Abstract
Introduction
The study was conducted to investigate the prevalence and genetic diversity of Chlamydia spp. in poultry in Poland and estimate possible transmission to humans.
Material and Methods
Molecular diagnostic methods followed by sequencing and strain isolation were used on cloacal/faecal swabs collected from 182 apparently healthy poultry flocks including chickens, turkeys, ducks, and geese. Serum samples obtained from people exposed (study group) and non-exposed (control group) to birds were tested by complement fixation test to acquire data on Chlamydia spp. antibody level.
Results
Overall, 15.9% of the tested flocks were Chlamydiaceae-positive and three Chlamydia spp. were identified. Predominant chlamydial agent found was C. gallinacea occurring in 65.5% of all positive poultry flocks and in 73.0% of positive chicken flocks. The sequences from four chicken flocks were assigned to C. abortus, whereas C. psittaci was confirmed in one duck and one goose flock. The analysis of ompA variable domains revealed at least nine genetic variants of C. gallinacea. Chlamydial antibodies were detected in 19.2% of human serum samples in the study group in comparison with 10.8% in the controls.
Conclusion
The obtained results confirm that chlamydiae are common among chicken flocks in Poland with C. gallinacea as a dominant species. Moreover, the presence of C. abortus in chickens is reported here for the first time. Further investigation should focus on possible zoonotic transmission of C. gallinacea and C. abortus as well as potential pathogenic effects on birds’ health and poultry production.
Keywords: poultry, Chlamydia gallinacea, Chlamydia abortus, chlamydiosis, Poland
Introduction
Chlamydiae are bacteria belonging to the family Chlamydiaceae that cause chlamydiosis in wild and domestic birds, mammals, and humans. The genus Chlamydia includes 11 recognised species (27). Three of them, namely C. psittaci, C. avium, and C. gallinacea, with the latter two brought into this classification recently, occur commonly in birds. Transient colonisations by C. abortus, C. pecorum, C. trachomatis,C. suis , and C. muridarum were also noted occasionally in Aves (8, 20, 25, 26). Additionally, an individual case of C. ibidis, with Candidatus status in the Chlamydia genus, was recorded in an African sacred ibis (34). It is commonly known that C. psittaci is widespread throughout the world and can infect more than 450 bird species, including chickens, turkeys, ducks, and geese. C. psittaci infection in birds can persist for months to years, often without causing obvious illness (11). Several chlamydia-specific proteins, which are the source of diversity among chlamydial genomes, have now been identified on the inclusion membrane: the group of polymorphic membrane proteins (pmps), inclusion member protein A (inc A), and outer membrane protein (ompA) (23, 33). Until recently, there were nine ompA genotypes described in C. psittaci (A–F, E/B, M56, and WC), along with a number of provisional genotypes (YP84, R54, 6N, CPX0308, I, and J) representing strains which are untypable so far (15, 24). Before the new emerging chlamydial agent – C. gallinacea – was described, C. psittaci was considered to be the dominant Chlamydia species in poultry. According to recent data, C. gallinacea was mostly found in asymptomatic poultry (8). However, a decrease in the rate of weight gain was reported in infected chickens (8). It should be highlighted that relatively little is known regarding chlamydial agents in poultry in many countries, including Poland, and screening surveys of Chlamydiaceae presence in poultry have not been performed so far. Moreover, data about the genetic diversity and pathogenicity of recently classified C. gallinacea strains are scarce. Taking into account the fact that C. gallinacea found in poultry in a slaughterhouse in France coincided with atypical pneumonia cases reported in employees, its zoonotic potential cannot be ruled out (14). Therefore, the aim of the study was not only the evaluation of Chlamydiaceae prevalence and genetic diversity in poultry but also the estimation of the possibility of chlamydia transmission to humans, based on serological tests.
Material and Methods
Sampling
Sampling was performed between March 2014 and June 2016, in different regions of Poland (Fig. 1). In total, 1,830 cloacal/faecal swabs were collected from 182 apparently healthy poultry flocks (details in Table 1 and Supplementary file S1). On average, ten random samples were taken from each flock. Dry swabs were stored at −20°C for DNA extraction. Swabs in Chlamydia stabilising medium stored at −80°C were used for culture.
Fig. 1.
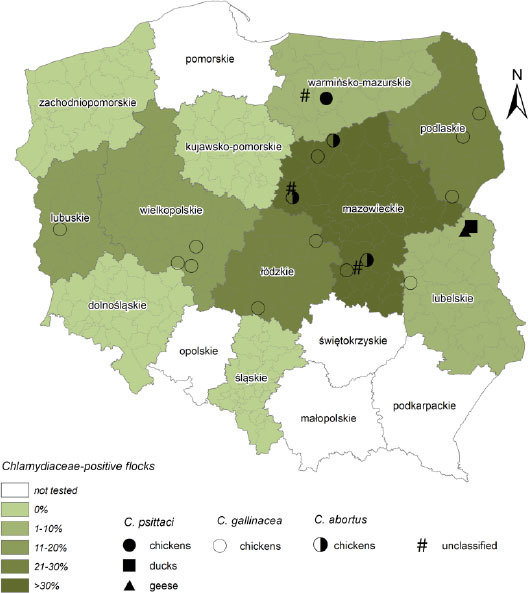
Area of bird sampling (ArcMap 10.4 software)
Table 1.
Prevalence of chlamydiae in different poultry flocks and host species in Poland
| Host species | Number of tested flocks | Number of samples | Number of Chlamydiaceae-positive flocks (%) | Species identification Number of positive flocks (%) | |||
|---|---|---|---|---|---|---|---|
| C. psittaci | C. gallinacea | C. abortus | Unclassified | ||||
| chicken | 113 | 1195 | 26 (23.0%) | 1 (3.8) | 19 (73.1) | 4 (15.4) | 2 (7.7) |
| duck | 23 | 225 | 1 (4.3%) | 1 | - | - | - |
| turkey | 28 | 230 | 1 (3.6%) | - | - | - | 1 |
| goose | 18 | 180 | 1 (5.6%) | 1 | - | - | - |
| Total | 182 | 1830 | 29 (15.9%) | 3 (10.3) | 19 (65.5) | 4 (13.8) | 3 (10.3) |
Blood samples (n = 500) were taken from humans in two sampling groups: control and study (see Supplementary file S2). The first group consisted of randomly selected blood donors (n = 250) and the second group consisted of individuals who declared contact with birds (n = 250), including persons with occupational exposure on poultry farms (n = 215), as well as veterinarians (n = 20) and ornithologists (n = 15) involved in handling wild birds (30). None of the persons tested reported any clinical symptoms at the time of interview. Whole blood and serum samples were collected into Vacutainers® containing no additives or only a preservative. Sampling was performed by qualified medical staff.
DNA extraction. A QIAamp DNA Mini Kit (Qiagen, France) was used for the DNA extraction from cloacal/faecal swabs according to the manufacturer's instructions, but with one modification. DNA extracts were eluted in 100 µl of elution buffer instead of 200 µl. DNA extracts were stored at −20°C before analysis.
Chlamydiaceae identification
All DNA extracts from birds (n = 1,830) were screened using a Chlamydiaceae-specific real-time PCR targeting a 23S rRNA gene fragment (111 bp) (4), which is conserved in all Chlamydiaceae. All analyses in the study were conducted on a 7500 Real-Time PCR System (Applied Biosystems, USA). Positive control using C. trachomatis (Genekam, Germany), and negative equivalents using DNase-RNase free water (Qiagen, Germany) were run with each assay. All sets of primers and probes used in this study are summarised in Supplementary file S3.
Microarrays
In total, 39 Chlamydiaceae-positive DNA samples (Ct < 36) from 16 poultry flocks were subjected to microarray tests performed by Alere Technologies GmbH, Germany.
Real-time PCR
All Chlamydiaceae-positive DNA extracts were included in further real-time PCR tests. Identification of C. psittaci based on the incA gene was conducted according to the protocol by Menard et al. (17), whereas C. gallinacea was detected by the amplification of an enoA gene fragment according to Laroucau et al. (14). Furthermore, specific real-time PCR assays were performed on selected samples to identify other Chlamydia species including C. abortus, C. pecorum, C. suis, and C. caviae (20), C. avium (39), and C. pneumoniae (12). In order to distinguish true target negatives from negatives due to PCR inhibition, an internal positive control (TaqMan Exogenous Internal Positive Control, Applied Biosystems, USA) was added to reaction mixtures according to the manufacturer’s instructions. Also, positive controls using C. abortus, C. pecorum, C. suis, and C. caviae, C. avium, and C. pneumoniae DNA, and negative equivalents using DNase-RNase free water (Qiagen, Germany) were run with each assay. All samples with a Ct value above 38.5 were considered negative.
Sequencing
Selected Chlamydiaceae-positive samples were used for DNA sequencing to confirm the identity of Chlamydia spp. Specific fragments of ompA, 16S rRNA, and 16S rRNA-23S rRNA intergenic spacer together with 23S rRNA domain I (IGS-23S rRNA) were amplified with previously published primer sets (5, 15, 22, 32) which are summarised in Supplementary file S3.
To investigate the diversity of the C. gallinacea ompA gene the amplification of variable domains (VD): VD 1–2 (435 bp) and VD 3–4 (421 bp) was performed according to the procedure proposed by Guo et al. (8). All PCR assays were performed on a Biometra thermocycler (Biometra, Germany). Amplified products were detected on ethidium-bromide-stained agarose gels with ultraviolet illumination and sent to Genomed (Poland) for sequencing.
Phylogenetic and ompA variability analysis
All dendrograms were constructed using neighbour joining (NJ) with the robustness of the clusters assessed by bootstrapping 1,000 replicates. One representative sequence from each individual flock was used (if available). PCR products of the ompA gene obtained for 13 Chlamydiaceae-positive poultry flocks were sequenced and the data were analysed using Geneious Pro 8.0 software (Biomatters, New Zealand). Amplicons were subjected to BLAST analysis against the GenBank database (NCBI) to identify related entries and aligned with a panel of Chlamydia reference strains including avian C. abortus genotypes G1 and G2. Phylogenetic trees were constructed based on alignments of 936 bp and 1,006 bp for 16S rRNA and IGS-23S rRNA, respectively.
OmpA (830 bp) as well as VD 1–2 (343 bp) and VD 3–4 (338 bp) fragments were aligned with sequences of C. gallinacea to build separate dendrograms, and the sequences included European and Chinese strains constituting different genetic variants.
Sequencing data from the present study were deposited in the GenBank database with the following accession numbers: MF140888–MF140897 (16S rRNA), MF140898–MF140900 (IGS-23S rRNA), MF140901–MF140919 (ompA), MF140920–MF140946 (VD 1–2 of ompA), and MF140872–MF140887 (VD 3–4 of ompA).
Isolation and propagation in cell culture
Buffalo green monkey (BGM) cells in minimal essential medium (MEM) (Lonza, Germany) with 5% serum were seeded into Trac bottles containing glass coverslips (Bibby Sterilin Ltd., UK) and incubated at 37°C with 5% CO2 in a fully humidified cabinet for four days. Swabs (with Ct value in Chlamydiaceae 23S real-time PCR <32) in 1–2 mL Chlamydia stabilising medium were ultrasonicated with a Branson 450D sonifier (ten 0.8 s pulses with 0.2 s pause between each pulse at an amplitude of 80%) (Branson Ultrasonics, USA) and 30–300 µL of medium were inoculated into six Trac bottles with confluent-grown BGM monolayers. After inoculation, the bottles were centrifuged at 3,000×g and at 37°C for 60 min and subsequently incubated for 2 h. The MEM was then replaced with serum-free medium UltraMDCK (Lonza, Germany) containing amphotericin (2.5 µg/mL), gentamicin (10 µg/mL), and vancomycin (25 µg/ mL). The medium was renewed after 18 h. Three days after inoculation, a single coverslip was fixed with methanol, and the monolayer was stained with IMAGEN Chlamydia (Oxoid Ltd., UK). A sample was considered positive when inclusions of typical chlamydial morphology appeared as bright apple-green spots after two passages.
Serological testing of human sera
In total, 500 human sera were tested by complement fixation test (CFT) in order to detect Chlamydia spp. antibodies. The CFT was performed according to the manufacturer’s protocol. The chlamydial antigen was obtained from Serion Immundiagnostica (Germany). A titre of 64 or higher was considered diagnostically significant and reported as a positive result.
Statistical analysis
All analyses were conducted using the programme STATISTICA ver. 10 (StatSoft, part of Dell Software, USA). The chi-squared test was carried out to calculate correlation among dependent variables of seropositivity level in study and control groups. The odds ratio (OR) was also calculated for assessment of the chance of chlamydial antibodies in both sampled populations.
Results
Bird survey
The Chlamydiaceae-specific real-time PCR (summary in Table 1 and Supplementary file S1) showed that 15.9% (29/182) of apparently healthy poultry flocks from different areas of Poland were positive (Fig. 1). Among 182 tested farms, the presence of Chlamydiaceae was molecularly confirmed mainly in chickens (26/113 farms; 23.0%). Only one turkey (1/28 farms; 3.6%), goose (1/18 farms; 5.6%), and duck (1/23 farms; 4.3%) flock was Chlamydiaceae-positive. In seven chicken flocks, the number of Chlamydiaceae-positive samples ranged between eight and ten (out of ten), whereas in the remaining 19 flocks, only one to five (out of ten) positive samples were detected. In the other tested hosts, no more than three positive samples were noted per flock.
Re-analysis of positive samples with a microarray assay revealed that C. gallinacea was present in most of the tested flocks. This result was confirmed by further molecular analysis. The majority of the Chlamydiaceae-positive chicken flocks (19/26, 73.1%) were positive in C. gallinacea qPCR with Ct values ranging from 21.8 to 38.3. Weak signals (average Ct value of 35.9 to 38.2) in C. abortus qPCR were obtained in four chicken flocks. Out of three C. psittaci-positive flocks, only in the duck flock (flock 15-63) was the shedding level high (Ct 23.8), while in the chicken (flock 15-2) and goose flocks (flock 15-41) average Ct values were 37.7 and 33.1, respectively. In a few flocks, microarrays and qPCR detected C. abortus and C. psittaci beside C. gallinacea. A co-infection of C. psittaci and C. gallinacea in chickens (flock 14-156) and geese (flock 15-41) was noted in qPCR tests. In chicken flock 14-67 species-specific qPCRs identified the three Chlamydia species: C. psittaci, C. gallinacea, and C. abortus, but further phylogenetic analysis confirmed only the presence of C. gallinacea. In two chicken and one turkey flocks (flocks 15-12, 14-157, and 14-200), species identification was unsuccessful.
Swabs sampled from chicken of flock 15-56 (samples 1 and 2) were inoculated onto BGM cell culture and two strains of C. gallinacea were successfully isolated from these samples. Partial ompA sequences were obtained from isolates and found to be 100% identical to those amplified directly from corresponding dry swabs.
Sequencing of 16S rRNA and IGS-23S rRNA was carried out to characterise studied DNA isolates. Successful amplification of the 16S rRNA gene was achieved for samples from eight poultry flocks, while IGS-23S rRNA amplicons were sequenced only for three flocks. Dendrograms constructed on the basis of these gene fragments and aligned with reference sequences of known Chlamydia species (Fig. 2 and 3) showed similar topology. All sequences from our study were positioned within the Chlamydiaceae family and were assigned to three Chlamydia species: C. psittaci, C. abortus, and C. gallinacea.
Fig. 2.
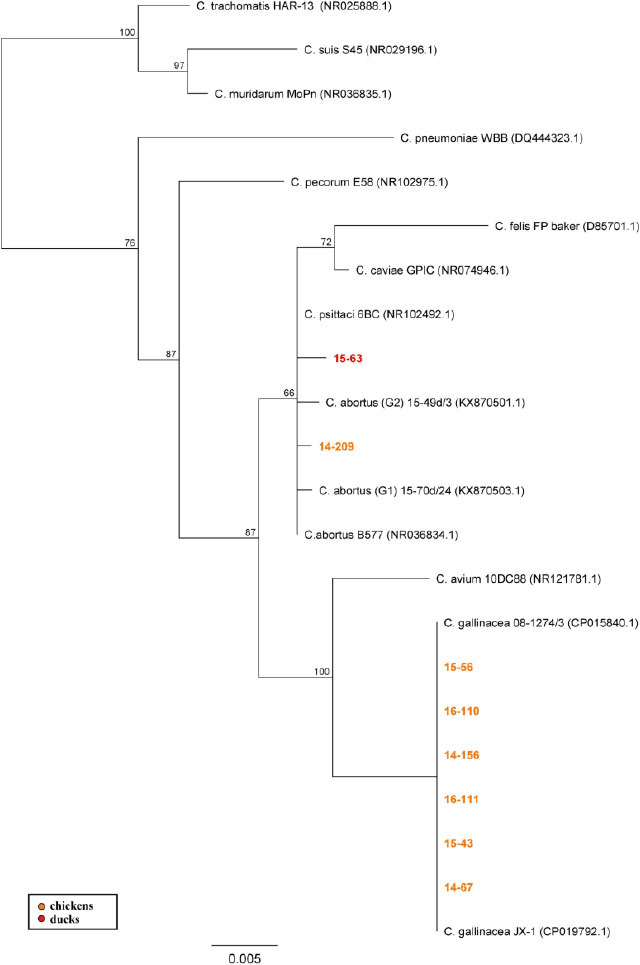
NJ dendrogram based on 16S rRNA gene fragment. Representative sequences of established Chlamydiaceae species were used; bar corresponds to the number of substitutions per site
Fig. 3.
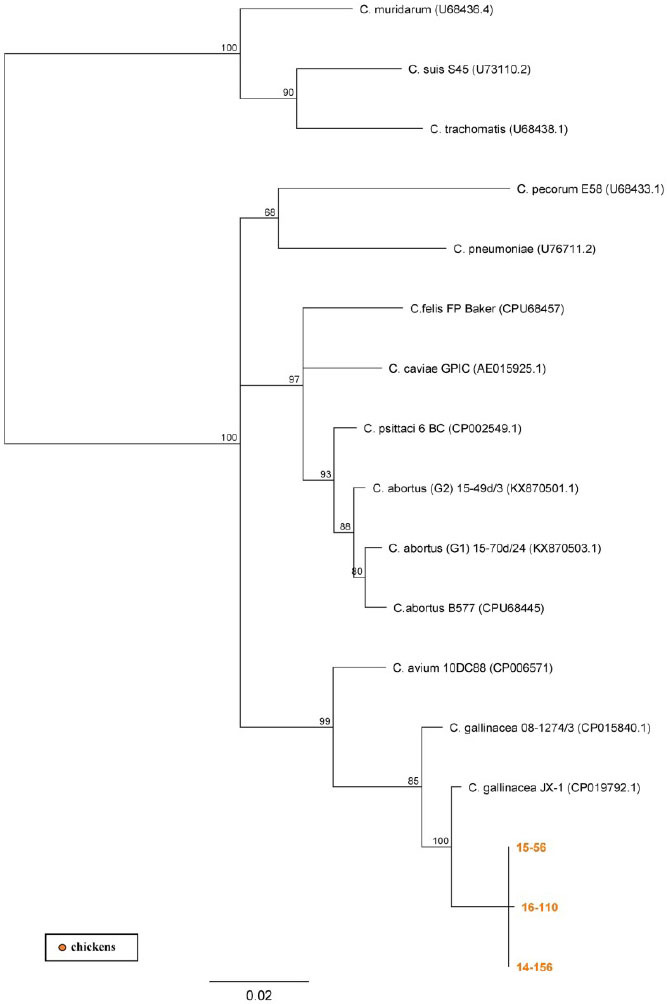
NJ dendrogram based on IGS-23S rRNA gene fragment. Representative sequences of established Chlamydiaceae species were used; bar corresponds to the number of substitutions per site
Phylogenetic analysis revealed that 2 out of 12 obtained ompA sequences (Fig. 4) from chicken (15-63) and goose flocks (15-41) were grouped with C. psittaci GD (AF26926.1) and C. psittaci (KX062086.1) with maximum 100% bootstrap support. A further three amplicons (14-204, 14-206, and 14-209) from chicken flocks were assigned to the C. abortus cluster with high bootstrap support and C. abortus B577 (M73036.1) as the closest relative. The remaining seven ompA sequences (14-67, 16-110, 14-205, 14-154, 14-156, 15-43, and 15-56) were grouped together with the C. gallinacea strains. The variability of this gene can be noted in the five subclades formed within the C. gallinacea group. The sequence 16-110 forms a subclade with Chlamydia spp. (HE660097.1) from a Slovenian isolate with 100% bootstrap support. Indicative of the heterogeneity, the sequence from flock 14-205 nevertheless was grouped together with Croatian Chlamydia sp. (HE660095.1) and a sister clade was formed by the 14-154 and 14-156 sequences. Both were supported by the highest bootstrap value. Sequence HE660099.1 obtained from Greek chickens was most similar to amplicons 15-43 and 15-56, while 14-67 from the present study did not group closely with any of the described strains. In-depth analysis of ompA variable domains was also performed. Sequences of VD 1–2 from 17 flocks were obtained but only 10 VD 3–4 fragments were sequenced successfully. Phylogenetic comparison of the resulting products identified nine genetic variants that were clearly separated, six of which were new (see Supplementary file S4).
Fig. 4.
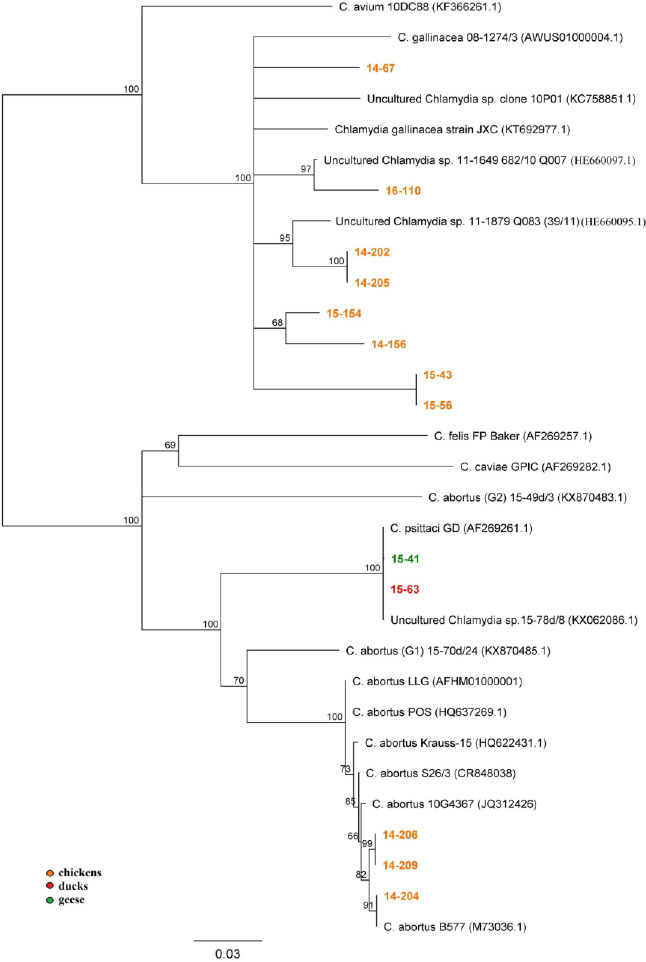
ompA-based NJ dendrogram (822 bp); bar corresponds to the number of substitutions per site
Human survey
Out of 250 human serum samples obtained from people declaring considerable exposure to birds (study group), 48 were positive in CFT for chlamydial antibodies (19.2%), whereas in the control group Chlamydia spp. antibodies were confirmed in 27 (10.8%) serum samples (see Supplementary file S2). Statistical analysis confirmed significant difference in seropositivity level between tested groups and odds ratio was calculated as 1.96. Specimens taken from workers employed on six C. gallinacea-positive farms (n = 50) were included in the study group, but in these samples the presence of antibodies specific to chlamydiae was not detected.
Discussion
A variety of Chlamydia species occurring in poultry hosts were detected in different European countries and China (8, 13, 38). C. psittaci has long been considered the main Chlamydia species in poultry, while recent scientific reports show that C. gallinacea, a newly emerging agent, is predominantly found in asymptomatic chickens (8). A monitoring survey of Chlamydia spp. dissemination in poultry in Poland has not been performed so far, and literature data are limited to description of the first case of C. gallinacea in hens (29). The present study was carried out on cloacal/faecal swabs sampled from poultry flocks in 11 out of 16 provinces to explore the prevalence of Chlamydiaceae shedders. Our results showed that 15.9% of tested flocks were Chlamydiaceae-positive. At the same time, it was revealed that Polish poultry, excluding chickens, are almost free from chlamydiae. However, within the chicken population the percentage of Chlamydiaceae-positive flocks was 23.0%. Chlamydia gallinacea was noted in 73.0% of Chlamydiaceae-positive chicken flocks, indicating this species to be the dominant chlamydial agent in this host. Available data show that C. gallinacea occurs in poultry flocks in at least five European countries: France, Greece, Slovenia, Croatia, and Poland, as well as in China (29, 38). In contrast, C. gallinacea was not found in a Belgian study (13). An extensive comparison of prevalence levels of C. gallinacea within Europe is impossible due to the lack of monitoring studies from other countries, but prevalence is probably lower than in China where C. gallinacea is endemic in chickens (8). Guo et al. (8) suggested that C. gallinacea is not commensal but a pathogen of moderate pathogenicity, and persistent infection can lead to reduced weight gain in poultry production. No clinical signs of chlamydia infection were observed in poultry included in our study.
In the presented study,C. psittaci and C. abortus were recorded less often in chicken flocks, with prevalence of 3.8% and 15.4% in Chlamydiaceae-positive flocks, respectively. The low rate of C. psittaci detection is not surprising as chickens are not a typical host of this chlamydial species.
The analysis of the ompA gene included here revealed that C. gallinacea strains encountered in Poland are diverse and different from previously known European and Chinese strains (8), but still share high sequence similarity with them (Fig. 4). Furthermore, none of the amplicons from the current study grouped closely with the C. gallinacea sequence (KC758851.1) obtained from strain 10P01 described in the first case report in Poland (29).
The two C. psittaci sequences from our study (15-41 and 15-63) represented genotype C that is typical for poultry. Interestingly, these sequences grouped together with a sequence obtained earlier by our research group from a swan in Poland (KX062086.1) (30). This is a clear indication of transmission of C. psittaci between wild and domestic birds.
It should be noted that three sequences from chickens (14-204, 14-206, and 14-209) were placed in the C. abortus clade together with typical mammalian strains. They share 100% sequence homology with C. abortus B577 (M73036.1) and C. abortus S26/3 (CR848038.1), both isolated from sheep, whereas up to seven single-nucleotide polymorphisms (SNPs) were present between them and the remaining C. abortus sequences. It is worth noting that this is the first report of natural infection with C. abortus in chickens. Previous data showed the presence of this pathogen in a few bird species, e.g. turkey, budgerigar, oriental white stork, and pigeon (3, 25, 28). C. abortus has attracted increasing scientific attention due to its pathogenicity and several events of systemic infection in humans (2, 9, 16, 21, 35, 36), as well as the isolation of the agent from new hosts including birds (30). Therefore, Pannekoek et al. (18) and ourselves, in our recent report on Chlamydiaceae prevalence in wild birds (30), propose a modification of the initial C. abortus species definition published by Everett et al. (5) and an expansion of this species to include not only the classical strains obtained from mammals but also avian isolates. Taking into account that the tested poultry was apparently healthy, it can be assumed that these birds were asymptomatic carriers of C. abortus. Crossing of the species barriers by chlamydiae is well known, with literature describing cases of C. trachomatis and C. pecorum as examples of a possible host change from mammals to birds (6, 19, 25) and C. psittaci as a vice versa example (10).
Avian chlamydiosis poses also a potential risk to humans. Four Chlamydia species, namely C. trachomatis, C. pneumoniae, C. abortus, and C. psittaci, are known to be able to infect humans. Outbreaks of the disease have been reported through direct contact with birds, both wild and breeding (31), though inadvertent exposure particularly in endemic areas also occurs (1). Human-to-human transmission has also been reported, raising the spectre of uncontrolled outbreaks (37). Description of C. gallinacea in poultry and C. avium in wild fowl by Sachse et al. (26) has raised the question of its aetiological role and possible zoonotic potential. Our survey in humans exposed to birds was based on available serological tools for detection of antibodies against Chlamydia spp. It should be noted that the lack of specific serological methods precluded the identification of any humoral immune response specific to C. gallinacea. Our results showed that the percentage of Chlamydia spp. seropositive samples is significantly higher in the study group than in the control. However, because CFT lacks specificity, it is unclear whether this discordance is due to different exposure to chlamydia-infected birds or to infections by the typical human pathogens, namely C. trachomatis and C. pneumoniae. Especially antibodies against C. pneumoniae are very common among the adult human population with seropositivity rising to 80% with age (7). Interestingly, 19.2% of tested individuals exposed to birds included in our survey were seropositive. All of them were bird breeders or poultry farm workers; however, no positive serological reactions were recorded among workers on some of C. gallinacea-positive farms, nor veterinarians and ornithologists who had had contact with wild birds. Taking into account the results of this study and the current state of knowledge, the zoonotic potential of the new Chlamydia players (C. gallinacea and C. avium) can be neither acknowledged nor excluded.
In conclusion, we have demonstrated that Chlamydiaceae are common in chicken flocks in Poland. The dominant species noted in this host is C. gallinacea, which exhibits different genetic variants. Moreover, this is the first report showing the occurrence of C. abortus in chickens. Further studies should focus on possible zoonotic transmission of C. gallinacea and C. abortus as well as potential pathogenic effects on poultry health and productivity. Moreover, a thorough molecular investigation of C. abortus and C. gallinacea strains isolated from production birds using next generation sequencing should be performed.
Acknowledgements
The authors are grateful to Elżbieta Anna Moniuszko, Mirosław Netczuk, Magdalena Szymańska-Krzywda, Ryszard Bartczak, Ewa Borzym, Grzegorz Tomczyk, Wojciech Kozdruń, and Michał Jóźwiak for their help with sample collection. Anna Wójcik, Agnieszka Jodełko, Sabine Scharf, Agnieszka Stolarek and Łukasz Bocian are acknowledged for their excellent technical assistance.
The online version of this article (DOI:10.1515/jvetres-2017-0072) offers the following supplementary material: 1) Supplementary file S1. Identity, origin and results obtained for Chlamydiaceae-positives samples; 2) Supplementary file S2. Summary of human survey results; 3) Supplementary file S3. Summary of primers used in the study; 4) Supplementary file S4. NJ dendrograms displaying variability of VD 1–2 (A) and VD 3–4 (B) ompA domains.
Footnotes
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: The scientific work was funded by the Polish Ministry of Science and Higher Education Iuventus Plus programme in the years 2015–2016 (project number IP2014047273).
Animal Rights Statement: Samples from birds were collected during clinical studies or routine activities following standard procedures. According to the Local Ethical Committee on Animal Testing at University of Life Sciences in Lublin (Poland) formal ethical approval is not required for this kind of study. Guidelines published by this ethics committee (Resolution No. 22/2006 of the National Ethic Committee for Experimentation of November 7 (2006) Poland. 2006) were used, which confirm that this work is acceptable without specific ethical approval. Moreover, consent of bird owners was obtained for sampling.
Human Rights Statement: The research related to human use has been complied with all the relevant national regulations, institutional policies and in accordance the tenets of the Helsinki Declaration. Sampling and laboratory testing of obtained specimens were approved by the Bioethics Committee at the Regional Chamber of Physicians Decision No. 224/2014/KB/V.
References
- 1.Branley J.M., Weston K.M., England J., Dwyer D.E., Sorrell T.C.. Clinical features of endemic community-acquired psittacosis. New Microbes New Infect. 2006;2:7–12. doi: 10.1002/2052-2975.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buxton D.. Potential danger to pregnant women of Chlamydia psittaci from sheep. Vet Rec. 1986;118:510–511. doi: 10.1136/vr.118.18.510. [DOI] [PubMed] [Google Scholar]
- 3.Chahota R., Ogawa H., Mitsuhashi Y., Ohya K., Yamaguchi T., Fukushi H.. Genetic diversity and epizootiology of Chlamydophila psittaci prevalent among the captive and feral avian species based on VD2 region of ompA gene. Microbiol Immunol. 2006;50:663–678. doi: 10.1111/j.1348-0421.2006.tb03839.x. [DOI] [PubMed] [Google Scholar]
- 4.Ehricht R., Slickers P., Goellner S., Hotzel H., Sachse K.. Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol Cell Probes. 2006;20:60–63. doi: 10.1016/j.mcp.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Everett K.D., Bush R.M., Andersen A.A.. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 6.Everett K.D.. Chlamydia and Chlamydiales: more than meets the eye. Vet Microbiol. 2000;75:109–26. doi: 10.1016/s0378-1135(00)00213-3. [DOI] [PubMed] [Google Scholar]
- 7.Grayston J.T.. Background and current knowledge of Chlamydia pneumoniae and atherosclerosis. J Inf Dis. 2000;181:402–410. doi: 10.1086/315596. [DOI] [PubMed] [Google Scholar]
- 8.Guo W., Li J., Kaltenboeck B., Gong J., Fan W., Wang C.. Chlamydia gallin acea, not C. psittaci, is the endemic chlamydial species in chicken Gallus gallus. Sci Rep. 2016;6:19638. doi: 10.1038/srep19638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyde S.R., Benirschke K.. Gestational psittacosis: case report and literature review. Mod Pathol. 1997;10:602–607. [PubMed] [Google Scholar]
- 10.Jelocnik M., Branley J., Heller J., Raidal S., Alderson S., Galea F., Gabor M., Polkinghorne A.. Multilocus sequence typing identifies an avian-like Chlamydia psittaci strain involved in equine placentitis and associated with subsequent human psittacosis. Nat Publ Gr. 2017;6:e7. doi: 10.1038/emi.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaleta E.F., Taday E.M.. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 2003;32:435–461. doi: 10.1080/03079450310001593613. [DOI] [PubMed] [Google Scholar]
- 12.Kohlhepp S.J., Hardick J., Gaydos C.. Chlamydia pneumoniae in peripheral blood mononuclear cells isolated from individuals younger than 20 years or older than 60 years. J Clin Microbiol. 2005;43:3030. doi: 10.1128/JCM.43.6.3030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagae S., Kalmar I., Laroucau K., Vorimore F., Vanrompay D.. Emerging Chlamydia psittaci infections in chickens and examination of transmission to humans. J Med Microbiol. 2014;63:399–407. doi: 10.1099/jmm.0.064675-0. [DOI] [PubMed] [Google Scholar]
- 14.Laroucau K., Aaziz R., Meurice L., Servas V., Chossat I., Royer H., De Barbeyrac B., Vaillant V., Moyen J., Meziani F., Sachse K., Rolland P.. Outbreak of psittacosis in a group of women exposed to Chlamydia psittaci-infected chickens. Eurosurveillance. 2015;20:1–6. doi: 10.2807/1560-7917.es2015.20.24.21155. [DOI] [PubMed] [Google Scholar]
- 15.Madani S.A., Peighambari S.M.. PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathol. 2013;42:38–44. doi: 10.1080/03079457.2012.757288. [DOI] [PubMed] [Google Scholar]
- 16.Meijer A., Brandenburg A., de Vries J., Beentjes J., Roholl P., Dercksen D.. Chlamydophila abortus infection in a pregnant woman associated with indirect contact with infected goats. Eur J Clin Microbiol Infect Dis. 2004;23:487–490. doi: 10.1007/s10096-004-1139-z. [DOI] [PubMed] [Google Scholar]
- 17.Menard A., Clerc M., Subtil A., Megraud F., Bebear C., de Barbeyrac B.. Development of a real-time PCR for the detection of Chlamydia psittaci. J Med Microbiol. 2006;55:471–473. doi: 10.1099/jmm.0.46335-0. [DOI] [PubMed] [Google Scholar]
- 18.Pannekoek Y., Dickx V., Beeckman D.S.A.B., Jolley K.A., Keijzers W.C., Vretou E., Maiden M.C.J., Vanrompay D., van derEnde A.. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS One. 2010;5:e14179. doi: 10.1371/journal.pone.0014179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K.. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet J. 2009;181:145–150. doi: 10.1016/j.tvjl.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 20.Pantchev A., Sting R., Bauerfeind R., Tyczka J., Sachse K.. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis. 2010;33:473–484. doi: 10.1016/j.cimid.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Pospischil A., Thoma R., Hilbe M., Grest P., Gebbers J.O. Abortion in woman caused by caprine Chlamydophila abortus (Chlamydia psittaci serovar 1) Swiss Med Wkly. 2002;132:64–66. doi: 10.4414/smw.2002.09911. [DOI] [PubMed] [Google Scholar]
- 22.Pudjiatmoko Fukushi H., Ochiai Y., Yamaguchi T., Hirai K.. Phylogenetic analysis of the genus Chlamydia based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1997;47:425–431. doi: 10.1099/00207713-47-2-425. [DOI] [PubMed] [Google Scholar]
- 23.Rockey D.D., Heinzen R.A., Hackstadt T.. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 24.Sachse K., Laroucau K., Hotzel H., Schubert E., Ehricht R., Slickers P.. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008;8:63. doi: 10.1186/1471-2180-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachse K., Kuehlewind S., Ruettger A., Schubert E., Rohde G.. More than classical Chlamydia psittaci in urban pigeons. Vet Microbiol. 2012;157:476–480. doi: 10.1016/j.vetmic.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Sachse K., Laroucau K., Riege K., Wehner S., Dilcher M., Creasy H.H., Weidmann M., Myers G., Vorimore F., Vicari N., Magnino S., Liebler-Tenorio E., Ruettger A., Bavoil PM., Hufert F.T., Rosselló-Móra R., Marz M.. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Sachse K., Bavoil P.M., Kaltenboeck B., Stephens R.S., Kuo C.C, Rosselló-Móra R., Horn M.. Emendation of the family Chlamydiaceae: Proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Sting R., Lerke E., Hotzel H., Jodas S., Popp C., Hafez H.M.. Comparative studies on detection of Chlamydophila psittaci and Chlamydophila abortus in meat turkey flocks using cell culture, ELISA, and PCR. Dtsch Tierarztl Wochenschr. 2006;113:50–54. [PubMed] [Google Scholar]
- 29.Szymańska-Czerwińska M., Niemczuk K., Sachse K., Mitura A., Karpińska T.A., Reichert M.. Detection of a new non-classified chlamydia species in hens in Poland. Bull Vet Inst Pulawy. 2013;57:25–28. [Google Scholar]
- 30.Szymańska-Czerwińska M., Mitura A., Niemczuk K., Zareba K., Jodełko A., Pluta A., Scharf S., Vitek B., Aaziz R., Vorimore F., Laroucau K., Schnee C.. Dissemination and genetic diversity of chlamydial agents in Polish wildfowl: Isolation and molecular characterisation of avian Chlamydia abortus strains. PLoS One. 2017;12:e0174599. doi: 10.1371/journal.pone.0174599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Telfer B.L., Moberley S.A., Hort K.P., Branley J.M., Dwyer D.E., Muscatello D.J., Correll P.K., England J., McAnulty J.M.. Probable psittacosis outbreak linked to wild birds. Emerg Infect Dis. 2005;11:391–397. doi: 10.3201/eid1103.040601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas V., Casson N., Greub G.. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture. Environ Microbiol. 2006;8:2125–2135. doi: 10.1111/j.1462-2920.2006.01094.x. [DOI] [PubMed] [Google Scholar]
- 33.Voigt A., Schöfl G., Saluz H.P. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One. 2012;7:e35097. doi: 10.1371/journal.pone.0035097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vorimore F., Hsia R., Huot-Creasy H., Bastian S., Deruyter L., Passet A., Sachse K., Bavoil P., Myers G., Laroucau K.. Isolation of a new Chlamydia species from the Feral Sacred Ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS One. 2013;8:e74823. doi: 10.1371/journal.pone.0074823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walder G., Meusburger H., Hotzel H., Oehme A., Neunteufel W., Dierich M.P., Würzner R.. Chlamydophila abortus pelvic inflammatory disease. Emerg Infect Dis. 2003;9:1642–1644. doi: 10.3201/eid0912.020566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walder G., Hotzel H., Brezinka C., Gritsch W., Tauber R., Würzner R., Ploner F.. An unusual cause of sepsis during pregnancy: recognizing infection with Chlamydophila abortus. Obstet Gynecol. 2005;106:1215–1217. doi: 10.1097/01.AOG.0000161060.69470.9c. [DOI] [PubMed] [Google Scholar]
- 37.Wallensten A., Fredlund H., Runehagen A.. Multiple human-to-human transmission from a severe case of psittacosis, Sweden, January-February 2013. Euro Surveill. 2014;19:20937. doi: 10.2807/1560-7917.es2014.19.42.20937. [DOI] [PubMed] [Google Scholar]
- 38.Zocevic A., Vorimore F., Marhold C., Horvatek D., Wang D., Slavec B., Prentza Z., Stavianis G., Prukner-Radovcic E., Dovc A.. Molecular characterization of atypical Chlamydia and evidence of their dissemination in different European and Asian chicken flocks by specific real-time PCR. Environ Microbiol. 2012;14:2212–2222. doi: 10.1111/j.1462-2920.2012.02800.x. [DOI] [PubMed] [Google Scholar]
- 39.Zocevic A., Vorimore F., Vicari N., Gasparini J., Jacquin L., Sachse K., Magnino S., Laroucau K.. A real-time PCR assay for the detection of atypical strains of Chlamydiaceae from pigeons. PLoS One. 2013;8:e58741. doi: 10.1371/journal.pone.0058741. [DOI] [PMC free article] [PubMed] [Google Scholar]


