Abstract
Increasing evidence indicates that molecular hydrogen-dissolved alkaline electrolyzed water (AEW) has various physiological activities such as antioxidative activity. Gut microbiota are deeply associated with our health through a symbiotic relationship. Recent reports have described that most gastrointestinal microbial species encode the genetic capacity to metabolize molecular hydrogen, meaning that molecular hydrogen might affect the gut microbial composition. Nevertheless, AEW effects on gut microbiota remain unknown. This study investigated AEW effects on the intestinal environment in mice, including microbial composition and short-chain fatty acid contents. After mice were administered AEW for 4 weeks, 16S rRNA gene sequencing analyses revealed their fecal microbiota profiles. Organic acid concentrations in cecal contents were measured using an HPLC system. Compared to the control group, AEW administration mice had significantly lower serum low-density lipoprotein cholesterol level and alanine aminotransferase activity. Organic acid concentrations of propionic, isobutyric, and isovaleric acids were higher in AEW-administered mice. Results of 16S rRNA gene sequencing analyses showed that the relative abundances of 20 taxa differed significantly in AEW-administered mice. Although the definitive role of gut microbes of AEW-administered mice remains unknown, our data demonstrate the possibility that AEW administration affects the gut microbial composition and that it has beneficial health effects in terms of cholesterol metabolism and liver protection.
Keywords: gut microbiota, propionic acid, low-density lipoprotein cholesterol, alanine aminotransferase, hydrogen, alkaline electrolyzed water, 16S rRNA, Clostridiaceae
INTRODUCTION
The large bowel is home to complex and diverse communities of microbiota that play important roles in health through a symbiotic relationship with the host. Disruption of this relationship under abnormal conditions is increasingly recognized as a major risk factor for various diseases such as metabolic disorders.1,2,3 The major function of microbiota in the intestinal lumen is the fermentation of dietary fiber and resistant starch,4 which generates short-chain fatty acids (SCFAs). Emerging evidence indicates that SCFAs have the most important physiological effects on the colonic mucosa, including stimulation of mucous secretion, increase of motility, and sodium and water absorption.5 Additionally, SCFAs can regulate systemic physiological and pathophysiological events, regulating the sympathetic nervous system, controlling body energy utilization, and differentiating immune cells.6,7,8 Changes of microbiota composition followed by alteration in SCFA contents play pivotal roles in host health and disease.
Some bioactivities of molecular hydrogen (H2)-dissolved alkaline electrolyzed water (AEW) have been reported. Koyama et al.9 reported for humans that AEW more strongly suppresses urinary excretion of exercise-induced 8-hydroxy-2'-deoxyguanosine, a biomarker of global DNA oxidation, than a placebo. Furthermore, a study using a hindlimb unloading model of rat has demonstrated that AEW ingestion decreases oxidative stress and attenuates muscle atrophy.10 Alkaline electrolyzed water was also used in some sterilization applications. Alkaline electrolyzed water showed a strong antimicrobial effect and served to reduce populations of pathogenic bacteria such as Escherichia coli O157 and Salmonella.11 However, molecular H2 is produced as an endproduct of carbohydrate fermentation. It is reoxidized primarily by sulfate-reduction, acetogenesis, and methanogenesis. A recent report described that 70% of gastrointestinal microbial species listed in the Human Microbiome Project encoded the genetic capacity to metabolize H2,12 meaning that molecular H2 might affect the gut microbial composition. Taken together, understanding AEW health benefits requires precise investigation of AEW effects on the intestinal microbiome. This report is the first of a study yielding insight into AEW effects on the intestinal environment, including alterations of SCFA contents in mice.
MATERIALS AND METHODS
Preparation of treatment solutions
The AEW used for this study was generated using an alkaline ionized water apparatus (Panasonic Corporation, Shiga, Japan), which have been approved as a medical device in Japan. The apparatus reduces water to produce molecular hydrogen. Drinking AEW has been found to be effective in relieving gastrointestinal symptoms for the patients with gastrointestinal symptoms.13 The H2 concentrations (pH) of normal water and AEW were 0.00 (6.83) and 0.32 (9.90) ppm, respectively. To reduce the loss of the H2, aluminum pouches were used to supply water. The pouches were kept pointing downward to prevent the ingress of air.
Animals
After 5-week-old male C57BL/6N mice used for this study were obtained (Shimizu Laboratory Supplies Co. Ltd., Kyoto, Japan), they were kept at 18–24°C and 40–70% relative humidity, with a 12-hour light/dark cycle. They were allowed free access to water and diet (CE-2; CLEA Japan Inc., Tokyo, Japan) for 1 week during their acclimatization period. Experimental procedures were conducted in accordance with NIH guidelines for the use of experiments animals. All experimental protocols were approved by the Animal Care Committee of Kyoto Prefectural University of Medicine (permission number M28-500).
After acclimatization, the mice were divided randomly into two groups of eight mice each. They were given either normal water (control) or AEW for 4 weeks. After the mice were killed under anesthesia, their blood, liver, epididymal adipose tissue, cecal content, and fresh stool samples were collected immediately.
Blood biochemical analysis
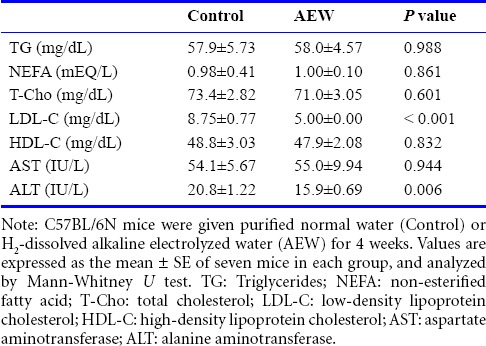
Blood supernatants were obtained as serum after centrifugation at 1,500 × g for 10 minutes. Concentrations of aspartate aminotransferase (AST), alanine aminotransferase (ALT), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), triglyceride (TG), total cholesterol (T-Cho), and non-esterified fatty acid (NEFA) in serum were measured at SRL Inc. (Tokyo, Japan).
Determination of organic acid concentrations in cecal contents
The organic acid concentrations in cecal contents were measured using a high performance liquid chromatography (HPLC) system according to the method described by Ushida et al.14 In brief, a portion of the cecal contents (0.3 mg) was suspended with 0.5 mL of 14% perchloric acid to eliminate protein. After centrifugation at 10,000 r/min at 4°C for 5 minutes, the supernatant was filtered through a cellulose acetate membrane filter with 0.45 µm pore size; then an aliquot of the resultant sample solution was injected into the HPLC system for analysis.
Microbiota analysis by 16S rRNA sequencing
Fecal samples were collected, placed into tubes, and kept at −80°C until further use. Bacterial genomic DNA was extracted from the fecal samples, as described in an earlier report.15 Preparation of the library for DNA sequencing using a MiSeq desktop sequencer (Illumina Inc., CA, USA) was conducted according to the protocol with minor modifications, as described in an earlier report.16 The V3-V4 regions of the 16S rRNA genes in each sample were amplified (KAPA HiFi HotStart Ready Mix; Kapa Biosystems Inc., MA, USA), with primers 341F and 805R that contained a 5'overhang adapter sequence. The amplicon was purified using NucleoFast 96 PCR plates (Takara Bio Inc., Shiga, Japan). A second PCR was conducted (KAPA HiFi HotStart Ready Mix) to attach a unique combination of dual indices (I5 and I7) and Illumina Inc. sequencing adapters to each sample. The amplicon of the second PCR was purified. Then the concentration was normalized (SequalPrep Normalization Plate Kit; Life Technologies Japan Ltd., Tokyo, Japan). Each of the normalized amplicons was then pooled evenly and concentrated using beads (AMPure XP; Beckman Coulter Inc., Tokyo, Japan). The size and quantity of the library were assessed respectively with a Bioanalyzer 2100 (Agilent Technologies Japan Ltd., Tokyo, Japan) and a Library Quantification Kit for Illumina (Kapa Biosystems Inc.). The library was denatured with 0.2 M NaOH (Sigma-Aldrich Japan K.K., Tokyo, Japan) and combined with phiX Control (v3, expected 20%; Illumina Inc.). From the library, 11 pM were combined with phiX Control and were heat-denatured at 96 °C for 2 minutes before sequencing using a 300 bp paired-end strategy on the MiSeq (Illumina Inc.), according to the manufacturer's instructions.
Sequence data analysis
Post-processing sequencing data were analyzed using software (Quantitative insights into Microbial Ecology (QIIME) pipeline ver. 1.8.0).17 Paired-end reads were assembled using the default parameters. Sequence reads with average quality < Q20 were removed. Chimeric sequences were removed using Usearch V6.1.544, based on the Uchime algorithm implemented in QIIME.18 Clean Fastq data were aligned into operational taxonomic units (OTUs) at 97% similarity, using open reference OTU picking against the 16S database (Greengenes database ver. 13.8).19 Taxonomic identification was performed at the phylum and genus levels. The representative (most abundant) sequence of each genus was investigated using basic local alignment search tool (BLAST) searches (http://blast. ncbi.nlm.nih.gov/Blast.cgi) to confirm the closest species. Alpha and beta diversity indices were computed at a sequence depth of 9,379 sequences for the sample of lowest sequence. Principal coordinate analysis (PCoA) was applied to plot the variation in the unweighted UniFrac distance between samples. In addition, unweighted-pair group method using arithmetic means (UPGMA) clustering was applied to unweighted UniFrac distance matrices to build an UPGMA tree.
Statistical analysis
Comparisons of two groups were done using Mann-Whitney U test. Statistical analyses were conducted using software (GraphPad Prism ver. 5.02; GraphPad Software Inc., San Diego, CA, USA). All results are expressed as the mean ± SE. Correlations between parameters were assessed by Pearson's correlation test. Differences for which P values of < 0.05 and < 0.01 were inferred as significant.
RESULTS
Effects of AEW on physical and serum biochemical parameters
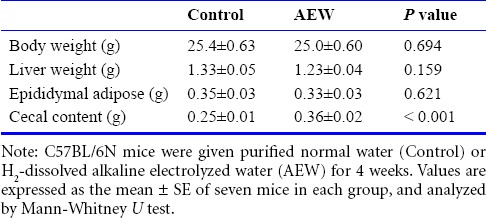
Six-week-old mice were administered AEW for 4 weeks. The AEW administration had no effect on body weight gain (Table 1). Similar results were obtained when liver weight and epididymal adipose weight were determined. However, the weight of cecal contents, which is a marker of intestinal fermentation, was significantly greater in AEW-treated mice than in control mice.
Table 1.
Effects of molecular hydrogen-dissolved alkaline electrolyzed water on physical parameters at 4 weeks of treatment
Biochemical parameters of the serum obtained from mice administered with AEW are presented in Table 2. The AEW-treated mice exhibited significantly lower serum LDL-C level and ALT activity. No appreciable difference in TG, NEFA, T-Cho, HDL-C level, or AST activity was found between the two groups.
Table 2.
Effects of molecular hydrogen-dissolved alkaline electrolyzed water on serum metabolic parameters at 4 weeks of treatment
Effects of AEW on composition of SCFAs in mouse cecum
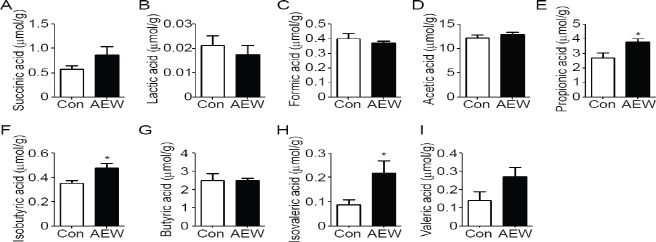
We calculated the concentrations of cecal organic acids as metabolites of microbiota using an HPLC system. Mice treated with AEW produced significantly more propionic acid, isobutyric acid, and isovaleric acid than control mice did (Figure 1). No difference was found in the production of succinic acid, lactic acid, formic acid, acetic acid, butyric acid, or valeric acid.
Figure 1.
Effects of AEW on short-chain fatty acids in mouse cecal contents.
Note: Concentrations of succinic (A), lactic (B), formic (C), acetic (D), propionic (E), isobutyric (F), butyric (G), isovaleric (H), and valeric acid (I) in the cecal are shown. Data represent the mean ± SE of seven mice. *P < 0.05, vs. control group (Con) (Mann-Whitney U test). AEW: Alkaline electrolyzed water group.
Effect of AEW administration on the gut microbiota
To assess changes in the microbial community induced by AEW, variable regions V3-V4 of 16S rRNA genes extracted from stool samples from mice in each group were sequenced using Illumina MiSeq platforms. The results are presented as OTUs using a 97% homology cutoff value. Phylogenic differences within gut microbiota were assessed using PCoA method (Figure 2A). The AEW-administered mice exhibited a distinct microbiota composition that clustered separately from that of the control mice. As depicted in Figure 2B, hierarchical clustering also showed that the microbial composition in the stool of AEW-administered mice differed from that of control mice. The AEW-administered mice exhibited significantly lower the number of Simpson and Shannon indices (P < 0.01, data not shown).
Figure 2.
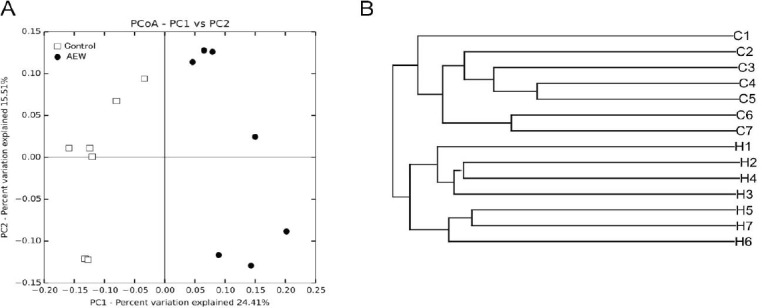
Modulation of molecular hydrogen-dissolved alkaline electrolyzed water (AEW) on the structure of gut microbiota.
Note: Principal coordinate analysis (PCoA) (A) and sample clustering results (B) of the unweighted UniFrac distances of microbial 16S rRNA sequences from the V3-V4 regions in the stool samples. C1 to C7 indicate individuals in the control group, and H1 to H7 indicate individuals in the AEW-administered group.
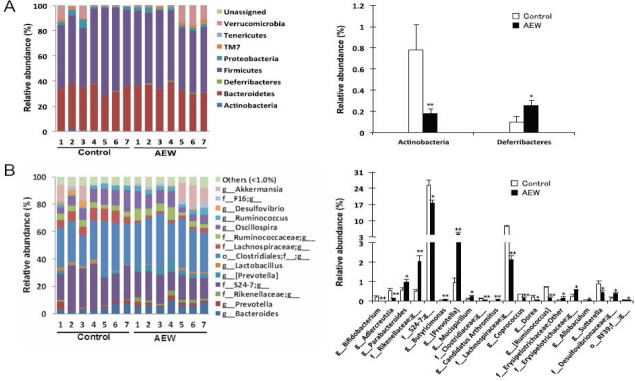
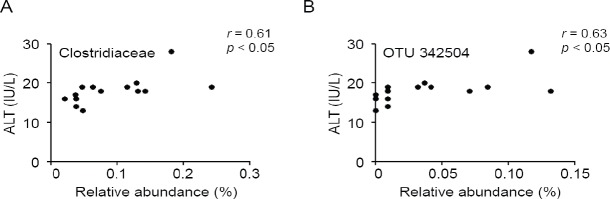
To evaluate AEW administration effects on gut microbial composition precisely, we compared the relative abundance of the entire detected taxa in each group (Figure 3). At the phylum level, the AEW administration led to significantly lower relative abundance of Actinobacteria than in control mice (Figure 3A). The relative abundance of Deferribacteres was higher in AEW-administered mice. At the genus level, the AEW-administered mice microbial community showed significantly greater abundance of Parabacteroides, Rikenellaceae, Butyricimonas, Prevotella, Mucispirillum, Candidatus arthromitus, Erysipelotrichaceae, Allobaculum, Desulfovibrionaceae, and RF39 (Figure 3B). The relative abundances of Bifidobacterium, Adlercreutzia, S24-7, Clostridiaceae, Lachnospiraceae, Coprococcus, Dorea, Ruminococcus, and Sutterella were lower in AEW-administered mice. Furthermore, Pearson correlation analysis revealed that the relative abundance of Clostridiaceae was positively correlated with serum ALT level (Figure 4A). To estimate which strain contributes to its positive correlation, Pearson correlation analysis with serum ALT level was performed for all sequences constituting Clostridiaceae. As shown in Figure 4B, the relative abundance of OTU 342504 (the most abundant sequence of Clostridiaceae) was positively correlated with serum ALT level. Taken together, although it is unclear whether the decrease in OTU 342504 leads to a decrease in serum ALT level, it is indicated that OTU 342504 is a bacteria strongly correlated with serum ALT level.
Figure 3.
Modulation of molecular hydrogen-dissolved alkaline electrolyzed water (AEW) on the composition of gut microbiota.
Note: Phylum level (A) and genus level (B) taxonomic distributions of the microbial communities in the stool samples ascertained from next-generation sequencing. The right graph portrays significantly different taxa. Data represent the means ± SE of seven mice. *P < 0.05, **P < 0.01, vs. control group (Mann-Whitney U test).
Figure 4.
Correlation between alanine aminotransferase (ALT) and gut microbiota.
Note: Correlation between the serum ALT level and the relative abundances of Clostridiaceae (A) and operational taxonomic units (out) 342504 (B) were calculated using Pearson correlation analysis.
DISCUSSION
Results of this study show that oral administration of AEW is associated with more cecal contents, indicating that AEW can facilitate intestinal fermentation. In fact, some SCFAs in the cecum were significantly more abundant in AEW-administered mice. Of these, propionic acid is known to have many physiological functions. Propionic acid lowers fatty acids contents in the liver and plasma, exerts immunosuppressive actions, and improves insulin sensitivity.20 Our previous study showed that propionic acid induced peroxisome proliferator-activated receptor α (PPARα) expression, and subsequently promoted fatty acid oxidation.21 It is particularly interesting that Hansen et al.22 reported that PPARα activation using agonist engenders a decrease in serum LDL-C levels in mice. Our data showed that the serum LDL-C level was significantly lower in AEW-administered mice than in control mice. These data suggest that alterations of propionic acid level are associated with LDL-C lowering effects of AEW. However, AEW-mediated lowering effects of LDL-C and ALT activity were very minor. To support a more definitive role for AEW for our health, further investigations using some disease model such as fatty liver and arteriosclerosis must be conducted.
According to data of the Human Microbiome Project, most intestinal bacteria can metabolize H2,12 suggesting that AEW might affect the gut microbial composition. Indeed, the relative abundances of 20 taxa differed considerably between AEW-administered mice and control mice. Results of 16S rRNA gene sequencing analyses showed significant association of AEW administration with higher relative abundances of Parabacteroides, Rikenellaceae, Butyricimonas, Prevotella, Mucispirillum, Candidatus arthromitus, Erysipelotrichaceae, Allobaculum, Desulfovibrionaceae, and RF39. After identifying the most abundant sequences in respective taxa, the closest species of these sequences were determined using BLAST for a homology search of the GenBank database. Butyricimonas virosa strain MT12 (the most abundant sequence of Butyricimonas) produces several SCFAs including isobutyric acid and propionic acid as metabolites from glucose and glycerol.23 Data from the SCFAs concentrations show that the concentrations of isobutyric acid and propionic acid were significantly higher in AEW-administered mice. These SCFAs increases might have been associated with the increase of the Butyricimonas. Candidatus arthromitus sp. SFB-mouse-NL (one of the closest species of the most abundant sequence of Candidatus arthromitus), also known as segmented filamentous bacteria, plays a key role in the maturation of gut innate and adaptive immune systems.24 Additionally, they can induce a strong IgA response and activation of immune cells such as CD8+ and CD4+ T lymphocytes. Regarding Parabacteroides, Rikenellaceae, Prevotella, Erysipelotrichaceae, Allobaculum, and RF39, their most abundant sequences were uncultured bacteria. It is particularly interesting that Allobaculum (the most abundant sequence: uncultured bacterium clone CRWD2) was not detected in feces of most control mice, meaning that this genus is highly susceptible to AEW and/or molecular H2. More recently, the relative abundance of Prevotella was found to be low in constipated patients. This genus has the positive effects on stool frequency and consistency,25 suggesting that the increase of Prevotella by AEW contributes to improvement of constipation. In fact, AEW treatment improved the reduction of feces induced by high fat diet treatment in mice (our unpublished data). Consequently, these clones might be deeply associated with several bioactivities of AEW.
The relative abundances of 9 taxa were significantly lower in AEW-administered mice than in control mice. Because the most abundant sequences of S24-7, Lachnospiraceae, Coprococcus, and Sutterella were uncultured clones, the physiological features of these genera have not been clarified. Therefore, the reasons for the lower abundance of these genera remain unclear. It is particularly interesting that the relative abundance of OTU 342504 (the most abundant sequence of Clostridiaceae) is correlated positively with serum ALT level (r = 0.63). The sequence of OTU 342504 is shown in Additional Table 1 (475.9KB, tif) . The closest species of this sequence is Eubacteriaceae bacterium Marseille-P2843. This anaerobic bacteria species was recently isolated from the human gut.26 Although the physiological features of this species have not yet been clarified, our data show the possibility that the relative abundance of this sequence is related to hepatocellular injury.
Sequence data of OTU 342504.
In conclusion, results presented herein indicate that AEW administration strongly affects the intestinal environment in mice, including the SCFA contents and the microbial composition in mice. Our previous report showed that the antioxidant activity of molecular hydrogen-dissolved alkaline electrolyzed water is enhanced with increasing hydrogen concentration even at the same pH condition.27 Therefore, we speculate that molecular hydrogen contribution is great for its beneficial effects. To elucidate the definitive role of gut microbes of AEW-administered mice, further investigations using fecal transplantation must be conducted. However, our data demonstrate the possibility that oral intake of AEW has beneficial roles for health in terms of cholesterol metabolism and liver protection.
Footnotes
Conflicts of interest
YT is a salaried employee of the Panasonic Corporation. This does not alter our adherence to MGR policies on sharing data and materials. The other authors have no conflict of interest, financial or otherwise, in relation to this study.
Financial support
This work was partly supported by Grants-in-Aid for Scientific Research (KAKENHI) to YH (No. 16K16281) and (B) to YN (No.16H05289) from the Japan Society for the Promotion of Science (JSPS), R& D Matching Funds on the Field for Knowledge Integration and Innovation, and scholarship funds for research from Functional Water Foundation. Funders had no involvement in the study design; data collection, management, analysis, and interpretation; paper writing; or decision to submit the paper for publication.
Research ethics
The study was performed in accordance with the Declaration of Helsinki and relevant ethical principles.
Data sharing statement
Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check
Checked twice by iThenticate.
Peer review
Externally peer reviewed.
Open peer reviewers
Lei Huang, Loma Linda University, USA; Robert Ostrowski, Polish Academy of Sciences Medical Research Centre, Poland.
Additional file
Additional Table 1 (475.9KB, tif) : Sequence data of OUT 342504.
Funding: This work was partly supported by Grants-in-Aid for Scientific Research (KAKENHI) to YH (No. 16K16281) and to YN (No.16H05289) from the Japan Society for the Promotion of Science (JSPS), R&D Matching Funds on the Field for Knowledge Integration and Innovation, and scholarship funds for research from Functional Water Foundation.
REFERENCES
- 1.Bajzer M, Seeley RJ. Physiology: obesity and gut flora. Nature. 2006;444:1009–1010. doi: 10.1038/4441009a. [DOI] [PubMed] [Google Scholar]
- 2.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Mol Aspects Med. 2013;34:39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 4.Cummings JH, Macfarlane GT. The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 5.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 6.Kimura I, Inoue D, Maeda T, et al. Short-chain fatty acids and ketones directly regulate sympathetic nervous system via G protein-coupled receptor 41 (GPR41) Proc Natl Acad Sci USA. 2011;108:8030–8035. doi: 10.1073/pnas.1016088108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura I, Ozawa K, Inoue D, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 9.Koyama K, Tanaka Y, Saihara Y, Ando D, Goto Y, Katayama A. Effect of hydrogen saturated alkaline electrolyzed water on urinary oxidative stress makers after an acute severe exercise: a randomized controlled trial. Anti Aging Med. 2008;4:117–122. [Google Scholar]
- 10.Fujita R, Tanaka Y, Saihara Y, Yamakita M, Ando D, Koyama K. Effect of molecular hydrogen saturated alkaline electrolyzed water on disuse muscle atrophy in gastrocnemius muscle. J Physiol Anthropol. 2011;30:195–201. doi: 10.2114/jpa2.30.195. [DOI] [PubMed] [Google Scholar]
- 11.Shimamura Y, Shinke M, Hiraishi M, Tsuchiya Y, Masuda S. The application of alkaline and acidic electrolyzed water in the sterilization of chicken breasts and beef liver. Food Sci Nutr. 2016;4:431–440. doi: 10.1002/fsn3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf PG, Biswas A, Morales SE, Greening C, Gaskins HR. H2 metabolism is widespread and diverse among human colonic microbes. Gut Microbes. 2016;7:235–245. doi: 10.1080/19490976.2016.1182288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka Y. Structure and function of alkaline ionized water apparatus. J Funct Water. 2017;12:29–34. [Google Scholar]
- 14.Ushida K, Sakata T. Effect of pH on oligosaccharide fermentation by porcine cecal digesta. Anim Sci Technol. 1998;69:100–107. [Google Scholar]
- 15.Matsumoto M, Inoue R, Tsuruta T, Hara H, Yajima T. Long-term oral administration of cows'milk improves insulin sensitivity in rats fed a high-sucrose diet. Br J Nutr. 2009;102:1324–1333. doi: 10.1017/S0007114509990365. [DOI] [PubMed] [Google Scholar]
- 16.Nishino K, Nishida A, Inoue R, et al. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2018;53:95–106. doi: 10.1007/s00535-017-1384-4. [DOI] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801:1175–1183. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Higashimura Y, Naito Y, Takagi T, Uchiyama K, Mizushima K, Yoshikawa T. Propionate promotes fatty acid oxidation through the up-regulation of peroxisome proliferator-activated receptor alpha in intestinal epithelial cells. J Nutr Sci Vitaminol (Tokyo) 2015;61:511–515. doi: 10.3177/jnsv.61.511. [DOI] [PubMed] [Google Scholar]
- 22.Hansen MK, McVey MJ, White RF, et al. Selective CETP inhibition and PPARalpha agonism increase HDL cholesterol and reduce LDL cholesterol in human ApoB100/human CETP transgenic mice. J Cardiovasc Pharmacol Ther. 2010;15:196–202. doi: 10.1177/1074248410362891. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto M, Takagaki A, Matsumoto K, Kato Y, Goto K, Benno Y. Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ‘Porphyromonadaceae’ isolated from rat faeces. Int J Syst Evol Microbiol. 2009;59:1748–1753. doi: 10.1099/ijs.0.007674-0. [DOI] [PubMed] [Google Scholar]
- 24.Bolotin A, de Wouters T, Schnupf P, et al. Genome sequence of "Candidatus Arthromitus" sp. Strain SFB-Mouse-NL, a commensal bacterium with a key eole in postnatal maturation of gut immune functions. Genome Announc. 2014;2:e00705–00714. doi: 10.1128/genomeA.00705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinheiro I, Robinson L, Verhelst A, et al. A yeast fermentate improves gastrointestinal discomfort and constipation by modulation of the gut microbiome: results from a randomized double-blind placebo-controlled pilot trial. BMC Complement Altern Med. 2017;17:441. doi: 10.1186/s12906-017-1948-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ndongo S, Lagier JC, Fournier PE, Raoult D, Khelaifia S. “Ihubacter massiliensis”: a new bacterium isolated from the human gut. New Microbes New Infect. 2016;13:104–105. doi: 10.1016/j.nmni.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue J, Shang G, Tanaka Y, et al. Dose-dependent inhibition of gastric injury by hydrogen in alkaline electrolyzed drinking water. BMC Complement Altern Med. 2014;14:81. doi: 10.1186/1472-6882-14-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence data of OTU 342504.