Abstract
Background
Renal cell carcinoma (RCC) is the most common cancer in kidney malignancies. UCA1 has been identified as an oncogenic lncRNA in multiple cancers, including RCC. However, the underlying molecular mechanism of UCA1 involved in RCC progression is far from being addressed.
Methods
Reverse-transcription quantitative polymerase chain reaction (RT-qPCR) assays were used to measure expressions of UCA1, miR129, and SOX4 mRNA. Western blot assays were employed to detect SOX4 protein expression. Cell proliferation, invasion, and apoptosis were assessed by CCK-8, Matrigel invasion, and annexin–fluorescein isothiocyanate (FITC) apoptosis-detection assays, respectively. The interaction between UCA1 and miR129 was demonstrated by luciferase, RNA pull-down, and RNA-immunoprecipitation (RIP) assays. Luciferase assays were also used to explore whether UCA1 was able to act as a molecular sponge of miR129 to affect the interplay of miR129 and SOX4.
Results
UCA1 expression was upregulated in RCC tissue and cells, and higher UCA1 expression was associated with advanced pathogenic status and poor prognosis of RCC patients. UCA1 knockdown suppressed proliferation and invasion and induced apoptosis in RCC cells. UCA1 inhibited miR129 expression by direct interaction in RCC cells. miR129 overexpression inhibited cell proliferation and invasion and promoted apoptosis. Moreover, miR129 downregulation abrogated UCA1 knockdown-mediated antiproliferation, anti-invasion, and proapoptosis effects in RCC cells. Furthermore, UCA1 acted as a ceRNA of miR129 to enhance target-gene SOX4 expression in RCC cells.
Conclusion
UCA1 promoted cell proliferation and invasion and inhibited apoptosis by regulating SOX4 via miR129 in RCC, offering a promising therapeutic target and prognosis marker for RCC patients.
Keywords: renal cell carcinoma, lncRNA, UCA1, miR129, SOX4, UCA1/miR129/SOX4 axis
Introduction
Originating from renal epithelium cells, renal cell carcinoma (RCC) accounts for 2%–3% of all cancers and approximately 90% of kidney cancers.1,2 Kidney cancer is the tenth most common cancer in women and the seventh most prevalent cancer in men, with an estimated 295,000 new cases and 134,000 deaths annually worldwide.3–5 Although considerable advances have been made in diagnosis, screening, prevention, and therapy for RCC, it is still a serious threat to human health and life.6,7 As such, it is imperative to explore the molecular basis underlying RCC progression to identify more effective intervention strategies.
lncRNAs, a class of noncoding RNAs with the length ranging from 200 nucleotides to almost 100 kilobases, play vital roles in cancer development.8 Moreover, accumulating lncRNAs have been identified as important regulators in the diagnosis, prognosis, carcinogenesis, and pathogenesis of RCC.9,10 For instance, MALAT1 lncRNA is highly expressed in RCC tissue, and MALAT1 silencing suppresses proliferation and invasion and facilitates apoptosis in RCC cells.11 GAS5 lncRNA is downregulated in RCC tissue and cells, and ectopic expression of GAS5 hampers proliferation, migration, and invasion, as well as induces cell-cycle arrest and apoptosis in RCC cells.12 UCA1 LncRNA has been found to be an oncogene in certain types of malignancies, such as colorectal cancer,13 breast cancer,14 and non-small-cell lung cancer.15 Moreover, a recent study showed that UCA1 expression was markedly increased in RCC tissue and cells and UCA1 overexpression promoted proliferation and migration, as well as inhibited apoptosis in RCC cells.16 Furthermore, Lu et al demonstrated that higher UCA1 expression was positively associated with poor prognosis and progressive tumor status of RCC patients, and UCA1 accelerated RCC-cell proliferation through negatively regulating miRNA495 and epigenetically silencing p21 expression.17 However, the molecular basis of UCA1 underlying RCC pathogenesis has not been fully elaborated.
In the present study, we found that UCA1 expression was upregulated in RCC tissue and cells. Moreover, UCA1 depletion suppressed proliferation and invasion and enhanced apoptosis through the miR129–SOX4 axis in RCC cells.
Methods
Tissue specimens and cell culture
RCC tissue and adjacent normal tissue were obtained from 40 RCC patients (median age 55 years, male:female 3:2) receiving nephrectomy at our hospital. The resected tumors were snap-frozen in liquid nitrogen and then stored at -80°C. Tumor stage was defined following the TNM classification standard of American Joint Committee on Cancer. Experimental protocols were approved by the ethics committees of Huaihe Hospital of Henan University. Written informed consent was obtained from all RCC patients prior to tissue use.
HK2 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in keratinocyte serum-free medium containing 0.05 mg/mL bovine pituitary extract and 5 ng/mL human recombinant EGF (Thermo Fisher Scientific, Waltham, MA, USA). Human RCC cells (769P, 786O, ACHN, A498, Caki2) were purchased from the China Center for Type Culture Collection (Wuhan, China). 769P and 786O cells were cultured in RPMI 1640 medium supplemented with 10% FBS (Thermo Fisher Scientific). ACHN cells were maintained in MEM-EBSS (HyClone, Logan, UT, USA) containing 10% FBS. A498 cells were maintained in the MEM-EBSS supplemented with 10% FBS and 1% inessential amino acids (Thermo Fisher Scientific). Caki2 cells were grown in modified McCoy’s 5A medium (Thermo Fisher Scientific) containing 10% FBS. All cultures were kept in a humidified atmosphere with 5% CO2 at 37°C.
Reagents and transfection
siRNAs targeting UCA1 (siUCA1-1) and siUCA-2 and its scrambled control (siCon), miR129 mimic and its scrambled control (miR-Con), miR129 inhibitor (anti-miR129), and anti-miR-Con were purchased from GenePharma (Shanghai, China). Full-length sequences of UCA1 were amplified by polymerase chain reaction (PCR) and inserted into pcDNA3.1 vector (Thermo Fisher Scientific) to gain the UCA1-overexpression plasmid. All oligonucleotides or plasmids were transfected into RCC cells using Lipofectamine 2000 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The target sequences of siUCA1 were siUCA1-1 (5′-GGACAACAGTACACGCATA-3′) and siUCA1-2 (5′-TCTTTGTCTCCTGGATTAAC-3′).
Reverse-transcription quantitative PCR assays
Total RNA was isolated from RCC tissue and cells using a mirVana miRNA-isolation kit and quantified using spectrophotometry (NanoDrop 2000c; Thermo Fisher Scientific). Then, cDNA was synthesized from 1 μg RNA using Moloney murine leukemia virus reverse transcriptase (Thermo Fisher Scientific) and a random primer or miRNA-specific reverse-transcription (RT) primer (miR129 or U6 snRNA RT primer). SYBR green PCR master mixes (Thermo Fisher Scientific) and quantitative primers were employed to detect expression patterns of UCA1, miR129, and SOX4 mRNA on LightCycler 480 real-time PCR system (Hoffman-La Roche, Basel, Switzerland). GAPDH acted as an endogenous control of UCA1 and SOX4 mRNA expression, and U6 snRNA was used to normalize the expression of miR129. RT primers and quantitative primers were miR129-RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAAGC-3′, U6-RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGG-3′, quantitative primers miR129 5′-CTTTTTGCGGTCTGGGCTTGC-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse), U6 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse), UCA1, 5′-CTCTCCATTGGGTTCACCATTC-3′ (forward) and 5′-GCGGCAGGTCTTAAGAGATGAG-3′ (reverse), SOX4, 5′-GGCCTCGAGCTGGGAATCGC-3′ (forward) and 5′-GCCCACTCGGGGTCTTGCAC-3′ (reverse), and GAPDH 5′-GGTGAAGGTCGGAGTCAACGG-3′ (forward) and GAGGTCAATGAAGGGGTCATTG-3′ (reverse). Relative expression was calculated by the 2−ΔΔCT method.18 All results were obtained from at least three experiments with triplicate reactions.
Western blot assays
Total proteins were extracted from 786O and ACHN cells using radioimmunoprecipitation-assay buffer (Beyotime, Haimen, China) supplemented with a protease-inhibitor cocktail (Hoffman-La Roche) and then quantified using Bradford reagent (Sigma-Aldrich, St Louis, MO, USA). Then, the same amount of proteins (40 μg) was resolved by sodium dodecyl sulfate polyacrylamide-gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Merck Millipore, Billerica, MA, USA). After immersion in 5% skimmed milk for 1 hour at room temperature, the membranes were incubated overnight at 4°C with appropriate primary antibodies against SOX4 (ab80261 1:500) and GAPDH (ab8245 1:2,000; Abcam, Cambridge, UK). Following probing with HRP-conjugated secondary antibody (ab205718 or ab6789 1:5,000; Abcam) for 1 hour at room temperature, specific protein bands were visualized using an electrochemiluminescence Western blotting substrate (Thermo Fisher Scientific).
Luciferase assays
Partial sequences of UCA1 and SOX4 3′UTR containing predicted miR129-binding sites were amplified by PCR and constructed into a pGL3 basic vector (Promega, Fitchburg, WI, USA) to generate wild-type (WT) UCA1 reporter and SOX4 3′UTR reporter. A KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan) was used to produce a mutant (Mut)-type UCA1 reporter containing miR129-Mut-binding sites. Then, constructed luciferase reporter and oligonucleotide/plasmid were cotransfected into RCC cells in combination with a pRL-TK vector (Promega). At 48 hours following transfection, luciferase activities in RCC cells were detected by a dual luciferase reporter-assay kit (Promega) with Renilla luciferase activity as an endogenous control.
CCK-8 assays
Cell-proliferation capacity was measured using CCK-8 (Dojindo, Kumamoto, Japan) following the manufacturer’s procedure. Generally, RCC cells were seeded into a 96-well plate at a density of 104 cells/well and incubated overnight at 37°C prior to transfection with oligonucleotides or plasmids. At 0, 24, 48, and 72 hours after transfection, 10 μL CCK-8 solution was added to each well for an additional 3 hours. Finally, optical density was determined at a wavelength of 450 nm by a microplate reader (model 680; Bio-Rad, Hercules, CA, USA).
Cell-apoptosis assays
The apoptosis rate of RCC cells was detected using an annexin V–fluorescein isothiocyanate (FITC) apoptosis-detection kit (Beyotime) referring to the manufacturer’s protocols. At 48 hours posttransfection, cells were washed with PBS, resuspended with annexin V–FITC binding solution, and stained with annexin V–FITC and propidium iodide at room temperature for 20 min at dark. Then, the cell-apoptosis rate was detected using flow cytometry (BD Biosciences, San Jose, CA, USA).
Matrigel invasion assays
RCC-cell-invasion ability was assessed using BioCoat Matrigel invasion chambers (BD Biosciences). Briefly, 2×105s RCC cells resuspended in 350 μL serum-free medium were plated in the upper chamber containing Matrigel-coated membrane, and 700 μL complete medium containing 10% FBS was added to the lower chamber. After incubation for 36 hours at 37°C, cells on the upper surface of the membrane were removed using a cotton swab. Cells on the lower side of the membrane were fixed with 100% methanol, stained with 0.1% crystal violet solution (Sigma-Aldrich) and counted under microscopy.
RNA-immunoprecipitation assays
RNA-immunoprecipitation (RIP) assays were performed in RCC cells using a Magna RIP RNA-binding protein-immunoprecipitation kit (Merck Millipore) following the manufacturer’s instructions. Briefly, 786O and ACHN cells were lysed using RIP lysis buffer containing RNase inhibitor (Thermo Fisher Scientific) and a protease-inhibitor cocktail (Hoffman-La Roche). Then, cells extracted were incubated with RIP buffer containing protein A/G magnetic beads coated with anti-Ago2 or negative control anti-IgG (Merck Millipore) antibody, followed by the isolation of RNA. Following this, RT quantitative PCR (qPCR) assays were used to test degrees of enrichment of UCA1 and miR129 in precipitates of 786O and ACHN cells.
RNA pull-down assays
RNA pull-down assays were performed to detect the potential binding ability of UCA1 and miR129 in 786O and ACHN cells, referring to previous research.19 A biotinylated UCA1 probe and biotinylated control probe were purchased from Sangon (Shanghai, China). Briefly, probes were dissolved in wash/binding buffer and then incubated with streptavidin-coupled magnetic beads (M-280 Dynabeads; Thermo Fisher Scientific) for 2 hours at room temperature, followed by the addition of cell lysates for an additional 2 hours at room temperature. Then, RNA complex conjugated with beads was eluted and miR129 expression quantified using RT-qPCR assays with U6 snRNA as an endogenous control.
Statistical analysis
All data are expressed as mean ± SD from over three independent experiments and were analyzed using Student’s t-test or one-way analysis of variance. Spearman’s rank correlation analysis was used to detect correlations between expression levels of UCA1 and miR129 or SOX4. Survival curves were generated by the Kaplan–Meier method and estimated using the log-rank test. Differences were regarded as statistically significant when P<0.05.
Results
UCA1 expression was upregulated in RCC tumor tissue and cells
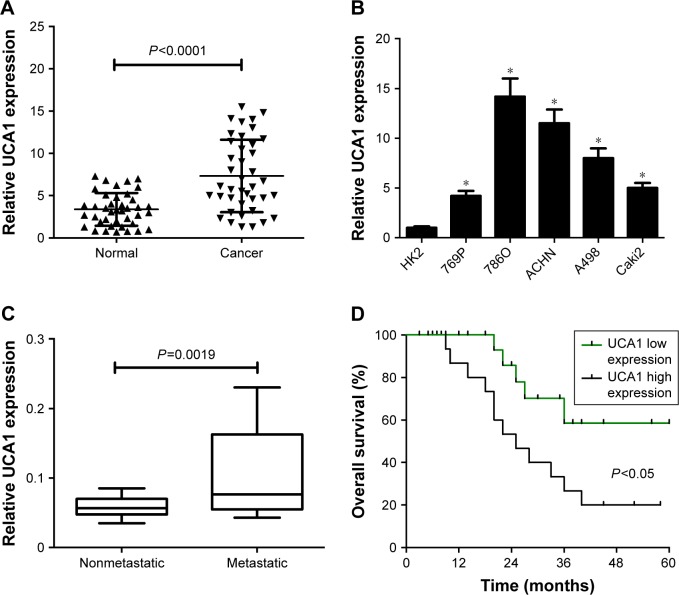
The expression pattern of UCA1 was explored in RCC tumor tissue and cells by RT-qPCR assays. Results showed that UCA1 expression was significantly increased in 40 cases of RCC tumor tissue compared with adjacent normal tissue (Figure 1A). Also, a notable upregulation in UCA1 expression was observed in a large panel of RCC cell lines (769P, 786O, ACHN, A498, Caki2) when compared to a normal renal proximal tubular epithelial cell line (HK2) (Figure 1B). Then, we further explored the association of UCA1 expression with clinical pathologic status and prognosis outcome. As displayed in Figure 1C, UCA1 level was markedly elevated in the advanced pathogenic stage (metastatic phase) patients when compared to earlier stage (nonmetastatic phase) patients. Moreover, overall survival was strikingly lower in the UCA1 high-expression group than in the UCA1 low-expression group (Figure 1D). As such, we concluded that dysregulation of UCA1 might be associated with RCC progression and prognosis.
Figure 1.
UCA1 was highly expressed in RCC tumor tissue and cells.
Notes: UCA1 expressions in 40 pairs of RCC tumor tissue and adjacent normal tissue (A), RCC cells (769P, 786O, ACHN, A498, Caki2) and normal proximal tubular epithelial cell line (HK2) (B), different pathogenic status (metastatic [n=22] and nonmetastatic [n=18] phases) (C). (D) Kaplan–Meier analysis of overall survival for RCC patients according to differences in UCA1 expression. Median UCA1 expression in RCC tumor tissue was used as a cutoff point to divide the UCA1 high-expression group and the UCA1 low-expression group. *P<0.05.
Abbreviation: RCC, renal cell carcinoma.
UCA1 silencing resulted in decreased proliferation and invasion and increased apoptosis in RCC cells
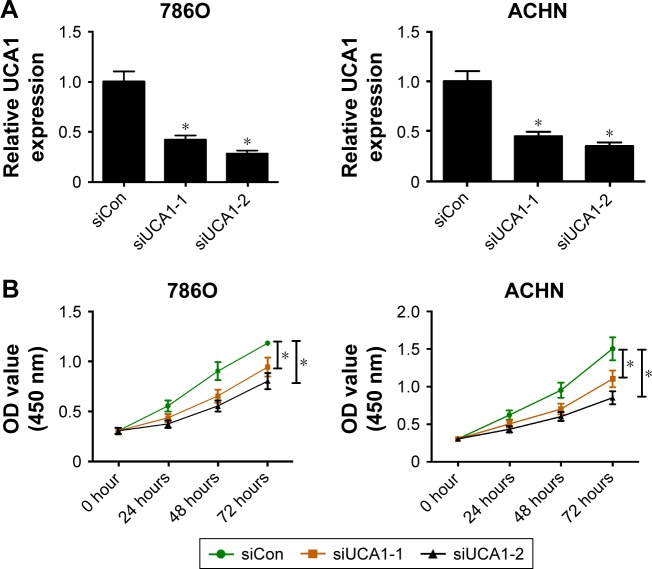
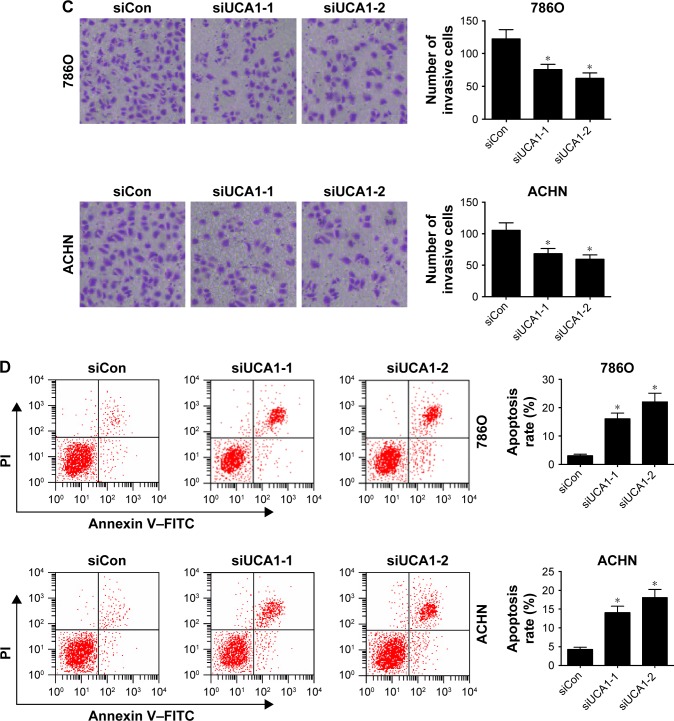
siRNAs targeting UCA1 (siUCA1-1 and siUCA1-2) and siCon were synthesized to investigate the role of UCA1 in RCC progression. Firstly, knockdown efficiency of siUCA1-1 and siUCA1-2 was detected in 786O and ACHN cells. As shown in Figure 2A, introduction of siUCA1-1 and siUCA1-2 triggered a marked decrease in UCA1 expression compared with siCon in 786O and ACHN cells, indicating that siUCA1-1 and siUCA1-2 could be used for subsequent loss-of-function experiments. CCK-8 assays showed that cell-proliferation ability was strikingly suppressed in UCA1-depleted cells in comparison with mock cells (Figure 2B). Also, Matrigel invasion assays revealed that RNAi-based silencing of UCA1 expression led to a substantial decline in invasion capacity of 786O and ACHN cells (Figure 2C). Furthermore, we demonstrated that UCA1 knockdown by siUCA1-1 and siUCA1-2 induced a significant enhancement in apoptosis rate in 786O and ACHN cells (Figure 2D). In a word, these results indicated the carcinogenic roles of UCA1 in RCC.
Figure 2.
UCA1 knockdown hampered proliferation and invasion and induced apoptosis in RCC cells.
Notes: 786O and ACHN cells were transfected with siRNAs targeting UCA1 (siUCA1-1 and siUCA1-2) and its scrambled control (siCon), followed by the detection of UCA1 expression at 48 hours posttransfection (A); assessment of proliferation ability after transfection at the time points indicated (B); evaluation of invasive capacity at 48 hours following siRNA treatment, magnification: 200× (C); measurement of apoptosis rate at 48 hours following siRNA treatment (D). *P<0.05.
Abbreviations: RCC, renal cell carcinoma; PI, propidium iodide; FITC, fluorescein isothiocyanate.
UCA1 inhibited miR129 expression by direct interaction in RCC cells
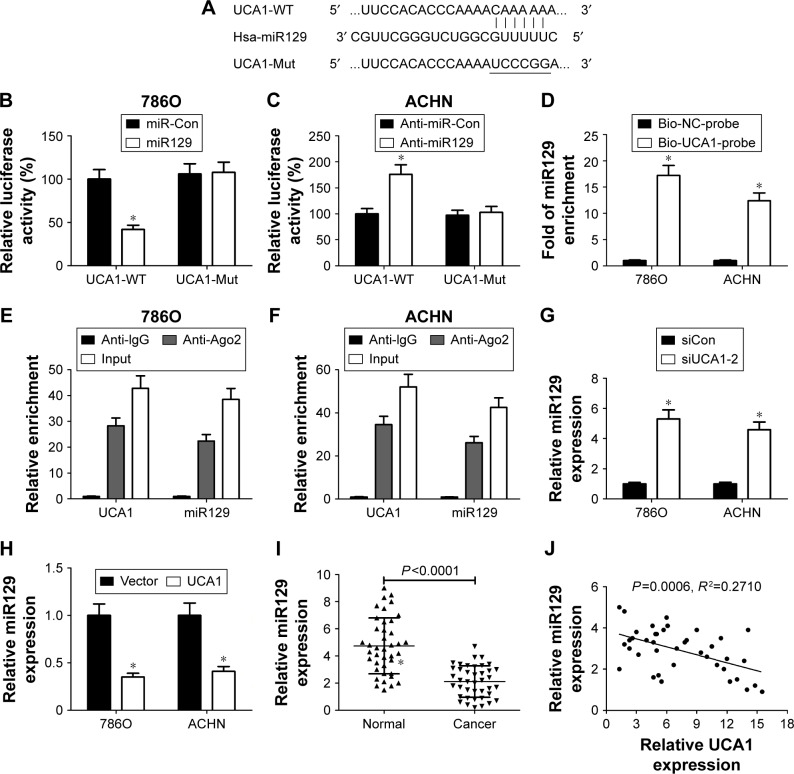
To gain further insight into the molecular mechanism of UCA1 in RCC progression, the miRcode website (http://www.mircode.org/index.php) was used to search for potential miRNAs that had a chance to interact with UCA1. Among candidate miRNAs, miR129 (Figure 3A) was chosen for further investigation, because of its tumor-suppressive effect in RCC.20,21 Then, luciferase, RNA pull-down, and RIP assays were performed further to explore whether UCA1 can interact with miR129 in RCC cells. Luciferase assays showed that the luciferase activity of UCA1-WT reporter was markedly decreased in miR129-overexpressed 786O cells (Figure 3B) but was notably increased in ACHN cells with reduced miR129 expression (Figure 3C). However, little change was observed in luciferase activity of UCA1-Mut reporter in 786O and ACHN cells after upregulating or suppressing miR129 expression (Figure 3B and C). Then, biotin-labeled RNA pull-down assays demonstrated that biotin-labeled specific UCA1 probes could pulled down abundant miR129 in 786O and ACHN cells (Figure 3D). Moreover, RIP assays disclosed that UCA1 and miR129 expressions were remarkably enriched by Ago2 antibody in 786O and ACHN cells (Figure 3E and F). In summary, these data revealed that UCA1 interacted directly with miR129 by putative binding sites in RCC cells. Subsequently, siUCA1-2 was selected for follow-up experiments, due to its better knockdown efficiency. RT-qPCR assays delineated that miR129 expression was markedly upregulated in UCA1- silenced RCC cells but greatly downregulated following UCA1 overexpression (Figure 3G and H). Additionally, miR129 expression was significantly lower in RCC tumor tissue (n=40) than in adjacent normal tissues (Figure 3I). Also, miR129 expression was negatively associated with UCA1 expression in 40 cases of RCC tumor tissue (Figure 3J). Taken together, these data suggested that UCA1 negatively regulated miR129 expression in RCC cells.
Figure 3.
UCA1 inhibited miR129 expression in RCC cells by direct interaction.
Notes: (A) Exhibition of putative binding sites between UCA1 and miR129 and mutant (Mut) sites in UCA1-Mut reporter. (B) 786O cells were cotransfected with miR control (Con) or miR129 mimic and wild-type (WT) UCA1 or UCA1-Mut reporter, followed by the detection of luciferase activity at 48 hours posttransfection. (C) Luciferase activity was measured in ACHN cells cotransfected with anti-miR-Con or miR129 inhibitor and UCA1-WT or UCA1-Mut reporter at 48 hours after transfection. (D) RNA pull-down assays were used to further validate the interaction between UCA1 and miR129 in 786O and ACHN cells. (E, F) RNA-immunoprecipitation assays were employed using Ago2 or IgG antibody to explore the enrichment degrees of UCA1 and miR129 in Ago2 or IgG immunoprecipitation complexes of 786O and ACHN cells. The input group acted as a positive control. (G, H) miR129 expression was determined in 786O and ACHN cells transfected with siCon (G), siUCA1-2 (G), UCA1 overexpression plasmid (H) or pcDNA3.1 empty vector (H). (I) miR129 expression analysis in 40 pairs of RCC tumor tissue and adjacent normal tissue. (J) Spearman’s correlation analysis of miR129 and UCA1 expression in 40 cases of RCC tumor tissue. *P<0.05.
Abbreviation: RCC, renal cell carcinoma.
MiR129 overexpression inhibited proliferation and invasion and induced apoptosis in RCC cells
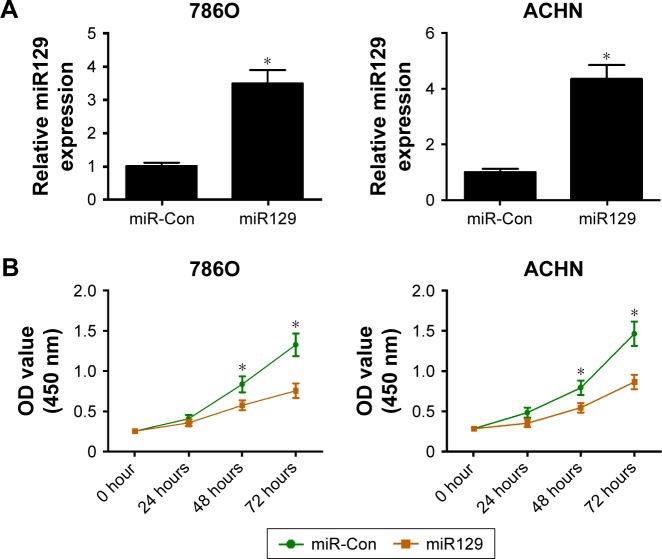
We further investigated the roles of miR129 in RCC cell proliferation, invasion, and apoptosis by transfection with miR129 mimic. As presented in Figure 4A, introduction of miR129 mimic triggered an obvious elevation in miR129 levels in 786O and ACHN cells. Further functional assays revealed that upregulation of miR129 resulted in a marked reduction in proliferation ability (Figure 4B) and invasion number (Figure 4C) in both 786O and ACHN cells. Moreover, a striking enhancement in apoptosis rate was observed in 786O and ACHN cells following increased miR129 expression (Figure 4D). In sum, enforced expression of miR129 suppressed proliferation and invasion and promoted apoptosis in RCC cells.
Figure 4.
miR129 overexpression inhibited proliferation and invasion and enhanced apoptosis in RCC cells.
Notes: 786O and ACHN cells were transfected with miR129 mimic and its scrambled control (miR-Con), followed by the detection of miR129 expression at 48 hours upon transfection (A); determination of proliferation ability posttransfection at the time points indicated (B); measurement of invasive cell numbers at 48 hours following transfection, magnification: 200× (C); examination of apoptosis rate at 48 hours after transfection (D). *P<0.05.
Abbreviations: RCC, renal cell carcinoma; PI, propidium iodide; FITC, fluorescein isothiocyanate.
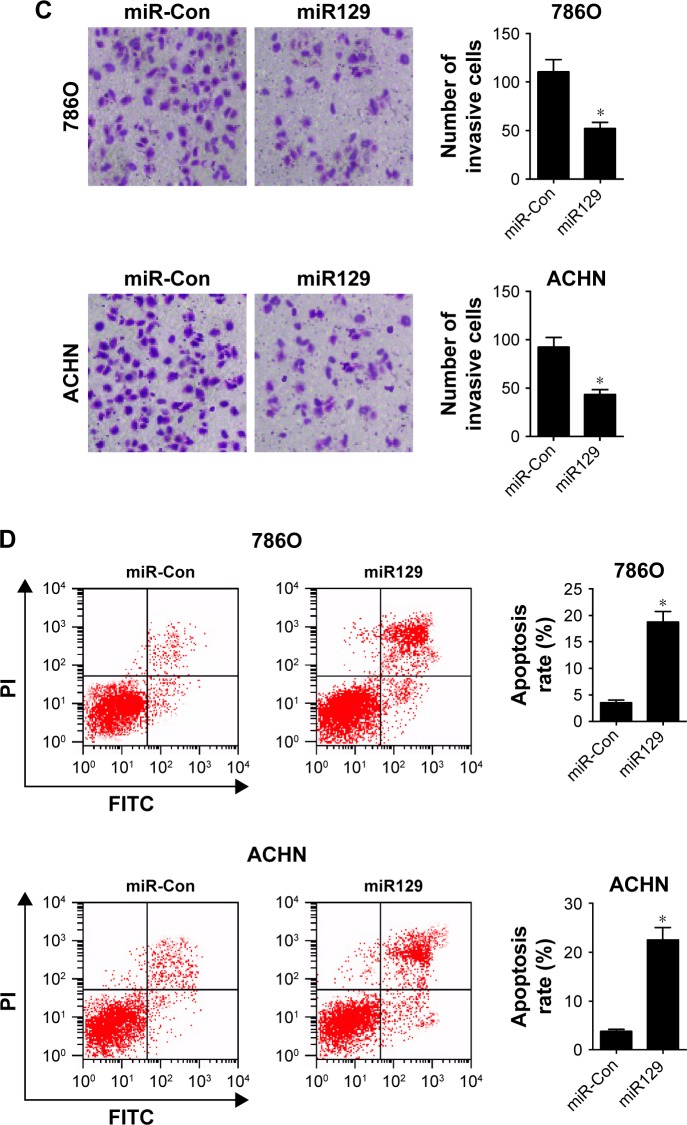
MiR129 downregulation abrogated UCA1-knockdown-mediated antiproliferation, anti-invasion, and proapoptosis in RCC cells
We further explored whether the carcinogenicity of UCA1 was mediated by miR129 in RCC cells. Results showed that introduction of miR129 inhibitor strikingly alleviated UCA1-knockdown inhibition effects on the proliferation (Figure 5A and B) and invasion (Figure 5C and D) of 786O and ACHN cells. Moreover, miR129 downregulation apparently weakened siUCA1-induced apoptosis in 786O and ACHN cells (Figure 5E and F). In a word, these results implicated that UCA1 deficiency repressed proliferation and invasion and enhanced apoptosis of RCC cells by increasing miR129 expression.
Figure 5.
miR129 downregulation attenuated UCA1 knockdown-mediated antiproliferation, anti-invasion, and proapoptosis effects in RCC cells.
Notes: 786O and ACHN cells were transfected with scrambled control (siCon), siUCA1-2, siUCA1-2 + anti-miR-Con, or siUCA1-2 + anti-miR129, followed by determination of cell proliferation (A, B), invasion (C, D), and apoptosis (E, F). *P<0.05.
Abbreviation: RCC, renal cell carcinoma.
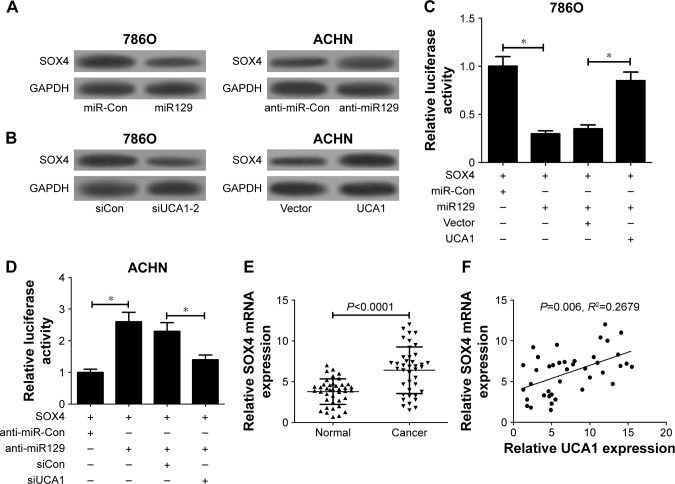
UCA1 acted as a ceRNA of miR129 to enhance target gene SOX4 expression in RCC cells
Previous studies have shown that miR129 exerts its antitumor effect by targeting SOX4 in several cancers, including RCC.20,22 We further discuss whether UCA1 can act as a ceRNA of miR129 to regulate SOX4 expression in RCC cells. At first, Western blot analysis displayed that SOX4 protein expression was decreased in miR129-overexpressed 786O cells but was increased in miR129-reduced ACHN cells (Figure 6A). Consistently, UCA1 silencing resulted in a downregulation in SOX4 expression in 786O cells, while enforced expression of UCA1 induced higher SOX4 expression in ACHN cells (Figure 6B). As demonstrated, UCA1 inhibited miR129 expression in RCC cells. We might come to the conclusion that UCA1 can act as a ceRNA of miR129 to regulate SOX4 expression. To validate this conclusion further, the effects of miR129 and UCA1 on luciferase activity of SOX4 reporter were determined in RCC cells. As exhibited in Figure 6C, miR129 upregulation inhibited luciferase activity of SOX4 reporter, while this effect was counteracted following UCA1 overexpression in 786O cells. On the contrary, miR129 deficiency induced an evident rise in luciferase activity of SOX4 reporter, and UCA1 knockdown undermined anti-miR129-mediated enhancement effect on luciferase activity in ACHN cells (Figure 6D). Moreover, SOX4 expression was obviously upregulated in RCC tumor tissue (n=40) in comparison with adjacent normal tissue (n=40; Figure 6E). Also, SOX4 mRNA expression was positively related to UCA1 expression in RCC tumor tissue (n=40; Figure 6F). Taken together, these results indicate that UCA1 performed as a molecular sponge of miR129 to promote target SOX4 gene expression in RCC cells.
Figure 6.
UCA1 acted as a ceRNA of miR129 to enhance target SOX4 gene expression in RCC cells.
Notes: (A) 786O cells were transfected with miR129 mimic or miR-Con, and ACHN cells were transfected with miR129 inhibitor or anti-miR-Con, followed by the detection of SOX4 protein expression. (B) 786O cells were transfected with siCon or siUCA1-2, and ACHN cells were transfected with pcDNA3.1 vector or pcDNA-UCA1 plasmid, followed by the determination of SOX4 protein expression. (C) Effects of miR129 and UCA1 overexpression on luciferase activity of SOX4 reporter were evaluated in 786O cells. (D) The effect of miR129 downregulation and UCA1 silencing on luciferase activity of SOX4 reporter was monitored in ACHN cells. (E) SOX4 mRNA expression in 40 pairs of RCC tumor tissue and adjacent normal tissue. (F) Spearman’s correlation analysis of SOX4 mRNA and UCA1 expressions in 40 cases of RCC tumor tissue. *P<0.05.
Abbreviation: RCC, renal cell carcinoma.
Discussion
RCC, a serious threat for human health and life with the highest mortality in urological malignancies,23 is characterized by the deficiency of early warning signs and the diversity of clinical manifestation.24 In China, RCC is the second-most prevalent major cancer in the urological system, with a mean 6% rise in incidence over the past two decades.25 A substantial amount of evidence shows that noncoding RNAs, including lncRNA and miRNA, play vital roles in the development and progression of RCC.26,27 UCA1 has been demonstrated as an oncogenic factor in multiple cancers. For instance, UCA1 enhances the metastatic ability of gastric cancer cells by affecting GRK2 protein stability via facilitating CblC-mediated GRK2 ubiquitination and degradation.28 Qian et al revealed that UCA1 facilitated proliferation and invasion and suppressed apoptosis in hypopharyngeal squamous cell carcinoma.29
In the present study, the role and molecular mechanism of UCA1 in RCC progression were further investigated. Firstly, we demonstrated that UCA1 was highly expressed in RCC tissue and cells, and higher UCA1 expression was associated with advanced clinical stage and lower overall survival of RCC patients, consistent with a previous report.17 Function analyses further displayed that UCA1 silencing resulted in a decrease in proliferation and invasion capacity and an increase in apoptosis rate in RCC cells, in line with an earlier study.16 lncRNAs can act as ceRNAs of miRNAs to regulate gene expression. miRNAs, a group of small noncoding RNAs of about 19–25 nucleotides in length, have been well documented to be closely associated with carcinogenesis, diagnosis, and prognosis in cancer, including RCC.26,30
The miRcode website was employed to search for potential miRNAs that had a chance of interacting with UCA1. Among candidate miRNAs, miR129 was chosen for further investigation, due to its antitumor effects in some cancers, such as esophageal carcinoma,22 gastric cancer,31 and bladder cancer.32 A recent study pointed out that miR129-3p expression was downregulated in RCC tissues and low miR129-3p expression was associated with poor prognosis in RCC patients.20 Moreover, miR129-3p weakened migration and invasion abilities of RCC cells by decreasing the levels of SOX4, MMP2, MMP9, and FAK phosphorylation.20 Xu et al also found that miR129-5p expression was remarkably downregulated in RCC tissue and cells and miR129-5p overexpression significantly inhibited proliferation, migration, and invasion of RCC cells by targeting IGF2BP1.21
In the present study, we further demonstrated that UCA1 interacted directly with miR129 by luciferase, RNA pull-down, and RIP assays. Moreover, ectopic expression of UCA1 suppressed miR129 expression in RCC cells. MiR129 expression was underexpressed and inversely associated with UCA1 levels in RCC tissue. Additionally, exogenous expression of miR129 inhibited proliferation and invasion and induced apoptosis in RCC cells. Restoration assays further revealed that miR129 downregulation abated UCA1 knockdown-mediated antiproliferation, anti-invasion, and proapoptosis effects in RCC cells. In other words, UCA1 exerted its oncogenic effect by decreasing miR129 expression in RCC.
Previous studies showed that miR129 suppressed tumorigenesis by downregulating SOX4 expression in several solid tumors, such as esophageal cancer,22 endometrial cancer,33 and RCC.20 SOX4, mapped to chromosome 6p22, has been identified as an oncogene and is highly expressed in a variety of cancers, such as breast cancer,34 hepatocellular carcinoma (HCC),35 and adenoid cystic carcinoma.36 Ruan et al also demonstrated that SOX4 expression was notably increased in RCC tissue and ectopic expression of SOX4 facilitated migration and invasion of RCC cells.37 Thereby, we further explored whether UCA1 acts as a ceRNA of miR129 to regulate SOX4 expression in RCC. Our study firstly exhibited that miR129 inhibited SOX4 expression and UCA1 promoted SOX4 expression in RCC cells. Moreover, luciferase assays elucidated that UCA1 undermined the interaction between miR219 and SOX4 in RCC cells. Clinical assays further showed that SOX4 expression was prominently increased and positively associated with UCA1 expression in RCC tissues.
Conclusion
Taken together, our results suggest that UCA1 knockdown hinders cell proliferation and invasion and induces apoptosis by sequestering miR129 from SOX4 in RCC, indicating a new regulatory mechanism of UCA1 underlying RCC progression and a potential therapy target for RCC.
Acknowledgments
This work was supported by the Science and Technology Project of Henan Province (172102310153).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ljungberg B, Hanbury DC, Kuczyk MA, et al. Renal cell carcinoma guideline. Eur Urol. 2007;51(6):1502–1510. doi: 10.1016/j.eururo.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 5.Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108(10):1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 7.Escudier B, Porta C, Schmidinger M. Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl 5):v58–v68. doi: 10.1093/annonc/mdw328. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Chen ZH, Wang XX, Huang ZN, He ZW, Chen YQ. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen JW, Chen YL, Gu LY, et al. lncRNAs act as prognostic and diagnostic biomarkers in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget. 2016;7(45):74325–74336. doi: 10.18632/oncotarget.11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blondeau JJ, Deng M, Syring I, et al. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics. 2015;7:10. doi: 10.1186/s13148-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14(2):1077–1082. doi: 10.7314/apjcp.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 13.Han Y, Yang YN, Yuan HH, et al. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46(5):396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 14.Tuo YL, Li XM, Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur Rev Med Pharmacol Sci. 2015;19(18):3403–3411. [PubMed] [Google Scholar]
- 15.Nie W, Ge HJ, Yang XQ, et al. lncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Wang T, Li Y, et al. Identification of long-non coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol Med Rep. 2016;13(4):3326–3334. doi: 10.3892/mmr.2016.4894. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Liu WG, Lu JH, et al. lncRNA UCA1 promotes renal cell carcinoma proliferation through epigenetically repressing p21 expression and negatively regulating miR-495. Tumour Biol. 2017;39(5) doi: 10.1177/1010428317701632. 1010428317701632. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Liu WH, Ma CL, Yang B, Yin CY, Zhang BN, Xiao YF. lncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun. 2017;493(3):1168–1175. doi: 10.1016/j.bbrc.2017.09.114. [DOI] [PubMed] [Google Scholar]
- 20.Chen XY, Ruan AM, Wang XG, et al. miR-129-3p, as a diagnostic and prognostic biomarker for renal cell carcinoma, attenuates cell migration and invasion via downregulating multiple metastasis-related genes. J Cancer Res Clin Oncol. 2014;140(8):1295–1304. doi: 10.1007/s00432-014-1690-7. [DOI] [PubMed] [Google Scholar]
- 21.Xu HL, Feng YY, Jia ZK, Zhang YG, Li J, Xia ML. MicroRNA-129-5p inhibits the proliferation and migration in renal cell carcinoma via targeting IGF2BP1. Int J Clin Exp Pathol. 2016;9(8):8254–8260. [Google Scholar]
- 22.Kang M, Li YM, Liu WQ, et al. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32(1):51–58. doi: 10.3892/ijmm.2013.1384. [DOI] [PubMed] [Google Scholar]
- 23.Slabý O, Svoboda M, Michálek J, Vyzula R. DNA a mikroRNA čipové technologie v diagnostice a predikci upacientů s renálním karcinomem [DNA and microRNA microarray technologies in diagnostics and prediction for patients with renal cell carcinoma] Klin Onkol. 2009;22(5):202–209. Czech. [PubMed] [Google Scholar]
- 24.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhao P, Dai M, Chen WQ, Li N. Cancer trends in China. Jpn J Clin Oncol. 2010;40(4):281–285. doi: 10.1093/jjco/hyp187. [DOI] [PubMed] [Google Scholar]
- 26.Redova M, Svoboda M, Slabý O. MicroRNAs and their target gene networks in renal cell carcinoma. Biochem Biophys Res Commun. 2011;405(2):153–156. doi: 10.1016/j.bbrc.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 27.Zhou SG, Wang JD, Zhang ZG. An emerging understanding of long noncoding RNAs in kidney cancer. J Cancer Res Clin Oncol. 2014;140(12):1989–1995. doi: 10.1007/s00432-014-1699-y. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZQ, He CY, Hu L, et al. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi: 10.1016/j.canlet.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Qian Y, Liu D, Cao S, et al. Upregulation of the long noncoding RNA UCA1 affects the proliferation, invasion, and survival of hypopharyngeal carcinoma. Mol Cancer. 2017;16(1):68. doi: 10.1186/s12943-017-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguiari G. MicroRNAs in clear cell renal cell carcinoma: biological functions and applications. J Kidney Cancer VHL. 2015;2(4):140–152. doi: 10.15586/jkcvhl.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu XC, Song HJ, Xia T, et al. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532(1):87–93. doi: 10.1016/j.gene.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 32.Dyrskjøt L, Ostenfeld MS, Bramsen JB, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69(11):4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 33.Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69(69):9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang JC, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–4608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 35.Liao YL, Sun YM, Chau GY, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–5589. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 36.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25(41):5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 37.Ruan H, Yang H, Wei H, et al. Overexpression of SOX4 promotes cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition. Int J Oncol. 2017;51(1):336–346. doi: 10.3892/ijo.2017.4010. [DOI] [PubMed] [Google Scholar]