Abstract
The unicellular metazoan zygote undergoes a series of cell divisions that are central to its development into an embryo. Differentiation of embryonic cells leads eventually to the development of a functional adult. Fate specification of pluripotent embryonic cells occurs during the early embryonic cleavage divisions in several animals. Early development is characterized by well-known stages of embryogenesis documented across animals—morulation, blastulation, and morphogenetic processes such as gastrulation, all of which contribute to differentiation and tissue specification. Despite this broad conservation, there exist clearly discernible morphological and functional differences across early embryonic stages in metazoans. Variations in the mitotic mechanisms of early embryonic cell divisions play key roles in governing these gross differences that eventually encode developmental patterns. In this review, we discuss molecular mechanisms of both karyokinesis (nuclear division) and cytokinesis (cytoplasmic separation) during early embryonic divisions. We outline the broadly conserved molecular pathways that operate in these two stages in early embryonic mitoses. In addition, we highlight mechanistic variations in these two stages across different organisms. We finally discuss outstanding questions of interest, answers to which would illuminate the role of divergent mitotic mechanisms in shaping early animal embryogenesis.
Keywords: mitosis, metaphase, cytokinesis, spindle orientation, early embryonic development, fate specification, metazoa
Introduction
Cell division is a key process shaping normal embryonic development. Mitosis involves the segregation of the replicated genome (karyokinesis) and separation of the cytoplasmic content (cytokinesis). These two key steps are tightly regulated in space and time during normal embryogenesis. Understanding cell division is thus crucial to understand developmental processes, leading to tissue and organ formation. The development of a single cell zygote into a multicellular functional adult involves multiple rounds of cell division. Early embryonic cleavage divisions, unlike later divisions, lack G1-G2 checkpoints (1) and depend heavily on maternal factors that continue dividing the egg cytoplasm into smaller cells, often employing the asymmetric mode of division (1). The molecular regulation of early embryonic divisions is an area of intense study. In this review, we discuss the broadly conserved mechanisms involved in nuclear and cytoplasmic divisions during early embryogenesis across different metazoan species, emphasizing species-specific differences.
Early Mitosis
The entry of embryonic cells into mitosis is characterized by chromosome condensation and nuclear envelope disintegration during prometaphase, mixing nuclear and cytoplasmic contents. This stage is signified by dramatic changes to both microtubule and actin cytoskeletal networks. Dynamic cytoskeletal reorganization leads to the hallmark spherical architecture of mitotic cells and a concomitant loss of cell polarity. The conserved mitotic spindle apparatus consisting of the centrosomes and the reorganized microtubules is formed and serves as a scaffold on which most events in metaphase are orchestrated. This phase culminates in equal segregation of sister chromatids to the daughter cells.
Mitotic Rounding and Formation of the Mitotic Spindle
During early mitosis, as chromatin undergoes condensation, major cytoskeletal rearrangements lead to cells attaining a typical spherical shape (Fig. 1A). This “mitotic rounding” is seen across metazoa—Dictyostelium, Drosophila melanogaster, mouse, and cultured human cells. The round shape confines the chromosomes to a limited volume to facilitate kinetochore-microtubule attachment. Mitotic rounding is crucial for spindle formation, spindle pole stability, chromosome capture and correct chromosome segregation (2).
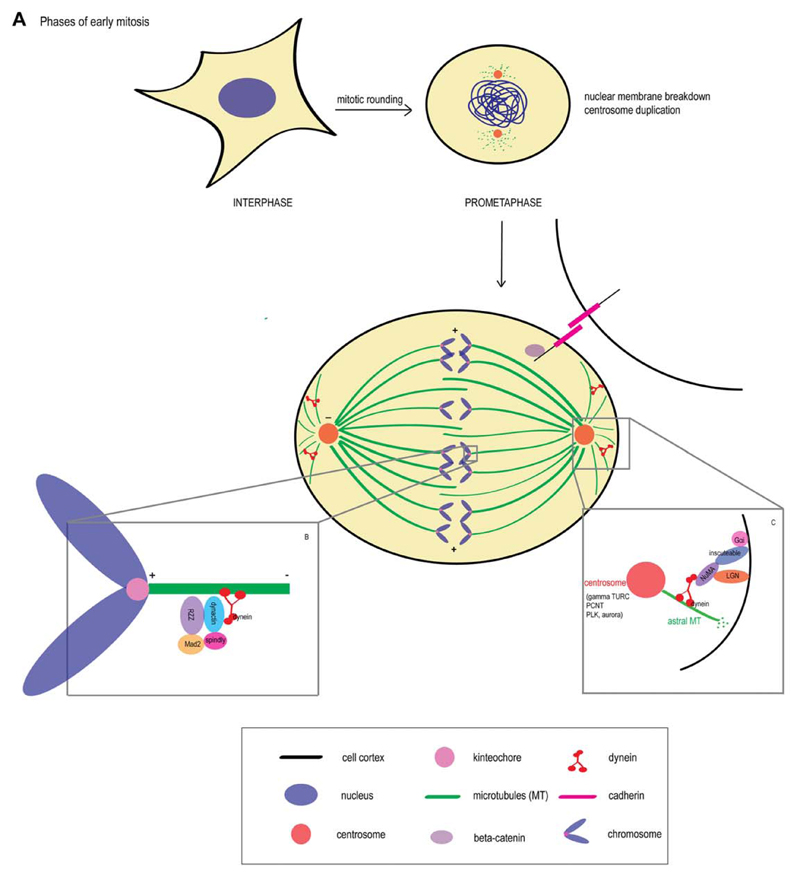
Fig 1. Phases of early mitosis.
A: Upon entry into the mitotic phase of cell cycle, an irregular shaped metazoan cell undergoes mitotic rounding and acquires a spherical shape coupled with drastic cytoskeletal reorganization. By prometaphase, the nuclear membrane disintegrates, the centrosomes undergo duplication and migrate to opposite ends, forming the spindle poles. At metaphase, the chromosomes lie in the center of the mitotic spindle. The chromosomes are held at the metaphase plate by the kinetochore microtubules, while the spindle itself is anchored to the cell cortex by astral microtubules. B: At the kinetochore, the minus-end directed dynein-dynactin motor complex transports cargo such as checkpoint proteins (e.g., Mad2) toward the spindle poles, initiating silencing of the checkpoint and entry into anaphase. C: At the polar cell cortex, dynein and the NuMA–LGN–Gαi complex are required for correct positioning and orientation of the mitotic spindle.
The mitotic spindle, consisting of an array of microtubules nucleating from opposite centrosomes, (microtubule organizing centres—MTOCs, also called spindle poles) and numerous associated proteins, ensures equal separation of the genetic material and also regulates the position of the cytokinetic furrow (Fig. 1A) (3). Formation of the spindle apparatus begins during prophase with chromatin condensation. The mammalian centrosome consists of mother and daughter centrioles surrounded by the electron dense pericentriolar material (PCM). Centrosomes are essential for cells to complete cytokinesis, undergo mitotic exit and enter into the S phase (4).
During S phase, centrosomes undergo semi-conservative duplication along with the genomic DNA and by early mitosis (prometaphase), reach the opposite sides of the nucleus, a process mediated by the microtubule-based motors kinesin Eg5 and dynein (5). After nuclear membrane breakdown, each centrosome nucleates an aster of microtubules, which form the spindle poles (Fig. 1A). Microtubules originating from the centrosomes grow and establish connections with the kinetochores on the centromeric DNA, finally organizing to form the “fusiform” bipolar spindle apparatus (Fig. 1A) (6). The spindle is composed of three kinds of microtubules—kinetochore, interpolar, and astral microtubules. The kinetochore microtubules attach to the chromosomes and separate them at anaphase. Interpolar microtubules form antiparallel arrays between the spindle poles and regulate the position of the cytokinetic furrow. Astral microtubules anchor the spindle to the cortex and regulate the orientation of the division axis and the position of cytokinesis (Fig. 1A) (3). Spindle microtubules undergo dynamic turnover, allowing rapid attachment to the chromosomes during spindle assembly. Microtubule dynamics is regulated by the activity of several microtubule associated proteins (MAPs) that are required for maintaining microtubular homeostasis (4).
There are two accepted models of spindle formation. The “search and capture” model suggests that the bipolar spindle is formed by mediating chromosome and microtubule attachments via pulling and pushing forces derived from molecular motors cytoplasmic dyneins and kinesins (7). The “self-assembly model” postulates that kinetochore fibers form spontaneously and interact with the centrosomal microtubules. In most cases, including mammalian cells, a combination of these two models has been reported (7). In most cells, centrosomes are required for spindle assembly. However, bipolar spindles can form even in the absence of centrosomes (4). Germ cells, like oocytes of Xenopus and mammals, lack centrioles and centrosomes and follow the acentriolar pathway of spindle assembly wherein spindles are assembled by nucleation of microtubules adjacent to the chromosomes (8). The breakdown of the germinal vesicle in the oocyte results in the formation of cytoplasmic MTOCs which move toward the chromosomes with the help of dyneins. Thus, a ball of microtubules is formed at the site of chromosomes. The kinetochores mediate attachment of the chromosomes to the outer surface of this ball, giving a “belt like” appearance of chromosomes around the ball. The MTOCs become spatially organized to two opposite poles of the ball, and the belt of chromosomes forms the future metaphase plate. The kinesin 5 motor pushes the two MTOC poles apart, thus giving rise to the bipolar spindle. This acentriolar mechanism of spindle assembly by cytoplasmic MTOCs is also employed in early embryonic divisions in the mouse (6,9).
Spindle Orientation During Embryogenesis
Orientation of the mitotic spindle regulates the positioning of the cell division axis. During early cleavage divisions, the spindle axis lies along the longest axis of the cytoplasm, commonly referred to as the Hertwig rule (10,11). During embryogenesis, the orientation of cell division regulates the content, position, and fate specification of cells, which along with other events, influences the formation of different tissues and organs. For example, in Xenopus, cell divisions upto the 32-cell stage are parallel to the surface but later become perpendicular to give rise to deeper cells that undergo differentiation (Fig. 2A) (11). In zebrafish embryos, cell divisions in the dorsal tissue are oriented along the long animal-vegetal axis of the cell, resulting in the elongation of the body axis (Fig. 2C) (10,11). As neurulation begins, the orientation of cell division shifts to the mediolateral axis (12). In Drosophila wing imaginal discs, dividing cells orient along the proximal–distal axis (11). Orientation also affects the spatial relationship between the daughter cells. For example, during neurogenesis in Drosophila embryos, spindles oriented parallel to the epithelium generate daughter cells with epithelial fate while those oriented perpendicular generate daughter cells with neuronal fate (13).
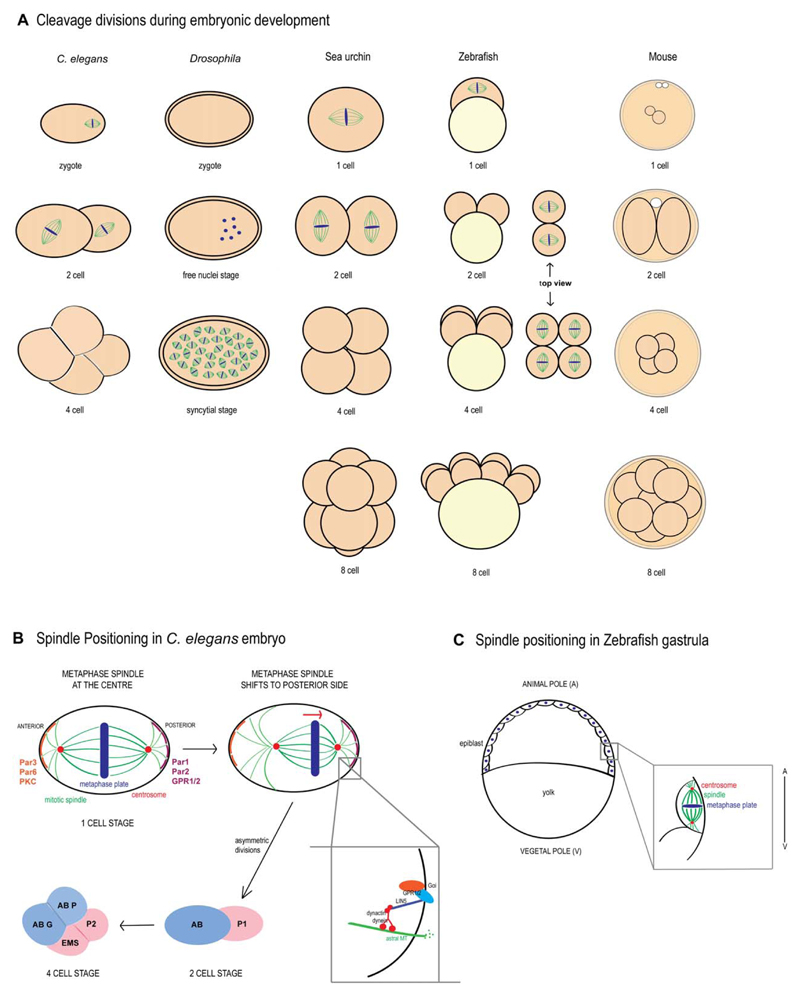
Fig 2. Cleavage divisions across metazoa.
A: Representation of spindle positioning from zygote (1 cell) to 8 cell stage in various metazoans. In the one-cell stage C. elegans embryo, the spindle is positioned asymmetrically toward the posterior end, giving rise to daughter cells with different fates. In Drosophila, successive nuclear divisions coupled with the absence of cytokinesis give rise to a syncytial embryo. Sea urchin embryos undergo asymmetric divisions giving rise to micromeres and macromeres. In contrast, early divisions are symmetric in zebrafish embryos. Mouse embryos also undergo asymmetric divisions, giving rise to daughter cells with different cell fates. B: In the one-cell stage C. elegans embryo, the mitotic spindle shifts to the posterior end, giving rise to AB and P1 cells, which again undergo asymmetric divisions. C: During gastrulation in zebrafish, spindles are positioned along the animal-vegetal axis.
Spindle orientation is also affected by physical constraints of the cell. For example, sea urchin eggs, when experimentally forced into different shapes, resulted in some cells not following the Hertwig rule. The division axis was along the largest axis of symmetry. Further, the nucleus was repositioned to the center of that specific shape and underwent elongation according to the future spindle axis. Manipulation of cell shape in developing mouse embryos also results in changes in the division plane (11). According to the “centriolic principle of spindle orientation,” centrioles migrate equally during spindle formation resulting in each division occurring perpendicular to the previous one (10), as seen in shrimp embryos.
At the molecular level, spindle orientation is regulated primarily by actomyosin contractility and spatially restricted polarity cues (10,14). In Caenorhabditis elegans, sperm entry results in the establishment of anterior–posterior polarity in the fertilized egg. The paternal centrosome initiates nucleation of microtubules and asymmetric contraction of the actomyosin cytoskeleton in the cell cortex. This leads to distribution of various PAR proteins to specific domains (Fig. 2B) (15). The PAR proteins play a major role in regulation of spindle positioning. PAR1 and PAR2 accumulate toward the posterior of the cell whereas PAR3 and PAR6 localize to the anterior. During the first division, the spindle aligns with the anterior–posterior axis and gives rise to two distinct asymmetric daughter cells. The anterior cell “AB” generates somatic cells while the posterior cell “P1” generates germ line cells (Fig. 2B) (15). In the nascent P1 cell, the spindle rotates by 90 degrees to again align along the anterior–posterior axis (15).
In mammalian cells, the alignment of the spindle is controlled by dynein–NuMA (Nuclear Mitotic Apparatus) complex (Fig. 1C). Lin5 in C. elegans and Mud in Drosophila are orthologs of vertebrate NuMA. NuMA, a nuclear protein in interphase, localizes to spindle poles and at the polar cell cortex in mitosis (16). NuMA interacts with cortical proteins LGN, Inscuteable, and Par3 and p150glued subunit of the dynactin complex at the polar cell cortex (Fig. 1C). The LGN-NuMA-Gα and PCP (planar cell polarity) pathways are evolutionarily conserved mechanisms regulating spindle orientation across metazoa (11). Yet, the mechanism of formation of the cortical NuMA–dynein–dynactin complex is not completely understood. The mechanism of regulation of microtubule depolymerization and cortical tension by this complex also remains an open question. In mammalian cells, Abelson kinase 1 (Abl1) and Polo like kinase 1 (Plk1) also play important roles in spindle orientation. Abl1 promotes an increase in the amount of LGN at the cell cortex, thus inducing formation of the NuMA–LGN complex. In contrast, Plk1, which is enriched at spindle poles, inhibits cortical dynein. However, the mechanism of Plk1 in the regulation of spindle positioning is unknown (17). In some cases, cell–cell adhesion also plays an important role in centrosome positioning and spindle orientation (Fig. 1A). In germ cells of Drosophila, E-cadherin imparts polarity cues to position the centrosomes and orient the mitotic spindle (18). In addition, extracellular signals affect spindle orientation. At the 4-cell stage in C. elegans embryos, the endomesodermal cell (EMS) and P2 cell (germline precursor cell) orient their division planes toward the shared cell–cell contact interface. This phenomenon is mediated by dynactin recruited to the cell surface by the MES-Wnt pathway (14). In zebrafish gastrula, disruption of Dishevelled (Dsh), a Wnt and PCP signaling factor, results in random cell divisions in the epiblast (11). The PCP pathway involving Wnt5 and Wnt11 is known to play key roles in oriented cell division in the fish embryo with dynein and NuMA acting as downstream effectors. Randomized spindle orientation was seen in zebrafish epiblast cells upon NuMA knockdown (3). In the developing fly wing as well, loss of PCP pathway members Fat and Dachsous showed mis-oriented cell division (11).
Positioning of the Mitotic Spindle
Spindle positioning is primarily mediated by astral microtubules and cortical polarity cues. The astral microtubules arise from the spindle poles and extend their plus ends toward the cell cortex. One of the most studied models of spindle positioning is the “cortical pulling mechanism.” Here, cortical dynein exerts pulling forces on the plus ends of astral microtubules. Dynein provides the pulling force and binds to Lin5 (NuMA in vertebrates), which in turn is associated with the cortically localized GPR1/2 (G protein regulator, LGN in vertebrates) and G α (G-protein α) subunit (Fig. 2B). In C. elegans, GPR, NuMA, and the dynein–dynactin–Lis1 complex, localized asymmetrically during anaphase, exert a greater pulling force at the posterior end of the cell as compared to the anterior (Fig. 2B). This pulling results in displacement of the spindle to the posterior and generates daughter cells of different sizes (11,17,19). RNAi mediated knockdown of dynein results in delay in alignment of the spindle along the long axis in the C. elegans zygote (20).
In the Xenopus embryo, mitotic spindles are oriented by an intricate balance of apico-basal forces of the cell during early divisions. Actin and myosin II primarily generate the apical forces. Basally directed forces are generated by astral microtubules and myosin10 (21). In snail embryos, the asymmetrical positioning of the spindle activates nodal signaling, resulting in initiation of asymmetry in the embryo (22). In Chaetopterus oocytes and sea urchin embryos, when the spindle is experimentally pulled away from the cortex, it re-positions to specific regions of the cortex. This movement is also driven by microtubule depolymerization and molecular motors (17,23).
Spindle Positioning: Basis for Asymmetric Divisions
The position of the mitotic spindle dictates the site of contractile ring assembly and the size of daughter cells, which form the basis for asymmetric divisions (19). Asymmetric divisions are well studied in germ cells, since a mammalian oocyte with 2N chromosomes undergoes asymmetric divisions to achieve genome reduction before fertilization. Due to the dramatic asymmetrical distribution of cytoplasmic material between the egg and the polar body, the egg is equipped with a majority of the cellular components to sustain the impending embryonic development. In C. elegans oocytes, meiotic spindles are positioned by nuclear localization to the cortical region by microtubular force generated by kinesin 1 (15,24). Murine oocytes require actin nucleators Formin2, Spire2 and ARP2/3 to position the spindle (19).
Asymmetric division or unequal portioning of cell fate determinants serves as a key mechanism to initiate cell fate differentiation in the developing embryo. Asymmetric division can be due to extracellular signals, polarity cues, and the intrinsic program of the cell (25). In sea urchin embryos, asymmetric divisions begin at the 16-cell stage. Animal pole-proximal cells undergo equal divisions while vegetal pole-proximal ones undergo unequal divisions, giving rise to micromeres and macromeres (Fig. 2A). The centrosomes in the micromeres are unique, as they do not generate asters (26). In mouse embryos, the first division is asymmetric, resulting in two cells with different fates (Fig. 2A). The first cell forms the embryo proper whereas the second cell gives rise to extraembryonic tissues. Here, the orientation of the spindle is determined by the site of the last meiotic division. As the two daughter cells have different fates, cell fate determinants appear to be partitioned unequally to the two daughters at the one-cell stage. This asymmetric division is not determinative in nature, but regulative as each resulting blastomere can give rise to a complete organism (27). At the 9-cell stage, blastomere compaction causes cell shape changes and cell polarization. Compaction also initiates the accumulation of PAR proteins in the cortical region, thus initiating asymmetric divisions based on the position of the mitotic spindle in the context of the apico-basal axis (10,28).
Asymmetric divisions also form the basis for establishment of left–right body asymmetry in the embryo, which results in the asymmetric positioning of organs in the body. During early divisions in Xenopus, cytoskeletal motor proteins such as Dnah1 and KIF3B localize fate determinants asymmetrically in the embryo, which results in a pH gradient and transfer of serotonin to the right side of the embryo. In contrast, nodal signaling is inhibited on the right side of the embryo by serotonin, but remains activated on the left, forming the basis for left–right asymmetry positioning of various organs (22).
Centrosomes play key roles during asymmetric divisions. Even in “seemingly” symmetrical divisions, centrosomal material is always portioned asymmetrically as one daughter cell receives the mother centrosome and the other receives the daughter (26). Many species have evolved with a complete lack of centrosomes. Cells of Planaria contain centrioles but do not form functional centrosomes (18). Mouse embryos too lack centrosomes and the cell divisions are random during early stages. The gamma tubulin centric foci can be first seen in interphase cells at the morula stage, but by the blastocyst stage (64–128 cells) centrioles can be seen in these foci. The biological significance of this phenomenon is not yet clearly understood. In the absence of centrosomes, spindles are assembled like meiotic spindles by a kinesin 5 mediated pathway. At blastula, this function of kinesin 5 is taken over by the centrosomes (6). Centrosomes are usually required for oriented cell divisions; however, exceptional cases such as mouse embryos lack centrosomes during the early stages and do not require oriented cell divisions (6). This phenomenon signifies an evolutionary shift from “mosaic” embryonic development where spindle orientation is tightly regulated, to “regulative” development in which spindle orientations are random (18).
Chromosome Segregation
The bipolar spindle also ensures correct segregation of chromosomes, failure of which could result in aneuploidy or embryonic lethality (6,29). In order to achieve faithful segregation, chromosomes are captured by kinetochore microtubules and arrayed to form the bipolar spindle. Kinetochores establish and maintain this attachment and facilitate separation of the chromosomes (30). Proper kinetochore–microtubule interaction is facilitated by the Spindle Assembly Checkpoint (SAC) signaling pathway. At metaphase, the SAC ensures correct chromosome orientation and stalls anaphase onset until all sister kinetochore pairs are attached to microtubules originating from opposite centrosomes (5,31). Key SAC proteins including kinetochore–microtubule attachment sensing Mad proteins (Mad1, Mad2) and inter-kinetochore tension sensing Bub proteins (Bub1, Bub3, BubR1) inhibit Cdc20-dependent activation of the anaphase promoting complex (APC), which promotes anaphase entry. Currently, there are two models of checkpoint function—the relay model and the mitotic checkpoint complex (MCC) model. According to the relay model, activated Mad2 binds to Cdc20 and transfers it to a Bub1–Bub3 complex in a relay-like manner; the Bub1–Bub3–Cdc20 complex then binds to APC. In the MCC model, activated Mad2 binds to a Bub1–Bub3 dimer and Cdc20 to form a tetrameric MCC complex, which then inhibits APC activation. Once all kinetochores stably attach to microtubules in a bipolar manner, the conserved Mad and Bub proteins along with key accessory proteins like Spindly and Rod–Zwilch–Zw10 complex (RZZ complex), are removed from kinetochores primarily by the cytoplasmic dynein motor (Fig. 1B) (5). The checkpoint is thus inactivated, resulting in APC activation followed by ubiquitylation-mediated proteasomal degradation of cyclin B and securin to permit anaphase onset (29,32).
During anaphase, depolymerization of kinetochore microtubules and simultaneous spindle elongation pulls segregated sister-chromatids apart toward the poles, eventually culminating in decondensation of the chromatin and reorganization into the new daughter nuclei. Concomitantly, cytoplasmic partitioning (cytokinesis) initiates at late anaphase through a signaling network emanating from the anaphase spindle, as discussed below.
Cytokinesis
Cytokinesis is initiated during anaphase when Cdk1 activity diminishes, allowing stabilization of microtubules and major cortical rearrangements to form an actomyosin ring perpendicular to the axis of chromosome separation. The ring constricts like a draw-string purse to form a cleavage furrow resulting ultimately in a narrow cytoplasmic bridge between the daughter cells that eventually abscises. Proper positioning of the cytokinetic plane and cytoplasmic segregation is as critical for daughter cell viability as correct DNA segregation. Faulty cytokinesis may lead to improper segregation of DNA, resulting in aneuploidy or anucleate cells as also to improper partitioning of key cytoplasmic factors. In the last few decades, a surge in mechanistic investigations using multiple model systems has provided deep insights into molecular mechanisms governing cytokinesis. Almost half the mitotic timeframe of a cell is occupied by cytokinesis, which is typified by four major steps—positioning of the division plane, assembling the constriction machinery, ingression of the furrow, and finally membrane abscission to separate the daughter cells.
Positioning the Division Plane
Cytokinesis usually forms equally sized daughter cells. In specialized cases however, cytokinesis can result in unequal divisions, generating specific tissue morphology. Improper positioning of the cytokinetic plane can lead to mis-segregation of chromatin and cell fate determinants resulting in defective morphogenesis (33). The nature and source of factors that position the cytokinetic plane are unclear. The way the cytokinetic plane is decided in metazoans is different from bacteria, fungi, and plants. Unlike in fungi and plants where the site is selected before commencing karyokinesis, animals select the furrow site after separating DNA. It has been hypothesized that animal cells prefer to adjust the cytokinetic plane depending on the position and orientation of the mitotic spindle in response to mechanical, geometrical, and biochemical signals from neighboring cells. This plasticity of deciding the division plane is fundamental to the asymmetric divisions during early embryonic development (34).
Landmark studies in echinoderm eggs (35) demonstrated the importance of the spindle in specification of the division site. Both chromosomes and centrioles are dispensable for furrow initiation (36,37). However, inhibition of microtubule polymerization during early, but not the later stages of mitosis prevents furrowing (38). Studies in different organisms have contested the role of the spindle midzone (antiparallel microtubules between the separating chromosomes) and astral microtubules in cytokinetic site positioning (Fig. 3A) (34).
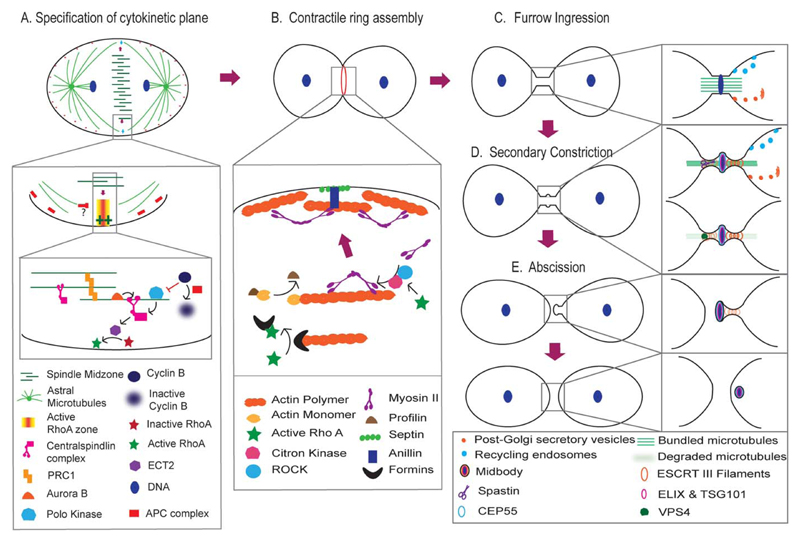
Fig 3. Mechanism of cytokinesis in animal cells.
A: Specification of the cytokinetic plane by cues from the mitotic spindle. Aster density limits cortical contractility only to the equator while inhibiting it at the polar cortex. The spindle midzone activates RhoA at the equator upon decline in Cdk1 activity via the Centralspindlin-ECT1 pathway. B: RhoA assembles the actomyosin based contractile ring by activating both actin and myosin at the equatorial plane. C: Furrow contraction and membrane insertion by vesicular trafficking forms a narrow cytoplasmic bridge between the daughter cells, at the center of which lies an electron-dense midbody. D: A secondary constriction mediated by the ESCRT complex appears on either side of the midbody. The microtubule bundle is disassembled by Spastin. E: Finally the ESCRT complex disappears and membrane abscission occurs, separating the two daughter cells.
The precise molecular mechanisms that position the furrow are debated. Multiple models agree on key roles for microtubules in initiating the furrow. The “astral stimulation” model proposes that astral microtubules stimulate furrowing by transporting an unknown factor preferentially to the equatorial cortex because it is influenced by asters from both the poles. The “astral relaxation” model asserts that high concentration of asters at the polar cortices causes cortical stiffness, thereby reducing its contractility leading to furrow induction at the more supple equatorial membrane (37). A third model emphasizes the role of the central spindle in deciding the division plane (33,37).
Classical experiments in echinoderm embryos showed that opposing asters alone in the absence of the central spindle can lead to furrow specification (39). However, evidence from multiple other organisms elucidates the importance of the central spindle in specifying the furrow plane. “Rapport experiments” in cultured mammalian cells showed that it is the central spindle and not the astral microtubules that initiate furrowing (40). Sea urchin, Xenopus and Drosophila embryos initiate furrowing even in the absence of close contact between astral microtubules with the cell cortex. These results question the astral stimulation model; rather they propose an inhibitory role of asters on the initiation signal, spatially restricting it to the equator (41).
A recent report in Drosophila demonstrated that the furrow can be initiated just by inhibiting Cdk1 activity even in the absence of organized microtubules (42). In C. elegans embryos, the central spindle was initially shown to be dispensable for furrow initiation (43). However, further genetic and laser ablation experiments demonstrated the existence of parallel and consecutive signals from astral tubules (that sense cell geometry) and the central spindle (that senses chromosome segregation) for furrow positioning. Interestingly, signaling from the central spindle supersedes that from astral microtubules (44).
In summary, animal cells position the division plane using multiple spatial cues to ensure error-free cell division. Both the spindle midzone and astral microtubules cooperate in this process and can each have different levels of contribution depending on organism and cell type. However, once initiated, the furrow is strengthened by persistent signaling from the spindle midzone (34).
Assembling the Cytokinetic Machinery
A key feature of cytokinesis is the contractile ring made of actin, non-muscle myosin, formins, and other associated proteins that assembles at the equatorial cortex. Molecular features of the contractile ring are similar in yeast and animals (37). Activation of the small GTPase RhoA is central to contractile ring assembly in C. elegans, Drosophila, sea urchins, echinoderms, and Xenopus embryos as well as mammalian cells (Fig. 3A) (34). The spindle midzone acts as a platform for key cytokinetic regulators for RhoA activation, which is limited to a narrow equatorial band through negative signaling from astral tubules (Fig. 3A). It is debatable whether signals from the spindle reach the cortex via active transport along microtubules or through cytoplasmic diffusion (34,45).
Like other small GTPases, RhoA exists in a GTP-bound (active) and GDP-bound (inactive) form. At the center of RhoA activation is the evolutionarily conserved centralspindlin complex, containing GTPase activating protein CYK-4/MgcRacGAP and mitotic kinesin-like protein MKLP1/ZEN-4. The complex localizes to the plus-ends of microtubules both at the spindle midzone and at peripheral asters near the equator (45). The centralspindlin complex recruits and activates ECT-2, a GTP exchange factor (GEF) that activates RhoA at the equatorial cortex. Fly Cyk-4 has been shown to physically bind and activate ECT2 (46), while MKLP1 may merely act as a localization factor as demonstrated in worms and mammalian cells that allow furrow formation even in the absence of MKLP1 (47,48). The spatiotemporal mechanism for RhoA activation is still unclear. It is hypothesized that Cyk-4 maintains a constant flux of RhoA through constant phosphorylation–dephosphorylation cycles, limiting its activity to a narrow contractile ring (Fig. 3A). Curiously, Cyk-4 has never been shown to be a RhoA-GAP. Recently in C. elegans embryos, a M-phase GAP (MP-GAP) was demonstrated as the primary GAP for RhoA (49). In addition to activating RhoA, Cyk-4 inactivates Rac, another Rho family GTPase, to stop actin filament branching in the contractile ring (50).
It is also unclear how actin and myosin are targeted to the cell cortex. Active RhoA initiates polymerization of equatorial actin by relieving self-inhibition of formins, inhibitors of actin polymerization and activates nucleation via profilins, the transporters of actin monomers (Fig. 3B). Excess of actin is cleared by cofilin, which destabilizes the redundant actin filaments. Thus, active assembly and disassembly of F-actin are required for proper furrow constriction. In different animal systems, myosin reaches the cortex independent of actin, either by random diffusion or via cortical flow (51). Myosin is stabilized at the equator by RhoA effectors rock and citron kinases (52) that phosphorylate myosin regulatory chain to activate its motor activity, making it competent to bind actin and assemble into myosin filaments (Fig. 3B). Alternatively, in mammalian cells recombinant myosin filaments lacking regulatory subunits can also assemble at the furrow and drive cytokinesis, suggesting a RhoA-independent mechanism of assembly at the cortex probably via cortical flow and interactions with heterologous protein partners (51). Myosin recruitment to the polar cortex is inhibited by astral microtubules, which further increases the contractility of the equatorial cortex (53).
The contractile ring is linked with the plasma membrane via anillin, a conserved scaffolding protein considered to be a key organizer of the cytokinetic machinery (Fig. 3B) (54). In the absence of anillin, the furrow initiates and ingresses but does not complete cytokinesis. Anillin is localized in the nucleus during S-phase and recruited to the cortex during cytokinesis, most likely by the RhoA/ECT2 pathway as demonstrated in human cells and Drosophila (55,56). Anillin bundles F-actin filaments, stabilizes myosin at the equator, and also interacts with multiple other furrow proteins. Anillin is proposed to link the contractile ring with the plasma membrane via its membrane interacting PH-domain and by recruiting septins, conserved membrane associated GTP-binding proteins, to the furrow (Fig. 3B) (54). Moreover, the furrow membrane has a specific phosphotidylinositol lipid composition. Both, phosphotidylinositol lipids and lipid kinases have been shown to regulate cytokinesis (57). Further work is required to elucidate the exact temporal mechanisms of actomyosin ring assembly.
Constriction and Furrow Ingression
Persistent signaling from the spindle midzone and inhibition of polar contractility by the dense asters is required for furrow ingression. Mathematical models suggest that line tension of the contractile ring and resistance to polar contractility (58) propel furrow ingression. How the actomyosin ring actually generates the force for constriction is still an open field for future biophysical investigations. High resolution microscopy and mutant analysis suggest that myosin filaments slide along actin filaments analogous to the force-generating machinery in muscles. The ring is constantly remodeled during constriction (59). In sea urchins and C. elegans, the ring disassembles and contracts at the same rate indicating that the concentration of its components per unit length remains constant during constriction. The time taken for this process to complete seems to be independent of the initial cell size at least in C. elegans embryos, perhaps for maintaining synchronous divisions (60).
Increase in surface area at the time of constriction is contributed by polarized vesicular trafficking that inserts additional membrane at the furrow. Several vesicular trafficking components localize to the leading edge of the ingressing furrow (Fig. 3C) (61). Factors that initiate ring constriction, regulate ring disassembly, and clearing of the components as it contracts need to be elucidated.
Abscission
Actomyosin ring constriction compacts the spindle midzone forming a narrow (1–2 μm diameter) intercellular cytoplasmic bridge. At the center of this bridge is an electron-dense structure called the midbody, made-up of condensed midzonal antiparallel microtubules traversing through a dense midbody ring made of several proteins (Figs. 3C and 3D) (62,63). The midbody serves as a scaffold for the assembly of the abscission machinery and anchors the ingressed furrow.
Several spindle midzone proteins remain associated with the midbody core (e.g., PRC1, KLF4) while some (e.g., Aurora B) relocalize to adjacent microtubules and a few others (e.g., Centralspindlin) concentrate on the midbody ring (59,64). The midbody ring contains several contractile ring components like anillins, septins, and RhoA (59,64). Anillins and septins anchor the ingressed furrow to the midbody ring. Recently, centralspindlin was also implicated in furrow anchorage via a membrane interacting domain of Cyk-4 and physical interaction of MKLP 1 with a membrane associated small GTPase Arf 6 (45).
Abscission is a two-step process requiring disassembly of the cytoskeletal material followed by membrane fusion (Figs. 3C and 3D). The bridge narrows and the actomyosin ring is disassembled by regulating RhoA activity. Later, cortical rearrangements form a secondary constriction on either side of the midbody ring. Both post-Golgi and endosomal vesicles are targeted to the midbody and are thought to fuse with the plasma membrane allowing narrowing of the intercellular bridge (65) (Fig. 3D). Endosomal vesicles also help in disassembly of cortical actin by delivering p50RhoGAP to the bridge (66). The exact roles of vesicular fusion, mechanism of targeting to the midbody, and the cargo that they deliver remain elusive. It has been suggested that membrane vesicles are required to set the stage for abscission but are not required for the terminal cut (59).
The secondary constriction contains membrane associated ESCRT complex (Endosomal Complexes Required for Transport) filaments encircling the bridge perpendicular to its length (Fig. 3D) (59). Studies in mammalian cells propose that MKLP1 recruits CEP55 to the midbody, which further brings ESCRT I (ALIX and TSG101) and ESCRT III components to the prospective constriction sites on either side of the midbody. Microtubules underneath the constriction sites are disassembled by AAA-ATPase spastins recruited by the ESCRT III complex (67). Further ESCRT III filaments likely get disassembled by another AAA-ATPase VPS4 recruited to the intercellular bridge by the ESCRT complex itself (Fig. 3D) (59). Finally, the membrane stochastically fuses at one or both sides of the midbody, splitting the cell (Fig. 3E). The exact details of how ESCRTs bring about the cut remain a blur and their roles in different animal models still need to be explored (59).
Variations in the Theme
Organisms have evolved several variations to conventional cytokinesis initiated by different molecular programs during the course of development. The contractile machinery of these variations is similar to the cytokinetic ring, but its assembly varies temporally and spatially.
Incomplete Cytokinesis
Karyokinesis is not always followed by cytokinesis. During insect embryogenesis, multiple rounds of rapid nuclear divisions occur without completing cytokinesis, forming a syncytium (Fig. 4B) (1). In sea urchins, the first few cleavages are incomplete, allowing sister blastomeres to remain connected via cytoplasmic bridges that may be required for intercellular communication in the absence of conventional gap junctions (68). Similar intercellular bridges are present in squid embryos where the midbody ring components disappear once the furrow ingresses, enabling vesicular and organelle transport between cells (69).
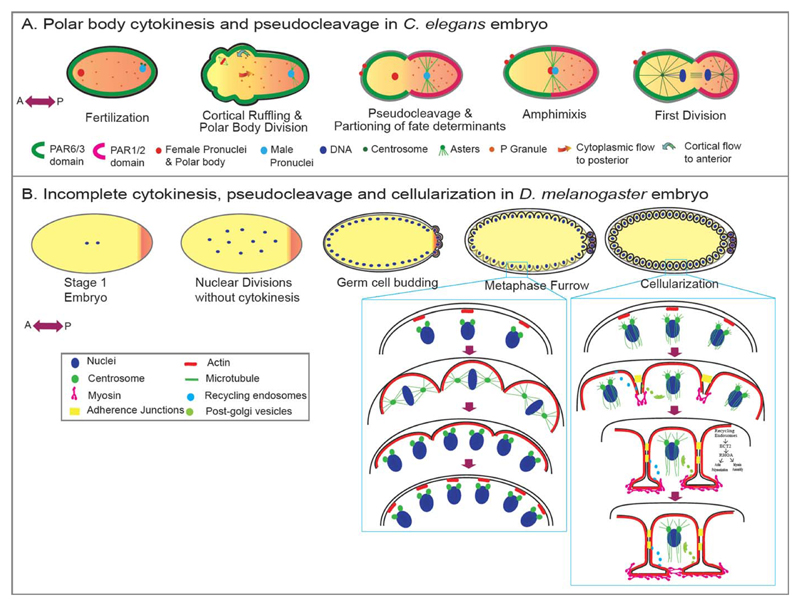
Fig 4. Variations in cytokinesis during metazoan development.
A: Fertilization-triggered meiotic divisions of the oocyte in C. elegans are extremely asymmetric resulting in polar bodies devoid of cytoplasm and the haploid oocyte retaining most of the cytoplasm. Soon after fertilization, cortical contraction in the C. elegans embryo forms a pseudocleavage that segregates the fate determinants in the zygote, a process that is essential for antero-posterior axis specification. B: Drosophila embryos initially demonstrate a cytokinesis-free nuclear division upto 8 nuclear cycles. At the 9th nuclear division, these nuclei migrate to the periphery and continue to divide without undergoing cytoplasmic divisions for another three cycles. During these peripheral divisions, they form pseudocleavages/partial furrows to shield the chromatin from the influence of neighboring asters. The nuclei finally undergo cellularization at interphase of the 14th nuclear division, utilizing an actomyosin-based contractile machinery similar to conventional cytokinesis.
Pseudocleavage
Partial furrows that regress with time represent another variation in cytokinesis. In Drosophila, pseudo-cleavages ensure proper segregation of chromosomes during peripheral nuclear divisions of stage 10–13 (Fig. 4B) (70). As the centrosomes migrate to opposite poles in early mitosis, they reorganize cortical actin, which encircles the dividing chromatin to shield them from neighboring asters, before finally regressing during telophase (Fig. 4B inset) (70). Drosophila partial furrows are stabilized by anillin and septins in a RanGTPase dependent pathway (70). Right after fertilization in C. elegans embryos, the anterior cortex contracts and forms a transient pseudocleavage directed by the sperm aster (Fig. 4A), which helps in establishing the division asymmetry to specify the anteroposterior axis and to segregate fate determinants (Fig. 4A) (71). As opposed to anaphase-initiated cytokinetic furrow, a contractile ring similar to conventional cytokinesis assembles during prophase to metaphase (70).
Cellularization
Cellularization of nuclei is mechanistically similar to cytokinesis. In Drosophila embryos, cellularization is initiated during the 14th nuclear division. Astral mictotubules direct remodeling of actin and ingression of the furrow through a RhoA-dependent actomyosin network, as seen in cytokinesis. As the furrow invaginates, post-Golgi and recycling endosome vesicles fuse with the leading edge of the furrow forming a double-membrane (Fig. 4B) (70). Similar to pseudocleavage, the cellularization furrow is initiated before chromatin separation in contrast to the late anaphase-initiated cytokinetic furrows signifying a markedly different method of furrow positioning in these unconventional forms of cytokinesis.
Polar Body Extrusion
Cytokinesis during meiotic division of the oocyte produces a large haploid egg and two haploid nuclei almost devoid of cytoplasm, called the polar body (1). In most animals, polar body cytokinesis happens right before amphimixis. Faulty polar body extrusion can lead to lethality due to increased ploidy. Unlike mitotic cytokinesis, the cleavage plane in polar body cytokinesis is determined by chromatin before anaphase onset via Ran GTP signaling without the help of astral or spindle midzone microtubules. Chromatin induces meiotic spindle migration, localization, and anchoring closer to the cortex in a perpendicular orientation. Final extrusion is mediated by the RhoA-dependent actomyosin machinery similar to cytokinesis (Fig. 4A) (72).
Coordinating Nuclear and Cytoplasmic Divisions
Cytoplasmic divisions typically begin at the end of nuclear divisions. Several factors that are required for proper chromosome segregation are also later required for cytokinesis. Tight spatio-temporal regulation of cytokinetic initiation by karyokinesis ensures high fidelity of cell division. The metaphase to anaphase transition is orchestrated by the APC that targets Cdk1 for degradation. The same molecular switch also leads to activation of the spindle midzone proteins to initiate cytokinesis (Fig. 3A). Although the switch for chromosome separation and cytokinesis remains the same, they are temporally regulated by a relative delay in activation of mitotic exit phosphatase PP2A-B55 (73).
The spindle midzone contains three major groups of proteins—centralspindlin complex (MKLP1/MgcRacGAP), Aurora B/INCENP (Inner-centromeric protein)/CPC (chromosomal-passenger complex), and PRC1 (protein regulating cytokinesis) (74). Proteins from each of these groups are phosphorylated by Cdk1 in early mitosis, leading to their inactivation until chromatin separation. APC releases this inhibition resulting in spindle midzone assembly that further signals actomyosin ring formation (74). Decline in Cdk1 activity concurrent with phoshphorylation of PRC1 by CPC allows it to dimerize and bind the antiparallel microtubule interface, spatially organizing them into bundles to form the spindle midzone (Fig. 3A). Additionally, Cdk1 inhibits the motor activity of MKLP1 that is relieved by Aurora B in animal cells and C. elegans, making it competent to bind the spindle (75). This allows CPC translocation from chromosomes to the spindle midzone by MKLP motor-driven transport in animal cells (59). However in C. elegans embryos, CPC can also reach the midzone in a MKLP-independent manner (75). Another key regulator of cytokinesis in multiple organisms is Polo-like kinase (Plk1). Plk1 phosphorylates Cyk-4 allowing it to interact with ECT2 in human cells (Fig. 3A) (76). Additionally, flies mutant for another CPC component Survivin show defects in central spindle formation and MKLP recruitment (77). A yeast-like “No cut” pathway has also been observed in animal cells and C. elegans embryos wherein both Plk1 and Aurora B kinases delay abscission by negatively regulating the ESCRT III complex until segregation of chromosomes is complete (59).
Cytokinetic midbody remnants have also been proposed to regulate spindle orientation and spindle tethering during the second embryonic division of C. elegans. The midbody remnant of the first embryonic division is inherited by the smaller posterior cell P1. During subsequent division of P1, the midbody remnant directs spindle rotation along the shorter axis of the cell making an exception to the Hertwig long-axis rule. The midbody also helps in the anchoring of the spindle and skews it ventrally, an event that is essential for dorso-ventral embryonic patterning (78).
Conclusions and Questions
The molecular mechanisms of early embryonic divisions are fundamentally conserved across metazoa. During karyokinesis, after nuclear envelope disintegration, condensed chromosomes of the mother cell organize around the equator of the cell with the help of the mitotic spindle and segregate equally to daughter cells, after which they are repackaged into new nuclei. In close orchestration with karyokinesis, the cytoplasmic contents of the mother cell start partitioning toward the two future daughter cells and dictate the timing and positioning of cleavage furrow ingression, eventually leading to the formation of euploid daughter cells. The entire division process involves cells going from a polarized to an unpolarized state during early mitosis, with polarity re-established by cytokinesis. Even seemingly symmetric embryonic divisions are biochemically asymmetric, with the programmed unequal partitioning of several cellular components shaping the course of embryonic development. Despite the inherent similarities, mechanistic variations in these two stages of early divisions across different animals play key roles in shaping their divergent developmental patterns. Mitotic spindle positioning as well as specification of the division plane axis are differentially governed in different species. Moreover, while most nuclear divisions are followed by faithful separation of cytoplasm, some animals have evolved to undergo multiple rounds of nuclear division without cytokinesis.
Despite intense research on early embryonic divisions, there are several intriguing questions that remain unanswered. The role of centrosomes in cell-fate specification in the early embryo is poorly understood. How does the paternal centrosome in the one-celled embryo govern the sanctity of the first cleavage division? How do the mother and daughter centrosomes of blastomeres govern the subsequent cleavage planes? During cytoplasmic division, how do molecular signals from the midzone and astral microtubules physically transmit to the midzonal cortex, especially in large embryonic cells, where the mitotic spindle occupies a relatively minor volume at the center of the cytoplasm? Further, how are actin and myosin recruited specifically to the equatorial plane? Finally, what is the molecular crosstalk between extracellular cues and intracellular division mechanisms in regulating mitosis during embryogenesis? Research on such questions across different metazoan models will also illuminate how different species have evolved to make mechanistic alterations to fundamental processes such as embryonic cell divisions. Such studies will also have a significant impact on cancer research and would open avenues for therapeutic intervention.
Acknowledgements
The authors thank all members of the Laboratory of Cellular Dynamics, Regional Centre for Biotechnology for critical reading and comments. All relevant literature could not be cited due to space constraints. The authors acknowledge funding from the Regional Centre for Biotechnology, the Department of Biotechnology of the Government of India, and the Wellcome Trust-DBT India Alliance.
Footnotes
The authors declare that there are no conflicts of interest.
References
- [1].Gilbert SF, editor. Developmental Biology. 6th edn. Sinauer Associates; Sunderland (MA): 2000. [Google Scholar]
- [2].Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, et al. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell. 2013;25:270–283. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- [3].Morin X, Bellaiche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- [4].Cassimeris L, Skibbens RV. Regulated assembly of the mitotic spindle: a perspective from two ends. Curr Issues Mol Biol. 2003;5:99–112. [PubMed] [Google Scholar]
- [5].Raaijmakers JA, Medema RH. Function and regulation of dynein in mitotic chromosome segregation. Chromosoma. 2014;123:407–422. doi: 10.1007/s00412-014-0468-7. [DOI] [PubMed] [Google Scholar]
- [6].Howe K, FitzHarris G. Recent insights into spindle function in mammalian oocytes and early embryos. Biol Reprod. 2013;89:71. doi: 10.1095/biolreprod.113.112151. [DOI] [PubMed] [Google Scholar]
- [7].Gadde S, Heald R. Mechanisms and molecules of the mitotic spindle. Curr Biol. 2004;14:R797–R805. doi: 10.1016/j.cub.2004.09.021. [DOI] [PubMed] [Google Scholar]
- [8].Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;198:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thery M, Bornens M. Cell shape and cell division. Curr Opin Cell Biol. 2006;18:648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- [11].Gillies TE, Cabernard C. Cell division orientation in animals. Curr Biol. 2011;21:R599–R609. doi: 10.1016/j.cub.2011.06.055. [DOI] [PubMed] [Google Scholar]
- [12].Castanon I, Gonzalez-Gaitan M. Oriented cell division in vertebrate embryogenesis. Curr Opin Cell Biol. 2011;23:697–704. doi: 10.1016/j.ceb.2011.09.009. [DOI] [PubMed] [Google Scholar]
- [13].Gonczy P. Mechanisms of spindle positioning: focus on flies and worms. Trends Cell Biol. 2002;12:332–339. doi: 10.1016/s0962-8924(02)02306-1. [DOI] [PubMed] [Google Scholar]
- [14].Werts AD, Goldstein B. How signaling between cells can orient a mitotic spindle. Semin Cell Dev Biol. 2011;22:842–849. doi: 10.1016/j.semcdb.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li R. The art of choreographing asymmetric cell division. Dev Cell. 2013;25:439–450. doi: 10.1016/j.devcel.2013.05.003. [DOI] [PubMed] [Google Scholar]
- [16].Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kotak S, Gonczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 2013;25:741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- [18].Tang N, Marshall WF. Centrosome positioning in vertebrate development. J Cell Sci. 2012;125:4951–4961. doi: 10.1242/jcs.038083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Moore JK, Cooper JA. Coordinating mitosis with cell polarity: molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lancaster OM, Baum B. Shaping up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol. 2014;34:109–115. doi: 10.1016/j.semcdb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- [22].Blum M, Schweickert A, Vick P, Wright CV, Danilchik MV. Symmetry breakage in the vertebrate embryo: when does it happen and how does it work? Dev Biol. 2014;393:109–123. doi: 10.1016/j.ydbio.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McCarthy EK, Goldstein B. Asymmetric spindle positioning. Curr Opin Cell Biol. 2006;18:79–85. doi: 10.1016/j.ceb.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McNally KL, Martin JL, Ellefson M, McNally FJ. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Dev Biol. 2010;339:126–140. doi: 10.1016/j.ydbio.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schatten H, Sun QY. The role of centrosomes in fertilization, cell division and establishment of asymmetry during embryo development. Semin Cell Dev Biol. 2010;21:174–184. doi: 10.1016/j.semcdb.2010.01.012. [DOI] [PubMed] [Google Scholar]
- [27].Ahringer J. Control of cell polarity and mitotic spindle positioning in animal cells. Curr Opin Cell Biol. 2003;15:73–81. doi: 10.1016/s0955-0674(02)00018-2. [DOI] [PubMed] [Google Scholar]
- [28].Muller HA. Of mice, frogs and flies: generation of membrane asymmetries in early development. Dev Growth Differ. 2001;43:327–342. doi: 10.1046/j.1440-169x.2001.00587.x. [DOI] [PubMed] [Google Scholar]
- [29].Kops GJ. Dividing the goods: co-ordination of chromosome biorientation and mitotic checkpoint signalling by mitotic kinases. Biochem Soc Trans. 2009;37:971–975. doi: 10.1042/BST0370971. [DOI] [PubMed] [Google Scholar]
- [30].Rago F, Cheeseman IM. Review series: the functions and consequences of force at kinetochores. J Cell Biol. 2013;200:557–565. doi: 10.1083/jcb.201211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pereira AJ, Maiato H. Maturation of the kinetochore-microtubule interface and the meaning of metaphase. Chromosome Res. 2012;20:563–577. doi: 10.1007/s10577-012-9298-8. [DOI] [PubMed] [Google Scholar]
- [32].Vleugel M, Hoogendoorn E, Snel B, Kops GJ. Evolution and function of the mitotic checkpoint. Dev Cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- [33].Oliferenko S, Chew TG, Balasubramanian MK. Positioning cytokinesis. Genes Dev. 2009;23:660–674. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- [34].von Dassow G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009;19:165–173. doi: 10.1016/j.tcb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- [35].Rappaport R, Rappaport BN. Duration of division-related events in cleaving sand dollar eggs. Dev Biol. 1993;158:265–273. doi: 10.1006/dbio.1993.1185. [DOI] [PubMed] [Google Scholar]
- [36].Hiramoto Y. Mechanics of cleavage in the sea urchin egg. Jpn J Med Sci Biol. 1965;18:320–321. [PubMed] [Google Scholar]
- [37].Glotzer M. Cleavage furrow positioning. J Cell Biol. 2004;164:347–351. doi: 10.1083/jcb.200310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hamaguchi Y. Microinjection of colchicine into sea urchin eggs. Dev Growth Diff. 1975;17:111–117. doi: 10.1111/j.1440-169X.1975.00111.x. [DOI] [PubMed] [Google Scholar]
- [39].Rappaport R. Experiments concerning the cleavage stimulus in sand dollar eggs. J Exp Zool. 1961;148:81–89. doi: 10.1002/jez.1401480107. [DOI] [PubMed] [Google Scholar]
- [40].Cao LG, Wang YL. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].von Dassow G, Verbrugghe KJ, Miller AL, Sider JR, Bement WM. Action at a distance during cytokinesis. J Cell Biol. 2009;187:831–845. doi: 10.1083/jcb.200907090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Menant A, Karess RE. Inducing “cytokinesis” without mitosis in unfertilized Drosophila eggs. Cell Cycle. 2012;11:2856–2863. doi: 10.4161/cc.21190. [DOI] [PubMed] [Google Scholar]
- [43].Verbrugghe KJ, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14:1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- [44].Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- [45].White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- [47].Dechant R, Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- [48].Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zanin E, Desai A, Poser I, Toyoda Y, Andree C, et al. A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell. 2013;26:496–510. doi: 10.1016/j.devcel.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Canman JC. Cytokinetic astralogy. J Cell Biol. 2009;187:757–759. doi: 10.1083/jcb.200911084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Beach JR, Egelhoff TT. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J Biol Chem. 2009;284:27377–27383. doi: 10.1074/jbc.M109.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Uehara R, Goshima G, Mabuchi I, Vale RD, Spudich JA, et al. Determinants of myosin II cortical localization during cytokinesis. Curr Biol. 2010;20:1080–1085. doi: 10.1016/j.cub.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Werner M, Munro E, Glotzer M. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Piekny AJ, Maddox AS. The myriad roles of Anillin during cytokinesis. Semin Cell Dev Biol. 2010;21:881–891. doi: 10.1016/j.semcdb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- [55].Somma MP, Fasulo B, Cenci G, Cundari E, Gatti M. Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol Biol Cell. 2002;13:2448–2460. doi: 10.1091/mbc.01-12-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- [57].Echard A. Phosphoinositides and cytokinesis: the “PIP” of the iceberg. Cytoskeleton (Hoboken) 2012;69:893–912. doi: 10.1002/cm.21067. [DOI] [PubMed] [Google Scholar]
- [58].Turlier H, Audoly B, Prost J, Joanny JF. Furrow constriction in animal cell cytokinesis. Biophys J. 2014;106:114–123. doi: 10.1016/j.bpj.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- [60].Carvalho A, Desai A, Oegema K. Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell. 2009;137:926–937. doi: 10.1016/j.cell.2009.03.021. [DOI] [PubMed] [Google Scholar]
- [61].McKay HF, Burgess DR. ‘Life is a highway’: membrane trafficking during cytokinesis. Traffic. 2011;12:247–251. doi: 10.1111/j.1600-0854.2010.01139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-98s cells. J Cell Biol. 1977;73:672–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Skop AR, Liu H, Yates J, III, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Ann Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- [65].Chen CT, Hehnly H, Doxsey SJ. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat Rev Mol Cell Biol. 2012;13:483–488. doi: 10.1038/nrm3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schiel JA, Prekeris R. Membrane dynamics during cytokinesis. Curr Opin Cell Biol. 2013;25:92–98. doi: 10.1016/j.ceb.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Reid E, Connell J, Edwards TL, Duley S, Brown SE, et al. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet. 2005;14:19–38. doi: 10.1093/hmg/ddi003. [DOI] [PubMed] [Google Scholar]
- [68].Andreuccetti P, Barone Lumaga MR, Cafiero G, Filosa S, Parisi E. Cell junctions during the early development of the sea urchin embryo (Paracentrotus lividus) Cell Differ. 1987;20:137–146. doi: 10.1016/0045-6039(87)90427-1. [DOI] [PubMed] [Google Scholar]
- [69].Cartwright J, Jr, Arnold JM. Intercellular bridges in the embryo of the Atlantic squid, Loligo pealei. II. Formation of the bridge. Cell Motil. 1981;1:455–468. doi: 10.1002/cm.970010406. [DOI] [PubMed] [Google Scholar]
- [70].Lee DM, Harris TJ. Coordinating the cytoskeleton and endocytosis for regulated plasma membrane growth in the early Drosophila embryo. Bioarchitecture. 2014;4:68–74. doi: 10.4161/bioa.28949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Maddox AS, Azoury J, Dumont J. Polar body cytokinesis. Cytoskeleton (Hoboken) 2012;69:855–868. doi: 10.1002/cm.21064. [DOI] [PubMed] [Google Scholar]
- [73].Cundell MJ, Bastos RN, Zhang T, Holder J, Gruneberg U, et al. The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol Cell. 2013;52:393–405. doi: 10.1016/j.molcel.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Berridge MJ. Cell Signalling Biology. Portland Press Limited; 2012. [Google Scholar]
- [75].Douglas ME, Davies T, Joseph N, Mishima M. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr Biol. 2010;20:927–933. doi: 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Wolfe BA, Takaki T, Petronczki M, Glotzer M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009;7:e1000110. doi: 10.1371/journal.pbio.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cabernard C. Cytokinesis in Drosophila melanogaster. Cytoskeleton (Hoboken) 2012;69:791–809. doi: 10.1002/cm.21060. [DOI] [PubMed] [Google Scholar]
- [78].Singh D, Pohl C. A function for the midbody remnant in embryonic patterning. Commun Integr Biol. 2014;7:e28533. doi: 10.4161/cib.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]