Abstract
Purpose
A significant increase in the incidence of fungal infections and drug resistance has been observed in the past decades due to limited availability of broad-spectrum antifungal drugs. Nanomedicines have shown significant antimicrobial potential against various drug-resistant microbes. Silver nanoparticles (AgNps) are known for their antimicrobial properties and lower host toxicity; however, for clinical applications, evaluation of their impact at cellular and molecular levels is essential. The present study aims to understand the cellular and molecular mechanisms of AgNp-induced toxicity in a common fungal pathogen, Candida albicans.
Methods
AgNps were synthesized by chemical reduction method and characterized using UV–visible spectroscopy, X-ray powder diffraction, transmission electron microscopy, scanning electron microscopy–energy dispersive X-ray spectroscopy, energy dispersive X-ray fluorescence, and zeta potential. The anti-Candida activity of AgNps was assessed by broth microdilution and spot assays. Effects of AgNps on cellular and molecular targets were assessed by monitoring the intracellular reactive oxygen species (ROS) production in the absence and presence of natural antioxidant, changes in surface morphology, cellular ultrastructure, membrane microenvironment, membrane fluidity, membrane ergosterol, and fatty acids.
Results
Spherical AgNps (10–30 nm) showed minimum inhibitory concentration (minimum concentration required to inhibit the growth of 90% of organisms) at 40 μg/mL. Our results demonstrated that AgNps induced dose-dependent intracellular ROS which exerted antifungal effects; however, even scavenging ROS by antioxidant could not offer protection from AgNp mediated killing. Treatment with AgNps altered surface morphology, cellular ultrastructure, membrane microenvironment, membrane fluidity, ergosterol content, and fatty acid composition, especially oleic acid.
Conclusion
To summarize, AgNps affected multiple cellular targets crucial for drug resistance and pathogenicity in the fungal cells. The study revealed new cellular targets of AgNps which include fatty acids like oleic acid, vital for hyphal morphogenesis (a pathogenic trait of Candida). Yeast to hypha transition being pivotal for virulence and biofilm formation, targeting virulence might emerge as a new paradigm for developing nano silver-based therapy for clinical applications in fungal therapeutics.
Keywords: AgNps, nanomedicine, drug resistance, membrane fluidity, antifungals, reactive oxygen species, ROS
Introduction
In recent years, the emergence of microbes resistant to conventionally used antimicrobials (antibacterial and antifungal drugs) has been observed.1 Antibacterial resistance attracts greater attention from the research community but fungal drug resistance is kept at bay, although it poses an equally challenging threat to global public health.2 Higher prevalence of fungal infections, increasing incidence of drug resistance, and lower availability of antifungal drugs with minimal adverse side effects have led to a significant rise in morbidity and mortality in immunocompromised individuals and patients hospitalized with serious diseases.3–8 Fungal infections result in 1,350,000 deaths annually and affect millions of individuals.9,10 Candida species are major fungal pathogens causing both superficial and life-threatening systemic mycosis. The growth of fungal biofilms on medical devices is also a major challenge in hospital settings where commonly found Candida species become the cause of severe secondary fungal infections in patients.4–6 Out of more than 17 species of Candida known to cause fungal infections, Candida albicans is one of the most prevalent fungal pathogens responsible for 90% of invasive fungal infections worldwide.11,12 The observed drug resistance and pathogenicity in C. albicans have been attributed to several cellular factors and a multitude of molecular mechanisms, which include alterations in expression and efficacy of drug efflux pump proteins, membrane fluidity, membrane lipid composition (sterols and fatty acids), ability to evade host defenses, adherence, biofilm formation on deep-seated host tissues and medical devices, hyphal morphogenesis, and production of tissue damaging hydrolytic enzymes.3,4,7,13–15
Currently, four main classes of antifungal drugs, namely, azoles, polyenes, allylamines, and echinocandins, are available for the treatment of candidiasis and each class of drug has a particular mode of action and definite cellular target.12 The extensive deployment and indiscriminate use of these antifungal drugs have led to the emergence of more drug-resistant isolates.5,12 Although, antifungals and the increasing incidence of multidrug resistance (MDR) have been a critical issue of patient care and therapeutics, less attention has, however, been given toward the development of new and effective antifungals.6 The continuous evolution of drug-resistant fungal isolates constantly demands the development of newer and safer antifungal agents with broad-spectrum activity and minimal host toxicity. Recently, nanomedicines, comprising various nanoparticles and a combination of nanomaterials with existing drugs, have shown promising potential to combat microbial drug resistance.16 Various nanoparticles such as silver, zinc, gold, zinc oxide (ZnO), iron oxide (Fe3O4), titanium oxide (TiO2), zirconium oxide (ZrO2), and combinations of nanomaterials like chitosan/guanidine-functionalized graphene oxide, chitosan–iron oxide-coated graphene oxide have exhibited antimicrobial activity.17–24 However, the mechanism of antimicrobial activity of these nanomaterials or nanomedicines is still debatable and their safe medical adoption requires detailed investigations on the biological entities.25 Among many nanomaterials, silver nanoparticles (AgNps), known for their antimicrobial properties and higher microbial toxicity, have gained more impetus in biomedical and industrial applications than other metal nanoparticles.26,27
Furthermore, AgNps have been approved as a bactericidal agent for burn wound dressings (Acticoat™; Smith & Nephew, London, UK)28 by the US Food and Drug Administration. According to the latest information of Nanotechnology Consumer Products Inventory, >400 (nearly half of the total) registered “nano-enabled” consumer products claim to contain AgNps.27
Silver and nano silver have been utilized in many consumer products such as wound dressings, disinfectants, soaps, detergents, dental alloys, cosmetics, and many more products with high commercial potential.28 However, safer clinical applications of nano silver still require more clinical trials and research at the level of molecular biology to delineate the intracellular pathways involved before evaluating its potential host toxicity. The understanding of physiochemical interactions between the surfaces of nanomaterials and biomolecules like protein, lipids, biological fluids, and genetic material is most likely to shed light on the underlying mechanisms of its action.25
We have recently reported that AgNps exhibit both reactive oxygen species (ROS)-dependent and ROS-independent anti-Candida activity,29 indicating that some other antimicrobial mechanisms are active in addition to the already known ROS-mediated microbial toxicity of nanomaterials.30 However, a complete understanding of the cellular and molecular mechanisms of action of AgNps against fungal pathogens certainly merits investigation to devise better strategies for MDR reversal.29 A systematic analysis of the mechanisms involved in AgNp-induced fungal cell toxicity may lead to the identification of new drug targets or functional links directly related to MDR, pathogenicity, and virulence.
In the present study, we have investigated the cellular and molecular mechanisms associated with AgNp-mediated cell toxicity in the opportunistic fungal pathogen, C. albicans. Candida serves as the representative mode for the study of fungal pathogens31 and the results obtained from this study can be extended to other fungal pathogens of clinical relevance.
Materials and methods
Materials
Media chemicals (yeast extract, peptone, glucose, and agar) for culture of the microbial cells were procured from Hi-Media (Mumbai, India) and Fisher Scientific (Hampton, NH, USA). Analytical grade sodium borohydrate, silver nitrate, sodium citrate, EDTA, sorbitol, Tris-Cl, and MgSO4 obtained from Qualigens (Mumbai, India) were used for preparing buffers. All solvents including n-heptane, methanol, ethanol, and water were of high performance liquid chromatography grade, high purity, and procured from Merck Millipore (Burlington, MA, USA). Lyticase enzyme, pyrogallol salts, ascorbic acid (AA), solvents such as dimethyl sulfoxide and tetrahydrofuran, ergosterol standards, and fluorescent probes 1,6-diphenyl-1,3, 5-hexatriene (DPH) and 2,7-dichlorofluorescein diacetate (DCFH-DA) were obtained from Sigma-Aldrich Co. (St Louis, MO, USA).
Strains and growth media
Wild-type C. albicans SC5314 strain was used for this study; the strain description has been reported elsewhere.32 The fungal strain was maintained on liquid broth containing 2% (w/v) dextrose, 1% (w/v) yeast extract, and 2% (w/v) peptone (YEPD medium) at 30°C and for solid YEPD plates, 2.5% (w/v) agar was added to the YEPD media. Strains were preserved in 15% (v/v) glycerol at −80°C. Cells from frozen glycerol stocks were revived on YEPD plates at 30°C and maintained at 4°C. Liquid media cell cultures were grown at 30°C with continuous shaking at 140–150 rpm for 14–16 hours and the exponentially growing cells were used for all the experiments. All experiments were performed in triplicate and the average of three independent experiments along with the standard deviation was calculated for each assay. Student’s t-test and p-value were calculated using GraphPad Prism version 5.0 (GraphPad Software Inc, La Jolla, CA, USA) to show the significance and reproducibility of experiments. A p-value <0.05 was considered significant.
Synthesis of AgNps
AgNps were prepared by a two-step process – seed preparation and nanoparticles formation. Silver seed was prepared as described in an earlier publication33 and the seed solution was stirred well for 5 minutes and kept for aging for 2 hours. The 100 mL aqueous solution of 1 mM silver nitrate was heated to near boiling temperature, and then 3 mL of silver seed and sodium citrate were added in such a way that the final concentration of sodium citrate was 1 mM. The solution was heated till it turned greenish yellow in color and cooled to room temperature indicating the formation of AgNps. Particles were then separated by centrifugation at 40,000 rpm. The obtained particles were washed with deionized water thrice to remove impurities by centrifugation at 40,000 rpm and freeze dried for further experiments.
Characterization of AgNps
Optical properties of AgNp were recorded with a UV–visible spectrophotometer as described elsewhere (UV–VIS Spectrophotometer Lambda 35; PerkinElmer Inc, Waltham, MA, USA).16,34 Size and shape of AgNps were analyzed using a 200 kV transmission electron microscope (JEM 2100F; JEOL, Tokyo, Japan) and mean particle size was determined using Image Tool Software (multipoint image database software for grain and particle analysis; Dietermann & Heuser Solution GmbH, Greifenstein, Germany). Surface morphology of AgNps was analyzed by a 20 kV scanning electron microscope (AG-EVO®40; Carl Zeiss AG, Jena, Germany) under high vacuum and elemental composition by energy dispersive X-ray spectroscopy (EDS; E-sprit 1.8 X-ray microanalysis, Quantax 200; Bruker Nano GmbH, Berlin, Germany). The presence of elemental silver was further confirmed through an energy dispersive X-ray fluorescence (EDXRF) spectrometer equipped with a 600 W Gd anode X-ray tube, 100 kV generator (EDXRF Epsilon 5; PANalytical, Almelo, the Netherlands), and auto quantified AgNps were analyzed using Epsilon 5 wizard software. The crystalline structure of AgNps was determined using an X-ray powder diffractometer (PANalytical X’Pert PRO diffractometer) using Cu Kα radiation (λ=1.5418 Å) in the scan range of 30°–80° diffraction angle, with a step size of 0.016°. The results were analyzed using X’Pert data collector software (PANalytical).34 Stability of AgNp in aqueous solution was examined through zeta potential measurements as described elsewhere.35 The zeta potential of AgNp in aqueous solution was recorded by a Malvern Zetasizer Nano-ZS (Malvern Instruments Ltd., Malvern, UK).
Assessment of antifungal activity of AgNp
Broth microdilution and spot assays were used to determine the antifungal activity of AgNps.
Broth microdilution for determination of minimum inhibitory concentration (MIC)
MIC was determined by the broth microdilution method in accordance with the recommendations of the Clinical and Laboratory Standard Institute as described elsewhere.36,37 Briefly, the cells were grown for 14–16 hours on agar plates (exponentially growing log phase cells) and resuspended in 0.9% saline to give an optical density of 0.1 at 600 nm (OD600), which corresponds to cell number 0.5–1×106 cells/mL. Further, the cells were diluted 100-fold in YEPD medium to make the final concentration of cells 0.5–1×104 cells/mL. The cells were allowed to grow at 30°C with continuous shaking in the presence of various concentrations of AgNps. In this method, growth of the cells was evaluated both visually and by recording OD600 nm in a microplate reader. Readings were recorded by a UV–visible spectrometer at 600 nm after every 4-hour interval and compared with growth control (no AgNps).
Spot assay
In this assay, cells from YEPD agar plate grown overnight at 30°C were resuspended in 0.9% saline and OD600 of the suspension was adjusted to 0.1. Then, 5 μL of 5-fold serial dilutions of each yeast culture was spotted onto YEPD plates in the absence (growth control) and presence of various concentrations of AgNps followed by incubation of the plates for 48 hours at 30°C as described elsewhere.38 The plates were then imaged and growth differences were evaluated by comparing the growth of the cells with AgNp-free controls.
Intracellular ROS
Intracellular ROS levels of C. albicans were detected using a fluorogenic cell-permeant dye DCFH-DA as previously reported with slight modifications.39 This fluorescent probe measures hydroxyl, peroxyl, and other ROS activity within the cell. Cells were grown until the mid-exponential phase in the presence of AgNps (5 and 40 μg/mL), absence of AgNps (growth control), and presence of antioxidant, AA (5 mM/mL). Cells were then harvested by centrifugation at 5,000 rpm for 10 minutes at 4°C, followed by washing twice with PBS buffer (pH 7.4) to remove the media. Then, 107 cells were resuspended in 3 mL PBS (pH 8.4). The fluorescent probe DCFH-DA (final concentration 10 μM) was added to each cell suspension and incubated at 30°C for 1 hour. Relative fluorescence (Rf) intensity was measured in a PerkinElmer L55 spectrofluorimeter at respective excitation and emission wavelengths of 488 and 540 nm with slit widths of 5 and 10 nm, respectively. The blank (absence of fluorescent probe) was maintained separately to avoid autofluorescence. To confirm the presence of ROS, the images of the fluorescent cells were observed using a confocal microscope FluoView™ FV1000 (Olympus Corporation, Tokyo, Japan).
Antifungal activity of AgNp in the presence of antioxidant AA
Spot assay was performed as described above to assess the antifungal activity of different concentrations of AgNps in the presence of antioxidant AA (5 mM/mL). Two growth controls were maintained separately – one without AgNps and antioxidant and another without AgNps but with the antioxidant. The plates were imaged and growth differences were evaluated by comparing the growth of the cells with controls after incubation of the plates for 48 h at 30°C.
Assessing the physical state of the membrane
Fluorescence polarization measurements
Membrane fluidity was assessed through fluorescence polarization measurements using fluorescent probe DPH. A method described earlier39 was followed with slight modifications. AgNp-treated and untreated (no AgNps) cells were harvested and washed to remove the media contents. Spheroplasts were prepared by incubating the cells with lyticase (10 U for 107 cells/mL) at 30°C for 3–4 hours. The prepared spheroplasts were resuspended in a labeling buffer with pH 7.4 (0.6 M sorbitol, 10 mM MgSO4, and 20 mM Tris-Cl) and incubated with 2 μM DPH for 1 hour at 30°C. Fluorescence polarization (p) was measured using a PerkinElmer LS55 spectrofluorimeter (excitation 360 nm, emission 426 nm, and slit size 10 nm for both excitation and emission) as follows:40
where, IVV is the corrected fluorescence intensity obtained with excitation by vertically polarized light and emission detected by the analyzer oriented vertically to the direction of polarized excitation light, IVH is the corrected fluorescence intensity obtained with excitation by vertically polarized light and emission detected by the analyzer oriented horizontally to the direction of polarized excitation light. Grating factor (G), the correction for optical components of the instrument, is calculated as IHV/IHH where subscripts HV and HH indicate the corrected fluorescence intensity values obtained with horizontal–vertical and horizontal–horizontal orientations for the polarizer and analyzer in that order, respectively.
Time-resolved fluorescence spectroscopy
Fluorescence lifetime decays of DPH in nanosecond timescales were measured in Time-Correlated Single Photon Counting (TCSPC) setup (FL920; Edinburgh Instruments, Livingston, UK). AgNp-treated and untreated (no AgNps) cells were harvested and then washed to remove the media. Further, the spheroplasts were prepared and resuspended in labeling buffer followed by incubation with 2 μM DPH for 1 hour at 30°C. DPH-labeled cells were placed in a quartz cuvette for measuring the fluorescence decay of DPH. Samples were excited at 375 nm using a picosecond diode laser (pulse width ~100 ps). Fluorescence was dispersed in a monochromator and then collected by a multichannel plate in a photomultiplier tube detector. The time resolution of TCSPC setup of ~100 ps was determined by measuring the instrument response function using LUDOX solution. Fluorescence was emitted at 426 nm and the 100-ns time was split into 4,096 channels. Decays were measured at a magic angle for 5,000 peak counts. The obtained results were plotted using IGOR6 software (WaveMetrics, Inc., Portland, OR, USA)41,42 to obtain the decay lifetime for DPH in AgNp-treated and untreated cells.
Sterol quantitation
Sterols were extracted as described earlier43 with slight modifications from cells grown overnight at 30°C in the absence (control) and presence of AgNps. Cell pellets were resuspended in 2.5 mL methanol, 1.5 mL potassium hydroxide (60% [wt/v]), and 1 mL pyrogallol dissolved in methanol (0.5% [wt/v]). The suspension was kept for refluxing at 80°C for 2 hours and allowed to cool. The sterol was extracted with the help of n-heptane and extraction was repeated by adding 3 mL of n-heptane two to three times. The ergosterol content was calculated as a percentage of the wet weight of the cells using the following equations:39
where, F is the factor for dilution in hexane, A281.5 nm is the absorbance of both ergosterol and 24(28)-DHE at 281.5 nm, A230 nm is the absorbance of 24(28)-DHE at 230 nm, and 290 and 518 are the extinction coefficient values (in percent per centimeter) for ergosterol and 24(28)-DHE, respectively.
Fatty acid analysis
Total lipids were extracted from Candida cells with slight modifications of the method used by Bligh and Dyer.44 Briefly, the AgNp-treated and untreated (no AgNps) Candida cells were harvested at the exponential phase and resuspended in 10 mL methanol. The cell suspension was sonicated for 15 minutes to rupture the cells and total lipids were extracted using 2:1 chloroform:methanol. The lower organic layer was collected in a separate tube, dried under N2 flush, and stored at −20°C for further analysis. Fatty acids were then extracted by alcoholic saponification of the total lipids as described earlier.39 Extracted fatty acids were derivatized by O-Bis(trimethylsilyl)trifluoroacetamide with trimethyl-chlorosilane (Sigma-Aldrich Co) using injection port derivatization and analyzed by gas chromatography (GC)–mass spectrometry (MS) (Trace GC Ultra coupled with TSQ quantum; Thermo Fisher Scientific, Waltham, MA, USA). Fatty acids were separated using DB5-MS column (30 m ×0.2 mm, film thickness 0.25 μm). Helium was used as the carrier gas with a constant flow rate of 1.2 mL/min. Initial oven temperature was set at 80°C for 1 minute and then increased up to 320°C at 2°C/min and held for 20 min. An aliquot of 2 μL was injected into GC-MS for analysis. GC injector temperature was set at 300°C. The total ion mass spectra were recorded in the mass range of 20–500 (m/z). Each peak was identified based on the retention times of peaks obtained with that of standard fatty acids and by comparing the mass spectral data of peak with that of NIST mass spectral data available with the instrument (NIST, Gaithersburg, MD, USA).
Overall distribution of the monounsaturated and polyunsaturated fatty acids was determined by calculation of the unsaturation index (UI) using the following formula45 by Stubbs and Smith, which states,
where % monoenes, % dienes, and % trienes represent the percentages of monounsaturated, di-unsaturated, and tri-unsaturated fatty acids, respectively.
Surface morphology and ultrastructure of AgNp-treated Candida cells
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were used to study the alterations in surface morphology and cellular ultrastructure, respectively,46 after treatment with AgNps. Cells grown till the mid-exponential phase without (control) and with AgNp at the concentration of 40 μg/mL were harvested, washed with PBS buffer to remove media, and fixed with 2% glutaraldehyde in 0.1 M phosphate buffer for 3 hours at 25°C. After washing two to three times with phosphate buffer (pH 7.2), cells were postfixed with 1% OsO4 in 0.1 M phosphate buffer for 1 hour at 4°C for further study under SEM and TEM. For SEM, an aliquot of the above treated cells was dehydrated in acetone and dropped onto a round glass coverslip with hexamethyldisilazane and dried, followed by sputter coating with gold for observation under SEM (Carl Zeiss EVO®40 with resolution: 3.0 nm at 30 kV (SE and W) and magnification 7–1,000,000×). For TEM, samples were dehydrated with graded acetone, cleared with toluene, infiltrated with toluene and araldite mixture at 25°C, and finally put overnight in pure araldite at 50°C. The samples were then embedded in 1.5 mL Eppendorf tube with pure araldite mixture at 60°C for semi-thin and ultrathin section cutting with an ultramicrotome (Leica EM UC6 Ultramicrotome; Leica Microsystems, Wetzlar, Germany). Sections taken on 3.05 mm diameter and 200-mesh copper grid were stained with uranyl acetate and observed at 120 kV under TEM (model name JEOL 2100F with accelerating voltage 80–200 kV and magnification 50–1,500,000×).
Results and discussion
Physical characterization of AgNps
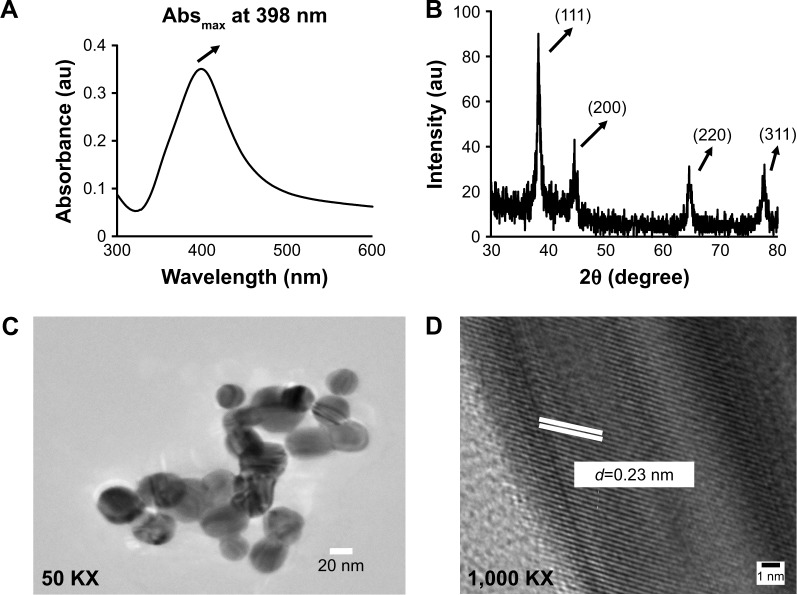
Characterization of the physicochemical properties (chemical composition, crystallinity, roughness/smoothness, size, shape, zeta potential, dispersion status, stability, etc) was carried out according to the International Organization for Standardization (ISO) guidelines ISO/TR 13014 and 13329, prior to nano–bio interactions studies.25,47 The formation of AgNps was confirmed by UV–visible absorption spectroscopy, where a strong single peak was observed at 398 nm (Figure 1A), related to the characteristic surface plasmon resonance (SPR) band of silver.48 A single SPR band was observed suggesting the formation of spherical AgNps and was in agreement with Mie’s theory.49 The synthesized AgNps were found to be stable for 3–4 months as revealed by absorption spectra taken at different time intervals.33 Furthermore, the X-ray diffraction (XRD) pattern was collected for AgNps to elucidate the crystalline nature of synthesized nanoparticles. XRD pattern (Figure 1B) showed distinct broad diffraction peaks at 2θ values of 38.1°, 44.5°, 64.6°, and 77.5° corresponding to (111), (200), (220), and (311) planes, respectively, suggesting the cubic phase (Joint Committee on Powder Diffraction Standards no 03-0921) of AgNps. The lattice spacing d111 for diffraction plane (111) is calculated to be 0.2355 nm, which is typically observed for crystalline AgNps.
Figure 1.
Characterization of optical and physical properties of AgNps synthesized by chemical reduction method. (A) Absorbance spectra for AgNps in the UV–visible spectrum range between 380 and 420 nm with an absorbance maxima at 398 nm. (B) X-ray diffraction spectrum of AgNps in the range 30°–80° showing four peaks at 2θ values of 38.1°, 44.5°, 64.6°, and 77.5° corresponding to (111), (200), (220), and (311) planes, respectively. (C) TEM image showing spherical AgNps. (D) High-resolution TEM image for AgNps showing lattice spacing (d=0.23 nm, d represents the distance between two lattice fringes).
Abbreviations: Abs, Absorbance maximum; AgNps, silver nanoparticles; KX, magnification; TEM, transmission electron microscopy.
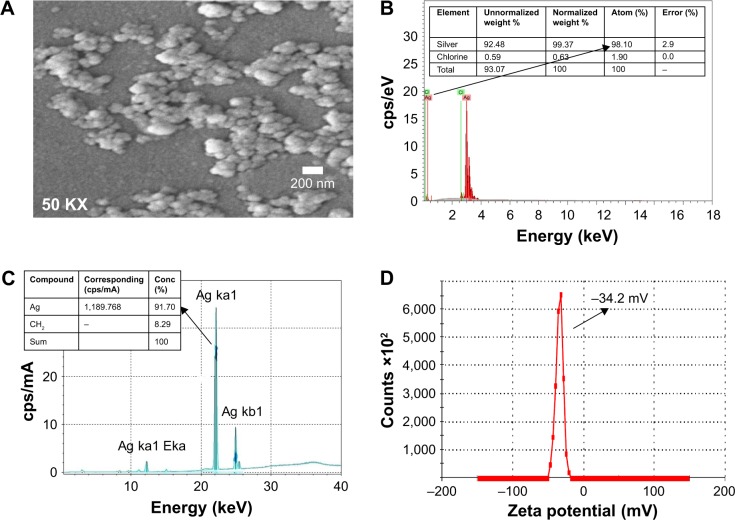
The TEM results (Figure 1C) revealed the synthesis of spherical AgNps in the size range of 10–30 nm. An interplanar distance “d” of 0.23 nm related to the (111) plane of silver is clearly visible in the high-resolution TEM image (Figure 1D, wherein d represents the distance between two lattice fringes) and corroborates the XRD results, thus confirming the formation of AgNps. The SEM images of AgNps also exhibited spherical morphology (Figure 2A) and the EDS spectrum shows the presence of a strong peak at 3 keV (Figure 2B), typical for crystalline AgNps. Throughout the EDS spectrum, no other peak belonging to any impurity was observed indicating the formation of impurity-free AgNps. EDXRF spectra also confirmed the presence of only silver (Figure 2C) suggesting that AgNps did not have any kind of contaminant. The zeta potential measurements displayed a strong peak at −34.2 mV (Figure 2D), confirming the stability of AgNps.
Figure 2.
Characterization of physicochemical properties of AgNps. (A) SEM image for AgNps. (B) SEM/EDS spectrum for AgNps with a strong peak at 3 keV. (C) EDXRF spectrum of AgNps showing the elemental composition of the prominent peak. (D) Zeta potential for AgNps.
Abbreviations: AgNps, silver nanoparticles; cps, counts per second; EDS, energy dispersive X-ray spectroscopy; EDXRF, energy dispersive X-ray fluorescence; KX, magnification; SEM, scanning electron microscopy.
Antifungal property of AgNps
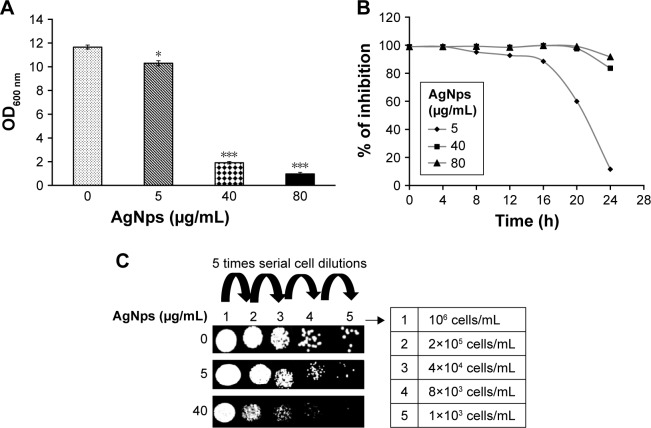
Silver and silver compounds are known for their effective use in the treatment of burns and chronic wounds because of their antimicrobial properties.16,50,51 The mechanism for fungicidal activity of AgNps is still debatable, though they are known to be detrimental to the fungal cells.30 AgNps prepared by different methods show varying extents of antifungal activity which has been attributed to their size, shape, and surface modification.2,30,52,53 In this study, antifungal activity of citrate-reduced AgNps was investigated in C. albicans, as a model of opportunistic common fungal pathogens by using broth microdilution and spot assays (Figure 3). Broth microdilution assay revealed that AgNps reduced the growth of Candida at lower concentration (5 μg/mL) whereas they almost completely inhibited their growth at 40 μg/mL (MIC90 for AgNps) (Figure 3A and B). Furthermore, the spot assay (Figure 3C) also displayed lower sensitivity of Candida cells toward AgNps at 5 μg/mL and enhanced sensitivity at 40 μg/mL, confirming the results of the broth microdilution. The results obtained for the antifungal activity of AgNps were interpreted by comparing the growth of the cells with the control (no AgNps). The high surface to volume ratio and presence of {111} facet in AgNps enhance their direct interaction with the fungal cells as compared to bulk silver, resulting in their improved antifungal efficacy.33,54 Delineation of the intracellular pathways involved and elucidation of the cellular and molecular mechanisms associated with the antifungal activity of AgNps remain indispensable to establish their safe medical adoption. We therefore systematically investigated the cellular and molecular mechanisms involved in the antifungal activity of AgNps in the subsequent sections.
Figure 3.
Antifungal activity of AgNps. (A) Determination of OD600 nm by broth microdilution method for Candida cells grown at 30°C in the absence (control) and presence of 5, 40, and 80 μg/mL AgNps, respectively. The values are mean ± SD of three independent sets of experiments. The * represents p<0.05 and *** represents p<0.001 with respect to control. (B) Dose-dependent percentage inhibition determined by broth microdilution method for Candida cells grown at 30°C in the presence of 5 μg/mL (♦), 40 μg/mL (■), and 80 μg/mL (▲) AgNp, respectively. (C) AgNp susceptibility profile determined by spot assay for Candida cells grown in the absence and presence of 5 and 40 μg/mL AgNp, respectively.
Abbreviations: AgNp, silver nanoparticle; OD, optical density.
ROS-mediated cell toxicity
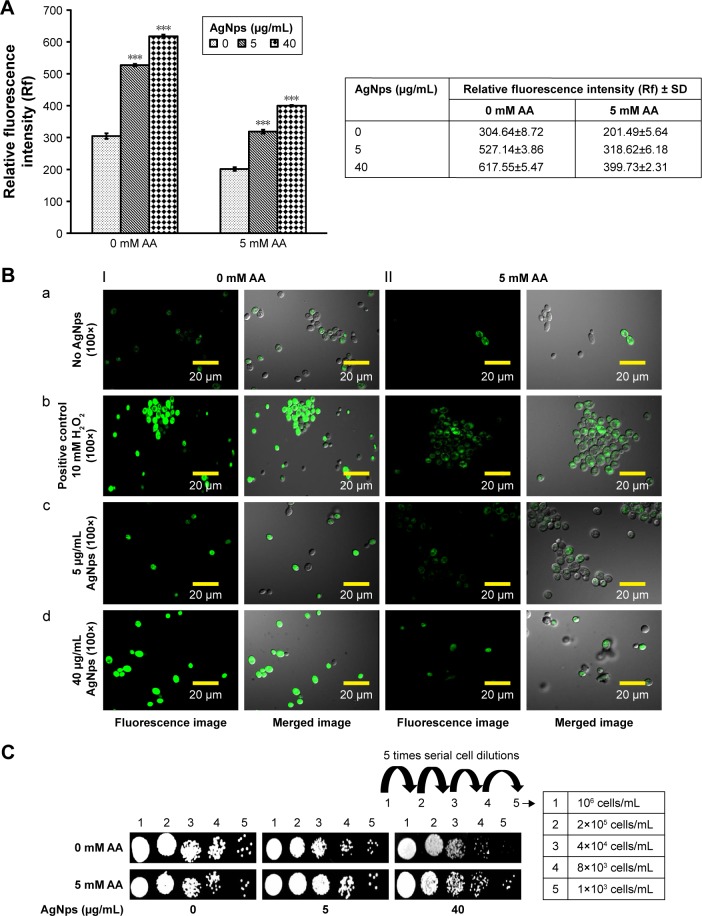
The action of several antifungals leads to augmentation of intracellular ROS, an important mediator for exerting antifungal effects.55 Also, the antifungal activity of nano silver is associated with induction of mitochondrial dysfunctional apoptosis through increase in oxidative stress via ROS generation especially hydroxyl radicals.30 In this study, we measured the endogenous ROS augmented by AgNps using DCFH-DA, a fluorogenic dye that measures hydroxyl, peroxyl, and other ROS activity within the cell. This cell-permeant dye diffuses into the cells and is deacetylated by cellular esterases to stable and colorless dichlorofluorescein, which subsequently gets oxidized by ROS to produce fluorescent 2,7-dichlorofluorescein. Here, we observed that compared to the basal ROS levels in control (untreated cells), AgNp-treated cells exhibited increased fluorescence within the cells implying enhanced intracellular ROS, which is depicted as Rf intensity in Figure 4A. This was further corroborated by the results obtained through confocal microscopy (Figure 4B; panels a, c, and d), wherein increased green fluorescence was observed due to elevated ROS levels within the AgNp-treated cells. The H2O2-treated cells were used as positive control (Figure 4B; panel b). Our results demonstrated a dose-dependent increase in endogenous ROS levels which were significantly elevated by around 73% in the presence of 5 μg/mL (subinhibitory concentration) AgNps and by almost more than double by 40 μg/mL (MIC90) AgNps.
Figure 4.
Intracellular ROS production in the Candida cells grown in the absence and presence of 5 μg/mL (subinhibitory) and 40 μg/mL (MIC90) AgNps estimated using the fluorescent dye DCFDA. (A) Quantitative measurement of endogenous ROS in AgNp-treated and untreated (control) Candida cells without addition of antioxidant and with addition of 5 mM antioxidant, AA. ROS levels are measured as Rf intensity and y-axis depicts the mean Rf values ± SD of three sets of independent experiments. Rf intensity was calculated by subtracting the Rf intensity for cells incubated without DCFDA (blank) from Rf intensity for cells incubated with DCFDA.55 *** represents p<0.001 calculated with respect to “no AgNps” (control). (B) Confocal images showing green fluorescence for endogenous ROS in Candida cells grown in the absence (B, panel a) and presence of 5 and 40 μg/mL AgNps (B, panels c and d), respectively, without antioxidant (I, left two panels) and with 5 mM AA (II, right two panels). Each left panel of both I and II depicts the fluorescence intensity (higher fluorescence indicates higher ROS production) measured by a confocal microscope and each right panel of both I and II shows the merge for the phase-contrast micrographs and fluorescence images at magnification 100×. The obtained results were compared with untreated cells (no AgNps; B, panel a) and positive control (cells treated with 10 mM hydrogen peroxide, H2O2; B, panel b). (C) AgNp susceptibility profiling of Candida cells grown in the absence and presence of 5 mM AA using spot assay. AgNps were used at respective concentrations of 5 μg/mL (subinhibitory) and 40 μg/mL AgNp (MIC90).
Abbreviations: AA, ascorbic acid; AgNps, silver nanoparticles; DCFDA, 2,7,dichlorofluorescein diacetate; MIC, minimum inhibitory concentration; Rf, relative fluorescence; ROS, reactive oxygen species.
Dose-dependent antifungal effects were exerted by AgNps which is evident from the spot assays in Figure 4C (upper panel) and it is possible that endogenous ROS could contribute to these AgNp-induced antifungal effects. The direct involvement of ROS in cell toxicity was investigated in the presence of natural antioxidant AA to elucidate the fungicidal effect of AgNps. It was found that without AgNp treatment, AA inhibited the basal ROS levels by around 33% (Figures 4A); in addition, ROS levels in AgNp-treated cells could also be restored to almost basal levels by 5 mM AA (Figure 4A and B). It may be noted that 5 mM AA itself did not exert any inhibitory effect on the growth of the cells (Figure 4C, lower panel). However, restoration of basal levels of endogenous ROS by antioxidant could not completely reverse the AgNp-induced sensitivity of treated fungal cells (Figure 4C, lower panel, extreme right) and restore their growth identical to that of the cells grown without AgNps (Figure 4C, upper left panel). The results thus implied that scavenging ROS through antioxidant could not provide complete protection against the killing action of AgNps and generation of intracellular ROS does not appear to be the sole major mechanism for AgNp-mediated Candida cell toxicity. It is envisioned that endogenous ROS acts either in parallel or in concerted action along with other cellular mechanisms paving the way for further experimental investigations to elucidate the underlying mechanisms for AgNp-mediated cell toxicity. It has been shown that AgNps exert antifungal activity through targeting cell membranes.56 Physiological changes have also been observed in AgNp-treated Candida cells showing dissipation of membrane potential, normal bud growth, and partly inhibited cell cycle.2 The physiological changes induced by AgNps on Candida were, therefore, observed through SEM and TEM investigations in the present study.
Cellular and intracellular imaging
SEM was used to examine the effect of 40 μg/mL (MIC90) AgNps on the surface morphology of C. albicans cells. The corresponding SEM micrographs showed a smooth outer surface (Figure 5A) in untreated cells whereas AgNp-treated cells showed distinct surface alterations and rough, wrinkled outer cell wall (Figure 5B). The observed surface morphological changes in the fungal cells may be due to the damage in cell wall and cell membrane.
Figure 5.
SEM micrographs (at 5 KX and 20 KX) for (A) untreated (no AgNps) Candida cells showing smooth surface of cell wall and (B) Candida cells treated with 40 μg/mL (MIC90) AgNps; the red arrows point at the rough outer cell wall after treatment with AgNps.
Abbreviations: AgNps, silver nanoparticles; KX, magnification; MIC, minimum inhibitory concentration; SEM, scanning electron microscopy.
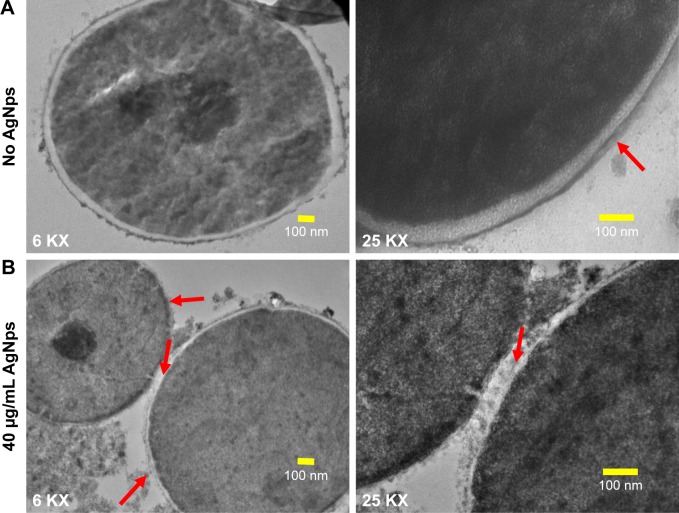
Few studies reported the detrimental effects of AgNps on Candida cells53,57–59 through disturbances in membrane potential,2 cellular ultrastructure,31,60 and triggering of apoptosis.30 We performed an ultrastructural analysis using TEM for AgNp-treated C. albicans cells. Candida cells in the absence of AgNp treatment (control) exhibited an unaltered cellular structure with well conserved and distinct cell wall and cell membrane (Figure 6A). On the other hand, Candida cells treated with 40 μg/mL (MIC90) AgNps displayed altered cellular ultrastructure with compromised cell wall and cell membrane integrity (Figure 6B), corroborating our SEM observations and in consistence with previous reports.31,60 The cell envelope (cell wall and cell membrane together) provides stability, rigidity, and resistance to physical stresses and plays a crucial role in pathogenicity and virulence of pathogenic fungi.61 It is possible that the effects exerted by AgNps on the cell wall and cell membrane might alter the physical state and, therefore, membrane fluidity.
Figure 6.
TEM images (at 6 KX and 25 KX) for Candida cells grown in the absence (no AgNps) and presence of AgNps. (A) Ultrastructure of cells grown without AgNps; cells without treatment revealed well-conserved ultrastructural features with a distinctive cell wall and cytoplasmic membrane and (B) cellular ultrastructure after treatment with 40 μg/mL (MIC90) AgNps; the red arrows indicate damage to the integrity of the cell wall and cytoplasmic membrane after treatment with AgNps.
Abbreviations: AgNps, silver nanoparticles; KX, magnification; MIC, minimum inhibitory concentration; TEM, transmission electron microscopy.
Membrane fluidity
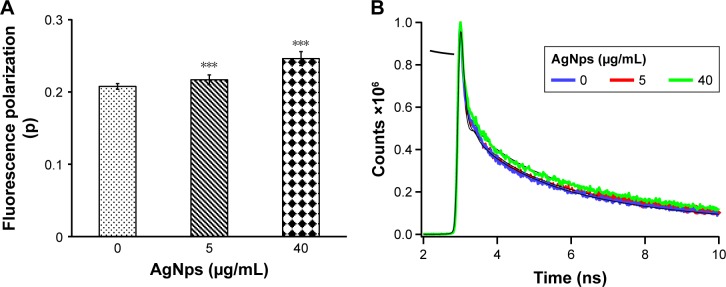
We have demonstrated an intricate relationship between membrane fluidity, lipid composition, and drug susceptibilities in C. albicans cells in our previous studies.7,36,38,39,62 Herein, the effect of AgNps on the physical state and membrane fluidity of Candida cells was characterized after treatment with 5 and 40 μg/mL AgNps. Fluorochrome, DPH was used to study the changes in the organizational dynamics of the hydrophobic interior of the membrane through steadystate fluorescence polarization (measured by “p”-values, Figure 7A) and time-resolved fluorescence measurements (Figure 7B; Table 1). Fluorescence being sensitive to the cellular physicochemical environment, the physical state of the membrane is reflected in the spectroscopic response of the fluorescence probes used.63 Fluorescence polarization is inversely proportional to membrane fluidity and an increase in the p-values implies an increase in membrane order and a reduction in membrane fluidity, typically due to a reduction in rotational mobility of the fluorophore. Candida cells treated with AgNps showed a dose-dependent reduction in membrane fluidity as evident from Figure 7A and it was observed that treatment with 5 μg/mL (subinhibitory concentration) AgNps and 40 μg/mL (MIC90) AgNps resulted in an increase in fluorescence polarization p-values by 4.3% (p<0.001) and 19% (p<0.001), respectively. The observed differences and reduced membrane fluidity may be due to the changes in lipid composition resulting in differences in the levels of compaction of cell membrane and altered lateral organization on the plane of the membrane.
Figure 7.
Fluorescence polarization “p” values and fluorescence decay curves for the fluorophore, DPH in Candida cells grown in the absence and presence of 5 μg/mL (subinhibitory) and 40 μg/mL (MIC90) AgNps. (A) Mean fluorescence polarization “p” values (fluorescence values being inversely proportional to membrane fluidity) ± SD of three sets of experiments shown on the y-axis. *** represents test of significance p<0.001 calculated with respect to no AgNps (control). (B) Normalized lifetime decay curves for DPH showing time-dependent fluorescence decay intensity (with fits) in linear scale with respect to time in nanoseconds for cells grown in the absence (control indicated by blue curve) and presence of 5 μg/mL (shown by red curve) and 40 μg/mL (depicted by green curve) AgNps. The lifetime decay time for DPH in nanoseconds is given in Table 1.
Abbreviations: AgNps, silver nanoparticles; DPH, 1,6-diphenyl-1,3,5-hexatriene; MIC, minimum inhibitory concentration.
Table 1.
Lifetime decay for DPH in nanoseconds in Candida cells grown in the absence (control) and presence of AgNps
| Sample description | Lifetime decay (ns) mean ± SD |
|---|---|
| No AgNps | 0.93±0.12 |
| 5 μg/mL AgNps | 1.11±0.21 |
| 40 μg/mL AgNps | 1.19±0.04 |
Abbreviations: AgNps, silver nanoparticles; DPH, 1,6-diphenyl-1,3,5-hexatriene; SD, standard deviation.
The polarity changes in local environment of the DPH are reliably indicated by the fluorescence lifetime of DPH. The mean fluorescent lifetimes as indicated in Table 1 were calculated by fitting tri-exponential fluorescent decay curves.41 The data revealed that there is a statistically significant, AgNp dose-dependent increase in DPH lifetime in Candida cells treated with AgNps. DPH decay time of 0.93 ns in untreated cells increased to decay time of 1.11 and 1.19 ns in the presence of 5 and 40 μg/mL (MIC90) AgNps, respectively. Fluorescence decay patterns of DPH represented the changes in cellular microenvironment/micro-viscosity due to AgNp treatment (Figure 7B). Thus, the observed longer decay time for DPH in AgNp-treated cells corroborated with their higher fluorescence polarization p-values implying a decrease in membrane fluidity due to AgNp treatment. Since, cell envelope functions as a barrier for the entry and exit of biomolecules across the cells, therefore, the reduced fluidity of the membrane may be associated with a specific secondary adaptation in response to AgNp treatment or linked to changes in lipid composition and/or membrane functions.
Membrane ergosterol levels and fatty acids
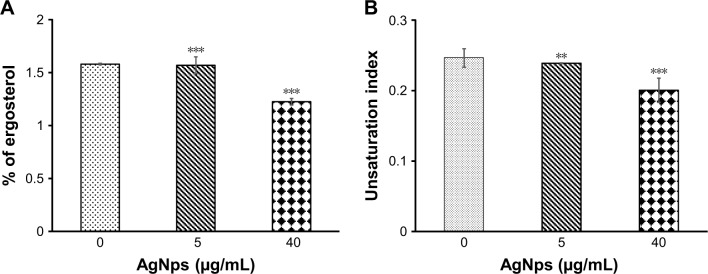
Lipids are essential components of membranes and perform a variety of functions that modulate fluidity and permeability. Among various classes of fungal lipids, membrane ergosterol is one of the most important constituents of fungal cell membrane.38 Ergosterol plays a vital role in imparting stability to the cell and loss of sterol leads to destabilization of membrane resulting in enhanced permeability and thus increase in sensitivity.38,62 Ergosterol also acquires importance because of the fact that the ergosterol biosynthetic pathway is the cellular target for most of the common antifungals (azoles, polyenes, and allylamines). Upregulation of sterol-rich microdomains has also been reported in drug-resistant cancer cell lines.64,65 In this study, AgNps were found to affect cell membrane integrity and the physical state of the membrane of Candida cells. Hence, it was pertinent to explore the effect of AgNps on membrane lipid composition of the cells, which might be responsible for the altered physical state of the membrane. We checked the ergosterol contents of the AgNp-treated cells in order to examine the effect of AgNps on the lipid composition. Although there was a reduction in ergosterol content by only 1.3% (p<0.001) in the presence of 5 μg/mL (subinhibitory) AgNps, there was a marked reduction by 22.8% (p<0.001) in the fungal cells treated with 40 μg/mL (MIC90) AgNps (Figure 8A). Ergosterol, an important constituent of the membrane microdomains, plays a critical role in cell physiology and cellular processes including cell to cell signaling, protein secretion, translocation of small molecules, and host pathogen interactions. Depletion of ergosterol after AgNp treatment might be responsible for the observed sensitivity of fungal cells, the compromised membrane integrity, and changes in cellular microenvironment.
Figure 8.
Ergosterol contents and UI for fatty acids of Candida cells grown in the absence (control) and presence of 5 μg/mL (subinhibitory) and 40 μg/mL (MIC90) AgNps. (A) Relative percentages of ergosterol contents in control and AgNp-treated cells calculated as mentioned in the “Material and methods” section; values on the y-axis are mean (the mean ergosterol content of cells is expressed as a percentage of the wet weight of the cells) ± SD of three independent sets of experiments. *** represents p<0.001 calculated with respect to no AgNps (control). (B) The degree of fatty acid unsaturation of the Candida cells grown in the absence (control) and presence of AgNps was calculated from the data in Table 2 and expressed as UI as described in the “Materials and methods” section. UI for unsaturated fatty acids represent mean values ± SD of three independent sets of experiments. ** represents p<0.01 and *** represents p<0.001, respectively, calculated with respect to no AgNps (control).
Abbreviations: AgNps, silver nanoparticles; MIC, minimum inhibitory concentration; SD, standard deviation; UI, unsaturation indices.
Normally, the cells are expected to cope up with the modulations in ergosterol levels by compensatory alterations in other membrane lipids to maintain optimal membrane fluidity, and this explains why the observed reduction in ergosterol levels in this study did not result in an increase in membrane fluidity. Saturated and unsaturated fatty acids are also critical determinants of membrane fluidity. The status of fatty acids in AgNp-treated cells was assessed using GC-MS and our results showed a significant increase in the percentage of saturated fatty acids and a simultaneous decrease in unsaturated fatty acids in AgNp-treated Candida cells (Table 2). The unsaturated fatty acids in fungal cells mainly are palmitoleic acid (16:1), oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3). The unsaturated fatty acids present in the cells help to determine the UI. In this study, it was observed that the UI decreased by only 3.2% (p<0.01) when cells were grown in the presence of 5 μg/mL (subinhibitory) AgNps but showed a marked reduction by 20% (p<0.001) in the presence of 40 μg/mL (MIC90) AgNps (Figure 8B). Therefore, it appears that the increase in saturated fatty acids and simultaneous decrease in unsaturated fatty acids compensated for the reduced ergosterol levels, resulting in a net decrease in membrane fluidity after AgNp treatment.
Table 2.
Relative percentages of fatty acids and values for UI in Candida cells (grown in the absence and presence of AgNps)
| Fatty acids | No AgNps (area %) | 5 µg/mL AgNps (area %) | 40 µg/mL AgNps (area %) |
|---|---|---|---|
| C10:0 | 0.34±0.1 | 0.34±0.02 | 0.43±0.13 |
| C12:0 | 0.705±0.30 | 0.46±0.03 | 0.58±0.05 |
| C14:0 | 14.25±2.80 | 10.75±0.89 | 9.87±1.43 |
| C16:0 | 9.97±0.36 | 11.9±1.01 | 12.19±0.26 |
| C16:1 | 5.3±0.16 | 7.5±0.28 | 8.09±0.34 |
| C17:0 | 0.28±0.04 | 0.6±0.14 | 0.54±0.19 |
| C18:0 | 1.67±0.06 | 1.81±0.02 | 2.36±0.21 |
| C18:1 | 11.5±0.49 | 9.47±0.59 | 5.94±0.77 |
| C18:2 | 2.92±0.91 | 2.28±0.15 | 1.91±0.82 |
| C18:3 | 0.7±0.04 | 0.79±0.02 | 0.74±0.12 |
| UI | 0.25±0.01 | 0.24±0.0006** | 0.20±0.01*** |
Notes: Values are mean ± SD of three independent sets of experiments.
represents p<0.01 and
represents p<0.001, calculated with respect to “no AgNps.”
Abbreviations: AgNps, silver nanoparticles; SD, standard deviation; UI, unsaturation index.
Close observation of the individual fatty acids revealed that among the unsaturated fatty acids, it was mainly the levels of C18:1 and C18:2 which were lowered after AgNp treatment. Interestingly, we found a significant simultaneous reduction in 18:1 unsaturated fatty acid (oleic acid) by 17.7% (p<0.01) and 48.3% (p<0.001) and in unsaturated fatty acid C18:2 by 21.9% (p<0.01) and 34.6% (p<0.001) in the presence of 5 and 40 μg/mL AgNps, respectively (Table 2). A direct functional consequence of the marked decrease in these unsaturated fatty acids of the membrane may be responsible for the observed reduction in membrane fluidity (Figure 7) in AgNp-treated cells. It is expected that the simultaneous decrease in unsaturated fatty acids is compensated by a small relative increase in unsaturated fatty acid C18:3 by 12.9% (p<0.01) and 6% (p<0.001) in cells treated with 5 and 40 μg/mL AgNps, respectively (Table 2). The observed decrease in levels of short chain C14:0 saturated fatty acid also reflects a compensatory reaction of cells due to low amounts of unsaturated fatty acids. The degree of fatty acid unsaturation is shown to be related to altered defense signaling pathways in plants.66
The ratio of saturated and unsaturated fatty acid is a key determinant of temperature sensing in Candida which is critical for pathogenesis and virulence.67 It has been shown that membrane fluidity and temperature sensing are interdependent in C. albicans via oleic acid.68 Oleic acid levels are important for hyphal morphogenesis,68 a striking feature of Candida which is defined as the transition from budding yeast to filamentous forms (hyphae and pseudo-hyphae). Hyphae formation is pivotal for biofilm development and causes systemic mycosis in immunocompromised patients. Candida biofilms are composed of a mixture of yeast cells and hyphal elements.69–71 This yeast to hypha transition contributes to the overall virulence of Candida cells and may constitute a significant target for the development of antifungal drugs.71 Impairing morphogenesis by targeting fatty acid may constitute a strategy to treat Candida infections. Small molecule inhibitors of fatty acid biosynthetic pathways in bacteria are already in clinical use for treatment as antibacterial agents, for example, tuberculosis.72 Fungal fatty acid biosynthesis is still emerging as an attractive target for the development of antifungal agents. This study reveals that fatty acids in Candida cells appear to be a potential cellular target of AgNps.
Conclusion
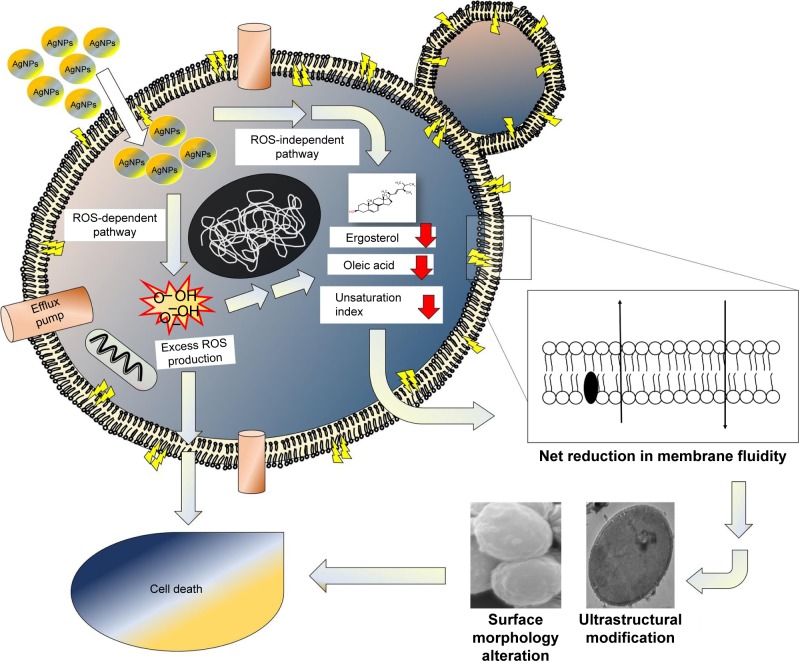
We have demonstrated that AgNps affect drug susceptibilities significantly in C. albicans through multiple cellular targets leading to altered membrane microenvironment, physical state of the membrane, lipid composition, cellular ultrastructure, and surface morphology (Figure 9). The observed AgNp-induced alteration in cellular targets can potentially affect the drug resistance, pathogenesis, and virulence of fungal pathogens.73 Among cellular targets of AgNps, fungal fatty acids, especially oleic acid, appear to be quite significant; therefore, targeting morphogenesis and virulence through fatty acids might devise a strategy for the treatment of Candida infections. Nano silver-based drug combinations could, thus, be ideal candidates to address the challenges of MDR and fungal therapeutics by favoring broad-spectrum activity, multiple cellular targets, and minimal host toxicity. Our results could be correlated and translated to nano silver-based drug therapy for other pathogenic fungi of medical importance.
Figure 9.
Schematic representation for the proposed model for mechanism of action of AgNps against Candida cells, depicting possible cellular targets and existence of ROS-dependent and ROS-independent pathways for fungicidal action of AgNps.
Abbreviations: AgNps, silver nanoparticles; ROS, reactive oxygen species.
Acknowledgments
We gratefully acknowledge the GC-MS facility of Council for Scientific and Industrial Research-Indian Institute of Toxicology Research (CSIR-IITR), Lucknow. This work has been supported by grants to TP from Indian funding agencies: Department of Biotechnology (DBT) (BT/PR5110/MED/29/497/2012 and BT/BI/12/045/2008), CSIR (OSDD/HCP001/11FYP/2010-11/134), University Grants Commission (UGC) (Jawaharlal Nehru University [JNU]-UPOEII), and Department of Science and Technology (DST) (JNU-DST-PURSE). We gratefully acknowledge JNU for providing infrastructural support, and DBT, India for the award of Senior Research Fellowship to VSR.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Goffeau A. Drug resistance: the fight against fungi. Nature. 2008;452(7187):541–542. doi: 10.1038/452541a. [DOI] [PubMed] [Google Scholar]
- 2.Kim KJ, Sung WS, Suh BK, et al. Antifungal activity and mode of action of silver nano-particles on Candida albicans. Biometals. 2009;22(2):235–242. doi: 10.1007/s10534-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 3.Kanafani ZA, Perfect JR. Resistance to antifungal agents: mechanisms and clinical impact. Clin Infect Dis. 2008;46(1):120–128. doi: 10.1086/524071. [DOI] [PubMed] [Google Scholar]
- 4.Sardi JCO, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJS. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol. 2013;62(1):10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 5.Orasch C, Marchetti O, Garbino J, et al. Candida species distribution and antifungal susceptibility testing according to European Committee on Antimicrobial Susceptibility Testing and new vs. old Clinical and Laboratory Standards Institute clinical breakpoints: a 6-year prospective candidaemia survey from the fungal infection network of Switzerland. Clin Microbiol Infect. 2014;20(7):698–705. doi: 10.1111/1469-0691.12440. [DOI] [PubMed] [Google Scholar]
- 6.Perlin DS. Echinocandin resistance in Candida. Clin Infect Dis. 2015;61(Suppl 6):S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad T, Sethumadhavan S, Fatima Z. Altered ergosterol biosynthetic pathway – an alternate multidrug resistance mechanism independent of drug efflux pump in human pathogenic fungi C. albicans. In: Méndez-Vilas A, editor. Science Against Microbial Pathogens: Communicating Current Research and Technological Advances. Spain: Formatex Research Center; 2011. pp. 757–768. [Google Scholar]
- 8.Hirakawa MP, Martinez DA, Sakthikumar S, et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 2015;25(3):413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlin DS, Shor E, Zhao Y. Update on antifungal drug resistance. Curr Clin Microbiol Reports. 2015;2(2):84–95. doi: 10.1007/s40588-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336(6082):647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 11.Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine and voriconazole against Candida spp. J Clin Microbiol. 2007;45(11):3522–3528. doi: 10.1128/JCM.00403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Hou Y, Chen X, Gao Y, Li H, Sun S. Combination of fluconazole with non-antifungal agents: a promising approach to cope with resistant Candida albicans infections and insight into new antifungal agent discovery. Int J Antimicrob Agents. 2014;43(5):395–402. doi: 10.1016/j.ijantimicag.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Kumamoto CA, Pierce JV. Immunosensing during colonization by Candida albicans: does it take a village to colonize the intestine? Trends Microbiol. 2011;19(6):263–267. doi: 10.1016/j.tim.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 2012;36(2):288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Wagh P, Wadhwani S, et al. Synthesis, optimization, and characterization of silver nanoparticles from Acinetobacter calcoaceticus and their enhanced antibacterial activity when combined with antibiotics. Int J Nanomedicine. 2013;8:4277–4290. doi: 10.2147/IJN.S48913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsueh Y, Lin K, Ke W, et al. The antimicrobial properties of silver nanoparticles in Bacillus subtilis are mediated by released Ag+ ions. PLoS One. 2015;10(12):1–17. doi: 10.1371/journal.pone.0144306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salem W, Leitner DR, Zingl FG, et al. Antibacterial activity of silver and zinc nanoparticles against Vibrio cholerae and enterotoxic Escherichia coli. Int J Med Microbiol. 2015;305(1):85–95. doi: 10.1016/j.ijmm.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohamed MM, Fouad SA, Elshoky HA, Mohammed GM, Salaheldin TA. Antibacterial effect of gold nanoparticles against Corynebacterium pseudotuberculosis. Int J Vet Sci Med. 2017;5(1):23–29. doi: 10.1016/j.ijvsm.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sirelkhatim A, Mahmud S, Seeni A, et al. Review on zinc oxide nano-particles: antibacterial activity and toxicity mechanism. Nano-Micro Lett. 2015;7(3):219–242. doi: 10.1007/s40820-015-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arakha M, Pal S, Samantarrai D, et al. Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle–bacteria interface. Sci Rep. 2015;5(14813):1–12. doi: 10.1038/srep14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubacka A, Suarez DM, Rojo D, et al. Understanding the antimicrobial mechanism of TiO2 based nanocomposite films in a pathogenic bacterium. Sci Rep. 2014;4(4134):1–9. doi: 10.1038/srep04134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, Gao Y, Sun Z, Chang D, Gao G, Dong A. Synthesis, characterization, and bactericidal evaluation of chitosan/guanidine functionalized graphene oxide composites. Molecules. 2017;22(12):1–15. doi: 10.3390/molecules22010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konwar A, Kalita S, Kotoky J, Chowdhury D. Chitosan-iron oxide coated graphene oxide nanocomposite hydrogel: a robust and soft antimicrobial biofilm. ACS Appl Mater Interfaces. 2016;8(32):20625–20634. doi: 10.1021/acsami.6b07510. [DOI] [PubMed] [Google Scholar]
- 25.Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano–bio interface. Nat Mater. 2009;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 26.Ge L, Li Q, Wang M, Ouyang J, Li X, Xing MMQ. Nanosilver particles in medical applications: synthesis, performance, and toxicity. Int J Nanomedicine. 2014;9(1):2399–2407. doi: 10.2147/IJN.S55015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mao BH, Tsai JC, Chen CW, Yan SJ, Wang YJ. Mechanisms of silver nanoparticle-induced toxicity and important role of autophagy. Nanotoxicology. 2016;10(8):1021–1040. doi: 10.1080/17435390.2016.1189614. [DOI] [PubMed] [Google Scholar]
- 28.Melaiye A, Youngs WJ. Silver and its application as an antimicrobial agent. Expert Opin Ther Pat. 2005;15(2):125–130. [Google Scholar]
- 29.Radhakrishnan VS, Dwivedi SP, Siddiqui MH, Prasad T. In vitro studies on oxidative stress-independent, Ag nanoparticles-induced cell toxicity of Candida albicans, an opportunistic pathogen. Int J Nanomedicine. 2018;13:91–96. doi: 10.2147/IJN.S125010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang IS, Lee J, Hwang JH, Kim KJ, Lee DG. Silver nanoparticles induce apoptotic cell death in Candida albicans through the increase of hydroxyl radicals. FEBS J. 2012;279(7):1327–1338. doi: 10.1111/j.1742-4658.2012.08527.x. [DOI] [PubMed] [Google Scholar]
- 31.Vazquez-Muñoz R, Avalos-Borja M, Castro-Longoria E. Ultrastructural analysis of Candida albicans when exposed to silver nanoparticles. PLoS One. 2014;9(10):1–10. doi: 10.1371/journal.pone.0108876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134(3):717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal S, Tak YK, Song JM. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the Gram-negative bacterium Escherichia coli. Appl Environ Microbiol. 2007;73(6):1712–1720. doi: 10.1128/AEM.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalimuthu K, Suresh Babu R, Venkataraman D, Bilal M, Gurunathan S. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf B Biointerfaces. 2008;65(1):150–153. doi: 10.1016/j.colsurfb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 35.Murdock RC, Braydich-Stolle L, Schrand AM, Schlager JJ, Hussain SM. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol Sci. 2008;101(2):239–253. doi: 10.1093/toxsci/kfm240. [DOI] [PubMed] [Google Scholar]
- 36.Prasad T, Chandra A, Mukhopadhyay CK, Prasad R. Unexpected link between iron and drug resistance of Candida spp: iron depletion enhances membrane fluidity and drug diffusion, leading to drug-susceptible cells. Antimicrob Agents Chemother. 2006;50(11):3597–3606. doi: 10.1128/AAC.00653-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rex JH, Alexander BD, Andes D, et al. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard – Third Edition. Vol. 28. Pennsylvania, USA: Clinical and Laboratory Standards Institute (CLSI); 2008. [Google Scholar]
- 38.Mukhopadhyay K, Prasad T, Saini P, Puoadyil TJ, Chattopadhyay A, Prasad R. Membrane sphingolipid–ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob Agents Chemother. 2004;48(5):1778–1787. doi: 10.1128/AAC.48.5.1778-1787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad T, Hameed S, Manoharlal R, et al. Morphogenic regulator EFG1 affects the drug susceptibilities of pathogenic Candida albicans. FEMS Yeast Res. 2010;10(5):587–596. doi: 10.1111/j.1567-1364.2010.00639.x. [DOI] [PubMed] [Google Scholar]
- 40.Shinitzky M, Barenholz Y. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim Biophys Acta. 1978;515(4):367–394. doi: 10.1016/0304-4157(78)90010-2. [DOI] [PubMed] [Google Scholar]
- 41.Verma SD, Pal N, Singh MK, Sen S. Probe position-dependent counterion dynamics in DNA: comparison of time-resolved Stokes shift of groove-bound to base-stacked probes in the presence of different monovalent counterions. J Phys Chem Lett. 2012;3(18):2621–2626. doi: 10.1021/jz300934x. [DOI] [PubMed] [Google Scholar]
- 42.Abe F, Hiraki T. Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim Biophys Acta. 2009;1788(3):743–752. doi: 10.1016/j.bbamem.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Martel CM, Parker JE, Bader O, et al. A clinical isolate of Candida albicans with mutations in ERG11 (encoding sterol 14-demethylase) and ERG5 (encoding C22 desaturase) is cross resistant to azoles and amphotericin B. Antimicrob Agents Chemother. 2010;54(9):3578–3583. doi: 10.1128/AAC.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 45.Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim Biophys Acta. 1984;779(1):89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 46.Walker LA, Munro CA, De Bruijn I, Lenardon MD, McKinnon A, Gow NAR. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4(4):e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng LC, Jiang X, Wang J, Chen C, Liu RS. Nano-bio effects: interaction of nanomaterials with cells. Nanoscale. 2013;5(9):3547–3569. doi: 10.1039/c3nr34276j. [DOI] [PubMed] [Google Scholar]
- 48.Link S, El-Sayed MA. Optical properties and ultrafast dynamics of metallic nanoparticles. Annu Rev Phys Chem. 2003;54(1):331–366. doi: 10.1146/annurev.physchem.54.011002.103759. [DOI] [PubMed] [Google Scholar]
- 49.Mie G. Contributions to the optics of turbid media, especially colloidal metal solutions. Ann Phys. 1908;330(3):377–445. [Google Scholar]
- 50.Franci G, Falanga A, Galdiero S, et al. Silver nanoparticles as potential antibacterial agents. Molecules. 2015;20(5):8856–8874. doi: 10.3390/molecules20058856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You C, Han C, Wang X, et al. The progress of silver nanoparticles in the antibacterial mechanism, clinical application and cytotoxicity. Mol Biol Rep. 2012;39(9):9193–9201. doi: 10.1007/s11033-012-1792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh P, Kim YJ, Singh H, et al. Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles. Int J Nanomedicine. 2015;10:2567–2577. doi: 10.2147/IJN.S72313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panáček A, Kolář M, Večeřová R, et al. Antifungal activity of silver nanoparticles against Candida spp. Biomaterials. 2009;30(31):6333–6340. doi: 10.1016/j.biomaterials.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 54.Morones JR, Elechiguerra JL, Camacho A, et al. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16(10):2346–2353. doi: 10.1088/0957-4484/16/10/059. [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi D, Kondo K, Uehara N, et al. Endogenous reactive oxygen species is an important mediator of miconazole antifungal effect. Antimicrob Agents Chemother. 2002;46(10):3113–3117. doi: 10.1128/AAC.46.10.3113-3117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swathy JR, Sankar MU, Chaudhary A, Aigal S, Anshup, Pradeep T. Antimicrobial silver: an unprecedented anion effect. Sci Rep. 2014;4:7161. doi: 10.1038/srep07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selvaraj M, Pandurangan P, Ramasami N, Rajendran SB, Sangilimuthu SN, Perumal P. Highly potential antifungal activity of quantum-sized silver nanoparticles against Candida albicans. Appl Biochem Biotechnol. 2014;173(1):55–66. doi: 10.1007/s12010-014-0782-9. [DOI] [PubMed] [Google Scholar]
- 58.Wady AF, Machado AL, Zucolotto V, Zamperini CA, Berni E, Vergani CE. Evaluation of Candida albicans adhesion and biofilm formation on a denture base acrylic resin containing silver nanoparticles. J Appl Microbiol. 2012;112(6):1163–1172. doi: 10.1111/j.1365-2672.2012.05293.x. [DOI] [PubMed] [Google Scholar]
- 59.Monteiro DR, Takamiya AS, Feresin LP, et al. Susceptibility of Candida albicans and Candida glabrata biofilms to silver nanoparticles in intermediate and mature development phases. J Prosthodont Res. 2015;59(1):42–48. doi: 10.1016/j.jpor.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Lara HH, Romero-Urbina DG, Pierce C, Lopez-Ribot JL, Arellano-Jiménez MJ, Jose-Yacaman M. Effect of silver nanoparticles on Candida albicans biofilms: an ultrastructural study. J Nanobiotechnology. 2015;13(91):1–12. doi: 10.1186/s12951-015-0147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaffin WL, López-Ribot JL, Casanova M, Gozalbo D, Martínez JP. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev. 1998;62(1):130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prasad T, Saini P, Gaur NA, Vishwakarma RA, Khan LA, Haq QMR. Functional analysis of CaIPT1, a sphingolipid biosynthetic gene involved in multidrug resistance and morphogenesis of Candida albicans. Antimicrob Agents Chemother. 2005;49(8):3442–3452. doi: 10.1128/AAC.49.8.3442-3452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar A, Radhakrishnan VS, Singh R, Kumar M, Mishra NN, Prasad T. A clinical resistant isolate of opportunistic fungal pathogen, Candida albicans revealed more rigid membrane than its isogenic sensitive isolate. In: Mendiz-Vilas A, editor. Multidisciplinary Approaches for Studying and Combating Microbial Pathogens. Florida, USA: Brown Walker Press; 2015. pp. 1–5. [Google Scholar]
- 64.Lavie Y, Fiucci G, Liscovitch M. Up-regulation of caveolae and caveolar constituents in multidrug-resistant cancer cells. J Biol Chem. 1998;273(49):32380–32383. doi: 10.1074/jbc.273.49.32380. [DOI] [PubMed] [Google Scholar]
- 65.Lavie Y, Liscovitch M. Changes in lipid and protein constituents of rafts and caveolae in multidrug resistant cancer cells and their functional consequences. Glycoconj J. 2000;17(3–4):253–259. doi: 10.1023/a:1026553626537. [DOI] [PubMed] [Google Scholar]
- 66.Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc Natl Acad Sci U S A. 2001;98(16):9448–9453. doi: 10.1073/pnas.151258398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leach MD, Cowen LE. Membrane fluidity and temperature sensing are coupled via circuitry comprised of Ole1, Rsp5, and Hsf1 in Candida albicans. Eukaryot Cell. 2014;13(8):1077–1084. doi: 10.1128/EC.00138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krishnamurthy S, Plaine A, Albert J, Prasad T, Prasad R, Ernst JF. Dosage-dependent functions of fatty acid desaturase Ole1p in growth and morphogenesis of Candida albicans. Microbiology. 2004;150(Pt 6):1991–2003. doi: 10.1099/mic.0.27029-0. [DOI] [PubMed] [Google Scholar]
- 69.Douglas L. Candida biofilms and their role in infection. Trends Microbiol. 2003;11(1):30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 70.Ramage G, VandeWalle K, Lopez-Ribot J, Wickes B. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214(1):95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 71.Shareck J, Belhumeur P. Modulation of morphogenesis in Candida albicans by various small molecules. Eukaryot Cell. 2011;10(8):1004–1012. doi: 10.1128/EC.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heath RJ, White SW, Rock CO. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl Microbiol Biotechnol. 2002;58(6):695–703. doi: 10.1007/s00253-001-0918-z. [DOI] [PubMed] [Google Scholar]
- 73.Maseet M, Khan N, Basir SF. Ergosterol biosynthesis and pathogenicity markers inhibition of Candida albicans by fungus mediated silver nanoparticles. World J Pharm Pharm Sci. 2017;6(2):600–618. [Google Scholar]