Abstract
Lysine deacetylases (KDACs) are enzymes that reverse the post-translational modification of lysine acetylation. Thousands of potential substrates, acetylated protein sequences, have been identified in mammalian cells. Properly regulated acetylation and deacetylation have been linked to many biological processes, while aberrant KDAC activity has also been linked to numerous diseases. Commercially available peptide substrates that are conjugated to fluorescent dye molecules, such as 7-amino-4-methylcoumarin (AMC), are commonly used to monitor deacetylation in studies addressing both substrate specificity and small molecule modulators of activity. Here, we have compared the activity of several KDACs, representing all major classes of KDACs, with substrates in the presence and absence of AMC as well as peptides for which tryptophan has been substituted for AMC. Our results unequivocally demonstrate that AMC has a significant effect on activity for all KDACs tested. Furthermore, the effect is not consistent across KDACs, neither in nature of the effect nor magnitude, making it impossible to predict the effect of AMC on a particular enzyme-substrate pair. AMC did not affect acetyllysine preference in a multiply acetylated substrate. In contrast, AMC significantly enhanced KDAC6 substrate affinity, greatly reduced Sirt1 activity, eliminated substrate sequence specificity of KDAC4, and had no consistent effect with KDAC8 substrates. These results indicate that profiling of KDAC activity with labeled peptides is unlikely to produce biologically relevant data.
TABLE OF CONTENTS
Introduction
Lysine deacetylases (KDACs) are enzymes that reverse the post-translational modification of lysine acetylation, by catalyzing the hydrolysis of ε-N-acetyllysine residues in proteins. The eighteen human KDACs are divided into several classes. Classes I, IIa, and IIb are metal-dependent enzymes that share a conserved reaction mechanism, and are often referred to as histone deacetylases. 1-3 Class III KDACs, or sirtuins, are NAD+-dependent enzymes that catalyze the same net reaction but are unrelated to the metal-dependent KDACs. The metal-dependent KDAC11 is sometimes considered to be its own class, class IV.4,5 Thousands of acetylated protein sequences have been identified in mammalian cells, and thus are potentially subject to deacetylation by KDACs.6-11 However, direct evidence for deactylation of acetylated proteins by a specific KDAC exists for only a small number of potential non-histone KDAC substrates.12-15 Properly regulated acetylation and deacetylation have been linked to many biological processes, while aberrant KDAC activity has also been linked to numerous diseases.4,5 Based on the therapeutic potential of regulating KDACs in vivo, research efforts are focused on identifying molecules that inhibit or activate these enzymes,5,16-18 as well as identifying substrates of specific KDACs.19-23
To address these research questions, several in vitro assays have been developed to measure KDAC activity with a variety of substrates. One common method utilizes peptide substrates, which are often derived from acetylated human proteins, that are conjugated to fluorescent dye molecules, such as 7-amino-4-methylcoumarin (AMC). In these assays, fluorescence of AMC occurs only when the dye molecule is cleaved from the peptide by trypsin, which is dependent on deacetylation.20 As several of these substrates are commercially available, they are heavily utilized for determining the effects of small molecules on enzyme activity and characterizing substrate interactions.24,25 In addition, this method has been used to investigate substrate preferences for specific KDACs, although it can only be used to investigate the residues on the N-terminal side of the acetylated lysine. In one instance, to address substrate specificity of KDACs, activity was determined for several KDACs against peptides containing different combinations of amino acids at the -2 and -1 positions relative to the acetylated lysine and with AMC conjugated to the C-terminal side.22 While convenient, these assays assume that the presence of AMC does not affect the enzyme’s function or substrate preferences. Such effects, especially any that is not consistent for multiple enzymes and substrates, would greatly impact how the results of such experiments can be interpreted, even when the peptide to which the AMC is conjugated is derived from a known substrate protein sequence.
Recently, we developed an assay which enables monitoring the activity of KDACs without use of a conjugated dye or other unnatural modification of substrate.26 This method relies on fluorescamine, a molecule that reacts with primary amines, resulting in fluorescence.27 Fluorescamine is particularly suited for this application, as it specifically reacts with the product of deacetylation (i.e. an unacetylated lysine) and therefore monitors deacetylation of unlabeled substrates. Notably, we found that normalizing the activity from a library of AMC conjugated peptides did not correlate with the activity obtained with unlabeled 5-mer peptides with the same amino acids at the -2 and -1 positions relative to the acetylated lysine.26 In contrast, our results did correlate with an alternate assay for label-free substrates based on detection of acetate.28 Therefore, the AMC conjugated peptides did not serve as reliable predictors for substrates of KDAC8. While this discrepancy could be due, at least in part, to the contribution of the amino acids in the +1 and +2 positions relative to the acetylated lysine, it may also be that the presence of AMC adjacent to the acetylated lysine affects the activity of the enzyme in a manner that is not biologically relevant. Notably, other groups have reported isolated instances where AMC conjugation to specific peptides led to increased activity with KDACs on specific substrates when compared to the use of other methods for detection of deacetylation.23,28,29 In addition, small molecule activators of some KDACs have been reported to be sequence-specific, and activators identified using AMC-conjugated substrates have not demonstrated activation with unlabeled substrates.30-33 However, to date no work has been reported that systematically probes the underlying mechanism of activity changes caused by AMC conjugation across multiple substrates and classes of KDACs.
To directly determine whether AMC affects KDAC activity when conjugated to peptide substrates in a more comprehensive way, we assayed the activity of several KDACs with three different commercially available AMC-conjugated substrates and compared the activity to the activity with the corresponding non-AMC conjugated peptides. The results demonstrated a clear, significant effect of the dye on activity for all classes of KDACs. Furthermore, the effect was not consistent for the different enzymes, making interpretation of previously collected data more challenging.
Methods
KDAC expression and purification
pFastbac1 (Life Technologies) containing human KDAC8 (Uniprot Q9BY41), fused to a tobacco-etch virus (TEV) protease cleavage site and His6 tag, was transformed into DH10Bac E. coli cells to produce bacmids containing KDAC8.34 Bacmids were purified and transfected into Sf9 cells using Cellfectin II (Life Technologies) as described elsewhere.35 Baculovirus from these transfections was then used to infect High Five insect cells in suspension. At 72 hours post-infection, cells were pelleted and frozen at -20 °C until lysis. Cells were resuspended in lysis buffer (30 mM MOPS pH 8.0, 150 mM KCl, 5% glycerol, 5 mM imidazole, 2 mM MgCl2, 1X HALT protease inhibitor [Thermo Scientific]). Purification was then performed as described previously.26 KDAC6 (Uniprot Q9UBN7, a gift from Eric Verdin, Addgene plasmid #13823)36 was cloned into pFastBac1 with the TEV protease cleavage site and His6 tag, then expressed and purified in insect cells using the same protocol as for KDAC8 except that the His6 tag was retained. Sirt1(193-741)-GST (Uniprot Q96EB6) was obtained from BPS Bioscience and dialyzed into KDAC storage buffer (30 mM MOPS pH 8.0, 150 mM KCl, 25% glycerol, 1 mM tris(2-carboxyethyl)phosphine). KDAC4(648-1057) (Uniprot P56524) was synthesized as a codon-optimized gene in pJExpress401 (DNA 2.0), then expressed and purified as previously described for KDAC8.26 Site-directed mutagenesis was used to create KDAC4 H976Y (KDAC4HY), which was then expressed and purified identically.
KDAC substrates
All peptides were N-terminally acetylated, and unlabeled peptides C-terminally amidated. {K-ac} was synthesized by adding ten-fold molar excess anhydrous acetic anhydride to acetyl-L-lysine amide hydrochloride (Chem-Impex International) in anhydrous DMSO. The sealed reaction was allowed to proceed for 3 days at room temperature. After confirmation of complete acetylation using fluorescamine, 2.5% dH2O was added to the reaction and it was dried using a vacuum concentrator until all solvent evaporated. Purity and complete acetylation were confirmed by NMR (Table S1). Fluorophore-coupled substrates were obtained commercially (Enzo Life Sciences, R&D Systems, or BPS Bioscience). All other substrates were commercial custom peptide syntheses purified to > 95% (Genscript).
Activity assays
Assays were performed as described previously.26 For endpoint assays, 100 μM substrate was incubated with KDAC8 (200 nM), Sirt1 (200 nM), KDAC6 (50 nM), KDAC4 (200 nM), or KDAC4HY (200 nM for unlabeled peptides, 10 nM for labeled substrates) for 1 hour at 25 °C. For kinetic analyses, at least eight concentrations of each peptide substrate were incubated with 10-50 nM KDAC6 or 150 nM KDAC8 in reaction buffer at 25 °C. Aliquots were taken at five timepoints and added to SAHA (suberoylanilide hydroxamic acid) at a final concentration of 100 μM. Timepoints for each substrate were selected to ensure that the data reflected the initial velocity of the reaction and varied between 1 and 60 minutes in increments of 0.25 to 15 minutes. Michealis-Menten steady state parameters were calculated as described previously,26 using at least three consecutive and linear data points for each concentration, again to ensure that calculations reflected initial velocity.
KDAC reactions assayed by MALDI MS
Several concentrations of KDAC6 and KDAC8 were incubated with 50 μM substrate for either 10 min (KDAC6) or 60 min (KDAC8) at 25 °C. Reactions were stopped by adding SAHA to a final concentration of 100 μM. The reaction was diluted 1:100 in TA85 (85% acetonitrile, 15% water, 0.1% trifluoroacetic acid). 0.5 μl was spotted onto a MTP Anchorchip target plate (Bruker Daltonics) and allowed to dry. 0.5 μl matrix (1.4 mg/ml α-cyano-4-hydroxycinnamic acid in TA85) was spotted on top of each sample. Samples were analyzed by matrix assisted laser desorption/ionization (MALDI) time-of-flight (TOF) mass spectrometry (MS) on an Autoflex Speed MALDI TOF/TOF (Bruker Daltonics) in positive reflector mode and masses were assigned to peaks using flexanalysis software (Bruker Daltonics). For each enzyme-substrate pair containing the highest enzyme concentration, samples were also analyzed in LIFT mode to obtain MS/MS spectra for the substrate and products. Spectra were analyzed in flexanalysis for the presence or absence of diagnostic y-ion peaks to distinguish between deacetylation at lysine positions 3 and 4, and the presence of expected a-ion, b-ion, and y-ion peaks confirmed by sequencing analysis using Biotools (Bruker Daltonics).
Results
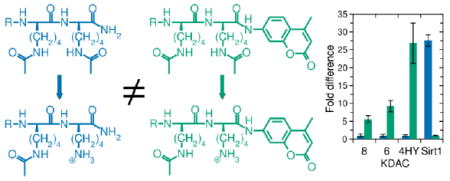
To investigate whether the presence of AMC in substrates affected KDAC activity, we performed quantitative assays with metal-dependent KDACs (KDAC4, KDAC6, and KDAC8) as well as a NAD+-dependent KDAC (Sirt1) as representative members of each major class of KDAC (class IIa, IIb, I, and III, respectively). Each KDAC was allowed to react with three sets of peptide substrates. Each substrate set included a base sequence where the acetylated lysine was at the C-terminus, a peptide that was identical except AMC was conjugated to the C-terminal side of the acetylated lysine, and a third peptide where a tryptophan (W) residue was included instead of the dye molecule at the +1 position (Figure 1). We hypothesized that the tryptophan side chain would at least partially mimic the effect of AMC, as both are large, hydrophobic molecules. The three AMC-containing peptides were obtained from commercial sources and activity was assessed by measuring the fluorescence of free AMC after treatment with trypsin to liberate AMC from deacetylated peptides. Activity against the remaining peptides was quantified using the fluorescamine assay. Unfortunately, we found that free AMC interferes with the fluorescamine signal such that we could not assess deacetylation of AMC-containing peptides using the fluorescamine assay; however, the resulting activity measurements can be directly compared, as specific activity was calculated in each case. We have previously determined that specific activity from the fluorescamine assay is comparable to measurements obtained using other methods.26
Figure 1. Substrate structures.
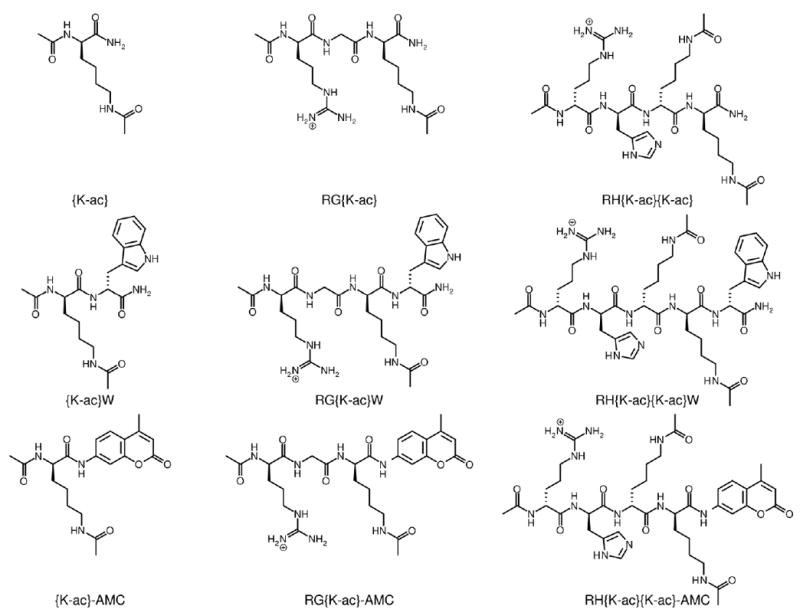
RG{K-ac} has been reported within several proteins, including as RG{K-ac}W.6,8-11 RH{K-ac}{K-ac} has been observed within the sequence of cytochrome p53;8 although tryptophan does not follow this sequence in any previously reported acetylated protein, we utilize it as the closest amino acid mimic of AMC.
Strikingly, the presence of AMC in the peptide greatly affected the activity of all KDACs tested, resulting in a significant difference between deacetylation of the peptides with and without AMC in every case where activity was observed (Figure 2 and Table S2). As expected, KDAC4 did not demonstrate activity with any of the peptides tested; however, it was active with a previously reported class IIa fluorogenic substrate that is not biologically relevant,37 indicating that it was a functional enzyme (data not shown). Thus, we used a previously reported KDAC4 gain-of-function variant (KDAC4HY),37 which allowed us to determine the effect of the fluorophore on a class IIa KDAC. The overall effects of the dye were not consistent across KDACs. The dye increased activity for the metal-dependent KDACs, but not equivalently. In particular, KDAC4HY activity increased up to 300-fold with the AMC-containing peptides, compared to approximately 10-fold for KDAC6 and a widely variable amount for KDAC8. In addition, KDAC4HY was equivalently active with all three AMC-containing peptides, although there were clear differences in activity with the non-AMC containing peptides. In contrast, Sirt1 demonstrated negligible activity when AMC was present despite having significant activity with the unlabeled substrates. In all cases except KDAC4HY, the presence of tryptophan substantially increased activity over the base peptides. For KDAC6 and KDAC8, this resulted in an intermediate level of activity, suggesting that the tryptophan may be partially compensating for the AMC dye. However, this pattern was not apparent for Sirt1 or KDAC4HY, where inclusion of the tryptophan resulted in no significant change in activity compared to the base peptide (KDAC4HY) or the highest level of activity (Sirt1). Taken together, there was no discernible pattern for the effect of either tryptophan or AMC on KDAC activity across enzyme classes.
Figure 2. Endpoint activity for KDACs with peptide substrates.
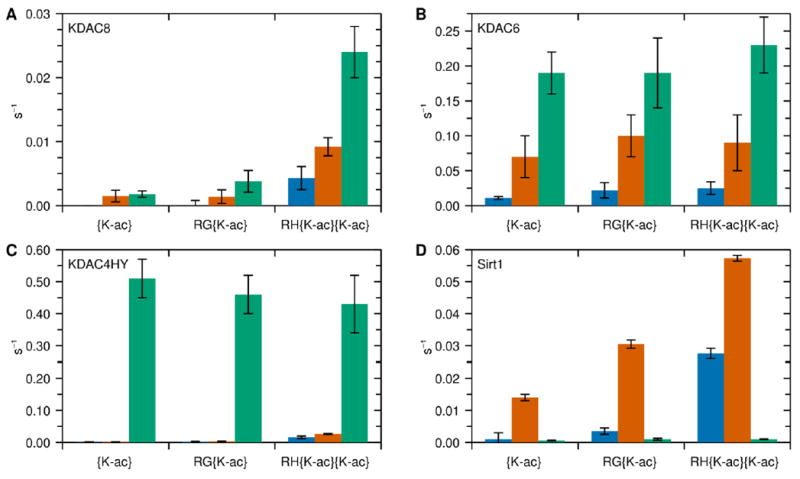
Three sets of peptide substrates where the C-terminus ends in an acetylated lysine (blue), tryptophan (orange), or AMC (green). Each peptide reacted with KDAC8 (A), KDAC6 (B), KDAC4HY (C), or Sirt1 (D) as described in the methods. The average specific activity and standard deviation for at least three replicates is shown for each enzyme-substrate pair. The differences in trends for each KDAC indicate that AMC conjugation has an effect on enzyme activity distinct from tryptophan, the amino acid most similar in structure.
To determine whether the apparently similar effect of AMC on the activity of KDAC6 and KDAC8 shared an underlying mechanism, we determined the steady-state kinetic parameters for both KDAC6 and KDAC8 to determine whether the dye affected catalytic activity, substrate affinity, or both. We hypothesized that tryptophan and AMC both elicited their effects by influencing the affinity of the enzyme for each substrate. Reactions were set up such that several concentrations of substrate were incubated with each KDAC and aliquots were taken at several different timepoints. We noticed that while KDAC6 deacetylated all substrates tested, for most substrates the reaction only proceeded at the initial velocity for a short time before slowing down and in many cases halting entirely, even though there was still a large excess of substrate present in the reaction (data not shown). The decrease in velocity was substrate-dependent rather than time-dependent, observed with some substrates in under 3 minutes; in contrast, KDAC6 maintained a constant velocity for at least one hour with other substrates. Therefore, we adjusted the timescale of each experiment to include only timepoints that reflected the true initial velocity. Steady-state kinetic parameters (kcat and KM) for each peptide with KDAC6 are shown in Table 1. While there were only modest differences in the rate of catalysis (kcat) between the peptides (only a 3.5 fold difference between the best and worst substrates), the substrate affinity varied dramatically. As with the endpoint data, a clear pattern emerged where the KM for each AMC-containing peptide was approximately 50-fold lower than the corresponding peptide without AMC. As expected, activity differences could not be attributed to differential effects of the inhibitor in the presence of AMC, as SAHA inhibition of KDAC6 was comparable in the presence and absence of AMC (Figure S1). Also consistent with the endpoint data, KDAC6 demonstrated intermediate affinity for the tryptophan-containing peptides in each case. The kinetic parameters for KDAC8 could not be definitively determined in all cases due to the relatively low activity of KDAC8 with some of the tested substrates (Table 1). However, it is apparent that KDAC8 does not follow a consistent trend in the same way that KDAC6 does, as the AMC-containing peptides increase or decrease kcat up to 10-fold depending on peptide sequence. Affinity does tend to increase (lower KM) with AMC-containing peptides, but to a much smaller degree than with KDAC6. Moreover, the tryptophan-containing peptides do not follow neatly in affinity between the other two substrates in each set. Similarly, the catalytic efficiency of KDAC6 is at least 4-fold greater for tryptophan-containing peptides and at least 25-fold greater for AMC-containing peptides compared to substrates with neither, while KDAC8 catalytic efficiency tends to increase in the same general trend but with greatly reduced magnitude of effect.
Table 1.
Steady state kinetic parameters for peptides with KDAC6 and KDAC8
| KDAC6 | KDAC8 | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Substrate | kcat (s-1) | KM (μM) | kcat/KM (M-1 s-1) | kcat (s-1) | KM (μM) | kcat/KM (M-1 s-1) |
| {K-ac} | 0.123 ± 0.018 | 800 ± 300 | 150 ± 30 | 0.011 ± 0.003 | 800 ± 400 | 1.4 ± 0.3 |
| {K-ac}W | 0.19 ± 0.02 | 70 ± 30 | 2500 ± 1000 | > 0.02 | > 5000 | 3.5 ± 0.6 |
| {K-ac}-AMC | 0.27 ± 0.04 | 15 ± 7 | 19000 ± 7000 | 0.07 ± 0.02 | 4200 ± 1700 | 16.3 ± 1.5 |
| RG{K-ac} | 0.32 ± 0.06 | 600 ± 200 | 530 ± 130 | 0.09 ± 0.05 | > 5000 | 4.4 ± 0.4 |
| RG{K-ac}W | 0.25 ± 0.04 | 60 ± 30 | 4300 ± 1800 | 0.041 ± 0.014 | 2700 ± 1700 | 15 ± 4 |
| RG{K-ac}-AMC | 0.40 ± 0.02 | 11 ± 3 | 36000 ± 7000 | 0.011 ± 0.003 | 500 ± 200 | 23 ± 6 |
| RH{K-ac}{K-ac} | 0.23 ± 0.03 | 370 ± 150 | 620 ± 180 | 0.10 ± 0.03 | 3100 ± 1100 | 34 ± 4 |
| RH{K-ac}{K-ac}W | 0.24 ± 0.02 | 90 ± 40 | 2600 ± 900 | 0.131 ± 0.014 | 1200 ± 300 | 106 ± 13 |
| RH{K-ac){K-ac}-AMC | 0.148 ± 0.016 | 9 ± 3 | 17000 ± 5000 | 0.34 ± 0.06 | 1300 ± 300 | 259 ± 19 |
We recognized that the interpretation of the data generated for the RH{K-ac}{K-ac} peptide series may not be straightforward because it contains two acetylated lysines, and we are unaware of any report describing the ability of KDACs to deacetylate one position versus the other for this sequence. To determine whether one or both acetyllysines were substrates for KDAC6 and/or KDAC8, we used MALDI mass spectrometry to determine how many deacetylation events were occurring on both the RH{K-ac}{K-ac}W and RH{K-ac}{K-ac}-AMC peptides. In all cases, only a singly deacetylated product appeared under conditions of lower KDAC concentration. When more enzyme was added, a doubly deacetylated product was observed in addition to the singly deacetylated product (Figure 3). Because the singly deacetylated product peak could be due to deacetylation at position 3, position 4, or a combination of two products, we further analyzed the peaks by tandem mass spectrometry. The lysine at position 4 was always deacetylated first, as the singly deacetylated products were identified as RH{K-ac}KW and RH{K-ac}K-AMC, indicating that both KDAC6 and KDAC8 demonstrate a preference for the second acetyllysine (Figure 4). Based on these observations, we can conclude that the Michaelis-Menten kinetic parameters calculated are valid, as the reaction conditions used in the kinetic experiments were far below the threshold conditions necessary to observe the second deacetylation event and the qualitative trend of the MS data confirms that percent product formation was very low during the time scale used for our initial velocity.
Figure 3. Deacetylation of doubly acetylated peptide substrates monitored by MALDI MS.
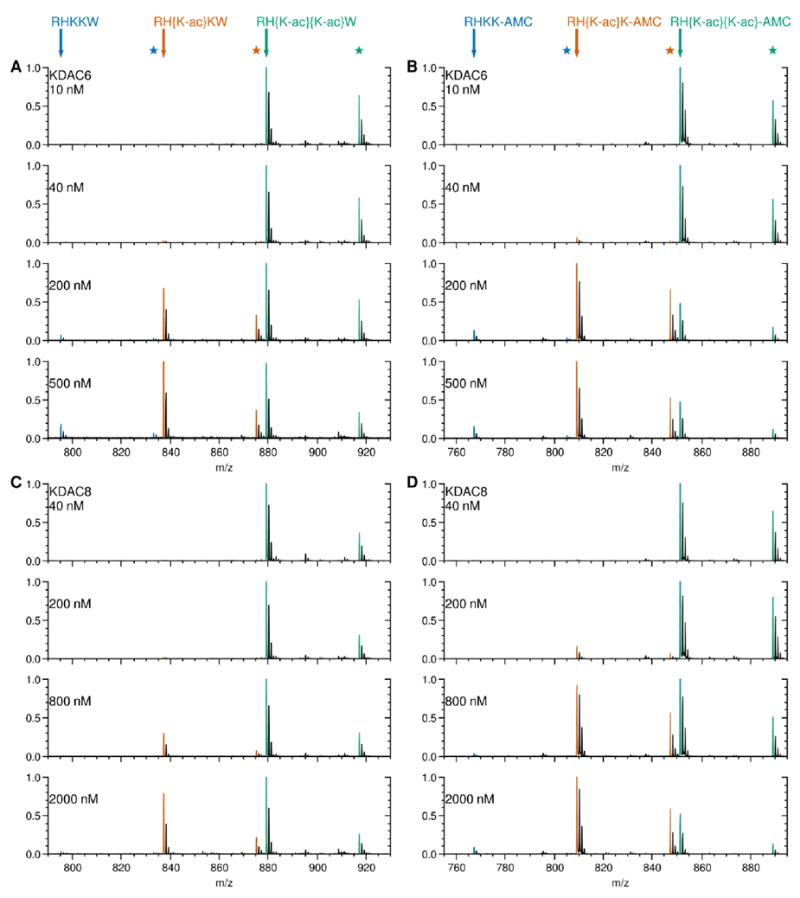
Several concentrations of KDAC were reacted with 50 μM doubly acetylated peptides in the following pairs: KDAC6 with RH{K-ac}{K-ac}W (A), KDAC6 with RH{K-ac}{K-ac}-AMC (B), KDAC8 with RH{K-ac}{K-ac}W (C), and KDAC8 with RH{K-ac}{K-ac}-AMC (D). After reactions were stopped, they were subjected to MADLI mass spectrometry as described in the methods. Peptides were detected based on m/z and identified based on theoretical masses of the M+H+ ion for each. Peaks representing the masses of the monoisotopic peptides are highlighted for the substrate (green), singly deacetylated product (orange), and doubly deacetylated product (blue). The theoretical peptide mass for each is indicated by an arrow. Stars represent the mass of the corresponding potassium adducts, observed due to the high salt concentration of the samples from the reaction buffer. For each reaction, the most intense signal was set to 1.0, and all other signals were normalized to it. The progression of the singly acetylated product matches that expected from fluorescence data. The doubly deacetylated product is present for all pairs at the highest concentration of KDAC, albeit at very low levels compared to the other peptides.
Figure 4. Sequence identification for products of doubly acetylated peptide reactions by tandem mass spectroscopy.
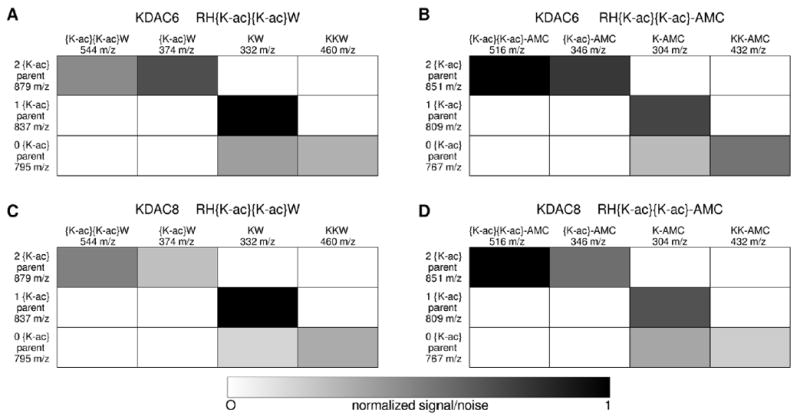
Reactions containing the highest concentration of enzyme from each of the pairs analyzed in Figure 3 were subjected to tandem mass spectrometry: KDAC6 with RH{K-ac}{K-ac}W (A), KDAC6 with RH{K-ac}{K-ac}-AMC (B), KDAC8 with RH{K-ac}{K-ac}W (C), and KDAC8 with RH{K-ac}{K-ac}-AMC (D). Substrate and product peptides (y-axis) were subjected to further fragmentation and the resulting y-ion fragments were analyzed. The signal/noise ratio for the diagnostic fragments (x-axis) are shown for each parent peptide, where white indicates a fragment that was not detected and black indicates high signal/noise ratios. All detected fragments were found to have at least 20% of the maximum signal/noise ratio, whereas all undetected fragments (white) were entirely absent. Diagnostic fragments are those that distinguish between deacetylation at lysine positions 3 and 4; other expected y-ion, a-ion, and b-ion peaks were observed but are not as useful for determining the acetylation pattern. Note that for the singly deacetylated peptide (1 {K-ac}) from each reaction pair, the diagnostic peptide fragment corresponding to deacetylation at position 4 (KW or K-AMC) was strongly detected, while the fragment corresponding to deacetylation at position 3 ({K-ac}W or {K-ac}-AMC) was always absent.
Discussion
Prior to this report, limited data suggested that conjugated fluorophores may affect KDAC6 and KDAC8 activity against model substrates.23,28,29 In one instance, AMC-based activity enhancement was previously observed for KDAC8 with the RH{K-ac}{K-ac} peptide using a mass spectrometry method.23 Our experiments demonstrate that this result is not substrate-specific, as all three sets of peptides tested here display very clear effects of AMC incorporation. For the metal-dependent KDACs tested, the presence of AMC led to a large increase in activity (Figure 2 and Table S2). These results are consistent with previously reported data for KDAC8 with another set of peptides assayed using a distinct method,28 and the values of kcat and KM we determined are consistent with available prior data for some of the AMC-containing peptides,3,38-40 with variation in kcat related to known effects of different reaction conditions that we have previously reported on.26,41 KDAC6 was previously shown to exhibit greater endpoint activity with a peptide containing C-terminal acetyllysine compared to a corresponding peptide with AMC in the C-terminal position.29 This result is not directly comparable to ours as we did not investigate any peptides with acetyllysine in the extreme C-terminal position and a free C-terminal carboxylate; however, it does reinforce that inclusion of AMC affects KDAC behavior.
Of particular interest is the fact that although KDAC6 and KDAC8 exhibited similar overall rate enhancement due to the AMC in endpoint assays, the underlying mechanism is entirely due to enhanced substrate affinity (i.e. lower KM values) in KDAC6, whereas KDAC8 demonstrated variation in both substrate affinity and catalytic ability (Table 1). We are unaware of any direct comparison of peptides with and without the AMC fluorophore for KDAC6, but prior data characterizing steady-state kinetics of KDAC6 resulted in KM values similar to those we observed, with AMC-containing peptides consistently much lower than unlabeled peptides and with relatively little variation in kcat.29,42-44 We did note a significant difference in magnitude between some previously reported kcat values for KDAC6 and those generated here, where our values were 3- to 5-fold higher, especially for the AMC-containing substrates.29,44 These differences can be attributed to the abrupt deceleration of KDAC6 that we observed, which occurs long before the substrate is significantly depleted. If the activity is calculated from a reaction which was allowed to proceed past this point, the kcat will be significantly underestimated. This also explains why our steady-state parameters predict endpoint velocities approximately 2-fold higher than actually observed for KDAC6, whereas the steady-state parameters reliably predict endpoint activity values for KDAC8. This behavior of KDAC6 must be considered when evaluating endpoint data, such as in inhibitor screens, because the assumption of constant velocity throughout the reaction over an extended period is invalid for this enzyme under typical in vitro reaction conditions. The underlying mechanism of this behavior of KDAC6 is an area of future investigation, potentially related to the fact that class IIb KDACs contain two catalytic domains.
The largest AMC-dependent activity enhancement was observed for KDAC4HY, which increased activity 100-fold in some cases compared to the non-AMC containing peptide (Figure 2C and Table S2). Notably, while KDAC4HY showed distinct preferences among the non-AMC containing peptides, this specificity was lost upon the addition of AMC, as the activity with all three of the AMC-containing peptides was very similar. The trend was opposite for Sirt1, which is not metal dependent, as Sirt1 demonstrated negligible reactivity with AMC-containing peptide derivatives, but showed clear activity with each non-AMC containing counterpart (Figure 2D and Table S2). A previous report with a related peptide also demonstrated reduced activity of Sirt1 when AMC is conjugated.32 Overall, it is clear that AMC does not affect all KDACs in the same manner, and the activity of each KDAC is uniquely impacted.
While limited observations have previously suggested that conjugated dyes might enhance KDAC activity with peptide substrates, AMC and other dye conjugated substrates continue to be widely used to study KDACs, including substrate specificity and small molecule inhibitors and activators. This report directly demonstrates that AMC affects activity of KDACs of all classes in a manner independent of the substrate sequence. Together, the results presented here call into question the conclusions drawn from experiments using AMC-modified substrates. Certainly, KDAC activity with an AMC-conjugated substrate does not necessarily correlate to activity with a corresponding natural substrate, even for AMC-conjugated peptides derived from known substrates such as those utilized here, as the KDACs interact very differently with peptides in the presence of the dye. We have previously demonstrated that libraries of AMC-containing peptides did not reliably predict activity of KDAC8 with unlabeled substrates,26 and the results presented here indicate that the lack of correlation between labeled and unlabeled substrates applies to all KDACs. Therefore, profiling of KDAC activity with labeled peptides is unlikely to produce biologically relevant data.
In addition, a prior study suggested that the dye may have a differential effect on KDACs depending upon whether they are in a complex with other proteins. Specifically, KDAC3-SMRT activity against the RH{K-ac}{K-ac} peptide was not affected by the presence or absence of the AMC dye; however, KDAC3 alone demonstrated activity with another AMC-containing peptide but did not show activity with any non-dye containing peptides in a screen.23 This result is significant because in vivo KDAC3 exists in a complex containing SMRT, which has been reported to be essential for KDAC3’s activity,45,46 suggesting that the dye enhances activity in a manner that is not biologically relevant. All of these observations indicate that the effects of the dye are not consistent, neither in magnitude nor direction of effect, so it would not be possible to predict the effect of the dye for a particular substrate-KDAC pair.
While this report focused on the commercially-available AMC-containing substrates, several other fluorogenic substrates have been recently developed including those for removal of other acyl modifications by sirtuins.47-49 For example, “green” substrates based on fluorescien are now available and should also be used with caution, as these substrates utilize a larger fluorophore and are often dual substrates conjugated to a single fluorophore, and therefore deviate more than AMC-containing substrates from a biological context. Other recently developed substrates allow deacetylation to be monitored in a single-step.24,50,51 For some of these substrates, the dyes are positioned further away from the acetylated lysine than for the AMC substrates;52,53 however, it will be important to determine how the configuration of these substrates affects KDAC activity. An assumption that AMC or other fluorophores might mimic large non-polar residues after the acetyllysine is clearly incorrect, as tryptophan in the +1 position causes distinctly different activity effects than AMC. Although fluorophore-conjugated substrates are convenient, the recent development of several alternative high-throughput assays utilizing fluorescence or mass spectrometry enables the use of unlabeled substrates for nearly all applications.26,28,54,55 Use of unlabeled peptides also ensures that acetyllysine preference in multiply-acetylated substrates is not skewed by use of the fluorophore. Although we did not observe any significant change in acetyllysine preference due to AMC-conjugation in the single peptide tested with KDAC6 and KDAC8 (Figures 3 and 4), such potential effects should be considered. Utilizing mass spectrometry to monitor deacetylation for multiply-acetylated substrates can definitively demonstrate whether one or more acetylated lysines are being targeted by the deacetylase. Protein acetylation frequently occurs on adjacent or nearby lysine residues (e.g., histones), and so this distinction is important both for understanding substrate preference and determining kinetic parameters.
While the data presented here did not directly address whether conjugated fluorophores modulate the effects of small molecules on KDAC activity, the possibility must be considered. Consistent with our data, AMC was previously found to decrease affinity of Sirt1 for a peptide substrate.30 Interestingly, this decreased affinity was alleviated by resveratrol through a conformational change, and resveratrol had no effect on the activity of Sirt1 against the same peptide without AMC.30,31,56 Because the fluorophore specific nature of the activation was discovered well after the initial observation, several “activators” of Sirt1 were reported, leading to the proposal of an erroneous mechanism to explain how the sirtuins extend lifespan in model organisms.17 Subsequent work examining the activation of sirtuins has concluded that activating molecules all function allosterically and are substrate-specific, and therefore AMC-conjugated peptides are not reliable tools for screening of potential activators.30-33,57 Additionally, N-acetylthioureas that were reported to activate KDAC8 were later found to merely restore basal activity levels for enzymes that were functioning well below expected levels,18,41 suggesting that in vitro “activation” of KDACs should be treated with skepticism regarding any biological relevance when identified using AMC-conjugated substrates. These results may also shed light on the observation that most KDAC inhibitors identified in vitro do not have the same effect in cells, as many of the screens identifying novel inhibitors utilize AMC-containing peptides. Because AMC changes substrate affinity and/or catalytic rate, inhibitors that bind non-competitively (i.e., allosterically) may be especially affected by conjugation of these molecules to model substrates, as allosteric interactions are most likely to be sensitive to substrate-specific events due to the concurrent binding of substrate and modulator. Our observations directly demonstrate that KDACs react differently and unpredictably with AMC-containing model substrates compared to more biologically relevant substrates. Thus, substrate-specific artifacts must be carefully considered and explicitly controlled for when identifying small molecule modulators of KDAC activity. When possible, it would be advantageous to utilize KDAC assays that do not rely on fluorophore-conjugated substrates.
Supplementary Material
Acknowledgments
We thank Dr. Florastina Payton-Stewart and Dr. Subramanya Pingali for assistance with obtaining NMR spectra, Dr. Melyssa R. Bratton and Elena Y. Glotser for expressing KDAC6 and KDAC8 in insect cells, and Asia N. Matthew for cloning of KDAC4HY.
Funding
This material is based upon work supported by, or in part by, the U. S. Army Research Laboratory and the U. S. Army Research Office under contract/grant number W911NF1310129. This publication was also made possible in part by support from the Louisiana Cancer Research Consortium, the NIH-RCMI grant #5G12MD007595 from the National Institute of Minority Health and Health Disparities, awards #TL4GM118968 and #UL1GM118967 from the National Institute of General Medical Sciences, and the National Science Foundation grant CHE 1625993. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the Louisiana Cancer Research Consortium, the National Institutes of Health, or the National Science Foundation.
Abbreviations
- AMC
7-amino-4-methylcoumarin
- DMSO
dimethyl sulfoxide
- GST
glutathione S-transferase
- KDAC
lysine deacetylase
- MALDI
matrix assisted laser desorption/ionization
- MOPS
3-(N-morpholino)propanesulfonic acid
- MS
mass spectrometry
- NAD+
nicotinamide adenine dinucleotide
- NMR
nuclear magnetic resonance
- SAHA
suberoylanilide hydroxamic acid
- Sirt
sirtuin
- TEV
tobacco etch virus
- TOF
time-of-flight
Footnotes
NMR peaks for {K-ac} (Table S1), endpoint specific activity values (Table S2), and SAHA titrations (Figure S1). The Supporting Information is available free of charge on the ACS Publications website at DOI:
Notes
The authors declare no competing financial interest.
References
- 1.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 2.Chen K, Zhang X, Wu Y-D, Wiest O. Inhibition and mechanism of HDAC8 revisited. J Am Chem Soc. 2014;136:11636–11643. doi: 10.1021/ja501548p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gantt SL, Gattis SG, Fierke CA. Catalytic activity and inhibition of human histone deacetylase 8 is dependent on the identity of the active site metal ion. Biochemistry. 2006;45:6170–6178. doi: 10.1021/bi060212u. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y-L, Yang W-M. Beyond histone and deacetylase: an overview of cytoplasmic histone deacetylases and their nonhistone substrates. J Biomed Biotechnol. 2011;2011:146493. doi: 10.1155/2011/146493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci. 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang X-J, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, Allis CD, Hake SB. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci U S A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan K-L. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, Olsen JV. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012;2:419–431. doi: 10.1016/j.celrep.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schölz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, Lain S, Cox J, Matthias P, Mann M, Bradner JE, Choudhary C. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 12.Bheda P, Jing H, Wolberger C, Lin H. The substrate specificity of sirtuins. Annu Rev Biochem. 2016;85:405–429. doi: 10.1146/annurev-biochem-060815-014537. [DOI] [PubMed] [Google Scholar]
- 13.López JE, Sullivan ED, Fierke CA. Metal-dependent deacetylases: cancer and epigenetic regulators. ACS Chem Biol. 2016;11:706–716. doi: 10.1021/acschembio.5b01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra M. Class IIa HDACs - new insights into their functions in physiology and pathology. FEBS J. 2015;282:1736–1744. doi: 10.1111/febs.13061. [DOI] [PubMed] [Google Scholar]
- 15.Wolfson NA, Ann Pitcairn C, Fierke CA. HDAC8 substrates: histones and beyond. Biopolymers. 2013;99:112–126. doi: 10.1002/bip.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murugan K, Sangeetha S, Ranjitha S, Vimala A, Al-Sohaibani S, Rameshkumar G. HDACiDB: a database for histone deacetylase inhibitors. Drug Des Devel Ther. 2015;9:2257–2264. doi: 10.2147/DDDT.S78276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang L-L, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 18.Singh RK, Mandal T, Balsubramanian N, Viaene T, Leedahl T, Sule N, Cook G, Srivastava DK. Histone deacetylase activators: N-acetylthioureas serve as highly potent and isozyme selective activators for human histone deacetylase-8 on a fluorescent substrate. Bioorg Med Chem Lett. 2011;21:5920–5923. doi: 10.1016/j.bmcl.2011.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson DE, Udeshi ND, Wolfson NA, Pitcairn CA, Sullivan ED, Jaffe JD, Svinkina T, Natoli T, Lu X, Paulk J, McCarren P, Wagner FF, Barker D, Howe E, Lazzaro F, Gale JP, Zhang Y-L, Subramanian A, Fierke CA, Carr SA, Holson EB. An unbiased approach to identify endogenous substrates of “histone” deacetylase 8. ACS Chem Biol. 2014;2014:2210–2216. doi: 10.1021/cb500492r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegener D, Wirsching F, Riester D, Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem Biol. 2003;10:61–68. doi: 10.1016/s1074-5521(02)00305-8. [DOI] [PubMed] [Google Scholar]
- 21.Gurard-Levin ZA, Kilian KA, Kim J, Bahr K, Mrksich M. Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem Biol. 2010;5:863–873. doi: 10.1021/cb100088g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riester D, Hildmann C, Grunewald S, Beckers T, Schwienhorst A. Factors affecting the substrate specificity of histone deacetylases. Biochem Biophys Res Commun. 2007;357:439–445. doi: 10.1016/j.bbrc.2007.03.158. [DOI] [PubMed] [Google Scholar]
- 23.Gurard-Levin ZA, Kim J, Mrksich M. Combining mass spectrometry and peptide arrays to profile the specificities of histone deacetylases. Chembiochem. 2009;10:2159–2161. doi: 10.1002/cbic.200900417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minoshima M, Kikuchi K. Chemical tools for probing histone deacetylase (HDAC) activity. Anal Sci. 2015;31:287–292. doi: 10.2116/analsci.31.287. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Liu T, Liao S, Li Y, Lan Y, Wang A, Wang Y, He B. A mini-review on sirtuin activity assays. Biochem Biophys Res Commun. 2015;467:459–466. doi: 10.1016/j.bbrc.2015.09.172. [DOI] [PubMed] [Google Scholar]
- 26.Toro TB, Watt TJ. KDAC8 substrate specificity quantified by a biologically-relevant, label-free deacetylation assay. Protein Sci. 2015;24:2020–2032. doi: 10.1002/pro.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Udenfriend S, Stein S, Bohlen P, Dairman W, Leimgruber W, Weigele M. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science. 1972;178:871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 28.Wolfson NA, Pitcairn CA, Sullivan ED, Joseph CG, Fierke CA. An enzyme-coupled assay measuring acetate production for profiling histone deacetylase specificity. Anal Biochem. 2014;456:61–69. doi: 10.1016/j.ab.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, Napper A, Curtis R, DiStefano PS, Fields S, Bedalov A, Kennedy BK. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 32.Dai H, Kustigian L, Carney D, Case A, Considine T, Hubbard BP, Perni RB, Riera TV, Szczepankiewicz B, Vlasuk GP, Stein RL. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, Pentelute BL, Schuman ER, Stevens LA, Ling AJY, Armour SM, Michan S, Zhao H, Jiang Y, Sweitzer SM, Blum CA, Disch JS, Ng PY, Howitz KT, Rolo AP, Hamuro Y, Moss J, Perni RB, Ellis JL, Vlasuk GP, Sinclair DA. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciccarone VC, Polayes DA, Luckow VA. Molecular Diagnosis of Infectious Diseases. Humana Press; New Jersey: 1997. Generation of recombinant baculovirus DNA in E. coli using a baculovirus shuttle vector; pp. 213–236. [DOI] [PubMed] [Google Scholar]
- 35.Hawley-Nelson P, Ciccarone V, Moore ML. Transfection of cultured eukaryotic cells using cationic lipid reagents. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2008. [DOI] [PubMed] [Google Scholar]
- 36.Fischle W, Emiliani S, Hendzel MJ, Nagase T, Nomura N, Voelter W, Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- 37.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc Natl Acad Sci U S A. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh RK, Mandal T, Balasubramanian N, Cook G, Srivastava DK. Coumarin-suberoylanilide hydroxamic acid as a fluorescent probe for determining binding affinities and off-rates of histone deacetylase inhibitors. Anal Biochem. 2011;408:309–315. doi: 10.1016/j.ab.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porter NJ, Christianson NH, Decroos C, Christianson DW. Structural and functional influence of the glycine-rich loop G(302)GGGY on the catalytic tyrosine of histone deacetylase 8. Biochemistry. 2016;55:6718–6729. doi: 10.1021/acs.biochem.6b01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toro TB, Pingali S, Nguyen TP, Garrett DS, Dodson KA, Nichols KA, Haynes RA, Payton-Stewart F, Watt TJ. KDAC8 with high basal velocity is not activated by N-acetylthioureas. PloS One. 2016;11:e0146900. doi: 10.1371/journal.pone.0146900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–91. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- 43.Zou H, Wu Y, Navre M, Sang B-C. Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun. 2006;341:45–50. doi: 10.1016/j.bbrc.2005.12.144. [DOI] [PubMed] [Google Scholar]
- 44.Miyake Y, Keusch JJ, Wang L, Saito M, Hess D, Wang X, Melancon BJ, Helquist P, Gut H, Matthias P. Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat Chem Biol. 2016;12:748–754. doi: 10.1038/nchembio.2140. [DOI] [PubMed] [Google Scholar]
- 45.Li J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You S-H, Lim H-W, Sun Z, Broache M, Won K-J, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba R, Hori Y, Kikuchi K. Intramolecular long-distance nucleophilic reactions as a rapid fluorogenic switch applicable to the detection of enzymatic activity. Chem. 2015;21:4695–4702. doi: 10.1002/chem.201406093. [DOI] [PubMed] [Google Scholar]
- 48.Chiang Y-L, Lin H. An improved fluorogenic assay for SIRT1, SIRT2, and SIRT3. Org Biomol Chem. 2016;14:2186–2190. doi: 10.1039/c5ob02609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galleano I, Schiedel M, Jung M, Madsen AS, Olsen CA. A continuous, fluorogenic sirtuin 2 deacylase assay: substrate screening and inhibitor evaluation. J Med Chem. 2016;59:1021–1031. doi: 10.1021/acs.jmedchem.5b01532. [DOI] [PubMed] [Google Scholar]
- 50.Xie Y, Ge J, Lei H, Peng B, Zhang H, Wang D, Pan S, Chen G, Chen L, Wang Y, Hao Q, Yao SQ, Sun H. Fluorescent probes for single-step detection and proteomic profiling of histone deacetylases. J Am Chem Soc. 2016;138:15596–15604. doi: 10.1021/jacs.6b07334. [DOI] [PubMed] [Google Scholar]
- 51.Schuster S, Roessler C, Meleshin M, Zimmermann P, Simic Z, Kambach C, Schiene-Fischer C, Steegborn C, Hottiger MO, Schutkowski M. A continuous sirtuin activity assay without any coupling to enzymatic or chemical reactions. Sci Rep. 2016;6:22643. doi: 10.1038/srep22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baba R, Hori Y, Mizukami S, Kikuchi K. Development of a fluorogenic probe with a transesterification switch for detection of histone deacetylase activity. J Am Chem Soc. 2012;134:14310–14313. doi: 10.1021/ja306045j. [DOI] [PubMed] [Google Scholar]
- 53.Minoshima M, Matsumoto T, Kikuchi K. Development of a fluorogenic probe based on a DNA staining dye for continuous monitoring of the histone deacetylase reaction. Anal Chem. 2014;86:7925–7930. doi: 10.1021/ac501881s. [DOI] [PubMed] [Google Scholar]
- 54.Hubbard BP, Sinclair DA. Measurement of sirtuin enzyme activity using a substrate-agnostic fluorometric nicotinamide assay. Methods Mol Biol. 2013;1077:167–177. doi: 10.1007/978-1-62703-637-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun S, Buer BC, Marsh ENG, Kennedy RT. A label-free sirtuin 1 assay based on droplet-electrospray ionization mass spectrometry. Anal Methods Adv Methods Appl. 2016;8:3458–3465. doi: 10.1039/C6AY00698A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao D, Wang M, Qiu X, Liu D, Jiang H, Yang N, Xu R-M. Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol. Genes Dev. 2015;29:1316–1325. doi: 10.1101/gad.265462.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittenhafer-Reed KE, Feldman JL, Denu JM. Catalysis and mechanistic insights into sirtuin activation. Chembiochem. 2011;12:281–289. doi: 10.1002/cbic.201000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


