Abstract
The murine pancreas buds from the ventral embryonic endoderm at approximately 8.75 dpc and a second pancreas bud emerges from the dorsal endoderm by 9.0 dpc. Although it is clear that secreted signals from adjacent mesoderm-derived sources are required for both the appropriate emergence and further refinement of the pancreatic endoderm, neither the exact signals nor the requisite tissue sources have been defined in mammalian systems. Herein we use DiI fate mapping of cultured murine embryos to identify the embryonic sources of both the early inductive and later condensed pancreatic mesenchyme. Despite being capable of supporting pancreas induction from dorsal endoderm in co-culture experiments, we find that in the context of the developing embryo, the dorsal aortae as well as the paraxial, intermediate and lateral mesoderm derivatives only transiently associate with the dorsal pancreas bud, producing descendants that are decidedly anterior to the pancreas bud. Unlike these other mesoderm derivatives, the axial (notochord) descendants maintain association with the dorsal pre-pancreatic and early pancreas bud. This fate mapping data points to the notochord as the likely inductive source in vivo while also revealing dynamic morphogenetic movements displayed by individual mesodermal subtypes. Because none of the mesoderm examined above produced the pancreatic mesenchyme that condenses around the induced bud to support exocrine and endocrine differentiation, we also sought to identify the mesodermal origins of this mesenchyme. We identify a portion of the coelomic mesoderm that contributes to the condensed pancreatic mesenchyme. In conclusion, we identify a portion of the notochord as a likely source of the signals required to induce and maintain the early dorsal pancreas bud, demonstrate that the coelomic mesothelium contributes to the dorsal and ventral pancreatic mesenchyme and provide insight into the dynamic morphological rearrangements of mesoderm-derived tissues during early organogenesis stages of mammalian development.
INTRODUCTION
The pancreas arises from naïve foregut endoderm and is induced, patterned and undergoes further differentiation in response to signals provided by adjacent mesoderm derived tissues. Distinct ventral and a dorsal pancreas buds each arise independently from the foregut endoderm by 9.0 days post coitum (dpc) in mouse and day 26 in human [reviewed in (Gittes, 2009)]. Although each bud can be identified by 9.0 dpc as Pdx1 expressing evaginations of stratified epithelium, they have different molecular requirement to form pancreatic endoderm from the foregut endoderm (Ahlgren et al., 1996; Ahlgren et al., 1997; Fujitani et al., 2006; Harrison et al., 1999; Jacquemin et al., 2003; Li et al., 1999; Sherwood et al., 2009; Yoshitomi and Zaret, 2004).
We previously used a fate-mapping approach to identify the dorsal and ventral pre-pancreatic endoderm in the early endodermal epithelium (Angelo et al., 2012; Tremblay and Zaret, 2005). The dorsal pre-pancreatic endoderm overlies the dorsal midline between somites 2–4 prior to induction, which we identify as the onset of Pdx1 expression in the dorsal pancreas and is generally thought to occur between the 10–12 somite stage (S) in mouse, while the ventral pre-pancreatic endoderm is lateral and anterior to the dorsal, demonstrating that the progenitors interact with distinct mesoderm-derived tissue. Dorsal progenitors can be induced by adjacent mesoderm-derived tissues to initiate the appropriate pancreatic program (Kim et al., 1997; Lammert et al., 2001). In contrast rather than being induced, the ventral foregut endoderm appears to have a default pancreatic state (Deutsch et al., 2001). A variety of experimental data from mouse indicates that the ventral pancreas is restricted by FGF and BMP signals that promote formation of the adjacent endoderm-derived organ, the liver bud (Chung et al., 2008; Deutsch et al., 2001; Palaria et al., 2018; Spence et al., 2009). Thus the differences in the induction of the pancreatic program between the dorsal and ventral pre-pancreatic endoderm underscores the distinct transcriptional requirements noted above.
Numerous studies have demonstrated that signals secreted from adjacent mesoderm are sufficient to induce pancreatic gene expression from competent endoderm (Dessimoz et al., 2006; Hebrok et al., 1998; Kim et al., 1997; Kumar et al., 2003; Wells and Melton, 2000). Several tissues, including the dorsal aortae, lateral plate mesenchyme, and notochord, are sufficient in the context of tissue recombination experiments in chick, to induce pancreas-specific gene expression from pre-pancreatic endoderm, and thus all have been hypothesized to be endogenous inducers of pancreas development (Apelqvist et al., 1997; Hebrok et al., 1998; Kim et al., 1997; Kumar et al., 2003). For example, recombination experiments demonstrate that lateral plate mesoderm induces PDX1 from anterior foregut endoderm. Additionally, secreted factors normally expressed from lateral plate mesoderm such as Activin A, BMP or Retinoic Acid are each sufficient to induce pancreatic markers from anterior endoderm/mesoderm explants. Conversely, inhibition of Activin or BMP, is sufficient to inhibit Pdx1 expression in pre-pancreatic endoderm/mesoderm explants (Kumar et al., 2003). Furthermore, either Activin βB, produced by the notochord, or FGF2, produced by lateral plate mesoderm, is sufficient to induce Pdx1-expression in isolated endoderm (Hebrok et al., 1998). Given that all current clinical work designed to create functional pancreatic cells from stem cells in vitro is focused on mammalian tissues, it is important that the signal or signals involved in pancreas induction are identified in mammalian systems.
After pancreas bud induction and emergence, the nascent buds are associated with another mesoderm-derivative, the condensed pancreatic mesenchyme, which guides pancreatic epithelial proliferation, differentiation and morphogenesis. Early studies demonstrated that the pancreatic epithelium is responsive to cues from pancreatic and non-pancreatic mesenchyme, influencing cytodifferentiation towards more endocrine or exocrine fate (Golosow and Grobstein, 1962). Later co-culture studies refined this idea, demonstrating that pancreas mesenchyme induced acinar fate, basement membrane rich conditions led to ductal fate, while endoderm cultured without added mesenchyme promotes islet growth (Gittes et al., 1996; Miralles et al., 1998). Furthermore, when co-cultures are performed with the condensed pancreatic mesenchyme and pancreatic epithelium, both the maturity and proximity of that mesenchyme to endoderm affect exocrine/endocrine differentiation. Proximity of mesenchyme to pancreatic epithelium dictates the proportion of exocrine to endocrine cells (Li et al., 2004). Furthermore, the maturity of pancreatic mesenchyme dictates its ability to pattern competent epithelium. Recombination of 11.5 dpc pancreatic epithelium with 10.5 dpc mesenchyme results in a less mature epithelium when compared to 10.5 dpc pancreatic epithelium/11.5 mesenchyme co-cultures (Rose et al., 1999). More recent explant studies suggest that at 13.5 dpc, pancreatic mesenchyme may be responsible for maintaining the balance between the PDX1+ progenitor population and differentiation into a terminal endocrine fate (Attali et al., 2007). Finally in vivo ablation of pancreatic mesenchyme highlights its role in directing early pancreatic endoderm growth and differentiation and in the acquisition of appropriate morphogenesis during later pancreatic development (Landsman et al., 2011). These data indicate a plasticity of the pancreatic epithelium and underscore its dose-dependent sensitivity to signals. While the importance of the pancreatic mesenchyme has long been appreciated, the embryonic origin of the pancreatic mesenchyme is unknown. For example, it is not known whether the mesoderm adjacent to the dorsal pre-pancreatic endoderm is the same as that which later condenses around it or if a separate mesenchyme migrates to surround and guide pancreatic morphogenesis and differentiation.
To identify which of the adjacent mesoderm-derived tissues exhibit a sustained interaction with the pre-pancreatic endoderm and early-specified pancreas thus meeting our criteria as an inductive tissue, we created a DiI fate map of the mesodermal tissues proximal to the dorsal pancreatic progenitors immediately prior to and at the onset of induction as indicated by the onset of Pdx1 expression and bud formation. Although it is not possible to discount a step-wise specification of the dorsal pancreas via multiple transient interactions, the co-culture and explant data demonstrates that the induction of a pancreatic program from competent dorsal endoderm requires at least 18 hours of stable interaction, thus we reasoned that identification of a sustained mesoderm-pre/pancreatic endoderm interaction in vivo would highlight a likely inductive event. We find that the majority of mesoderm adjacent to the dorsal pre–pancreatic endoderm at 8.5 dpc are substantially displaced from the nascent pancreas buds by 9.5 dpc while a defined portion of the labeled notochord remains adjacent to the dorsal bud during and shortly after the induction period. Because none of this mesoderm gave rise to the condensed dorsal pancreas mesenchyme, we extensively fate mapped the mesoderm lateral and posterior to that described above. Towards this goal, we identify a discreet portion of the 8.5 dpc coelomic mesothelium as contributing descendants to both the dorsal and ventral pancreatic mesenchyme. These data shed light on potential pancreas-inductive tissue in the mammalian embryo and help tease apart the body of pancreas literature gathered from diverse species while highlighting an unanticipated complexity in mesoderm-endoderm interactions during post-gastrulation mammalian development.
MATERIAL AND METHODS
1.1 Embryo dissection, culture and DiI-labeling
Dissections, embryo collection, culture and DiI-labeling was performed as previously described (Angelo et al., 2012) except that mesenchyme directly underlying the endoderm label was also injected. To accomplish this, a pulled pipette with an outer tip diameter of approximately 10 μm, was frontloaded with DiI and inserted into the targeted mesenchyme. After manual ejection of the dye, the needle was removed and additional DiI applied to the endoderm directly over the labeled mesenchyme.
1.2 Fate map construction
Initial labels were mapped onto illustrations representing the ventral surface of the 8.5 dpc embryo as described (Angelo et al., 2012). Labels that gave rise to multiple tissue populations were assumed to lie on the interface of these tissues and analysis in combination with similarly located labels on additional embryos used to identify each mesodermal population. Post-culture maps were produced by identifying labeled tissues in sections of each embryo and using readily identifiable structures such as the first and second branchial arches, thyroid bud, otic pit, atrium and ventricle of the heart, liver bud and limb buds to determine the anterior-posterior position of the dye. The relative location of ventral and dorsal structures within each image and the number of embryo sections on each slide between each image was recorded to ensure that position of the embryo upon sectioning did not obscure analysis. These labels were then mapped onto sagittal illustrations traced from an image of a 9.5 dpc embryo.
1.5 Immunofluorescence
Immunofluorescence was performed as described (Angelo and Tremblay, 2013). Primary antibodies included: rabbit anti-PDX1 (1:1000, [Abcam, 47267)], guinea pig anti-PDX1 (1:1000, [Abcam, 47308)], and mouse anti-ISL1 [1:200, (DSHB 40.2D6)].
RESULTS
Fate mapping of tissues associated with the dorsal pancreas progenitors
We previously demonstrated that the dorsal pancreas progenitors are localized to the definitive endoderm that overlies somites 2–4 of the 3–11S mouse embryo (Angelo and Tremblay, 2013). During these stages, the dorsal pancreas progenitors lie directly over the somites, lateral plate mesenchyme and dorsal aortae and are directly apposed to or contiguous with the medially located notochord. To further explore the interaction of the pre-pancreatic endoderm and adjacent tissues, we labeled discrete portions of pre-pancreatic endoderm, which lies on the surface of the embryo, as well as proximal underlying mesoderm. To insure that the two labeled tissues were proximal, we plunged a DiI-filled pipette through the pre-pancreatic endoderm into the underlying mesoderm, ejected a small bolus of DiI to label this mesoderm and then withdrew to label the overlying endoderm. Figure 1 illustrates the immediate outcome of such a label. This 7S embryo was labeled, fixed and sectioned sagittally to reveal DiI in the pre-pancreatic endoderm and ventral half of the second somite (Fig. 1A,B,B′; somites are numbered). These structures are easily identified in the false-colored enlargement, (Fig. 1B″; green endoderm, orange somites, arrow points to injection site). It is notable that in this example, the injection itself caused some aberrant scattering of cells between somite and endoderm. While this figure illustrates that a limited amount of cell damage occurs at the onset of the experiment, we find that in a typical labeling experiment, where embryos are cultured for at least 24 hours, the post-culture mesoderm derived structures appear morphologically normal and structures containing labeled cells appear symmetric with their unlabeled lateral counterpart. Finally we should note that in this and other whole mount images throughout this manuscript, the visible DiI is usually that on surface endoderm while overlying tissues tend to occlude any underlying mesoderm label. It was, however, possible to see the point of injection during the labeling process, thus directing our fate mapping procedure.
Fig. 1. Example of a DiI labeled Embryo.
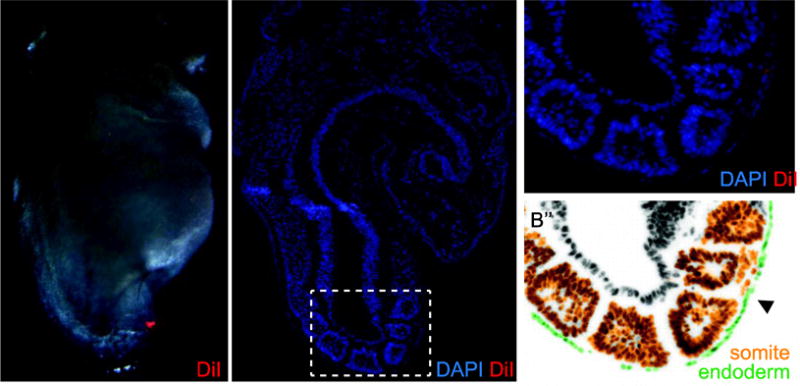
A) A 7-somite (S) embryo labeled over the second somite on the right side with DiI (red). B) A DAPI stained sagittal section of the same embryo shows DiI (red) in the pre-pancreatic endoderm and in the underlying somitic mesoderm, better seen in enlargement (B′), and site of DiI release (arrowhead). C) False color rendition of the same section highlights endoderm (green) and somites (orange). Hf, head fold; h, heart; the somites are numbered.
These data demonstrate that we are able to label discrete, internal portions of the mesoderm adjacent to pre-pancreatic endoderm. Because the mesoderm-derived tissues are closely juxtaposed and because DiI quickly but not instantaneously precipitates out of solution, rarely is the initial label restricted to a single mesoderm lineage and thus precision is limited. However, because DiI only fluoresces when incorporated into membranes and because membranes are divided between daughter cells, such limitations are overcome by imaging the embryo at the onset of labeling and by careful identification the labeled descendants at the end of culture in section analysis. Finally we should note that prior to 8S the notochord has not yet submerged and is contiguous with the pre-pancreatic endoderm (Balmer et al., 2016) and thus during these stages, midline endoderm labels may also include notochord.
We next examined the association of the pre-pancreatic endoderm and underlying mesoderm during development. Because the dorsal pre-pancreatic endoderm overlies somites 2–4 of the 3–11S embryo, we chose fate map the mesoderm underlying and proximal to somites 2–4 at these stages and slightly later using DiI (3–14S; illustrated in Fig. 2A). After DiI-labeling, embryos are cultured for at least 24 hours, through the early stages of dorsal pancreas development (illustrated in Fig. 2B). This strategy ensures that the DiI-labeled mesoderm is proximal to the pre-pancreatic endoderm at the start of culture and allows us to determine how these tissue populations are ultimately arranged not only after the onset of pancreas development but also after embryonic turning, gut tube formation and germ-layer inversion.
Fig. 2. Mesenchyme Fate Mapping Summary.
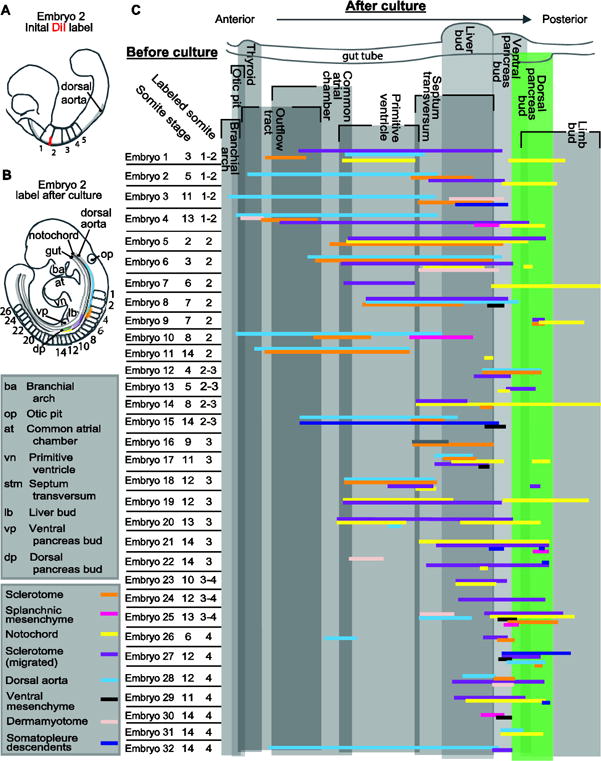
A–B) A schematized version of DiI-labeled Embryo 2 before (A) and after (B) culture. A) The initial DiI label (red) between somites 1 and 2 is indicated on this 5-somite embryo. B) After culture, the DiI labeled descendants are indicated by the colors on this E9.5 embryo. Each color on the embryo represents a different mesodermal-derivative (see the color key) and the position of each represents the position of each descendant population. C) Individual embryo data from all 32 fate-mapped embryos is presented vertically in the chart. Each embryo was given a number (Embryo1–32, for reference purposes here and throughout subsequent figures), the somite stage of each embryo at the onset of culture is indicated and the location of the initial DiI-label listed. The initial label was limited to the mesoderm proximal to the dorsal pancreas progenitors, which lie within the endoderm overlying somites 2–4. At the end of culture each embryo was sectioned transversely and the location of the DiI-labeled mesoderm-derivative mapped along the anterior/posterior axis relative to other easily identifiable embryonic structures also present in that section. The vertical grey boxes delimit the boundary of each listed embryonic structure along the anterior/posterior axis and overlap between boxes, resulting in a darker shade, indicates that those structures are present on a transverse section at that relative anterior/posterior position. Each cluster of horizontal colored lines represents data from one embryo, and the color of each line indicates the structure to which labeled cells contributed (see color key). The presence of stacked horizontal lines indicated that multiple tissue types were simultaneously labeled at the onset of culture.
The extent, location and types of mesoderm labeled in each of the 32 DiI-labeled embryos (E1–E32) are summarized in Figure 2. To discern the identity of the labeled mesoderm at the end of culture, each embryo was sectioned transversely and fluorescence used to identify the DiI-labeled descendants and the immunolabeled PDX1+ pancreas buds. We should note that although all embryos presented herein were DiI-labeled in a portion of the pre-pancreatic dorsal endoderm and underlying associated mesoderm, the information in the summaries includes only information on the mesoderm.
Several general trends can be appreciated from the mesoderm fate-mapping summary (Fig. 2). The first is the dramatic separation of initially contiguous mesoderm sub-types. Because each label was discrete and localized, we were surprised to find such a wide distribution of mesoderm derivatives that were initially apposed. The second is that each mesoderm sub-type contributed descendants that were anterior to the dorsal pancreas bud in a pattern unique to each tissue-type and is discussed below.
Most mesoderm surrounding the dorsal pre-pancreatic endoderm and early pancreas bud interact transiently
The separation of labeled mesoderm from the dorsal pancreatic endoderm, is illustrated by the representative embryo labeled at the lateral edge of the second somite (Fig. 3B–C; herein referred to as lateral labels) at 5S and cultured through 9.5 dpc (Fig. 3D). In this embryo the anterior most DiI-labeled cells are in the dorsal aorta (blue asterisk) and adjacent mesenchyme. These labeled descendants stretch from the caudal most region of the head (marked by the otic placode and thyroid; Fig. 3E, blue arrowhead, op and th respectively) to regions containing caudal aspects of the heart (Fig. 3F, between blue arrowheads). As the aortic label ends, a separate medial-lateral population becomes apparent within the somitic sclerotome and dermamyotome (Fig. 3F–G′, orange and peach arrowheads, respectively). Both aortae and somite, which are proximal to the pre-pancreatic endoderm at 8.5 dpc, are located anterior to the dorsal pancreas bud at 9.5 dpc. The caudal extent of the labeled mesoderm is located in sections containing the posterior liver bud (Fig. 3G, dashed line, lb) and thus labeled mesoderm is absent from sections containing ventral pancreas (Fig. 3H, dotted line, vp) as well as those containing DiI-labeled dorsal pancreas (Fig. 3I dotted line, dp, red arrow). A representative older (11S) embryo illustrates that the pattern of anterior aorta label followed by medial-lateral somite label is consistent throughout the stages examined (2–15S, compare Fig. 3B–I to Fig. 3J–Q). In this older embryo, DiI-labeled aortic endothelium and adjacent mesenchyme are present anteriorly, near the level of the otic placode and the branchial arches (Fig. 3M,N). We find that mesoderm that is initially proximal to the first and second somites produce descendants that are surprisingly anterior to descendants of the pancreatic endoderm. Not only are mesoderm and endoderm displaced from one another but the direction and extent of displacement is unique to each mesodermal subtype. An example of this can be seen in whole mount in Figure 3L where the labeled dorsal aortae stretches along the rostral/caudal axis while labeled somitic mesenchyme has spread dorsoventrally (Fig. 3L, enlarged in L′).
Fig. 3. Most mesoderm/pancreatic endoderm interactions are transient.
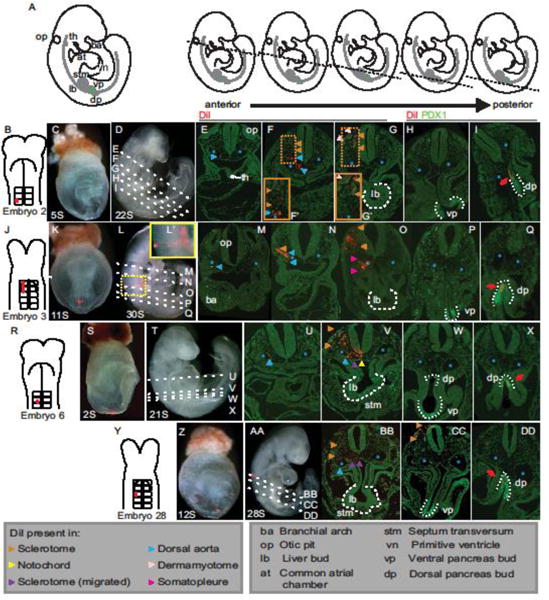
A) Illustrations representing post-culture embryos with a portion of the morphological structures used as landmarks indicated on the first illustration and deciphered in the box below. The dashed line on each of the smaller illustrations indicates the approximate location of the transverse plane exhibited by the sections below it. B–I) An illustration of Embryo 2 that was DiI labeled at the border of somites 1–2 of the 5S embryo (B) and shown in whole mount before (C) and after (D) culture to 22S. Analysis of transverse sections through this embryo (planes represented by dashed lines through D) reveals the anterior-most DiI in the dorsal aorta (structures indicated by blue asterisk, blue arrowheads indicate DiI labeled dorsal aorta; E), more posteriorly DiI-labeled cells are found in dorsal aorta, adjacent mesenchyme and sclerotome (orange arrowheads; F). Sections containing liver bud (white dashed outline, lb) show DiI-labeled cells in sclerotome and dermamyotome (peach arrowheads; G), while DiI is absent from mesoderm-derivatives in more posterior sections containing the ventral and dorsal pancreas buds labeled immunofluorescently for PDX1 in green (white dotted outline, vp and dp, respectively; H,I). As expected DiI-labeled dorsal pancreatic endoderm is present (I). J–Q) A 1–2 somite label in an older 11S embryo (J, K) cultured to 30S (L; inset in L′ shows a magnified view of the DiI label) shows a similar spread of DiI-labeled mesoderm. In anterior most sections DiI is found in in dorsal aorta (M), followed more posteriorly by labeled sclerotome (N) and finally in somatopleure descendants (pink arrowheads, O). The dorsal pancreatic endoderm was also DiI+ (P,Q). R–X) Embryo 6 was labeled at the presumptive third somite (R–S) and cultured through 21S (T), producing DiI descendants that are found more posteriorly in the embryo at the end of culture than those in which the initial label was more anterior (U–V, compare to E–F). However no DiI-labeled mesoderm-derivative is observed proximal to the ventral (W) or dorsal pancreas (X). Y–DD) In this 12 S embryo labeled over the lateral portion of the third somite (Y–Z), after culture through 28S (AA) the DiI-label is more condensed and is present in a limited area around the liver bud (BB) and ventral pancreas (CC) but absent from the mesoderm in sections containing dorsal pancreas (DD). The embryo number under B, J, R, Y corresponds to those listed in Figure 2. th=thyroid; op=otic pit; ba=branchial arch; at=common atrial chamber; vn=primitive ventricle; stm=septum transversum mesenchyme; lb=liver bud; vp, ventral pancreas; dp, dorsal pancreas. Arrowheads indicate DiI in structures with color-codes as listed. Blue asterisks demarcate dorsal aortae. Red arrows indicate DiI-labeled dorsal pancreas bud.
Because our fate-mapping strategy produces multiple mesoderm-derived labeled tissues in each embryo, an analysis of the pattern of descendants produced by multiple embryos allows us to determine the borders between the antecedents of these tissues. For example, a comparison of lateral labeled mesoderm in young embryos (2–6S; Fig. 3B–I) to middle of a somite (also referred to as mid-somite) mesoderm labels in comparably staged embryos (Fig. 3R–X) demonstrates that the antecedents of the dorsal aortae and medial-lateral somitic mesoderm extend from the middle to the lateral of a somite. Mid-somite labeling of younger embryos (Fig. 3R–S) cultured to ~9.5 dpc (Fig. 3T) not only results in labeling of the dorsal aortae and medial-lateral somite (Fig. 3U–V, blue and orange arrowheads, respectively), but also notochord and migrated sclerotome (Fig. 3V, yellow and purple arrowheads, respectively). As both lateral and mid-somite labels give rise to dorsal aortae and medial-lateral sclerotome/dermamyotome, we conclude that their antecedents are within the area that the initial labels overlap.
The area occupied by each somite proximal mesoderm domain narrows medio-laterally as the embryo develops from a relatively simple, and fairly flat, structure to one that has differentiated and expanded dorsoventrally. This observation is most apparent when comparing the descendants produced by similar labels performed on embryos at different stages. For example, mid-somite labels in older embryos (6–15S; Fig. 3Y, Z) cultured to ~9.5 dpc (Fig. 3AA) produces descendants within the dorsal aortae (blue arrowheads), medial-lateral somite (orange arrowheads) and mesenchyme dorsal to the gut tube (purple arrowheads, Fig. 3BB–CC) but no notochord label. This is in contrast to the younger embryo described above (Fig. 3R–X), which is a similar label at a younger initial age. Additionally, lateral labels in younger embryos gave rise to midline sclerotome (Fig. 3B–I) while similar labels in older embryos did not (Fig. 3J–Q). A role of initial embryo age in extent of anterior migration of tissues is also notable. For example, when the mesoderm is similarly labeled on the third somite in younger (2S; Fig. 3R–X, Embryo 6) and older embryos (11S; Fig. 3J–Q, Embryo 28), the aortic label extends further anterior in the younger embryo.
Sustained interaction of the notochord and dorsal pre-pancreatic endoderm
Embryonic midline (hereafter referred to as midline) labels performed at all stages examined (2–15S) produced DiI-positive notochord that often remained in the same A/P domain as the dorsal pancreas bud (n=15/20), suggesting that a regional domain of the notochord consistently interacts with the dorsal pancreas from the progenitor (until 10–12S) through early budding stages (Fig. 2) Several trends are observed in embryos with labeled notochord. Unlike other labeled mesoderm-derivatives, the notochord is the only one that repeatedly displays a discontinuous distribution of labeled cells (n=5/20) and is the only labeled mesoderm-derivative that is repeatedly observed posterior to the dorsal pancreas (n=7/20; Fig. 2). Furthermore, notochord labeled embryos often include the presence of labeled midline mesenchyme, presumably migrated scleretome. For example a 3S embryo labeled at the midline of the second somite pair (Fig. 4A,B) was cultured through 24S. DiI-labeled descendants are present in the notochord and in the dorsal mesenchyme around the notochord in section containing rostral portions of the developing ventricles of the heart (Fig. 4C,D). In sections containing the anterior liver bud, DiI+ notochord-proximal mesenchyme remains, while DiI is notably absent from the notochord (Fig. 4E, purple arrowhead). In more caudal sections that also contain the dorsal pancreas, labeled notochord is present (Fig. 4F, red arrow indicates DiI-labeled pancreas) and the labeled notochord extends slightly caudal to the pancreas (Fig. 4G). A similar pattern of DiI-labeled notochord and notochord proximal mesoderm is found upon labeling the medial-lateral of the third somite in a slightly older embryo (Fig. 4 H–I, 13S). Labeled notochord and adjacent mesenchyme are apparent in sections anterior to the liver bud, while only the notochord is labeled in sections containing the dorsal pancreas bud. DiI is absent from the notochord in sections between the liver and dorsal pancreas buds (Fig. 4K–N). Very small labels in other tissues resulted in scattered cells sparsely labeled throughout a portion of that tissue, appearing to be a result of dilution rather than separation of groups of labeled cells. When a gap in labeling is observed in notochord, DiI is robust rostral and caudal to the portion of unlabeled cells, making it unlikely that this is an artifact of dilution (compare Fig. 4E to D, F, and L, M to K, N). Taken together these observations suggest that the notochord that is displaced posteriorly may be distinct from the anteriorly extending portion (Cambray and Wilson, 2007; Le Douarin, 2001; Yamanaka et al., 2007).
Fig. 4. The notochord maintains its proximity to the dorsal pancreas.
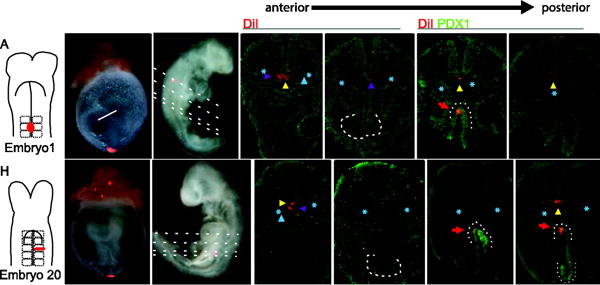
A–G) A midline DiI-label at somite 2 of the 3S embryo depicted in the illustration (A) and photographed in whole mount before (B) and after (C) culture to 24S. Analysis of transverse sections (plane indicated by dashed lines in C) reveals DiI in the notochord (yellow arrowhead) and adjacent cells, presumably migrated sclerotome (purple arrowhead), in sections rostral to (D) and containing the liver bud (lb, white dashed outline; E). DiI is present in notochord at the level of the dorsal pancreas bud, labeled immunofluorescently for PDX1 in green, (white dotted line, dp; F) and extending slightly more posterior (G). H–N) Similar initial midline labels produced on a 13S embryo (H–I) results in DiI-labeled descendants in cells around the notochord in more anterior sections (K), and in the notochord in sections that contain dorsal pancreas (M–N). lb, liver bud; vp-ventral pancreas bud; dp, dorsal pancreas bud. Purple arrowhead indicates DiI in sclerotome, yellow arrowhead indicates DiI-labeled notochord, blue arrowhead indicates DiI-labeled dorsal aortae, blue asterisk demarcates the dorsal aortae, red arrows point to DiI-labeled dorsal pancreas bud.
In conclusion, although the pre-pancreatic endoderm is surrounded by a variety of mesoderm, many of which have been shown to support pancreas induction in explant assays, the notochord is the one tissue that exhibits a sustained regional interaction with the dorsal pancreatic endoderm.
Coelomic mesothelium contributes to the dorsal pancreatic mesenchyme
An important step in the patterning and differentiation of the initially uniform pancreatic bud is the accumulation and interaction of the dorsal pancreatic epithelium with a condensed ISL+ mesenchyme (Ahlgren et al., 1997; Attali et al., 2007; Li et al., 2004). Because this mesenchyme, hereafter referred to as the dorsal pancreatic mesenchyme (DPM), directs the differentiation from naïve pancreas epithelium to endocrine and exocrine cell lineages, we sought to determine its mesodermal source and the time at which it becomes proximal to the dorsal pancreas bud. Based on the amount of anterior displacement displayed by mesoderm-derivatives proximal to the pre-pancreatic/early pancreatic endoderm, we focused these studies on mesoderm-derived tissue lateral and posterior to those examined above.
To identify the DPM, we DiI-labeled discrete portions of 6–14S embryonic mesoderm, cultured the embryos for at least 24 hours and sectioned to identify the labeled mesoderm descendants. DiI-labeling of midline cells (n=9, SFig. 1A–C), somatopleure (n=23, SFig. 1D–F), and splanchnopleure, (n=91, SFig. 1G–I) never resulted in labeled DPM. The only tissue we identified that contributes to the DPM is the ISL1+ dorsal coelomic mesothelium adjacent to the 7th–9th somite pairs in the 8–13S embryo. Figure 5A summarizes the coelomic mesothelium data and highlights an 8S embryo labeled at the level of the 8th somite (Fig. 5B,C) that was cultured to 26S (Fig. 5D) and produced DiI-labeled descendants in both the ISL1+ coelomic mesothelium adjacent to the dorsal pancreas bud and in ISL1+ DPM that appears to be migrating from the coelomic mesothelium (Fig.5 E,E′). Because the dorsal pancreas progenitors are located over the 2nd–4th somites in 8–13S embryos, our data suggests the coelomic-derived DPM is not in direct contact with the pancreatic progenitors until after 13S. Interestingly, although the numbers are limited, we find that the ventral coelomic mesothelium can also contribute to the mesenchyme surrounding the ventral pancreas bud (n=3, SFig. 2). Intriguingly, and unlike the early inductive mesoderm, these results suggest that the ventral pancreatic mesenchyme and DMP share a common mesoderm-derived source.
Fig. 5. Coelomic mesothelium gives rise to dorsal pancreas mesenchyme.
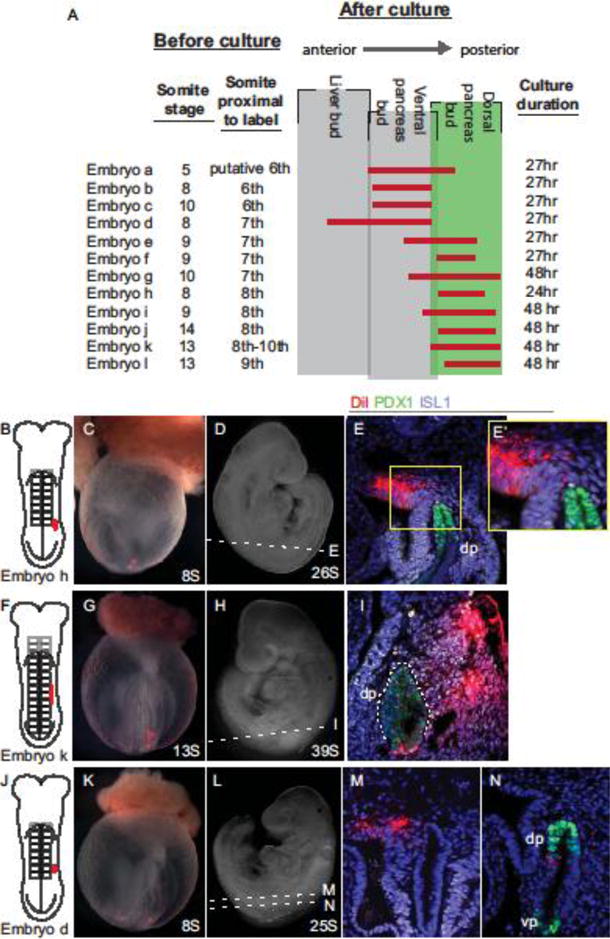
A) Summary of dorsal coelomic mesothelium labels. Each embryo is assigned a letter (Embryos a-l) and the initial somite stage and anterior/posterior level of the initial label provided. The vertical grey boxes represent the boundary of each listed embryonic structure, red lines indicate the anterior/posterior span of DiI-labeled coelomic cells as observed in transverse sections after culture of each embryo through the indicated stage. B–E) DiI was used to label the coelomic cavity adjacent to the eighth somite of an 8S embryo, as illustrated (B) and photographed in whole mount prior to (C) and after (D) culture through26S. Immunofluorescently labeled sections for ISL1 (white) and PDX1 (green) show DiI (red) in coelomic mesothelium and in ISL1+ mesenchyme dorsal to the dorsal pancreas bud (E, E′). F–I) A similar initially labeled embryo (F, G) cultured through 39S (H) shows DiI-labeled cells throughout ISL1+ mesenchyme adjacent to the dorsal pancreas (I). J–N) A more anterior label directed into the coelomic cavity at the seventh somite of an 8S embryo (J, K) and cultured through 25S (L) reveals DiI in the coelomic mesothelium and adjacent mesenchyme anterior to the dorsal pancreas (M) but absent in the pancreatic endoderm (N).
Dorsal coelomic labels at the 6–7th somite level produced labeled dorsal mesenchyme immediately anterior to and sometimes at the level of the dorsal pancreas (Fig. 5A; Embryos a–g), indicating that the anterior boundary of the presumptive DPM is approximately at the 6th somite pair. An example embryo labeled within the coelomic cavity at the 7th somite (Fig. 5J,K) and cultured to 25S (Fig. 5L) displays DiI+ cells in in coelomic mesothelium anterior to the dorsal pancreas bud (Fig. 5M), but absent from sections containing pancreas buds (Fig. 5N).
Because much of the literature regarding the patterning role of pancreatic mesenchyme has focused on later stages of development (10.5 dpc), we sought to confirm that the mesenchyme adjacent to the dorsal pancreas bud at ~9.5 dpc is that which has been characterized at ~10.5 dpc. Thus we repeated the coelomic mesothelial labels with embryos that were cultured for up to 48 hours, through ~10.5 dpc. Such an embryo, labeled at 13S in the coelomic mesothelium proximal to the 8th–10th somites (Fig. 5F,G), and cultured to 39S (Fig. 5H) produces DiI-labeled descendants that are apparent throughout the ISL1+ DPM (Fig. 5I). At these later stages of development, both the pancreas and duodenum express PDX1 and thus the dorsal pancreas is outlined by the white dotted line. Although the duodenum is DiI+, indicating that this endoderm overlies the coelomic mesothelium that will contribute to the DPM, the dorsal pancreas itself is not, consistent with our previous data, identifying the dorsal pre-pancreatic endoderm as that overlaying somite pairs 2–4 (Angelo et al., 2012).
Finally it is important to note that while only a small number of DiI+ cells appear to be in the process of migrating out of the mesothelium at ~9.5 dpc, a larger proportion of labeled cells are apparent in the DPM at ~10.5 dpc. This supports the hypothesis that the DPM infiltrates the dorsal space around the bud between these time points. This is important, as one would expect the mesenchyme that queues endocrine and exocrine differentiation to be in contact with pancreatic epithelium after the onset of induction.
DISCUSSION
Several mesoderm-derived tissues including lateral plate mesoderm, endothelial cells of the dorsal aortae, and the notochord are each capable of inducing pancreatic gene expression from nascent chick endoderm (Apelqvist et al., 1997; Hebrok et al., 1998; Hebrok et al., 2000; Kim et al., 1997; Kumar et al., 2003; Lammert et al., 2001). However few of these data have been directly recapitulated in mouse. Furthermore, given the extensive amount of morphogenesis that occurs during early organogenesis in the mammalian embryo, an additional gap in the literature includes whether any of the candidate inductive tissues engage in stable interactions with the pre-pancreatic and early pancreatic endoderm. The fate mapping presented herein demonstrates that most mesoderm proximal to the dorsal pancreas progenitors immediately before or during specification is transiently associated with the pre-pancreatic endoderm and is displaced anterior to the nascent dorsal pancreas bud. Conversely, the same portion of the notochord remains associated with the dorsal pancreas prior to, at the onset of and slightly after the onset of pancreas induction. Thus, unlike any other potentially inductive interaction, the notochord and dorsal pancreatic endoderm maintain a relatively stable and sustained interaction, supporting data that has shown the notochord to be involved in pancreas induction.
Mesenchyme Fate Mapping Reveals Unique Tissue Migrations
An important aspect of this work is identifying a significant amount of displacement between initially contiguous mesoderm and endoderm beginning at early somite stages. Between 8.5–9.5 dpc a series of dynamic changes in morphology are grossly apparent. In the region of the developing foregut, a substantial amount of remodeling results from the closing of the gut tube, turning of the embryo, expansion of the dorsal-ventral axis and remodeling of developing structures such as the dorsal aortae, somites and gut itself (Garriock et al., 2010; Santibanez et al., 2011; Tam, 1981; Tam et al., 2007; Tam and Behringer, 1997; Tremblay and Zaret, 2005). Our data reveals the constant, dynamic flow of tissues prior to, during and immediately after embryonic turning. Specifically as the endoderm, and the mesoderm dorsal and lateral to it, expand and differentiate during this period of development, our data reveals that they are rapidly displaced from one another.
In general we find that mesoderm and endoderm that are initially adjacent during early somitogenesis produce descendants in which the endoderm is positioned posterior to mesoderm, with the exception of notochord, by 9.5 dpc (Fig. 2). Additionally we find that labeled mesoderm descendants produce continuous trails along the A/P axis while labeled endoderm remains in a relatively more intact domain (Fig. 2,3). This type of migration is not unprecedented; indeed at gastrulation stages mesoderm and endoderm appear to have divergent trajectories from the primitive streak to meet at the developing anterior. The endoderm moves longitudinally from the primitive streak across the distal tip of the embryo to the forming anterior while the mesoderm moves laterally from the primitive streak toward the anterior (Lawson and Pedersen, 1992; Tam and Behringer, 1997).
Labels made at the anterior most somites result in streaks of cells that spread over large portions of the embryo, both along the A/P axis and dorsoventrally as sclerotome moves toward to notochord and somites differentiate into sclerotome and dermamyotome (Fig. 2C Embryos 1–8, 10, 11, 14, 15, 19, 20, Fig. 3B–X). It should be noted that the size of labels after culture reflects the initial number of cells labeled (compare Fig. 2C Embryo 8 versus Embryo 9). Importantly and also apparent in Figures 2 and 3, these larger labels often involve two or more tissue types such that post-culture analysis reveals trails of labeled cells within multiple mesodermal tissues arranged along the A/P axis. The separation of these initially adjacent mesodermal tissues from each other and co-labeled endoderm reveals a pattern when multiple embryos are compared. For example, dorsal aortae moves most anteriorly and notochord relatively little when compared with the endoderm (summarized in Fig. 1), indicating orchestrated movement among mesodermal derivatives that are unique to each tissue-type.
Notochord morphogenesis and regionalization
While the notochord has previously been implicated in dorsal pancreas induction, the mechanism is unknown, particularly since no inductive factors are uniquely expressed in the domain that associates with the dorsal pancreas. In addition to remaining in the same A/P plane as the dorsal pancreas between 8.5–9.5 dpc, the notochord is the only tissue in our study that displayed two distributions that were not regularly observed in other labeled mesoderm. This includes prominent gaps in the A/P distribution of labeled cells (Fig. 2, n=5/20) and the appearance of labeled notochord descendants posterior to the dorsal pancreas bud (Fig. 2, n=7/20).
The gap in labeled notochord could result from a dilution of the membrane labeled DiI via localized proliferation, the addition of otherwise sequestered cells to the region of the gap, or may be indicative of altered cell movements by notochord descendants in that region. If increased proliferation produces a gap in DiI intensity then we might expect to see a gradient of label on either end of the gap. Instead we see robust DiI intensity on both the anterior and posterior region of the gap, suggesting that a regional increase in proliferation is likely not the cause. Others have demonstrated that the notochord is composed of 3 populations of cells. (Yamanaka et al., 2007). The notochord anterior to somite 2, termed the anterior head process (AHP) arises from a dispersed cell population anterior to the node, utilizing direct convergence. In contrast, the trunk notochord, defined as that posterior to somite 2, and the tail notochord, defined as that posterior to the hindlimb buds, are both node-derived although the former utilizes convergence extension and the latter posterior migration to form. It is notable that when we observe a gap in labeled notochord the excluded region includes the caudal-most portion of the AHP notochord, and suggests that the progenitors of the notochord in the unlabeled gap are distinct from the remainder of the AHP notochord. Furthermore, given that the trunk notochord utilizes convergent extension to form, it is also reasonable to attribute the slight posterior displacement of the trunk notochord to convergent-extension movements.
We find it interesting that the border between the AHP and trunk notochord overlies the dorsal pancreatic endoderm progenitors. Our data shows two distinctly labeled sections of notochord, one anterior and one posterior, presumably descendants of AHP and trunk notochord, respectively. Upon pancreas bud formation (~9.5 dpc), the anterior portion of labeled notochord stretches from the anterior liver bud towards the common atrial chamber (anterior heart, Fig.2 Embryo 1, 6, 7, 19, 20). In juxtaposition, the notochord adjacent to the dorsal pancreas bud is that which appears to have been posteriorly displaced, suggesting the posterior directed notochord is the portion poised to provide inductive signals to dorsal pancreas. Furthermore, these fate-mapping results are consistent with the regionalized gene expression of the notochord, noted below, and further suggest a mechanism of regionalization where the domain of notochord capable of induction is in contact with the portion of endoderm competent to respond. Studies in chick indicate that foregut endoderm is competent to express Pdx1, but not the hindgut endoderm (Kumar et al., 2003). This could explain the formation of one dorsal organ, at the border of the fore- and midgut, adjacent to a posteriorly extending portion of notochord.
There are multiple examples of genes expressed in notochord anterior to the dorsal pancreatic endoderm (Ding et al., 1998; Failli et al., 2002; Furuta et al., 1997; Rodriguez-Magadan et al., 2008; Tamplin et al., 2011; Tamplin et al., 2008) or posterior to it (Borello et al., 1999; Danesh et al., 2009; Lyons et al., 1995; Medioni et al., 2010; Tamplin et al., 2011; Tamplin et al., 2008) at specific time points before, during and soon after induction. The hypothesis that the notochord is also partitioned by patterns of discrete gene expression, much like the gut tube which is regionalized by similar domains of gene expression (Sherwood et al., 2009), may also explain the presence of a single dorsal endoderm organ bud arising at the junction of these expression domains.
Mesothelial origin of the condensed pancreatic mesenchyme
In addition to the initial inductive events that specify the dorsal pancreas bud, a second mesoderm derivative, the condensed pancreatic mesenchyme, directs exocrine and endocrine differentiation and is vital for the creation of a functional pancreas. Both the presence and proportion of exocrine and endocrine cell types in the developing epithelium is dictated by the amount of appropriate mesenchyme and its proximity (Attali et al., 2007; Li et al., 2004). Because the fate of the pancreatic parenchyma appears to be entirely dictated by the mesenchyme, an endoderm/mesenchyme interaction must be tightly regulated for proper differentiation to occur.
One way to control the dose of potent patterning signals could be the slow infiltration of mesenchymal cells from the coelomic mesothelium via an epithelial to mesenchymal transition (EMT). Such a transition would allow for the slow accumulation of mesenchyme and patterning signals, guarding against premature differentiation and exhaustion of the PDX1+ progenitor pool (Attali et al., 2007). Our fate mapping supports the hypothesis of a slow infiltration of mesenchyme from the coelomic mesothelium beginning at ~9.5 dpc and extending through at least 10.5 dpc (Fig. 5D). These data suggest that mesothelial cells robustly populate the DPM by ~10.5 dpc and are then poised to pattern the pancreatic epithelium.
The coelomic mesothelium has been shown to differentiate into diverse cell lineages, often through an EMT, and to contribute to multiple developing organs [reviewed in (Ariza et al., 2016)], thus our results are not without precedent. Genetic lineage tracing, utilizing several Cre-lines with expression within portions of coelomic mesothelium, have been used to demonstrate this dynamic disbursement. For example, Mesothelin-expressing coelomic mesothelium produces alveolar myofibroblasts, intestinal fibroblasts and smooth muscle cells of the trunk (Rinkevich et al., 2012) and GATA4-expressing mesothelium contributes to portions of the liver, aorta, body wall, esophagus, adrenals, and genital ridge lineages (Delgado et al., 2014; Rojas et al., 2005). Although a similar EMT has not been previously demonstrated in the pancreas, the prevalence of coelomic mesothelium contribution and the diversity of resulting cell lineages supports our novel observation that a specific portion of the coelomic mesothelium contributes to the pancreatic mesenchyme.
While our data demonstrates that the coelomic mesothelium contributes to the DPM, it does not exclude the possibility of an additional mesenchymal source. In other organs contribution from coelom and additional mesenchyme has been documented. For example, the lung pulmonary mesenchyme is derived from splanchnopleural mesoderm that is infiltrated by WT1+ coelomic mesothelium (Cano et al., 2013). However, no other labeled mesoderm gave rise to the DPM (n= 0/114). Extensive labeling was performed directly posterior to the pancreas progenitors, targeting the few loose midline dorsal mesenchyme cells seen in more anterior midline labels (Fig. 4D,K), and lateral to the somites, targeting splanchnopleure. Midline labels resulted in a small population of ISL1− cells close to the neural tube and not the dorsal pancreas bud (SFig. 1A–C). Splanchnic mesenchyme seemed a plausible source of DPM as it encases the gut tube (Matsushita and Matsushita, 1995; Wells and Spence, 2014). After extensive labeling of somatopleure or splanchnopleure no DiI+ DPM cells was observed (SFig1 D–I). Labeled splanchnic mesenchyme remained adjacent to the lateral portions of the gut tube and did not migrate dorsally. Descendants of somatopleure extended dorsolateral to the gut and yielded neither ISL1+ cells or any cells near the dorsal pancreas bud. The DPM remained illusively between the labeled clones of these two tissues. The resulting descendants of labeled splanchnopleure, somatopleure and coelomic mesothelium appear mutually exclusive.
CONCLUSIONS
Herein we present a fate map of the mesoderm derivatives that are proximal to the pancreatic endoderm prior to and shortly after the onset of Pdx1 expression and bud formation. Overall, the descendants of each labeled tissue type displays its own pattern of distribution that is particularly obvious in the aggregate and are ultimately located anterior to the early dorsal pancreas bud at the end of culture. Furthermore, these data suggest that the interaction of the dorsal pancreas progenitors/early dorsal pancreas bud with much of the initially proximal mesoderm derived tissue is transient, excluding the possibility that such mesoderm acts as a stable source of signals required for pancreas induction in vivo. The notochord is the sole mesoderm derivative that maintains its proximity to dorsal pancreatic epithelium during and after induction, supporting the hypothesis that the notochord is the tissue that governs pancreatic induction in vivo. As development proceeds the notochord is dorsally displaced from the pancreas [reviewed in (Gittes, 2009)] and we demonstrate that dorsal mesenchyme originating from the coelomic mesothelium gradually infiltrates the dorsal region surrounding the nascent bud. This fate mapping documents tissue movement in the developing embryo from before induction to the time when the first differentiation will take place within the dorsal pancreas bud and presents a platform for further studies to understand the embryo as a whole and how the signaling within may be translated into differentiation protocols to lead to further advances in stem cell and tissue replacement therapies.
Supplementary Material
SFig. 1. Midline mesenchyme and lateral plate labels do not give rise to pancreatic mesenchyme. A–C′) A representative midline labeled 12S embryo in illustration (A) and whole mount view (B) cultured to 26S and sectioned shows DiI (red) in a few scattered midline cells between neural tube and gut. D–F) Labels within somatopleuric mesenchyme (D, E) result in DiI labeled cells lateral to the dorsal pancreas bud and never proximal (F,F′). G–I) Labels of splanchnic mesenchyme (G, H) result in DiI labeled cells lateral to the gut and never proximal to either pancreas bud (I,I′).
SFig. 2. Ventral coelomic mesothelium contributes to ventral pancreas mesenchyme. A–C) An example of a 14S embryo labeled in the coelomic cavity at the seventh somite (A, B) and cultured to 27S. Sections of this embryo were labeled immunofluorescently for PDX1 (green) show DiI (red) in the ventral coelomic mesothelium and mesenchyme adjacent to the ventral pancreas bud (C, C′).
Highlights.
Most proposed inductive tissues move away from dorsal pancreas precursors in vivo.
Only a portion of the notochord remains proximal to the dorsal pancreas.
The condensed pancreatic mesenchyme is derived from coelomic mesothelium.
Acknowledgments
We would like to thank members of the Tremblay and Mager labs for their support and stimulating discussion of this work, Dr. Jesse Mager, Amrita Palaria, Chelsea Marcho, and Gabriel El Sebae for their helpful critiques, and the mice that made this work possible. This work was supported by and NIH grant RO1DK87753 and basic science award from the American Diabetes Association (1-10BS-178 to KDT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Ahlgren U, Pfaff SL, Jessell TM, Edlund T, Edlund H. Independent requirement for ISL1 in formation of pancreatic mesenchyme and islet cells. Nature. 1997;385:257–260. doi: 10.1038/385257a0. [DOI] [PubMed] [Google Scholar]
- Angelo JR, Guerrero-Zayas MI, Tremblay KD. A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS One. 2012;7:e40707. doi: 10.1371/journal.pone.0040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo JR, Tremblay KD. Laser-mediated cell ablation during post-implantation mouse development. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:1202–1209. doi: 10.1002/dvdy.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Current biology : CB. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Ariza L, Carmona R, Canete A, Cano E, Munoz-Chapuli R. Coelomic epithelium-derived cells in visceral morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2016;245:307–322. doi: 10.1002/dvdy.24373. [DOI] [PubMed] [Google Scholar]
- Attali M, Stetsyuk V, Basmaciogullari A, Aiello V, Zanta-Boussif MA, Duvillie B, Scharfmann R. Control of beta-cell differentiation by the pancreatic mesenchyme. Diabetes. 2007;56:1248–1258. doi: 10.2337/db06-1307. [DOI] [PubMed] [Google Scholar]
- Balmer S, Nowotschin S, Hadjantonakis AK. Notochord morphogenesis in mice: Current understanding & open questions. Developmental dynamics : an official publication of the American Association of Anatomists. 2016;245:547–557. doi: 10.1002/dvdy.24392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Buffa V, Sonnino C, Melchionna R, Vivarelli E, Cossu G. Differential expression of the Wnt putative receptors Frizzled during mouse somitogenesis. Mech Dev. 1999;89:173–177. doi: 10.1016/s0925-4773(99)00205-1. [DOI] [PubMed] [Google Scholar]
- Cambray N, Wilson V. Two distinct sources for a population of maturing axial progenitors. Development. 2007;134:2829–2840. doi: 10.1242/dev.02877. [DOI] [PubMed] [Google Scholar]
- Cano E, Carmona R, Munoz-Chapuli R. Wt1-expressing progenitors contribute to multiple tissues in the developing lung. Am J Physiol Lung Cell Mol Physiol. 2013;305:L322–332. doi: 10.1152/ajplung.00424.2012. [DOI] [PubMed] [Google Scholar]
- Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danesh SM, Villasenor A, Chong D, Soukup C, Cleaver O. BMP and BMP receptor expression during murine organogenesis. Gene expression patterns : GEP. 2009;9:255–265. doi: 10.1016/j.gep.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I, Carrasco M, Cano E, Carmona R, Garcia-Carbonero R, Marin-Gomez LM, Soria B, Martin F, Cano DA, Munoz-Chapuli R, Rojas A. GATA4 loss in the septum transversum mesenchyme promotes liver fibrosis in mice. Hepatology. 2014;59:2358–2370. doi: 10.1002/hep.27005. [DOI] [PubMed] [Google Scholar]
- Dessimoz J, Opoka R, Kordich J, Grapin-Botton A, Wells J. FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mechanisms of development. 2006;123:42–55. doi: 10.1016/j.mod.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lóra J, Zaret K. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development (Cambridge, England) 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, Hui CC. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- Failli V, Bachy I, Retaux S. Expression of the LIM-homeodomain gene Lmx1a (dreher) during development of the mouse nervous system. Mech Dev. 2002;118:225–228. doi: 10.1016/s0925-4773(02)00254-x. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Piston DW, Hogan BL. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development. 1997;124:2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- Garriock R, Czeisler C, Ishii Y, Navetta A, Mikawa T. An anteroposterior wave of vascular inhibitor downregulation signals aortae fusion along the embryonic midline axis. Development (Cambridge, England) 2010;137:3697–3706. doi: 10.1242/dev.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes GK. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. doi: 10.1016/j.ydbio.2008.10.024. [DOI] [PubMed] [Google Scholar]
- Gittes GK, Galante PE, Hanahan D, Rutter WJ, Debase HT. Lineage-specific morphogenesis in the developing pancreas: role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- Golosow N, Grobstein C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Developmental Biology. 1962:242–255. doi: 10.1016/0012-1606(62)90042-8. [DOI] [PubMed] [Google Scholar]
- Harrison K, Thaler J, Pfaff S, Gu H, Kehrl J. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nature genetics. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim S, Melton D. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes & development. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M, Kim S, St Jacques B, McMahon A, Melton D. Regulation of pancreas development by hedgehog signaling. Development (Cambridge, England) 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Developmental Biology. 2003;258:105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- Kim S, Hebrok M, Melton D. Notochord to endoderm signaling is required for pancreas development. Development (Cambridge, England) 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Landsman L, Nijagal A, Whitchurch TJ, Vanderlaan RL, Zimmer WE, Mackenzie TC, Hebrok M. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS biology. 2011;9:e1001143. doi: 10.1371/journal.pbio.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Pedersen RA. Clonal analysis of cell fate during gastrulation and early neurulation in the mouse. Ciba Foundation Symposium. 1992;165:3–21. doi: 10.1002/9780470514221.ch2. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. Early neurogenesis in Amniote vertebrates. Int J Dev Biol. 2001;45:373–378. [PubMed] [Google Scholar]
- Li H, Arber S, Jessell TM, Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nature genetics. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- Li Z, Manna P, Kobayashi H, Spilde T, Bhatia A, Preuett B, Prasadan K, Hembree M, Gittes GK. Multifaceted pancreatic mesenchymal control of epithelial lineage selection. Dev Biol. 2004;269:252–263. doi: 10.1016/j.ydbio.2004.01.043. [DOI] [PubMed] [Google Scholar]
- Lyons KM, Hogan BL, Robertson EJ. Colocalization of BMP 7 and BMP 2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. Mech Dev. 1995;50:71–83. doi: 10.1016/0925-4773(94)00326-i. [DOI] [PubMed] [Google Scholar]
- Matsushita S, Matsushita S. Fate mapping study of the splanchnopleural mesoderm of the 1.5-day-old chick embryo. Roux Arch Dev Biol. 1995;204:392–399. doi: 10.1007/BF00360484. [DOI] [PubMed] [Google Scholar]
- Medioni C, Bertrand N, Mesbah K, Hudry B, Dupays L, Wolstein O, Washkowitz AJ, Papaioannou VE, Mohun TJ, Harvey RP, Zaffran S. Expression of Slit and Robo genes in the developing mouse heart. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:3303–3311. doi: 10.1002/dvdy.22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Czernichow P, Scharfmann R. Follistatin regulates the relative proportions of endocrine versus exocrine tissue during pancreatic development. Development. 1998;125:1017–1024. doi: 10.1242/dev.125.6.1017. [DOI] [PubMed] [Google Scholar]
- Palaria A, Angelo JR, Guertin T, Mager J, Tremblay K. Patterning of the hepatopancreatobiliary (HPB) boundary by BMP signaling reveals heterogeneity within the murine liver bud. Hepatology. 2018 doi: 10.1002/hep.29769. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Mori T, Sahoo D, Xu PX, Bermingham JR, Jr, Weissman IL. Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nat Cell Biol. 2012;14:1251–1260. doi: 10.1038/ncb2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Magadan H, Merino E, Schnabel D, Ramirez L, Lomeli H. Spatial and temporal expression of Zimp7 and Zimp10 PIAS-like proteins in the developing mouse embryo. Gene expression patterns : GEP. 2008;8:206–213. doi: 10.1016/j.modgep.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- Rose MI, Crisera CA, Colen KL, Connelly PR, Longaker MT, Gittes GK. Epithelio-mesenchymal interactions in the developing mouse pancreas: morphogenesis of the adult architecture. J Pediatr Surg. 1999;34:774–779. doi: 10.1016/s0022-3468(99)90372-x. discussion 780. [DOI] [PubMed] [Google Scholar]
- Santibanez JF, Quintanilla M, Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clinical science. 2011;121:233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin SCJC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Developmental cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. The control of somitogenesis in mouse embryos. J Embryol exp Morph. 1981;65:103–128. [PubMed] [Google Scholar]
- Tam P, Khoo P, Lewis S, Bildsoe H, Wong N, Tsang T, Gad J, Lorraine R. Sequential allocation and global pattern of movement of the definitive endoderm in the mouse embryo during gastrulation. Development. 2007;134:251–260. doi: 10.1242/dev.02724. [DOI] [PubMed] [Google Scholar]
- Tam PP, Behringer RR. Mouse gastrulation: the formation of a mammalian body plan. Mech Dev. 1997;68:3–25. doi: 10.1016/s0925-4773(97)00123-8. [DOI] [PubMed] [Google Scholar]
- Tamplin OJ, Cox BJ, Rossant J. Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord. Dev Biol. 2011;360:415–425. doi: 10.1016/j.ydbio.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Tamplin OJ, Kinzel D, Cox BJ, Bell CE, Rossant J, Lickert H. Microarray analysis of Foxa2 mutant mouse embryos reveals novel gene expression and inductive roles for the gastrula organizer and its derivatives. BMC genomics. 2008;9:511. doi: 10.1186/1471-2164-9-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay K, Zaret K. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Developmental Biology. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Wells J, Melton D. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development (Cambridge, England) 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- Wells JM, Spence JR. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SFig. 1. Midline mesenchyme and lateral plate labels do not give rise to pancreatic mesenchyme. A–C′) A representative midline labeled 12S embryo in illustration (A) and whole mount view (B) cultured to 26S and sectioned shows DiI (red) in a few scattered midline cells between neural tube and gut. D–F) Labels within somatopleuric mesenchyme (D, E) result in DiI labeled cells lateral to the dorsal pancreas bud and never proximal (F,F′). G–I) Labels of splanchnic mesenchyme (G, H) result in DiI labeled cells lateral to the gut and never proximal to either pancreas bud (I,I′).
SFig. 2. Ventral coelomic mesothelium contributes to ventral pancreas mesenchyme. A–C) An example of a 14S embryo labeled in the coelomic cavity at the seventh somite (A, B) and cultured to 27S. Sections of this embryo were labeled immunofluorescently for PDX1 (green) show DiI (red) in the ventral coelomic mesothelium and mesenchyme adjacent to the ventral pancreas bud (C, C′).


