Abstract
Cellular membranes are laterally organized into domains of distinct structures and compositions by the differential interaction affinities between various membrane lipids and proteins. A prominent example of such structures are lipid rafts, which are ordered, tightly packed domains that have been widely implicated in cellular processes. The functionality of raft domains is driven by their selective recruitment of specific membrane proteins to regulate their interactions and functions; however, there have been few general insights into the factors that determine the partitioning of membrane proteins between coexisting liquid domains. In this work, we used extensive coarse-grained and atomistic molecular dynamics simulations, potential of mean force calculations, and conceptual models to describe the partitioning dynamics and energetics of a model transmembrane domain from the linker of activation of T cells. We find that partitioning between domains is determined by an interplay between protein-lipid interactions and differential lipid packing between raft and nonraft domains. Specifically, we show that partitioning into ordered domains is promoted by preferential interactions between peptides and ordered lipids, mediated in large part by modification of the peptides by saturated fatty acids (i.e., palmitoylation). Ordered phase affinity is also promoted by elastic effects, specifically hydrophobic matching between the membrane and the peptide. Conversely, ordered domain partitioning is disfavored by the tight molecular packing of the lipids therein. The balance of these dominant drivers determines partitioning. In the case of the wild-type linker of activation of T cells transmembrane domain, these factors combine to yield enrichment of the peptide at Lo/Ld interfaces. These results define some of the general principles governing protein partitioning between coexisting membrane domains and potentially explain previous disparities among experiments and simulations across model systems.
Introduction
Mammalian membranes are laterally heterogeneous, being partitioned into a variety of lateral domains by a myriad of protein- and lipid-mediated interactions. One example of such partitioning is the proposed self-aggregation of certain membrane lipids into transient, relatively ordered membrane domains, termed “lipid rafts.” (1, 2) The prevailing model for the physical and chemical principles underlying raft formation is the coexistence of Lo (raft-analogous) and Ld (nonraft) domains in various model membranes, including planar lipid bilayers (3, 4, 5), lipid monolayers (6, 7), giant unilamellar vesicles (GUVs) (4, 8, 9), and giant plasma membrane vesicles (GPMVs) (10, 11). The composition of the former three synthetic model membranes can be precisely tuned for studying the roles of specific membrane components. In contrast, GPMVs are isolated directly from cellular membranes, providing a more physiological, though less defined, model to study the behavior of biological assemblies. These investigations have converged on a common picture of Lo domains that enrich sphingolipids, saturated phospholipids, and cholesterol and exclude more unsaturated lipids (1, 2, 12) and most proteins (13). These compositional differences lead to differences in lipid acyl chain order, which in turn result in significant differences in lipid packing (14). These general trends are maintained across model systems, though the differences between phases appear to generally be smaller in GPMVs compared to synthetic bilayers (1, 11, 15, 16).
Because of the differences in lipid compositions and packing of Lo and Ld domains, it should be expected that transmembrane proteins will interact differently with Lo versus Ld membranes, leading to preferential domain affinities for membrane proteins. Although there have been sporadic investigations into the question of membrane protein partitioning between coexisting domains (13, 17, 18, 19, 20), there remain few generalizable insights. A common model system for studying protein partitioning between coexisting membrane domains is the single-pass transmembrane adaptor protein linker for activation of T cells (LAT), which is critical for T-cell-antigen-receptor-mediated signaling (21, 22). The palmitoylated transmembrane domain (TMD) of LAT (tLAT) partitions into the ordered (raft) phase of cell-derived GPMVs as a function of its palmitoylation, hydrophobic length, and TMD surface area (13, 23, 24, 25, 26). Palmitoylation has been widely implicated in ordered domain affinity (27), whereas the other two structural factors have been characterized more recently but appear to be widely applicable to single-pass transmembrane proteins (25, 26). Interestingly, the TMD alone is sufficient to recapitulate the raft affinity of the whole protein, suggesting that at least for this protein, raft partitioning is determined entirely by the TMD (26). In contrast to findings in GPMVs, experiments with GUVs have found that tLAT is distinctly excluded from the ordered phase (28). Similarly, coarse-grained molecular dynamics (MD) simulations with planar lipid bilayers showed a consistent exclusion of the tLAT from the ordered domains but with a notable enrichment around the domain boundary (29, 30). The explanations for these disparities remain unclear, as does the overall energetics governing the domain partitioning of LAT and all other proteins.
In this work, we performed extensive free MD simulations and umbrella-sampling calculations to reveal the membrane partitioning energetics of tLAT in three-component biomimetic planar lipid bilayers. Free MD simulations revealed that palmitoylation and hydrophobic length were important for preferential interactions between the tLAT and ordered phase lipids, fully consistent with experiments in biological membranes (13, 23, 24, 26). However, we also observed enrichment of TMD peptides at the Lo/Ld interface. These observations were confirmed by umbrella sampling, which we used to calculate the overall free-energy profiles for partitioning. We synthesize these results into a conceptual model that posits that the relative balance between preferential protein-lipid interactions and interdomain packing differences is the critical driver of protein partitioning between coexisting domains. These observations may explain disparities between simulations and experimental measurements in various model systems in addition to defining the determinants of Lo/Ld partitioning energetics for transmembrane peptides.
Methods
Coarse-grained MD simulations
Coarse-grained models (31, 32, 33), which allow a much longer timescale and larger length-scale for MD simulations, have been widely used to study the dynamics of lipids and proteins in model membranes. In this work, we used one of these popular coarse-grained models, Martini (version 2.1) (33, 34), to investigate the membrane partitioning dynamics of tLAT in phase-separated membranes. 2000 lipids, including 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-diarachidonoyl-sn-glycero-3-phosphocholine (DAPC), and cholesterol (Chol), as well as water and salt ions (Na+ & Cl−), were used for simulations with the box dimension of 10.1 × 10.1 × 11.4 nm.
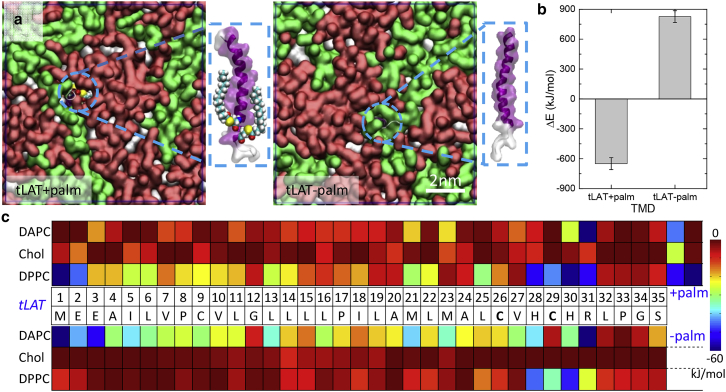
In this work, wild-type tLAT and a shortened mutant (Δ6endo) were used (Fig. 1 a). Martini simulations of the LAT transmembrane peptide and its behavior in Lo/Ld systems have been previously described (29, 30). In the Martini protein model, both bead types and bond/angle/dihedral parameters of each residue are determined by the secondary structures of the protein (34). Despite significant investigations into LAT (UniProt Knowledgebase: O43561, http://www.uniprot.org/uniprot/O43561), no crystal structures are available. To obtain Martini parameters for tLAT and its variants, we predicted their secondary structures in the following steps: 1) predict the peptide structure with the Robetta webserver (35) and 2) evaluate the structural variation of peptides in membranes using 100-ns-scale all-atom MD simulation with the CHARMM36 force field (36, 37, 38). We chose the highly conserved peptide structure (>80%) as well as “coil” for variable residues to build the coarse-grained models of tLAT and its shortened mutant (Fig. 1 b). To obtain palmitoylated tLAT, palmitoyl residues were added to two cysteine residues at the C-terminus (Fig. 1, a and b) using the same parameters as our previous work for H-Ras (17, 20, 39).
Figure 1.
Coarse-grained MD simulations of tLAT partitioning between coexisting liquid membrane phases. (a) The amino acid sequences of tLAT and its truncated mutant (Δ6endo, amino acids in blue were removed) are given. Underlined amino acids represent the positions for palmitoylation. (b) Martini configurations of TMDs studied in this work are shown, with +palm for palmitoyls and −palm without palmitoyls; the backbones of TMDs are colored in blue, side chains in yellow, and palmitoyls in tan. (c) The interaction energy differences between the various TMDs with Lo versus Ld domain lipids are shown; means that TMD interacts preferentially with Lo domain lipids. The mean ± SD is based on a 10-block average over the last 20 μs trajectories. (d) Snapshots of the first frame (t = 0 μs) and the last frame (t = 40 μs) for the coarse-grained systems are shown. DPPC is colored in red, cholesterol in white, and DAPC in green, and the TMD coloring style is the same as in (b). To see this figure in color, go online.
For all classical coarse-grained MD simulations, a cutoff of 1.2 nm was used for van der Waals interactions, and the Lennard-Jones potential was smoothly shifted to zero between 1.0 and 1.2 nm to reduce cutoff noise. For electrostatic interactions, the coulombic potential was smoothly shifted from 0 to 1.2 nm, with a cutoff at 1.2 nm. The default relative dielectric constant (15) of the force field was used in the simulations (33). Peptides, lipids, and water and ions were coupled separately to V-rescale heat baths (40) at T = 298 K (coupling constant τ = 1 ps). The systems were simulated at 1 bar pressure using a semiisotropic Parrinello-Rahman pressure coupling scheme (41) with a coupling constant τ = 5 ps and compressibility of 3 × 10−4 bar−1. The nonbonded interaction neighbor list was updated every 10 steps with a cutoff of 1.2 nm.
All-atom MD simulations
The CHARMM36 force field (36, 37, 38) was used to simulate the behavior of wild-type tLAT with and without palmitoylation in a DPPC/DAPC/Chol bilayer. Grid-based energy correction maps were used to refine conformational properties of protein backbones (42). The initial conformation of wild-type tLAT without palmitoylation was obtained via the Robetta webserver (35), and then the two all-atom MD simulation systems were generated using CHARMM-GUI (43, 44). The Lennard-Jones potential was smoothly shifted to zero between 1.0 and 1.2 nm, with a cutoff of 1.2 nm to reduce cutoff noise. Particle mesh Ewald electrostatics (45) with a real-space cutoff of 1.2 nm was used. Peptides, lipids, and water and ions were coupled separately to Nose-Hoover heat baths (46, 47) at T = 298 K (coupling constant τ = 1 ps). The systems were simulated at 1 bar pressure using a semiisotropic Parrinello-Rahman pressure coupling scheme (41) with a coupling constant τ = 5 ps and compressibility of 4.5 × 10−5 bar−1. Bonds with H-atoms were constrained with the LINCS algorithm (48). The nonbonded interaction neighbor list was updated every 20 steps with a cutoff of 1.2 nm.
In this work, all MD simulations were run with constant pressure and temperature, as well as leap-frog Verlet algorithm and periodic boundary conditions, using GROMACS 5.0.4 (49). Coarse-grained MD simulations were run for 10 μs (effective time (33) is around 40 μs) with a time step of 20 fs and a trajectory-saving frequency of 200 ps (effective time is used for description of coarse-grained MD simulations in this work). All-atom MD simulations were run for 2 μs with a time step of 2 fs and a trajectory-saving frequency of 20 ps. System snapshots were rendered by Visual Molecular Dynamics (Theoretical and Computational Biophysics Group, University of Illinois at Urbana-Champaign, IL) (50).
To evaluate the interaction polarity of LAT with Lo versus Ld domain lipids, we calculated the residue-based interaction energy (E, containing van der Waals and coulombic interactions) between the peptide and DPPC + Chol (Lo) compared to DAPC (Ld). Then, the relative Lo interaction preference for each amino acid was described by the following equation:
where R = 1 means full Lo domain preference and R = 0 full Ld domain preference.
Umbrella sampling
Umbrella sampling is an enhanced sampling method to calculate relative free energy (formally potential of mean force, PMF) along certain reaction coordinates (51). To get one-dimensional PMF, three steps are needed: 1) a reaction coordinate is chosen; 2) biased simulations are run at different , with constrained by a harmonic potential; and 3) the mean force at each is calculated, and is obtained by integrating along according to . Using the Martini force field, the DPPC/DAPC/Chol bilayer shows very stable phase separation (52), which makes it suitable for the umbrella-sampling simulations performed in this work. First, we constructed a sandwich-model membrane composed of a stripe of Lo domain (DPPC and Chol, ratio: 2:1, initial width: 6.6 nm) at the center, flanked by Ld domains (DAPC, initial width: 11 nm, which is wide enough to avoid peptides’ crossing the box boundary). After a 400-ns equilibrium simulation, the box dimensions of this model membrane are 27.0 × 12.1 × 10.2 nm. As shown in Fig. 3 a, the striped domains are parallel to the y axis of the simulation box. Hence, we chose the center-of-mass (COM) distance between the peptide and the Lo domain as the reaction coordinate for umbrella-sampling simulations. Because cholesterol molecules can flip-flop between the two lipid leaflets and exchange between Lo and Ld domains, only DPPC molecules were used for the calculation of the COM of the Lo domain. 34 independent simulation systems with different (, interval of 0.2 nm) were built for each peptide. Each system was run for 3.2 μs with the pull code of GROMACS 5.0.4 (49), with the first 0.8 μs reserved for the equilibrium (total simulation time: 34 × 3 × 3.2 μs = 326.4 μs). The final free-energy profiles were obtained from analysis of the last 2.4 μs trajectories.
Figure 3.
Energetics of tLAT partitioning via umbrella sampling simulations. (a) The COM distance between a TMD and the Lo domain was chosen as the reaction coordinate for umbrella sampling (upper). Three representative initial configurations for the umbrella sampling (DPPC = red, Chol = white, DAPC = green, TMD same as Fig. 1b; initial x-width of Lo domain is ∼6.6 nm) are shown. (b) Potential of mean force (PMF) profiles for tLAT peptides in Lo domain (0 < < 3.3 nm; shaded red) and Ld domain ( > 3.3 nm; shaded green) are shown. The mean ± SD is from a six-block average over the last 2.4 μs of 3.2 μs. (c) The heat map shows a two-dimensional DPPC localization probability map (deep red = max probability, deep blue = zero probability) based on the last 2.4 μs trajectory of the umbrella-sampling simulation at = 3.2 nm. The black dots represent COM locations of tLAT+palm during this period. The narrow domain boundary and stable location of the peptide is the evidence for good sampling. This particular system is shown because the peptide is at the Lo/Ld interface, which is the most dynamic sample among all umbrella-sampling windows. (d) Top-view snapshots of the 40-μs-scale coarse-grained MD simulation system of tLAT at much higher peptide/lipid ratio (60/1440) are shown. The colors of the molecules are the same as those in Fig. 1d. To see this figure in color, go online.
Results and Discussion
Interfacial accumulation of tLAT in coarse-grained free MD simulations
To investigate the partitioning of LAT in phase-separated model membranes, nine copies of a peptide based on the human tLAT were uniformly placed in randomly distributed phospholipid bilayers (2000 lipids, DPPC/DAPC/Chol: 5/3/2). As previously described (52), this lipid system rapidly (within a few hundred nanoseconds) segregates into stable membrane domains, which also makes it feasible for further investigation with all-atom MD simulations (see below). 40-μs coarse-grained MD simulations indeed yielded the expected separation into stable liquid membrane domains without any notable effect of the presence of peptides (Fig. 1 d). The peptides also adopted a nonrandom distribution, as all three peptides showed an obvious preference for the interface between Lo and Ld domains, fully consistent with previous work from the Marrink group (29, 30).
Previous experimental work has implicated TMD length (26) and palmitoylation (23) as important determinants of ordered phase affinity for tLAT. We probed the effect of these modifications by either shortening the TMD (i.e., removing six amino acids from the cytoplasmic side of the TMD-Δ6endo) or removing the palmitoyl moieties (tLAT−palm). Surprisingly, neither of these major structural changes produced a notable effect on overall peptide localization, as both Δ6endo and tLAT−palm enriched at the domain interface (Fig. 1 d). To ensure that this effect was robust, we also tested peptide partitioning in another membrane model system, DPPC/DLiPC/Chol (DLiPC: 1,2-dilinoleoyl-sn-glycero-3-phosphocholine), which forms much less stable membrane domains (52). Despite the significant differences between the two membrane systems (22), all three peptides tested showed the same interfacial accumulation in both bilayers (Fig. S3). This accumulation of tLAT at the Lo/Ld interface seems to conflict with observations in GUVs (28) and GPMVs (26), which reported preference for Ld or Lo domains, respectively, without any noted enrichment at the domain interface. The energetics of the unexpected interfacial accumulation in MD simulations are discussed below.
Peptide-lipid interactions in simulations are consistent with partitioning in biological membranes
Because the interfacial accumulation of all peptides prevented direct assessment of partitioning by localization, we instead determined the relative affinity of the peptides for ordered versus disordered phase lipids by calculating the interaction energy between the TMDs and Lo versus Ld domain lipids (Fig. 1 c). We observed that the palmitoylated tLAT indeed preferentially interacts with Lo domain lipids (i.e., DPPC and Chol), whereas depalmitoylated tLAT (tLAT−palm) had stronger interactions with Ld domain lipids (DAPC). Similarly, reducing the hydrophobic length of tLAT also significantly decreased the relative affinity for Lo domain lipids (Fig. 1 c). These observations show impressive qualitative agreement to results in isolated biological membranes (GPMVs), wherein the tLAT partitions approximately equally between coexisting domains, whereas both palmitoylation (23) and truncation mutants (26) lead to significant enrichment in the disordered phase.
To support the inferences from the coarse-grained MD simulations and obtain more detailed insights into the molecular interactions guiding protein partitioning, we performed 2 μs all-atom MD simulations. The same membrane systems (DPPC/DAPC/Chol: 5/3/2) were assembled by CHARMM-GUI (46, 47), with an initial configuration of 400 evenly distributed lipids. The first 1 μs was reserved for equilibration into relatively stable membrane domains (clear phase separation was observed within 500 ns.). The last 1 μs trajectory was used for data analysis. No significant structural changes were observed for either peptide in our all-atom MD simulations. As in the coarse-grained observations, the atomistic simulations revealed that tLAT, both with and without palmitoylations, preferentially localized at the domain boundaries (Fig. 2 a; Fig. S4). Despite the overall accumulation at the domain interface, the peptides had preferential interactions with certain membrane lipids. Specifically, palmitoylated tLAT showed significantly higher interaction energies with Lo domain lipids, and these trends were reversed for depalmitoylated tLAT (Fig. 2 b). These observations are fully consistent with the results of the coarse-grained MD simulations, validating the coarse-grained approach for detailed analysis of protein interactions with phase separated membranes. More importantly, both the coarse-grained and all-atom models reproduced key features of experimental measurements in biological membranes (26), in that both suggested preferred interactions between tLAT and ordered phase lipids and an inversion of this preference without palmitate moieties.
Figure 2.
All-atom MD simulations of tLAT partitioning between coexisting liquid phases. (a) Snapshots of the last frame (t = 2 μs) for the all-atom systems tLAT+palm and tLAT−palm are shown; red = DPPC, white = cholesterol, and green = DAPC (50). (b) The interaction energy differences between TMDs and Lo versus Ld domain lipids are shown; means that TMD prefers to interact with Lo domain lipids; the mean ± SD is of a five-block average of the last 1 μs trajectories. (c) TMD-lipid interaction contributions from each amino acid and palmitoyl moieties for the two systems based on the analysis of the last 1 μs trajectories are shown. The upper heat map represents tLAT+palm; the lower is for tLAT−palm. For palmitoylated tLAT, most of the preferential interactions with Lo phase lipids come from the palmitate moieties. To see this figure in color, go online.
The atomistic resolution afforded by all-atom MD simulations allowed us to analyze the detailed residue-level interactions between LAT peptides and surrounding lipids (Fig. 2 c). The most striking observation was that both palmitoyl groups showed strong attractive interactions with DPPC, suggesting that these interactions may be responsible for the enhanced Lo-lipid affinity observed in both coarse-grained and all-atom MD simulations of the palmitoylated LAT peptide. These inferences are consistent with the preferential interactions between saturated lipid fatty acyl chains (53, 54, 55), which are likely responsible for the palmitoylation-dependent raft affinity previously observed in isolated plasma membranes (20). The differences in the pattern of lipid interaction energies between the two peptides (Fig. 2 c) are driven largely by the partitioning effect of palmitoylation. That is, palmitoylation increases the affinity of the peptide for the ordered phase, increasing the likelihood that peptide residues will interact with DPPC. In contrast, the depalmitoylated peptide preferentially enriches near disordered lipids and therefore has more interactions with DAPC.
Energetics of tLAT partitioning from umbrella sampling
MD simulations of both coarse-grained and all-atom models confirmed the critical role of palmitoylations in regulating the raft affinity of transmembrane peptides. Further, coarse-grained MD simulations recapitulated experimental observations (26) on the important role of TMD hydrophobic length in determining protein partitioning between membrane domains. However, several discrepancies remain between MD simulations, GUV experiments, and GPMV experiments. Specifically, they are different with respect to overall peptide distributions: in GUV experiments, peptides are enriched in the Ld phase (28); in GPMVs, peptides are slightly enriched in Lo phase (26), whereas peptides in MD simulations show interfacial accumulation (Figs. 1 and 2). To gain insight into these effects, we investigated the detailed energetics of domain affinity by coarse-grained umbrella-sampling simulations. For these calculations, we used the DPPC/DAPC/Chol system, which forms stable striped domains (52). We constructed sandwich-style phase-separated membrane systems with an Lo domain in the middle flanked by two Ld domains and chose the COM distance between the TMD and the Lo domain along the axis perpendicular to the domain stripes as the reaction coordinate for umbrella-sampling simulations (Fig. 3 a). PMF or relative free energy profiles were derived by combining results from 34 systems with different initial reaction coordinates ranging from 0 to 6.6 nm ( = 0.2 nm) (Fig. 3 a). Umbrella-sampling simulations were run for 3.2 μs, with the first 0.8 μs reserved for equilibrium and the last 2.4 μs for analysis. We calculated the mean force at each individual reaction coordinate and then obtained the PMF/ profiles by integrating mean forces along the reaction coordinate .
Fig. 3 b shows the PMF profiles for the three peptides, with each displaying a prominent energy minimum near the membrane domain interface (∼3.2 nm), fully consistent with preferences for the membrane domain interface observed in coarse-grained (Fig. 1) and all-atom (Fig. 2) simulations. Importantly, we observed good sampling of the peptide at the interface without any notable disruption of the interface itself by the presence of the peptide (Fig. 3 c). To ensure this result was robust, we constructed an “inverted” sandwiched membrane with an Ld domain at the center and a new reaction coordinate being the lateral COM distance between the Ld domain and the peptide. The PMF profile in this system was in excellent quantitative consistency with the original PMF data (Fig. S6). All three peptides had a lower free energy in the Ld domain; this effect is discussed below. However, the relative difference between the free energy in the Lo versus the Ld domain (; the partitioning free energy) was lowest for the tLAT peptide and higher for both the depalmitoyled and the truncated variants. These trends from the free-energy calculations are consistent with the energetics shown in Fig. 1, but more importantly are consistent with experimental partitioning measurements of tLAT in biomembranes (i.e., GPMVs).
According to the PMF profile in Fig. 3 b, the energy minimum for tLAT is the domain interface, with the next lowest energy state being the Ld domain. This observation could elucidate the disparity between GUV experiments (Ld domain preference) (28) and MD simulations (domain interface preference) based on the following hypothesis: if the domain interface in GUVs was fully occupied by tLAT, the excess tLAT would prefer Ld domain because of the lower energy difference. To test this hypothesis, we performed an additional 40 μs coarse-grained MD simulation with a high abundance of tLAT (peptide/lipid = 60/1440) using the same bilayer system as in Fig. 1. As shown in Fig. 3 d, saturation of the domain interface with peptides resulted in the remaining peptides partitioning into the Ld domain. This observation suggests that the apparent mismatch between GUV experiments and MD simulation may be that the relative peptide enrichment at the domain interface in GUVs cannot be resolved by light microscopy because of the molecular length-scale of this interface (i.e., >3 nm). Further, because of limited box size, the interface in simulations comprises a much larger proportion of the overall membrane surface than in GUVs, in which the interface is of molecular scale (as in simulations), whereas the bulk phases are macroscopic. Such an effect would tend to overemphasize the interfacial localization in simulations.
Thus, we believe this saturation of the interface may explain the apparent discrepancy between partitioning in MD simulations (preference for the domain interface) and GUVs (preference for the Ld domain). However, we believe our simulations and experimental observations may also inform the discrepancies between observations in model membranes (MD and GUVs show preference for Ld domains) (28) and GPMV experiments (slight preference for Lo domains) (26). Our previous observations suggest that the lipid packing differences between Lo and Ld domains in GPMVs are notably smaller than those in GUVs (15, 56, 57). Thus, even peptides with potential affinity for ordered domains (e.g., palmitoylated LAT) are excluded from the artificially high packing of the Lo domains in model membranes. Thus, artificial model membranes do not quantitatively reproduce the absolute partitioning of peptides between ordered and disordered membrane domains. However, our observations reveal that relative differences between peptides are indeed consistent between MD simulations and experiments in isolated plasma membranes, as shorter or depalmitoylated TMDs had lower ordered domain affinity and DPPC/Chol interactions than wild-type tLAT. In other words, the domain-partitioning energetics of TMDs is determined by two major inputs: 1) protein-lipid interactions, with both simulations and GPMV experiments revealing preferential interactions of LAT-TMD with Lo domain lipids; and 2) interdomain lipid packing differences, which are highly dependent on the specifics of the model system. Below, we formalize this hypothesis with a simplified conceptual model to elucidate how lipid packing differences determine transmembrane peptide partitioning between coexisting phases.
In this model, the interaction between a TMD peptide and a phase-separated membrane is parameterized by which defines the relative contact area between the peptide and a given membrane domain. As shown in the Fig. 4 b, when the peptide is entirely located in the Lo domain and in the Ld domain. Between those values, the peptide is located at the interdomain boundary. In this model, TMD partitioning free energy mainly consists of two contributions: 1) interactions between the peptide and Lo versus Ld lipids , as measured in Figs. 1 c and 2 b, which are determined by the chemical properties of the TMD peptide; and 2) steric effects , which are determined by lipid packing differences between Lo and Ld domains as well as the TMD size. If these two components have different profiles with respect to , with one being asymmetric about (e.g., red curves in Fig. 4 c), the overall free-energy profile should be nonmonotonic, including potentially a minimum at the Lo/Ld interface (as observed in the PMF calculations, Fig. 3 b). A possible explanation for such an asymmetric profile is an inherent polarity of the peptides, with one side preferring the Lo environment and vice versa. To evaluate this possibility, we analyzed the relative preference of each amino acid side chain for DPPC versus DAPC in the atomistic simulations of the tLAT peptide. We indeed observed a distinct asymmetry in peptide interactions with lipids, with one face of the helix having notably higher interactions with Lo phase lipids than the other (Fig. S7). This polarity in the LAT peptide could certainly account for the interfacial preference we observed in all our simulations. Notably, not all peptides show this behavior in simulations. Previous observations from the Marrink group (see Schäfer et al. (29) and de Jong et al. (30)) show that most peptides generally accumulate in Ld domains, consistent with many experiments in GUVs (28, 29, 58) and GPMVs (59, 60, 61).
Figure 4.
Conceptual model for contributions of TMD partitioning free energy. (a) The schematics of translocating TMDs between Ld and Lo domains are shown. (b) A schematic of TMD at membrane domain interface (top view) is shown. : the peptide is completely in Lo domain; : the peptide is fully in Ld domain. (c) A schematic for domain-partitioning free-energy contributions from protein-lipid interactions (red) and steric effects (blue) is shown. Hypothetical curves are shown for partitioning in GUVs (solid) compared to GPMVs (dashed). Note that although the interface is represented as a sharp boundary in these schemata, real interfaces feature a transitional zone with an effective width and a distinct compositional profile. To see this figure in color, go online.
Our hypothetical model (Fig. 4 c) also suggests how differences in and between domains in various systems could contribute to partitioning, which helps explain the disparity in the preferred partitioning of palmitoylated tLAT between GPMVs (11, 15, 56) and GUVs (28). The most notable difference between these systems is the much smaller packing differences between Lo and Ld domains in GPMVs (15, 16, 56), which generates a much smaller energy cost for translocating palmitoylated tLAT from Ld to Lo domains (dashed blue line in Fig. 4 c). This effect may account for the different partitioning behaviors observed between these two systems (black lines in Fig. 4 c).
Conclusions
In this work, we used molecular simulations to elucidate the energetics of transmembrane protein partitioning between coexisting fluid membrane domains, using tLAT as a model. Coarse-grained MD simulations reproduced observations from experiments in isolated biological membranes, as long, palmitoylated peptides had preferred interactions with Lo domain lipids compared to shorter and depalmitoylated peptides. These observations were also validated by corresponding all-atom MD simulations. In all simulations, we observed enrichment of peptides at the Lo/Ld interface, which was further confirmed by free-energy profiles calculated using umbrella-sampling simulations. Atomistic simulations suggest that this effect may arise from an inherent polarity of the peptide with respect to its interaction with the coexisting phases. The partitioning free-energy profiles produce a comprehensive picture of peptide partitioning and clarified disparities between experiments in various model membranes and MD simulations. These results prompt a conceptual model wherein TMD partitioning free energy is driven by a balance between protein-lipid interactions and steric effects, the former mainly determined by chemical properties of the TMD peptide components and the latter dominated by lipid packing differences between Lo and Ld domains and the physical features of the TMD. The insights in this work may explain inconsistences in protein partitioning experiments with different model systems and provide guidelines for designing peptides to target specific membrane domains.
Acknowledgments
The authors thank Dr. Joseph Lorent, Dr. Hualin Li, and members of the Siewert-Jan Marrink group (Groningen University) for helpful discussions on membrane partitioning dynamics of tLAT, and Dr. Hongyin Wang and Dr. Priyanka Prakash for the help in the data analysis of the revised manuscript.
This work was supported by the National Institutes of Health (grant nos. R01GM100078, R01GM114282, and R01GM124072), the Volkswagen Foundation (grant no. 93091), and the Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator Recruitment Grant (R1215). We thank the Extreme Science and Engineering Discovery Environment (project: TG-MCB150054) and the Texas Advanced Computing Center for generous computational resources.
Editor: Markus Deserno.
Footnotes
Supporting Materials and Methods and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30387-4.
Contributor Information
Alemayehu A. Gorfe, Email: alemayehu.g.abebe@uth.tmc.edu.
Ilya Levental, Email: ilya.levental@uth.tmc.edu.
Supporting Material
References
- 1.Levental I., Veatch S. The continuing mystery of lipid rafts. J. Mol. Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sezgin E., Levental I., Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozano M.M., Hovis J.S., Boxer S.G. Dynamic reorganization and correlation among lipid raft components. J. Am. Chem. Soc. 2016;138:9996–10001. doi: 10.1021/jacs.6b05540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Risselada H.J., Marrink S.J. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 2008;105:17367–17372. doi: 10.1073/pnas.0807527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X., Zhang S., Gorfe A.A. The aliphatic chain of cholesterol modulates bilayer interleaflet coupling and domain registration. FEBS Lett. 2016;590:3368–3374. doi: 10.1002/1873-3468.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan C., Johnston L.J. Distribution of ganglioside GM1 in L-α-dipalmitoylphosphatidylcholine/cholesterol monolayers: a model for lipid rafts. Biophys. J. 2000;79:2768–2781. doi: 10.1016/S0006-3495(00)76516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baoukina S., Mendez-Villuendas E., Tieleman D.P. Molecular view of phase coexistence in lipid monolayers. J. Am. Chem. Soc. 2012;134:17543–17553. doi: 10.1021/ja304792p. [DOI] [PubMed] [Google Scholar]
- 8.Veatch S.L., Keller S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veatch S.L., Keller S.L. Miscibility phase diagrams of giant vesicles containing sphingomyelin. Phys. Rev. Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 10.Baumgart T., Hammond A.T., Webb W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sezgin E., Kaiser H.J., Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 12.Simons K., Sampaio J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorent J.H., Levental I. Structural determinants of protein partitioning into ordered membrane domains and lipid rafts. Chem. Phys. Lipids. 2015;192:23–32. doi: 10.1016/j.chemphyslip.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 14.Ollila S., Hyvönen M.T., Vattulainen I. Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J. Phys. Chem. B. 2007;111:3139–3150. doi: 10.1021/jp065424f. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser H.J., Lingwood D., Simons K. Order of lipid phases in model and plasma membranes. Proc. Natl. Acad. Sci. USA. 2009;106:16645–16650. doi: 10.1073/pnas.0908987106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sezgin E., Levental I., Schwille P. Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim. Biophys. Acta. 2012;1818:1777–1784. doi: 10.1016/j.bbamem.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Li Z., Janosi L., Gorfe A.A. Formation and domain partitioning of H-ras peptide nanoclusters: effects of peptide concentration and lipid composition. J. Am. Chem. Soc. 2012;134:17278–17285. doi: 10.1021/ja307716z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun A.R., Lacy M.M., Sachs J.N. α-Synuclein-induced membrane remodeling is driven by binding affinity, partition depth, and interleaflet order asymmetry. J. Am. Chem. Soc. 2014;136:9962–9972. doi: 10.1021/ja5016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronchi P., Colombo S., Borgese N. Transmembrane domain-dependent partitioning of membrane proteins within the endoplasmic reticulum. J. Cell Biol. 2008;181:105–118. doi: 10.1083/jcb.200710093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X., Li Z., Gorfe A.A. Reversible effects of peptide concentration and lipid composition on H-Ras lipid anchor clustering. Biophys. J. 2015;109:2467–2470. doi: 10.1016/j.bpj.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wange R.L. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci. STKE. 2000;2000:re1. doi: 10.1126/stke.2000.63.re1. [DOI] [PubMed] [Google Scholar]
- 22.Su X., Ditlev J.A., Vale R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levental I., Lingwood D., Simons K. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc. Natl. Acad. Sci. USA. 2010;107:22050–22054. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Resh M.D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 25.Lorent J.H., Diaz-Rohrer B., Levental I. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat. Commun. 2017;8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-Rohrer B.B., Levental K.R., Levental I. Membrane raft association is a determinant of plasma membrane localization. Proc. Natl. Acad. Sci. USA. 2014;111:8500–8505. doi: 10.1073/pnas.1404582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levental I., Grzybek M., Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 28.Shogomori H., Hammond A.T., Brown D.A. Palmitoylation and intracellular domain interactions both contribute to raft targeting of linker for activation of T cells. J. Biol. Chem. 2005;280:18931–18942. doi: 10.1074/jbc.M500247200. [DOI] [PubMed] [Google Scholar]
- 29.Schäfer L.V., de Jong D.H., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. USA. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Jong D.H., Lopez C.A., Marrink S.J. Molecular view on protein sorting into liquid-ordered membrane domains mediated by gangliosides and lipid anchors. Faraday Discuss. 2013;161:347–363. doi: 10.1039/c2fd20086d. discussion 419–459. [DOI] [PubMed] [Google Scholar]
- 31.Izvekov S., Voth G.A. A multiscale coarse-graining method for biomolecular systems. J. Phys. Chem. B. 2005;109:2469–2473. doi: 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z.J., Deserno M. A systematically coarse-grained solvent-free model for quantitative phospholipid bilayer simulations. J. Phys. Chem. B. 2010;114:11207–11220. doi: 10.1021/jp102543j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 34.Monticelli L., Kandasamy S.K., Marrink S.J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 35.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klauda J.B., Monje V., Im W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 39.Janosi L., Li Z., Gorfe A.A. Organization, dynamics, and segregation of Ras nanoclusters in membrane domains. Proc. Natl. Acad. Sci. USA. 2012;109:8097–8102. doi: 10.1073/pnas.1200773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 41.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 42.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J., Cheng X., Im W. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jo S., Kim T., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 45.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 46.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 47.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 48.Hess B., Bekker H., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 49.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 51.Kästner J. Umbrella sampling. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011;1:932–942. [Google Scholar]
- 52.Lin X., Lorent J.H., Levental I. Domain stability in biomimetic membranes driven by lipid polyunsaturation. J. Phys. Chem. B. 2016;120:11930–11941. doi: 10.1021/acs.jpcb.6b06815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sodt A.J., Sandar M.L., Lyman E. The molecular structure of the liquid-ordered phase of lipid bilayers. J. Am. Chem. Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sodt A.J., Pastor R.W., Lyman E. Hexagonal substructure and hydrogen bonding in liquid-ordered phases containing palmitoyl sphingomyelin. Biophys. J. 2015;109:948–955. doi: 10.1016/j.bpj.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uline M.J., Longo G.S., Szleifer I. Calculating partition coefficients of chain anchors in liquid-ordered and liquid-disordered phases. Biophys. J. 2010;98:1883–1892. doi: 10.1016/j.bpj.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sezgin E., Gutmann T., Schwille P. Adaptive lipid packing and bioactivity in membrane domains. PLoS One. 2015;10:e0123930. doi: 10.1371/journal.pone.0123930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levental K.R., Levental I. Isolation of giant plasma membrane vesicles for evaluation of plasma membrane structure and protein partitioning. Methods Mol. Biol. 2015;1232:65–77. doi: 10.1007/978-1-4939-1752-5_6. [DOI] [PubMed] [Google Scholar]
- 58.Bacia K., Schuette C.G., Schwille P. SNAREs prefer liquid-disordered over “raft” (liquid-ordered) domains when reconstituted into giant unilamellar vesicles. J. Biol. Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 59.Levental I., Grzybek M., Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2011;108:11411–11416. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson S.A., Stinson B.M., Baumgart T. Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim. Biophys. Acta. 2010;1798:1427–1435. doi: 10.1016/j.bbamem.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sengupta P., Hammond A., Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim. Biophys. Acta. 2008;1778:20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.