Abstract
Purpose
The aim of this study was to identify the molecular genetic basis of cone-rod dystrophy in 18 unrelated families of Polish origin. Cone-rod dystrophy is one of the inherited retinal dystrophies, which constitute a highly heterogeneous group of disorders characterized by progressive dysfunction of photoreceptors and retinal pigment epithelium (RPE) cells.
Methods
The study group was composed of four groups of patients representing different Mendelian inheritance of the disease: autosomal dominant (AD), autosomal recessive (AR), X-linked recessive (XL), and autosomal recessive or X-linked recessive (AR/XL). The combined molecular strategy included Sanger sequencing of the RPGR-ORF15 gene (three families with XL and three families with the AR/XL mode of inheritance), mutation-specific microarray analysis of the ABCA4 gene (five families with the AR mode of inheritance and two families with the AR/XL mode of inheritance), targeted next-generation sequencing (NGS) of inherited retinal disease–associated (IRD) genes (seven families with the AD mode of inheritance and five families with the AR mode of inheritance), and whole exome sequencing, performed in select families who had been mutation-negative in the analysis with the targeted NGS panel (one family with the AD mode of inheritance, one family with the AR mode of inheritance, and two families with the AR/XL mode of inheritance).
Results
Based on this combined strategy, we managed to identify potentially causative variants in seven out of 18 families with CRD. Five of these variants are novel: c.3142_3143dupAA, p.(Glu1049Argfs*41) in the RPGR-ORF15 gene, two variants: c.1612delT, p.(Trp538Glyfs*15) and c.2389dupG, p.(Ile798Hisfs*20) in the PROM1 gene in one family, c.592A>C, p.(Ser198Arg) in the PRPH2 gene and the variant c.1691A>G, p.(Asp564Gly) in the ATF6 gene that we have already reported to be pathogenic. NGS on the IRD panel allowed the molecular basis of CRD to be identified in four out of 14 families with a total detection rate of 38%. WES allowed identification of the molecular genetic basis of CRD in one family.
Conclusions
This is the first report on the spectrum of disease genes and pathogenic variants causing CRD in the Polish population. The study presents five novel variants identified in four genes and therefore, broadens the spectrum of probable pathogenic variants associated with CRD.
Introduction
Inherited retinal dystrophies (IRDs) are a highly heterogeneous group of disorders characterized by progressive dysfunction of photoreceptors and RPE cells. They represent the major cause of familial blindness, with more than 2 million people affected worldwide [1]. In general, IRDs are classified based on the type of retinal cells (cones or rods) that are primarily affected, the age of onset, and the progression of degeneration [2].
Cone-rod dystrophy (CRD; OMIM 120970) and cone dystrophy (CD; OMIM 602093) are progressive forms of IRDs with a prevalence ranging from 1/30,000 to 1/40,000 [3]. Whereas CRD is characterized by the progressive loss of cone function, followed by loss of rod photoreceptor function, in CD cone function decreases progressively, while rod function is normal until the late stages of the disorder. The main symptoms of CRD are decreased visual acuity, color vision defects, and photophobia, sometimes followed by the progressive loss of peripheral vision and night blindness [3-5]. Perifoveal atrophy of the outer retina and a “bull’s eye” appearance of the retina are typically observed on fundus examination and optical coherence tomography (OCT) in patients with CRD [6].
Cone-rod dystrophies are genetically heterogeneous with all modes of inheritance documented. To date, ten genes for the autosomal dominant forms of CRD/CD are known: AIPL1 (ID: 23746, OMIM: 604392), CRX (ID: 1406, OMIM: 602225), GUCA1A (ID: 2978, OMIM: 600364), GUCY2D (ID: 3000, OMIM: 600179), PITPNM3 (ID: 83394, OMIM: 608921), PROM1 (ID: 8842, OMIM: 604365), PRPH2 (ID: 5961, OMIM: 179605), RIMS1 (ID: 22999, OMIM: 606629) SEMA4A (ID: 64218, OMIM: 607292), UNC119 (ID: 9094, OMIM: 604011). Twenty two genes are known to be implicated in the autosomal recessive forms: ABCA4 (ID: 24, OMIM: 601691), ADAM9 (ID: 8754, OMIM: 602713), ATF6 (ID: 22926, OMIM: 605537), C21ORF2 (ID: 755, OMIM: 603191), C8ORF37 (ID: 157657, OMIM: 614477), CACNA2D4 (ID: 93589 OMIM: 608171), CDHR1 (ID: 92211, OMIM: 609502), CERKL (ID: 375298, OMIM: 608381) CNGA3 (ID: 1261, OMIM: 600053), CNGB3 (ID: 54714, OMIM: 605080), CNNM4 (ID: 26504, OMIM: 607805), GNAT2 (ID: 2780, OMIM: 139340), IFT81 (ID: 28981, OMIM: 605489), KCNV2 (ID: 169522, OMIM: 607604), PDE6C (ID: 5146, OMIM: 600827), PDE6H (ID: 5149, OMIM: 601190), POC1B (ID: 282809, OMIM: 614784), RAB28 (ID: 9364, OMIM: 612994), RAX2 (ID: 84839, OMIM: 610362), RDH5 (ID: 5959, OMIM: 601617) RPGRIP1 (ID: 57096 OMIM: 605446), TTLL5 (ID: 23093, OMIM: 612268). The X-linked form of CRD is associated with mutations in RPGR (ID: 6103, OMIM: 312610) and CACNA1F (ID: 778, OMIM: 300110; RetNet).
Most of the previously reported molecular analyses of patients affected with CRD were based on targeted Sanger sequencing of selected genes. This approach yields a low detection rate that is further confounded due to the clinical and genetic overlap between CRD and other retinal dystrophies. The detection rate (or diagnostic yield) in CRD has increased since the introduction of next-generation sequencing (NGS) technologies, including the targeted capture of known disease genes (“disease panels”), whole exome sequencing (WES), and whole genome sequencing (WGS). Apart from searching for new mutations in the known genes, WES and WGS give the opportunity to discover novel genes associated with CRD [7]. It has recently been discovered that some unsolved cases with IRD can be attributed to copy number variations (CNVs) and noncoding variants. Moreover, some IRD genes appeared to be CNV-prone [8,9]. NGS-based technologies, especially WGS, but also disease panels and WES, can be applied as a useful tool for detecting CNVs [10-12]. The aim of this study was to determine the underlying genetic defect of the disease in 18 Polish families with CRD (37 patients).
Methods
Selection of patients
A total of 37 patients from 18 families of Polish origin were recruited for this study, having previously been clinically diagnosed with CRD based on the following criteria: early childhood decrease of visual acuity, photophobia, decreasing scotopic electroretinogram (ERG) amplitudes, color vision defects, and progressive loss of central vision (central scotoma). All patients had a positive family history compatible with autosomal dominant inheritance (AD, seven families), autosomal recessive inheritance (AR, five families), X-linked recessive inheritance (XL, three families), or either autosomal recessive or X-linked recessive inheritance (AR/XL, three families).
The study complied with the tenets of the Declaration of Helsinki and the Association for Research in Vision and Ophthalmology (ARVO) statement on human subjects, it was also approved by the Poznan University of Medical Sciences Institutional Review Board. Written informed consent was obtained from all participants or their legal guardians.
Molecular genetic analysis
Venous blood was collected from all index cases, as well as from affected and unaffected available family members (the total number of samples analyzed was 75). Genomic DNA was isolated from leukocytes according to the conventional salting-out procedure [13].
A total of six families, including three families with X-linked recessive inheritance and three families with either autosomal recessive or X-linked recessive inheritance, were screened for mutations in the RPGR-ORF15 (NM_001331041) gene using Sanger sequencing (Asper Biotech, Tartu, Estonia).
APEX-arrayed primer extension for ABCA4 (NM_000350) gene variants was performed in five families (Families 3, 5, 6, 12, and 17) with an AR mode of inheritance and in two families (Families 11 and 15) with either the AR or XL mode of inheritance (Asper Biotech). The version of the APEX array applied was designed to cover 558 variants in the ABCA4 gene.
A patient from Family 3 carrying a single heterozygous variant in the ABCA4 gene was screened further for possible structural rearrangements using multiple ligation probe analysis (MLPA; SALSA MLPA P151 ABCA4 mix-1 probemix and SALSA MLPA P152 ABCA4 mix-2 probemix) according to the manufacturer’s instructions (MRC-Holland, Amsterdam, Netherlands). Sanger sequencing for the most common deep intronic variants V1-V7 in the ABCA4 gene (V1: c.5196+1137G>A, V2: c.5196+1216C>A, V3: c.5196+1056A>G, V4: 4539+2001G>A, V5: 4539+2028C>T, V6: 6342G>A, V7: 4773+3A>G) which may affect the correct splicing was performed as described earlier [14]. The primer sequences are available upon request.
PCR products were purified using ExoSAP-IT (Exonuclease I and Shrimp Alkaline Phosphatase Cleanup for PCR products, Affymetrix, ThermoFisher) and directly sequenced using Dye Terminator chemistry (v3.1 BigDye®Terminator, Life Technologies). For verification of variants identified by targeted NGS and WES, as well as for amplification of intronic variants V1-V7 in the ABCA4 gene the following PCR conditions were applied: 95 °C, 3 min (preliminary denaturation); 40 cycles of denaturation at 94 °C for 30 s, annealing for 30 s with temperature starting from 63 °C, decreasing to 55 °C (touchdown PCR −0.2 °C per cycle), elongation at 72 °C for 45 s; and the final synthesis at 72 °C, 10 min. PCR was run in a Mastercycler pro thermocycler (Eppendorf, Hamburg, Germany) using HiFiTaq polymerase (Novazym, Poznan, Poland)The sequencing products were separated on an ABI 3130xl capillary sequencer Genetic Analyzer (Applied Biosystems).
Seven families with the AD mode of inheritance, five families with the AR mode of inheritance, and two families with either the AR or X-linked recessive (one patient from each family) mode of inheritance were analyzed using a targeted NGS diagnostic panel for eye diseases. Targeted NGS of retinal disease–associated genes was performed at CeGaT (Center for Genomics and Transcriptomics, Tuebingen, Germany). A capture panel of 105 inherited retinal disease–associated genes (IRD panel) was used.
Details of the panel design, library preparation, and capture sequencing have been published [15]. Visualization of the CNV variants was performed using the Integrative Genomics Viewer (IGV) software [16]. All putative disease-causing variants were validated with Sanger sequencing. The sequence variants identified were then cross-checked to the Human Gene Mutation Database (HGMD), the Exome Variant Server (NHLBI Exome Sequencing Project ESP), the 1000 Genomes Project database (1000 Genomes Project Consortium 2012) and gnomAD (genome Aggregation Database). In silico analysis using Sorting Intolerant from Tolerant (SIFT), PolyPhen-2, Panther, and MutationTaster software was performed to predict the possible effect of the novel missense mutations. NetGene2 and ESE software packages were used to predict the outcome of splice-site mutations. Novel variants identified in this study were classified according to American College of Medical Genetics and Genomics (ACMG) guidelines [17]. Segregation analysis for the presence and independent inheritance of altered alleles was performed in the families with potentially causative variants in PROM1 (NM_006017), CNGB3 (NM_019098), PRPH2 (NM_000322), and GUCA1A (NM_001319061) genes with Sanger sequencing of the appropriate exons.
Due to financial constraints, and based on patient consent, only four families who lacked disease-causing mutations after the IRD panel analysis were selected for WES. A duo-based WES (two affected family members) was performed in one family with the AD mode of inheritance (Family 4). Exomes were enriched using the SureSelect XT Human All Exon 50 Mb kit, version 4 or 5 (Agilent Technologies, Santa Clara, CA), and sequenced on the HiSeq 2500 system (Illumina, San Diego, CA). Sequencing data were analyzed as previously described [7]. ExomeDepth version 1.0.0 was used for CNV calling [18]. WES was also conducted in one family with the AR mode of inheritance (Family 17) and two families with the AR/XL mode of inheritance (Families 11 and 15), but in contrast to the family with the AD mode of inheritance, only one affected patient was analyzed per family. WES in families with AR and AR/XL CRD was performed using the Nextera Rapid Capture Exome Kit (Illumina) and paired-end sequencing (2 × 100 bp) on an Illumina HiSeq 1500 instrument. Sequencing data were analyzed according to the previously described procedure in agreement with the Broad Institute recommendations [19]. CNVkit was used to search for CNVs in whole exome Illumina data; 42 reference samples had been preanalyzed for statistics of target (exome) bins of size 300 bp (or single exon size if shorter) and of antitarget bins of size up to 100,000 bp [20]. Each sample alignment .bam file, hg19 realigned, was analyzed with CNVkit using the CBS segmentation algorithm with the p value threshold set to 0.0001 (default) [21,22]. CNV segments were reported if their real-value copy number computed from the log2 ratio was 0.2 higher or lower than neutral. Usually, the segments were subjected to further analysis only if the difference was greater than 0.5.
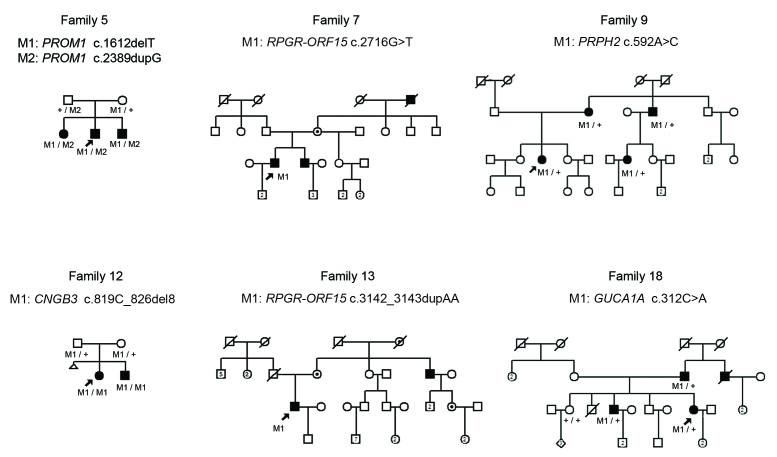
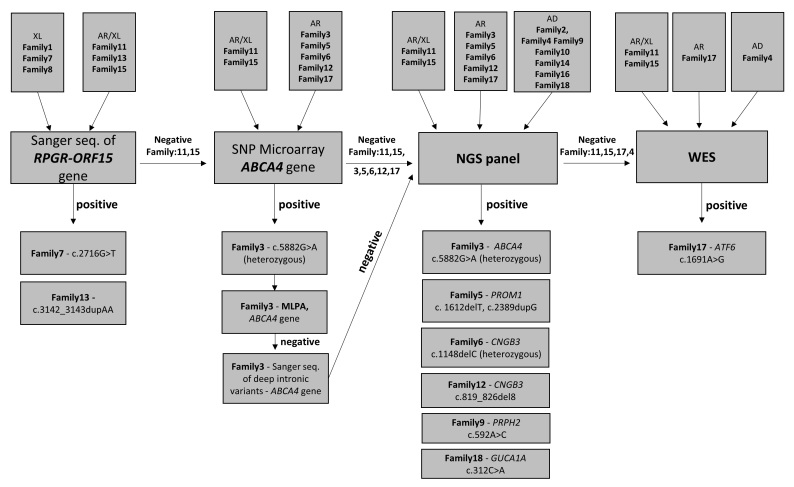
Candidate variants were validated and tested for cosegregation within a family using Sanger sequencing. The pedigrees of the families with CRD are presented in Figure 1 and Appendix 1. The diagnostic strategy used in patients with CRD is presented in Figure 2.
Figure 1.
Pedigrees and genotyping results of families with CRD whose genetic background of the disease has been identified. The genotypes are provided for all subjects available for molecular genetic analysis. Family number and disease-causing variant(s) are noted above each pedigree. Wild-type variants are indicated with +, while disease-causing variant(s) are indicated with M1 and M2.
Figure 2.
Diagnostic strategy used in patients with CRD.
Results
Clinical features
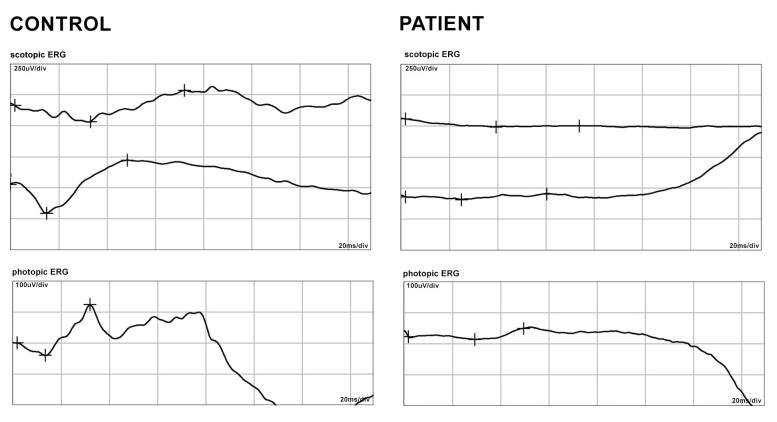
All patients showed typical CRD signs and symptoms: photophobia, decreased visual acuity, and color vision defects. The flash ERG scotopic and photopic responses of a typical patient are shown in Figure 3. Ophthalmological findings of the indexes are summarized in Table 1.
Figure 3.
ERG data from a patient with CRD (Family 9). The left panel shows the electroretinograms (ERGs) of a normal healthy individual; the right panel shows the ERGs of the patient, which show the extinguished rod and cone response.
Table 1. Ophthalmological findings of CRD patients.
| Family | Gender | Age (yrs) | Inheritance | BCVA right-left | Fundus/AF | Color Vision disturbances | Nyctalopia | Nystagmus | Photophobia | Perimetry | ERG |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
M |
29 |
XL |
0.1–0.1 |
pale optic disc |
+ |
- |
+ |
+ |
ND |
photopic and scotopic diminished |
| 2 |
F |
53 |
AD |
0.02- 0.02 |
normal |
+ |
- |
- |
- |
concentric narrowed |
ND |
| 3 |
F |
33 |
AR |
0.2–0.3 |
bull's eye maculopathy |
+ |
- |
- |
+ |
normal |
photopic diminished, scotopic slightly diminished |
| 4 |
M |
27 |
AD |
0.2–0.2 |
bilateral mild changes |
+ |
- |
- |
+ |
visual field deficit |
photopic and scotopic diminished |
| 5 |
M |
17 |
AR |
0.4- 0.4 |
bilateral macular granular appearance |
+ |
+ |
- |
+ |
concentric narrowed |
photopic extinguished, scotopic 50% |
| 6 |
F |
16 |
AR |
0.1–0.1 |
ND |
+ |
- |
+ |
+ |
ND |
photopic 15%, scotopic 5% |
| 7 |
M |
50 |
XL |
0.05- 0.1 |
bull's eye maculopathy |
+ |
- |
- |
+ |
central scotoma |
photopic extinguished, scotopic diminished |
| 8 |
M |
24 |
XL |
0.1–0.1 |
normal |
- |
- |
+ |
+ |
ND |
photopic and scotopic diminished |
| 9 |
F |
36 |
AD |
0.3–0.3 |
bull’s eye maculopathy |
+ |
+ |
- |
- |
irregular scotomas |
photopic and scotopic diminished |
| 10 |
F |
35 |
AD |
0.1- 0.1 |
bilateral pale optic disc |
+ |
- |
+ |
+ |
ND |
photopic and scotopic extinguished |
| 11 |
M |
43 |
AR,XL |
0.06- 0.1 |
loss of macular reflex/ RPE atrophy |
+ |
- |
+ |
+ |
visual field deficit, mostly peripheral |
ND |
| 12 |
F |
20 |
AR |
0.2–0.3 |
normal |
+ |
- |
+ |
+ |
ND |
photopic diminished, scotopic 50% |
| 13 |
M |
38 |
AR,XL |
0.6- 0.9 |
bull's eye maculopathy, narrowing of the retinal blood vessels |
+ |
+ |
- |
+ |
irregular scotomas |
photopic and scotopic diminished |
| 14 |
F |
13 |
AD |
0.1- 0.1 |
loss of macular reflex, pale optic disc |
- |
+ |
+ |
- |
ND |
photopic and scotopic diminished |
| 15 |
M |
20 |
AR,XL |
0.2- 0.2 |
macula with reflex, normal optic disc and blood vessels |
- |
- |
- |
- |
normal |
photopic residual, scotopic subnormal |
| 16 |
F |
45 |
AD |
0.1- 0.1 |
normal/ RPE atrophy, macular granular appearance |
+ |
+ |
+ |
+ |
concentric narrowed (5°) |
photopic and scotopic diminished |
| 17 |
M |
9 |
AR |
0.1–0.2 |
loss of macular reflex, foveal hypoplasia |
+/− |
- |
- |
+ |
ND |
extinguished photopic and diminished scotopic |
| 18 | F | 34 | AD | 0.1- 0.1 | bilateral macular granular appearance | + | - | - | + | central irregular scotomas | photopic diminished, scotopic 50%–80% |
ND – no data. Grey color indicates the first symptom reported by the patient
Molecular genetic findings
Patients with the AD mode of inheritance
Molecular results for the patients with CRD are summarized in Table 2. The IRD panel was performed in seven families with the AD mode of inheritance.
Table 2. Variants identified in CRD patients together with the analyses performed.
| Family no. |
Analysis |
Gene | Variant classification | Genotype | Reference | |||
|---|---|---|---|---|---|---|---|---|
| RPGR-ORF15 sequencing | SNP microarray of ABCA4 | NGS RD panel | WES | |||||
|
Dominant inheritance | ||||||||
| 9 |
- |
- |
+ |
- |
PRPH2 |
c.592A>C, p.(Ser198Arg; pathogenic II) |
heterozygous |
[a] |
| 18 |
- |
- |
+ |
- |
GUCA1A |
c.312C>A, p.(Asn104Lys) |
heterozygous |
Jiang et al., 2008 |
|
Recessive inheritance | ||||||||
| 5 |
- |
+ |
+ |
- |
PROM1 |
c.1612delT, p.(Trp538Glyfs*15; pathogenic Ia)
c.2389dupG, p.(Ile798Hisfs*20; pathogenic Ia) |
heterozygous
heterozygous |
[a]
[a] |
| 12 |
- |
+ |
+ |
- |
CNGB3 |
c.819_826del8, p.(Arg274Valfs*13)
gnomAD browser prevalence: 0.0001185 |
homozygous |
Sundin et al. 2000 |
| 17 |
- |
+ |
+ |
+ |
ATF6 |
c.1691A>G, p.(Asp564Gly; pathogenic II) |
homozygous |
[Skorczyk-Werner et al., 2017] 1 |
|
Recessive inheritance – single mutation identified | ||||||||
| 3 |
- |
+ |
+ |
- |
ABCA4 |
c.5882G>A, p.(Gly1961Glu)
gnomAD browser prevalence: 0.003931 |
heterozygous |
Allikmets et al. 1997 |
| 6 |
- |
+ |
+ |
- |
CNGB3 |
c.1148delC, p.(Thr383Ilefs*13)
gnomAD browser prevalence: 0.002810 |
heterozygous |
Sundin et al. 2000 |
|
X – linked inheritance | ||||||||
| 13 |
+ |
- |
- |
- |
RPGR
-ORF15 |
c.3142_3143dupAA, p.(Glu1049Argfs*41; pathogenic Ic) |
hemizygous |
[a] |
| 7 | + | - | - | - | RPGR -ORF15 | c.2716G>T, p.(Glu906*) | hemizygous | Sharon et al. 2003 |
[a] – this study; 1– identified in this study, but already published; ; + molecular analysis was performed, - molecular analysis was not performed; Variants designation is based on NM_000322 for PRPH2, NM_001319061 for GUCA1A, NM_006017 for PROM1, NM_019098 for CNGB3, NM_007348 for ATF6, NM_000350 for ABCA4 and NM_001331041 for RPGR-ORF15 (GRCh38). Classification of novel variants according to American College of Medical Genetics and Genomics (ACMG) guidelines. The gnomAD browser prevalences are provided for mutated alleles in non-Finnish Europeans.
NGS revealed potentially causative variants in genes associated with the AD form of CRD in Families 9 and 18. In Family 9, a novel nucleotide variant c.592A>C, p.(Ser198Arg) in exon 2 of the PRPH2 gene was identified, verified with Sanger sequencing (Figure 4A), and validated with segregation analysis (Figure 1). In silico analyses indicated that the substitution is probably damaging.
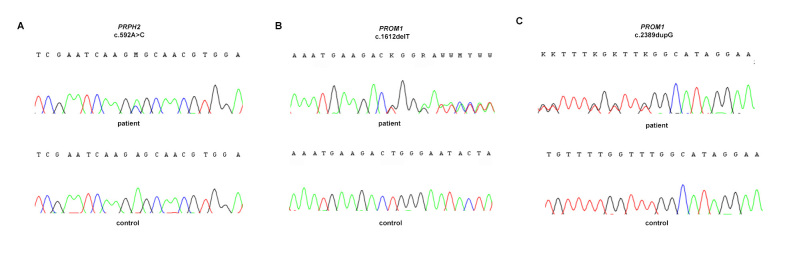
Figure 4.
Chromatograms showing three novel variants identified in patients with CRD. A: Sequence trace of the PRPH2 gene (fragment of exon 2) in the affected individual (Family 9) carrying a heterozygous missense variant c.592A>C (upper panel) and a normal control individual (lower panel). B: Sequence trace of the PROM1 gene (fragment of exon 14) in the affected individual (Family 5) carrying the heterozygous frameshift change c.1612delT (upper panel) and a normal control individual (lower panel). C: Sequence trace of the PROM1 gene (fragment of exon 23) in the affected individual (Family 5) carrying the heterozygous frameshift change c.2389dupG (upper panel) and a normal control individual (lower panel). Because the RPGR-ORF15 gene was sequenced commercially by Asper Biotech (Tartu, Estonia), the chromatogram is not available. The chromatogram showing the mutation c.1691A>G in the ATF6 gene was included in our previous report [19].
One previously described substitution c.312C>A, p.(Asn104Lys) in the GUCA1A gene was discovered in Family 18 [23]. WES analysis performed in Family 4 did not allow identification of potentially causative variants that segregate with disease.
Patients with the XL and XL/AR modes of inheritance
Sanger sequencing of the RPGR-ORF15 was performed in three families (Families 1, 7, and 8) presenting with an XL mode of inheritance and in three additional families (Families 11, 13, and 15) with the AR/XL mode of inheritance. Sequence analysis of the RPGR-ORF15 revealed two hemizygous variants in two families (7 and 13). In Family 13, we identified a novel indel variant c.3142_3143dupAA, p.(Glu1049Argfs*41) resulting in a frameshift and premature termination of translation. Unfortunately, parental genomic DNA was unavailable for allelic segregation analysis. The known nonsense change c.2716G>T, p.(Glu906*) was identified in Family 7 [24]. Commercial sequencing electropherograms from Asper Biotech were not available for inclusion in Figure 4.
Patients with the AR mode of inheritance
Five families with AR CRD (Families 3, 5, 6, 12, and 17) and two families with XL/AR CRD negative for variants in the RPGR-ORF15 gene (Families 11 and 15) were screened for variants in the ABCA4 gene using APEX analysis. The APEX analysis revealed an alteration in ABCA4 in only one family (Family 3). A previously described missense variant, c.5882G>A, p.(Gly1961Glu), was present in the heterozygous state in the female index patient [25]. To exclude any copy number variations, such as large deletions/duplications, MLPA was performed within the ABCA4 gene in this family. However, no genomic rearrangements were found in the ABCA4 gene. In addition, several deep intronic variants (V1–V7) in the ABCA4 gene, which may affect gene splicing, have been reported to segregate with the disease [14]. The analysis of these sites did not reveal the presence of any of these intronic variants. As the absence of an alteration on a second allele of the ABCA4 gene did not allow the molecular diagnosis, the proband was referred for a further molecular analysis.
All families with the AR mode of inheritance (3, 5, 6, 12, and 17) and the families with the AR/XL mode of inheritance (11 and 15) were submitted for IRD panel–based NGS. NGS allowed the identification of putative sequence variants in three families (5, 6, and 12) and confirmed the previously identified ABCA4 heterozygous variant in Family 3. In Family 5, two novel frameshift variants in the PROM1 gene were identified: c. 1612delT, p.(Trp538Glyfs*15) in exon 14 (Figure 4B) and c.2389dupG, p.(Ile798Hisfs*20) in exon 23 (Figure 4C) in the form of a compound heterozygote (Figure 1). Both variants had not been described in scientific literature previously and were not functionally tested for potential pathogenicity.
In Family 12, a previously reported homozygous frameshift variant c.819_826del8, p.(Arg274Valfs*13) in exon 6 of the CNGB3 gene was detected (Figure 1) [26].
In Family 6, NGS revealed a previously described heterozygous frameshift variant c.1148delC, p.(Thr383Ilefs*13) in exon 10 of the CNGB3 gene [26].
Three families who remained unsolved with targeted NGS including one with the AR mode of inheritance (Family 17) and two with the AR/XL mode of inheritance (Families 11 and 15) were subject to WES. WES allowed the genetic basis of the retinal dystrophy in one family to be identified (Family 17): a novel variant c.1691A>G, p.(Asp564Gly) in the ATF6 gene, which we reported to be pathogenic [19]. None of the novel variants identified based on the combined strategy of Sanger sequencing and the NGS technique was found in a control cohort annotated in the Exome Variant Server (EVS) database (Exome Variant Server 2015) or in the 1000 Genomes Project database (1000 Genomes Project Consortium 2012), ExAC Browser Beta (Exome Aggregation Consortium 2015), and gnomAD browser beta (genome Aggregation database).
Discussion
In this study, we present the results of molecular screening in 18 Polish families who have CRD. Notably, this is the first report on the spectrum of disease genes and pathogenic variants causing CRD in a Polish population. In total, we identified the most likely genetic cause of disease in seven families (Table 2).
Sequencing of the RPGR-ORF15 gene, performed in families with the XL and AR/XL modes of inheritance, resulted in the identification of two variants in two families (Families 7 and 13). Interestingly, the patient from Family 7, with a known nonsense change c.2716G>T, p.(Glu906*) presented a more severe course of the disease (refer to the earlier age of onset and worse visual acuity) than the patient from Family 13, with a frameshift variant.
Depending on the population and its ethnic background, mutations in the ABCA4 gene have been shown to be the most prevalent cause of AR CRD, accounting for 30–60% cases [27]. The carrier frequency is estimated at around 2% in the general population, although some studies suggest that the frequency might be considerably higher and account for even 5–20% of individuals [27-31]. The APEX analysis revealed an ABCA4 alteration in only one family (Family 3) [25]. Taking into consideration the high ABCA4 mutation carrier frequency, the presence of the variant on one allele is not sufficient to confirm the molecular diagnosis. This finding suggests that ABCA4 plays a minor role in Polish patients with AR CRD. One of the possible reasons for these discrepancies and the main limitation of the study was the small size of the study group. Fourteen families involved in this study were examined with targeted NGS with the application of a custom IRD panel, which allowed a possible genetic cause to be identified in four out of 14 families.
The variant reported in Family 12, c.819_826del8, p.(Arg274Valfs*13) in the CNGB3 gene, has been previously reported as a recurrent mutation in families with achromatopsia, although only in a compound heterozygous state with another, usually missense variant [9,32-36]. Mutations in the CNGB3 gene are also known to be a rare cause of AR CRD, described thus far in only six families [37-39]. All the reported AR CRD cases have been found to be homozygous for frameshift or splice-site mutations. Therefore, it is possible that c.819_826del8, p.(Arg274Valfs*13) in a homozygous state is the cause of AR CRD in Family 12.
The missense variant c.592A>C identified in the PRPH2 gene of the patient from Family 9 has not been reported before, but the substitution in the same codon (AGC>AGG) c.594C>G resulting in the same amino acid change p.(Ser198Arg) has been previously identified as a likely pathogenic in two families who have an autosomal dominant form of retinitis pigmentosa (RP) [40,41]. The missense substitution p.(Ser198Arg) is localized in the large intradiscal loop domain (D2) of the peripherin-2 protein, at an evolutionary conserved amino acid position. The D2 loop plays a crucial role in the dimerization of homo- and heterotetramers with ROM1, which is critical for PRPH2-ROM1 interactions important for the formation and stabilization of photoreceptor outer segments [41,42]. The previously reported results of in silico analyses (PolyPhen and SIFT software) for the p.(Ser198Arg) substitution, as well as the analysis (Mutation Taster software) for the variant c.592A>C (this study) indicates that the substitution is probably damaging. However, additional functional analyses are required to confirm the pathogenicity of the p.(Ser198Arg) variant. Notably, the substitution c.594C>G, p.(Ser198Arg) has been identified to date only in families affected by AD RP [40,41].
In Family 6, NGS on the IRD panel revealed a previously described heterozygous frameshift variant c.1148delC, p.(Thr383Ilefs*13) in exon 10 of the CNGB3 gene [26]. The lack of a second allele in this patient might be explained in several ways. First, NGS on the retinal panel does not allow large genomic rearrangements in the heterozygous state to be identified. Recently, the prevalence of CNVs in the CNGB3 gene was determined to be approximately 2% of all cases [9]. Moreover, according to Mayer et al., CNVs can account for more than one-third of unknown mutations in patients with single heterozygous mutations upon Sanger sequencing [8,9]. There is also a possibility that the missing variant is located deep inside the intron, disturbing the proper splicing or expression of the CNGB3 gene. Finally, taking into consideration that the carrier frequency for CNGB3 mutations is estimated at about 1:200 births, the members of Family 6 could possibly be only carriers, and a causative mutation is located in another gene [32]. Due to the lack of color vision defects, clinical diagnosis of achromatopsia was excluded.
In one family with AR CRD (Family 5), we identified two novel variants in the PROM1 gene. Missense mutations in the PROM1 gene have mostly been associated with autosomal dominant forms of Stargardt-like macular dystrophy, bull’s eye macular dystrophy, and cone-rod dystrophy. To date, only a few cases of nonsense, frameshift, and intronic mutations have been described in patients with severe forms of autosomal recessive CRD and retinitis pigmentosa [43-49]. Both variants identified in this family result in frameshift mutations and the occurrence of a premature termination codon. The derived proteins are substantially shortened, lacking the important transmembrane domain. Therefore, the pathogenicity of these mutations is highly probable.
The mutation detection rate for the IRD panel applied was 38% (8/21 alleles), which is lower in comparison with previous studies that used the same IRD panel in a larger cohort of patients with various retinal dystrophies, where the overall detection rates were 55% and 50% for the group of patients with CRD [7,15]. The lower detection rate for the panel-based analysis can be explained in several ways. The present study group was smaller than the cohorts analyzed in previous reports. Moreover, the spectrum of CRD disease genes and mutations may differ from that of Western European countries and the United States, which was the basis for the preparation of the custom IRD NGS panel. In addition, the version of the IRD panel applied for the families in the present study encompassed only CRD genes that were known at that time (22 genes). To date, 12 more genes and loci that had not been incorporated in that version of the panel have been found to be associated with CRD: ATF6, CNGA3, CNNM4, C21ORF2, C8ORF37, GNAT2, ITF81, PDE6H, POC1B, RAB28, and TTLL5 (RetNet). Therefore, we cannot exclude variants in these genes in families who have not been examined with WES.
Four families of the families who had been mutation-negative upon analysis with the IRD panel were selected for WES, which allowed the causative change in one family with the autosomal recessive mode of inheritance to be identified [19]. However, it is not reasonable to assess the mutation detection rate for WES, as this analysis was not performed in all the families.
Searches for CNVs in WES data have not revealed any potentially causative variations. As this analysis was performed only for WES data and not a targeted NGS panel, we cannot exclude CNVs in families who have not undergone WES. The presence of mutations in non-coding regions, or deep intronic mutations, which have been reported to date as a cause of retinal dystrophies cannot be excluded in the group as a whole, either [9,14,50]. As these types of mutations are not possible to detect with the use of WES, the families who are negative for WES should be further analyzed with whole genome sequencing. These studies confirm that the NGS on the retinal panel seems to be a more effective method for diagnosis of the molecular basis of CRD, compared to other methods, including Sanger sequencing [7,15].
Acknowledgments
This study was supported by a grant from the Polish Ministry of Science and Higher Education (806/N-NIEMCY/ 2010/0) to MK and a grant from the German Ministry of Education and Science (BMBF - HOPE2: 01GM1108A) to BW.
Appendix 1. Supplemental material.
Pedigrees of the CRD families whose genetic background of the disease remained unexplained, with the exception of Family 17 (molecular results were reported previously). To access the data, click or select the words “Appendix 1.”
References
- 1.Sahel JA, Marazova K, Audo I. Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb Perspect Med. 2014;5:a017111. doi: 10.1101/cshperspect.a017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.den Hollander AI, Black A, Bennett J, Cremers FP. Lighting a candle in the dark: advances in genetics and gene therapy of recessive retinal dystrophies. J Clin Invest. 2010;120:3042–53. doi: 10.1172/JCI42258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamel CP. Cone rod dystrophies. Orphanet J Rare Dis. 2007;2:7. doi: 10.1186/1750-1172-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moore AT. Cone and cone-rod dystrophies. J Med Genet. 1992;29:289–90. doi: 10.1136/jmg.29.5.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaelides M, Hardcastle AJ, Hunt DM, Moore AT. Progressive cone and cone-rod dystrophies: phenotypes and underlying molecular genetic basis. Surv Ophthalmol. 2006;51:232–58. doi: 10.1016/j.survophthal.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Fishman GA, Stone EM, Eliason DA, Taylor CM, Lindeman M, Derlacki DJ. ABCA4 gene sequence variations in patients with autosomal recessive cone-rod dystrophy. Arch Ophthalmol. 2003;121:851–5. doi: 10.1001/archopht.121.6.851. [DOI] [PubMed] [Google Scholar]
- 7.Weisschuh N, Mayer AK, Strom TM, Kohl S, Glockle N, Schubach M, Andreasson S, Bernd A, Birch DG, Hamel CP, Heckenlively JR, Jacobson SG, Kamme C, Kellner U, Kunstmann E, Maffei P, Reiff CM, Rohrschneider K, Rosenberg T, Rudolph G, Vamos R, Varsanyi B, Weleber RG, Wissinger B. Mutation Detection in Patients with Retinal Dystrophies Using Targeted Next Generation Sequencing. PLoS One. 2016;11:e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Schil K, Naessens S, Van de Sompele S, Carron M, Aslanidis A, Van Cauwenbergh C, Mayer AK, Van Heetvelde M, Bauwens M, Verdin H, Coppieters F, Greenberg ME, Yang MG, Karlstetter M, Langmann T, De Preter K, Kohl S, Cherry TJ, Leroy BP, De Baere E. Mapping the genomic landscape of inherited retinal disease genes prioritizes genes prone to coding and noncoding copy-number variations. Genet Med. 2018;20:202–13. doi: 10.1038/gim.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer AK, Van Cauwenbergh C, Rother C, Baumann B, Reuter P, De Baere E, Wissinger B, Kohl S, Group AS. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum Mutat. 2017;38:1579–91. doi: 10.1002/humu.23311. [DOI] [PubMed] [Google Scholar]
- 10.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, Megy K, Grozeva D, Dewhurst E, Malka S, Plagnol V, Penkett C, Stirrups K, Rizzo R, Wright G, Josifova D, Bitner-Glindzicz M, Scott RH, Clement E, Allen L, Armstrong R, Brady AF, Carmichael J, Chitre M, Henderson RHH, Hurst J, MacLaren RE, Murphy E, Paterson J, Rosser E, Thompson DA, Wakeling E, Ouwehand WH, Michaelides M, Moore AT. Consortium NI-BRD, Webster AR, Raymond FL. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am J Hum Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellingford JM, Horn B, Campbell C, Arno G, Barton S, Tate C, Bhaskar S, Sergouniotis PI, Taylor RL, Carss KJ, Raymond LFL, Michaelides M, Ramsden SC, Webster AR, Black GCM. Assessment of the incorporation of CNV surveillance into gene panel next-generation sequencing testing for inherited retinal diseases. J Med Genet. 2018;55:114–21. doi: 10.1136/jmedgenet-2017-104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, Jiang Q, Allen AS, Zhu M. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35:899–907. doi: 10.1002/humu.22537. [DOI] [PubMed] [Google Scholar]
- 13.Lahiri DK, Bye S, Nurnberger JI, Jr, Hodes ME, Crisp M. A non-organic and non-enzymatic extraction method gives higher yields of genomic DNA from whole-blood samples than do nine other methods tested. J Biochem Biophys Methods. 1992;25:193–205. doi: 10.1016/0165-022x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 14.Braun TA, Mullins RF, Wagner AH, Andorf JL, Johnston RM, Bakall BB, Deluca AP, Fishman GA, Lam BL, Weleber RG, Cideciyan AV, Jacobson SG, Sheffield VC, Tucker BA, Stone EM. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–45. doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glockle N, Kohl S, Mohr J, Scheurenbrand T, Sprecher A, Weisschuh N, Bernd A, Rudolph G, Schubach M, Poloschek C, Zrenner E, Biskup S, Berger W, Wissinger B, Neidhardt J. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet. 2014;22:99–104. doi: 10.1038/ejhg.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, Wood NW, Hambleton S, Burns SO, Thrasher AJ, Kumararatne D, Doffinger R, Nejentsev S. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–54. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skorczyk-Werner A, Chiang WC, Wawrocka A, Wicher K, Jarmuz-Szymczak M, Kostrzewska-Poczekaj M, Jamsheer A, Ploski R, Rydzanicz M, Pojda-Wilczek D, Weisschuh N, Wissinger B, Kohl S, Lin JH, Krawczynski MR. Autosomal recessive cone-rod dystrophy can be caused by mutations in the ATF6 gene. Eur J Hum Genet. 2017;25:1210–6. doi: 10.1038/ejhg.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLOS Comput Biol. 2016;12:e1004873. doi: 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olshen AB, Bengtsson H, Neuvial P, Spellman PT, Olshen RA, Seshan VE. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation. Bioinformatics. 2011;27:2038–46. doi: 10.1093/bioinformatics/btr329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Wheaton D, Bereta G, Zhang K, Palczewski K, Birch DG, Baehr W. A novel GCAP1(N104K) mutation in EF-hand 3 (EF3) linked to autosomal dominant cone dystrophy. Vision Res. 2008;48:2425–32. doi: 10.1016/j.visres.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharon D, Sandberg MA, Rabe VW, Stillberger M, Dryja TP, Berson EL. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003;73:1131–46. doi: 10.1086/379379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–46. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- 26.Sundin OH, Yang JM, Li Y, Zhu D, Hurd JN, Mitchell TN, Silva ED, Maumenee IH. Genetic basis of total colourblindness among the Pingelapese islanders. Nat Genet. 2000;25:289–93. doi: 10.1038/77162. [DOI] [PubMed] [Google Scholar]
- 27.Manitto MP, Roosing S, Boon CJ, Souied EH, Bandello F, Querques G. Clinical Utility Gene Card for: autosomal recessive cone-rod dystrophy. Eur J Hum Genet. 2015;23 doi: 10.1038/ejhg.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yatsenko AN, Shroyer NF, Lewis RA, Lupski JR. Late-onset Stargardt disease is associated with missense mutations that map outside known functional regions of ABCR (ABCA4). Hum Genet. 2001;108:346–55. doi: 10.1007/s004390100493. [DOI] [PubMed] [Google Scholar]
- 29.Riveiro-Alvarez R, Aguirre-Lamban J, Lopez-Martinez MA, Trujillo-Tiebas MJ, Cantalapiedra D, Vallespin E, Avila-Fernandez A, Ramos C, Ayuso C. Frequency of ABCA4 mutations in 278 Spanish controls: an insight into the prevalence of autosomal recessive Stargardt disease. Br J Ophthalmol. 2009;93:1359–64. doi: 10.1136/bjo.2008.148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaakson K, Zernant J, Kulm M, Hutchinson A, Tonisson N, Glavac D, Ravnik-Glavac M, Hawlina M, Meltzer MR, Caruso RC, Testa F, Maugeri A, Hoyng CB, Gouras P, Simonelli F, Lewis RA, Lupski JR, Cremers FP, Allikmets R. Genotyping microarray (gene chip) for the ABCR (ABCA4) gene. Hum Mutat. 2003;22:395–403. doi: 10.1002/humu.10263. [DOI] [PubMed] [Google Scholar]
- 31.Maugeri A, Flothmann K, Hemmrich N, Ingvast S, Jorge P, Paloma E, Patel R, Rozet JM, Tammur J, Testa F, Balcells S, Bird AC, Brunner HG, Hoyng CB, Metspalu A, Simonelli F, Allikmets R, Bhattacharya SS, D’Urso M, Gonzalez-Duarte R, Kaplan J, te Meerman GJ, Santos R, Schwartz M, Van Camp G, Wadelius C, Weber BH, Cremers FP. The ABCA4 2588G>C Stargardt mutation: single origin and increasing frequency from South-West to North-East Europe. Eur J Hum Genet. 2002;10:197–203. doi: 10.1038/sj.ejhg.5200784. [DOI] [PubMed] [Google Scholar]
- 32.Kohl S, Varsanyi B, Antunes GA, Baumann B, Hoyng CB, Jagle H, Rosenberg T, Kellner U, Lorenz B, Salati R, Jurklies B, Farkas A, Andreasson S, Weleber RG, Jacobson SG, Rudolph G, Castellan C, Dollfus H, Legius E, Anastasi M, Bitoun P, Lev D, Sieving PA, Munier FL, Zrenner E, Sharpe LT, Cremers FP, Wissinger B. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet. 2005;13:302–8. doi: 10.1038/sj.ejhg.5201269. [DOI] [PubMed] [Google Scholar]
- 33.Nishiguchi KM, Sandberg MA, Gorji N, Berson EL, Dryja TP. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mutat. 2005;25:248–58. doi: 10.1002/humu.20142. [DOI] [PubMed] [Google Scholar]
- 34.Varsanyi B, Wissinger B, Kohl S, Koeppen K, Farkas A. Clinical and genetic features of Hungarian achromatopsia patients. Mol Vis. 2005;11:996–1001. [PubMed] [Google Scholar]
- 35.Thiadens AA, Slingerland NW, Roosing S, van Schooneveld MJ, van Lith-Verhoeven JJ, van Moll-Ramirez N, van den Born LI, Hoyng CB, Cremers FP, Klaver CC. Genetic etiology and clinical consequences of complete and incomplete achromatopsia. Ophthalmology. 2009;116:1984–9. doi: 10.1016/j.ophtha.2009.03.053. [DOI] [PubMed] [Google Scholar]
- 36.Wawrocka A, Kohl S, Baumann B, Walczak-Sztulpa J, Wicher K, Skorczyk-Werner A, Krawczynski MR. Five novel CNGB3 gene mutations in Polish patients with achromatopsia. Mol Vis. 2014;20:1732–9. [PMC free article] [PubMed] [Google Scholar]
- 37.Roosing S, Thiadens AA, Hoyng CB, Klaver CC, den Hollander AI, Cremers FP. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014;42:1–26. doi: 10.1016/j.preteyeres.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Thiadens AA, Phan TM, Zekveld-Vroon RC, Leroy BP, van den Born LI, Hoyng CB, Klaver CC, Roosing S, Pott JW, van Schooneveld MJ, van Moll-Ramirez N, van Genderen MM, Boon CJ, den Hollander AI, Bergen AA, De Baere E, Cremers FP, Lotery AJ. Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology. 2012;119:819–26. doi: 10.1016/j.ophtha.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Huang L, Zhang Q, Li S, Guan L, Xiao X, Zhang J, Jia X, Sun W, Zhu Z, Gao Y, Yin Y, Wang P, Guo X, Wang J. Exome sequencing of 47 chinese families with cone-rod dystrophy: mutations in 25 known causative genes. PLoS One. 2013;8:e65546. doi: 10.1371/journal.pone.0065546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, Garcia CA, Ruiz RS, Blanton SH, Northrup H, Gire AI, Seaman R, Duzkale H, Spellicy CJ, Zhu J, Shankar SP, Daiger SP. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47:3052–64. doi: 10.1167/iovs.05-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manes G, Guillaumie T, Vos WL, Devos A, Audo I, Zeitz C, Marquette V, Zanlonghi X, Defoort-Dhellemmes S, Puech B, Said SM, Sahel JA, Odent S, Dollfus H, Kaplan J, Dufier JL, Le Meur G, Weber M, Faivre L, Cohen FB, Beroud C, Picot MC, Verdier C, Senechal A, Baudoin C, Bocquet B, Findlay JB, Meunier I, Dhaenens CM, Hamel CP. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in france and characterization of biochemical and clinical features. Am J Ophthalmol. 2015;159:302–14. doi: 10.1016/j.ajo.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 42.Loewen CJ, Moritz OL, Molday RS. Molecular characterization of peripherin-2 and rom-1 mutants responsible for digenic retinitis pigmentosa. J Biol Chem. 2001;276:22388–96. doi: 10.1074/jbc.M011710200. [DOI] [PubMed] [Google Scholar]
- 43.Boulanger-Scemama E, El Shamieh S, Demontant V, Condroyer C, Antonio A, Michiels C, Boyard F, Saraiva JP, Letexier M, Souied E, Mohand-Said S, Sahel JA, Zeitz C, Audo I. Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: mutation spectrum and new genotype-phenotype correlation. Orphanet J Rare Dis. 2015;10:85. doi: 10.1186/s13023-015-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eidinger O, Leibu R, Newman H, Rizel L, Perlman I, Ben-Yosef T. An intronic deletion in the PROM1 gene leads to autosomal recessive cone-rod dystrophy. Mol Vis. 2015;21:1295–306. [PMC free article] [PubMed] [Google Scholar]
- 45.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Roper K, Weigmann A, Huttner WB, Denton MJ. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet. 2000;9:27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- 46.Michaelides M, Gaillard MC, Escher P, Tiab L, Bedell M, Borruat FX, Barthelmes D, Carmona R, Zhang K, White E, McClements M, Robson AG, Holder GE, Bradshaw K, Hunt DM, Webster AR, Moore AT, Schorderet DF, Munier FL. The PROM1 mutation p.R373C causes an autosomal dominant bull’s eye maculopathy associated with rod, rod-cone, and macular dystrophy. Invest Ophthalmol Vis Sci. 2010;51:4771–80. doi: 10.1167/iovs.09-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Permanyer J, Navarro R, Friedman J, Pomares E, Castro-Navarro J, Marfany G, Swaroop A, Gonzalez-Duarte R. Autosomal recessive retinitis pigmentosa with early macular affectation caused by premature truncation in PROM1. Invest Ophthalmol Vis Sci. 2010;51:2656–63. doi: 10.1167/iovs.09-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pras E, Abu A, Rotenstreich Y, Avni I, Reish O, Morad Y, Reznik-Wolf H. Cone-rod dystrophy and a frameshift mutation in the PROM1 gene. Mol Vis. 2009;15:1709–16. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Zulfiqar F, Xiao X, Riazuddin SA, Ahmad Z, Caruso R, MacDonald I, Sieving P, Riazuddin S, Hejtmancik JF. Severe retinitis pigmentosa mapped to 4p15 and associated with a novel mutation in the PROM1 gene. Hum Genet. 2007;122:293–9. doi: 10.1007/s00439-007-0395-2. [DOI] [PubMed] [Google Scholar]
- 50.Mayer AK, Rohrschneider K, Strom TM, Glockle N, Kohl S, Wissinger B, Weisschuh N. Homozygosity mapping and whole-genome sequencing reveals a deep intronic PROM1 mutation causing cone-rod dystrophy by pseudoexon activation. Eur J Hum Genet. 2016;24:459–62. doi: 10.1038/ejhg.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]