Abstract
We combined recognition imaging and force spectroscopy to study the interactions between receptors and ligands on the single molecule level. This method allowed the selection of a single receptor molecule reconstituted in a supported lipid membrane at low density, with the subsequent quantification of the receptor-ligand unbinding force. Based on atomic force microscopy (AFM) tapping mode, a cantilever tip carrying a ligand molecule was oscillated across a membrane. Topography and recognition images of reconstituted receptors were recorded simultaneously by analyzing the downward and upward parts of the oscillation, respectively. Functional receptor molecules were selected from the recognition image with nanometer resolution before the AFM was switched to the force spectroscopy mode, using positional feedback control. The combined mode allowed for dynamic force probing on different pre-selected molecules, resulting in higher throughput when compared with force mapping. We applied this method for a quantitative characterization of the binding mechanism between mitochondrial uncoupling protein 1 (UCP1) and its inhibitor adenosine triphosphate (ATP). Moreover the dynamics of force loading was varied to elucidate the binding dynamics and map the interaction energy landscape.
Keywords: Atomic Force Microscopy, Single Molecule Force Spectroscopy, Recognition Imaging, Membrane, Uncoupling Protein (UCP), Adenosine Triphosphate (ATP)
1. Introduction
Ligand binding to receptors is one of the most important regulatory elements in biology as the initiating step in signaling pathways and cascades. Measuring the interaction forces between cognate receptor-ligand pairs leads to new insights into the recognition process between the interaction partners.1, 2 Because of its piconewton (pN, 10−12 N) force sensitivity and nanometer positional precision, as well as its possibility to be operated under nearly physiological conditions, the atomic force microscope (AFM) has proven to be a powerful tool for exploring such forces on the single molecule level by employing the force spectroscopy mode.3 This technique has been widely used to gain new insights into the molecular principles of many biological processes and phenomena, such as protein folding and unfolding,4, 5 DNA mechanics,6 cell adhesion7 and molecular recognition.8–10 For the detection of receptor-ligand forces, ligands are bound to the outer apex of the AFM tip and receptors are coupled to the probe surface, or vice versa. In the force measurements, the tip first approaches (termed ‘trace’) the surface, whereupon a receptor-ligand complex is possibly formed due to specific ligand-receptor recognition. Thereafter the tip is retracted (termed ‘retrace’) from the probe surface. These tip movements are repeated many times in a cyclic fashion at a fixed lateral position. Provided that a receptor ligand-complex has formed in the trace, a temporarily increasing force is applied to the receptor-ligand bond during retrace. Finally the bond breaks at a certain measurable force (termed ‘unbinding force’). The force recordings are depicted in force-distance cycles (FDCs). Varying the dynamics of the experiment changes the loading rate (rate of increasing force) applied to the receptorligand bond. Data extracted from such studies (termed ‘force spectroscopy,’ ‘FS’) yield information about the binding pocket, binding energy barriers and chemical reaction rates.8–10
For practical purposes a dense receptor distribution on the probe surface is preferred to ensure for a high probability in receptor-ligand complex formation during FDCs. Thus conventional FS is not well applicable for very low receptor densities, as is often the case for membrane proteins reconstituted in artificial lipid membranes. A possible application of FS on such delicate samples is performing FDCs on different positions during simultaneous lateral scans. This mode, found in the literature as force volume or force mapping, was originally proposed by Cleveland et al.11 The first proof of concept on biological samples has been realized by visualizing streptavidin patterns using a biotinylated tip.12 In the meantime this technique has been applied on a variety of organic materials,13, 14 on single receptor molecules15, 16 and on cells17, 18 to map the arrangement of chemical groups. Force mapping mode provides a quantitative analysis of unbinding forces in the investigated scan area, but it is limited by its time resolution. The recoding time of a force map depends on acquisition parameters like pixel resolution, sweep duration or number of FDCs per pixel, and can range up to several hours for high lateral resolution maps. Although a lot of progress has been made in terms of performance and speed,19–22 it is still considerably slower than many conventional imaging modes.
A faster method for mapping molecular recognition sites is TREC (topography and recognition) imaging, which has developed into a versatile tool in biophysical research, molecular biology and bionanotechnology.23–25 The advantages of this non-optical recognition microscopy technique are the following:
-
(i)
The topography of the sample surface (topography image) is obtained simultaneously with the localization of the recognition sites (recognition image), both with nanometer lateral resolution.
-
(ii)
Neither labeling of the sample nor a demanding probe preparation is needed.
-
(iii)
TREC is fast in imaging binding sites even with a lateral resolution comparable to other imaging modes.
The disadvantage of TREC is that it is a pure imaging technique and does not quantify receptor-ligand interaction forces.
TREC is based on a dynamic AFM mode in fluid, in which the tip is magnetically oscillated close to its fundamental resonance frequency and scanned across the sample surface. Additionally, the AFM cantilever carries a ligand on the outer tip apex that is tethered via a flexible polyethylene glycol (PEG) linker. The tip-bound ligand is capable of binding to the cognate receptors on the sample surface.26–28 With this configuration and a modified feedback loop,29 two independent images are recorded simultaneously: A topography image of the sample surface and a lateral map of the corresponding receptor recognition sites. The separation into topographical and recognition signals is achieved by splitting the cantilever’s oscillation amplitude into downward and upward parts with respect to the cantilever’s zero position. The maxima (or peaks) of these two parts are the parameters of choice for recording topography and recognition images independently and at the same time:
-
(i)
topographic elevations reduce the downward amplitude peaks of the tip oscillation, while topographic depressions increase them.
-
(ii)
recognition of a target receptor on the sample surface by a tip-bound ligand causes reduction of the upward amplitude peaks of the tip oscillation due to the physical connection of the tip to the surface.
Consequently, the envelope of the lower amplitudes generates the topographic image, while the envelope of the upper amplitudes yields the map of the recognition sites.
To overcome limits of the force-volume technique with respect to time and spatial resolution and to fulfill the demand of measuring unbinding forces between ligands and receptors at low receptor densities on the surface, we here combined TREC imaging and force spectroscopy. This method enabled us the selection of
-
(i)
a single, functional receptor molecule reconstituted in a supported lipid membrane from the TREC recognition image, and
-
(ii)
the subsequent quantification of the receptor-ligand unbinding force in the FS mode.
As a proof of principle, we studied the interactions between uncoupling protein 1 (UCP1) and adenosine triphosphate (ATP).
UCP1 is expressed in brown adipose tissue (BAT) and is involved in non-shivering thermogenesis under cold-acclimating conditions. After the discovery of BAT in adults the role of UCP1 as a therapeutic target for the obesity treatment has increased.30 Thereby a tight and specific regulation of proton transport through the mitochondrial membrane is essential. We and other research groups have demonstrated that proton transport, which is mediated by UCP1, is inhibited by PN.31–36 Several theories have been published to explain the inhibitory effect of ATP, even though the crystallographic structure of UCP1 is 37, 38 We thus developed a measuring mode for quantitatively characterizing the interactions between ATP bound to AFM tips and UCPs present on probe surfaces at low density. This mode is capable of localizing single molecule recognition events and proving the UCP1’s functionality, as well as quantifying the interaction force with ATP using the AFM.
2. Experimental Details
2.1. Proteoliposome Preparation
If not written otherwise, all chemical were obtained from Sigma GmbH (Munich, Germany). Proteoliposomes containing UCP1 were prepared as previously described.39 In short, murine UCP1 (mUCP1) was cloned in a pET-24a (Novagen, Germany) expression vector, and expressed in E. coli strain Rosetta (DE3) (Novagen, Germany). Isolated inclusion bodies containing mUCP1 were solubilized in TE/G-buffer (100 mM Tris, 5 mM EDTA, 10% glycerin, pH 7.5) with 2% SLS and 1 mM DTT. 50 mg of E. coli polar lipid (Avanti Polar Lipids, Alabaster, AL), dissolved in TE/G-buffer with 2% Triton X-100 and 0.5% octyl-polyoxyethylene, was slowly added to 1 mg of solubilized mUCP1 in the presence of 2 mM GTP and 1 mM DTT. Detergents and GTP were removed by dialysis against assay-buffer (50 mM Na2SO4, 10 mM MES, 10 mM Tris; pH 7.35) and Bio-Beads SM-2 (Bio-Rad, Germany) were used. By centrifugation and application of hydroxyapatite (Bio-Rad, Munich, Germany) aggregates and unfolded proteins were eliminated. The protein content of proteoliposomes was determined using the Micro BCA Protein Assay (Perbio Science, Bonn, Germany). Proteoliposomes were stored at −80 °C until used.
2.2. Surface Chemistry
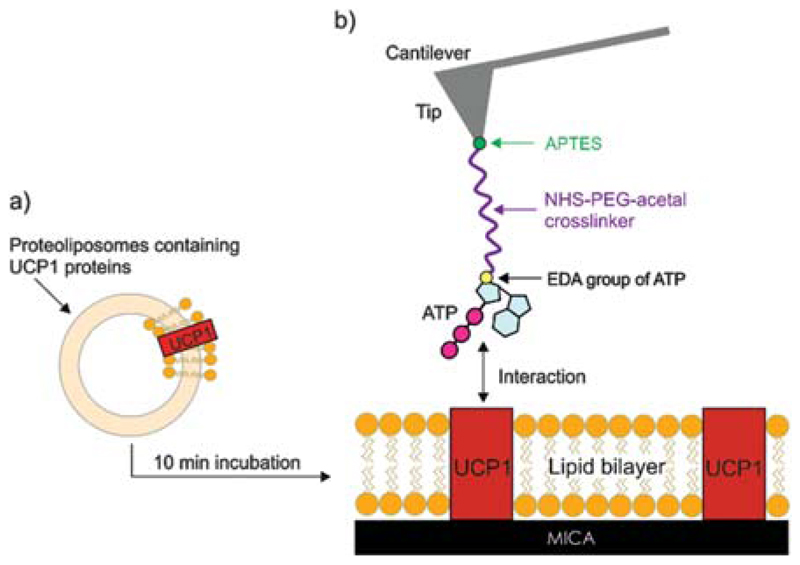
Freshly cleaved mica was placed on the AFM sample plate and mounted with a flow-through fluid cell. Then 20 μl of UCP1 stock solution of proteoliposomes (with a lipid concentration of ~5 mg/ml and protein concentration of ~125.0 μg/ml) was diluted to a final lipid concentration of ~1 mg/ml with assay buffer consisting of 50 mM NaSO4, 10 mM MES, 10 mM TRIS, 0.6 mM EDTA at pH 7.2. After short vortexing, the solution was pipetted into the fluid cell onto the mica surface. After incubation for about 10 min lipid bilayer batches containing UCP1 molecules were formed on the mica surface (shown in Figs. 1 and 4(a)). Thereafter, the sample was thoroughly washed with the assay buffer. Finally, 600–700 μl assay buffer was added to the fluid cell for AFM measurements.35
Fig. 1.
Tip and surface chemistry. (a) Formation of a UCP1 containing lipid bilayer from proteoliposomes after their incubation on mica surface. (b) Tip functionalization with NHS-PEG-acetal and EDA-ATP.
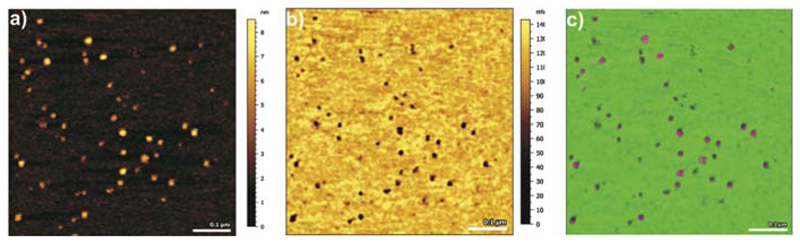
Fig. 4.
TREC imaging: UCP1 distribution and functionality. (a) Topography image. UCP1 molecules imaged with an ATP-tethered tip. (b) Corresponding recognition image. Black dots of the recognition image arise from the decrease of the oscillation upwards peaks as a result from UCP1-ATP binding during recognition. (c) Superposition of recognition map of UCP1-ATP binding domains (in pink) onto the corresponding topography image. Color scale (green to pink) is 0–8 nm. Image sizes are 600 × 600 nm.
2.3. Tip Chemistry
Commercial silicon-nitride AFM cantilevers (MSNL levers, Bruker) were functionalized with the ethylene diamine derivative of ATP (EDA-ATP, 2′-/3′-O-(2-aminoethylcarbamoyl)-adenosine-5′-O-triphosphate, BioLog) by a well-established three-step procedure:
-
(i)
amino-functionalization of the cantilevers by gas phase silanization with (3-aminopropyl)-triethoxysilane (APTES) to convert the tip surface into a chemically addressable surface,40
-
(ii)
attachment of a distensible heterobifunctional polyethylene glycol crosslinker (acetal-PEG-NHS), and
-
(iii)
coupling of EDA-ATP to the free end of the PEG chain.41
The APTES coating was performed exactly according to Ebner et al.40 For coupling of the acetal-PEG-NHS crosslinker, the APTES-functionalized AFM tips were incubated in 0.5 ml of a 1 mg/ml solution of acetal-PEG-NHS in chloroform containing 30 μl of triethylamine (TEA) as catalyst for 2 h. Subsequently, the tips were washed in chloroform (3 × 10 min) and dried in a gentle stream of nitrogen. Immediately before ligand coupling, the acetal group was deprotected by incubation of the acetal-PEG-NHS functionalized tips in 1% citric acid (in water) solution for 10 minutes, followed by rinsing in water (3 × 5 min) and drying as described above. For linking of the EDA-ATP to the free end of the PEG chain, one lyophilized portion of EDA-ATP (10 mM) was redissolved in 9 μl of strong buffer A (1 mM EDTA and 300 mM NaH2PO4, pH adjusted to 7.4 with NaOH) and 1 μl of 200 mM solution of NaCNBH3 (freshly prepared by dissolving 16 mg NaCNBH3 in a mixture of 25 μl 100 mM NaOH plus 225 μl H2O and diluting with 2 ml strong buffer A) was added. After mixing, the cantilevers were placed into a 10 μl droplet and incubated for 1 h. Afterwards 0.5 μl 1 M ethanolamine (pH 9.6) was added for about 10 min in order to passivate unreached aldehyde groups (see Fig. 1(b)). Finally the tips were washed 3 times and stored in strong buffer A at 4 °C.41
2.4. AFM Measurements
All measurements were performed using a PicoPlus 5500 AFM (Keysight, formerly Agilent) and ATP-functionalized cantilever. Experiments were carried out in assay buffer to ensure physiological conditions.
2.4.1. TREC Imaging
Topography and recognition images were simultaneously performed by oscillating an ATP-functionalized cantilever at the 2nd level of its resonance frequency through an alternating voltage-carrying piezo. Images were recorded at an amplitude set point of 23.2 nm, comparable to 90–95% of the amplitude detected before the cantilever touched the sample surface. The scanning speed for imaging was 1.5 line/s, the resolution was set to 256 × 256–512 × 512 pixel, and the scan area varied between 0.3 and 5 μm. Experiments for blocking ATP recognition were conducted by injecting ATP stock solution into the measurement solution to a final concentration of ~8.3 mM.
2.4.2. Force Spectroscopy
For probing the unbinding forces between UCP1 and ATP, as well as their kinetic off rate (koff) and the xβ value, force distance cycles (FDCs) were recorded at different loading rates. FDCs were recorded many times (500–1000) for each loading rate. The loading rate was varied via the pulling speed in z-direction, which was adjusted by means of sweep duration time t [in s] and scan range [in nm], resulting in pulling speeds from 100 nm/s–3000 nm/s. In order to increase the probability for ATP binding, a hold time of 1 s was applied, so as to leave the cantilever on the sample surface for 1s before it was retracted. After recording FDCs the binding probability of ATP binding was calculated according to the ratio of the number of FDCs showing a binding event to the whole number of acquired FDCs. For the spring constant determination the thermal noise method was applied.42 The spring constant determination of each used tip was repeated five times.
3. Results and Discussion
3.1. Experimental Design
Combining the techniques of TREC imaging24 and force spectroscopy43 faces three major challenges:
-
(i)
A common technical issue during AFM measurements is thermal drift, which can lead to the loss of the protein under investigation between two imaging scans or during force spectroscopy experiments. This issue can be significantly improved by using a closed loop scanner (N9524A, Keysight) in which sensors detect the deviations from the ideal movement and apply appropriate corrections to the piezo drive signal.
-
(ii)
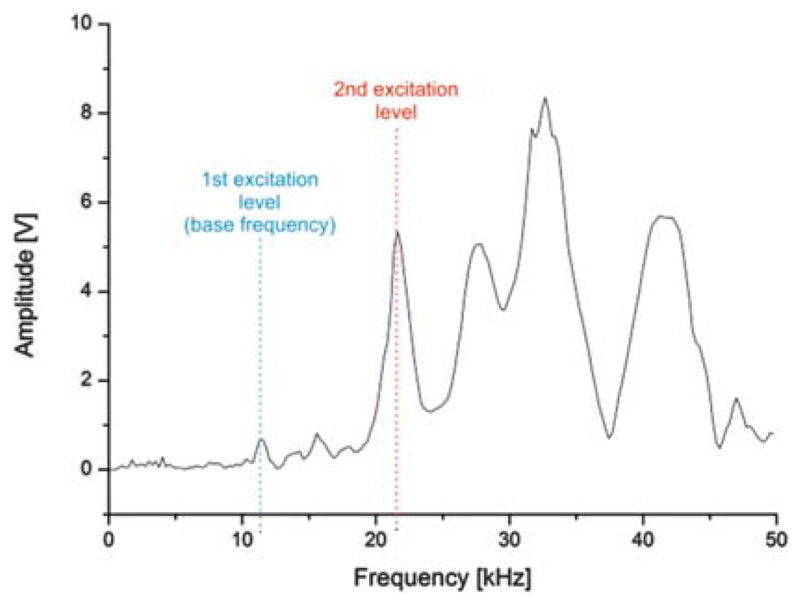
It is mandatory to use the same cantilever for the entire combined TREC and force spectroscopy experiment. For an optimal force sensitivity a rather soft spring is required (spring constant 0.01–0.03 N/m)44 in conventional force spectroscopy, whereas for high-resolution dynamic imaging a harder spring (0.14–0.30 N/m) is preferred.24 This basically means identifying an acceptable compromise which satisfies both needs. This issue was solved by using a silicon nitride tip with a spring constant of 0.03 N/m (MSNL levers, Bruker). Since such tips are not available with a magnetic coating (as it is standard for the magnetic excitation commonly used in TREC24), an acoustic excitation of the cantilever was applied instead. We thus optimized an alternative TREC mode for cantilevers with a small spring constant.
-
(iii)
In order to avoid the remaining drift that still occurred despite using the closed loop scanner, constant environmental conditions were ensured.
We strictly maintained a lab temperature of 21 °C ± 1 °C and did not start the measurements before the system (sample, measurement buffer and cantilever) was completely equilibrated.
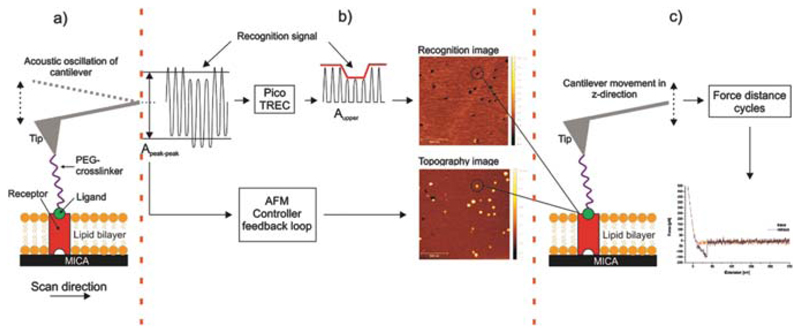
Figure 2 shows the experimental setup. Based on the AFM tapping mode, a cantilever tip carrying a ligand molecule is acoustically oscillated at its second excitation level (see Fig. 3) to gain an adequate amplitude. During scanning across a membrane into which UCPs are reconstituted (Fig. 2(a)), topography and recognition images are recorded simultaneously by analyzing the downwards and upwards peaks of the oscillation (Fig. 2(b)), respectively. Functionally active receptor molecules were selected from the recognition image with nanometer accuracy before the AFM was switched into the force spectroscopy mode using positional feedback control (Fig. 2(c)). Thereafter force distance cycles were recorded at different pulling speeds. The combined mode allowed for dynamic force probing of pre-selected molecules, resulting in a higher throughput when compared with force mapping.
Fig. 2.
Experimental setup of combined TREC imaging and force spectroscopy. (a) Receptors (UCPs) reconstituted into a lipid bilayer formed on a mica surface and a single ligand (ATP) conjugated on a cantilever tip via a PEG crosslinker. (b) TREC mode: The cantilevers oscillation signal is split into two parts. (i) Aupper, the upwards oscillation peaks yields a map of recognition sites which are shown as dark dots in the recognition image. Molecules which did not bind to the ligand on the tip are invisible in the recognition image. (ii) Apeak–peak, downwards amplitude peaks lead to the topography image. (c) Switching into the force spectroscopy mode: repeatedly approaching and withdrawing of the ligand-carrying AFM tip on the previously selected receptor molecule to measure the interaction forces between these two molecules. Force versus distance data of force distance cycles, shown in red for the approaching period (trace) and in black for the retraction (retrace). In the latter a typical receptor-ligand unbinding event is visible as parabolic shaped downwards signal.
Fig. 3.
AC tuning curve for cantilevers during acoustical excitation. The 2nd frequency level (f~21 kHz) was chosen for imaging instead of the base resonance frequency level (f~10.5 kHz) so as to achieve an adequate driving amplitude.
3.2. TREC Imaging: UCP1 Distribution and Functionality
For visualization of the UCP1 molecules we fused proteoliposomes containing purified UCP1 onto mica, as depicted above (Fig. 2). The correct folding and activity of the protein was tested in electrophysiological experiments, as previously described.39 In large area scans membrane batches containing UCP1 molecules and membranes holes were visible in the topography image (data not shown). After zooming onto an area covered by complete membrane batch, UCP1 molecules were visualized as round shaped structures (Fig. 4(a)). The black dots visible in the corresponding recognition image reveal the existence of ATP binding sites on UCP1 that were accessible by the nucleotide coupled to the cantilever (Fig. 4(b)). Almost all UCPs that were detected in the topography image (Fig. 4(a)) were recognized by the ATP-functionalized tip (Fig. 4(b)). The recognition efficiency of close to 100% at the applied pulling speed is clearly visible in the overlay of the recognition image with the topographical image (Fig. 4(c)).
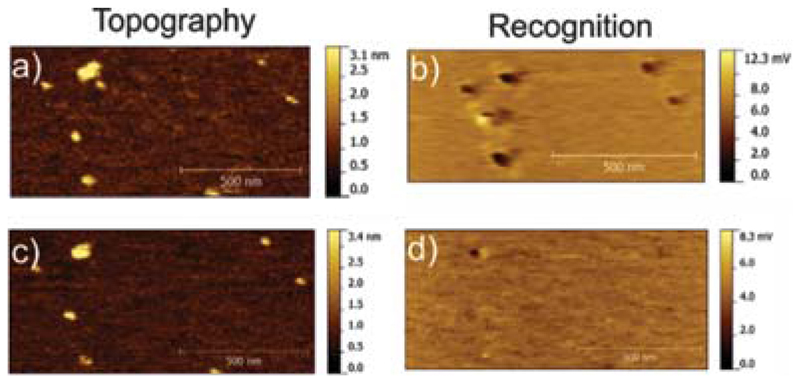
The specificity of the recognition signals was proven by a block experiment. Here ~8.6 mM of free ATP was injected into the buffer solution and effectively blocked the binding sites for ATP in the UCP1 binding pocket (Figs. 5(b, d)). The nearly complete loss of the recognition spots after addition of free ATP (Figs. 5(b, d)), while leaving the topography image unchanged (Figs. 5(a, c)), clearly confirms that most of the recognition spots were caused by specific binding of tip-linked ATP to UCP1 molecules in the planar lipid membrane. This coincides with already published results for the UCP1-ATP interaction.35, 41
Fig. 5.
Block experiment. (a and b) Topography and recognition images of UCP1 molecules in the lipid membrane acquired with an ATP-tethered tip, prior to blocking. (c and d) Topography and recognition images after blocking by addition of free ATP into the solution while scanning the same position. Almost all recognition spots (black) disappeared, demonstrating the specificity of the UCP1-ATP interaction.
After recording both topography and recognition images were analyzed after levelling by mean plane subtraction and scanning line correction (using Gwyddion 2.41). Polynomial background was removed as well. Membrane regions lacking protein were used to evaluate the threshold for recognition (using PicoImage ©Keysight). Only proteins identified with a recognition signal above the threshold were considered as being recognized by the functionalized tip.
3.3. Force Spectroscopy
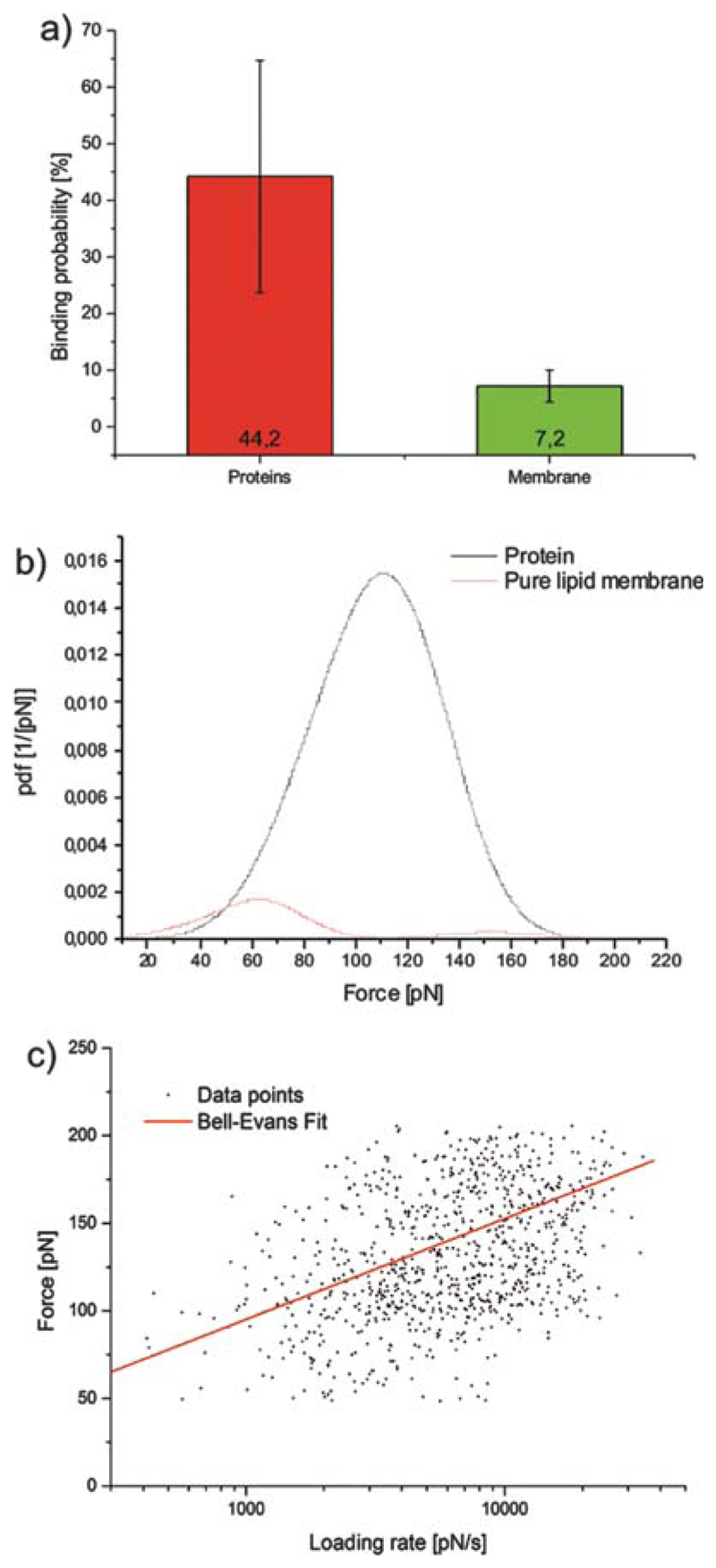
After identifying ATP-binding UCP1 molecules, we switched from tapping to contact mode and positioned the cantilever on a selected recognition spot using positional feedback control. Force spectroscopy experiments were started (Fig. 6(a)) and the binding activity was probed by calculating the ratio of FDCs carrying a binding event over the number of totally recorded FDCs (termed as ‘binding probability’). The binding probability recorded on different UCP1 proteins (Fig. 6(a)) amounted to 44.2% ± 20.5% (n = 7). In contrast, when FDCs were taken on pure lipid membranes lacking UCP1 molecules, binding probabilities (Fig. 6(a)) were significantly lower, 7.2% ± 2.8% (n = 5). This is a clear indication that the unbinding forces detected between UCP1 and ATP (Fig. 6(a)) were of specific nature.
Fig. 6.
Force spectroscopy of the UCP1-ATP interaction. (a) Comparison of the binding probabilities of the interaction between ATP and UCP1 molecules, and between ATP and pure lipid membrane. (b) Experimental probability density functions (pdf) of the unbinding forces were generated from binding experiments on UCP1 (black) and on the pure lipid membrane (red). (c) Loading rate dependence of the most probable unbinding force for the UCP1-ATP interaction.
From FDCs recorded on UCP proteins, distributions of unbinding forces were obtained by constructing empirical probability density functions (pdf) using a MATLAB© routine. The maximum of such pdfs reflects the most probable unbinding force, whereas the broadness of this distribution corresponds to the standard derivation.45 Two examples of force pdfs are shown in Figure 6(b). The force pdf gained from the binding experiment carried out on the pure lipid membrane was normalized to the binding on UCP1 to demonstrate the specificity of the experiment (see binding probabilities above). The maximum force for the FDCs recorded on UCP molecules amounts to 105 pN ± 11 pN, with a clear maximum peak in the force pdf. In contrast the force pdf for the experiment on the pure lipid membrane (maximum force = 62 pN ± 24 pN) appears more randomly distributed around lower forces.
Then we applied different loading rates to the molecular bonds by varying the pulling velocities in FDCs. The loading rate r of every unbinding event was calculated by multiplying the pulling speed with the effective spring constant of the system (keff). The latter depends on the nominal spring of the cantilever, the spring of the PEG crosslinker and the stiffness of the reconstituted proteins in the lipid bilayer. Using an established maximum likelihood approach, koff and xβ were determined as described in Wilding et al.46 and fitted according to the single barrier model.47 In this model the rupture force F (as a function of r) amounts to:
| (1) |
koff represents the kinetic off-rate in solution, and xβ is a characteristic length-scale in the interaction energy potential.
In accordance with the Eq. (1), the force of every single unbinding event was plotted against its corresponding loading rate on a semi-logarithmic scale (Fig. 6(c)). Data analysis revealed details of the energy landscape of UCP1-ATP binding: the characteristic length-scale of the energy barrier was xβ ~ 1.65 ± 0.01 Å, the kinetic off-sate koff = 0.89 ± 0.11 s−1, and the corresponding bond lifetime τ, given by the inverse kinetic off-rate (τ = 1/koff), amounted to 1.12 ± 0.14 s.
4. Conclusions
Our combined mode offers several advantages that are essential for measuring the unbinding forces between a ligand on the AFM tip and receptors reconstituted in an artificial lipid membrane at low density. A major benefit is the possibility to perform topography imaging, recognition imaging and force spectroscopy in one selected area. This results in a high throughput and renders carrying out dynamic force probing possible on different pre-selected molecules. Parameter adjustment for this combined method and data analysis are straightforward. One drawback is that the experiments are sensitive to changes in environmental conditions, such as temperature fluctuations. This could lead to the loss of the protein during force spectroscopy and requires switching back to the recognition imaging mode and readjusting the cantilever onto the molecule of interest. However, by using a positional feedback control, the measurements are reasonably stable at thermal equilibrium. As a proof of principle the binding of ATP to the uncoupling protein 1 was studied. With the combination of the two techniques presented in this paper we were able to measure the interaction forces between these two interaction partners and yield details about the interaction energy landscape.
Acknowledgments
This work was supported by the Austrian Research Fund (FWF, P25357). We thank Sarah Bardakji for excellent technical assistance and Quentina Beatty for editing.
References and Notes
- 1.Hinterdorfer P, Dufrêne YF. Nature Methods. 2006;3:347. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 2.Hinterdorfer P. Methods in Cell Biology. 2002;68:115. doi: 10.1016/s0091-679x(02)68007-0. [DOI] [PubMed] [Google Scholar]
- 3.Clausen-Schaumann H, Seitz M, Krautbauer R, Gaub HE. Current Opinion in Chemical Biology. 2000;4:524. doi: 10.1016/s1367-5931(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 4.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 5.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. Nature. 1998;393:181. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 6.Rief M, Clausen-Schaumann H, Gaub HE. Nature Structural and Molecular Biology. 1999;6:346. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 7.Benoit M, Gabriel D, Gerisch G, Gaub HE. Nature Cell Biology. 2000;2:313. doi: 10.1038/35014000. [DOI] [PubMed] [Google Scholar]
- 8.Florin E-L, Moy VT, Gaub HE. Science. 1994;264:415. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- 9.Lee GU, Chrisey LA, Colton RJ. Science. 1994;266:771. doi: 10.1126/science.7973628. [DOI] [PubMed] [Google Scholar]
- 10.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. Proceedings of the National Academy of Sciences. 1996;93:3477. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleveland JP, Radmacher M, Hansma PK. Atomic scale force mapping with the atomic force microscope, Forces in Scanning Probe Methods. Springer; Netherlands: 1995. pp. 543–549. [Google Scholar]
- 12.Manne S, Gaub HE. Current Opinion in Colloid and Interface Science. 1997;2:145. [Google Scholar]
- 13.Frisbie CD, Rozsnyai LF, Noy A, Wrighton MS, Lieber CM. Science. 1994;265:2071. doi: 10.1126/science.265.5181.2071. [DOI] [PubMed] [Google Scholar]
- 14.Noy A, Frisbie CD, Rozsnyai LF, Wrighton MS, Lieber CM. Journal of the American Chemical Society. 1995;117:7943. [Google Scholar]
- 15.Kim IH, Lee MN, Ryu SH, Park JW. Analytical Chemistry. 2011;83:1500. doi: 10.1021/ac102695e. [DOI] [PubMed] [Google Scholar]
- 16.Kim DH, Lee JE, Xu ZY, Geem KR, Kwon Y, Park JW, Hwang I. Nature Communications. 2015;6:6843. doi: 10.1038/ncomms7843. [DOI] [PubMed] [Google Scholar]
- 17.Alsteens D, Dague E, Rouxhet PG, Baulard AR, Dufrêne YF. Langmuir. 2007;23:11977. doi: 10.1021/la702765c. [DOI] [PubMed] [Google Scholar]
- 18.Dague E, Alsteens D, Latgé JP, Verbelen C, Raze D, Baulard AR, Dufrêne YF. Nano Letters. 2007;7:3026. doi: 10.1021/nl071476k. [DOI] [PubMed] [Google Scholar]
- 19.Alsteens D, Dupres V, Yunus S, Latgé JP, Heinisch JJ, Dufrêne YF. Langmuir. 2012;28:16738. doi: 10.1021/la303891j. [DOI] [PubMed] [Google Scholar]
- 20.Adamcik J, Berquand A, Mezzenga R. Appl Phys Lett. 2011;98:3701. [Google Scholar]
- 21.Rico F, Su C, Scheuring S. Nano Letters. 2011;11:3983. doi: 10.1021/nl202351t. [DOI] [PubMed] [Google Scholar]
- 22.Heu C, Berquand A, Elie-Caille C, Nicod L. Journal of Structural Biology. 2012;178:1. doi: 10.1016/j.jsb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Stroh C, Wang H, Bash R, Ashcroft B, Nelson J, Gruber H, Lohr D, Lindsay SM, Hinterdorfer P. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12503. doi: 10.1073/pnas.0403538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroh CM, Ebner A, Geretschläger M, Freudenthaler G, Kienberger F, Kamruzzahan ASM, Smith-Gill SJ, Gruber HJ, Hinterdorfer P. Biophysical Journal. 2004;87:1981. doi: 10.1529/biophysj.104.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebner A, Kienberger F, Kada G, Stroh CM, Geretschläger M, Kamruzzahan ASM, Wilding L, Johnson TW, Ashcroft B, Nelson J, Lindsay SM, et al. ChemPhysChem. 2005;6:897. doi: 10.1002/cphc.200400545. [DOI] [PubMed] [Google Scholar]
- 26.Ebner A, Wildling L, Kamruzzahan ASM, Rankl C, Wruss J, Hahn CD, Hölzl M, Zhu R, Kienberger F, Blaas D, Hinterdorfer P, et al. Bioconjugate Chemistry. 2007;18:1176. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- 27.Kamruzzahan ASM, Ebner A, Wildling L, Kienberger F, Riener CK, Hahn CD, Pollheimer PD, Winklehner P, Hölzl M, Lackner B, Schörkl DM, et al. Bioconjugate Chemistry. 2006;17:1473. doi: 10.1021/bc060252a. [DOI] [PubMed] [Google Scholar]
- 28.Ebner A, Wildling L, Zhu R, Rankl C, Haselgrübler T, Hinterdorfer P, Gruber HJ. Functionalization of probe tips and supports for single-molecule recognition force microscopy, STM and AFM Studies on (Bio) Molecular Systems: Unravelling the Nanoworld. Springer; Berlin, Heidelberg: 2008. pp. 29–76. [DOI] [PubMed] [Google Scholar]
- 29.Preiner J, Ebner A, Chtcheglova L, Zhu R, Hinterdorfer P. Nanotechnology. 2009;20:215103. doi: 10.1088/0957-4484/20/21/215103. [DOI] [PubMed] [Google Scholar]
- 30.Nedergaard J, Bengtsson T, Cannon B. American Journal of Physiology-Endocrinology and Metabolism. 2007;293:E444. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls DG, Bernson VS, Heaton GM. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation, Effectors of Thermogenesis. Birkhäuser; Basel: 1978. pp. 89–93. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, Klingenberg M. FEBS Letters. 1980;113:299. doi: 10.1016/0014-5793(80)80613-2. [DOI] [PubMed] [Google Scholar]
- 33.Urbánková E, Voltchenko A, Pohl P, Ježek P, Pohl EE. Journal of Biological Chemistry. 2003;278:32497. doi: 10.1074/jbc.M303721200. [DOI] [PubMed] [Google Scholar]
- 34.Beck V, Jaburek M, Demina T, Rupprecht A, Porter RK, Ježek P, Pohl EE. The FASEB Journal. 2007;21:1137. doi: 10.1096/fj.06-7489com. [DOI] [PubMed] [Google Scholar]
- 35.Zhu R, Rupprecht A, Ebner A, Haselgrübler T, Gruber HJ, Hinterdorfer P, Pohl EE. Journal of the American Chemical Society. 2013;135:3640. doi: 10.1021/ja312550k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholls DG. European Journal of Biochemistry. 1976;62:223. doi: 10.1111/j.1432-1033.1976.tb10151.x. [DOI] [PubMed] [Google Scholar]
- 37.Krauss S, Zhang C-Y, Lowell BB. Nature Reviews Molecular Cell Biology. 2005;6:248. doi: 10.1038/nrm1592. [DOI] [PubMed] [Google Scholar]
- 38.Klingenberg M. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2010;1797:579. doi: 10.1016/j.bbabio.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 39.Rupprecht A, Sokolenko EA, Beck V, Ninnemann O, Jaburek M, Trimbuch T, Klishin SS, Jezek P, Skulachev VP, Pohl EE. Biophysical Journal. 2010;98:1503. doi: 10.1016/j.bpj.2009.12.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebner A, Hinterdorfer P, Gruber HJ. Ultramicroscopy. 2007;107:922. doi: 10.1016/j.ultramic.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 41.Wildling L, Unterauer B, Zhu R, Rupprecht A, Haselgrübler T, Rankl C, Ebner A, Vater D, Pollheimer P, Pohl EE, Hinterdorfer P, et al. Bioconjugate Chemistry. 2011;22:1239. doi: 10.1021/bc200099t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutter JL, Bechhoefer J. Review of Scientific Instruments. 1993;64:1868. [Google Scholar]
- 43.Ebner A, Nevo R, Rankl C, Preiner J, Gruber HJ, Kapon R, Reich Z, Hinterdorfer P. In: Handbook of Single-Molecule Biophysics. Hinterdorfer P, Oijen A, editors. Springer; US: 2009. pp. 407–447. [Google Scholar]
- 44.Zlatanova J, Lindsay SM, Leuba SH. Progress in Biophysics and Molecular Biology. 2000;74:37. doi: 10.1016/s0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- 45.Baumgartner W, Hinterdorfer P, Schindler H. Ultramicroscopy. 2000;82:85. doi: 10.1016/s0304-3991(99)00154-0. [DOI] [PubMed] [Google Scholar]
- 46.Wildling L, Rankl C, Haselgrübler T, Gruber HJ, Holy M, Newman AH, Zou M-F, Zhu R, Freissmuth M, Sitte HH, Hinterdorfer P. Journal of Biological Chemistry. 2012;287:105. doi: 10.1074/jbc.M111.304873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans E. Faraday Discuss. 1999;111:1. doi: 10.1039/a809884k. [DOI] [PubMed] [Google Scholar]