Abstract
Background
Candida albicans is a major opportunistic fungal pathogen. One of the most important virulence factors that contribute to the pathogenesis of candidiasis is its ability to form biofilms. A key characteristic of Candida biofilms is their resistance to antifungal agents. Due to significant morbidity and mortality rates related to biofilm-associated drug resistance, there is an urgency to develop novel nanotechnology-based approaches preventing biofilm-related infections.
Methods
In this study, we report, for the first time, the synthesis of selenium nanoparticles by irradiating selenium pellets by nanosecond pulsed laser ablation in liquid chitosan as a capping agent. Synergy of the fungicidal effect of selenium nanoparticles and chitosan was quantified by the combination index theorem of Chou–Talalay.
Results
This drug combination resulted in a potent fungicidal effect against a preformed C. albicans biofilm in a dose–response manner. By advanced electron microscopy techniques, we documented the adhesive and permeabilizing properties of chitosan, therefore allowing selenium nanoparticles to enter as the cell wall of the yeast became disrupted and distorted. Most importantly, we demonstrated a potent quantitative synergistic effect when compounds such as selenium and chitosan are combined.
Conclusion
These chitosan-stabilized selenium nanoparticles could be used for ex vivo applications such as sterilizers for surfaces and biomedical devices.
Keywords: selenium nanoparticles, laser ablation in liquids, Candida albicans, biofilm, chitosan, synergy
Introduction
Candida albicans biofilm
C. albicans is a major opportunistic fungus capable of causing a broad spectrum of diseases. Candidiasis is the fourth most common hospital-acquired bloodstream infection (BSI) in the USA causing severe sepsis.1 One of the most important factors that contribute to the pathogenesis of candidiasis is its ability to form biofilm formation.2 Indwelling prosthetic materials and most catheters represent ideal surfaces for the adherence and growth of Candida biofilms. Therefore, Candida is the most frequently isolated fungal pathogen in catheter-related BSIs (CRBSIs) resulting in high morbidity and mortality rates in hospitalized patients.3 Candida biofilms are enclosed by an exopolymeric substance or extracellular polymeric substance (EPS) matrix which protects the pathogen from adverse exposure to environmental conditions,4 the host’s immune defense and fungicides. Furthermore, detached yeasts from the biofilm are the source of infection by spreading the BSI.5 Morphogenetic conversions between yeast and hyphae have a key role in biofilm formation and represent an important virulence factor for disease pathogenesis.6 The main characteristic of Candida biofilm is the high level of drug resistance that can endure up to 1,000-fold as compared to their planktonic (freely suspended) counterparts.7 As Candida biofilms have reduced susceptibility against currently existing antifungals,7,8 there is an urgent need to develop novel combination nanotherapeutic approaches for Candida biofilm treatment.9
Combination drug therapy and synergy
Combination drug therapy is the use of two or more pharmacological agents; this is a standard clinical practice in the treatment of antibiotic-resistant infections.10 This combination therapy is designed to achieve synergy,11 defined as the interaction of two or more drugs to accomplish a combined effect greater than the sum of their separate effects.12 Synergy can be demonstrated doing an analysis of the inhibitory assay by the CompuSyn software showing the combination index (CI) with a quantitative definition for additive effect (CI=1), synergism (CI<1), and antagonism (CI>1). The theorem of Chou–Talalay also provides algorithms for automated computer simulation for synergism, as shown in the CI plot.13 The mass–action law13–15-based software CompuSyn16 is one of the most cited software packages.17
Selenium nanoparticles (SeNPs)
As selenium (Se) sulfide is used in the treatment of superficial mycoses in the form of a topical antifungal shampoo18 and SeNPs have demonstrated fungicidal effects against C. albicans biofilms,19 we decided to test pure SeNPs in combination with chitosan (CS) or alone against a preformed C. albicans biofilm. Se is a naturally occurring metalloid trace element that is vital as nutrient with important human health benefits due to its dynamic role inhibiting the formation of free radicals; therefore, Se prevents oxidative stress, a major source of age-related diseases;20 there are approximately 25 known selenoproteins incorporated in the human genome;21 however, in high quantities, Se becomes toxic showing a narrow margin between beneficial and toxic effects.22 Yeasts of the genus Candida have the ability to accumulate (within the cell) extensive quantities of trace elements21 incorporated as organic compounds.23 The mechanisms of accumulation and transformation of Se into the cell wall (CW) architecture of C. albicans remain elusive.23 Se enters the yeast by chemisorption with the formation of ionic bonds by the CW polymers.23–25 The fungicidal effect may be due to the mixing of Se with cell proteins in which Se displaces sulfur (due to the chemical analogy of both elements), for sulfur-containing amino acids such as cysteine (Cys) and methionine (Met).23 Yeasts absorb Se into the cytosol, using transporters such as sulfate permeases Sul1 and Sul2.26 Selenoproteins in excess generate reactive oxygen species (ROS) causing DNA strand breaks;27 this process can lead to changes in protein misfolding, stability, structural changes, and enzyme dysfunction.21,23–25 Toxic activity of inorganic Se compounds in the yeasts involves the reaction of selenites with thiol-containing compounds.25 Therefore, SeNPs have demonstrated an anti-Candida biofilm effect.19
CS
Another compound tested in this study alone or in combination with SeNPs was CS, an effective capping and stabilizing agent of SeNPs28,29 with fungicidal effects.30–32 CS is a polysaccharide derivative of chitin, extracted from fungi, arthropods such as crustaceans and insects. It is a biocompatible, natural, biodegradable, bioadhesive, and positively charged polymer with low cytotoxicity.33 Microbicidal and fungicidal properties of CS are well established.30 The polycationic agents of the polymer interact with anionic components of the yeast reducing the negative surface charge of the outer CW ensuing in a strong attachment of the biopolymer, permeabilizing the CW as a result.30,34 CS-decorated SeNPs (CS-SeNPs) synthesized by chemical methods have been previously studied in cancer research.28,29
Nanotechnology
Nanotechnologies have demonstrated a remarkable potential against infections;35 therefore, there is an urgency to develop novel nanotechnology-based drugs alone and in combination to overcome biofilm drug resistance.36,37 C. albicans biofilm inhibition by silver nanoparticles (AgNPs)38,39 or SeNPs has been previously reported;40–42 our group previously described the inhibition of a preformed biofilm by SeNPs synthesized by a femtosecond laser, and we demonstrated that crystallinity and size are the two main physical parameters of SeNPs that affect the viability of C. albicans biofilms.19
CS-SeNPs synthesized by nanosecond laser ablation in liquids
In this study, we report, for the first time, the synthesis of SeNPs by irradiating Se pellets submerged in deionized (DI) water consisting of CS at 0.25% (CS-SeNPs). The irradiation was achieved by using a pulsed nanosecond neodymium-doped yttrium aluminum garnet (Nd:YAG) laser. The ability to synthesize contamination-free pure nanoparticles is essential for biomedical applications. To the best of our knowledge, this is the first time that CS-SeNPs were tested against a preformed mature C. albicans biofilm, also showing that the combination of SeNPs and CS has a potent fungicidal effect in a dose-dependent manner measured by a well-established phenotypic assay,43–45 and most importantly, we demonstrated a potent synergistic fungicidal effect with the combination of CS with SeNPs against the C. albicans biofilms. Synergy was analyzed by the CompuSyn software46 to quantitatively determine the synergistic effect;13,15 we analyzed the CI with the data of the nonlinear dose–effect curves (effect-based approaches).12 Finally, we showed by advanced electron microscopy (EM) that CS-SeNPs permeabilize the outer CW due to CS properties, allowing Se to enter. The change of the characteristic spherical structure of the Candida cells is due to protein misfolding that causes a change of structure induced by Se toxicity.21,23–26 We conclude that CS-SeNPs have a synergistic effect with an effective dose–response inhibition of the preformed mature biofilm in vitro. More studies are warranted to understand the synergistic effect of the combination of Se and CS against the Candida biofilm.
Materials and methods
Materials
Se pellets (Se, <5 mm, ≤99.999% trace metals), CS (low molecular weight), sodium hydroxide (NaOH, American Chemical Society [ACS] reagent, ≥97.0%, pellets), and acetic acid (ACS reagent, ≥99.7%) were purchased from Sigma-Aldrich (St Louis, MO, USA). Molecular biology grade water (MT46000CM) was purchased from Thermo Fisher Scientific (Waltham, MA, USA).
Nanoparticle synthesis
The synthesis of SeNPs and CS-SeNPs was performed using a nanosecond laser source by placing 0.35 g of Se pellets (purity 99.9%) deposited as target at the bottom of a 1.7 mL microcentrifuge tube that contained 0.5 mL of DI water or 0.25% (wt/vol) CS solution. The laser used for the irradiation (Ekspla, NT342A, Vilnius, Lithuania) was the Nd:YAG with a laser pulse duration of 3.6 ns at a wavelength of 1,064 nm and a repetitive rate of 20 Hz. The laser beam was aimed at the target vertically from above into the open microcentrifuge tube where the beam focused onto the pure Se pellets placed at the bottom. The target was irradiated for 15 min producing a red–orange color in the solution that was extracted for further analysis. The SeNPs produced in the 0.25% (wt/vol) CS solution were washed to remove the excess CS from the sample. An acid wash was performed using a 50/50 solution of acetic acid and DI water which was added to the sample and then centrifuged to produce a pellet resuspended in phosphate-buffered saline (PBS) solution.
Characterization
The SeNPs were produced by irradiating Se pellets using the nanosecond laser pumped by a 20 Hz Q-switched Nd:YAG laser powered at 20 mJ. The determination of the concentration of the solution was performed using atomic absorption spectroscopy (AA-6200; Shimadzu, Kyoto, Japan) with an Se lamp (L2433-34NQ; Hamamatsu, Boston, MA, USA). Hydrodynamic size and zeta potential (ZP) of SeNPs were characterized using the dynamic light scattering (DLS) system (Zetasizer Nano ZS; Malvern Instruments, Malvern, UK) at 25°C. The high-resolution transmission electron microscope (HRTEM) (JOEL 2010F) and atomic resolution microscope (ARM, JOEL ARM 200F) were used to acquire images of the SeNPs to determine the size and shape of the nanoparticles using the Cs probe at a voltage of 200 kV with a spatial resolution of 0.75 Å. The specially resolved elemental analysis was performed by X-ray emission spectroscopy attachment to the HRTEM.
Strain, media, and culture conditions
The wild-type C. albicans clinical strain SC531447 was used in all experiments as a control standard. In addition, we used TW148 and two C. albicans clinical isolates resistant to fluconazole, named TW1748,49 and 648650,51 strains as previously described. Frozen cells from stocks stored at −80°C were propagated overnight in yeast–peptone–dextrose (YPD) agar plates. Flasks of YPD liquid media were inoculated with a loopful of Candida growth and incubated in an orbital shaker (180 rpm) at 30°C and grown for 14–16 h. Biofilms were assessed using the 96-well microtiter plate-based method as previously reported.43
Activity against a preformed C. albicans biofilms
Biofilms were assessed using a well-known 96-well microtiter plate-based method.43 Briefly, yeast cells collected from overnight cultures were washed in sterile PBS and resuspended at a final concentration of 1.0×106 cells/mL in Roswell Park Memorial Institute (RPMI) medium-1640. Biofilms were formed on tissue culture-treated, 96-well microtiter plates (Corning Incorporated) incubated at 37°C for 24 h. The biofilms formed and attached to the flat bottom of the wells were washed twice with PBS. CS-SeNPs, SeNPs, and CS were assessed against the preformed biofilm for their fungicidal activity at concentrations ranging from 5 to 2,500 and 0.05 to 25 ppm for CS and SeNPs, respectively, in serial twofold dilutions as previously described.43,44,58 The plates were covered with parafilm and incubated for another 24 h. Then, plates were carefully washed twice, and the biofilms were quantified using the tetrazolium salt (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide [XTT]) reduction assay to test the efficacy of the nanoparticle preparations. All tests were performed in duplicate and were repeated at least three times in independent experiments. The IC50 was performed with the dose–response fits calculated using Origin 9 software (OriginLab Corporation, Northampton, MA, USA).59
Cytotoxicity assay
A stock solution of CS-SeNPs was diluted to the desired concentrations ranging from 50 to 1 ppm in growth medium and subsequently added into a 96-well plate containing human retinal pigment epithelial cell line ARPE-19 (5×104 cells/well). Microtiter plates were incubated at 37°C in a 5% CO2 air humidified atmosphere for 24 h. The cell experiments were performed in triplicate, and readings of the cell viability using a luminescent cell viability assay (CellTiter-Glo; Promega Corporation, Madison, WI, USA) were performed after 24 h.
Visualization by SEM of the effects of nanoparticles on preformed C. albicans biofilms
For SEM ultrastructural observation of the mature biofilms cultured on 48-well plates for 24 h treated with CS-SeNPs (3.5 ppm), SeNPs (21.7 ppm), and CS (25,000 ppm), we gently washed the treated biofilm twice with PBS and fixed with 4% formaldehyde and 1% glutaraldehyde (GA) for 1 h at room temperature. The fixed samples were washed with PBS and then stained in 1% osmium tetroxide (OsO4) solution buffered with PBS for 1 h. The samples were dehydrated with a series of ethanol solutions (25% for 10 min, 50% for 10 min, 70% for 10 min, 95% for 10 min, and absolute alcohol for 20 min).60 The dehydrated specimens were then transferred to 300-mesh carbon-coated copper grids to be observed by SEM in a Hitachi S-5500 (Hitachi Ltd., Tokyo, Japan).
Visualization by TEM of the effects of nanoparticles on preformed C. albicans biofilms
For ultrastructural TEM imaging, Candida biofilms were grown for 24 h and then treated with CS-SeNPs (3.5 ppm). The treated mature biofilm was then centrifuged at 3,500 rpm for 10 min. After washing two times with PBS, cells were fixed in 1 mL of 4% formaldehyde and 1% GA for 2 h. The fixed samples were stained with 1% OsO4 for 1 h. After washing the Candida biofilms with PBS to eliminate the heavy metal stain, a dehydration series was performed with 25, 50, 75, 95, and 100% ethanol diluted in dH2O. The absolute dehydration was assured with propylene oxide before embedding in an epoxy resin LX-112 (Ladd Research Industries, Williston, VT, USA), and the resin was left 48 h at 60°C to harden. The epoxy resin-embedded sections were cut (90 nm thick) using an ultramicrotome (Leica Microsystems, Wetzlar, Germany) and a 45° angle diamond knife as previously described.61 Ultrathin sections were mounted on an uncoated copper mesh grid and visualized using JEOL JEM-2010F.
CI analysis
CI theorem of Chou–Talalay analyses13,14 was calculated using the computer software CalcuSyn15,16 version 2.0 (Biosoft, Cambridge, UK).46 We analyzed the CI based on the nonlinear dose–effect curves (effect-based approaches).12 To analyze the dose–effect parameters (% inhibition of the biofilm) of each compound alone (CS and SeNPs) as well as in combination (CS-SeNPs) and calculate the CI value, the parameters can be automatically determined from the median-effect equation.13 Comprehensive procedures of automated dose–effect dynamic analysis via mathematical induction for quantization of synergism in drug combination studies are given in the user’s manual for CompuSyn software.14 Briefly, the CI-isobologram equation was used, where CI values around 1 demonstrate additive effects of the two drugs tested. CI<1 indicates a synergistic effect of the two drugs combined, and CI>1 indicates an antagonistic effect. We need to input each dose–response result as shown in Table 1.
Table 1.
Synergy based on the nonlinear dose–effect curves
| Dose (ppm), CS | Biofilm inhibition (%)a | Dose (ppm), SeNPs | Biofilm inhibition (%)a | Dose combination; 2,500+25 ppm; CS + SeNPs | Biofilm inhibition (%)a | Fa | CIb |
|---|---|---|---|---|---|---|---|
| Synergism (CI<1) | |||||||
| 2,500 | 45±1 | 25 | 59±7 | 2,525 | 80±2 | 0.97 | 2.53E-4 |
| 1,250 | 41±1 | 12.5 | 24±3 | 1,262 | 73±3 | 0.908 | 2.91E-4 |
| 630 | 26±1 | 6.3 | 23±2 | 636 | 71±1 | 0.887 | 2.45E-4 |
| 310 | 24±1 | 3.1 | 20±2 | 313 | 46±7 | 0.898 | 9.45E-5 |
| 160 | 22±1 | 1.6 | 16±4 | 161 | 13±1 | 0.635 | 0.00194 |
| Antagonism (CI>1) | |||||||
| 80 | 20±2 | 0.8 | 11±5 | 80.8 | 10±1 | 0.095 | 2.32 |
| 40 | 19±1 | 0.4 | 8±4 | 40.4 | 3±1 | 0.063 | 6.25 |
| 20 | 7±1 | 0.2 | 3±3 | 20.2 | 1±1 | 0.03 | 65.46 |
| 10 | 5±1 | 0.1 | 6±3 | 10.1 | 0±0 | 0.01 | 2,910 |
| 5 | 4±1 | 0.05 | 5±5 | 5.05 | 0±0 | 0.006 | 7,448 |
| 2.5 | 3±1 | 0.02 | 1±1 | 2.02 | 0±0 | 0.004 | 24,384 |
| 1.2 | 0±1 | 0.01 | 0±1 | 1.01 | 0±0 | 0.003 | 32,414 |
Notes: This table is constructed from the data generated by CompuSyn report.
The data of the nonlinear dose–effect curves were obtained from the XTT assay and were analyzed by the CompuSyn software. Data are presented as the mean (±SEM) of three independent experiments performed on different days.
CI was calculated from the CI equation and algorithms using CompuSyn software. When the CI is 1, the interaction for the inhibitory effect is additive, CI >1 is antagonistic, and CI <1 means a synergistic effect. Bold values are synergistic.
Abbreviations: CI, combination index; CS, chitosan; Fa, fraction affected; SEM, standard error of the mean; SeNPs, selenium nanoparticle; XTT, 2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide.
Statistical analyses
The IC50 was calculated by dose–response fitting using Origin 9 software. The graphs show the mean ± standard error of the mean from three separate experiments performed in duplicate. For quantitative assays, statistical analyses were performed with Student’s t-test. Statistical significance was accepted at a P-value of <0.05.
Results
Characterization
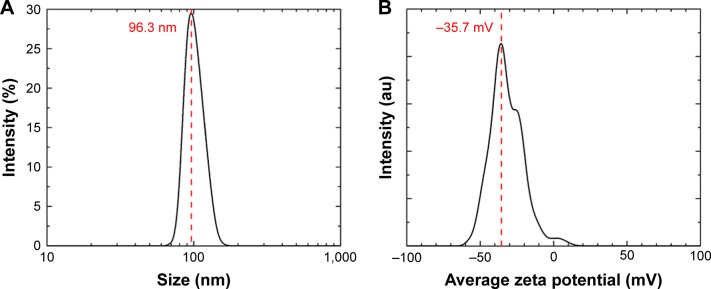
The average hydrodynamic diameter of the CS-SeNPs dispersed in DI water was around 96 nm determined by DLS (Figure 1A). The ZP had a negative charge of about −35.7 mV (Figure 1B). The concentration of the solution determined by atomic absorption spectroscopy (AAS) was 25.5±0.5 ppm. Energy-dispersive spectroscopy (EDS) spectra of CS-SeNPs demonstrate the presence of Se signal in the sample by spectral mapping acquisition (Figure 2). SeNPs were detected on the outer CW of the C. albicans (red dots) by EDS mapping. For the nanoparticles produced in the presence of CS (CS-SeNPs), HRTEM images show spherical shape of the nanoparticles, and the diffraction pattern of small and large nanoparticles indicates that both are crystalline in nature. The CS matrix surrounding the nanoparticles can be observed (Figure S1A–D); this CS matrix enhances the crystalline nature of the nanoparticle compared to the amorphous nature of SeNPs produced in DI water. The stability of the nanoparticles due to the CS was demonstrated by ZP which had a negative charge of about −35.7 mV, where the binding effects of the CS to the nanoparticle maintain the stability of the nanoparticles.
Figure 1.
DLS and ZP.
Notes: (A) Hydrodynamic diameters of the synthesized SeNPs stabilized in bovine serum albumin and dispersed in DI water; the average size of the nanoparticles is 96.3 nm. (B) The ZP showed a negative charge of −35.7 mV.
Abbreviations: DI, deionized; DLS, dynamic light scattering; SeNPs, selenium nanoparticles; ZP, zeta potential.
Figure 2.

EDS spectrum of SeNPs.
Notes: Se EDS peaks are labeled. Strong signals from the atoms in the SeNPs observed in the spectrum confirm Se metalloid nanoparticles on the yeast outer CW by spectral mapping acquisition.
Abbreviations: CW, cell wall; EDS, energy-dispersive spectroscopy; Se, selenium; SeNPs, selenium nanoparticles; UTSA, University of Texas at San Antonio.
Inhibition of preformed biofilms
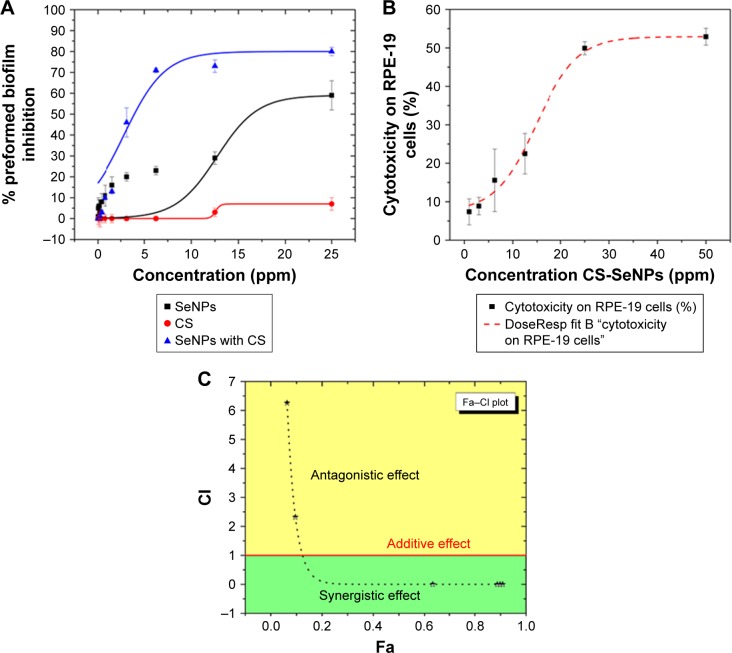
In this study, we used a well-established phenotypic assay as reported by Pierce et al44 and Pierce and Lopez-Ribot.45 Results indicated an inhibitory effect of SeNPs in a dose-dependent manner on the preformed C. albicans biofilm, with a calculated IC50 of 21.7 ppm. CS alone had an inhibitory effect on the biofilm obtaining a 7% inhibition at 25 ppm. CS-SeNPs showed the most potent inhibition against preformed biofilms in a dose–response manner with an IC50 of 3.5 ppm, which pointed to a strong synergistic effect when compared to both compounds alone (Figure 3A). We also compared the activity of CS-SeNPs on preformed biofilms of two drug-sensitive strains (SC531447 and TW148) and two drug-resistant C. albicans strains (TW1748,49 and 648650,51). No statistical difference in dose–response curves was observed between sensitive and resistant clinical strains. Statistical significance was accepted at a P-value of <0.05.
Figure 3.
Dose–response curves.
Notes: (A) Dose–response curves for the activity of SeNPs, CS, and CS-SeNPs against preformed C. albicans biofilms. The half maximal inhibitory concentration (IC50) values were calculated as SeNPs (21.7 ppm) and CS-SeNPs (3.52 ppm). Values are the mean ± SEM (error bars) from independent experiments performed in duplicate. (B) Cytotoxicity of CS-SeNPs in RPE-19 cells, the 50% cytotoxic concentration (CC50) is 26.3 ppm. Cytotoxicity of CS-SeNPs was quantified by a Luciferase assay after 24 h of compound incubation. X-axis, concentration in ppm; Y-axis, percentage of cell cytotoxicity. Dose–response curves were plotted using Origin 9 curve fit. Each data point represents the mean ± SEM of three independent experiments performed on different days. (C) The CI was determined. A horizontal red line marks CI=1. The data are the mean values from three independent experiments. Combined doses of CS and SeNPs from 160–1.6 to 25,000–25 ppm, respectively, resulted in Fa from 0.6 to 0.9 with a CI<1 (synergy). Doses under 3.5–35 ppm of CS-SeNPs, respectively, were antagonistic.
Abbreviations: C. albicans, Candida albicans; CI, combination index; CS, chitosan; CS-SeNPs, CS-decorated SeNPs; Fa, fraction affected; SEM, standard error of the mean; SeNPs, selenium nanoparticles.
Cytotoxicity assay
The CS-SeNP concentrations showing potent fungicidal effects against mature C. albicans biofilms (3.5 ppm) are lower than those at which they exhibit cytotoxicity, as demonstrated by a cytotoxicity assay using human ARPE cells (obtained from the American Type Culture Collection, Manassas, VA, USA), resulting in a 50% cytotoxicity concentration (CC50) of 26.3 ppm (Figure 3B).
Visualization by scanning EM (SEM) of the effects of nanoparticles on preformed C. albicans biofilms
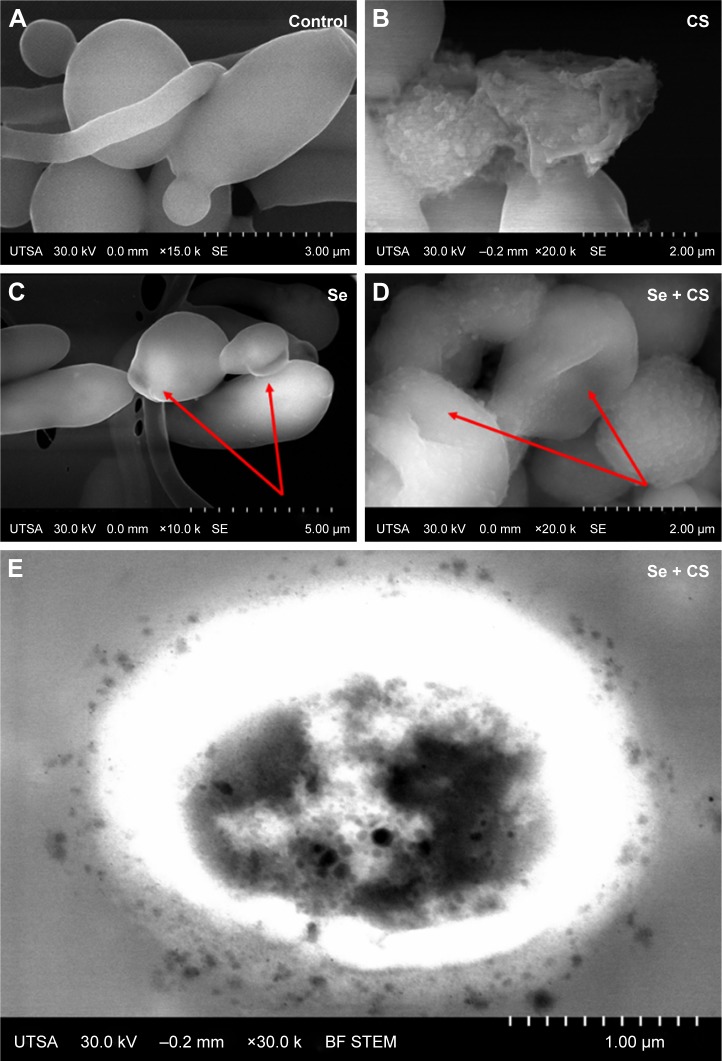
The ultrastructural changes were imaged under SEM after treatment with CS, SeNPs, and CS-SeNPs. Figure 4A shows untreated C. albicans SC5314 biofilms with a dense network of yeast cells and hyphae; yeast cells display a characteristic ovoid morphology. Figure 4B shows C. albicans biofilms treated with CS at 25,000 ppm; yeast cells became enlarged and deformed; CS adheres to the outer CW of the cells. C. albicans treated with SeNPs at 21.7 ppm (Figure 4C) has changes in morphology (red arrows). Yeasts treated with CS-SeNPs at 3,500–3.5 ppm, respectively, show major changes in morphology and structure. Red arrows show major deformation of the yeast cells and the polycationic polymer adhered to the negatively charged outer CW (Figure 4D). Bright-field (BF) scanning transmission electron microscopy (STEM) image shows CS-SeNPs entering the yeast (Figure 4E), CS adhered to the outer CW, with SeNPs (darker dots).
Figure 4.
SEM.
Notes: (A) Untreated C. albicans biofilms show a characteristic ovoid morphology, and a dense network of yeast cells and hyphae (B) biofilms treated with CS at 25,000 ppm showed enlarged and deformed yeast cell, (C) C. albicans treated with SeNPs at 21.7 ppm shows changes in morphology (red arrows) and (D) C. albicans treated with CS-SeNPs at 3.5 ppm has major changes in morphology and structure (red arrows), the polymer adheres to the CW. (E) BF STEM image of a C. albicans biofilm treated with CS-SeNPs. Throughout the CW, nanoparticles can be observed entering and the CS biopolymer containing SeNPs adheres to the outer CW. CS-SeNPs, CS-decorated SeNPs.
Abbreviations: BF, bright-field; C. albicans, Candida albicans; CS, chitosan; CW, cell wall; SEM, scanning electron microscopy; SeNPs, selenium nanoparticles; STEM, scanning transmission electron microscopy; UTSA, University of Texas at San Antonio.
Visualization by transmission EM (TEM) of the effects of nanoparticles on preformed C. albicans biofilms
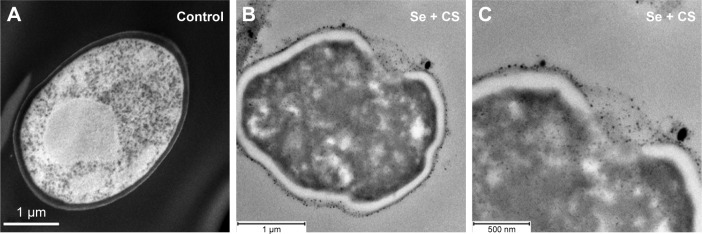
In TEM image, we observed the C. albicans yeast cell after thin sectioning; cells without treatment showed a characteristic ovoid morphology and intact CW (Figure 5A). After 24 h treatment with CS-SeNPs (3.5 ppm), a major distortion of the ovoid morphology of the cell is shown and CS-SeNPs are observed adhering to the outer CW (Figure 5B and C), metalloid nanoparticles enter the cell by disruption of the CW and the cell membrane. Ultrathin-section electron micrographs of the yeast cells document intracellular accumulation of the nanoparticles.
Figure 5.
TEM.
Notes: (A) Ultrastructure of untreated C. albicans yeast cell after thin sectioning shows a characteristic ovoid morphology. (B and C) After 24 h treatment with CS-SeNPs (3.5 ppm), the yeast cell loses the characteristic morphology as the cell distorts, the CW disrupts, and the CS-SeNPs enter the cell. CS-SeNPs, CS-decorated SeNPs.
Abbreviations: C. albicans, Candida albicans; CS, chitosan; CW, cell wall; SeNPs, selenium nanoparticles; TEM, transmission electron microscopy.
Synergy
CalcuSyn16 generated CI values according to the Chou–Talalay method.15 The CS-SeNP combination treatment was synergistic when CS (2,500–160 ppm) combined with SeNPs (25–1.6 ppm), resulting in CI values of <1; for lower doses, the effect was antagonistic with CI values of >1 as shown in Table 1. The additive effect of drug combination depends on the individual dose–effect curves and enables the formulation of synergy, additivity, or antagonism. The dose–effect-based methods depend on the Loewe additivity model.12
Discussion
A wild-type C. albicans strain SC5314 was used in this study; although this is a “susceptible” strain under normal planktonic (free-floating) growing conditions, it has been demonstrated that biofilms formed by this strain display high level of resistance against most clinically used antifungals.43 The antifungal properties of CS30 and SeNPs alone had been previously reported;40 our group demonstrated that SeNPs bind and get adsorbed into the yeast cell, and the main physical properties of SeNPs against the Candida biofilm are size and crystallinity of the nanoparticles.19
In this study, we demonstrated the first synthesis of pure, stable, and crystalline nanoparticles by pulsed laser-ablation in liquids (PLAL) with a nanosecond laser. This laser synthesis is well adapted for biomedical applications because the surface of the produced nanoparticles is free of any chemical byproducts or surfactants.52 Therefore, the biological interaction between the yeast cell and the nanoparticle is not altered by any contaminants. To achieve this, the CS-SeNPs were synthesized by PLAL in DI water containing 0.25% of CS resulting in SeNPs wrapped within a CS shell, shown by HRTEM;53 this CS wrapping is most likely due to the amino acid groups in CS interacting with the surface of the SeNPs, where the coordination behaviors of the nanoparticles maintain the nanoparticle stability and enhance the crystallinity.54 The average size of these wrapped nanoparticles and the electric potential surrounding the particle, called ZP, were measured by DLS. The particles were around ~100 nm in size with an ZP equal to −35.7 mV; this large negative value confirms that the CS-SeNPs are stable in solution due to the polycationic surface charge of the CS30,32,55 (Figure 1A and B). Furthermore, the slight increase in the value of the ZP of the CS-SeNPs when compared to the SeNPs can be attributed to the polycationic surface charge of the CS and suggests that CS is coating the SeNPs.
The CS and SeNPs alone or combined were tested by a standardized method for in vitro antifungal susceptibility43,56 against mature C. albicans biofilms which are known to be very difficult to inhibit because sessile cells become shielded inside an exopolymeric matrix of the biofilm displaying intrinsic resistance to most conventional antifungals.4,36 Taken separately, SeNPs (IC50 21.7 ppm) and CS have shown fungicidal properties, but when combined (CS-SeNPs) achieved a potent inhibitory effect (IC50 3.5 ppm) against the mature biofilm in a dose–response manner (Figure 3A). To quantify the synergistic effect of the synthesized CS-SeNPs, we used the Chou–Talalay method15 specifically designed for drug combination. This theory uses algorithms developed by CompuSyn software13 to demonstrate synergy, additivity, or antagonism, as shown in the fraction affected (Fa)–CI plot57 (Figure 3C). We analyzed the dose–effect parameters of each compound alone (CS and SeNPs) as well as in combination, and thus obtained the CI value.12,13 With higher doses of CS-SeNPs, a stronger synergistic interaction was obtained as expected, and doses below the IC50 became antagonistic (Table 1).
This is the first reported case, to the best of our knowledge, that proved the synergistic fungicidal effect of CS-SeNPs against preformed C. albicans biofilms.
We theorize that as the bioadhesive polycationic polymer CS strongly binds and permeates the CW of the C. albicans yeast (by reducing the negative surface charge of the CW30,34) this permeabilization enables SeNPs to straightforwardly enter the yeast in excess. Inside the cell, Se competes by affinity with sulfur21,23–25 producing an oxidative stress23 resulting in DNA strand breaks,25 therefore, creating changes in the morphology of the yeast cell19 as documented by advanced EM (Figures 4D and 5C).
The dose of CS-SeNPs required to inhibit the mature biofilm (IC50 3.5 ppm) is within the noncytotoxic range on human cells (CC50 26.3 ppm); thus CS-SeNPs could be a good choice for antibiofilm coatings. Future research is warranted to determine the complete mechanism of action and toxicity.
Conclusion
Applying nanosecond PLAL, we obtained, for the first time, the synthesis of CS-stabilized pure SeNPs free of contaminants. To the best of our knowledge, this is the first time that CS-SeNPs have demonstrated efficient dose-dependent activity against preformed C. albicans biofilms with an IC50 of 3.5 ppm of CS-SeNPs. By EM, we observed the fungicidal effect of CS-SeNPs by permeabilization of the outer CW due to the CS properties, and the changing of the structure of C. albicans was due to SeNP effect, documenting the entry of SeNPs in excess into the yeast cell. Most importantly, the results of the CI show a potent synergistic effect.
Supplementary material
HRTEM images of the CS-SeNPs.
Notes: Electron diffraction pattern of (A and B) large and (C and D) small nanoparticles indicates that both are crystalline in nature. CS-SeNPs, CS-decorated SeNPs.
Abbreviations: CS, chitosan; HRTEM, high-resolution transmission electron microscope; SeNPs, selenium nanoparticles.
Acknowledgments
HHL and JLLR would like to acknowledge the National Institute on Minority Health and Health Disparities (G12MD007591) from the National Institutes of Health, the National Science Foundation Partnerships for Research and Education in Materials (NSF-PREM) (DMR-0934218), and the Welch Foundation (AX-1615). GG and KLN would like to acknowledge grant support from the Biomedical Research Grants Program of the San Antonio Area Foundation. BAV, LCM, and JM would like to acknowledge support from the Air Force Office of Scientific Research (FA9550-15-1-0109).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Delaloye J, Calandra T. Invasive candidiasis as a cause of sepsis in the critically ill patient. Virulence. 2014;5(1):161–169. doi: 10.4161/viru.26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harriott MM, Noverr MC. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19(11):557–563. doi: 10.1016/j.tim.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gahlot R, Nigam C, Kumar V, Yadav G, Anupurba S. Catheter-related bloodstream infections. Int J Crit Illn Inj Sci. 2014;4(2):162–167. doi: 10.4103/2229-5151.134184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathé L, Van Dijck P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 2013;59(4):251–264. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee S, Maiti P, Dey R, Kundu A, Dey R. Biofilms on indwelling urologic devices: microbes and antimicrobial management prospect. Ann Med Health Sci Res. 2014;4(1):100–104. doi: 10.4103/2141-9248.126612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taff HT, Mitchell KF, Edward JA, Andes DR. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013;8(10):1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn DM, George T, Chandra J, Mukherjee PK, Ghannoum MA. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother. 2002;46(6):1773–1780. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Yue L, Gu W, Li X, Zhang L, Sun S. Synergistic effect of fluconazole and calcium channel blockers against resistant Candida albicans. PLoS One. 2016;11(3):e0150859. doi: 10.1371/journal.pone.0150859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leekha S, Terrell CL, Edson RS. General principles of antimicrobial therapy. Mayo Clin Proc. 2011;86(2):156–167. doi: 10.4065/mcp.2010.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L, Kohli M, Smith A. Nanoparticles for combination drug therapy. ACS Nano. 2013;7(11):9518–9525. doi: 10.1021/nn405674m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3(3):e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou T-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 14.Chou T-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 15.Chang TT, Chou TC. Rational approach to the clinical protocol design for drug combinations: a review. Acta Paediatr Taiwan. 2000;41(6):294–302. [PubMed] [Google Scholar]
- 16.Bijnsdorp IV, Giovannetti E, Peters GJ. Analysis of drug interactions. Methods Mol Biol. 2011;731:421–434. doi: 10.1007/978-1-61779-080-5_34. [DOI] [PubMed] [Google Scholar]
- 17.Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias MF, Bernardes-Filho F, Quaresma-Santos MV, Amorim AG, Schechtman RC, Azulay DR. Treatment of superficial mycoses: review. Part II. An Bras Dermatol. 2013;88(6):937–944. doi: 10.1590/abd1806-4841.20132018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guisbiers G, Lara HH, Mendoza-Cruz R, et al. Inhibition of Candida albicans biofilm by pure selenium nanoparticles synthesized by pulsed laser ablation in liquids. Nanomedicine. 2017;13(3):1095–1103. doi: 10.1016/j.nano.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tinggi U. Selenium: its role as antioxidant in human health. Environ Health Prev Med. 2008;13(2):102–108. doi: 10.1007/s12199-007-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kieliszek M, Błażejak S, Kurek E. Binding and conversion of selenium in Candida utilis ATCC 9950 yeasts in bioreactor culture. Molecules. 2017;22(3):352. doi: 10.3390/molecules22030352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao L, Ma Y, Zhuang M, Luo T, Wang Y, Hong A. Chitosan-decorated selenium nanoparticles as protein carriers to improve the in vivo half-life of the peptide therapeutic BAY 55-9837 for type 2 diabetes mellitus. Int J Nanomedicine. 2014;9:4819–4828. doi: 10.2147/IJN.S67871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kieliszek M, Błażejak S, Gientka I, Bzducha-Wróbel A. Accumulation and metabolism of selenium by yeast cells. Appl Microbiol Biotechnol. 2015;99(13):5373–5382. doi: 10.1007/s00253-015-6650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieliszek M, Błażejak S, Płaczek M. Spectrophotometric evaluation of selenium binding by Saccharomyces cerevisiae ATCC MYA-2200 and Candida utilis ATCC 9950 yeast. J Trace Elem Med Biol. 2016;35:90–96. doi: 10.1016/j.jtemb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Kieliszek M, Błażejak S, Bzducha-Wróbel A, Kurcz A. Effects of selenium on morphological changes in Candida utilis ATCC 9950 yeast cells. Biol Trace Elem Res. 2016;169(2):387–393. doi: 10.1007/s12011-015-0415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrero E, Wellinger RE. Yeast as a model system to study metabolic impact of selenium compounds. Microb Cell. 2015;2(5):139–149. doi: 10.15698/mic2015.05.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letavayová L, Vlasáková D, Spallholz JE, Brozmanová J, Chovanec M. Toxicity and mutagenicity of selenium compounds in Saccharomyces cerevisiae. Mutat Res. 2008;638(1):1–10. doi: 10.1016/j.mrfmmm.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhai X, Zhang C, Zhao G, Stoll S, Ren F, Leng X. Antioxidant capacities of the selenium nanoparticles stabilized by chitosan. J Nanobiotechnology. 2017;15(1):4. doi: 10.1186/s12951-016-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S, Luo Y, Zeng H, et al. Encapsulation of selenium in chitosan nanoparticles improves selenium availability and protects cells from selenium-induced DNA damage response. J Nutr Biochem. 2011;22(12):1137–1142. doi: 10.1016/j.jnutbio.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Peña A, Sánchez NS, Calahorra M. Effects of chitosan on Candida albicans: conditions for its antifungal activity. Biomed Res Int. 2013;2013:527549. doi: 10.1155/2013/527549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kvasničková E, Paulíček V, Paldrychová M, Ježdík R, Maťátková O, Masák J. Aspergillus fumigatus DBM 4057 biofilm formation is inhibited by chitosan, in contrast to baicalein and rhamnolipid. World J Microbiol Biotechnol. 2016;32(11):187. doi: 10.1007/s11274-016-2146-9. [DOI] [PubMed] [Google Scholar]
- 32.Pu Y, Liu A, Zheng Y, Ye B. In vitro damage of Candida albicans biofilms by chitosan. Exp Ther Med. 2014;8(3):929–934. doi: 10.3892/etm.2014.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheung RCF, Ng TB, Wong JH, Chan WY. Chitosan: an update on potential biomedical and pharmaceutical applications. Mar Drugs. 2015;13(8):5156–5186. doi: 10.3390/md13085156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai T, Tanaka M, Huang Y-Y, Hamblin MR. Chitosan preparations for wounds and burns: antimicrobial and wound-healing effects. Expert Rev Anti Infect Ther. 2011;9(7):857–879. doi: 10.1586/eri.11.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X, Radovic-Moreno AF, Wu J, Langer R, Shi J. Nanomedicine in the management of microbial infection – overview and perspectives. Nano Today. 2014;9(4):478–498. doi: 10.1016/j.nantod.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierce CG, Srinivasan A, Uppuluri P, Ramasubramanian AK, López-Ribot JL. Antifungal therapy with an emphasis on biofilms. Curr Opin Pharmacol. 2013;13(5):726–730. doi: 10.1016/j.coph.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh R, Smitha MS, Singh SP. The role of nanotechnology in combating multi-drug resistant bacteria. J Nanosci Nanotechnol. 2014;14(7):4745–4756. doi: 10.1166/jnn.2014.9527. [DOI] [PubMed] [Google Scholar]
- 38.Monteiro DR, Gorup LF, Silva S, et al. Silver colloidal nanoparticles: antifungal effect against adhered cells and biofilms of Candida albicans and Candida glabrata. Biofouling. 2011;27(7):711–719. doi: 10.1080/08927014.2011.599101. [DOI] [PubMed] [Google Scholar]
- 39.Silva S, Pires P, Monteiro DR, et al. The effect of silver nanoparticles and nystatin on mixed biofilms of Candida glabrata and Candida albicans on acrylic. Med Mycol. 2013;51(2):178–184. doi: 10.3109/13693786.2012.700492. [DOI] [PubMed] [Google Scholar]
- 40.Shakibaie M, Salari Mohazab N, Ayatollahi Mousavi SA. Antifungal activity of selenium nanoparticles synthesized by Bacillus species Msh-1 against Aspergillus fumigatus and Candida albicans. Jundishapur J Microbiol. 2015;8(9):e26381. doi: 10.5812/jjm.26381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kheradmand E, Rafii F, Yazdi MH, Sepahi AA, Shahverdi AR, Oveisi MR. The antimicrobial effects of selenium nanoparticle-enriched probiotics and their fermented broth against Candida albicans. Daru. 2014;22:48. doi: 10.1186/2008-2231-22-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cremonini E, Zonaro E, Donini M, et al. Biogenic selenium nanoparticles: characterization, antimicrobial activity and effects on human dendritic cells and fibroblasts. Microb Biotechnol. 2016;9(6):758–771. doi: 10.1111/1751-7915.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce CG, Uppuluri P, Tristan AR, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3(9):1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce CG, Uppuluri P, Tummala S, Lopez-Ribot JL. A 96 well microtiter plate-based method for monitoring formation and antifungal susceptibility testing of Candida albicans biofilms. J Vis Exp. 2010 Oct 21; doi: 10.3791/2287. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pierce CG, Lopez-Ribot JL. Candidiasis drug discovery and development: new approaches targeting virulence for discovering and identifying new drugs. Expert Opin Drug Discov. 2013;8(9):1117–1126. doi: 10.1517/17460441.2013.807245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bijnsdorp IV, Giovannetti E, Peters GJ. Analysis of drug interactions. Methods Mol Biol. 2011;731:421–434. doi: 10.1007/978-1-61779-080-5_34. [DOI] [PubMed] [Google Scholar]
- 47.Gillum AM, Tsay EY, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198(2):179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 48.Singh A, Yadav V, Prasad R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS One. 2012;7(6):e39812. doi: 10.1371/journal.pone.0039812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohberger A, Coste AT, Sanglard D. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot Cell. 2014;13(1):127–142. doi: 10.1128/EC.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pierce CG, Chaturvedi AK, Lazzell AL, et al. A novel small molecule inhibitor of Candida albicans biofilm formation, filamentation and virulence with low potential for the development of resistance. NPJ Biofilms Microbiomes. 2015;1(1):15012. doi: 10.1038/npjbiofilms.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vila TVM, Chaturvedi AK, Rozental S, Lopez-Ribot JL. In vitro activity of miltefosine against Candida albicans under planktonic and biofilm growth conditions and in vivo efficacy in a murine model of oral candidiasis. Antimicrob Agents Chemother. 2015;59(12):7611–7620. doi: 10.1128/AAC.01890-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto A, Tamura A, Honda T, et al. Transfer of the species dissolved in a liquid into laser ablation plasma: an approach using emission spectroscopy. J Phys Chem. 2015;119(47):26506–26511. [Google Scholar]
- 53.Guisbiers G, Wang Q, Khachatryan E, et al. Anti-bacterial selenium nanoparticles produced by UV/VIS/NIR pulsed nanosecond laser ablation in liquids. Laser Phys Lett. 2015;12(1):16003. [Google Scholar]
- 54.Yu B, Zhang Y, Zheng W, Fan C, Chen T. Positive surface charge enhances selective cellular uptake and anticancer efficacy of selenium nanoparticles. Inorg Chem. 2012;51(16):8956–8963. doi: 10.1021/ic301050v. [DOI] [PubMed] [Google Scholar]
- 55.Martinez LR, Mihu MR, Tar M, et al. Demonstration of antibiofilm and antifungal efficacy of chitosan against candidal biofilms, using an in vivo central venous catheter model. J Infect Dis. 2010;201(9):1436–1440. doi: 10.1086/651558. [DOI] [PubMed] [Google Scholar]
- 56.Siles SA, Srinivasan A, Pierce CG, Lopez-Ribot JL, Ramasubramanian AK. High-throughput screening of a collection of known pharmacologically active small compounds for identification of Candida albicans biofilm inhibitors. Antimicrob Agents Chemother. 2013;57(8):3681–3687. doi: 10.1128/AAC.00680-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu D, Kahen E, Cubitt CL, et al. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci Rep. 2015;5:16991. doi: 10.1038/srep16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lara HH, Romero-Urbina DG, Pierce C, Lopez-Ribot JL, Arellano-Jiménez MJ, Jose-Yacaman M. Effect of silver nanoparticles on Candida albicans biofilms: an ultrastructural study. J Nanobiotechnology. 2015;13:91. doi: 10.1186/s12951-015-0147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Veroli GY, Fornari C, Goldlust I, et al. An automated fitting procedure and software for dose-response curves with multiphasic features. Sci Rep. 2015;5:14701. doi: 10.1038/srep14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer ER, Hansen BT, Nair V, Hoyt FH, Dorward DW. Scanning electron microscopy. Curr Protoc Microbiol. 2012 doi: 10.1002/9780471729259.mc02b02s25. Chapter 2:Unit2B.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuwajima M, Mendenhall JM, Lindsey LF, Harris KM. Automated transmission-mode scanning electron microscopy (tSEM) for large volume analysis at nanoscale resolution. PLoS One. 2013;8(3):e59573. doi: 10.1371/journal.pone.0059573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HRTEM images of the CS-SeNPs.
Notes: Electron diffraction pattern of (A and B) large and (C and D) small nanoparticles indicates that both are crystalline in nature. CS-SeNPs, CS-decorated SeNPs.
Abbreviations: CS, chitosan; HRTEM, high-resolution transmission electron microscope; SeNPs, selenium nanoparticles.