Abstract
The majority of gene loci that have been associated with type 2 diabetes play a role in pancreatic islet function. To evaluate the role of islet gene expression in the etiology of diabetes, we sensitized a genetically diverse mouse population with a Western diet high in fat (45% kcal) and sucrose (34%) and carried out genome-wide association mapping of diabetes-related phenotypes. We quantified mRNA abundance in the islets and identified 18,820 expression QTL. We applied mediation analysis to identify candidate causal driver genes at loci that affect the abundance of numerous transcripts. These include two genes previously associated with monogenic diabetes (PDX1 and HNF4A), as well as three genes with nominal association with diabetes-related traits in humans (FAM83E, IL6ST, and SAT2). We grouped transcripts into gene modules and mapped regulatory loci for modules enriched with transcripts specific for α-cells, and another specific for δ-cells. However, no single module enriched for β-cell-specific transcripts, suggesting heterogeneity of gene expression patterns within the β-cell population. A module enriched in transcripts associated with branched-chain amino acid metabolism was the most strongly correlated with physiological traits that reflect insulin resistance. Although the mice in this study were not overtly diabetic, the analysis of pancreatic islet gene expression under dietary-induced stress enabled us to identify correlated variation in groups of genes that are functionally linked to diabetes-associated physiological traits. Our analysis suggests an expected degree of concordance between diabetes-associated loci in the mouse and those found in human populations, and demonstrates how the mouse can provide evidence to support nominal associations found in human genome-wide association mapping.
Keywords: mediation analysis, eQTL, module-QTL, T2D, β-cell, genome-wide association (GWA)
TYPE 2 diabetes (T2D) is a highly heritable disease (h2 ≈ 0.5) (Sanghera and Blackett 2012). More than 100 gene loci associated with diabetes or diabetes-related phenotypes have been identified through genome-wide association studies (GWAS) (Morris et al. 2012; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium et al. 2014; Ng et al. 2014; Flannick and Florez 2016; Fuchsberger et al. 2016; Jason et al. 2017). However, the effect size of each locus is small; their odds ratios are typically ∼1.05–1.10, and most of the heritability of T2D in human populations remains to be elucidated. In addition, much remains to be discovered about the regulatory mechanisms responsible for the wide range in susceptibility to diabetes.
The risk of T2D is quite low in the absence of obesity. Prior to the start of the obesity epidemic, ∼60 years ago, the incidence of T2D was < 1%. Today, the incidence is > 9%. Thus, the gene loci responsible for the susceptibility to T2D act primarily in the context of obesity. Obesity usually leads to insulin resistance, resulting in an increased demand for insulin production to maintain normal glucose levels. Although the etiology of T2D involves interactions among multiple organ systems, one concept that has emerged from GWAS is that, for the most part, the causal genetic factors leading to T2D exert their effect by limiting the capacity of pancreatic β-cells to secrete sufficient insulin to maintain normal glucose levels. A substantial proportion of the candidate genes that have emerged from genetic studies in humans and model organisms affect β-cell function or β-cell mass (Billings and Florez 2010; Mohlke and Boehnke 2015; Prasad and Groop 2015). In the case of monogenic diabetes syndromes, essentially all the causal genes are expressed in β-cells (Fajans et al. 2001; Shih and Stoffel 2001; Taneera et al. 2014).
For purposes of genetic analysis, treating T2D as a binary disease is clearly inadequate. Thus, human and model organism studies focus on quantitative diabetes-related traits, including plasma glucose and insulin levels. However, normal blood glucose levels could equally reflect a healthy, or compensatory, state that is near the breakpoint. One way to confront this complexity is to examine the compensatory mechanisms in genetically diverse individuals that display a wide range of compensatory responses to an environmental stressor.
We hypothesized that the biochemical reactions and cellular signaling pathways that constitute the range of stress-induced responses across individuals would be observed as correlated changes in the patterns of gene expression in key organ systems. By summarizing expression patterns within groups of mRNAs as meta-traits, we can achieve a large dimension reduction, enabling a clearer understanding of the molecular functions involved in the disease process. To translate findings from mouse to human, an understanding of the processes involved in the disease is perhaps more relevant than the identification of causal variants.
In the context of a genetic study, mRNA abundance can be mapped in much the same way as a physiological trait. The relationship between genotype and mRNA abundance involves a unidirectional line of causality (Schadt et al. 2005; Millstein et al. 2009; Neto et al. 2010, 2013). Anchoring mRNA abundance and other phenotypes to genetic variation provides a powerful means to reveal causal drivers: genes that harbor genetic variants that influence disease-associated phenotypes. Often, multiple mRNA abundance traits map to the same locus and are influenced by common genetic drivers (Albert and Kruglyak 2015; Yao et al. 2017). When these comapping mRNAs encode proteins that are associated with common physiological functions this can shed light on potential biological functions of the driver gene(s). These connections evoke testable hypotheses whereby variation in the expression of a driver gene, rather than a genetic variant, can be established as a more proximal cause for a disease-related phenotype. The association between driver genes and their downstream effects can unveil novel pathways as well as the tissue sites of their action (Franzen et al. 2016).
T2D is a disorder of relative insulin deficiency. Pancreatic β-cells are challenged by an increased demand for insulin resulting from insulin resistance. A deficiency of β-cell mass or β-cell function usually does not result in diabetes; however, in the context of insulin resistance, it can lead to an insulin shortfall and diabetes. Because pancreatic islets are not accessible for detailed experimental study in humans, it is not feasible to directly interrogate β-cell function in the context of a properly-powered human GWAS. The repertoire of available mouse strains, harboring the same degree of genetic variability in human populations, displays a wide range in β-cell function. This range can be interrogated in nondiabetic mice where diet treatment is used to challenge the β-cells to increase their insulin secretion.
We conducted a study to map diabetes-related traits in Diversity Outbred (DO) mice. The DO is an outbred mouse population derived from eight inbred strains, including three wild-derived strains. The DO captures a broad range of genetic variation comparable to that found in human populations (Svenson et al. 2012) and presents a similarly broad range of individual susceptibility to T2D. We metabolically challenged 500 DO mice with a high-fat/high-sucrose (HF/HS) Western-style diet, measured diabetes-related physiological phenotypes (e.g., plasma glucose and insulin), and isolated their pancreatic islets for molecular characterization using RNA sequencing (RNA-seq) to reveal variation in patterns of gene expression in islets. Previously, this diet was used as a metabolic challenge to evoke broad phenotypic responses in 43 inbred mouse strains and to genetically map several metabolic phenotypes, including obesity, gut microbial composition, glucose homeostasis, dyslipidemia, hepatic steatosis, atherosclerosis, and energy balance (Svenson et al. 2007; Parks et al. 2013; Spiezio et al. 2014; Sinasac et al. 2016). A recent study by Threadgill and colleagues compared the metabolic effects of the Western-style diet to a control diet, and three additional diets (Mediterranean, Japanese, and Maasai/ketogenic) in four mouse strains; A/J, C57BL/6J, FVB/NJ, and NOD/ShiltJ (Barrington et al. 2018). The Western-style diet, compared to a control diet, had detrimental health effects, such as increased body fat, low-density lipoprotein cholesterol, and liver triglyceride (TG) in all strains, but the effect size varied across stains. In response to the other diets, improvements in these metabolic effects was also strain-dependent. These strain-dependent effects underscore the importance of studying the metabolic impact of diet on diverse genetic backgrounds, making the DO panel a particularly relevant population for this study.
Previous studies that used human islets have identified cis-regulatory maps of gene regulation and overlaid these maps with marks of chromatin accessibility and association with diabetes-related traits from human GWAS (van de Bunt et al. 2015; Varshney et al. 2017). Herein, we present a comprehensive picture of the genetically driven variation of > 21,000 transcripts expressed in mouse islets. We show how these transcripts are coregulated and associated with local and distal expression QTL (eQTL). We demonstrate that the genetic drivers behind this variability in the DO mice correspond to genetic effects at human GWAS loci. Thus, in addition to the conservation of the physiological and biochemical pathways involved in diabetes, genetically variable driver genes appear to be conserved across mammals. Our data provide a resource that can be integrated with other molecular and genetic data, both human and model organism, to identify causal genes and explore the mechanisms involved in the initial stages of T2D pathogenesis.
Materials and Methods
Animal husbandry and measurement of physiological phenotypes
DO mice were obtained from the Jackson Laboratories (stock no. 009376) at 4 weeks of age and maintained within the Department of Biochemistry animal vivarium at the University of Wisconsin. Waves of 100 DO mice, half for each sex, were obtained three times per year, until 500 DO mice were surveyed. DO generations 18, 19, and 21 were included in the cohort. Upon arrival, all DO mice were maintained on a HF/HS diet (44.6% kcal fat, 34% carbohydrate, and 17.3% protein) from Envigo Teklad (catalog number TD.08811). We maintained all DO mice on the HF/HS diet to provide an environmental stressor, sensitizing the mice to the development of diabetes. Our study does not provide information about specific dietary effects, as a control diet was not included in the study design. To measure food intake, all DO mice were singly housed. Although there is no biological replication of genetically identical animals in an outcross population, there is replication of genotypes at specific loci. This local genetic replication enables one to link phenotype with genotype, as in a human GWAS or QTL mapping studies in DO mice.
Body weight was measured biweekly, and 4-hr fasting plasma samples were collected for insulin, glucose, and TG measurements. At ∼18 weeks of age, an oral glucose tolerance test (oGTT) was performed on 4-hr fasted mice to evaluate dynamic changes in plasma insulin and glucose. Glucose (2 g/kg) was administered via oral gavage. Blood was collected from a retro-orbital bleed before glucose administration, and at 5, 15, 30, 60, and 120 min. Area under the curve (AUC) was determined from these time points for glucose and insulin. Glucose was measured by the glucose oxidase method using a commercially available kit (TR15221; Thermo Scientific). Insulin was measured by radioimmunoassay (SRI-13K; Millipore, Bedford, MA). Pancreas weight was not recorded, as it was inflated in situ with collagenase as the first step to islet isolation.
HOMA-IR and HOMA-B, homeostatic model assessment (HOMA) of insulin resistance (IR) and pancreatic islet function (B), were determined using fasting plasma values of glucose and insulin at the time the oGTT was administered (time = 0 values). HOMA-IR is equal to (glucose × insulin) / 405 and HOMA-B = (360 × insulin) / (glucose – 63). Units for plasma glucose and insulin are milligram/deciliter and milliunits/liter, respectively.
Islet RNA profiling
Whole-islet RNA was isolated and evaluated for quality. The average RNA-integrity number for all DO islet samples was 9.24 ± 0.04. RNA samples from 96 animals from each of the first four waves of mice (384 in total) were submitted to the Jackson Laboratory high-throughput sequencing core facility. RNAs were sheared using the E220 Focused-ultrasonicator (Covaris). The Jackson Laboratory core generated whole genome islet mRNA libraries using the KAPA Hyper Prep Kit for Illumina Sequencing (KAPA Biosystems), targeting an insert size of 300 bp using magnetic bead-based size selection. Libraries from waves 1, 2, and 3 were constructed at the Jackson Laboratory and randomized into pools of 24 samples with TruSeq RNA indices (6 bp). Libraries from wave 4 were constructed at the New York Genome Center and were pooled in sets of 24 using 8-bp dual index TruSeq RNA indices. Each pooled RNA sample was sequenced across four randomly assigned lanes on a HiSeq2500 (Illumina) at 1 × 100 bp at the New York Genome Center. To control for potential batch effects, we incorporated wave as a covariate in all subsequent analyses.
Reads were aligned to eight strain-specific transcriptomes of the DO founders (Munger et al. 2014). We used an expectation maximization algorithm (https://github.com/churchill-lab/emase) to obtain estimated total read counts for each gene as a sum across alleles and isoforms (Raghupathy et al. 2018). We normalized read counts in each sample using upper-quantile normalization.
Mouse genotyping and haplotype reconstruction
Genotyping was performed on tail biopsies as described in Svenson et al. (2012), using the Mouse Universal Genotyping Array (GigaMUGA) [143,259 markers (Morgan et al. 2015)] at Neogen (Lincoln, NE). Genotypes were converted to founder strain–haplotype reconstructions using R/DOQTL software (Gatti et al. 2014). We interpolated the GigaMUGA markers onto an evenly spaced grid with 0.02-cM spacing and added markers to fill in regions with sparse physical representation, resulting in 69,005 pseudomarkers. We also reconstructed individual chromosome (Chr) haplotypes from the RNA-seq data using a hidden Markov model (GBRS, https://github.com/churchill-lab/gbrs). We identified three samples with inconsistent genotypes between the GigaMUGA and RNA-seq-based haplotype reconstructions. Three additional samples were determined to have poor quality RNA-seq data. These six samples were excluded, leaving a total of 378 samples for subsequent analyses.
Genome scans to identify QTL for physiological and gene expression traits
Genetic mapping analysis was carried out with the qtl2 R package (http://kbroman.org/qtl2) using the founder haplotype regression method with the Leave One Chr Out option for kinship correction (Gatti et al. 2014) (http://kbroman.org/qtl2). Phenotype data were log transformed. We performed genome scans for each phenotype with sex and experimental cohort (wave) as covariates. We estimated significance thresholds by permuting the phenotype values 1000 times while holding the genotypes fixed, mapping the permuted trait, and retaining the maximum LOD score from each permutation (Churchill and Doerge 1994). Complete details of our analyses are provided in R Markdown files that can be used to reproduce our work in RStudio (Supplemental Material, File S1, File S2, and File S3). A guide to the contents and data type analyzed in each Markdown file [e.g., physiological traits, eQTL hotspots, and “module eigengenes” (MEs)] is provided in File S4.
Normalized RNA-seq data were transformed to normal scores [van der Waerden’s method, (Conover 1999)] and we carried out QTL mapping as described above. We called an eQTL a “local eQTL” if the marker with the maximum LOD score was within ± 4 Mb of the transcriptional start site (TSS) and called the remaining eQTL “distal eQTL.” We identified eQTL hotspots by counting the number of eQTL with LOD > 7.2 that occurred in a sliding 4-Mb window with a 1-Mb overlap between windows. We retained all genes with LOD > 6 within ± 2 Mb (4-Mb width) of the center of each hotspot and calculated the first principal component (PC1) of the genes in each hotspot.
Mediation analysis to identify causal drivers
To identify candidate driver genes for distal eQTL, we applied mediation analysis (MacKinnon et al. 2007). We denote the genotype at the QTL locus as Q, the gene expression of the distal eQTL transcript as Y, and the gene expression of the candidate driver gene as M. We hypothesize a causal pathway model Q → M → Y in which the genetic variation on the distal eQTL transcript is fully or partially mediated by variation in the expression of the driver gene. To evaluate this model, we applied the causal steps method and verified each of four conditions:
The distal transcript Y should have a significant LOD score at the locus Q.
The candidate mediator M should have a significant LOD score at Q. Typically, the LOD score of the mediator will be greater than the distal eQTL. In addition, we require that the transcript M should derive from a gene at the locus Q, i.e., that the mediator has a local eQTL, and that the allele effects pattern matches that of the distal eQTL.
Adding the mediator M to the regression of Y on Q should cause a drop in the distal eQTL LOD score. To determine if the drop in LOD score is significant, we tested every transcript genome-wide as a candidate mediator and computed a Z-score. We identified candidate mediators with a Z-score < 6.
Adding the distal transcript Y as a covariate in the regression of M on Q, we require that the conditional LOD score remains significant at a nominal (not genome-wide adjusted) level of 0.01.
We evaluated genes with local eQTL at hotspot loci as candidate mediators; these genes satisfy condition 2. We evaluated the LOD drop for the PC1 of the hotspot genes and for each individual gene with a distant eQTL at the hotspot. The logic of mediation analysis allows us to rule out candidate mediators, but not to prove them. There are some potential pitfalls to genetic mediation analysis (Didelez and Sheehan 2007). A transcript with a strong local eQTL (typically with LOD > 100) can satisfy conditions 1 through 4 because it can act as surrogate for the local genotype. If the distal eQTL is mediated by a mechanism other than a change in gene expression of a local transcript, mediation on RNA abundance will fail to detect this.
Weighted gene coexpression network analysis
Coexpression gene modules were computed using Weighted Gene Coexpression Network Analysis (WGCNA) (Zhang and Horvath 2005), which performs network construction and module detection based on correlation between traits. For the WGCNA analysis, we included all expression traits (21,771 transcripts) that were measured in 378 DO islet samples. We used a signed WGCNA network with minimum module size of 30 and a soft thresholding power of 12. A first PC (PC1) was calculated for each module, which we refer to as the ME, and used for module–trait correlation and module QTL analyses. The ME for each module was computed from the islet RNA-seq measurements for all DO mice, with no distinction made for sex. Thus, MEs are based purely on the correlation structure for all transcripts within a module for all mice. To identify which modules showed a sex difference in the expression pattern of the transcripts, we performed a Student’s t-test using the ME value between males and females (Table S5). Module enrichment analysis was done using allez (Newton et al. 2007) to identify enriched categories in the Gene Ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG). This analysis identifies differential enrichment of a functional category among traits in a module, compared to traits not in the module. Of the ∼22,000 transcripts used for module calculation, 13,412 (∼62%) were assigned to a module. The average number of transcripts per module was 327 ± 51 transcripts and ranged from 42 (lightsteelblue1) to 1494 (turquoise).
Identification of human loci syntenic to mouse QTL and integration with diabetes-associated GWAS
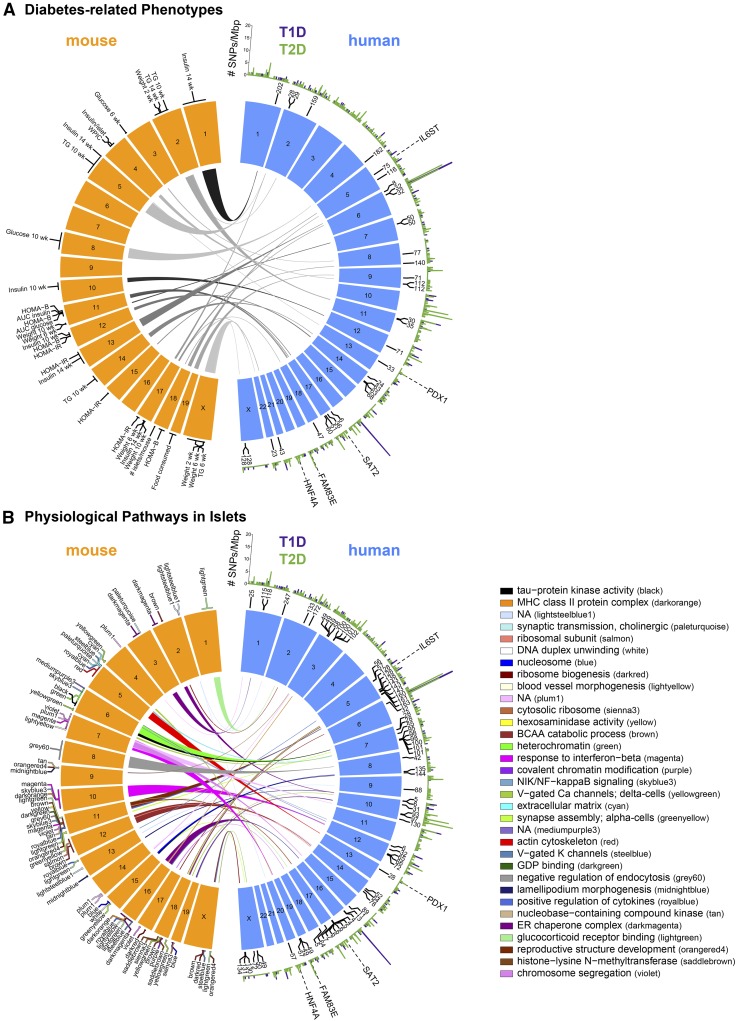
The R/Bioconductor package ChIPseeker (v1.13.1) (Yu et al. 2015) was used to first annotate the QTL locations to the nearest annotated gene in the University of California, Santa Cruz R/Bioconductor package TxDb.Mmusculus.UCSC.mm10.knownGene (v3.4.0) (Lawrence et al. 2013) using the function annotatePeak with options tssRegion = c(−1, 1), TxDb = TxDb.Mmusculus.UCSC.mm10.knownGene, annoDb = “org.Mm.eg.db,” overlap = “all,” and all other options set to default. Then, the syntenic regions in human (hg19) to each of the nearest QTL genes were mapped using the R/Bioconductor package biomaRt (v2.33.4) (Durinck et al. 2009). The locations of the syntenic human genes were rounded to the nearest megabase pair. The circle plots were made using the R package “circlize” (v0.4.1) (Gu et al. 2014) and initialized using the provided hg19 and mm10 cytobands. The circular layouts of the mouse and human genome (Figure 9) were made in R version 3.4.1 (https://www.r-project.org).
Figure 9.
Architectural view of diabetes-related physiological phenotypes and islet-based pathways in mouse and human. QTL for diabetes-related phenotypes (A) and physiological pathways (B) are illustrated for the mouse genome, which are connected to their syntenic regions in human. The gene nearest to the peak for the QTL in mouse was used to identify the corresponding locus in human. The thickness of the arc originating from the mouse QTL is proportional to the 95% C.I. for the QTL. Arcs interconnect mouse QTL to syntenic regions in human; numbers listed next to humans Chrs are megabase pair position. The color of the arcs in (B) correspond to the module color; top-enriched physiological pathways for modules are illustrated in the legend for (B). Chr numbers are shown; the Y-Chr is excluded. The number of SNPs from human GWAS (http://www.gwascentral.org/) with nominal or higher associations (−log10 P ≥ 4) to T2D (green) or T1D (purple) that occur within a 1-Mbp interval are shown for each human Chr, resulting in the identification of diabetes GWAS loci. The genomic location is shown for the five candidate gene drivers predicted from our mediation analysis of the eQTL hotspots; IL6ST, PDX1, SAT2, FAM83E, and HNF4A. BCAA, branched-chain amino acid; Chr, chromosome; eQTL, enhanced QTL; GWAS, genome-wide association study; T1D, type 1 diabetes; T2D, type 2 diabetes.
We used GWAS Central (http://www.gwascentral.org/) to download all SNPs that are associated with type 1 diabetes (T1D) and T2D above a threshold of −log10 P-value ≥ 4, resulting in 666 and 509 unique SNPs, respectively (Beck et al. 2014); these SNPs are contained within Table S8. We then determined the number of T1D- or T2D-associated SNPs occurring within a 1-Mbp genomic window, yielding the histograms shown for each Chr in Figure 9. The number of T1D-associated SNPs at the HLA locus on Chr 6 (∼30–33 Mbp) was 344. For display purposes of the T1D GWAS histogram shown in Figure 9, the maximum value for the 1-Mbp bins was set to 20 at the HLA locus. All analyses used to integrate the mouse QTL with human GWAS are reproduced in an R Markdown document, accompanying data files, and guide (File S5, File S6, and File S7, respectively).
We identified 67 module QTL and 30 physiological QTL for a total of 97; 49 of these QTL are syntenic to human loci that are within 1 Mbp of the T1D- and T2D-related GWAS SNPs that we obtained from GWAS Central. We performed a test of significance by randomly selecting 97 human genes from a restricted set of genes located on Chr 1–22 and X. The human genes were restricted to those from biomaRt that have syntenic regions in hg19 and were mappable to mm10. Additionally, we only included genes whose expression mean was greater than the fifth percentile of mean expression in our mouse islet RNA-seq data. For these 97 randomly selected genes, we then counted how many were within 1 Mbp of the diabetes-related SNPs from GWAS Central. We repeated this sampling 1000 times to derive an empirical P-value of enrichment.
Data availability
Raw sequence reads have been deposited in the Sequence Read Archive (SRP125176). Phenotypes, genotypes, and quantified gene expression data have been deposited with Dryad (doi:10.5061/dryad.pj105; data files: Attie Islet eQTL data). The data are also accessible through an interactive web-based analysis tool that will allow users to replicate the analyses reported here, including genetic mapping and mediation analysis (http://churchill-lab.jax.org/qtl/islet/DO378). Software for this interactive tool is maintained at https://github.com/churchill-lab/qtlviewer and https://github.com/churchill-lab/qtlapi. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5977459.
Results
Wide range of diabetes-related phenotypes in DO mice maintained on a Western-style diet
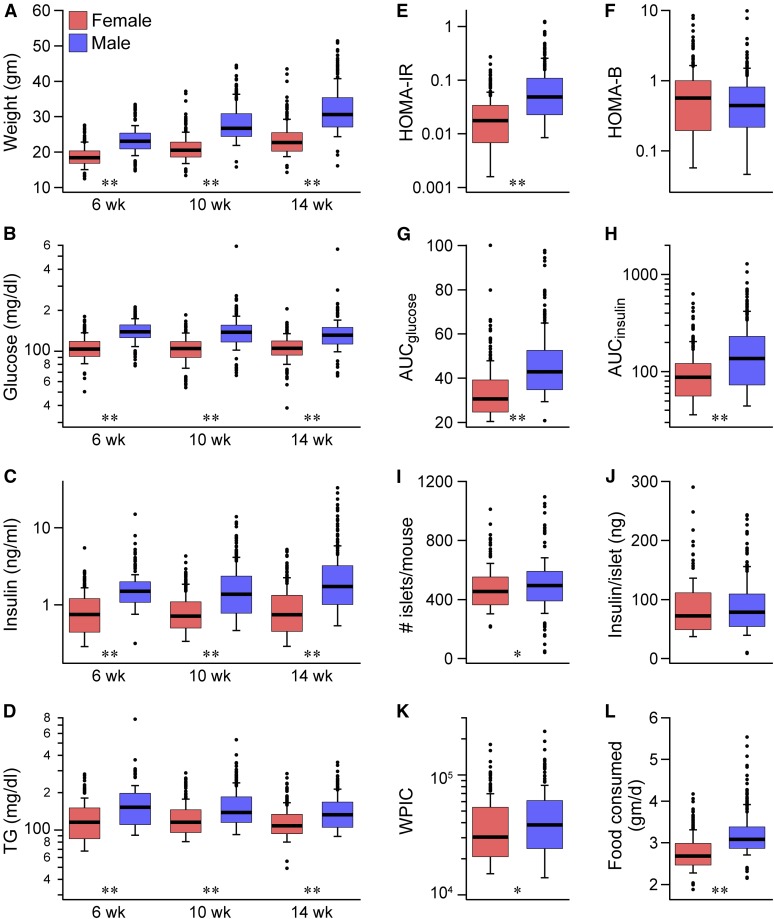
To better understand the impact of a Western-style diet on physiological traits related to diabetes, we generated a cohort of ∼500 DO mice that were metabolically challenged with a HF/HS diet (45% kcal fat and 34% sucrose). We measured body weight, fasting plasma glucose, insulin, and TGs at regular intervals (Figure 1, A–D). There was a large spread in phenotype values, due in part to the genetic diversity of DO mice. For example, body weight at 14 weeks of age ranged from ∼15 to > 50 g (Figure 1A) and fasting plasma insulin showed a ∼100-fold range (Figure 1C), consistent with significant differences in peripheral insulin resistance among the mice. Only two of the ∼500 DO mice evaluated became diabetic (blood glucose > 300 mg/dliter) (Figure 1B), suggesting that the HF/HS diet evoked a compensatory increase in insulin production to offset diet-induced insulin resistance. All mice were individually housed throughout the course of our study, allowing us to measure daily food consumption for individual mice. As with the other phenotypes, there was a broad range in the amount of food consumed (1.9–5.5 g/day) and was on average, higher in males than females (Figure 1L), reflecting differences in body weight between the sexes. At all ages, male DO mice demonstrated higher values for body weight (P < 2.2e−16), plasma glucose (P < 2.2e−16), insulin (P < 2.0e−12), and TGs (P < 6.5e−10), compared to female DO mice.
Figure 1.
Diabetes-related physiological phenotypes in Diversity Outbred (DO) mice. Body weight (A) and fasting plasma glucose (B), insulin (C), and triglycerides (TG) (D) are shown for female and male high-fat and high-sucrose (HF/HS) diet-fed DO mice at 6, 10, and 14 weeks (wk) of age. HOMA-IR (E) and HOMA-B (F) measures of insulin resistance and β-cell function, respectively, were computed from fasting plasma insulin and glucose measurements at 18 wk of age. Area under the curve (AUC) for glucose (G) and insulin (H) were determined from an oral glucose tolerance test (oGTT) performed at 18 wk of age. The number of islets per mouse (I), and the insulin content per islet (J) were determined at kill at 22 wk. Whole-pancreas insulin content (WPIC), a surrogate measure of pancreatic β-cell mass, was computed from islet number and insulin content per islet (K). Average food consumption (gram/day) was computed over the course of the 4 months the mice were maintained on the HF/HS diet (L). * P < 0.05 for male vs. female and ** P < 0.01. HOMA-IR, homeostatic model assessment of insulin resistance; HOMA-B, homeostatic model assessment of pancreatic beta-cell function.
To interrogate islet function, we performed an oGTT at ∼18 weeks of age. Prior to the oGTT, all mice were fasted for 4 hr and a blood sample was collected, from which we computed HOMA-IR and HOMA-B, homeostatic measures that are related to peripheral insulin resistance and β-cell function, respectively (Matthews et al. 1985). HOMA-IR (Figure 1E) showed a > 100-fold range and was higher in males than females (P = 1.0e−11), whereas HOMA-B was not different between the sexes (Figure 1F). The glucose (Figure 1G) and insulin (Figure 1H) responses during the oGTT (AUCglucose and AUCinsulin, respectively) were significantly higher in males than females (P = 2.0e−11) and showed a large dynamic range.
At ∼22 weeks of age, all DO mice were sacrificed and their islets isolated by hand-picking. The average number of islets per DO mouse was 483, ranging from 42 to 1096 (Figure 1I). The average amount of insulin per islet was 85 ng/islet and ranged from ∼9 to 290 ng/islet (Figure 1J). The whole-pancreas insulin content (WPIC), a surrogate measure of β-cell mass per pancreas, was 42.7 ± 1.4 µg/pancreas (Figure 1K). The number of islets per pancreas (P = 0.03) and WPIC (P = 0.01) were significantly elevated in male mice, whereas the insulin content per islet was not significantly different between the sexes (P ∼0.2). All mice were individually housed throughout the course of our study, allowing us to measure daily food consumption for individual mice. As with the other phenotypes, there was a broad range in the amount of food consumed (1.9–5.5 g/day) and was, on average, higher in males than females (Figure 1L). However, this difference is largely driven by the lower body weight of female mice (Figure 1A). Food consumption as a function of weight is nearly identical for male and female DO mice (Figure S1). Some of the physiological traits are highly intercorrelated (Figure S2). The correlation patterns are largely concordant between the sexes except for HOMA-B, which is more strongly correlated in males, and AUCglucose, which has stronger correlations with other traits in female mice.
Diabetes-related phenotypes map to multiple QTL loci
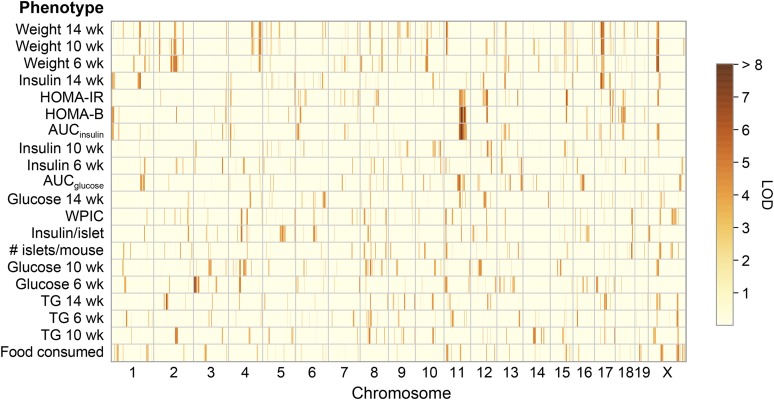
We next asked if the physiological phenotypes map to QTL. We identified a total of 34 physiological QTL (Figure 2) that were significant, or suggestive. Among the physiological traits, AUCinsulin and HOMA-B showed the strongest QTL, and comapped to a locus on Chr 11 at ∼85 Mbp with LOD scores of 11.3 and 10.3, respectively. HOMA-B has a second QTL on Chr 18 at ∼47 Mbp. Body weight at 6 and 10 weeks of age mapped to Chr 11 at ∼12 Mbp and to Chr 17 at ∼35 Mbp. Plasma insulin at 14 weeks comapped with the body weight QTL to Chr 17, consistent with a model whereby variation in body weight results in changes in peripheral insulin resistance, leading to increased plasma insulin to maintain euglycemia. However, plasma insulin at 14 weeks also mapped to Chr 1, Chr 5, and Chr 13, indicating that genetic factors acting through mechanisms independent of body weight influence circulating insulin levels. Other physiological QTL included WPIC, a surrogate for pancreatic β-cell mass, and insulin per islet on Chr 4 at ∼60 Mbp. HOMA-IR, HOMA-B, and plasma insulin at 10 weeks mapped to a locus on Chr 12. In summary, these results demonstrate that the complex nature of diabetes-related physiological phenotypes results from genetic regulation at multiple loci.
Figure 2.
Genetic architecture of diabetes-related phenotypes. QTL for diabetes-related physiological traits, colored by LOD score (red = high and yellow = low). Traits are ordered by unsupervised clustering such that traits with similar LOD profiles are grouped together. Table S6 lists all physiological QTL, their LOD scores, and their genomic positions in mouse and human. AUC, area under the curve; HOMA-B, homeostatic model assessment of pancreatic beta-cell function; HOMA-IR, homeostatic model assessment of insulin resistance; wk, week; WPIC, whole-pancreas insulin content; TG, triglyceride.
Genetic architecture of the transcriptome reveals hotspots in pancreatic islets
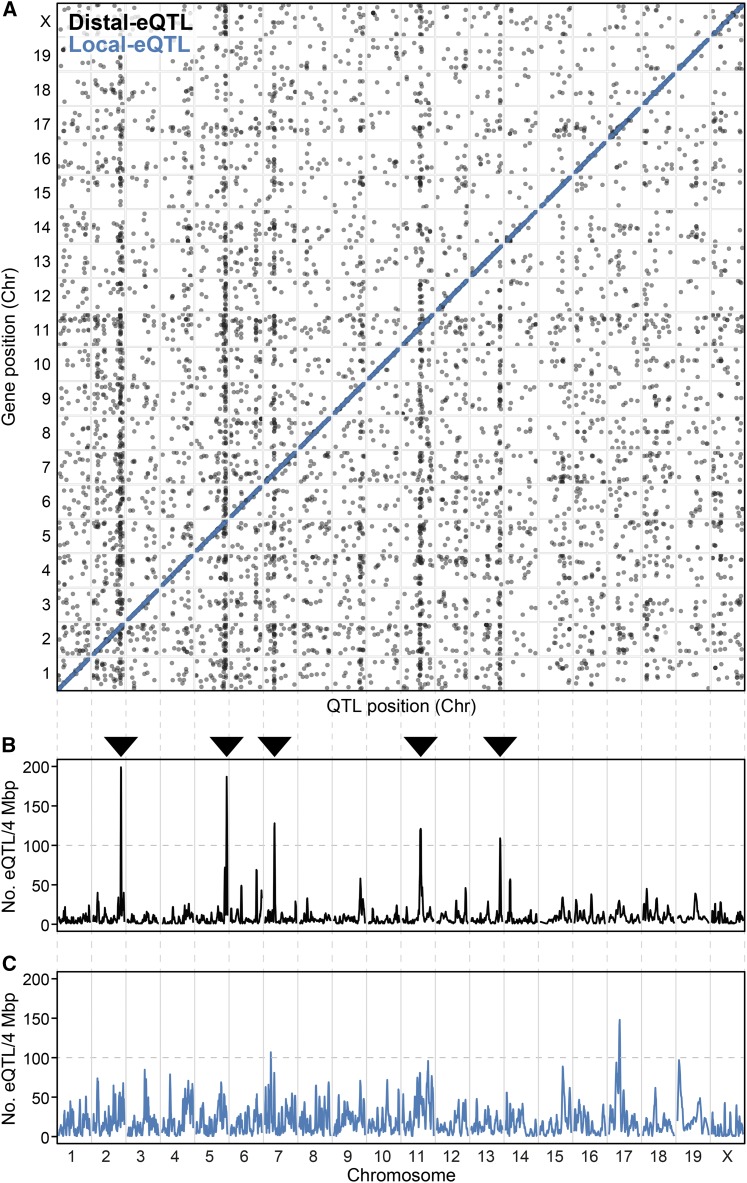
We sequenced whole-islet mRNA from 378 HF/HS-fed DO mice (188 females and 190 males) at an average depth of ∼36 million reads/sample and obtained quantitative estimates of abundance for 21,771 islet transcripts. RNA abundance was used to perform whole-genome scans, resulting in the identification of 18,820 significant eQTL [LOD > 7.2, corresponding to a permutation-based genome-wide P < 0.05; (Churchill and Doerge 1994)] (Figure 3A). We classified eQTL as either local (peak LOD within 4 Mbp of gene locus; blue dots) or distal (black dots). At this LOD threshold, 68% of the eQTL were local, suggesting that genetic variation proximal to the gene locus plays a key role in regulating transcript abundance among DO mice. LOD scores for local eQTL tended to be higher than for distal eQTL, consistent with what is normally observed in eQTL studies, and thus the proportion of local eQTL increases with higher LOD thresholds (West et al. 2007; Munger et al. 2014).
Figure 3.
Genetic architecture of gene regulation in pancreatic islets. (A) Inferred expression QTL (eQTL) with LOD ≥ 7.18 (P < 0.05) identified in islets from 378 Diversity Outbred mice maintained on a high-fat and high-sucrose diet. Black, distal eQTL (5669); blue, local eQTL (12,802). Data points correspond to the peak position of the eQTL. y-axis shows position of gene, while x-axis shows the genomic position of the eQTL. Local eQTL follow a diagonal pattern, whereas the distal eQTL form vertical bands. Profile illustrating the number of distal (B) or local (C) eQTL occurring within a 4-Mbp genomic window. Distal eQTL hotspots with > 100 comapping eQTL were identified in chromosomes (Chr) 2, 5, 7, 11, and 13 (see ▼).
As we have previously shown (Tian et al. 2015), islet distal eQTL can form clusters at genomic hotspots, where many expression traits comap to the same locus (Breitling et al. 2008). We used a 4-Mbp sliding window, with 1 Mbp steps, and defined windows with 100 or more distant eQTL that comap to be a hotspot. In DO islets, we identified five distal eQTL hotspots, on Chrs 2, 5, 7, 11, and 13 (Figure 3B and Table S1). Figure 3C illustrates the genomic profile for the number of comapping local eQTL, identifying several loci with ∼100 comapping local eQTL (e.g., Chrs, 7, 17, and 19). These local eQTL hotspots are all in regions of high gene density.
Mediation analysis predicts candidate drivers of eQTL hotspots
One possible explanation for comapping of many expression traits is the shared influence of polymorphic genetic factors present at the hotspot locus that directly or indirectly influence the distal eQTL genes. The effect of genetic variation on expression of the distal eQTL genes must be mediated through one or more trans-acting factors encoded at the hotspot locus. If the mediation occurs through transcriptional variation in regulator genes, we can identify candidate regulators by applying mediation analysis (Chick et al. 2016). A key step in mediation analysis looks for a substantial drop in the LOD score at the hotspot after conditioning on the expression of a candidate regulator gene. A candidate gene is the driver of the hotspot. We evaluated the conditions for mediation using PC1 at each of the five hotspots. In addition, we evaluated each of the distal eQTL genes to identify individual genes that share the same mediator as the PC1.
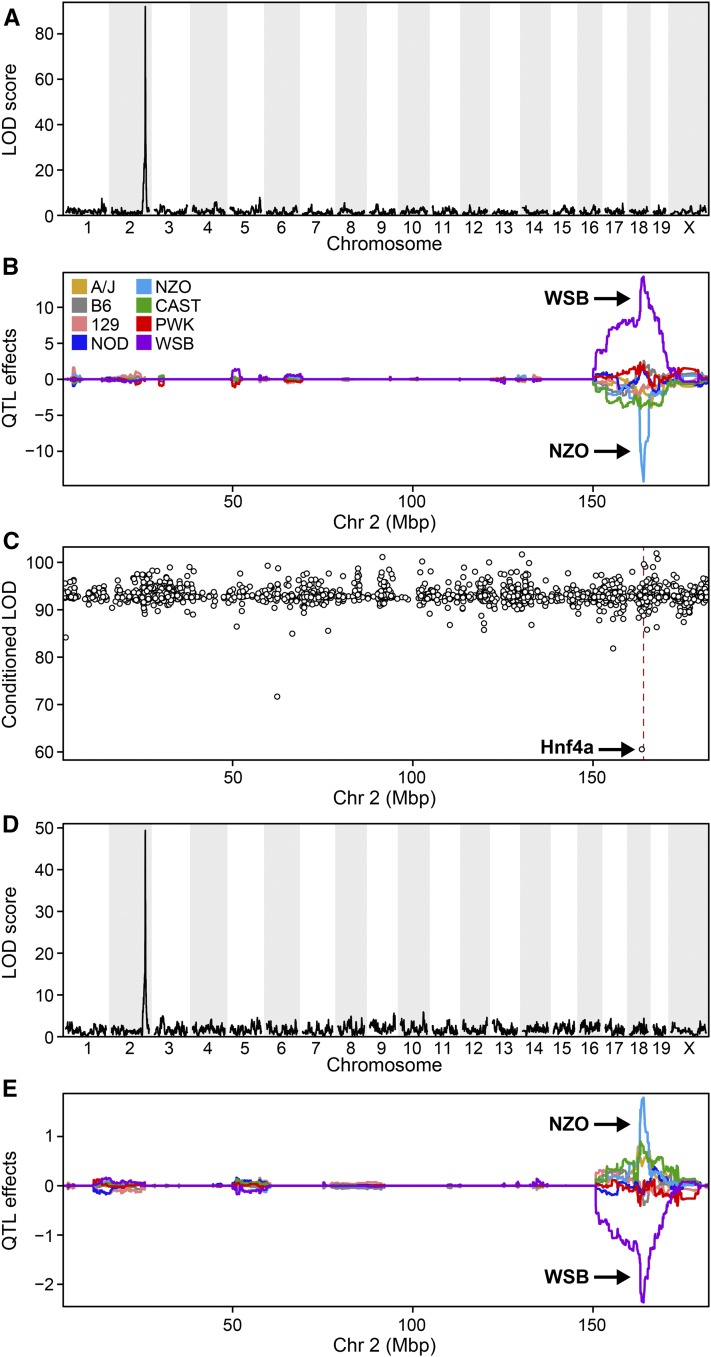
The PC1 for the Chr 2 hotspot at ∼164.0 Mbp (PC1Chr2) had a single significant QTL with a LOD score of 92.4 (Figure 4A). The WSB and NZO alleles were associated with low and high expression of PC1Chr2, respectively (Figure 4B). When conditioned on the expression of each of the genes on Chr 2, the LOD profile for PC1Chr2 was largely unchanged for most genes. However, when conditioned on Hnf4a, located on Chr 2 at ∼163.5 Mbp, the LOD profile for PC1Chr2 dropped significantly (Figure 4C), indicating that Hnf4a is a candidate mediator. The LOD profile (Figure 4D) and allele dependence (Figure 4E) for the local eQTL for Hnf4a closely matches that for PC1Chr2 (Figure 4, A and B), consistent with the results from conditioning PC1Chr2 on Hnf4a. We evaluated each of the distal eQTL genes, conditioning on Hnf4a, and found that 88 of the 147 genes in the hotspot had an LOD drop of ≥ 1.5 (Table S1). Hnf4a is a transcription factor. Using PSCAN (Zambelli et al. 2009), we asked if the promoters of transcripts comapping to the Hnf4a gene locus are enriched with a motif associated with Hnf4a binding. Known motifs for 635 transcription factors were surveyed within −950 to +50 bp of the transcription start sites for the 206 transcripts that map to the Chr 2 hotspot. Remarkably, the most significantly enriched (P = 3.6e−9) motif was for Hnf4a (Figure S3). Our results suggest that Hnf4a is a driver that mediates the distal eQTL for the Chr 2 hotspot expression traits.
Figure 4.
Mediation predicts Hnf4a as a driver at the chromosome (Chr) 2 expression QTL (eQTL) hotspot. (A) Genome-wide LOD profile for first principal component (PC1) of eQTL hotspot at ∼165 Mbp on Chr 2. (B) Allele dependence of Chr 2 hotspot, showing that WSB and NZO are the high and low alleles, respectively. (C) LOD score for Chr 2 eQTL hotspot after conditioning, one at a time, on the expression of 1892 genes that were located on Chr 2. Conditioning on Hnf4a local eQTL resulted in the largest drop in the LOD profile for the hotspot. (D) LOD profile for Hnf4a local eQTL with a peak at 163.9 Mbp on Chr 2. (E) Allele dependence for Hnf4a local eQTL demonstrates the same genetic architecture as the eQTL hotspot; WSB and NZO are the low and high allele, respectively.
We performed mediation analysis for the PC1s at the other four distal eQTL hotspots. At the Chr 13 hotspot, we identified Il6st as a candidate driver for 82 of 104 genes that map to this hotspot (Figure S4 and Table S1). When the PC1Chr13 was conditioned on the expression of Il6st, the LOD profile for the hotspot was essentially reduced to zero, suggesting that Il6st explains all of the genetic variation in PC1Chr13. Mediation analysis of the hotspots at Chrs 5, 7, and 11 identified Pdx-1 (mediates 77 of 182 genes; Figure S5), Fam83e (mediates 96 of 123 genes; Figure S6), and Sat2 (mediates 115 of 126 genes; Figure S7) as potential drivers of their respective hotspot eQTL (Table S1). The Chr 5 hotspot is complex and there may be additional driver genes at this locus.
We found concordance between QTL for physiological traits and four of the five eQTL hotspots. Fasting plasma insulin at 14 weeks of age mapped to the eQTL hotspot on Chr 5 and showed the same allele effects pattern. AUCglucose mapped to the Chr 11 eQTL hotspot and HOMA-IR mapped to the eQTL hotspot on Chr 13. Despite the weaker genetic signal and broad confidence intervals associated with many of the physiological QTL, their allelic effects indicate that these traits are responding to the same genetic drivers at the eQTL hotspots. Importantly, mediation analysis can nominate candidate genes with variants that influence diabetes-related phenotypes through their broad effects on pancreatic islet gene regulation.
Candidate drivers are associated with diabetes traits in human GWAS
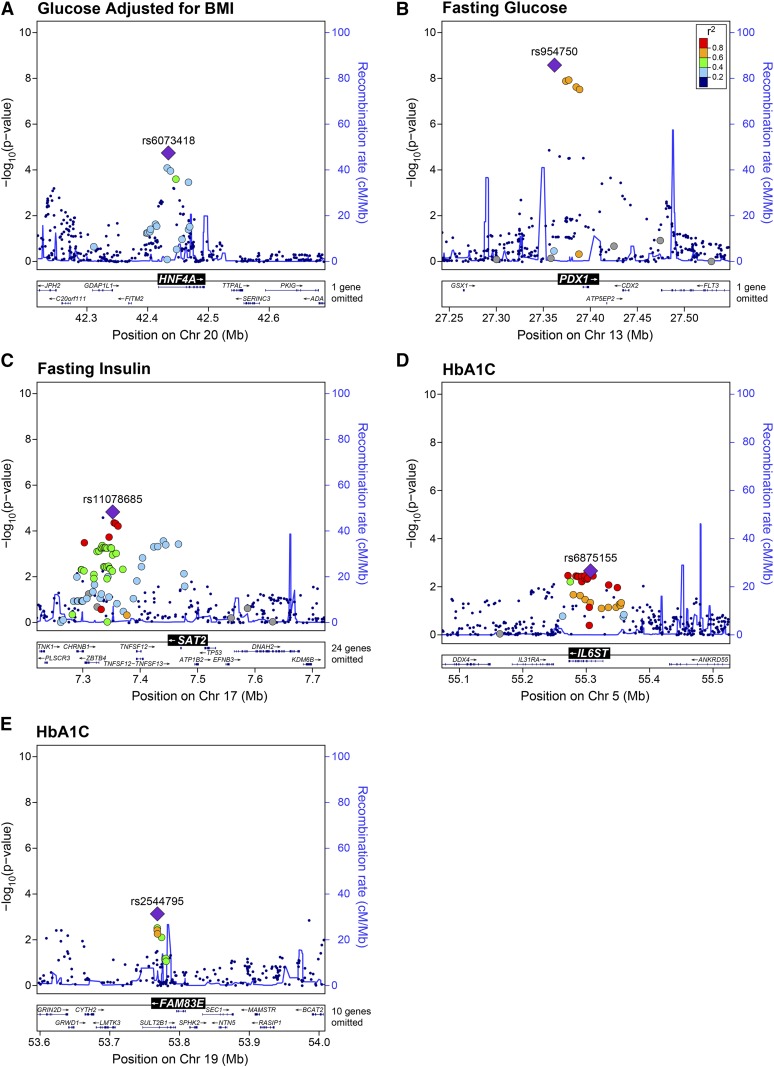
We next asked if genetic variation at the gene loci for the five candidate hotspot drivers is associated with diabetes-related phenotypes in human GWAS. We used LocusZoom (Pruim et al. 2010) to generate regional association plots at each of the candidate gene loci for diabetes-associated phenotypes measured in the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) studies, e.g., fasting glucose, insulin, or HbA1c (Dupuis et al. 2010; Soranzo et al. 2010; Manning et al. 2012; Scott et al. 2012a,b). While many of the SNPs associated with these phenotypes are subthreshold for a genome-wide query, the relatively small number of SNPs interrogated in our analysis (< 400) greatly reduces the multiple-testing penalty for these single-gene searches. We identified SNPs with significant association to one or more diabetes-related phenotypes at three of the five driver gene loci.
Genetic variation at HNF4A (Figure 5A) and PDX1 (Figure 5B), also known as MODY1 and MODY4, yielded SNPs associated with body mass index (BMI)-adjusted 2-hr glucose (P < 10−5) and fasting glucose (P < 10−8), respectively. SAT2, which has been linked to polyamine (Hyvonen et al. 2013) as well as thialysine (Coleman et al. 2004) metabolism, was associated with fasting insulin (P < 10−5) (Figure 5C). IL6ST (the β-subunit of the IL6 cytokine receptor) and FAM83E [a newly discovered gene that may be involved in MAPK signaling (Cipriano et al. 2014)] each were associated with HbA1c, albeit with marginal significance (P < 10−3) compared to the other loci (Figure 5, D and E, respectively). In summary, these results demonstrate that by integrating our conditional analyses at the eQTL hotspots to predict causal gene drivers, we performed single-gene queries in human GWAS that support a role for these genes in islet function and, potentially, diabetes risk.
Figure 5.
Candidate driver gene loci are associated with diabetes traits in human genome-wide association studies (GWAS). Association (−log10 P-value, left margin) to diabetes-related phenotypes for SNPs near candidate gene drivers of eQTL hotspots. (A) 2-hr glucose adjusted for body mass index (BMI) at HNF4A gene locus. (B) Fasting plasma glucose at PDX1 gene locus. (C) Fasting plasma insulin at SAT2 gene locus. Hemoglobin A1c (HbA1C) at the IL6ST (D) and FAM83E (E) gene loci. Plots were generated using LocusZoom (Pruim et al. 2010) from publicly available GWAS data. For each plot, the index SNP (purple diamond) is identified by Rs number, along with SNPs that are correlated to the index SNP (color scale shows correlation, r2), defining a haplotype block. The total number of SNPs plotted are 359, 324, 289, 315, and 217 for (A–E), respectively. Recombination frequencies (centimorgan/megabase) are plotted as blue traces and are shown along right margins for each locus. Legend for r2 values is shown in (B) and corresponds to all panels. Chr, chromosome.
Coexpression modules highlight biological processes related to diabetes
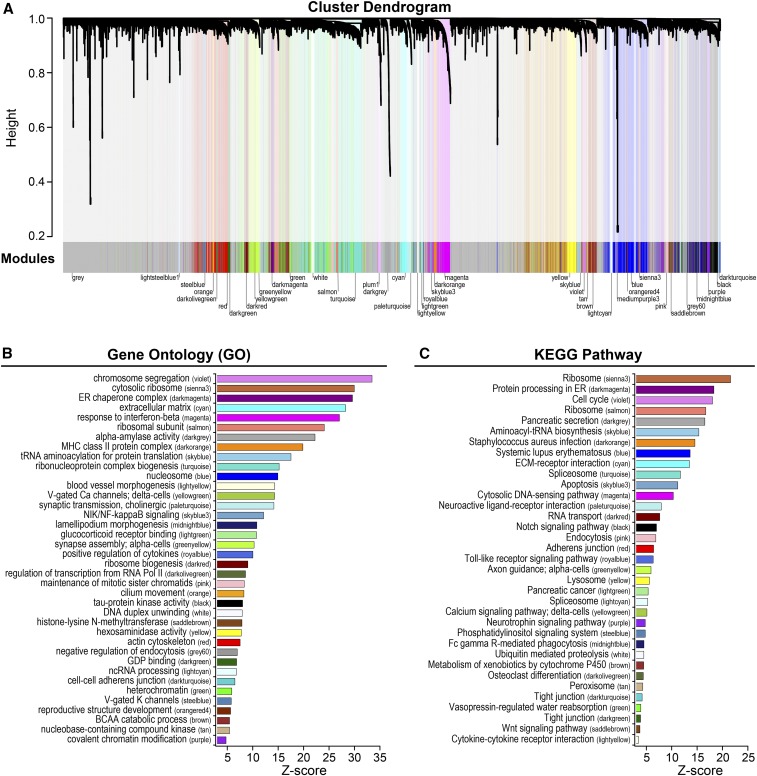
We employed cluster analysis to identify groups of genes with highly correlated expression patterns in the islet transcriptome (Zhang and Horvath 2005; Langfelder and Horvath 2008). Grouping of transcripts into modules provides a useful summary of the complex correlation structure that is typical of whole-transcriptome data. When grouping the transcripts into coexpression modules, we did not utilize information about functional annotations or whether the transcripts were subject to genetic regulation. Among the 21,771 transcripts from our whole-islet RNA-seq, 62% were assigned to a coexpression gene module. We identified a total of 41 modules with varying numbers of transcripts, ranging from 42 to 1494 (Table S2). The average number of transcripts per module was 327. Modules are depicted as the downward branches in the cluster dendrogram (Figure 6A), the length of which is proportional to the average gene–gene correlation within each module.
Figure 6.
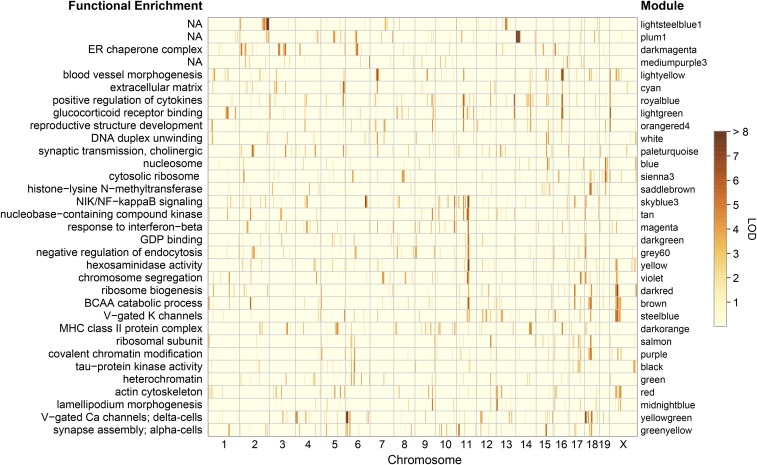
Coexpression gene modules in islets from Diversity Outbred (DO) mice. Cluster dendrogram illustrates modules (denoted by color and shown as downward branches) of highly correlated transcripts in islets from 378 DO mice that were maintained on a high-fat and high-sucrose diet (A). Transcripts falling within gray-colored areas were not sufficiently correlated to be included in a module; ∼62% of 21,771 transcripts were assigned to a module. Gene ontology (GO) (B) and Kyoto encyclopedia of genes and genomes (KEGG) pathway (C) enrichments for all modules. Modules are ranked by Z-score, highlighting those with the most significant enrichment. Top GO/KEGG terms are listed for each module. Table S2 provides a list of transcript membership for each module. Table S3 contains all significantly enriched (Z-score > 3) GO/KEGG terms for the modules. BCAA, branched-chain amino acid; ECM, extracellular matrix.
Highly correlated transcripts are often associated with common physiological or biochemical processes (Carlson et al. 2006; Gargalovic et al. 2006; Ghazalpour et al. 2006; Horvath et al. 2006; Keller et al. 2008). We performed gene set enrichment analysis (Newton et al. 2007) on each module to determine if any genes in the modules are associated with shared functional annotations. We will refer to modules by both a color key identifier and by the most common GO and/or KEGG functional annotations (Figure S8). Among the 41 modules identified, all but three were significantly enriched with one or more GO terms (Figure 6B) and/or KEGG pathways (Figure 6C). Table S3 lists the top enrichment terms for all modules; however, we note that not all genes in modules are associated with the functional annotations.
To determine the potential physiological significance of the islet modules, we asked if the modules were correlated with the diabetes-related phenotypes measured in the DO mice. For each module, we first computed a ME, in a similar fashion to PC1, to describe the pattern of transcript abundance among all the DO mice. The average percent variance explained by the MEs was 38 and ranged from ∼70 (mediumpurple3) to ∼24 (blue) (Table S4). The percent variance explained by the ME for transcripts that were not assigned to a module (transcripts in gray) was ∼2, demonstrating the highly coordinate nature of transcripts within the gene modules. Transcript abundance within 16 modules showed a highly significant difference between the sexes (Figure S9 and Table S5).
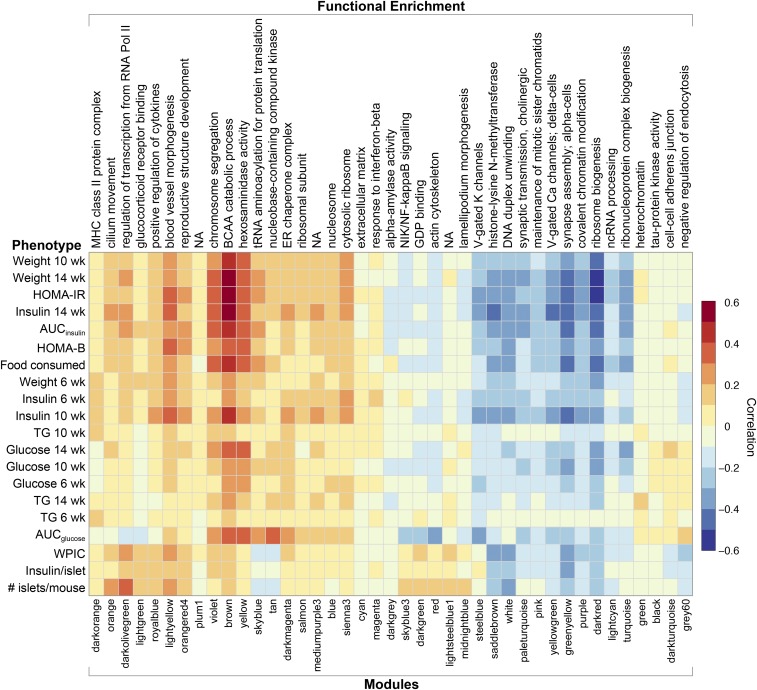
Due to the strong influence of sex on many of the physiological phenotypes (Figure 1) and module transcripts, we computed the Spearman’s nonparametric correlation between modules and traits, after adjusting for sex and DO breeding generation as covariates. This allowed us to determine which modules correlate with specific physiological phenotypes without sex as a confounding variable. Some of the physiological traits display significant correlations among themselves (Figure S2) and, as a result, several modules were significantly correlated with more than one physiological trait (Figure 7). We adjusted the P-value for the Spearman correlations using the Benjamini and Yekutieli method (Reiner et al. 2003) and found that an adjusted P-value of 0.05 corresponded to a correlation of ∼0.15. Here, we highlight features of the most relevant modules and for each, ask: (1) are the modules correlated with physiological phenotypes, (2) are they enriched for annotations suggesting that (some of) the genes may be functionally related to diabetes, and (3) do the modules demonstrate genetic regulation?
Figure 7.
Correlation between islet gene modules and physiological phenotypes. Heat map illustrates the Spearman’s nonparametric correlation between islet MEs and diabetes-related physiological phenotypes, after adjustment for sex and batch. Module names are shown along the bottom axis, and top-enriched GO terms along the top axis. Modules are ordered by unsupervised hierarchical clustering, resulting in them grouping by correlation patterns to the physiological traits. Red, positive correlation; blue negative correlation. AUC, area under the curve; GO, gene ontology; HOMA-B, homeostatic model assessment of pancreatic beta-cell function; HOMA-IR, homeostatic model assessment of insulin resistance; MEs, module eigengenes; wk, week; WPIC, whole-pancreas insulin content; NA, not applicable.
Modules associated with ribosome biogenesis (darkred), branched-chain amino acid (BCAA) catabolism (brown), cell cycle (violet), and hexosaminidase activity (yellow) were most strongly correlated with body weight, plasma insulin, AUCinsulin, HOMA-B, and HOMA-IR (Figure 7). The δ-cell module (yellowgreen) module, also correlated with these traits, is discussed below. The correlations are strongest between these modules and the later time points of physiological traits that were measured at multiple times (e.g., insulin at 14 weeks). The brown, violet, and yellow modules all share a significant QTL on Chr 11 at ∼70 Mb that is concordant with the Chr 11 hotspot and with a QTL for HOMA-IR (Figure 8). The brown module is enriched for BCAA catabolism genes including Bckhda, Bckhdb, and Bckdk. The latter is one of several genes in the module that are mediated by Sat2. The violet module is enriched for the GO term “chromosome segregation” and the KEGG pathway term “cell cycle.” It contains 145 transcripts, including many that are known to play a role in the regulation of the cell cycle: the cyclins A2, B2, and B1, Asf1b, Pbk, Aurkb, Cdk1, Bub1, Bub1b, Mcm3, Mcm5, Plk1, Top2a, and Ube2c, among others. Transcripts in the violet module largely overlap with an islet cell cycle-related module that we previously identified when comparing diabetes-resistant (B6) and diabetes-susceptible (BTBR) mouse strains (Keller et al. 2008). The violet module has a second QTL on Chr 17 at ∼70 Mb, which is concordant with the cell cycle module discovered in the B6 × BTBR cross. The darkred module has a distinct QTL on Chr 17 at ∼34 Mb, which is concordant with QTL for weight at 6 and 10 weeks of age.
Figure 8.
Genetic architecture of islet gene modules. Mod-QTL are illustrated for all modules, colored by LOD score (red = high and yellow = low). Top-enriched GO term for each module is shown; three modules did not significantly enrich for any terms (plum1, lightsteelblue1, and mediumpurple3). Modules are ordered by unsupervised clustering such that traits with similar LOD profiles are grouped together (e.g., Chr 11). Table S7 lists all mod-QTL, their LOD scores, and genomic positions in mouse and human. BCAA, branched-chain amino acid; Chr, chromosome; GO, gene ontology; mod-QTL, module-QTL; NA, not applicable.
Several modules show enrichment for genes with physiological functions that are relevant to the pathology of T2D but did not comap to hotspots or correlate with physiological traits. The darkmagenta module is enriched for the GO term “ER chaperone complex” and the KEGG pathway “protein processing in ER.” It includes transcripts for Pdia4, Calr, Pdia3, Hspa5, P4hb, Hsp90b1, Sec23b, Sec61a1, Dnajb11, and Pdia6, all of which are critical for ER homeostasis. The cyan module contained 261 transcripts and was enriched for the GO “extracellular matrix” and KEGG “ECM-receptor interaction” terms. The skyblue3 module is enriched for GO term “NIK/NF-κ B signaling” and the KEGG pathway term “apoptosis.” It contains, Nfkb1, Nfkb2, Tnf, Traf2, Bid, Casp3, Fas, Il1a, Nfkb1, Nfkbia, and Trp53. The lightgreen module is enriched for “glucocorticoid receptor binding” and “Pancreatic cancer”; Nr4a1, Nr4a3, Nr4a2, Stat3, Bcl2l1, Jak1, Pik3r1, Rac1, and Tgfb3.
Three modules (plum1, lightsteelblue, and mediumpurple3) with the strongest module-QTL (mod-QTL) did not enrich for functional annotations. The plum1 module consists of 91 transcripts, all but one of which correspond to genes located on proximal Chr 14, and mapped to Chr 14 at ∼7.5 Mbp, LOD > 90 (Figure 8). Transcripts within plum1 derive from a large region of the Chr (3–7.5 Mb) that is rich in repeated gene families, harbors segmental variations across the DO founder strains, and is a recombination coldspot (Morgan et al. 2017). Eighty-two of the 91 transcripts in the plum1 module are gene models (e.g., GM10409, GM3020, and GM26552, etc.) or predicted genes that do not have associated physiological annotations. The correlated expression of these transcripts reflects an unusually elevated level of linkage disequilibrium across the region. The lightsteelblue1 module consists of 42 transcripts, 40 of which are clustered in another recombination cold spot with segmental variations located on Chr 2 at 177 Mbp; lightsteelblue1 mapped to Chr 2 at ∼175 Mbp, LOD = 89. The genes in the lightsteelblue module are mostly gene models with little functional annotation. The MEs for either plum1 or lightsteelblue1 were not significantly correlated with any of the physiological traits (Figure 7). The mediumpurple3 module also maps to spatially clustered genes, most of which are located on distal Chr 6; mod-QTL at ∼150 Mbp, LOD ∼9. The mediumpurple3 gene clusters are not in recombination coldspots and encode functional RNAs, including the nuclear 5S ribosomal RNA and several small nuclear RNAs.
There were nine modules with functional enrichment but no significant or suggestive mod-QTL. These include the largest module, turquoise, with 1494 transcripts, many involved in mRNA processing and ribosome function, as well as 12 of the 14 mitochondrial-encoded transcripts. The darkgrey module consists of 175 transcripts enriched for “α-amylase activity,” and “pancreatic secretion,” and includes Amy1, Amy2a5, Amy2a4, Amy2a3, and Amy2a2. These transcripts are typically expressed in acinar tissue and most likely reflect residual contamination in isolated islet preparations, and thus we would not expect to find it to be controlled by genetic factors.
Modules enriched in α-cell- and δ-cell-specific transcripts
Two modules were enriched with transcripts selectively expressed in α-cells or δ-cells. These could reflect either increased activity or increased cell numbers relative to the predominant β-cell component of islets. The greenyellow module consists of 387 transcripts, including Gcg, Irx1, Irx2, Arx, Mafb, Ttr, Gria3, and Sstr2. These transcripts have been previously associated with selective expression in α-cells (Dorrell et al. 2011; Ackermann et al. 2016; DiGruccio et al. 2016; Xin et al. 2016; Lawlor et al. 2017). The yellowgreen module contains 109 transcripts, including many that have been linked to selective expression in δ-cells; Sst, Hhex, Rbp4, and Ghsr (Dorrell et al. 2011; Ackermann et al. 2016; DiGruccio et al. 2016; Xin et al. 2016; Lawlor et al. 2017).
In addition to the cell type-specific nature of the yellowgreen (δ-cells) and greenyellow (α-cells) modules, both are significantly enriched with one or more GO terms and KEGG pathways (Figure 6, B and C, respectively). The yellowgreen module is enriched for “voltage-gated Ca2+ channel activity” and includes transcripts for several Ca2+ channels; Cacna1e, Cacna1g, Cacna2d3, Cacna2d2, Cacna1h, Cacna1i, and Trpa1. Greenyellow is enriched for “synapse assembly” (Cbln2, Ephb2, Lrrn3, Nrxn2, Nrxn3, Oxtr, Ptprd, Wnt5a, Flrt3, Lrrtm1, Slitrk1, Shank2, Nrg1, Lrrtm3, Lrp4, Adgrb2, and Lrrc4b), which may be related to the control of glucagon secretion by the central nervous system.
Next, we asked if a module is similarly enriched for known β-cell genes. In contrast to the α-cell- and δ-cell-enriched modules, key β-cell genes were identified in separate modules, including Ins1 and Ins2 (yellow), Nkx6.1 (black), Pdx-1 and Glp1r (grey60), Slc30a8 and Slc2a2 (pink), Mafa (lightcyan), Ucn3 (red), and G6pc2 and Iapp (brown). These results suggest that unlike known β-cell-associated transcripts, those in α-cells and δ-cells are highly coordinate in their expression pattern among the islets of the DO mice. The β-cell transcripts do not show this same coordinate regulation, suggesting that they may reflect a greater degree of heterogeneity within the β-cell population, as has been previously reported for islets collected from different regions of the pancreas in mice following dietary challenge (Ellenbroek et al. 2013), as well for islets collected from nondiabetic vs. T2D human subjects (Dorrell et al. 2016; Xin et al. 2016; Lawlor et al. 2017).
Transcripts within a module are highly correlated, suggesting that this coordinate regulation is maintained across the DO islets that were profiled. Further, the α-cell and δ-cell modules showed significant QTL, implying that, at least in part, some of this coordinate regulation is driven by factors present at these loci; e.g., Ptprz1 for the δ-cell mod-QTL on Chr 6. In contrast to the α-cell and δ-cell modules, β-cell-specific transcripts were scattered among several different modules, suggesting that the β-cell-specific transcripts are not as tightly coordinated. We speculate that the discordant regulation of these genes among the DO mice reflects different compensatory responses. There was a > 100-fold range of plasma insulin values among the DO mice, presumably reflecting differences in insulin production in response to diet-induced insulin resistance. It is possible that these differences reflect greater changes in the β-cell transcriptome, rather than in α-cells or δ-cells. Further, it may be possible that islets collected from a single mouse harbor greater variability in β-cell transcripts than non-β-cell transcripts.
The α-cell- (greenyellow) and δ-cell-enriched (yellowgreen) modules each showed a decline in transcript abundance in male vs. female mice (Figure S9). These results suggest that the cellular composition of the DO islets—or the proportion of α, δ, and β-cells—may be different between the sexes, with islets from males having fewer non-β-cell types than females. A similar sex difference in the proportion of α-cells and δ-cells was reported for islets evaluated from female vs. male a backcross progeny of C57BL/6J × Mus spretus-F1 × C57BL/6J; PMID: 16353357 (BSB) mice (Fisler et al., 1993 Slavin et al. 2010). A complete list of modules demonstrating differences between the sexes is provided in Table S5.
We identified distinct mod-QTL for the modules associated with α-cells (greenyellow) and δ-cells (yellowgreen) (Figure 8). The δ-cell module mapped to four distinct loci; Chr 4 at ∼3.2 Mbp, Chr 6 at ∼4.8, and Chr 18 at ∼5.4 Mbp and ∼44.2 Mbp. The α-cell module showed suggestive mod-QTL on Chr 11 at ∼16.8 Mbp and Chr 15 at 60 Mbp. Unlike the α-cell and δ-cell modules, we did not identify any mod-QTL for the acinar-enriched module (darkgrey). These results suggest that islet cell composition may be under distinct genetic regulation, whereas the acinar component of the isolated islets is driven by nongenetic influences; e.g., the efficiency of the collagenase digestion of the pancreas prior to islet isolation.
An interactive web interface enables browsing of mouse islet eQTL
We have constructed a web interface that allows the user to quickly determine if a transcript shows genetic regulation in the DO islets (http://churchill-lab.jax.org:21010/). For example, entering the gene symbol Tcf7l2, a locus that is strongly associated with diabetes in human GWAS, into the search window, results in a genome-wide LOD profile that identifies two significant (P < 0.05) eQTL; Chr 13 at ∼112.5 Mbp and Chr 19 at ∼55.4 Mbp (Figure S10). The gene for Tcf7l2 is located on Chr 19 at ∼55.7 Mbp. The allele dependence for the local eQTL (Figure S11A) is driven by alleles from CAST (low) and B6 (high), whereas the distal eQTL (Figure S11B) is associated with NZO (high), suggesting that the genetic dependence of each of the eQTL for Tcf7l2 is distinct.
Mediation shows that the local eQTL for Tcf7l2 is, as expected, driven by Tcf7l2 expression (Figure S12A), whereas the distal eQTL appears to be mediated by the expression of Il6st (Figure S12B), the driver predicted from our mediation analysis for the Chr 13 eQTL hotspot (Figure S4). The transcript for Tcf7l2 is included in the royalblue module, which was enriched for the GO “positive regulation of cytokine production” (Figure 6A) and KEGG “Toll-like receptor signaling pathway” terms (Figure 6B), was positively correlated with plasma insulin at 10 weeks of age (Figure 7) and mapped most strongly to Chr 13 at ∼111.6 Mbp (Figure 8). These tools demonstrate how the user can begin with a gene of interest, determine if it shows distal vs. local genetic regulation, and integrate the findings with the physiological pathways associated with the gene modules, as well as the genetic architecture of the modules and individual transcripts that we have identified. SNPs at each eQTL that are associated with the expression of Tcf7l2 can be downloaded and surveyed for potential consequences, e.g., missense, frameshift, intronic, splice junction, etc. [Figure S13A (Chr 19) and Figure S13B (Chr 13)]. In addition to eQTL for individual transcripts, the website allows the user to survey LOD summaries, allele effect and SNP association plots for physiological phenotypes (e.g., HOMA-B, number of islets per mouse), eQTL hotspots, and MEs. This interactive resource enables a user to discover regulatory loci linked to the expression of any gene expressed in pancreatic islets, and to explore potential candidate drivers and downstream physiological pathways.
Discussion
We present, for the first time, the genetic architecture of gene regulation in pancreatic islets from mice subjected to a Western-style HF/HS diet (45% kcal fat and 34% sucrose). We used DO mice, which contain a high level of genotypic and phenotypic diversity; a level comparable to that present in the human population. The study in humans most closely resembling our mouse experiment was reported by Groop and colleagues (Taneera et al. 2012). They obtained pancreatic islets from 63 human cadaveric donors, including six who had T2D, and interrogated 48 candidate genes located near SNPs associated with T2D in human GWAS. The expression of KCNJ11, WFS1, SLCA2A, JAZF1, and G6PC2 was decreased in islets from the T2D donors. They identified five cis-eQTL; however, none showed significant expression differences in islets from T2D vs. normal subjects.
We quantitated the abundance of > 21,000 mRNA transcripts and identified nearly 19,000 eQTL (genome-wide P < 0.05) (Figure 3). More than 70% of these are local eQTL. In a previous islet genetic study (Tian et al. 2015, 2016), we observed a preponderance of distal eQTL, and believe that the difference is related to a higher LOD threshold in our current study; local eQTL usually have higher LOD scores than distal eQTL. We identified five distal eQTL hotspots and applied mediation analysis to identify candidate causal drivers. Two of these drivers are genes known to be involved in monogenic forms of diabetes: HNF4A (MODY1) and PDX1 (MODY4) (Figure 5). Genetic variation in the MODY genes has also been shown to play a role in T2D (Ndiaye et al. 2017).
We chose Pdx1 as a candidate driver at the Chr 5 eQTL hotspot for several reasons: (1) The LOD drop upon mediation against Pdx1 ranked second among the other local eQTL at the locus (Figure S5C); (2) the allele effect pattern for the Pdx1 local eQTL (Figure S5E) was the closest match to that for the Chr5 hotspot PC1 (Figure S5B) with NOD and B6/129 as the high and low alleles, respectively; and (3) the other candidate mediators have very strong local eQTL LOD scores. This is one of the pitfalls of mediation; a strong local eQTL can act as a surrogate for genotype, making it difficult to rule out the eQTL gene as a candidate mediator. The logic of mediation analysis is designed to rule out candidate mediators, so when there are several candidates we consider other evidence, including the allele effects and functional plausibility of the candidate.
In addition to the MODY genes, we identified three novel drivers of eQTL hotspots, Il6st (Chr 13), Sat2 (Chr 11), and Fam83e (Chr 7), and demonstrated associations with diabetes-related traits in human GWAS when we restrict the association testing to the region defined by synteny with the mouse eQTL (Figure 5). Il6st, also known as glycoprotein 130 (gp130), is the β-subunit of the IL-6 cytokine receptor. A recent report showed that Il6st is required for IL-6-mediated activation of STAT3 and stimulation of glucagon secretion (Chow et al. 2014). Further, α-cell-specific deletion of Il6st resulted in protection from streptozotocin-induced diabetes (Chow et al. 2014), suggesting that, in the context of inflammation that accompanies T2D, Il6st may promote hyperglycemia by stimulating glucagon secretion from α-cells. Il6st is in the royalblue module, along with transcripts that are significantly enriched for the GO term “positive regulation of cytokine production.” The FAM83 family consists of eight members, several of which are increased in expression or copy number in various cancers, and are downstream of the phosphatidylinositol three-kinase and epidermal growth factor receptor pathways (Snijders et al. 2017).
Both HOMA-IR, a measure of insulin resistance, and HOMA-B, a measure of insulin production, map to a common locus on Chr 11 at ∼85 Mbp (Figure 2). Several modules also mapped to this locus, including those enriched in cell cycle, BCAA catabolism, and NIK-NF-κ B signaling (Figure 8). Mediation analysis consistently identified Sat2 as the driver gene for this locus (Figure S7). Sat2 encodes Spermidine/Spermine N1-acetyltransferase 2 and is included in the BCAA module. Indeed, Sat2 has been shown to function with p65 as a coactivator of NF-κB (Vogel et al. 2006). Despite its 61% homology with Sat1, Sat2 does not appear to have significant activity toward polyamines. Instead, it catalyzes the transfer of an acetyl group to thialysine residues, forming Nε-thialysine (Coleman et al. 2004).
We measured glucose tolerance in all mice, as well as insulin during the oGTT. Essentially all of the mice were nondiabetic throughout the course of our study, and yet showed a > 100-fold range in plasma insulin, suggesting that the dietary challenge elicited a large variation in the physiological responses required to maintain plasma glucose. Insulin, AUCinsulin, HOMA-IR, and body weight were all positively correlated with a module enriched in BCAA catabolism (Figure 7). These same phenotypes were negatively correlated with ribosome biogenesis. Thus, protein metabolism, synthesis, and turnover were the strongest correlates. These results are consistent with a growing body of evidence linking BCAAs with metabolic disease (Newgard 2012). In contrast, plasma glucose and TGs were not strongly correlated with any of the modules, suggesting that these traits are effectively buffered by compensatory variation in insulin levels.
Mod-QTL have been identified for hepatic gene expression networks that were shown to be highly correlated and to comap with body weight in mice (Ghazalpour et al. 2006; Fuller et al. 2007). Other groups have used the WGCNA approach to identify mod-QTL in liver that are associated with high-density lipoprotein cholesterol (Leduc et al. 2012), or adiposity and hepatic steatosis (Davis et al. 2012), and in heart for cardiac left ventricular mass (Scott-Boyer et al. 2014). Our study is the first to apply the WGCNA mod-QTL approach to pancreatic islets.
Two of the coexpression gene modules that we identified appear to reflect the activity, or relative proportion, of α-cells and δ-cells in the islets. In contrast, no single module was enriched for transcripts selectively expressed in β-cells. This could be an indication that there is more heterogeneity in the β-cell population of islets than there is for α- and δ-cells, which may have been triggered by the metabolic challenge imposed by the HF/HS diet, requiring increased insulin production in some, but not all, mice. The heterogeneity could be due to subtypes of β-cells (Dorrell et al. 2016) or to plasticity of the cells, i.e., their ability to transdifferentiate to other cell types (Jonas et al. 1999; Papizan et al. 2011; Talchai et al. 2012; Wang et al. 2014).
We identified mod-QTL for the δ-cell-enriched module (yellowgreen) (Figure 8) on Chrs 4, 6, and 18. None of the genes most strongly associated with selective expression in δ-cells and included in the yellowgreen module (Hhex, Sst, Rbp4, and Ghsr) are physically located at these QTL. However, mediation analysis for these genes consistently identified Ptprz1 as a candidate driver for the Chr 6 locus and Armc4 as a candidate driver for the Chr 18 locus, suggesting that these genes control the expression of the δ-cell identity module. Ptprz1 is included in the yellowgreen module and is expressed > 10 times higher in δ-cells than α-cells or β-cells (DiGruccio et al. 2016), consistent with it playing a key role δ-cells. Ptprz1 is a receptor for the heparin-binding glycoprotein pleiotrophin (PTN) and has been associated with several cancers, including glioblastoma (Shi et al. 2017) and pancreatic cancer (Xue et al. 2018). In addition to δ-cell-specific transcripts, six genes that encode Ca2+ channels were included in the yellowgreen module, including R-type Ca2+ channels (e.g., Cacna1e), which have been previously linked to glucose-stimulated somatostatin secretion (Zhang et al. 2007).
We identified mod-QTL for the α-cell-specific module (greenyellow) and, as for the δ-cell module, the mod-QTL loci were distinct from the genes that encode the α-cell-specific transcripts. Mediation analysis of the α-cell-specific transcripts did not reveal consistent candidates as driver genes. This α-cell module was enriched for synapse formation, usually associated with the central nervous system, and may reflect the strong neural connection with α-cell function or development.
We compared the chromosomal locations of the QTL for the diabetes-related phenotypes (Figure 9A) and islet module pathways (Figure 9B) with their corresponding locations in the human genome and observed a significant enrichment (P = 0.003) for diabetes-associated GWAS peaks at threshold of −log10 P ≥ 4. This observation supports the hypothesis that the genetic drivers of diabetes risk are concordant across mice and humans. In other words, not only are the same pathways involved, but our findings suggest that genetic variation in the same gene loci is the underlying cause for the difference in diabetes incidence and susceptibility.
Our project allows one to nominate novel loci in the human genome based on associations with diabetes-related phenotypes, or physiological pathways in pancreatic islets identified in mouse. For example, the mod-QTL we identified for the δ-cell module on Chr 6 at ∼4.8 Mbp is syntenic to Chr 7 at ∼94.7 Mbp in human (Figure 9B). Our mediation analysis strongly suggests that Ptprz1 is a driver of this mod-QTL. Using the gene search tool at GWAS Central (http://www.gwascentral.org/generegion), one can then determine if any SNPs have been identified that are proximal to PTPRZ1 and associated to diabetes-related traits. The results show that SNPs associated with HbA1c levels have been reported at this locus (Strachan et al. 2007). Similarly, the cell cycle module (violet) has a mod-QTL on Chr 11 at ∼69.8 Mbp. In human, this region is syntenic to Chr 17 at ∼7.3 Mbp, which is proximal to the gene TMEM102, where SNPs have been identified that are associated with T2D (Sladek et al. 2007). Finally, one can ask if QTL identified for a diabetes-related phenotype in our mouse study highlight a novel locus in human. For example, among the diabetes-related physiological traits in our study, HOMA-B and AUCinsulin demonstrated the strongest genetic signal, whereby both mapped to a locus on Chr 11 at ∼84 Mbp (Figure 9A) with LODs > 8. In human, this region is syntenic to Chr 17 at ∼60.1 Mbp. Interestingly, SNPs have been identified at this locus that are associated glucose levels 2 hr after an oral glucose challenge (Saxena et al. 2010).
We applied a systems genetics approach, which used gene expression traits to interrogate the function of pancreatic islets under a diabetogenic stressor, and identified groups of comapping transcripts as well as coexpression modules. When we performed mediation analysis on the comapping transcripts, we identified strong candidate genes as likely drivers. Modules provide an alternative view of the coordinated expression patterns in the islet cells; functional enrichment analysis attaches biological interpretation to these modules and is often correlated with physiological measures of diabetes-associated traits. Genetic mapping of the MEs reveals multiple QTL peaks with modest LOD scores reflecting heterogeneity in the genetic drivers of the module. Mediation analysis of module QTL was less successful at identifying candidate driver genes; however, mediation of individual genes within the module can be effective. Our web-based analysis tools can support this kind of data exploration.
Acknowledgments
We are grateful for graphical expertise provided by Laura Vanderploeg. The project was supported by grants from the National Institutes of Health (NIH) to A.D.A. (DK-101573 and DK-102948), C.K. (GM-102756 and U54 AI-117924), and G.A.C. and K.W.B. (GM-076483), and the Clinical and Translational Science Award program, through the NIH National Center for Advancing Translational Sciences (grant UL1 TR-000427).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5977459.
Communicating editor: L. McIntyre
Literature Cited
- Ackermann A. M., Wang Z., Schug J., Naji A., Kaestner K. H., 2016. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol. Metab. 5: 233–244. 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert F. W., Kruglyak L., 2015. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 16: 197–212. 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- Barrington W. T., Wulfridge P., Wells A. E., Rojas C. M., Howe S. Y. F., et al. , 2018. Improving metabolic health through precision dietetics in mice. Genetics 208: 399–417. 10.1534/genetics.117.300536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Hastings R. K., Gollapudi S., Free R. C., Brookes A. J., 2014. GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur. J. Hum. Genet. 22: 949–952. 10.1038/ejhg.2013.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings L. K., Florez J. C., 2010. The genetics of type 2 diabetes: what have we learned from GWAS? Ann. N. Y. Acad. Sci. 1212: 59–77. 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Li Y., Tesson B. M., Fu J., Wu C., et al. , 2008. Genetical genomics: spotlight on QTL hotspots. PLoS Genet. 4: e1000232 10.1371/journal.pgen.1000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. R., Zhang B., Fang Z., Mischel P. S., Horvath S., et al. , 2006. Gene connectivity, function, and sequence conservation: predictions from modular yeast co-expression networks. BMC Genomics 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick J. M., Munger S. C., Simecek P., Huttlin E. L., Choi K., et al. , 2016. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534: 500–505. 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow S. Z., Speck M., Yoganathan P., Nackiewicz D., Hansen A. M., et al. , 2014. Glycoprotein 130 receptor signaling mediates alpha-cell dysfunction in a rodent model of type 2 diabetes. Diabetes 63: 2984–2995. 10.2337/db13-1121. [DOI] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano R., Miskimen K. L., Bryson B. L., Foy C. R., Bartel C. A., et al. , 2014. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol. Cancer Res. 12: 1156–1165. 10.1158/1541-7786.MCR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C. S., Stanley B. A., Jones A. D., Pegg A. E., 2004. Spermidine/spermine-N1-acetyltransferase-2 (SSAT2) acetylates thialysine and is not involved in polyamine metabolism. Biochem. J. 384: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover W. J., 1999. Practical Nonparametric Statistics. Wiley, New York. [Google Scholar]
- Davis R. C., van Nas A., Castellani L. W., Zhao Y., Zhou Z., et al. , 2012. Systems genetics of susceptibility to obesity-induced diabetes in mice. Physiol. Genomics 44: 1–13. 10.1152/physiolgenomics.00003.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) Consortium et al. , 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelez V., Sheehan N., 2007. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 16: 309–330. [DOI] [PubMed] [Google Scholar]
- DiGruccio M. R., Mawla A. M., Donaldson C. J., Noguchi G. M., Vaughan J., et al. , 2016. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol. Metab. 5: 449–458. 10.1016/j.molmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Schug J., Lin C. F., Canaday P. S., Fox A. J., et al. , 2011. Transcriptomes of the major human pancreatic cell types. Diabetologia 54: 2832–2844. 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Schug J., Canaday P. S., Russ H. A., Tarlow B. D., et al. , 2016. Human islets contain four distinct subtypes of beta cells. Nat. Commun. 7: 11756 10.1038/ncomms11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., et al. , 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42: 105–116. 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P. T., Birney E., Huber W., 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4: 1184–1191. 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek J. H., Tons H. A., de Graaf N., Loomans C. J., Engelse M. A., et al. , 2013. Topologically heterogeneous beta cell adaptation in response to high-fat diet in mice. PLoS One 8: e56922 10.1371/journal.pone.0056922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajans S. S., Bell G. I., Polonsky K. S., 2001. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 345: 971–980. [DOI] [PubMed] [Google Scholar]
- Fisler J. S., Warden C. H., Pace M. J., Lusis A. J. 1993. BSB: a new mouse model of multigenic obesity. Obes Res. 1: 271–80. [DOI] [PubMed] [Google Scholar]
- Flannick J., Florez J. C., 2016. Type 2 diabetes: genetic data sharing to advance complex disease research. Nat. Rev. Genet. 17: 535–549. 10.1038/nrg.2016.56. [DOI] [PubMed] [Google Scholar]
- Franzen O., Ermel R., Cohain A., Akers N. K., Di Narzo A., et al. , 2016. Cardiometabolic risk loci share downstream cis- and trans-gene regulation across tissues and diseases. Science 353: 827–830. 10.1126/science.aad6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., et al. , 2016. The genetic architecture of type 2 diabetes. Nature 536: 41–47. 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller T. F., Ghazalpour A., Aten J. E., Drake T. A., Lusis A. J., et al. , 2007. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm. Genome 18: 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargalovic P. S., Imura M., Zhang B., Gharavi N. M., Clark M. J., et al. , 2006. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc. Natl. Acad. Sci. USA 103: 12741–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D. M., Svenson K. L., Shabalin A., Wu L. Y., Valdar W., et al. , 2014. Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 4: 1623–1633. 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A., Doss S., Zhang B., Wang S., Plaisier C., et al. , 2006. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B., 2014. Circlize implements and enhances circular visualization in R. Bioinformatics 30: 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Horvath S., Zhang B., Carlson M., Lu K. V., Zhu S., et al. , 2006. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc. Natl. Acad. Sci. USA 103: 17402–17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen M. T., Weisell J., Khomutov A. R., Alhonen L., Vepsalainen J., et al. , 2013. Metabolism of triethylenetetramine and 1,12-diamino-3,6,9-triazadodecane by the spermidine/spermine-N(1)-acetyltransferase and thialysine acetyltransferase. Drug Metab. Dispos. 41: 30–32. 10.1124/dmd.112.047274. [DOI] [PubMed] [Google Scholar]
- Jason F., Fuchsberger C., Mahajan A., Teslovich T. M., Agarwala V., et al. , 2017. Sequence data and association statistics from 12,940 type 2 diabetes cases and controls. Sci. Data 4: 170179 10.1038/sdata.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas J. C., Sharma A., Hasenkamp W., Ilkova H., Patane G., et al. , 1999. Chronic hyperglycemia triggers loss of pancreatic beta cell differentiation in an animal model of diabetes. J. Biol. Chem. 274: 14112–14121. [DOI] [PubMed] [Google Scholar]
- Keller M. P., Choi Y., Wang P., Davis D. B., Rabaglia M. E., et al. , 2008. A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Res. 18: 706–716. 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor N., George J., Bolisetty M., Kursawe R., Sun L., et al. , 2017. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 27: 208–222. 10.1101/gr.212720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., et al. , 2013. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9: e1003118 10.1371/journal.pcbi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc M. S., Blair R. H., Verdugo R. A., Tsaih S. W., Walsh K., et al. , 2012. Using bioinformatics and systems genetics to dissect HDL-cholesterol genetics in an MRL/MpJ x SM/J intercross. J. Lipid Res. 53: 1163–1175. 10.1194/jlr.M025833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon D. P., Fairchild A. J., Fritz M. S., 2007. Mediation analysis. Annu. Rev. Psychol. 58: 593–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A. K., Hivert M. F., Scott R. A., Grimsby J. L., Bouatia-Naji N., et al. , 2012. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44: 659–669. 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews D. R., Hosker J. P., Rudenski A. S., Naylor B. A., Treacher D. F., et al. , 1985. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419. [DOI] [PubMed] [Google Scholar]
- Millstein J., Zhang B., Zhu J., Schadt E. E., 2009. Disentangling molecular relationships with a causal inference test. BMC Genet. 10: 23 10.1186/1471-2156-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohlke K. L., Boehnke M., 2015. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum. Mol. Genet. 24: R85–R92. 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Fu C. P., Kao C. Y., Welsh C. E., Didion J. P., et al. , 2015. The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6: 263–279. 10.1534/g3.115.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A. P., Gatti D.M., Najarian M. L., Keane T. M., Galante R. J., et al. , 2017. Genetics. 206: 603–619. 10.1534/genetics.116.197988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segre A. V., et al. , 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger S. C., Raghupathy N., Choi K., Simons A. K., Gatti D. M., et al. , 2014. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics 198: 59–73. 10.1534/genetics.114.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndiaye F. K., Ortalli A., Canouil M., Huyvaert M., Salazar-Cardozo C., et al. , 2017. Expression and functional assessment of candidate type 2 diabetes susceptibility genes identify four new genes contributing to human insulin secretion. Mol. Metab. 6: 459–470. 10.1016/j.molmet.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto E. C., Keller M. P., Attie A. D., Yandell B. S., 2010. Causal graphical models in systems genetics: a unified framework for joint inference of causal network and genetic architecture for correlated phenotypes. Ann. Appl. Stat. 4: 320–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto E. C., Broman A. T., Keller M. P., Attie A. D., Zhang B., et al. , 2013. Modeling causality for pairs of phenotypes in system genetics. Genetics 193: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard C. B., 2012. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 15: 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M. A., Quintana F. A., Den Boon J. A., Sengupta S., Ahlquist P., 2007. Random-set methods identify distinct aspects of the enrichment signal in gene-set analysis. Ann. Appl. Stat. 1: 85–106. [Google Scholar]
- Ng M. C., Shriner D., Chen B. H., Li J., Chen W. M., et al. , 2014. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 10: e1004517 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizan J. B., Singer R. A., Tschen S. I., Dhawan S., Friel J. M., et al. , 2011. Nkx2.2 repressor complex regulates islet beta-cell specification and prevents beta-to-alpha-cell reprogramming. Genes Dev. 25: 2291–2305. 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks B. W., Nam E., Org E., Kostem E., Norheim F., et al. , 2013. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17: 141–152. 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R. B., Groop L., 2015. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 6: 87–123. 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim R. J., Welch R. P., Sanna S., Teslovich T. M., Chines P. S., et al. , 2010. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghupathy N., Choi K., Vincent M. J., Beane G. L., Sheppard K., et al. , 2018. Hierarchical analysis of RNA-seq reads improves the accuracy of allele-specific expression. Bioinformatics. 1–8. 10.1093/bioinformatics/bty078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Yekutieli D., Benjamini Y., 2003. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375. [DOI] [PubMed] [Google Scholar]
- Sanghera D. K., Blackett P. R., 2012. Type 2 diabetes genetics: beyond GWAS. J. Diabetes Metab. 3: 6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R., Hivert M. F., Langenberg C., Tanaka T., Pankow J. S., et al. , 2010. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 42: 142–148. 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E. E., Lamb J., Yang X., Zhu J., Edwards S., et al. , 2005. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37: 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. A., Chu A. Y., Grarup N., Manning A. K., Hivert M. F., et al. , 2012a No interactions between previously associated 2-hour glucose gene variants and physical activity or BMI on 2-hour glucose levels. Diabetes 61: 1291–1296. 10.2337/db11-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. A., Lagou V., Welch R. P., Wheeler E., Montasser M. E., et al. , 2012b Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 44: 991–1005. 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Boyer M. P., Praktiknjo S. D., Llamas B., Picard S., Deschepper C. F., 2014. Dual linkage of a locus to left ventricular mass and a cardiac gene co-expression network driven by a chromosome domain. Front. Cardiovasc. Med. 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ping Y. F., Zhou W., He Z. C., Chen C., et al. , 2017. Tumour-associated macrophages secrete pleiotrophin to promote PTPRZ1 signalling in glioblastoma stem cells for tumour growth. Nat. Commun. 8: 15080 10.1038/ncomms15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih D. Q., Stoffel M., 2001. Dissecting the transcriptional network of pancreatic islets during development and differentiation. Proc. Natl. Acad. Sci. USA 98: 14189–14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinasac D. S., Riordan J. D., Spiezio S. H., Yandell B. S., Croniger C. M., et al. , 2016. Genetic control of obesity, glucose homeostasis, dyslipidemia and fatty liver in a mouse model of diet-induced metabolic syndrome. Int. J. Obes. 40: 346–355. 10.1038/ijo.2015.184. [DOI] [PubMed] [Google Scholar]
- Sladek R., Rocheleau G., Rung J., Dina C., Shen L., et al. , 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885. [DOI] [PubMed] [Google Scholar]
- Slavin B. G., Zarow C., Warden C. H., Fisler J. S., 2010. Histological, immunocytochemical, and morphometrical analyses of pancreatic islets in the BSB mouse model of obesity. Anat. Rec. (Hoboken) 293: 108–116. 10.1002/ar.21019. [DOI] [PubMed] [Google Scholar]
- Snijders A. M., Lee S. Y., Hang B., Hao W., Bissell M. J., et al. , 2017. FAM83 family oncogenes are broadly involved in human cancers: an integrative multi-omics approach. Mol. Oncol. 11: 167–179. 10.1002/1878-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., et al. , 2010. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes 59: 3229–3239. 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiezio S. H., Amon L. M., McMillen T. S., Vick C. M., Houston B. A., et al. , 2014. Genetic determinants of atherosclerosis, obesity, and energy balance in consomic mice. Mamm. Genome 25: 549–563. 10.1007/s00335-014-9530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan D. P., Rudnicka A. R., Power C., Shepherd P., Fuller E., et al. , 2007. Lifecourse influences on health among British adults: effects of region of residence in childhood and adulthood. Int. J. Epidemiol. 36: 522–531. [DOI] [PubMed] [Google Scholar]
- Svenson K. L., Von Smith R., Magnani P. A., Suetin H. R., Paigen B., et al. , 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 102: 2369–2378. [DOI] [PubMed] [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talchai C., Xuan S., Lin H. V., Sussel L., Accili D., 2012. Pancreatic beta cell dedifferentiation as a mechanism of diabetic beta cell failure. Cell 150: 1223–1234. 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera J., Lang S., Sharma A., Fadista J., Zhou Y., et al. , 2012. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 16: 122–134. 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Taneera J., Storm P., Groop L., 2014. Downregulation of type II diabetes mellitus and maturity onset diabetes of young pathways in human pancreatic islets from hyperglycemic donors. J. Diabetes Res. 2014: 237535 10.1155/2014/237535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Keller M. P., Oler A. T., Rabaglia M. E., Scheuler K. L., et al. , 2015. Identification of the bile acid transporter Slco1a6 as a candidate gene that broadly affects gene expression in mouse pancreatic islets. Genetics 201: 1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Keller M. P., Broman A. T., Kendziorski C., Yandell B. S., et al. , 2016. The dissection of expression quantitative trait locus hotspots. Genetics 202: 1563–1574. 10.1534/genetics.115.183624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Bunt M., Manning Fox J. E., Dai X., Barrett A., Grey C., et al. , 2015. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 11: e1005694 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney A., Scott L. J., Welch R. P., Erdos M. R., Chines P. S., et al. , 2017. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl. Acad. Sci. USA 114: 2301–2306. 10.1073/pnas.1621192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel N. L., Boeke M., Ashburner B. P., 2006. Spermidine/spermine N1-acetyltransferase 2 (SSAT2) functions as a coactivator for NF-kappaB and cooperates with CBP and P/CAF to enhance NF-kappaB-dependent transcription. Biochim. Biophys. Acta 1759: 470–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., York N. W., Nichols C. G., Remedi M. S., 2014. Pancreatic beta cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 19: 872–882. 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M. A., Kim K., Kliebenstein D. J., van Leeuwen H., Michelmore R. W., et al. , 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Y., Kim J., Okamoto H., Ni M., Wei Y., et al. , 2016. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 24: 608–615. 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- Xue J., Zhu W., Song J., Jiao Y., Luo J., et al. , 2018. Activation of PPARα by clofibrate sensitizes pancreatic cancer cells to radiation through the Wnt/β-catenin pathway. Oncogene 37: 953–962. [DOI] [PubMed] [Google Scholar]
- Yao C., Joehanes R., Johnson A. D., Huan T., Liu C., et al. , 2017. Dynamic role of trans regulation of gene expression in relation to complex traits. Am. J. Hum. Genet. 100: 571–580. 10.1016/j.ajhg.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L. G., He Q. Y., 2015. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31: 2382–2383. 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zambelli F., Pesole G., Pavesi G., 2009. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 37: W247–W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Horvath S., 2005. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4: Article17. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bengtsson M., Partridge C., Salehi A., Braun M., et al. , 2007. R-type Ca(2+)-channel-evoked CICR regulates glucose-induced somatostatin secretion. Nat. Cell Biol. 9: 453–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequence reads have been deposited in the Sequence Read Archive (SRP125176). Phenotypes, genotypes, and quantified gene expression data have been deposited with Dryad (doi:10.5061/dryad.pj105; data files: Attie Islet eQTL data). The data are also accessible through an interactive web-based analysis tool that will allow users to replicate the analyses reported here, including genetic mapping and mediation analysis (http://churchill-lab.jax.org/qtl/islet/DO378). Software for this interactive tool is maintained at https://github.com/churchill-lab/qtlviewer and https://github.com/churchill-lab/qtlapi. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.5977459.