Abstract
The advent of ultrafast highly brilliant coherent X-ray Free Electron Laser sources has driven the development of novel structure determination approaches for proteins, and promises visualisation of protein dynamics on the fastest timescales with full atomic resolution. Significant efforts are being applied to the development of sample delivery systems that allow these unique sources to be most efficiently exploited for high throughput serial femtosecond crystallography. We present here the next generation of a fixed target crystallography chip designed for rapid and reliable delivery of up to 11,259 protein crystals with high spatial precision. An experimental scheme for predetermining the positions of crystals in the chip by means of in-situ spectroscopy using a fiducial system for rapid, precise alignment and registration of the crystal positions is presented. This delivers unprecedented performance in serial crystallography experiments at room temperature under atmospheric pressure with a raw hit rate approaching 100% with an effective indexing rate of approximately 50%, increasing the efficiency of beam usage, and allowing the method to be applied to systems where the number of crystals is limited.
Keywords: Femtosecond Time-Resolved Crystallography, High Throughput Serial Crystallography, Fixed Target Sample Delivery, Room Temperature Crystallography
1. Introduction
X-ray diffraction is currently the standard approach for high resolution structure determination of proteins, and is conventionally carried out at synchrotron light sources. Recent developments in X-ray sources have led to the realisation of hard X-ray Free Electron Lasers (XFEL). The advantage of these fourth-generation X-ray light sources is their high peak brilliance and improved beam coherence in contrast to synchrotron-generated X-ray radiation. Further, X-rays are generated as short pulses in the 10-100 femtosecond regime, enabling experiments to probe ultrafast chemical and biochemical dynamics (Kern et al., 2014,Tenboer et al., 2015,Barends et al., 2015).
Protein crystallography experiments at XFELs are based on the approach now known as Serial Femtosecond Crystallography (SFX), where the above beam properties allow single diffraction patterns to be recorded from micron-sized protein crystals in a few tens of femtoseconds (Chapman et al., 2011). Using this approach, it is possible to collect a large number of diffracted photons from each protein crystal before the onset of X-ray radiation damage (“diffraction before destruction”) (Boutet et al., 2012, Barty et al., 2011). However, peak brilliance 108 times higher than a synchrotron source and short duration of the pulse at an XFEL do result in severe radiation damage to each crystal and make a sample change between each X-ray shot necessary (Barty et al., 2013). This requires either a very large number of small protein crystals or the translation and exposure of multiple regions of larger crystals in order to obtain a sufficient sampling of reciprocal space to allow protein structure determination. Samples must be delivered into the X-ray beam with high spatial precision and, ideally, sufficient speed/repetition rate to fully use the X-ray beam flux and avoid misalignment or mistiming in presenting the target to the X-ray beam. Currently, SFX experiments predominantly use variations of a liquid jet (DePonte et al., 2008, Boutet et al., 2012, Sierra et al., 2012), LCP injectors (Weierstall et al., 2014) in vacuum, grease matrix (Sugahara et al., 2015), or an acoustic injector (Roessler et al., 2016) for sample delivery. In the liquid jet method, the crystal solution is ejected at high velocity (albeit with small net volume flow) in a stream onto which, for time-resolved experiments, the pump (laser) and probe (X-ray) pulses are incident. These approaches can support structure determination, but suffer from low hit rates since the random nature of the position of crystals within the jet means that while all X-ray pulses may intersect with the jet, few hit a crystal, wasting photon flux. Even worse for many experiments, where crystal production is difficult or expensive, the vast majority of the crystals are wasted, since to maintain the thin flow of crystals the jet velocity needs to be high, increasing the number of crystals which do not intersect an X-ray pulse. The hit rate has been estimated to be as low as one hit per 1,600 crystals in some cases (Weierstall et al., 2014). Furthermore, the maximum pressure the crystals can handle before flow-induced damage occurs limits the crystal flow. The flowing stream of sample also adds to the difficulty (compounded by the destructive nature of the X-ray pulse) of taking diffraction reference shots, which would simplify pump-probe experiment data analysis. It is therefore, highly desirable to develop sample delivery approaches with hit rates that approach 100%, and which provide the possibility of recording reference data. For time-resolved studies, the requirement for a high hit rate is particularly acute since sufficient data for structure analysis must be obtained for each and every time point. This significantly increases the volume of sample needed, bearing in mind the high signal to noise required to resolve the very small changes in structure that direct chemical and biological processes or other structural transitions.
Fixed targets provide an attractive solution to the problems described above, and in principle allow for the convenient harvesting of crystals and their precise presentation to laser and X-ray beams. Several fixed target systems have been developed. These include polydimethylsiloxane (PDMS) based chips (Guha et al., 2012, Lyubimov et al., 2015), for which the crystals sit on the substrate, resulting in noticeable background scatter from the substrate material. An alternate approach utilizes a thin film as a substrate for crystal sample deposition (Hunter et al., 2014, Murray et al., 2015), or patterned silicon substrates (Roedig et al., 2015) for rapid sample positioning and polycarbonate grids coupled with a goniometer (Baxter et al., 2016, Cohen et al., 2014). This contributes less background but eliminates the possibility of prior mapping out of crystal positions. Using a large single crystal as a fixed target has also been implemented (Chreifi et al., 2016, Hirata et al., 2014), but large crystals with the appropriate shape and large accessible surface area and thickness are not easily available for many samples of interest.
We have previously reported a crystallography chip, made by silicon nanofabrication methods (Zarrine-Afsar et al., 2012), for use at microfocus synchrotron beamlines and for time-resolved studies at XFELs that addresses all the above problems (Mueller et al., 2015). The chip represents a significant improvement over injector schemes, as it requires a smaller amount of sample, and effectively minimizes background scatter compared to other fixed target approaches, due to the removal of excess mother liquor/solvent; a feature specifically incorporated in the design of the chip loading process. The chip contains precisely defined features (pit or well-shaped) in which the crystals are trapped. The feature sizes and chip support layer match the crystal size distribution of the sample under study. The chip structure allows a large number of micro-crystals to be loaded, using turbulent fluid flow to introduce random crystal orientations to ensure sufficient sampling of reciprocal space (Mueller et al., 2015). The loaded chip can be mounted on a high speed translation stage for crystal positioning in the X-ray beam (Sherrell et al., 2015). The accuracy of the nanofabrication process allows for precise positioning of the crystals, increasing the hit rate tremendously while keeping sample waste to a minimum, due to both the relatively high occupancy and the possibility to recover crystals not trapped during the loading process as all eluent can be collected. The chip also enables the recording of reference diffraction patterns and thus – radiation damage and signal to noise permitting – use of the ratio method for differential detection of the very small early structural changes typically involved in time-resolved studies (Coppens et al., 2009, Mueller et al., 2015), thereby reducing the volume of data required for structural dynamics studies and further increasing overall efficiency with respect to sample consumption.
In our initial reports the chip could not be operated with the maximum possible hit rate as all features on the chip were probed sequentially, irrespective of the presence or absence of a crystal (Zarrine-Afsar et al., 2012, Mueller et al., 2015). In the present design, we have further improved the chip to reduce multiple crystal loading per features, and added a system of fiducial marks for rapid alignment with well-defined reference points and addressing of individual compartments in the chip. The fiducial system enables error-free position determination and identification of each crystal in the array. Finally, we have developed a high speed single crystal spectrometer to map the crystal positions prior to the diffraction experiment. High speed mapping is important for crystals with finite lifetimes once mounted, and increases hit rates substantially as only features containing crystals are sampled at the beamline.
As a proof of principle, we demonstrate here that this chip concept, with a fiducial system and mapping protocol, can achieve a raw hit rate of up to 85%. When inherent variations in crystal quality as well as intensity fluctuations due to the SASE profile of the XFEL beam are taken into account, this approaches the maximum achievable sampling efficiency. Furthermore, the method facilitates a data collection rate that comes close to the fully optimized use of the XFEL X-ray flux with low sample consumption, thus opening up the study of precious protein systems. Specifically, we demonstrate the efficacy of the chip concept using carboxy-myoglobin, as a prelude to femtosecond time-resolved studies of ligand dissociation from the heme binding site.
2. Results
2.1. Chip design
The crystallography chip consists of compartments with features designed specifically for trapping micrometer-sized crystals (Fig. 1). The current design is comprised of 9 × 9 compartments, where each compartment contains an array of 12 × 12 features (Fig. 2), resulting in 11,664 positions on the chip, 11,259 of those for trapping crystals. Each feature is square-shaped with a 100 μm wide top aperture and a bottom aperture size ranging in width from 5 μm to 30 μm.
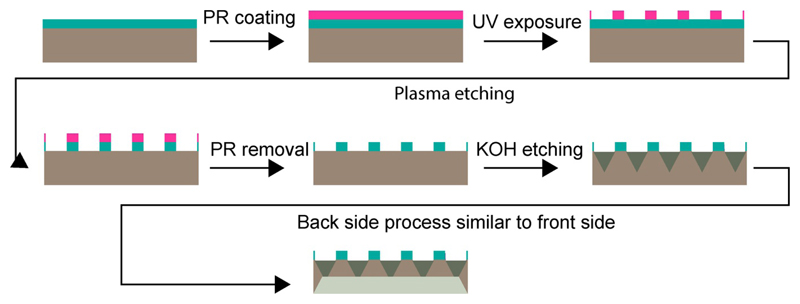
Figure 1.
Fabrication scheme for the crystallography chip. The process starts with conventional UV lithography to transfer the pattern of loading sites to a silicon wafer (gray, with a layer of silicon nitride, SiN, in cyan) coated with photoresist (pink). The pattern is then transferred to the SiN layer using reactive ion etching and the photoresist is stripped. The SiN layer plays the role of the mask for wet etching in the subsequent steps. KOH etching results in a trapezoidal shape that enhances loading efficiency. The same procedure is repeated for the back side of the chip to make it thin enough to fit a specific size of crystals.
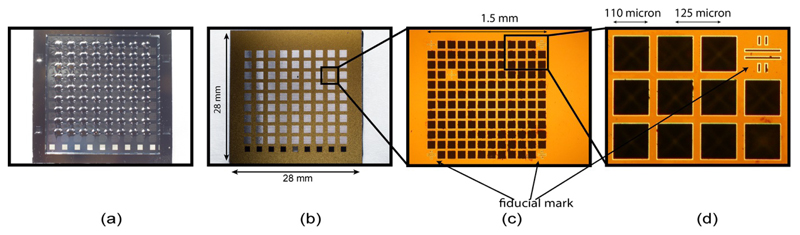
Figure 2.
Back (a) and front (b) of the chip, and a close up view of an individual compartment (c). Each chip comprises 9 × 9 compartments with 12 × 12 features per compartment. As each feature is designed to hold a single crystal the maximum capacity of the chip is 11,259 (139 × 81) crystals (based on 144 features minus 5 fiducials per compartment), (d) magnified portion of a compartment showing the crystal trapping features and a fiducial mark.
A key characteristic of the etched pattern on the silicon wafer is the trapezoidal geometry resulting from anisotropic etching of the different silicon crystal planes (Seidel et al., 1990). The aperture and depth of the resulting features can be optimized to match the average crystal size of the system of interest. The opening at the bottom of each feature allows for removal of excess solution to recover any untrapped smaller crystals, and to remove as much of the mother liquor remaining from the loading process as possible, dramatically reducing unwanted background scatter.
The thin regions of the chip designed for crystal trapping are easily deformable and, if not supported, can flex during loading and translation resulting in a loss of positioning accuracy. The chip therefore includes thick spacer bars between compartments to increase rigidity (Fig. 2a, b).
It should be noted that the thickness of the raw silicon wafer varies by 10 μm, which affects the final feature sizes, and the etching rate at the extremities of the individual compartments and the chip itself. The etching is slightly slower at the edge of the chip than in the centre. This leads to a radial distribution of feature sizes of about 5 μm over each individual compartment and over the whole chip. This non-uniformity means that a small number of features on the outermost part of the chip might not be fully open at the bottom and thus cannot hold a crystal. Nonetheless, this geometry and method of fabrication result in a chip capable of trapping up to 11,259 single crystals. The method of fabrication can be improved and easily scaled and thus allows adjustment of the number of compartments and features to further increase the chip capacity if desired.
Each compartment has five fiducials, three of them at the corners for alignment, and two additional serving as unique address, allowing precise and unambiguous positioning of every feature on the chip. The fiducial marks are designed to enable alignment of the chip using an inline imaging system with minimum aberrations to locate the centroid of the fiducial mark. The fiducial marks have a length of 100 μm and width of 10 μm and are in the form of a double cross or ‘#’ symbol to facilitate automated image analysis (Fig. 2c,d). The ‘#’ reduces edge confusion in the image processing/alignment algorithm, hence reducing the number of false positive matches. In principle, the corner features of every compartment could be used as intrinsic fiducial marks but as the feature dimensions have some irregularity, due to the silicon processing, this is less reliable.
2.2. Chip loading
Two auxiliary parts have been fabricated for use in the loading process to improve its ease and efficiency (Fig. 4). The first is an extremely flat silicon “bottom plate” (< 5 nm surface roughness over the whole contact area) with a hole through it, which lies underneath the crystallography chip. This very smooth surface provides a good seal for a closed flow system to direct the crystals through the features to trap the crystals. The second is a silicon chip with 9 linear channels etched across it, which span the length of the chip and with widths matching the size of one compartment. This part sits on top of the crystallography chip to direct the flow of liquid during sample loading, reducing sample waste by minimizing flow over dead space on the chip and maximizing the flow rate over the features. This avoids adsorption of crystals on to the chip at locations where they would not be sampled in the diffraction experiment. The whole assembly is stacked and held together accurately using a custom holder.
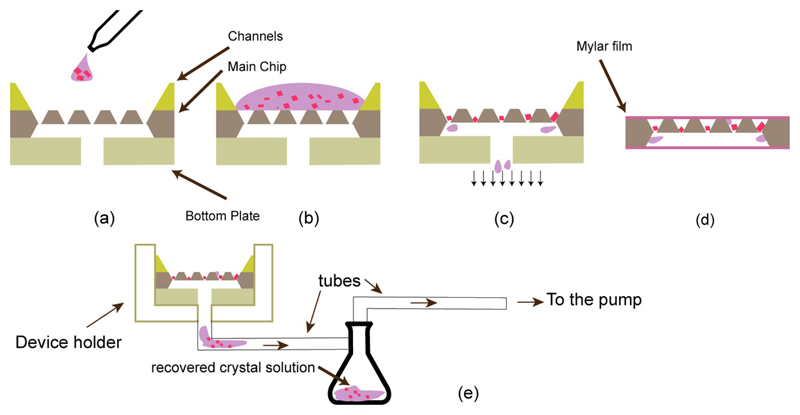
Figure 4.
(a, b, c) Chip loading scheme with chip components designed for crystal solution loading. The silicon piece with channels is placed on top of the chip for guiding crystal solution into the compartment rows. The bottom plate helps to apply effective negative pressure for trapping the crystals in features (c). (d) The loaded chip is wrapped in a thin film of mylar to avoid crystal dehydration. The crystal solution removed from the chip can be collected in an additional reservoir for use with a different chip size (e). All figures are schematics and are not to scale.
To minimize stress on the crystals, the loading process is performed at a controlled temperature, to match that of crystallization, and at controlled humidity to prevent salt crystal formation from mother liquor (see Materials and Methods section for details). First the crystals, in their mother liquor, are dispensed onto the chip using a pipette, using siliconated tips to minimize sticking of crystals to the pipette tip. Depending on crystal density volumes of 60 – 80 μl crystal slurry are sufficient to load a chip with approximately 40% occupancy. Negative pressure is applied to the under-side of the chip and the solution is sucked through the features, leaving the crystals trapped inside a feature with a minimal volume of mother liquor. The etched away area on the back-side of the chip creates a large evacuated volume for efficient removal of the mother liquor (Fig. 4c). The amount of negative pressure applied during the loading process needs to be carefully adjusted. Too much suction dehydrates the protein crystals too quickly, while too little suction leaves excess mother liquor on the surface of the chip, resulting in increased background scatter. Adjusting suction and humidity is important when high salt concentrations are used as precipitant to avoid formation of salt crystals. Next, the chip is removed from the closed flow system and rapidly sealed with 4 μm thick mylar film on each side. Finally, the chip is placed in a custom holder, which is specially designed to accommodate the two 4 μm films of stretched mylar.
The mylar film’s permeability is small enough to minimize protein dehydration and maintain the crystals in a humid environment long enough for data collection. The mylar introduces some diffuse background to the diffraction pattern (see Fig. 3), which is subtracted during standard data processing procedures. The feature’s bottom hole acts as an aperture, which helps to remove background scatter (see Fig. 3). In the future other, lower scatter, polymer films or 30 – 50 nm SiN films could be substituted to significantly further reduce background scatter. In addition, the silicon nitride and etched silicon surfaces, which are usually covered with their native oxides, are very hydrophilic, therefore it may be possible to improve loading efficiencies and removal of excess mother liquor by reducing the chip’s hydrophilicity using simple surface treatments.
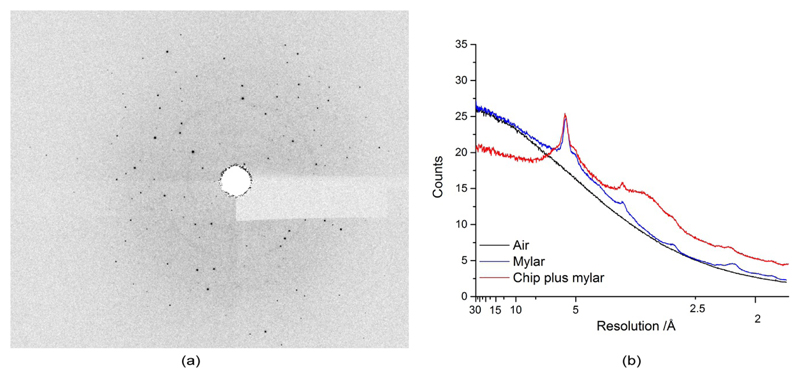
Figure 3.
(a) Example of diffraction image with mylar background, (b) comparison of diffuse scatter of air, air plus mylar and a loaded chip (empty feature) as a function of resolution. The feature acts as an aperture reducing background at low resolution.
2.3. Optical spectroscopy setup
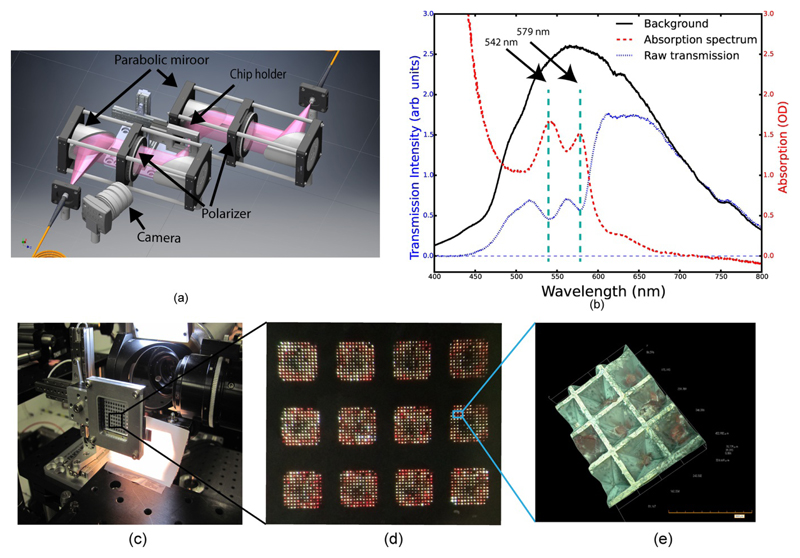
To increase the hit rate, we have developed a high-speed UV/Vis microspectrometer (adapted from Kirchner et al., 2014), with a focus spot size of 100 micron and covered wavelength range of 200 – 850 nm) to map crystal positions in the chip. Transmission spectra are recorded from each feature to determine whether or not it harbours a crystal. This information can be subsequently passed to the beamline data acquisition software so that empty features are not addressed during the SFX experiment. This approach also allows differentiation of protein and salt crystals, as salt crystals strongly scatter the light and can therefore be easily identified. The rapid filling of the chip relative to the rate of salt crystal formation greatly reduces the growth of salt crystals during chip loading (§2.2). However, for salt based crystallization conditions, some features can still be occupied with salt crystals and, in addition, salt crystals can grow on protein crystal surfaces.
To ensure complete compatibility between the chip mapping and positioning of the chip at the X-ray beamline, we use identical precision translation stages for the spectroscopy/mapping and for the X-ray beamline and the same coordinate system transform to convert feature positions into translation stage motions as previously described (Sherrell et al., 2015). Once mounted, the chip is translated along the spectroscopy probe beam axis until it is at the beam focus and the optical spot size is minimized. Chip tilt is compensated for in the coordinate system transform based on the position of the three outermost corner fiducial marks on the chip. The chip is then translated successively to each feature where a spectrum is recorded and named according to the feature address as defined by the coordinate system. The experimental setup is shown schematically in Fig. 5a, as well as representative spectra of carboxy-myoglobin crystals recorded from the chip (Fig. 5b). The time needed to acquire a spectrum with acceptable signal-to-noise limits the speed at which a chip can be scanned. In the experiments reported here the integration time for recording each feature spectrum was 250 ms, resulting in a 40 minute acquisition time for a whole chip map. However, improvements in integration time and the use of an external trigger have since been implemented, leading to an acquisition time of approximately 8 minutes per chip.
Figure 5.
(a) Schematic representation of the high-speed spectroscopy setup for mapping the loaded chip. (b) Example spectrum of a carboxy-myoglobin crystal recorded from a crystal containing feature. The characteristic absorption bands for CO-myoglobin (542, 579 nm) (Springer & Sligar, 1987) are indicated. (c) The chip mounted in the beam at the XPP instrument of LCLS. (d) A magnified picture of chip compartments loaded with crystals. The CO-myoglobin crystals can be easily identified due to their red colour. (e) 3D image of crystals trapped inside a feature (image taken using a Hirox RH2000 microscope).
After the chip has been fully scanned, the background-corrected transmission spectra are processed to determine the loaded feature positions. This is done by taking the ratio of the integral of a part of the transmission spectrum which is expected to show significant absorption (500 – 700 nm for CO-myoglobin) in comparison to the integral of a part of the spectrum where there is little absorption (700 – 850 nm). A feature is marked as containing a crystal if the resulting number is below the threshold ratio of an empty feature. A list is then generated containing each feature address and a binary value corresponding to whether or not the feature contains a crystal. This scheme is very fast and is generally effective, but suffers from a few drawbacks. Firstly, the effectiveness of the asymmetric filter described above depends on the sample’s specific absorption spectrum. Secondly, it is impossible, based on the spectroscopic data alone, to distinguish crystalline sample from amorphous sample. Subsequent diffraction data showed that 15% of features marked as containing a crystal did not yield Bragg peaks, indicating the presence of amorphous material in our crystal preparation. Nonetheless, we were able to consistently achieve 85% raw hit rates at the XPP instrument, LCLS (Chollet et al., 2015, §2.4). It is possible that a more robust way of correlating the spectral signal to that of crystalline protein can be found to increase the hit rate, however, there is likely to be some residual crystal-to-crystal variation in diffraction quality that will preclude reaching exactly 100% hit rates. Nevertheless, the present hit rate already allows for the highly efficient use of current XFEL pulse stream.
2.4. Data acquisition and analysis
For a proof of principle, we investigated the use of the crystallography chip with spectroscopic mapping during an experiment at the Linac Coherent Light Source (LCLS) XPP instrument, determining the structure of carboxy-myoglobin using the SFX approach. Spectroscopic mapping was carried out on six chips, which then underwent X-ray data collection. The use of a monochromator resulted in circa 1010 photons/image, considerably less than the 1012 photons/image typical in an SFX experiment. For this reason, in a typical ‘full throttle’ experiment, we would expect a useful data set to be collected from substantially fewer images, with the expected attendant improvements in signal to noise with more diffracted x-ray photons. The average number of X-ray hits recorded for a scanned chip was around 5,000 (Table 1). Here, an X-ray hit is defined as an image containing > 10 intensity maxima, as determined by a fast peak search using onDA(Mariani et al., 2015), which we report here as raw hit rates (hraw). This indicates that the crystal loading efficiency on the chip was approximately 50%. The use of a shot-list based on chip mapping resulted in an actual overall hit rate greater than 85%.
Table 1.
Data collection and processing
| Diffraction source | LCLS XFEL source (XPP Endstation) |
| Wavelength (Å) | 1.29 |
| Temperature (K) | 293 |
| Detector | Rayonix MX170-HS CCD |
| Crystal-detector distance (mm) | 340 |
| Exposure time per image (s) | 0.1 |
| Space group | P21 21 21 |
| a, b, c (Å) | 38.13, 46.48, 84.52 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 33.43-1.9 |
| Images merged | 8957 |
| No. of unique reflections | 12354 |
| Completeness (%) | 100 |
| 〈I/σ(I)〉 | 6.0 |
| Multiplicity | 51.6 |
| Rsplit(%) | 10.1(46.1) |
| Matthew’s coefficient, VM (Å3/Da) | 2.18 |
| Solvent content, Vs (%) | 43.5 |
| Observations per image to edge of detector | 166 ± 28 |
| CC1/2 (%) | 99.6 (42.6) |
A quantity that describes the success rate of diffraction events more reliably is the fraction of shots that lead to indexable diffraction patterns, hidx, which is available after data reduction and indexing of the entire dataset. The “indexable hit rate” hidx can never be higher than hraw, and will usually be considerably lower, as not every crystal diffraction pattern can be indexed. Obviously hidx depends not only on the efficiency of sample delivery, but also on the type and quality of the crystals, and on the performance of the indexing software. Utilizing CrystFEL (White et al., 2012) to index the diffraction data gives an hidx of 43%, which represents on average 2,000 successfully indexed images per chip. Finally, because of the stochastic nature of X-ray pulse generation, there is a shot-to-shot variation in both pulse intensity and spectral distribution. In our experiments an X-ray monochromator was used. As a consequence, the natural variation in the X-ray pulse intensity leads to a significant fraction of “empty” pulses. As a result, even features containing a “good” crystal may not result in a “hit”. To take this into account, corrected hit rates, h´raw and and h´idx, were calculated on the basis of the number of X-ray pulses observed with intensity greater than a certain threshold. This correction leads to an increase of the hidx rates to 46%, i.e. close to 50% indexable diffraction patterns. The total number of diffraction images (> 10 peaks, hraw) collected from the six chips was on the order of 20,000, of which half resulted in successfully indexed diffraction patterns (Table 2, exact number 10,751). Not all of these images have been used in final structure determination (see Table 3 and Materials and Methods section).
Table 2.
Hit rate statistics for mapped chips
| Chip Number | 1 | 2 | 3 | 4 | 5 | 6 | Average (%) |
| Positive Spectra | 6983 | 2237 | 9174 | 4325 | 8618 | 3782 | 52* (Fill-Factor) |
| Max No. diffraction images possible ** | 4467 | 1485 | 5993 | 3100 | 6064 | 2609 | 57 |
| Diffraction images with > 10 peaks | 4093 | 1432 | 5298 | 2790 | 4161 | 2148 | 84 (hraw) |
| Indexed images | 1802 | 825 | 2479 | 1611 | 2863 | 1171 | 46 (hidx) |
The number of features with a verified spectrum of CO-myoglobin.
The maximum expected number of diffraction images taking empty X-ray pulses into account.
Table 3.
Refinement statistics
| Resolution range (Å) | 42.26-1.9 |
| Completeness (%) | 100 |
| Rwork/ Rfree | 21.1/26.0 |
| No. of non-H atoms | |
| Protein | 1225 |
| Sulfate | 1 |
| Water | 20 |
| R.m.s. deviations | |
| Bonds (Å) | 0.0159 |
| Angles (°) | 1.76 |
| Ramachandran plot | |
| Most favoured (%) | 96.7 |
| Allowed (%) | 3.3 |
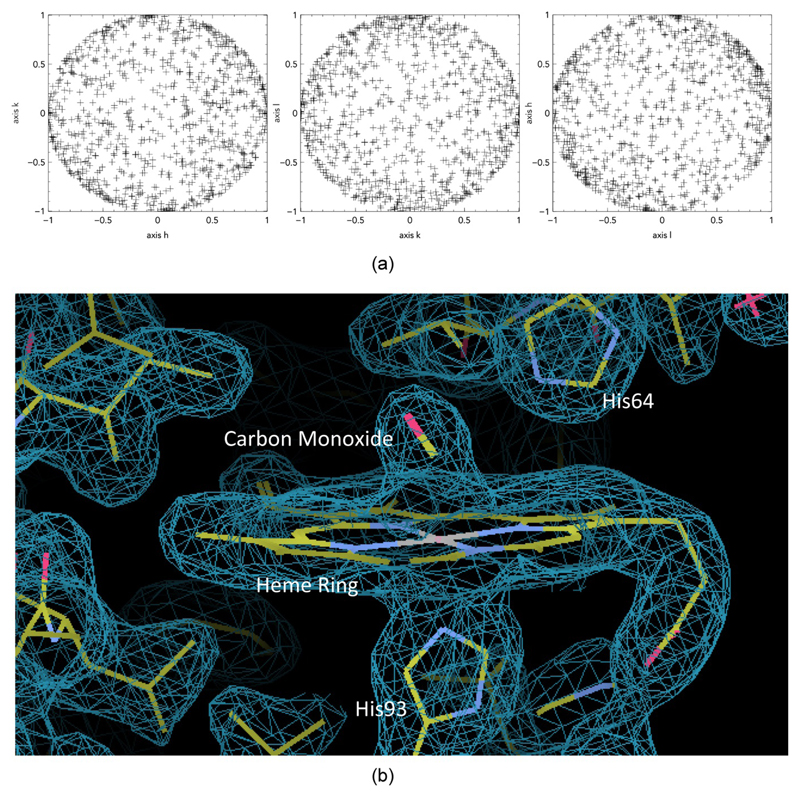
The dimensions of the crystals that were successfully indexed were 20 × 20 (40 – 70) μm. Analysis of the orientation matrices (Fig. 6a) confirms that the crystals are reasonably randomly oriented, with no evidence of preferred alignment within the chip features. The quality of the intensities derived by the application of cppxfel (Ginn et al., 2015a) was reasonable (Table 1), despite the heavy attenuation of the XFEL pulses (see Materials and Methods section). The TakeTwo algorithm within cppxfel increased the indexing rates to approximately 100% of images which registered more than 20 spots using DIALS spot-finding (Waterman et al., 2016). This data, when merged (with cppxfel), yielded a complete data set and supported model building and refinement to obtain a high resolution model capable of supporting the analysis of small conformational changes. Table 3 summarizes refinement statistics and Fig. 6b shows a representative portion of the electron density map.
Figure 6.
(a) The orientation matrices for different axis representing random orientation of crystals, (b) Representative electron density at 1.9 Å resolution, contoured at 1 σ for carbon monoxide (CO) bound to the heme iron of sperm whale (Physeter macrocephalus) myoglobin generated from data collected using the crystallography chip.
3. Discussion
The batch process for chip fabrication makes it possible to produce many chips at once. The tunability of the fabrication process enables development of different sized features to accommodate a range of crystal sizes up to hundreds of microns. This makes the crystallography chip well-suited for a large range of crystalline protein samples, down to dimensions of a few microns. This aspect of the chip is especially important for SFX experiments and the study of proteins for which large crystals are difficult to obtain. In addition, due to the well-shaped features, the loading leads to centred crystals, ensuring that the X-ray beam hits the centre of the crystal rather than an edge.
The minimal amount of mother liquor around each crystal and the thin mylar “windows” result in very low background, making the chip an attractive alternative compared to other sample delivery methods such as the liquid jet and LCP injectors, where the mother liquor or LCP carrier introduces significant background scatter to the diffraction data. An additional advantage of the silicon based chip is that it can be cleaned and reused. Finally, we have not observed any evidence of radiation damage to the chip itself at either microfocus synchrotron beamlines or when exposed to a XFEL beam.
One of the greatest advantages of the fixed-target chip, in combination with spectroscopic mapping, is the ability to define a spatial address for each protein crystal located in the chip and thus extract corresponding data selectively for a particular sample. This provides the possibility to perform additional diagnostics with the potential to improve SFX analysis, e.g. post-sorting of diffraction data based on optical absorption. In general, any measurable characteristic of an individual crystal can be recorded and referenced later for data analysis.
Another application of the optical spectroscopy setup is in-situ sample characterization. Unstable samples that require sealing from atmosphere or specific ambient conditions can be tested for longevity, e.g. in the case of carboxy-myoglobin the ligand binding state can be monitored for each individual crystal. In addition, chips can be scanned before and after X-ray exposure to observe the extent of X-ray and laser-induced damage (in case of time-resolved SFX experiments). This is important as femtosecond time-resolved experiments generally involve very small structural changes and thus there is a need to go to the maximum photoexcitation possible to ensure the majority of molecules in the crystal are reacting. This requires laser powers that are typically right at the limits of multiphoton processes unrelated to the photophysics and photochemistry of interest. This capability allows spectroscopic analysis to ensure proper excitation conditions below this limit and the onset of x-ray damage.
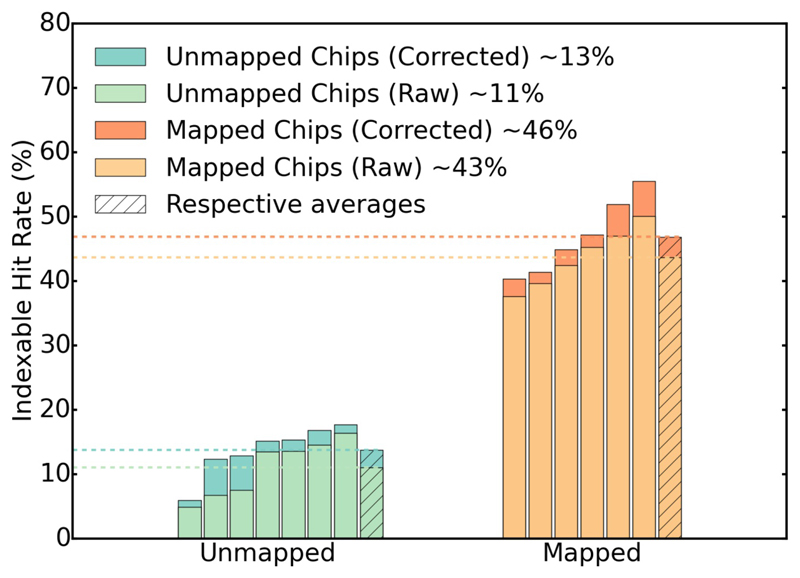
The improved jet-based sample delivery system utilizing LCP media has reported hit rates around 8% with an indexing success rate within this subset of images of 22%, resulting in an overall successful hidx rate of about 1.6% (Weierstall et al., 2014). Even allowing for possible differences in crystal quality, the hit rates achieved with this chip constitute an improvement of at least an order of magnitude over competing methods. The improved percentage of successfully indexed shots is due to a combination of reduced background scatter with the chip (see unmapped hit rates, Fig. 7) with a further gain in efficiency due to spectroscopic mapping (Table 2, and Fig. 7). Differential polarization analysis, using a simple polarizer between the sample and detector in the microspectrometer, or second harmonic generation based imaging to distinguish amorphous and crystalline protein will further improve this correlation to close to the 100% hraw limit.
Figure 7.
Percentage of indexed diffraction patterns for unmapped (left) and mapped chips (right); light colors: without correction for missing X-ray pulses, dark colours: after correction. Mapping the chip improves the success rate ~ three fold.
With improvements in correction to partialities (using a modified approach based on Ginn, H. M. et al., 2015), we find 8,957 indexable diffraction patterns to be sufficient to obtain a structure from monochromatic XFEL data. This is another development which is a significant improvement over previous methods, which required 20,000 or more indexed diffraction patterns (Kirian et al., 2010) with a pink beam. This means that in principle, with improved spectral mapping (including the differentiation between crystalline and amorphous material) and if optimal loading is achieved (> 50%), one or two chips could provide sufficient diffraction data for structure determination (ultimately depending on diffraction quality and the crystals’ parameters such as the space group). Data collection at XFELs is currently limited in principle by the repetition rate of the FEL (120 Hz for LCLS, US, 30 Hz for SACLA, Japan), but practically by the repetition rate of the available detectors. At the XPP instrument, the Rayonix detector used in these experiments runs at 10 Hz. Taking into account the mounting and alignment time, this would mean sufficient data for de novo structure determination could be collected within less than 20 minutes. For the chips described (feature spacing of 125 micron), the present translation stage system is capable of scanning at 107 Hz using the current approach of pausing at each feature for laser/X-Ray exposure.
Larger data sets will, of course, improve the overall signal to noise and 20,000 useful diffraction patterns could, in principle, be acquired at 100 Hz from an optimally loaded and mapped chip containing a larger number of features in less than 4 minutes. For time-resolved studies, this would mean that a full time series with up to 100 time points could be collected within a few hours. This would enable multiple systems to be studied for comparative studies of structural dynamics or studies over many decades in time in a single experimental visit to capture more fully relevant dynamics.
4. Conclusion
We present an improvement of a chip-based approach to sample delivery for SFX (extension of Mueller et al., 2015, Zarine-Asfar et al., 2012), which combines spectroscopic mapping of crystal positions with an innovative low background fixed target, to provide a versatile and highly efficient system with significant advantages over other sample delivery methods. In its current version, the chip is designed for time-resolved SFX experiments, but it can be used at virtually any synchrotron or XFEL beamline in combination with the dedicated mini-end station (Sherrell et al., 2015). The efficient process of loading and handling samples, in combination with spectral mapping, significantly increases productive data acquisition time. Further improvements in pre-mapping of the chip, in combination with fast read-out X-Ray detectors, will greatly facilitate high throughput serial crystallography at XFELs, potentially requiring only minutes of data collection for static structure determination. This chip concept is therefore an ideal approach for precious protein samples and oversubscribed XFEL beamlines. The most significant future improvement will be the identification of spectral features or other probes of single crystal character that are better correlated to X-ray diffraction quality so that the mapping procedure can lead to the maximum possible indexable hit rates for a given system.
5. Materials and Methods
5.1. Nanofabrication
The chip is made out of a single side polished silicon wafer (WRS material), with a thickness of 550 μm, coated with 50 nm Low Pressure Chemical Vapor Deposition (LPCVD) silicon nitride, applied as an etching mask for KOH etching. Conventional lithography is utilized to transfer the photomask pattern onto an organic photoresist (Shiply S1818), which is spin-coated onto the silicon wafer. After exposure to UV light, the pattern is developed and cleaned. In the next step, the silicon nitride under the photoresist is etched away using SF6 plasma reactive ion etching. The photoresist is used in the etching stage as an etch mask and must be stripped away afterwards. The wafer is then placed in a super-saturated aqueous KOH solution (40% w/w) at 80°C for etching away bulk silicon until the desired feature depth is reached. A similar process is carried out for developing the backside features until the desired hole size is reached. The wafer is then cleaned, using a standardized cleaning process by using H2O2:NH4OH:H2O solution at ratio 1:1:5 by volume at 70°C, which leaves the chip surface hydrophilic. Finally, the wafer is separated into individual chips by dicing.
5.2. Protein expression, purification and crystallization
Sperm whale myoglobin expression and purification
The sperm whale (Physeter catodon) myoglobin (SWMb) expression plasmid (pMb413a) with a D122N mutation was kindly provided by Professor John S. Olson (Rice University, USA) and the protein was expressed constitutively in E. coli BL21(DE3) cells for three days. SWMb with bound carbon monoxide (SWMb-CO) was purified as described previously, with some modifications (Springer & Sligar, 1987, Mueller et al., 2015). Briefly, i) all buffers used in purification were saturated with 1 atm of CO gas, ii) after ammonium sulfate precipitation, the collected precipitant was dissolved in a small volume of 20 mM Na-phosphate pH 6.0 and dialyzed overnight against this buffer, iii) the dialyzed protein was concentrated and applied onto a CM Sepharose (GE Healthcare Life Sciences) fast flow column, which was pre-equilibrated with the same buffer and a linear pH gradient: 20 mM Na-phosphate pH 6.0 to 50 mM Na-phosphate pH 9.0 was used for elution, iv) eluted protein fractions were concentrated and applied onto a Superdex 75 gel filtration column (GE Healthcare Life Sciences) in 20 mM Tris-HCl and 150 mM NaCl buffer (pH 8.0) to remove aggregates, v) the peak fractions were combined, solid sodium dithionite was added (20 mM final concentration) and, after 10 minute incubation, the protein was rapidly applied to a sephadex G-25 column equilibrated with 10 mM Tris-HCl buffer (pH 9.0) under 1 atm of CO (Li et al., 1994). The eluate containing SWMb-CO was concentrated for crystallization.
SWMb-CO crystallization
To ensure that the SWMb crystals were grown in the reduced CO-bound form, the crystallization buffers were saturated with CO gas. The SWMb-CO crystals were grown in 3 ml Monoject Blood Collection Tubes (Covidien, Mansfield, MA) prefilled with 1 atm of CO gas. 180 - 200 μl of crystallization solution containing 50-60 mg/ml protein in 10 mM Tris-HCl (pH 9.0) and 2.5 - 2.6 M ammonium sulfate were added into the Monoject tubes. Seeding was used to promote nucleation of large quantities of small crystals. The SWMb-CO crystals were crushed in 10 mM Tris-HCl (pH 9.0) and 3.2 M ammonium sulfate. The seeds were diluted 1:100 under the same buffer conditions and 10 μl was added to each tube. The size distribution of the final crystals was controlled by adding 180 μl of 10 mM Tris-HCl, pH 9.0 and 3.2 mM ammonium sulfate pre-saturated with CO gas to each tube after crystals appeared. This method produces crystals of appropriate sizes, corresponding to the chip parameters, as described previously (Mueller et al., 2015). Crystals were loaded onto the chip as shown in Fig. 4 To prevent salt crystal formation all experiments for chip loading have been performed in a portable glove bag (Sigma-Aldrich) with a temperature of 24°C and a humidity of 60%.
5.3. In-situ microspectrometer
A fiber-coupled microfocus absorption spectroscopy setup for sample characterization and chip mapping was developed (adapted from Kirchner et al., 2014). The setup utilizes a fiber-coupled broadband LED light source, off-axis parabolic mirrors, a fiber-coupled USB spectrometer, and a 3-axis translation stage (SmarAct) for accurate sample positioning (similar to the beamline end station described in Sherrell et al., 2015).
The LED light (OceanOptics) source outputs light within the wavelength range from 425 nm - 650 nm. It is fiber-coupled to a 200 μm core diameter fiber. The first off-axis parabolic mirror (OPAM) collimates and reflects the light by 90° onto a second OPAM with half the effective focal length of the first. This mirror focuses the light onto the sample. These two mirrors constitute an all-reflective 2:1 microscope, resulting in a focal spot size of nominally 100 μm at the sample. A third OPAM collimates the transmitted light onto a fourth OPAM that focuses the light into a fiber-coupled USB spectrometer (Ocean Optics, USB2000-UV-VIS). Absorption spectra were generated by taking the logarithm of the background subtracted transmission spectra divided by the light source spectrum.
The parabolic mirror pairs are placed in a rigid four bar optical cage system (Thorlabs, Fig. 5a) using a custom designed mount, which fixes the rotation of the mirrors. This ensures very easy and precise alignment, which does not need to be adjusted. A CMOS camera (Thorlabs) and a close-focus zoom lens (Edmond Optics), with a minimum field-of-view of approximately 6 mm and a large working distance, is used as an off-axis viewing system for chip alignment.
5.4. SFX Data Collection
X-ray data collection was performed at the XPP instrument of LCLS at room temperature with an X-ray beam size of 20 × 20 μm and 1010 X-ray photons per pulse with a photon energy of 9.58 keV and a nominal sample-to-detector distance of 67.8 mm, under proposal LF70. A diamond (111) monochromator was used, resulting in a narrow bandwidth, The diffraction data collection rate was 10 Hz, which was limited by detector readout speed. Data were recorded using a Rayonix MX 170-HS CCD detector.
In the manner described previously (Mueller et al., 2015) data collection was organized such that 10 shots of the XFEL were integrated into a single frame of the detector, providing the reference or “before” image, and in a second step a single shot of the same crystal was recorded. In a pump-probe experiment, this second image would be the only one with a possible pump interaction. We note that the first image integrated from ten individual XFEL shots was recorded with a beam attenuated roughly 30 times after the monochromator, ensuring that radiation damage is below detection threshold. The total absorbed X-ray dose per crystal was 5.7 kGy for the sum of 10 attenuated pulses plus 16.8 kGy without attenuator, using a 480 μm sapphire attenuator. These values are calculated using the software suite Raddose-3D (Zeldin et al., 2013), ignoring the effect of photoelectron escape. These are well below the damage threshold (Garman limit) at which noticeable spectroscopic changes are observed determined from previous studies (Owen et al., 2011). A broadly accepted rule of thumb in macromolecular crystallography is that a structural change of less than 20% does not contribute significantly to the final electron density (Pearson et al., 2007) (see supporting Information for Safe Dose calculation).
5.5. Data Analysis
Individual images were extracted from the XTC streams and stored in HDF5 format for indexing with Cheetah and indexamjig (CrystFEL). From the output of indexamajig information was extracted to calculate both raw hit rates (hraw) and fractions of indexed images (hidx). Spots were searched for on these extracted images using the DIALS spot-finding routine (Waterman et al., 2016), which is based on the XDS algorithm (Kabsch, 2010). For integration, data merging and structure determination, with an improved method based on post-refinement and partiality correction (Ginn et al., 2015a), images were indexed using the TakeTwo algorithm in cppxfel (Ginn et al., 2016, in press) and integrated using simple summation, with an integration box of a 5×5 foreground, one-pixel buffer and two layers of additional surrounding pixels for background subtraction. The detector distance was refined to 68.6 mm and the wavelength to 1.293 Å, in opposition to 67.8 mm and 1.312 Å, which were recorded during beam time. Crystal orientations were initially refined according to a previous protocol to minimize the spread of Ewald spheres on which the reflections are observed (Ginn et al., 2015b), with additional refinement of the three unit cell dimensions for the orthorhombic space group. For this purpose, predicted reflections were registered as strong reflections with an integrated intensity of 32 or greater. Post-refinement was carried out against a reference data set according to a previous protocol (Ginn et al., 2015a) using a binary partiality of 0 or 1. A partiality of 1 is assigned to reflections for which the conventionally calculated partiality is 0.2 or greater, and 0 if this is not the case. Only one cycle of post-refinement was performed after an initial Monte Carlo merge in order to refine which reflections were included and excluded in the merge, but partiality correction on this data was found to be unstable, likely due to the monochromatic beam. The reference data set was generated by an initial merge of the images and used to post-refine individual images to improve the merging statistics Rsplit and CC1/2. The bandwidth for refinement was set to 0.01% of the pulse wavelength, and mosaicity to 0.04°, leading to around 166 reflections accepted per image. Images were rejected below a correlation threshold of 0.8 and individual reflections were rejected beyond 1.8 standard deviations from the mean of all observation intensities for a given reflection (parameters defined in Ginn et al., 2015a). Linear scale factors were applied to each image during merging to best match the reference density. Intensities were converted to amplitudes using Truncate (Winn et al., 2011) and phases generated from a previously solved synchrotron data set using REFMAC5 (Murshudov et al., 2011).
The protein and heme group from the sperm whale myoglobin structure PDB ID = 1VXA was used as a starting model for molecular replacement. The atomic B factors were set to 20 using the Phenix software package (Adams et al., 2010). Phaser from CCP4 (Winn et al., 2011) was used for molecular replacement. Building and refinement were done using the COOT (Emsley & Lohkamp, 2010) and REFMAC5 programs. Following the placement of waters and a single sulfate atom, the carbon monoxide was built into the density and refined (Fig. 6b). The refinement values are presented in Table 3. Coordinates and structure factors for the Physeter macrocephalus myoglobin with carbon monoxide covalently bound have been deposited in the Protein Data Bank under the accession code 5JOM.
Acknowledgements
This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (R.J.D.M.), the Max Planck Society (R.J.D.M.), the Canadian Institute for Advanced Research (R.J.D.M., O.P.E., D.I.S.), the Canada Excellence Research Chair program (O.P.E.), O.P.E. is the Anne and Max Tanenbaum chair in Neuroscience at the University of Toronto.
The research leading to these results has received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement no. 623994 (H. M. -W.).
Use of the Linac Coherent Light Source (LCLS), SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science and Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515.
Footnotes
The use of a silicon based chip as a fixed target for serial crystallography using XFELs and synchrotrons is demonstrated. The ability to load more than 10,000 crystals and pre-map their positions using in-situ spectroscopy, with a system of fiducial marks for rapid and precise alignment, allows high hit rates to be achieved whilst minimizing the number of crystals required. This enables serial room temperature atmospheric pressure experiments at rates currently limited only by detector readout.
References
- Adams PD, Pavel V, Chen VB, Ian W, Echols N, Moriarty NW, Read RJ, Richardson DC, Jane S, Thomas C. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends TRM, Foucar L, Ardevol A, Nass K, Aquila A, Botha S, Doak RB, Falahati K, Hartmann E, Hilpert M, et al. Science. 2015;350:445–450. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- Barty A, Caleman C, Aquila A, Timneanu N, Lomb L, White TA, Andreasson J, Arnlund D, Bajt S, Barends TRM, et al. Nat Photonics. 2011;6:35–40. doi: 10.1038/nphoton.2011.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barty A, Küpper J, Chapman HN. Annu Rev Phys Chem. 2013;64:415–435. doi: 10.1146/annurev-physchem-032511-143708. [DOI] [PubMed] [Google Scholar]
- Baxter EL, Aguila L, Alonso-Mori R, Barnes CO, Bonagura CA, Brehmer W, Brunger AT, Calero G, Caradoc-Davies TT, Chatterjee R, et al. Acta Crystallogr Sect D Struct Biol. 2016;72:2–11. doi: 10.1107/S2059798315020847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet S, Lomb L, Williams GJ, Barends TRM, Aquila A, Doak RB, Weierstall U, Deponte DP, Steinbrener J, Shoeman RL, et al. Science. 2012;74:10–13. [Google Scholar]
- Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, et al. Nature. 2011;470:73–77. [Google Scholar]
- Chollet M, Alonso-Mori R, Cammarata M, Damiani D, Defever J, Delor JT, Feng Y, Glownia JM, Langton JB, Nelson S, et al. J Synchrotron Radiat. 2015;22:503–507. doi: 10.1107/S1600577515005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chreifi G, Baxter EL, Doukov T, Aina E, Mcphillips SE, Song J, Meharenna YT, Soltis SM, Poulos TL. Proc Natl Acad Sci. 2016;113:1226–1231. doi: 10.1073/pnas.1521664113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AE, Soltis SM, González A, Aguila L, Alonso-Mori R, Barnes CO, Baxter EL, Brehmer W, Brewster AS, Brunger AT, et al. Proc Natl Acad Sci. 2014;111:17122–17127. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens P, Pitak M, Gembicky M, Messerschmidt M, Scheins S, Benedict J, Adachi S, Sato T, Nozawa S, Ichiyanagi K, et al. J Synchrotron Radiat. 2009;2009:226–230. doi: 10.1107/S0909049508040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePonte DP, Weierstall U, Starodub D, Schmidt K, Spence JCH, Doak RB. J Phys D Appl Phys. 2008;41:7. [Google Scholar]
- Emsley P, Lohkamp B. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn HM, Brewster AS, Hattne J, Evans G, Wagner A, Grimes JM, Sauter NK, Sutton G, Stuart DI. Acta Crystallogr Sect D Biol Crystallogr. 2015a;71:1400–1410. doi: 10.1107/S1399004715006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn HM, Messerschmidt M, Ji X, Zhang H, Axford D, Gildea RJ, Winter G, Brewster AS, Hattne J, Wagner A, et al. Nat Commun. 2015b;6:6435. doi: 10.1038/ncomms7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha S, Perry SL, Pawate AS, Kenis PJA. Sensors Actuators, B Chem. 2012;174:1–9. doi: 10.1016/j.snb.2012.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K, Shinzawa-Itoh K, Yano N, Takemura S, Kato K, Hatanaka M, Muramoto K, Kawahara T, Tsukihara T, Yamashita E, et al. Nat Methods. 2014;11:734–736. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- Hunter MS, Segelke B, Messerschmidt M, Williams GJ, Zatsepin NA, Barty A, Benner WH, Carlson DB, Coleman M, Graf A, et al. Sci Rep. 2014;4:1–5. doi: 10.1038/srep06026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern J, Tran R, Alonso-Mori R, Koroidov S, Echols N, Hattne J, Ibrahim M, Gul S, Laksmono H, Sierra RG, et al. Nat Commun. 2014;5:4371. doi: 10.1038/ncomms5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner FO, Lahme S, Riedle E, Baum P. AIP Adv. 2014;4:077134. [Google Scholar]
- Kirian RA, Wang X, Weierstall U, Schmidt KE, Spence JCH, Hunter M, Fromme P, White T, Chapman HN, Holton J. Opt Express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Quillin ML, Phillips GN, Olson JS. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- Lyubimov AY, Murray TD, Koehl A, Araci IE, Uervirojnangkoorn M, Zeldin OB, Cohen AE, Soltis SM, Baxter EL, Brewster AS, et al. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:928–940. doi: 10.1107/S1399004715002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani V, Morgan A, Yoon CH, Lane TJ, White T, O’Grady C, Kuhn M, Aplin S, Koglin J, Chapman HN. 2015 doi: 10.1107/S1600576716007469. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, Marx A, Epp SW, Zhong Y, Kuo A, Balo AR, Soman J, Schotte F, Lemke HT, Owen RL, et al. Struct Dyn. 2015;2:054302. doi: 10.1063/1.4928706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray TD, Lyubimov AY, Ogata CM, Vo H, Brunger T, Berger JM. Acta Crystallogr Sect D Biol Crystallogr. 2015;71:1987–1997. doi: 10.1107/S1399004715015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RL, Yorke BA, Gowdy JA, Pearson AR. J Synchrotron Radiat. 2011;18:367–373. doi: 10.1107/S0909049511004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson AR, Pahl R, Kovaleva EG, Davidson L, Wilmot CM. J Synchrotron Radiat. 2007;14:92–98. doi: 10.1107/S0909049506051259. [DOI] [PubMed] [Google Scholar]
- Roedig P, Vartiainen I, Duman R, Panneerselvam S, Stübe N, Lorbeer O, Warmer M, Sutton G, Stuart DI, Weckert E, et al. Sci Rep. 2015;5:10451. doi: 10.1038/srep10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler CG, Agarwal R, Allaire M, Yukl ET, Zhu D, Bommer M, Brewster AS, Browne MC, Chatterjee R, Cho E, et al. Structure. 2016;24:1–10. [Google Scholar]
- Sherrell DA, Foster AJ, Hudson L, Nutter B, O’Hea J, Nelson S, Paré-Labrosse O, Oghbaey S, Miller RJD, Owen RL. J Synchrotron Radiat. 2015;22:1372–1378. doi: 10.1107/S1600577515016938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra RG, Laksmono H, Kern J, Tran R, Hattne J, Alonso-Mori R, Lassalle-Kaiser B, Glockner C, Hellmich J, Schafer DW, et al. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer BA, Sligar SG. Proc Natl Acad Sci U S A. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara M, Mizohata E, Nango E, Suzuki M, Tanaka T, Masuda T, Tanaka R, Shimamura T, Tanaka Y, Suno C, et al. Nat Methods. 2015;12 doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, et al. Science. 2015;346:1–6. [Google Scholar]
- Waterman DG, Winter G, Gildea RJ, Parkhurst JM, Brewster AS, Sauter K, Evans G. Acta Crystallogr Sect D Struct Biol. 2016;72:558–575. doi: 10.1107/S2059798316002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierstall U, James D, Wang C, White TA, Wang D, Liu W, Spence JCH, Bruce Doak R, Nelson G, Fromme P, et al. Nat Commun. 2014;5 doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TA, Kirian RA, Martin AV, Aquila A, Nass K, Barty A, Chapman HN. J Appl Crystallogr. 2012;45:335–341. [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrine-Afsar A, Barends TRM, Müller C, Fuchs MR, Lomb L, Schlichting I, Miller RJD. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:321–323. doi: 10.1107/S0907444911055296. [DOI] [PubMed] [Google Scholar]
- Zeldin OB, Gerstel M, Garman EF, Zeldin OB, Gerstel M, Garman EF. J Appl Crystallogr. 2013;46:1225–1230. [Google Scholar]