) were conducted on solid YPD media at 30° for 2 days. Diploids were selected by printing mated-cell mixtures onto SD media and grown at 30° for 1 day. To fix mitotypes, diploid strains were streaked for single colonies twice on SD media with 2 days of growth at 30°. Each strain containing putative recombinant mtDNAs resulted from independent matings. Two independent biological replicates of diploid parental control strains were created by mating congenic strains containing only one mitotype.
To look for evidence of mitochondrial recombination, restriction fragment length polymorphism (RFLP) assays were performed. Total DNA was isolated from 5 ml overnight cultures grown in YPD using a glass-bead cell disruption and phenol–chloroform extraction, as previously described (Hoffman and Winston 1987). Three mtDNA loci were amplified using the primers and amplification conditions listed in Table S4. Amplified products were digested using HinfI (ATP6), Ase1 (COX2), or HindIII (COX3), and separated on 2% agarose gels. Additionally, total mtDNA was isolated from 21 diploid strains according to Defontaine et al. (1991), including a DNase treatment (Wolters et al. 2015), and modified for smaller volumes. The total mtDNA was digested with EcoRV and separated by electrophoresis on 0.8% agarose gels. Mitochondrial recombination was inferred through nonparental haplotype and total mtDNA RFLP patterns.
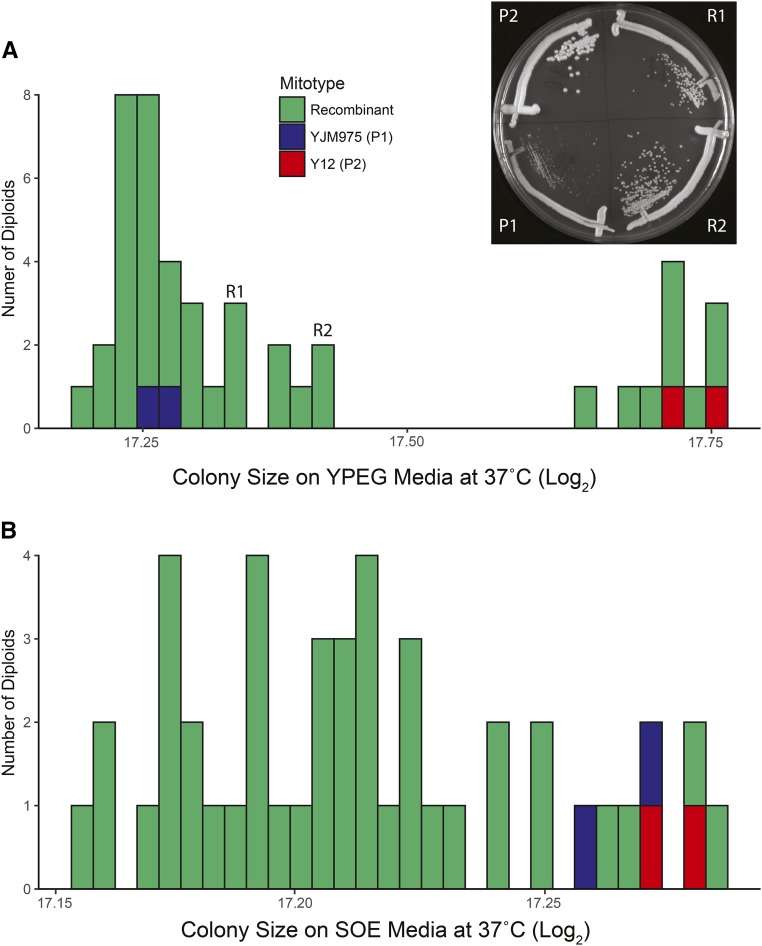
To compare colonies formed from single cells (Figure 5 inset), cells were streaked directly onto YPEG media and photographed following incubation at 37° for 4 days.
Figure 5.
Mitochondrial recombination generates novel phenotypes. Diploid strains containing isogenic homozygous nuclear genotypes and parental (Y12 (red) or YJM975 (blue) or recombinant (green) mitotypes were spotted via robotic transfer onto solid media, and colony sizes were quantified after 48 hrs. Two biological replicates for each parental control diploid were included. Colony sizes distributions are plotted as histograms. (A) YPEG, 37°. The intermediate phenotypes of representative recombinant strains (R1 and R2) are shown in the insert. (B) SOE, 37°. See Figure S1 for phenotypes for additional media and temperatures. SOE, ; YPEG, .
Statistics
All statistical analyses were performed in R (version 3.4.0). The differences in colony sizes from the arrays between 0 and 48 hr were used as a proxy for growth differences. Random effects ANOVAs (containing terms for nuclear genotype, mitotype, and nuclear × mitotype interaction) were performed using the lmer function of the lme4 package (Bates et al. 2014). Phenotypic variances were determined from each model and normalized by dividing the mean for a given condition. Direct effects of mitotypes were evaluated in each nuclear genotype by dividing the average growth of each strain by the average growth of all strains containing the identical nuclear genotype, and converting to percentages. To determine whether strains with potentially recombinant mtDNAs had phenotypes that differed from parental controls, individual ANOVAs were performed on each strain (fixed factor comparing parent 1, parent 2, and strain), followed by a post hoc analysis using the Tukey honest significant difference function to assess whether the recombinant strain was significantly different from both parental controls. A Bonferroni correction was applied to compare results under standard (α = 0.05) and stringent (α = 0.0012) significance thresholds (Table S5).
Estimating mito–mito epistasis
Epistasis between mitochondrial variants (mito–mito epistasis) was explicitly tested for these crosses using a variance partitioning approach developed for haploid organisms (Shaw et al. 1997). Recombinant mitotypes were nested within each parental cross and represented by multiple technical replicates. The variance attributed to parental crosses (PC) and putative recombinants were estimated from a nested random effects model using the lmer function in the lme4 package (Bates et al. 2014). Additive-by-additive epistasis (i.e., mito–mito epistasis) was estimated as: Parental cross, putative mitochondrial recombinant, and technical replicates correspond to the terms “diploids,” “haploids,” and “clones” as described by Shaw et al. (1997).
The significance of VAA was evaluated with several criteria. First, the significance of the putative recombinant term (nested within parental cross) was tested in the random effects model via a hierarchical comparison of models with and without that term. Second, the significance of the difference between the putative recombinant variance and the parental cross variance was tested by comparing the observed value to 1000 permutations of the data where individual data points were permuted (Anderson and Ter Braak 2003). Third, we determined whether 95% C.I.s of VAA were positive and did not overlap zero. C.I.s were calculated using a percentile bootstrap approach by resampling from within the technical replicates such that the overall structure of the putative recombinants and diploid crosses was maintained (Quinn and Keough 2002). VAA was considered significant if, and only if, all three criteria were met.
Data availability
All growth measures used for analyses are provided in Table S6, Table S7, and Table S8. Strains are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5970955.
Results
Respiratory growth is influenced by mtDNA haplotypes under stress conditions
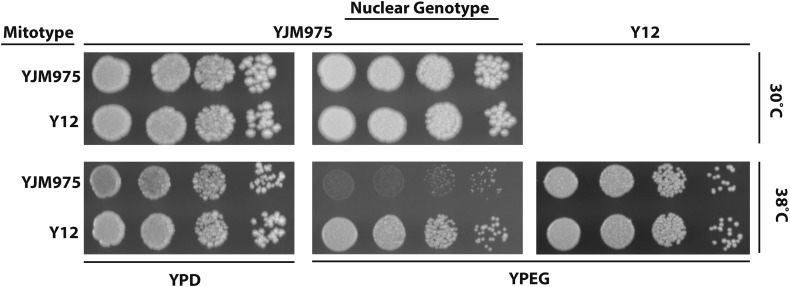
In S. cerevisiae, mito–nuclear interactions can explain the majority of mitotype-related growth differences (Paliwal et al. 2014). We previously observed that in yeast cells growing on fermentable sugars at high temperatures, mitotypes had direct effects on growth (Paliwal et al. 2014). Here, we hypothesized that temperature stress would reveal mitotype-specific effects on nonfermentable media where oxidative phosphorylation is essential for energy production. To test this, we plated S. cerevisiae cells containing identical nuclear genotypes and two divergent mitotypes onto solid media containing fermentable and nonfermentable sugars, and followed colony sizes as a proxy for mitochondrial genotype fitness (Figure 1). Cells containing the nuclear genotype from a strain of European origin (YJM975) and its own mitotype or that from a divergent Sake strain (Y12) showed no differences in fitness when grown on fermentable or nonfermentable sugars at standard temperature (30°). However, at high temperature (37°), growth differences due to mitotypes were observed. On nonfermentable media, where growth requires mitochondrial respiration, cells containing the Y12 mitotype formed larger colonies than isonuclear cells containing the YJM975 mitotype, while colony size differences on fermentable media were barely observable. To determine if these respiratory growth differences were due to direct effects of mitotypes or to mito–nuclear interactions, the mitotypes were introduced into a second nuclear background. In the Y12 nuclear background, no differences in fitness between mitotypes were observed although cells containing the Y12 nuclear genotype grew better on nonfermentable media than those with the YJM975 genotype (Figure 1). The variable effect of these mitotypes is consistent with mito–nuclear epistasis; however, nuclear alleles in the Y12 genotype with large effects on growth may have obscured independent effects of mitotypes.
Figure 1.
Mitochondrial genotype affected respiratory growth at elevated temperature. S. cerevisiae strains containing different combinations of nuclear and mitochondrial genomes were grown overnight in liquid fermentable media, serially diluted, and grown for 2 days on fermentable (YPD) or nonfermentable media (YPEG) media at 30 and 38°. Nonfermentable media requires mitochondrial respiration for growth. Nuclear and mitotype origins are indicated.
We expanded our assay to identify mitochondrial genotype-specific fitness differences between 81 strains containing unique combinations of nine divergent nuclear genotypes and mtDNAs (nine nuclear × nine mtDNA, Table S1). This large and systematic array of mito–nuclear combinations provides a powerful platform to determine whether phenotypic differences are due to genetic differences in nuclear genomes, mitotypes, or mito–nuclear interactions. Cells were grown on solid media under atmospheric oxygen levels using high-density colony arrays in 18 environmental conditions, including fermentable and nonfermentable sugars at three temperatures, and fitness differences were analyzed by ANOVAs (Table 1). Strains bearing the SK1 nuclear genotype failed to transfer or form uniform colonies due to severe flocculation and were omitted from further analysis. In all media and temperature conditions, mito–nuclear interactions and nuclear genotypes were highly significant contributors to fitness differences. Mitotypes showed significant independent fitness effects in each nonfermentable medium at elevated temperature. In the presence of strong interaction terms, interpretation of main effects is difficult. Still, the main effect for mitotype showed increasing χ2 statistics and decreasing P-values with increasing temperatures, suggesting that mitochondrial respiration was directly impacted by variation in mitotypes during temperature stress. Temperature-related mitotype effects were even stronger in media containing the exogenous oxidative stress agent menadione (YPEGM).
Table 1. Nuclear, mitotype, and mito–nuclear effects on growth.
| Condition | Nuclear | Mitotype | Mito–nuclear | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Media | ° | d.f. | χ2 | P | d.f. | χ2 | P | d.f. | χ2 | P |
| CSMEG | 30 | 5 | 42.87 | *** | 5 | 0.27 | ns | 5 | 81.69 | *** |
| 35 | 5 | 100.62 | *** | 5 | 7.58 | * (0.0059) | 5 | 261.39 | *** | |
| 37 | 5 | 69.39 | *** | 5 | 4.46 | * (0.0346) | 5 | 666.08 | *** | |
| YPEG | 30 | 5 | 79.04 | *** | 5 | 0 | ns | 5 | 638.80 | *** |
| 35 | 5 | 23.76 | *** | 5 | 0.12 | ns | 5 | 836.52 | *** | |
| 37 | 5 | 78.33 | *** | 5 | 10.06 | ** (0.0015) | 5 | 318.40 | *** | |
| YPEGM | 30 | 5 | 89.38 | *** | 5 | 0.08 | ns | 5 | 706.30 | *** |
| 35 | 5 | 32.09 | *** | 5 | 7.76 | * (0.0053) | 5 | 484.19 | *** | |
| 37 | 5 | 42.47 | *** | 5 | 24.21 | *** (8.65 × 10−7) | 5 | 219.67 | *** | |
| CSM | 30 | 5 | 127.82 | *** | 5 | 0 | ns | 5 | 363.13 | *** |
| 35 | 5 | 222.92 | *** | 5 | 0 | ns | 5 | 259.84 | *** | |
| 37 | 5 | 204.09 | *** | 5 | 0 | ns | 5 | 506.38 | *** | |
| SOE | 30 | 5 | 62.14 | *** | 5 | 0.30 | ns | 5 | 298.82 | *** |
| 35 | 5 | 45.21 | *** | 5 | 0 | ns | 5 | 659.51 | *** | |
| 37 | 5 | 80.46 | *** | 5 | 0.01 | ns | 5 | 799.73 | *** | |
| YPD | 30 | 5 | 87.81 | *** | 5 | 0 | ns | 5 | 398.46 | *** |
| 35 | 5 | 110.05 | *** | 5 | 2.37 | ns | 5 | 124.93 | *** | |
| 37 | 5 | 130.99 | *** | 5 | 0 | ns | 5 | 203.22 | *** | |
To determine the significance of each term, ANOVAs comparing the full model [nuclear + mitotype + (nuclear × mitotype)] with a model lacking the indicated term were evaluated. Factors were treated as random effects. Significance codes: ns P > 0.05, * P < 0.05, ** P < 0.005, and *** P < 5 × 10−6. CSM, ; CSMEG, ; ns, not significant; SOE, ; YPEG, ; YPEGM.
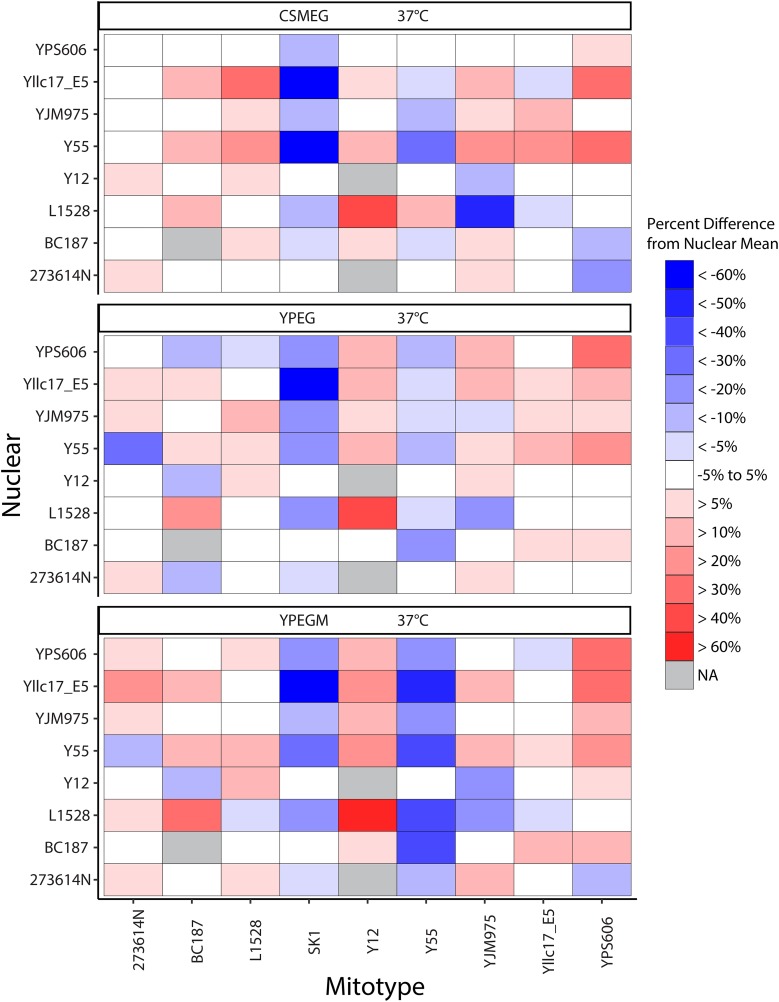
To more closely follow independent effects of mitotypes, we compared the respiratory growth of each strain with the average growth for all strains with the same nuclear genotype (Figure 2). The Y12 mitotype provided a respiratory growth advantage under temperature stress in the YJM975 nuclear background, and also in six of the seven nuclear backgrounds tested. Four additional mitotypes showed relative growth increases at high temperatures in each of the respiratory and oxidative stress conditions tested, while two mitotypes decreased growth (Table 2). Two mitotypes (including YJM975) showed both increased or decreased growth, depending on the nuclear backgrounds, although these fitness differences are relatively small compared to growth averages.
Figure 2.
Fitness effects of mitotypes in nonfermentable media. Heat maps show the relative increase (red) or decrease (blue) in fitness by specific mitotypes. Relative fitness is shown for each nuclear background in media where direct effects of mitotypes were statistically significant. Mitotypes had observable direct effects in nonfermentable media (CSMEG, YPEG, and YPEGM) at 37°. Percent changes in growth were determined by comparing the growth rate of each mito–nuclear genome combination with the average growth of all strains with the same nuclear genotype. Average percent increases and decreases are provided in Table 2. CSM, ; CSMEG, ; NA, not applicable; YPEG, ; YPEGM.
Table 2. Mitotype effects on growth.
| Mitotype | YPEG 37° (%) | CSMEG 37° (%) | YPEGM 37° (%) | Average (%) |
|---|---|---|---|---|
| Y12 | 15.5 | 11.1 | 23.8 | 16.8 |
| BC187 | 1.0 | 8.6 | 8.1 | 5.9 |
| YPS606 | 10.7 | 4.2 | 14.2 | 9.7 |
| L1528 | 2.5 | 11.2 | 5.4 | 6.4 |
| Yllc17_E5 | 4.3 | 3.5 | 1.2 | 3.0 |
| SK1 | −21.9 | −29.1 | −20.0 | −23.7 |
| Y55 | −8.3 | −6.2 | −31.7 | −15.4 |
| YJM975 | 0.8 | −2.4 | 0.3 | −0.4 |
| 273614N | −0.6 | 3.0 | 5.7 | 2.7 |
The percent increase or decrease in fitness by each mitotype (as shown in Figure 2) were averaged across all nuclear genotypes. Average fitness differences across three stress conditions are provided in the final column. CSMEG, ; YPEG, ; YPEGM.
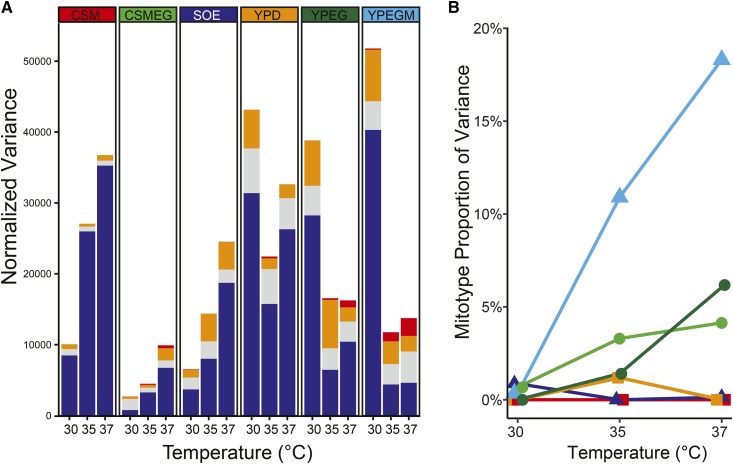
Direct effects from mitotypes can also be observed in colony size variances. Overall phenotypic variances in colony sizes increased with increasing temperatures in defined fermentable and nonfermentable media (Figure 3A). This is consistent with previously observed responses to temperature in yeasts grown in liquid culture (Paliwal et al. 2014) and on complex traits in Drosophila (Bubliy et al. 2001). In contrast, phenotypic variances were generally higher in rich media whose exact nutrient components are not defined (YPD, YPEG, and YPEGM) and decreased as cells were grown at higher temperatures. The contributions of the nuclear genome, mitotype, and mito–nuclear interactions to total phenotypic variances were estimated using variance component analyses of the full ANOVA models (Figure 3A and Table S9). Mitotypes did not explain any proportion of these variances at lower temperatures, but as temperatures increased, the proportions of variance due to mitotypes increased during respiratory growth, particularly in the presence of an oxidative stress agent (YPEGM, Figure 3B). The variance component due to mitotypes exceeded that of mito–nuclear interactions in this oxidative stress environment (18.3% vs. 16.0%, respectively, Table S9).
Figure 3.
Temperature increases phenotypic variances due to mitotypes under stress conditions. (A) Total normalized phenotypic variances and individual variance component analyses including nuclear (blue), mitotype (red), and mito–nuclear epistasis (orange), and residual (gray), are presented as stacked bar graphs. (B) The proportion of total phenotypic variance due to independent effects of mitotypes is plotted across increasing temperature for each media. Line colors match media as shown in (A). CSM, ; CSMEG, ; SOE, ; YPEG, ; YPEGM.
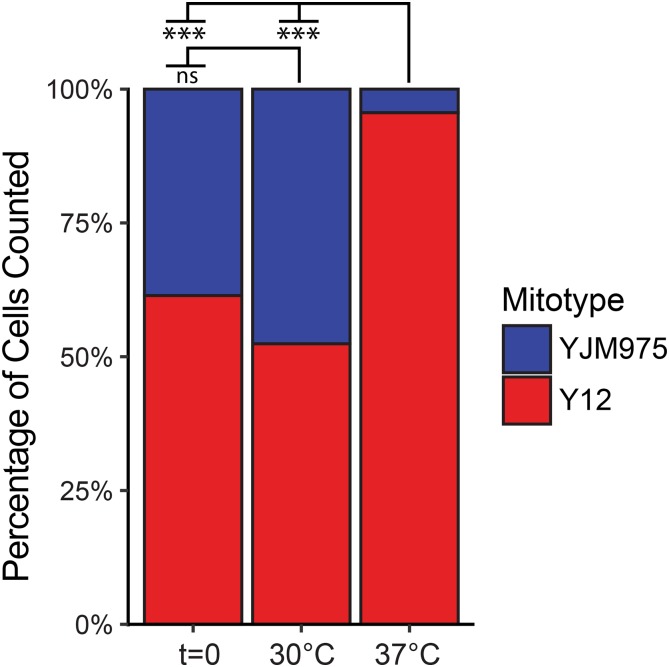
A competition experiment was performed to determine if the growth differences of strains grown in isolation on solid media corresponded to fitness advantages when grown in mixed cultures. Nonmating cells with isogenic nuclear genotypes, and either a neutral (YJM975) or high-fitness (Y12) mitotype, were pooled in equal amounts and grown in direct competition on nonfermentable media (Figure 4). The proportions of mitotypes were approximately equal before (61.4%) and after competition (52.4%) at 30° (nonsignificantly different with P = 0.03 vs. α of 0.017 after Bonferroni correction), but after competitive growth at the elevated temperature, the proportion of cells containing the high-fitness mtDNA increased substantially (95.6%, P < 2 × 10−16). This result demonstrates that larger colony sizes due to high-fitness mtDNAs reflect competitive advantages.
Figure 4.
High-fitness mitotypes provide competitive advantage. Overnight cultures of yeast strains containing the YJM975 nuclear genotype and YJM975 (blue) or Y12 (red) mitotypes were mixed and competitively grown on solid respiratory media (YPEG_R). The proportion of cells containing each mitotype was determined before (t = 0) and after competition on YPEG media for 2 days by counting numbers of large (Y12) and small (YJM975) colonies that formed on YPEG media at 37°. Proportions shown are based on total numbers of colonies counted before (n = 223) and after competition at 30° (n = 391) and 37° (n = 205). Significance codes following Bonferroni correction: ns (not significant) P > 0.05, * P < 0.05, ** P < 0.005, and *** P < 5 × 10−6.
Taken together, these results show that genetic variation in mitochondrial haplotypes directly impacts mitochondrial fitness. Even in the presence of strong mito–nuclear interactions, certain mitotypes affect yeast growth in consistent patterns and can be considered as high- and low-fitness mtDNAs. These effects are largely contingent on stress conditions that tax mitochondrial functions.
Mitochondrial recombination reveals functional interactions between mitochondrial loci
In nature, biparental inheritance of yeast mtDNAs during matings could introduce different mtDNAs into the same cell. Mitochondrial recombination would result in allele reorganization, revealing novel phenotypes due to direct effects and epistasis that may promote adaptive evolution.
Single or multiple alleles within the mitochondrial genome could be contributing to fitness differences. To test this, we created isonuclear diploid strains containing recombinant mtDNAs by mating haploid strains with congenic nuclear genotypes (differing by mating type and a selectable marker) and different mitotypes (Table S2). A total of 41 independent matings were performed, followed by clonal propagation of unique diploid strains to fix mitotypes. Restriction analysis of whole mtDNAs and three amplified mitochondrial markers showed evidence of recombination in 41% of tested diploids (Table S10), though the low resolution of these assays likely underestimates the number of diploids containing recombinant mtDNAs. If the high-fitness mtDNA was due to a single locus, then recombinant mtDNAs would have either the high- or low-fitness allele and would be expected to generate phenotypes identical to a parental mtDNA. We found that on rich nonfermentable media at high temperature (YPEG 37°), colony sizes from strains with recombinant mtDNAs were sometimes in between either control strain containing parental mitotypes (Figure 5A). Fitness differences between strains with recombinant vs. both parental mtDNAs were observed in 10 of the 18 environmental conditions examined (Table S5). These results indicate that multiple mitochondrial loci underlie mitotype-specific phenotypic differences.
Multiple loci could contribute to phenotypes through additive or epistatic interactions between mitochondrial alleles. For polygenic traits with additive alleles, recombination would produce mtDNAs that generate phenotypes both above and below those of the parental mtDNAs. If one parental mtDNA contained all positive alleles and the other none, then progeny containing recombinant mtDNA would only generate intermediate phenotypes. For epistatic loci, recombination would generate novel interacting allele combinations that could present phenotypic distributions that resemble additive alleles. However, if high-fitness epistatic allele combinations were interrupted, recombination could produce low-fitness mtDNAs that result in lower growth than parental mtDNAs.
The intermediate phenotypes observed in strains with recombinant mtDNAs on respiratory media are consistent with a model with multiple loci with additive effects (Figure 5A). However, when strains were grown at high temperature on a medium designed to emulate exudate from oak trees (SOE), a natural habitat for Saccharomyces, the phenotypic distribution was consistent with negative epistasis (Figure 5B). In this condition, growth rates of strains with parental mtDNAs were similar to each other, while 28 (of 41) strains containing recombinant mtDNAs grew worse than either parent (P < 0.05 for independent ANOVAs comparing growth of each recombinant strain to each parent; Table S5). It is likely that the interruption of one or more coadapted mito–mito allele combinations led to the observed negative epistasis in this ecologically relevant media.
To determine if mito–mito epistasis was a general feature among mtDNAs, we created additional recombinant mtDNAs between four different mitotypes (Table S2). Growth rates were collected for isonuclear strains containing fixed recombinant mtDNAs (mitotypes YJM975 × Y12, YJM975 × YPS606, YJM975 × Y55, and YPS606 × Y55). To explicitly test for mito–mito epistasis, we used a variance partitioning approach developed to test for epistasis in haploid genetic systems (Shaw et al. 1997). Specifically, additive by additive (i.e., mito–mito) epistasis (VAA) was estimated as twice the difference between the variances attributed to the PC and the putative recombinants (R), i.e., VAA = 2 * (σ2R−σ2PC) (see Materials and Methods). There was evidence for mito–mito epistasis (P < 0.001) in each environmental condition (Table 3). These analyses were repeated for balanced subsets of the data, such that each mitotype was only represented exactly once or twice (using data from mitotype crosses YJM975 × Y12 and YPS606 × Y55, or YJM975 × YPS606, YJM975 × Y55, and YPS606 × Y55, respectively). In both balanced designs, VAA estimates were significant and positive, with one exception (double representation, YPD at 37°). The negative VAA estimate in this exception suggests a violation of test assumptions and could be generated if the interacting loci were tightly linked. If mitochondrial recombination was not sufficient to interrupt linkage in these crosses, our power to observe mito–mito epistasis would be reduced. Still, we detected positive mito–mito epistasis in most cases.
Table 3. Mito–mito epistasis (VAA) estimates.
| Media: | SOE | YPD | YPEG | ||||
|---|---|---|---|---|---|---|---|
| Temperature (°C) | 30 | 37 | 30 | 37 | 30 | 37 | |
| All crosses | VAA | 0.0051 | 0.0360 | 0.0011 | 0.0028 | 0.0019 | 0.0042 |
| C.I. lower | 0.0046 | 0.0354 | 0.0009 | 0.0026 | 0.0017 | 0.0041 | |
| C.I. upper | 0.0078 | 0.0400 | 0.0014 | 0.0032 | 0.0022 | 0.0044 | |
| Single representation | VAA | 0.0112 | 0.0351 | 0.0010 | 0.0030 | 0.0010 | 0.0037 |
| C.I. lower | 0.0109 | 0.0345 | 0.0008 | 0.0027 | 0.0008 | 0.0036 | |
| C.I. upper | 0.0141 | 0.0394 | 0.0014 | 0.0033 | 0.00144 | 0.0039 | |
| Double representation | VAA | 0.0060 | 0.0413 | 0.0034 | −0.0030 | 0.0030 | 0.0037 |
| C.I. lower | 0.0045 | 0.0395 | 0.0033 | −0.0033 | 0.0029 | 0.0035 | |
| C.I. upper | 0.0091 | 0.0473 | 0.0037 | −0.0027 | 0.0033 | 0.0038 | |
Positive (nonzero) VAA estimates indicate mito–mito epistasis. The overall magnitudes of the VAA values do not provide information regarding their relative contribution to genetic variance. All estimates were significantly different from zero based on permutation analysis (see Materials and Methods). Positive estimates were confirmed based on upper and lower confidence intervals (C.I.s) that did not overlap zero estimated from bootstrapping analysis. Subsets of the data were analyzed for single representation (YJM975 × Y12 and YPS606 × Y55 crosses) or double representation (each mitotype represented twice, YJM975 × Y12 excluded). SOE, ; YPEG, ; YPEGM, .
In sum, mito–mito epistasis appears to be a common feature in yeast mtDNAs. Mito–mito epistasis estimates were highest for the ecologically relevant media (SOE) at high temperatures and suggest that selection for interacting mitochondrial alleles has influenced mtDNA evolution.
Discussion
The evolutionary significance of mitochondrial recombination is not well understood. In this work, we show that recombination can reveal interactions between mitochondrial loci that affect fitness. Our analyses were facilitated by using strains with isogenic nuclear genotypes and parental or recombinant mtDNAs in fixed environments, such that phenotypic variances could be directly attributed to mtDNAs. Because mtDNAs are homoplasmic in yeast, dominance effects (VD) could be eliminated, and the remaining phenotypic distributions must be due to additive (VA) and/or epistatic (VI) components within mtDNAs. In most cases, recombinant mtDNAs created distributions that were consistent with additive effects. However, negative interactions between mitochondrial alleles best explain the lowered fitness values of nearly all strains with recombinant mtDNAs shown in Figure 5B. Epistasis can be difficult to detect because, for most allele frequencies, the additive (main) effects of interacting loci will be greater than the total epistatic variance unless the epistatic loci have strong deleterious interactions (Mackay 2014). Still, statistically significant nonzero values of mito–mito epistasis (VAA) were estimated when assessing recombinant mtDNAs from four distinct parental mitotypes. Mito–mito epistasis is most likely a general feature underling fitness differences in yeast mtDNAs.
Given the large number of physical interactions between the products of mitochondrial genes, it is possible that multiple mito–mito interactions contribute to mtDNA fitness. In the simplest case where each parental mtDNA contains unique alleles for two interacting loci (e.g., X1Y1 and X2Y2), offspring with recombinant mtDNA should, at most, form two distinct phenotypic classes (representing X2Y1 and X1Y2). While our sample sizes are too low to robustly describe distribution shapes, the phenotypic distribution in Figure 5B appears broad, consistent with the interruption of multiple different interacting genes. Large deletions within mtDNAs arise in clonally replicating cells [resulting in “petites” (Euphrassi 1949)] and so we omitted nonrespiring diploid strains resulting from the crosses generating recombinant mtDNAs. Our observed numbers of petites varied between crosses (Table S11) and could reflect additional deleterious mito–mito interactions between certain mtDNAs that were not analyzed in this study.
We found that recombination between parental mtDNAs generated lower-fitness mtDNAs using laboratory crossing schemes, showing that mito–mito incompatibilities exist within species. In nature, mito–mito incompatibilities generated via mitochondrial recombination may lead to reduced fitness in hybrids. In S. paradoxus, recombination between mtDNAs from diverging populations generated hybrid diploids with lower fitness than hybrids with parental mtDNAs (Leducq et al. 2017). Given the heterozygous nuclear genotypes in these hybrids, dominant interactions between mitochondrial and nuclear alleles could not be ruled out entirely. However, it is likely that negative interactions between mitochondrial loci could be contributing to incipient speciation events. In natural hybridization zones, S. paradoxus hybrids contain mtDNAs with specific recombination patterns of mtDNAs (Leducq et al. 2017; Peris et al. 2017), consistent with selection for mito–mito (and possibly higher-order mito–mito–nuclear) interactions. Coevolved mito–mito interactions are likely to strengthen hybridization barriers already imposed by mitochondrial–nuclear incompatibilities in Saccharomyces yeasts (Sulo et al. 2003; Lee et al. 2008; Chou et al. 2010; Spirek et al. 2014).
Strains with recombinant mtDNAs had different phenotypic distributions when grown in different environmental conditions. Thus, selection potential for direct mitotype effects (including mito–mito epistasis) vs. mito–nuclear interactions will be altered in different environments. In the nine mitotype × nine nuclear genotype strain collection, we found that direct effects of mitotypes were significant only when respiring cells were grown in temperature and oxidative stress conditions. This is perhaps a reflection of increased ATP demands (Postmus et al. 2011) and a mitochondrial role in stress responses at elevated temperatures in cells (Knorre et al. 2016; Munro and Treberg 2017). In contrast, mito–nuclear interactions were highly significant in each condition tested, as shown here and in Paliwal et al. (2014), including fermentable media where mitochondrial metabolism is downregulated due to glucose repression. It is possible that mito–nuclear interactions play roles that are more related to cellular homeostasis rather than oxidative phosphorylation. In support of this, cosegregating mitochondrial and nuclear alleles in natural populations of fish mapped to genes with roles in mitochondrial protein regulation and translation, but not to genes encoding subunits of oxidative phosphorylation complexes (Baris et al. 2017).
While coevolved mito–nuclear genome combinations generally show higher fitness than synthetic genome combinations, exceptions are routinely observed, including in yeast [Figure 1A and Paliwal et al. (2014)], copepods, (Willett and Burton 2003), and flies (Rand et al. 2006). Mitochondrial alleles with strong direct effects may help to explain these observations. Without recombination, selection for beneficial mitochondrial alleles may be impeded by linkage to other mitochondrial alleles causing mito–nuclear incompatibilities. Recombination may promote adaptive evolution by separating beneficial alleles from linked deleterious alleles. Biparental inheritance of plastid DNA can lead to rescue of a cyto–nuclear incompatibility (Barnard-Kubow et al. 2017), suggesting that the same is possible with mitochondrial recombination. Low levels of recombination in natural populations may weaken mito–nuclear coevolution, especially if there is selection on direct effects that could promote admixture. However, strong mito–mito incompatibilities may reinforce mito–nuclear incompatibilities in the presence of mitochondrial recombination.
Mitochondrial recombination has played a role in the evolution of mtDNAs in yeasts (Wu et al. 2015; Leducq et al. 2017; Wang et al. 2017). High rates of recombination have been reported in laboratory conditions (Fritsch et al. 2014), although mitochondrial heteroplasmy does not necessarily occur in the diploid cells produced following matings (Zinn et al. 1987). We observed that ∼40% of mated cells contained recombinant mtDNAs as identified through low-resolution genotyping approaches. Additional mated strains had mtDNA genotypes that matched one parental mtDNA but phenotypes that matched the alternative mtDNA parental control strain, suggesting that many recombination events went undetected. A closer analysis of mitochondrial recombination is necessary to develop mapping approaches and to understand how functional variation is created in nature.
Mito–mito epistasis adds to the complexity of genetic interactions affecting life-history traits. In systems where mtDNA inheritance is uniparental and largely absent of recombination, mildly deleterious mutations are frequently maintained (Neiman and Taylor 2009). Secondary mutations that compensate for an interrupted epistatic interaction should be selected for and promote mito–mito coevolution. Selection for interacting mitochondrial loci may also contribute to unexpected maintenance of mtDNA heteroplasmy (Ye et al. 2014). In recombining systems, mitochondrial recombination will create new mito–mito allele combinations that could influence the removal of deleterious mutations, hybrid fitness, and coevolutionary processes. Generation and characterization of recombinant mtDNAs, as done here, and experimental evolution in yeast offer approaches to test such evolutionary mechanisms in the future.
Acknowledgments
This work was supported by awards to H.L.F. and A.C.F. from the National Institutes of Health (R01 GM-101320), and to H.L.F. and J.F.W. from the National Science Foundation (DEB-1601580). G.C. was supported by a Natural Sciences and Engineering Research Council (NSERC) graduate fellowship. C.R.L. holds the Canada Research Chair in cell and systems biology and was funded by an NSERC discovery grant for this work.
Author contributions: The project was developed by J.F.W., A.C.F., and H.L.F. Experimental work was performed by J.F.W., G.C., and A.G. Statistical analyses were done by J.F.W. and A.C.F. Resources, instrumentation, and research supervision were provided by C.R.L. and H.L.F. All authors contributed to manuscript preparation.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5970955.
Communicating editor: K. Peichel
Literature Cited
- Anderson M. J., Ter Braak C. J. F., 2003. Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Simul. 73: 85–113. 10.1080/00949650215733 [DOI] [Google Scholar]
- Azevedo L., Carneiro J., van Asch B., Moleirinho A., Pereira F., et al. , 2009. Epistatic interactions modulate the evolution of mammalian mitochondrial respiratory complex components. BMC Genomics 10: 266 10.1186/1471-2164-10-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baris T. Z., Wagner D. N., Dayan D. I., Du X., Blier P. U., et al. , 2017. Evolved genetic and phenotypic differences due to mitochondrial-nuclear interactions. PLoS Genet. 13: e1006517 10.1371/journal.pgen.1006517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard-Kubow K. B., McCoy M. A., Galloway L. F., 2017. Biparental chloroplast inheritance leads to rescue from cytonuclear incompatibility. New Phytol. 213: 1466–1476. 10.1111/nph.14222 [DOI] [PubMed] [Google Scholar]
- Barr C. M., Neiman M., Taylor D. R., 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168: 39–50. 10.1111/j.1469-8137.2005.01492.x [DOI] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B. M., Walker S., 2014. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67: 1–48. [Google Scholar]
- Berger K. H., Yaffe M. P., 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8: 508–513. 10.1016/S0966-842X(00)01862-X [DOI] [PubMed] [Google Scholar]
- Brankovics B., van Dam P., Rep M., de Hoog G. S., van der Lee T. A. J., et al. , 2017. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics 18: 735 10.1186/s12864-017-4116-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubliy O. A., Loeschcke V., Imasheva A. G., 2001. Genetic variation of morphological traits in Drosophila melanogaster under poor nutrition: isofemale lines and offspring–parent regression. Heredity (Edinb) 86: 363–369. [DOI] [PubMed] [Google Scholar]
- Chatelain E. H., Pichaud N., Ballard J. W., Tanguay R. M., Morrow G., et al. , 2011. Functional conservatism among Drosophila simulans flies experiencing different thermal regimes and mitochondrial DNA introgression. J. Exp. Zoolog. B Mol. Dev. Evol. 316B: 188–198. 10.1002/jez.b.21389 [DOI] [PubMed] [Google Scholar]
- Chou J. Y., Hung Y. S., Lin K. H., Lee H. Y., Leu J. Y., 2010. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 8: e1000432 10.1371/journal.pbio.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciborowski K. L., Consuegra S., Garcia de Leaniz C., Beaumont M. A., Wang J., et al. , 2007. Rare and fleeting: an example of interspecific recombination in animal mitochondrial DNA. Biol. Lett. 3: 554–557. 10.1098/rsbl.2007.0290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consuegra S., John E., Verspoor E., de Leaniz C. G., 2015. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 47: 58 10.1186/s12711-015-0138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahal S., Dubey S., Raghavan S. C., 2017. Homologous recombination-mediated repair of DNA double-strand breaks operates in mammalian mitochondria. Cell. Mol. Life Sci.. 10.1007/s00018-017-2702-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defontaine A., Lecocq F. M., Hallet J. N., 1991. A rapid miniprep method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res. 19: 185 10.1093/nar/19.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingley S. D., Polyak E., Ostrovsky J., Srinivasan S., Lee I., et al. , 2014. Mitochondrial DNA variant in COX1 subunit significantly alters energy metabolism of geographically divergent wild isolates in Caenorhabditis elegans. J. Mol. Biol. 426: 2199–2216. 10.1016/j.jmb.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Rago J. P., Hermann-Le Denmat S., Paques F., Risler F. P., Netter P., et al. , 1995. Genetic analysis of the folded structure of yeast mitochondrial cytochrome b by selection of intragenic second-site revertants. J. Mol. Biol. 248: 804–811. 10.1006/jmbi.1995.0261 [DOI] [PubMed] [Google Scholar]
- Diss G., Dube A. K., Boutin J., Gagnon-Arsenault I., Landry C. R., 2013. A systematic approach for the genetic dissection of protein complexes in living cells. Cell Rep. 3: 2155–2167. 10.1016/j.celrep.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Euphrassi B., 1949. Action de l acriflavine sur les levures, pp. 165–180 in Unites biologiques douees de continuite genetique. Editions du Centre National de la Recherche Scientifique, Paris. [Google Scholar]
- Fox T. D., Staempfli S., 1982. Suppressor of yeast mitochondrial ochre mutations that maps in or near the 15S ribosomal RNA gene of mtDNA. Proc. Natl. Acad. Sci. USA 79: 1583–1587. 10.1073/pnas.79.5.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch E. S., Chabbert C. D., Klaus B., Steinmetz L. M., 2014. A genome-wide map of mitochondrial DNA recombination in yeast. Genetics 198: 755–771. 10.1534/genetics.114.166637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto J. M., Newton K. J., 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 68: 225–252. 10.1146/annurev-arplant-043015-112232 [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F., 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272. 10.1016/0378-1119(87)90131-4 [DOI] [PubMed] [Google Scholar]
- Hou J., Friedrich A., Gounot J. S., Schacherer J., 2015. Comprehensive survey of condition-specific reproductive isolation reveals genetic incompatibility in yeast. Nat. Commun. 6: 7214 10.1038/ncomms8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. Y., Chou J. Y., 2017. Environmental factors can influence mitochondrial inheritance in the Saccharomyces yeast hybrids. PLoS One 12: e0169953 10.1371/journal.pone.0169953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorre D. A., Sokolov S. S., Zyrina A. N., Severin F. F., 2016. How do yeast sense mitochondrial dysfunction? Microb. Cell 3: 532–539. 10.15698/mic2016.11.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis E. D., Zouros E., 2001. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18: 1168–1175. 10.1093/oxfordjournals.molbev.a003904 [DOI] [PubMed] [Google Scholar]
- Lagisz M., Poulin R., Nakagawa S., 2013. You are where you live: parasitic nematode mitochondrial genome size is associated with the thermal environment generated by hosts. J. Evol. Biol. 26: 683–690. 10.1111/jeb.12068 [DOI] [PubMed] [Google Scholar]
- Leducq J. B., Henault M., Charron G., Nielly-Thibault L., Terrat Y., et al. , 2017. Mitochondrial recombination and introgression during speciation by hybridization. Mol. Biol. Evol. 34: 1947–1959. 10.1093/molbev/msx139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Y., Chou J. Y., Cheong L., Chang N. H., Yang S. Y., et al. , 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135: 1065–1073. 10.1016/j.cell.2008.10.047 [DOI] [PubMed] [Google Scholar]
- Li Q., Lin K., Sun H., Liu S., Huang K., et al. , 2016. Mitochondrial haplogroup M9a1a1c1b is associated with hypoxic adaptation in the Tibetans. J. Hum. Genet. 61: 1021–1026. 10.1038/jhg.2016.95 [DOI] [PubMed] [Google Scholar]
- Lucassen M., Koschnick N., Eckerle L. G., Portner H. O., 2006. Mitochondrial mechanisms of cold adaptation in cod (Gadus morhua L.) populations from different climatic zones. J. Exp. Biol. 209: 2462–2471. 10.1242/jeb.02268 [DOI] [PubMed] [Google Scholar]
- Mackay T. F., 2014. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat. Rev. Genet. 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masneuf I., Hansen J., Groth C., Piskur J., Dubourdieu D., 1998. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64: 3887–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Ferreira J., Vilela J., Fonseca M. M., da Fonseca R. R., Boursot P., et al. , 2014. The elusive nature of adaptive mitochondrial DNA evolution of an arctic lineage prone to frequent introgression. Genome Biol. Evol. 6: 886–896. 10.1093/gbe/evu059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D., Ruiz-Pesini E., Golik P., Macaulay V., Clark A. G., et al. , 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 100: 171–176. 10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D., Treberg J. R., 2017. A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J. Exp. Biol. 220: 1170–1180. 10.1242/jeb.132142 [DOI] [PubMed] [Google Scholar]
- Neiman M., Taylor D. R., 2009. The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 276: 1201–1209. 10.1098/rspb.2008.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliwal S., Fiumera A. C., Fiumera H. L., 2014. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics 198: 1251–1265. 10.1534/genetics.114.168575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. S., Kim M. J., Jeong S. Y., Kim S. S., Kim I., 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr. Genet. 62: 809–826. 10.1007/s00294-016-0585-3 [DOI] [PubMed] [Google Scholar]
- Passamonti M., Boore J. L., Scali V., 2003. Molecular evolution and recombination in gender-associated mitochondrial DNAs of the Manila clam Tapes philippinarum. Genetics 164: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris D., Sylvester K., Libkind D., Goncalves P., Sampaio J. P., et al. , 2014. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol. Ecol. 23: 2031–2045. 10.1111/mec.12702 [DOI] [PubMed] [Google Scholar]
- Peris D., Arias A., Orlic S., Belloch C., Perez-Traves L., et al. , 2017. Mitochondrial introgression suggests extensive ancestral hybridization events among Saccharomyces species. Mol. Phylogenet. Evol. 108: 49–60. 10.1016/j.ympev.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Piganeau G., Gardner M., Eyre-Walker A., 2004. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21: 2319–2325. 10.1093/molbev/msh244 [DOI] [PubMed] [Google Scholar]
- Postmus J., Tuzun I., Bekker M., Muller W. H., de Mattos M. J., et al. , 2011. Dynamic regulation of mitochondrial respiratory chain efficiency in Saccharomyces cerevisiae. Microbiology 157: 3500–3511. 10.1099/mic.0.050039-0 [DOI] [PubMed] [Google Scholar]
- Quinn G. P., Keough M. J., 2002. Experimental Design and Data Analysis for Biologists. Cambridge University Press, Cambridge, United Kingdom: 10.1017/CBO9780511806384 [DOI] [Google Scholar]
- Rainieri S., Kodama Y., Nakao Y., Pulvirenti A., Giudici P., 2008. The inheritance of mtDNA in lager brewing strains. FEMS Yeast Res. 8: 586–596. 10.1111/j.1567-1364.2008.00363.x [DOI] [PubMed] [Google Scholar]
- Rand D. M., Fry A., Sheldahl L., 2006. Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172: 329–341. 10.1534/genetics.105.046698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R., 1987. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell 48: 1047–1060. 10.1016/0092-8674(87)90712-4 [DOI] [PubMed] [Google Scholar]
- Sammler S., Bleidorn C., Tiedemann R., 2011. Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination. BMC Genomics 12: 35 10.1186/1471-2164-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott G. R., Schulte P. M., Egginton S., Scott A. L., Richards J. G., et al. , 2011. Molecular evolution of cytochrome C oxidase underlies high-altitude adaptation in the bar-headed goose. Mol. Biol. Evol. 28: 351–363. 10.1093/molbev/msq205 [DOI] [PubMed] [Google Scholar]
- Shaw A. J., Weir B. S., Shaw F. H., 1997. The occurrence and significance of epistatic variance for quantitative characters and its measurement in haploids. Evolution 51: 348–353. 10.1111/j.1558-5646.1997.tb02421.x [DOI] [PubMed] [Google Scholar]
- Silva G., Lima F. P., Martel P., Castilho R., 2014. Thermal adaptation and clinal mitochondrial DNA variation of European anchovy. Proc. Biol. Sci. 281: 20141093 10.1098/rspb.2014.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirek M., Polakova S., Jatzova K., Sulo P., 2014. Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5: 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., et al. , 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25: 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulo P., Spirek M., Soltesova A., Marinoni G., Piskur J., 2003. The efficiency of functional mitochondrial replacement in Saccharomyces species has directional character. FEMS Yeast Res. 4: 97–104. 10.1016/S1567-1356(03)00109-0 [DOI] [PubMed] [Google Scholar]
- Ujvari B., Dowton M., Madsen T., 2007. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 3: 189–192. 10.1098/rsbl.2006.0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspohl A., Pignedoli S., Giudici P., 2018. The inheritance of mitochondrial DNA in interspecific Saccharomyces hybrids and their properties in winemaking. Yeast 35: 173–187. 10.1002/yea.3288 [DOI] [PubMed] [Google Scholar]
- Wagih O., Parts L., 2014. gitter: a robust and accurate method for quantification of colony sizes from plate images. G3 (Bethesda) 4: 547–552. 10.1534/g3.113.009431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Xiang H., Liu L., Kong M., Yin T., et al. , 2017. Mitochondrial haplotypes influence metabolic traits across bovine inter- and intra-species cybrids. Sci. Rep. 7: 4179 10.1038/s41598-017-04457-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Huang Y., Liu N., Yang J., Lei F., 2015. Seven complete mitochondrial genome sequences of bushtits (Passeriformes, Aegithalidae, Aegithalos): the evolution pattern in duplicated control regions. Mitochondrial DNA 26: 350–356. 10.3109/19401736.2014.1003821 [DOI] [PubMed] [Google Scholar]
- Willett C. S., Burton R. S., 2003. Environmental influences on epistatic interactions: viabilities of cytochrome c genotypes in interpopulation crosses. Evolution 57: 2286–2292. 10.1111/j.0014-3820.2003.tb00240.x [DOI] [PubMed] [Google Scholar]
- Wolters J. F., Chiu K., Fiumera H. L., 2015. Population structure of mitochondrial genomes in Saccharomyces cerevisiae. BMC Genomics 16: 451 10.1186/s12864-015-1664-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Buljic A., Hao W., 2015. Extensive horizontal transfer and homologous recombination generate highly chimeric mitochondrial genomes in yeast. Mol. Biol. Evol. 32: 2559–2570. 10.1093/molbev/msv127 [DOI] [PubMed] [Google Scholar]
- Ye K., Lu J., Ma F., Keinan A., Gu Z., 2014. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. USA 111: 10654–10659. 10.1073/pnas.1403521111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A., 1987. Kinetic and segregational analysis of mitochondrial DNA recombination in yeast. Plasmid 17: 248–256. 10.1016/0147-619X(87)90033-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All growth measures used for analyses are provided in Table S6, Table S7, and Table S8. Strains are available upon request. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5970955.