Abstract
Access to DNA within nucleosomes is required for a variety of processes in cells including transcription, replication and repair. Consequently, cells encode multiple systems that remodel nucleosomes. These complexes can be simple, involving one or a few protein subunits, or more complicated multi-subunit machines1. Biochemical studies2–4 have placed the motor domains of several remodellers on the superhelical location (SHL) 2 region of the nucleosome. Structural studies on Chd1 and Snf2 (RSC) in complex with nucleosomes5–7 have provided insights into the basic mechanism of nucleosome sliding by these complexes. However, how larger, multi-subunit remodelling complexes, such as INO80, interact with nucleosomes or how remodellers carry out functions such as nucleosome sliding8, histone exchange9, and nucleosome spacing10–12 remains poorly understood. Although some remodellers work as monomers13, others work as highly cooperative dimers11,14,15. Here we present the structure of the INO80 chromatin remodeller with a bound nucleosome revealing that INO80 interacts with nucleosomes in a unique manner with the motor domains located at the entry point to the wrap around the histone core rather than at SHL2. The Arp5-Ies6 module of INO80 makes additional contacts on the opposite side of the nucleosome. This unique arrangement allows the H3 tails of the nucleosome to play a role in regulation, differing from other characterised remodellers.
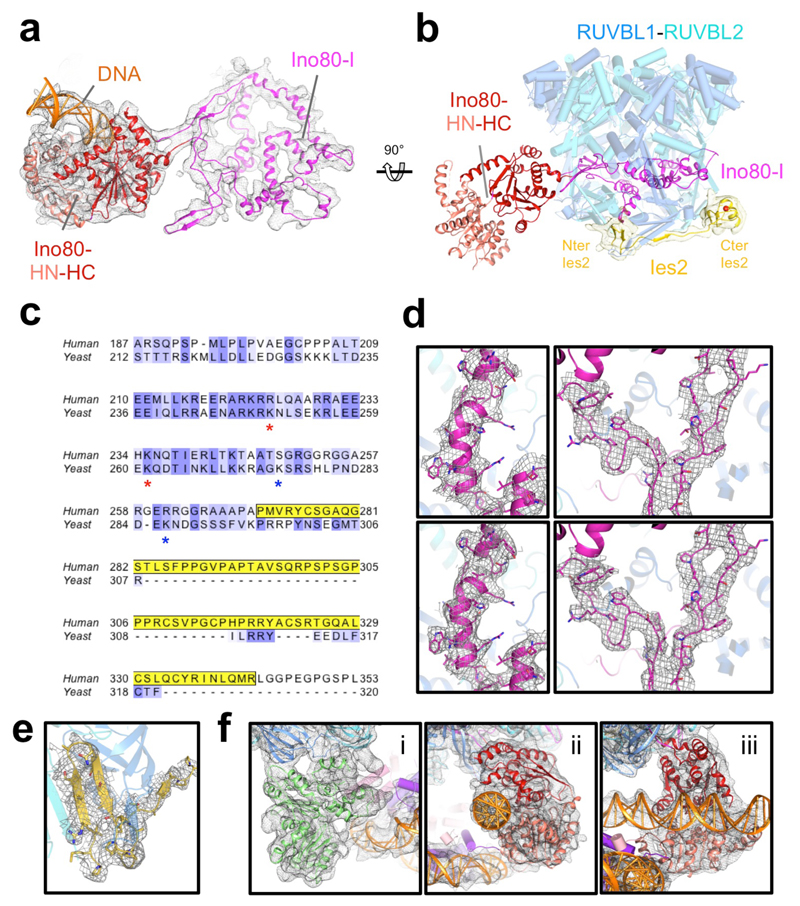
We prepared a complex between human INO80 core complex10 and human nucleosomes flanked by 52 and 25 base pair overhangs (Extended Data Fig. 1) in the presence of ADP·BeF3, that tightens nucleosome binding (Extended Data Fig. 1). Although prepared at a 2:1 molar ratio INO80:nucleosome, the majority of the particles on our EM grids contained either free INO80 complex or a 1:1 complex (Methods & Extended Data Fig. 2). We processed the data to obtain two different reconstructions (Methods and Extended Data Figure 2). One was selected to obtain nucleosome complexes (4.8 Å resolution, Fig. 1, Extended Data Figs. 2 & 3, Extended Data Table 1, Supplementary Video 1), while the other (3.8 Å resolution, Extended Data Figs. 2 & 3 and Extended Data Table 1) used all particles initially but during the final stages of processing, the region corresponding to the bound nucleosome was masked out to optimise fitting on the INO80 component (Methods and Extended Data Fig. 2). This map showed essentially the same features as our previous apo structure16 but with significant improvement in certain areas such as Ies2 and the RUVBL1-RUVBL2 hexamer16, allowing us to improve our model and assign sequence to the Ino80-I region (see Methods and Extended Data Fig. 4). Furthermore, this map allowed us to determine the location of a zinc-binding domain of Ies2. Parts of Ies2 track across the RUVBL1-2 hexamer and interact with the OB domains from adjacent RUVBL1 and RUVBL2 subunits (Extended Data Fig. 4). Interestingly, this part of human Ies2 corresponds to an extension at the C-terminus that is absent in the yeast protein. The density is running towards the motor domains but is disordered beyond the interface with the RUVBL subunits. Previous crosslinking data on the yeast apo INO80 complex indicated an interface between the Ies2 subunit and the motor domains17, and the Ies2 subunit regulates ATPase activity in both yeast and human INO80 complexes10,18–20. Crosslinks observed between yeast Ies2 and the Ino80 motor domains are located just beyond the ordered part of the human Ies2 structure but is close to the motor domains (Extended Data Fig. 4).
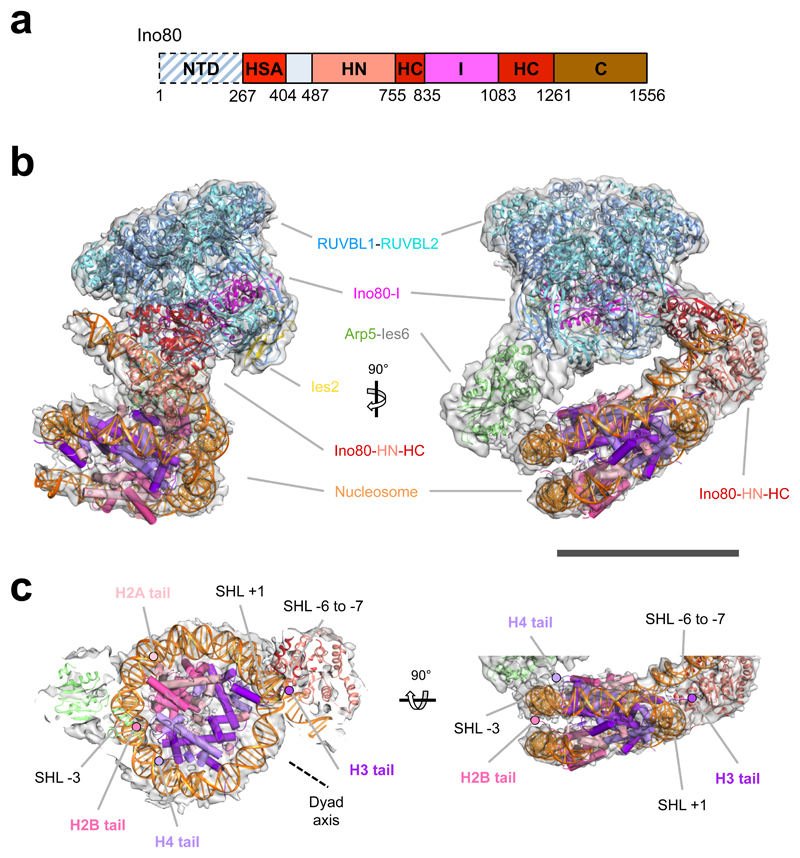
Fig. 1. Human INO80-nucleosome complex.
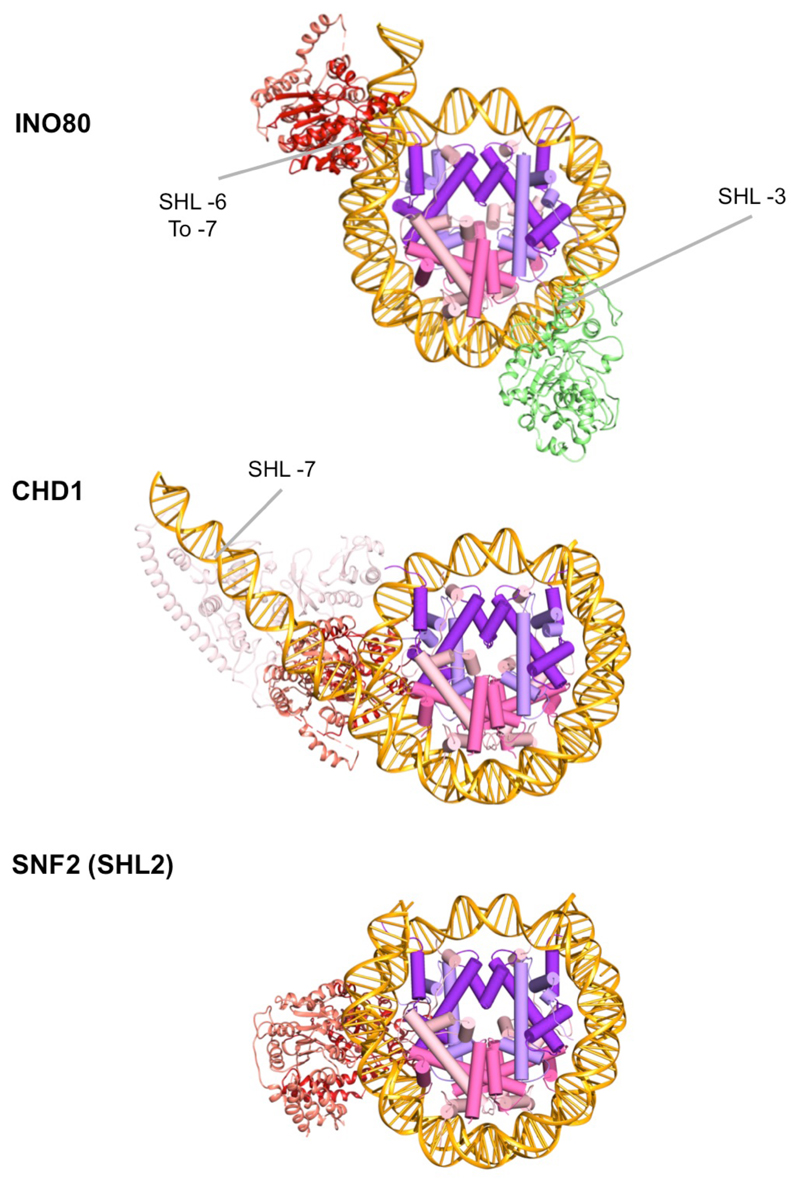
a, Ino80 subunit with functional domains labeled. b, 3D INO80-nucleosome complex reconstruction with RUVBL1-2, Ino80, Arp5, Ies2 and nucleosome structural models fitted. Scale bar, 100 Å. c, INO80 nucleosome interactions with histones and nucleosome positions labelled. INO80 contacts nucleosome at SHL-6 and SHL-3. Also shown are locations of the histone tails.
The INO80-nucleosome complex structure revealed protein secondary structural elements (Extended Data Fig. 3) and a bound nucleosome (Fig. 1a-c). The RUVBL1-2 heterohexamer encloses a large insertion in the C-terminal Ino80 motor domain (Figs. 1a, 1b, Extended Data Fig. 4) as seen in the INO80 apo structure16. This insertion region is connected to a region of density that is much better ordered than in the apo complex and fits the C-terminal motor domain of the Ino80 subunit, with density for the N-terminal domain alongside (Extended Data Fig. 4). Consistent with binding of ADP·BeF3, we observe the ATPase domains in the closed, nucleotide bound state5. However, rather than being located at SHL +2 of the nucleosome wrap (as observed in Chd1 and Snf25,6), the motor domains (as predicted from biochemical studies3), are instead located across SHL -6 to -7 in an orientation consistent with tracking along one strand of the DNA duplex in the anticipated 3’-5’ direction (Fig. 3a, Extended Data Fig. 5) when compared to other well characterised Superfamily 2 DNA translocases like NS321. This orientation would pump duplex DNA from the overhang onto the nucleosome towards the dyad axis. This contact region for the motor domains differs completely from all other characterised remodellers (Fig. 2a, Extended Data Fig. 5), but is consistent with footprinting and crosslinking studies on yeast INO80 complex3. The INO80 footprint spanning SHL -6 to -7 is due to contacts with the motor domains. The N-terminal motor domain also contacts across the gyres at SHL +1. Similar contacts across the gyres are observed in the Snf2 and Chd1 structures5,6 and are essential for nucleosome sliding4,6.
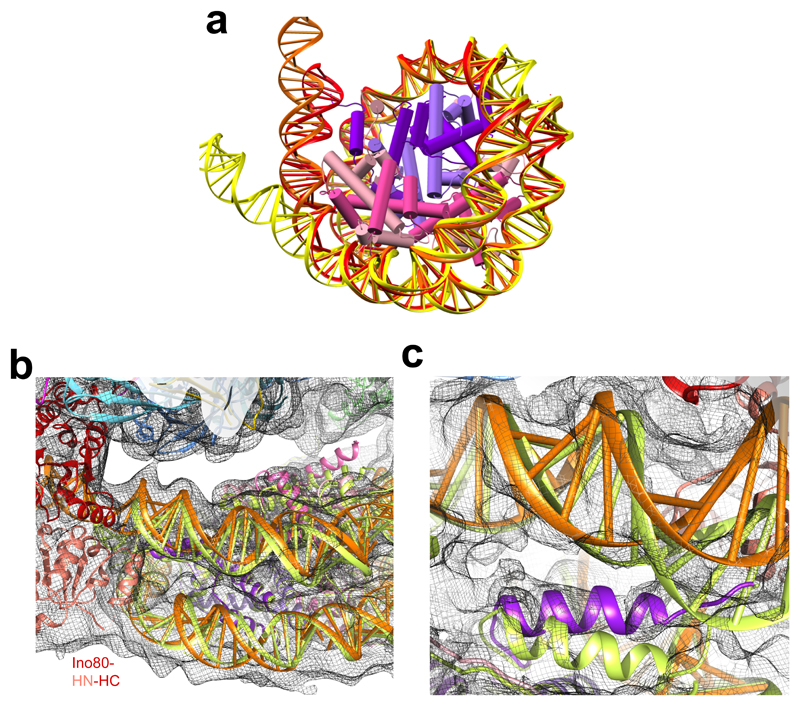
Fig. 3. Nucleosome distortion in the INO80-nucleosome complex.
a, DNA is peeled off in Ino80 (orange) and Chd1 (yellow) compared to the free nucleosome (red). In Ino80 this is due to motor domain interaction while in Chd1 this is due to the SANT and SLIDE domain interactions. b, DNA near to the motor domains in Ino80-nucleosome (orange) is lifted compared to a canonical nucleosome (lime green), which also causes slight rotation of H2A/H2B (pink). c, Lifting of the DNA in Ino80-nucleosome complex and movement of the H3 N-terminal helix (purple) near the motor domains.
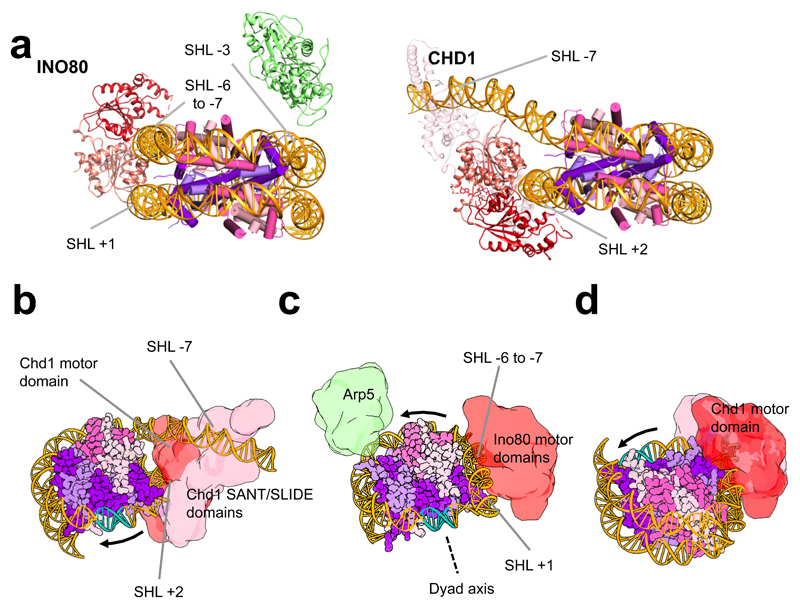
Fig. 2. Comparison of INO80 with Chd1 and a model for translocation by INO80.
a, INO80 and Chd1 nucleosome complexes viewed from the side of the nucleosome. b, Chd1 is proposed to push DNA towards the dyad axis (cyan). c, Ino80 (aligned on the nucleosome as in b) would push DNA past Arp5-Ies6 towards the dyad but from the opposite direction. d, Same as b but instead with the view aligned on the motor domains as in c.
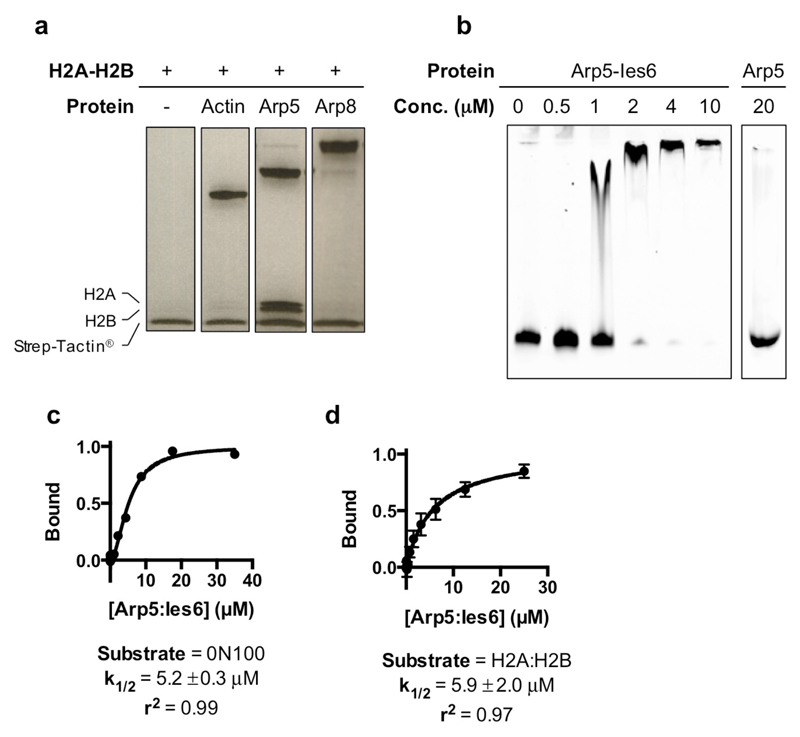
Significantly, footprinting studies also indicated contacts at SHL -2 to -33. Our structure reveals these to be due to Arp5-Ies6 and are proximal to H2A/H2B on almost the opposite side of the nucleosome to those made by the motor domains (Figs 1b & 1c, Fig. 2a). Consistent with these contacts, Arp5 binds to H2A/H2B dimers in solution and the Arp5-Ies6 complex binds to nucleosomes (Extended Data Fig. 6). The Arp5-Ies6 module also plays a key role in coupling ATPase and sliding activities10,19,20.
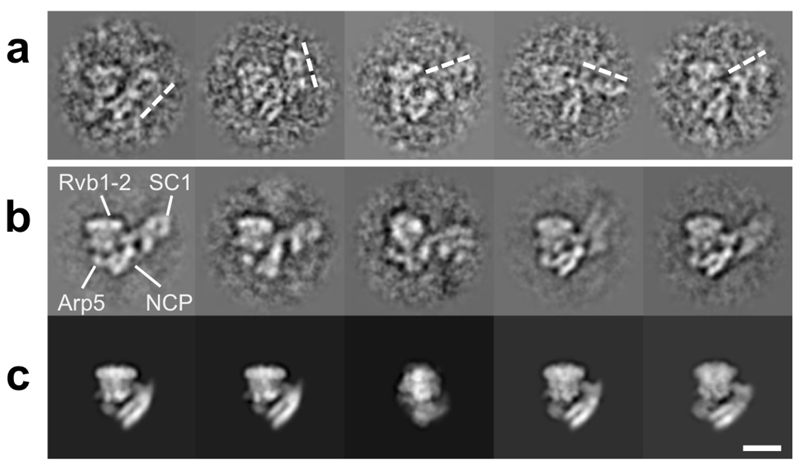
Although the structures reveal much detail about how INO80 contacts its nucleosome substrate, an obvious omission from our structure is the N-terminal region of the INO80 complex containing the actin, Arp4 and Arp8 subunits. Notably, this region (termed SC1) is flexible in the apo structure but careful selection of particles allowed us to locate this region of the complex16. This region remains flexible in the complex with nucleosomes resulting in it being averaged out in the structure. However, although visible in single particles and in carefully selected 2D class averages (Extended Data Fig. 7), it is too variable in location to be defined suggesting that it does not make extensive contacts with the nucleosome in this conformational state. The SC1 components have been shown to interact with histones22,23 and it may be that this component also interacts with the histone core in the active INO80 dimer or in a different functional state on the catalytic pathway.
Ino80, Chd1 and Snf2-like enzymes all translocate duplex DNA by tracking principally along one strand with a 3’-5’ directionality8,24,25 in a manner analogous to that employed by single-strand Superfamily 2 translocases like NS321. However, while INO80 and Chd1 slide nucleosomes away from DNA ends10,12, Snf2-like enzymes instead slide nucleosomes towards DNA ends25. Although similar regions of nucleosomal DNA are contacted, the structures place the motor domains of INO80 at a different location than in Chd1 and Snf2 (Fig. 2a, Extended Data Fig. 5). A consequence of this difference is that INO80 would pump DNA from the overhang towards the dyad whereas Chd1 and Snf2 would do this from the opposite direction5,6 (Fig. 2, Extended Data Fig. 5). This position of the motor domains of INO80 would move nucleosomes away from ends, consistent with biochemical observations10,12. The common directionality of sliding towards DNA ends suggested by the Chd1 and Snf2 structures raises a conundrum because Chd1 and Snf2-like enzymes have been shown to have opposing directional specificities for nucleosome sliding24,25.
The Snf2 and Chd1 nucleosome complexes show broadly similar contacts between the motor domains and the SHL +2 position of the DNA wrap. Both structures also show contacts across the DNA gyres to contact SHL -6 as predicted by biochemical studies4. Previous work showed that even the closely related SWR1 complex, which shares many subunits in common with INO80, is positioned at the SHL +2 to +3 location26. By contrast, the motor domains of INO80 bind at a completely different location but still contact the DNA across the gyres, albeit quite different parts of the nucleosome wrap.
The binding of INO80 induces unwrapping of the DNA at SHL -6 to -7, albeit to a lesser extent than Chd1 (Fig. 3a & Extended Data Fig. 5). However, the consequences are more dramatic because a more subtle distortion of the DNA wrap extends all the way from the motor domains round to the Arp5-Ies6 contact. The distortion lifts this DNA gyre away from the other. The associated H2A/H2B dimer moves along with the DNA causing it to lift away from the H3/H4 tetramer, presumably weakening this interface (Fig. 3b). Finally, as a consequence of the peeling back of the DNA at the entry site, the H3 histone tail remains associated with the DNA and alters conformation compared to the H3/H4 core (Fig. 3c). These conformational changes may have a role in histone exchange.
The Arp5-Ies6 subunits couple ATP hydrolysis to nucleosome sliding in INO8010,19,20. Furthermore, cyclic partial unwrapping of the DNA around the H2A/H2B interface is required for the histone exchange activity reported for INO803. The location of the DNA contacts we observe here suggests a simple mechanism for such a process. The directionality of translocation by the motor domains would push DNA towards the Arp5-Ies6 contact region (Fig. 2c). Unless released, this would result in a partial unwrapping of the DNA wrap, bulging out between these contacts across the H2A/H2B interface to facilitate H2A/H2B dimer exchange. Even though our structure has not undergone catalytic ATP turnover, the distortions induced by binding to INO80 appear to prepare the nucleosome for dimer exchange. Further ATP-dependent translocation by the motor domains would increase this effect by pushing DNA towards Arp5-Ies6. Interestingly, the SWR1 complex, that is related to INO80 and contains several subunits in common, facilitates histone exchange but is unable to slide nucleosomes27. Unlike INO80, the motor domains of SWR1 are located at the canonical SHL +2 position and the Swc2 subunit contacts the DNA overhang28. This two point contact, but with swapping of motor domain and DNA overhang contacts, raises the possibility of a similar mechanism for releasing the H2A/H2B dimer/DNA interface, with the motor domains pushing (or pulling) against a second contact to provide strain and lift the DNA wrap from the nucleosome surface.
INO80 slides nucleosomes from DNA ends and is able to sense flanking DNA length of up to 50-60 bp12,14. Like some other remodellers11,15, INO80 acts as a cooperative dimer in sliding14. Curiously, ATPase activity becomes uncoupled from sliding when INO80 has positioned nucleosomes at the centre of short DNA fragments but continues at the same rate as when the nucleosome is sliding12,14. The two contact points with the nucleosome in the structure suggests a basis for this behaviour. Since the motor domains pump DNA towards the dyad via the Arp5-Ies6 contact, they will also be underwinding the DNA as well as unwrapping it from the nucleosome surface. If the motor domains were to slip, then the DNA could simply re-associate with the nucleosome surface, resulting in a futile cycle of ATP hydrolysis. On the other hand, if the grip by Arp5-Ies6 were to slip instead, the DNA could be pushed forward across the surface resulting in sliding of the DNA wrap across the nucleosome surface. As a result, a translocation step size of one base per ATP, as shown for most SF1 and SF229, might build up tension in the DNA before being released in apparently larger step sizes as DNA slips past the Arp5-Ies6 grip point. Precisely such behaviour has been observed for nucleosome sliding at the single molecule level30,31 and may be an intrinsic part of nucleosome remodelling mechanisms. Such a mechanism would be coupled to sliding if it were to prevent “back-slippage”, thus providing directional translocation against a ratchet. For enzyme systems that require dimers, a mechanism that regulates the forward slippage between the partners could explain this behaviour, which presumably correlates with some form of regulation of activity, particularly for remodellers like INO80 that have higher order functions such as nucleosome spacing and phasing12,14,32.
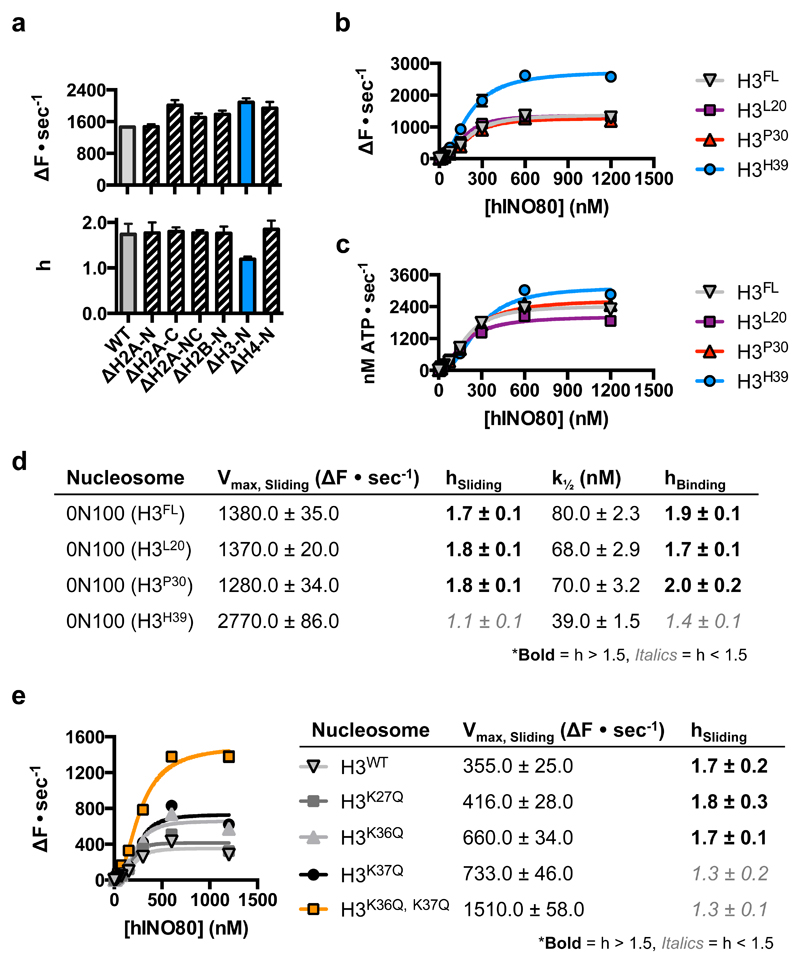
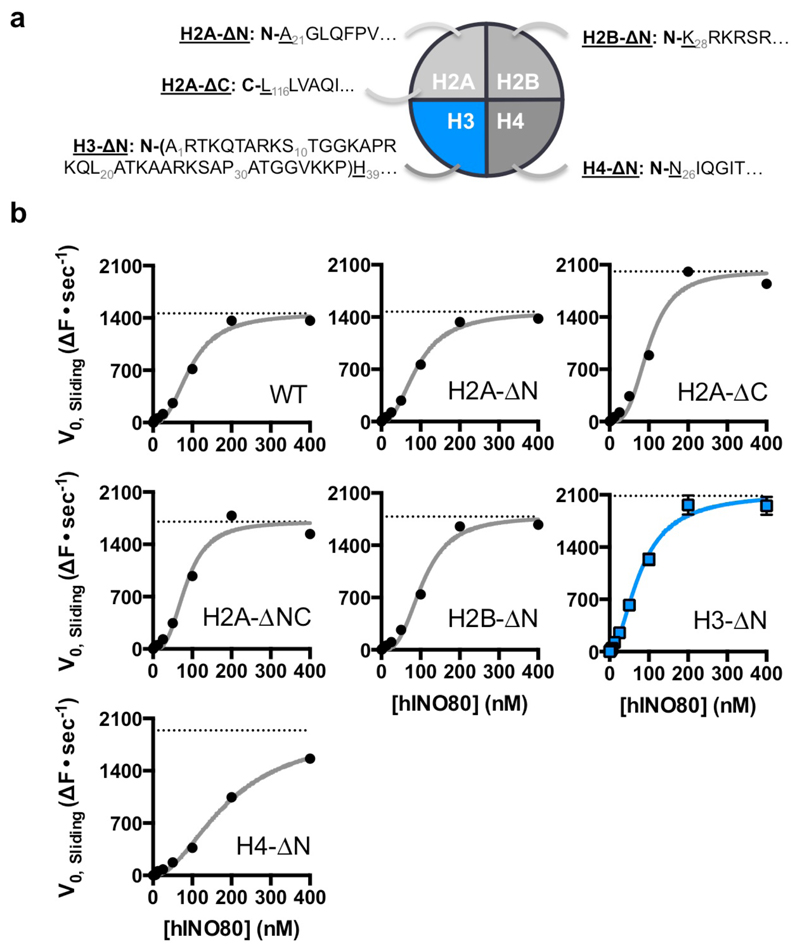
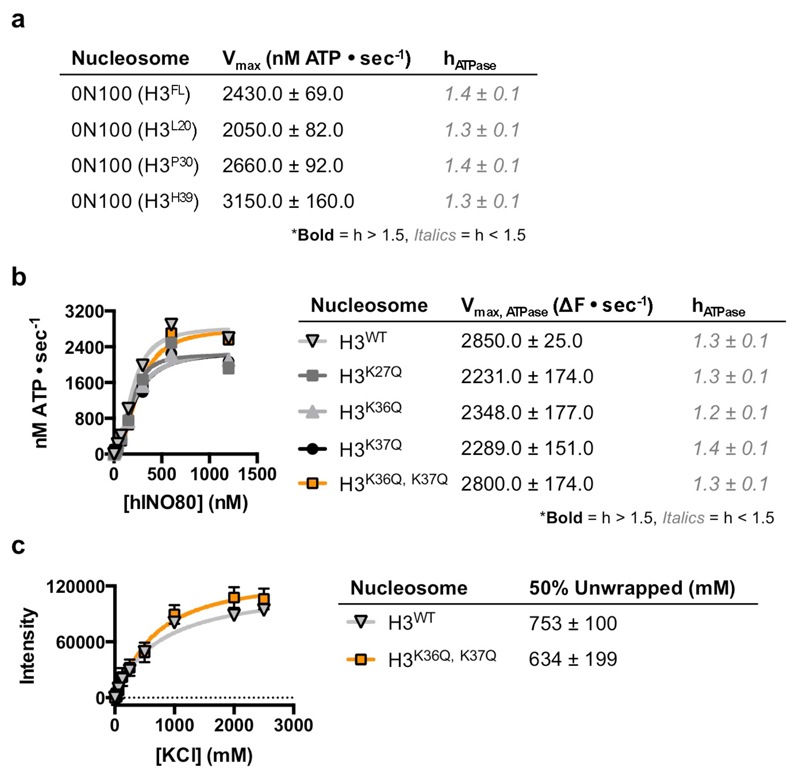
Several remodelling complexes are regulated by H4 tails33–35 through a complex interplay between regulatory components (AutoN and NegC) of the motor domains, that are missing in INO8014. The unique binding mode of INO80 raises questions about its regulation by histones because the H4 tails are too far away to interact with the motor domains (Fig. 1c), suggesting regulation by a different mechanism. We prepared a number of nucleosome variants in which the histone tails were individually deleted (Fig. 4 and Extended Data Fig. 8). For these tailless nucleosome substrates, sliding rates were comparable in all variants tested. Since INO80 functions as a dimer14, we also assessed the effects of the histone tails on the cooperativity. Interestingly for the H3 tail deletion, the Hill coefficient dropped significantly both for activity and binding (Fig. 4a), demonstrating a contribution of the H3 tail to INO80 dimer cooperativity that we localised to residues 31-39 (Fig. 4b). The ATPase and affinity of INO80 for tailless and cognate nucleosomes was similar (Figs. 4c & 4d), consistent with regulation being distinct from other remodellers that are regulated by H4 tails34. Individual mutations to mimic lysine acetylation (K36Q and K37Q) both showed a small, but reproducible, stimulation of sliding activity, but the K37Q mutation also showed the loss in cooperativity observed with the full H3 tail truncation (Fig. 4e and Extended Data Fig. 9). A double mutation showed a cooperative effect in sliding while retaining the loss in cooperativity. By contrast, a control substitution (K27Q) showed no effect on activity. These data support a role for H3 tails in regulating cooperativity in INO80 sliding and identify K37 as a key component in this process. The location of one H3 tail adjacent to the motor domains supports this idea (Fig. 1c) but, rather than being adjacent to the C-terminal motor domain as seen for remodellers regulated by H4 tails5,36, the H3 tail instead sits next to the N-terminal motor domain of the Ino80 subunit. The location of this H3 tail is normally between the DNA gyres as one end exits the nucleosome wrap37. However, the unwrapping of DNA from the nucleosome surface we observe in the structure breaks these contacts at the DNA entry site causing the H3 tail at that site to undergo a conformational change in response. Evidently, this unwrapping is required to initiate sliding by INO80 although the details about this process will require us to determine the structure of an INO80 dimer bound to a nucleosome.
Fig. 4. INO80 is regulated by H3 tails.
a, Initial nucleosome sliding rates of human nucleosomes lacking different histone tail, using a FRET-based assay25. b, Effect of increasing the extent of H3 tail truncation on nucleosome sliding. No effect is observed with 30 residues removed but a 39 residue truncation induces stimulation of sliding and, c, ATPase rates for H3 tail truncations, d, loss of cooperativity for both sliding and binding of nucleosomes, e, Lysine to glutamine mutations in the H3 tail affect both the rate and cooperativity of sliding. n=2 biologically independent experiments in all the graphs. Error bars represent SD from the mean values.
Our work reveals that INO80 adopts a unique mode of interaction with nucleosomes that permits, or possibly requires, regulation by a mechanism that also differs from other systems. However, further work will be required to determine details of these interactions and how these relate to the requirement of INO80 dimers for sliding activity.
Methods
Preparation of nucleosomes
For EM sample preparation, a 52N25 nucleosome was used, where N refers to the 147 bp nucleosome core. While we used the Widom-601 positioning sequence38 as the basis for this core, we introduced a point mutation within the sequence to remove a HinfI restriction site (GATTC to GATTG) to assist with sample preparation. The nucleosome was then prepared via the ligation method described in10. Tailless histones H2A–ΔN (A21-K130), H2A-ΔC (S1- L116), H2A–ΔNC (A21-L116), H2B-ΔN (K28-K125), H3-ΔN/H3H39 (H39-A135), H3P30 (P30-A135), H3L20 (L20-A135), H4-ΔN (N25-G102), additional mutations in H3 (K27Q, K36Q, K37Q and K36Q/K37Q) and H4 N25C mutation for labeling were introduced by standard mutagenesis methods. Tailless H3 nucleosomes and full-length were labeled on H4N25C. Human H2A, H2B, H3.1 and H4 were co-expressed in E. coli, lysed in buffer A (20 mM Tris pH 7.5, 400 mM NaCl, 0.1 mM EDTA, 1mM TCEP) and purified as soluble octamers on HiTrap Heparin HP in buffer A and eluted with a salt gradient, followed by Superdex S200 in buffer B (20 mM Tris pH 7.5, 2M NaCl, 0.1 mM EDTA, 1mM TCEP. Following labeling with Alexafluor 555 or 647 C2-maleimide, the octamer was re-purified by Superdex S200 in buffer B.
Preparation of hINO80-nucleosome-ADP·BeF3 complexes
hINO80 complex was prepared as described previously10. Nucleosomes were prepared as described above. hINO80-nucleosome-ADP·BeF3 complexes were prepared at a final concentration of 350 nM hINO80, 175 nM nucleosome, 3 mM ADP, 3 mM BeCl2, 15 mM NaF and 5 mM MgCl2. hINO80, nucleosomes, ADP and MgCl2 were prepared at 10x concentration in ‘EM Buffer’ (25 mM HEPES pH 8.0, 50 mM NaCl, 1 mM TCEP). BeCl2 and NaF were prepared at 10x concentration in water. The components were then mixed in the following order. First, hINO80 and nucleosomes were mixed together with the volume of ‘EM Buffer’ needed to obtain the final concentrations and incubated at 37 ºC for 15 min. This was followed by the addition of ADP and MgCl2 and a further 15 min incubation at 37ºC. Lastly, NaF and BeCl2 were added simultaneously.
Electron microscopy grid preparation
Grids for cryo-electron microscopy were prepared by depositing 3.5 μl of sample onto Quantifoil R2/2 copper grids. Samples were blotted before being flash-frozen in liquid ethane at liquid nitrogen temperature with a FEI Vitrobot Mark IV (waiting time 30 seconds, blotting time 0.5 seconds) at 4 °C and 100% humidity.
Data Collection
A set of 5,479 movies was collected at eBIC (Diamond Light Source, Didcot, UK) on a Titan Krios microscope operated at 300kV acceleration voltage. Images were recorded on a Falcon 3EC direct electron detector operating in linear mode at a magnification of 129,000 for a final pixel size of 1.09 Å/pixel with defocus range from -2.0 to -4.0 μm. The total dose was 80 e-/Å2 fractionated over 39 frames.
Image processing
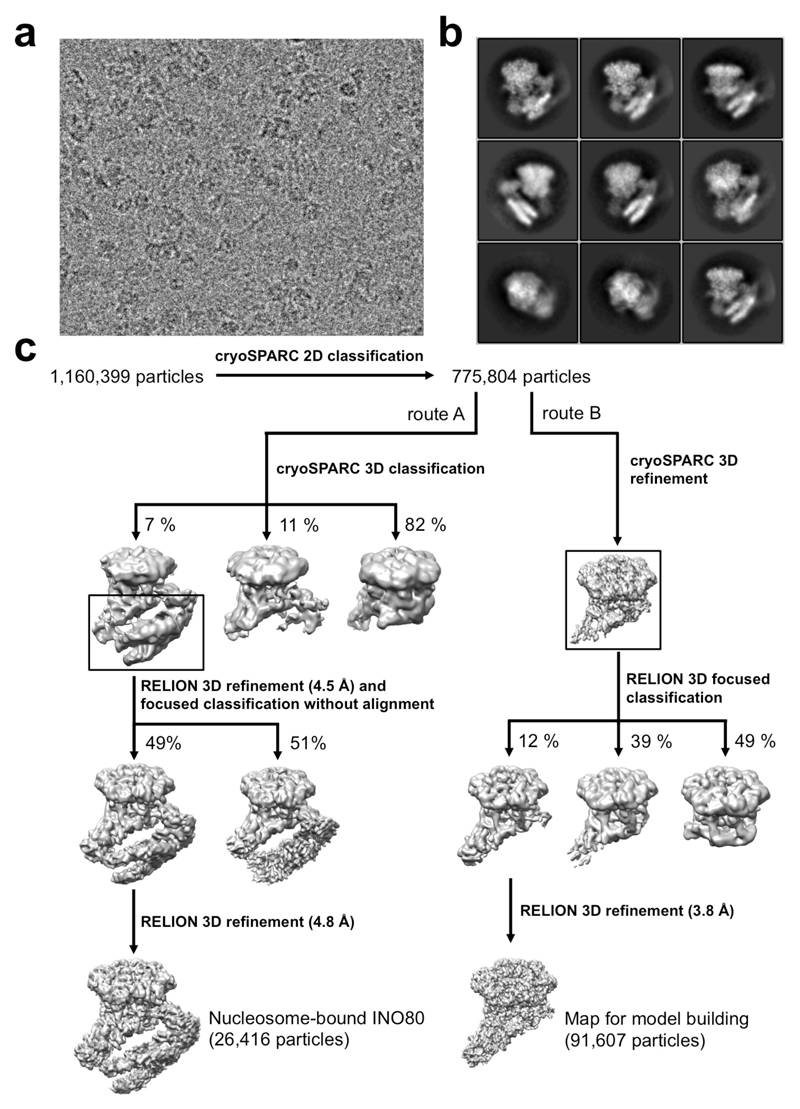
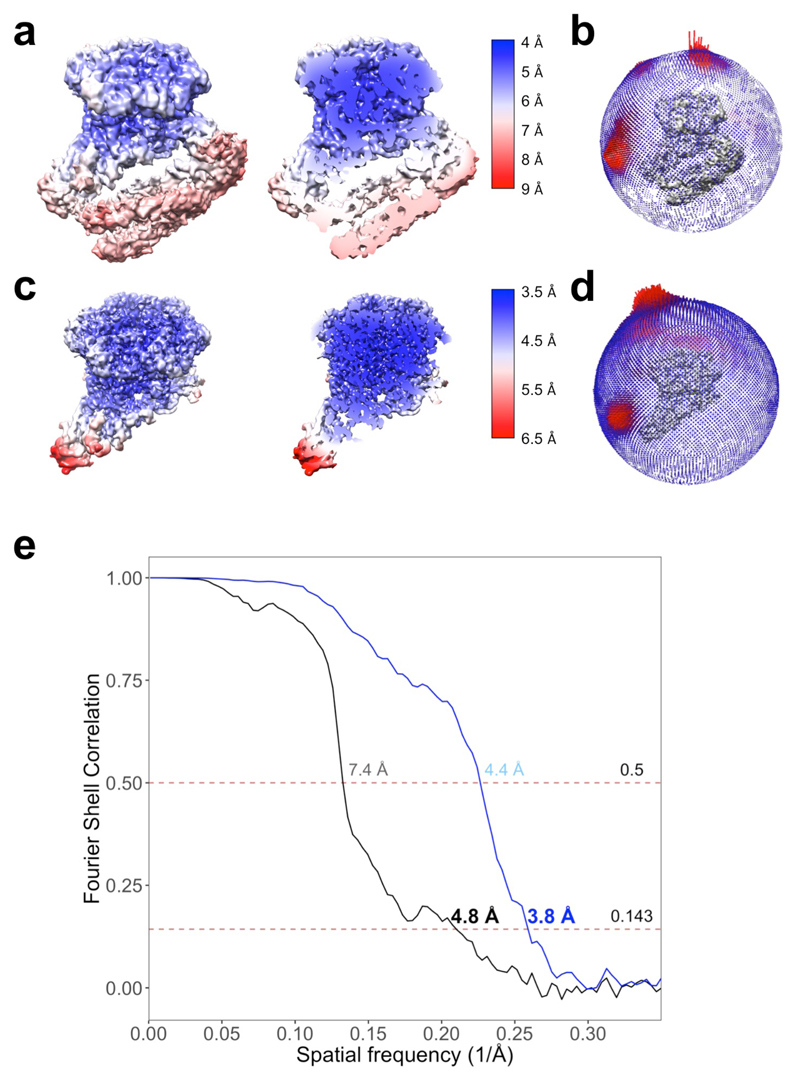
Individual movie frames were aligned using MotionCor239. CTF parameters were estimated using Gctf40. Particle picking was performed in Gautomatch using class averages obtained from a small dataset of the same sample previously collected in-house. Subsequent image processing was carried out in RELION 2.1.B141 and cryoSPARC 0.5.642. Global and local resolution estimates were calculated in RELION using the gold-standard Fourier Shell Correlation (FSC=0.143) criterion43. A total of 1,160,399 particles were extracted into boxes of 270 x 270 pixels. After 2D classification in cryoSPARC to remove false positives and noisy particles, a set of 775,804 particles was selected to perform downstream image processing, which is summarized in route A in Extended Data Figure 1. Briefly, nucleosome-bound particles were selected by a combination of 2D and 3D classifications in cryoSPARC and RELION. A final set of 26,416 homogeneous nucleosome-bound particles was selected to perform a final 3D refinement in RELION. The final model was refined to an overall resolution of 4.8 Å. FSC calculation was calculated after applying a mask generated by binarizing the map at a threshold of 0.012, extending the resulting mask by 6 pixels and adding a soft edge of 7 pixels. Statistics regarding the final model are presented in Extended Data Figure 2 and Extended Data Table 1.
A 3.8 Å map was generated with 91,607 particles selected by means of 3D classification in RELION with a mask which excluded the nucleosome (route B in Extended Data Figure 1). This map was used to aid model building.
Model building and refinement
Deposited coordinates for RUVBL1-2 for the apo INO80 structure (PDB ID 5OAF) were docked into the nucleosome-bound map. These were then adjusted and manually rebuilt in COOT44 with the aid of the 3.8 Å map. A homology model for Arp5 was generated by submitting the sequence to I-TASSER45, using a series of actin-fold proteins as templates. Well-resolved secondary structure was built according to the density in the 4.8 Å INO80-nucleosome map. After adjustments the model was trimmed of all side chains. The sequence for Ies2 was submitted to the PHYRE46 server, which yielded multiple results for the C-terminal zinc-binding domain. PDB ID 2YQQ was used as a starting model, and the coordinates were then manually extended towards the N-terminus in COOT. A homology model for the Ino80 motor domains was generated by threading the sequence into the structure of the Chd1 motor domain (PDB ID 5O9G) with SWISS-MODEL47. Side chains were removed and the domains were rigid body fit into the map, followed by a round of jellybody refinement in REFMAC48. The Ino80 insert domain was manually built in COOT and connected to the Ino80 motor domains using the 3.8 Å map. Coordinates for a human nucleosome core particle (PDB ID 5AV9) were fit into the density corresponding to the nucleosome. Keeping the position of the histone octamer fixed, a model for the nucleosome was built in COOT by combining the coordinates of the human histone octamer and a Widom 601 DNA wrap (PDB ID 3LZ0). The region of DNA bound to the motor domains was extended using linear B-form DNA, following the path of DNA through Chd1 (PDB ID 5O9G) where possible. Histones H2A and H2B with their complexed DNA, as well as the N-terminal helix of H3 were moved according to clear changes in the density from the canonical position. After completing model building the coordinates were subject to real-space refinement in Phenix49.
Purification of Actin and Actin-related proteins
Human Actin and Actin-related proteins (Arp5 and Arp8) were expressed in Hi5 insect cells with an N-terminal octahistidine and C-terminal double-Strep tag. All proteins were purified to near homogeneity using sequential affinity chromatography steps (HisTrap HP followed by StrepTactin HP (GE Healthcare)). This was followed by buffer exchange into a storage buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM TCEP and 10% glycerol using a spin concentrator. The concentrated sample was then flash frozen in liquid nitrogen in small aliquots until further use. Arp5-Ies6 was prepared as previously described10.
Actin and Actin-related protein pulldown assay
Purified human Actin or Actin-related proteins (bait) and recombinant human H2A/H2B dimers (prey) were prepared at concentrations of 20 and 40 μM, respectively, in ‘pulldown buffer’ (25 mM Tris-HCl, 250 mM NaCl, 1 mM TCEP, 0.05% NP40). For each pulldown condition, these 2x stocks were mixed in equal volumes and placed on a roller at room temperature for 30 minutes to equilibrate. In preparation for the pulldown, 50 μL of Strep-Tactin Magnetic Beads slurry (Qiagen) was washed with ‘pulldown buffer’ on a magnetised support stand. After incubation, the protein mixture was added to the washed magnetic beads and incubated for a further 30 min at room temperature. The resin was then washed extensively with ‘pulldown buffer’ (at least ten 1 mL washes) to remove any unbound products before adding 50 μL of SDS-containing loading dye and boiling the sample. The bound products (i.e. those eluted from the resin following addition of loading dye) were then resolved by SDS-PAGE.
Arp5 and Arp5-Ies6 electrophoretic mobility shift assay (EMSA) with nucleosomes
Purified Arp5 and Arp5-Ies6 were incubated for 30 minutes at room temperature with 2 μM human 167 nucleosomes in buffer containing 25 mM Tris-HCl, 100 mM NaCl and 1 mM DTT. Final concentrations of Arp5 and Arp5-Ies6 were as indicated in Extended Data Figure 5. Equilibrated samples were then resolved by native PAGE on 6% acrylamide gels prepared and run in 0.5x TBE (Tris-Borate-EDTA) buffer.
MST with Arp5-Ies6 and various substrates
MST experiments were carried out similarly to those described previously14 for the interaction of the hINO80 core complex and nucleosomes. Briefly, Arp5-Ies6 was assayed for interaction with 0N100 nucleosomes, DNA (100 bp) and H2A:H2B histone dimers. Arp5:Ies6 was incubated at the appropriate concentrations with 40 nM fluorescently-labelled substrate for 30 min at room temperature in buffer containing 25 mM HEPES pH 8.0, 50 mM NaCl, 1 mM TCEP, 10% Glycerol, 0.1 mg/mL BSA and 0.01% Tween-20 Reactions were loaded into Premium Coated Capillaries (Nanotemper) and analysed using a Monolith NT.115 (Nanotemper). Thermophoresis data were extracted from the companion software and analysed in Prism 6 (Graphpad) graphing software with a “One Site – Specific binding with Hill slope” model. Nucleosomes were labelled on H4N25C as described previously10.
Nucleosome sliding assays
Increasing concentrations of hINO80 were incubated with 6 or 18 pmol end-positioned nucleosomes with 100 bp flanking DNA for 15 min at 37°C in a 54 μL volume in buffer containing 25 mM HEPES pH 8.0, 50 mM NaCl and 1 mM TCEP. Following incubation, 45 μL of these reaction mixes were transferred into a 384-well microtitre plate. Reactions were initiated by injection of 5 μL ATP and MgCl2 to a final concentration of 1 and 2 mM, respectively. Initial rate comparisons between full length and tailless nucleosomes were made by monitoring a change in FRET between AlexaFluor® 647 (Thermo Fisher Scientific) on the short-end of the DNA wrap, and AlexaFluor® 555 C2-maleimide (Thermo Fisher Scientific) on N25 of H4 (via an H4N25C mutation). Nucleosomes for the comparison of H3 acetylation mimics against wild type H3-containing nucleosomes were labelled on H3R2C instead of H4N25C. Initial rates for each concentration of hINO80 were plotted and analysed in GraphPad Prism 6.0f with an ‘Allosteric sigmoidal’ model; Hill coefficients were determined manually through a log-conversion of the data.
Nucleosome stability assays
Salt-stability assays were carried out on centrally-positioned nucleosomes with Cy5 and Cy3 fluorescent labels on opposite ends of flanking DNA. Stocks of nucleosomes with WT or mutant histones were mixed with increasing concentrations of KCl and aliquoted into a 384-well microtitre plate. The intensity of the Cy3 donor label was then measured across different KCl concentrations, with higher intensity corresponding to decreased quenching and therefore unwrapping of the DNA tails from the nucleosome core.
Extended Data
Extended Data Fig. 1. Analysis of INO80-nucleosome complex sample.
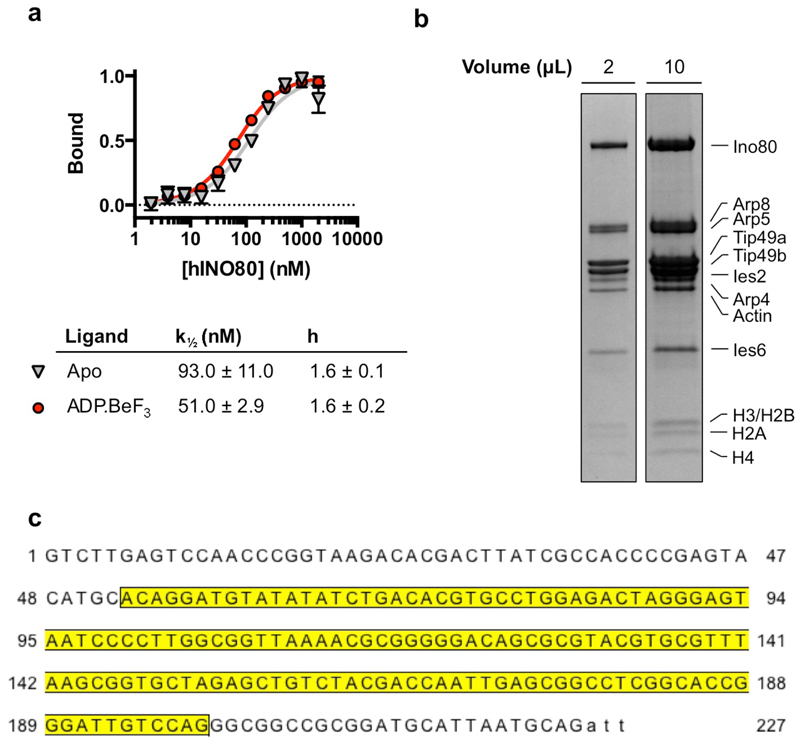
a, MST experiment of hINO80 with 60N12 nucleosome (+/- 3mM ADP•BeF3). Raw data (above) were processed to analyse binding and cooperativity (below). Data points represent mean values with SD, where n = 3 experimentally independent replicates. b, Gel of EM sample (hINO80 + nucleosome). Two loadings are shown to allow assessment of INO80 stoichiometry (left) or histones (right). n=3 independent experimental measurements. c, DNA sequence of the nucleosome (50N25) used for the structure determination. The Widom sequence (yellow) is flanked by 50 base pairs on one side and 25 base pairs on the other. An additional 3 base single strand overhang left over from restriction cleavage site is depicted in lower case. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 2. Cryo-EM data processing of INO80-nucleosome complex.
a, A typical micrograph out of 5,479. b, Representative 2D classes (out of 100) obtained with RELION from 775,804 particles. c, Image processing scheme. Data were processed by two parallel pathways to obtain maps for model building.
Extended Data Fig. 3. Quality of the structures.
a, Local resolution map of the INO80-nucleosome complex (4.8 Å) (left) and cut away (right). b, Angular distribution of these particles. c, Local resolution map of the INO80 complex (3.8 Å) (left) and cut away (right). d, Angular distribution of these particles. e, Corrected FSC curves of the reconstructions.
Extended Data Fig. 4. Assessment of various structural features in the INO80-nucleosome complex.
a, Overall fold of the Ino80-I and motor domains. b, Locations of the Ino80-I, motor domains and Ies2 regions relative to the RUVBL1-RUVBL2 hexamer. c, Sequence alignment of the C-terminal regions of human and yeast Ies2. The built part of the human Ies2 structure is indicated by a yellow bar. Asterisks indicate lysine residues in yeast Ies2 that crosslink to Ino80-HN (red) or Ino80-HC (blue). d, Representative density from two regions of the Ino80 insert. (Top) Density in the deposited 4.8 Å Ino80-nucleosome map (Bottom) Improvement in density in the 3.8 Å map, which facilitated model building. e, Coordinates of Ies2 showing formation of beta-sheet secondary structure with RUVBL1 (chain E) and RUVBL2 (chain D) within the 3.8 Å map. f, (i) Fit of Arp5 into 4.8 Å map. (ii) DNA and motor domains fit into the 4.8 Å map. (iii) Perpendicular view of (ii), showing the DNA crossing the motor domains.
Extended Data Fig. 5.
Comparisons of INO80-nucleosome interactions with those of Chd1 and Snf2. Viewed from the top of nucleosome, showing that all the motor domains are located on one side while Arp5-Ies6 (green) contacts the other side of the DNA wrap. Chd1 induces an unwrapping of the DNA at the SHL -7 position due in a large part to interactions with the accessory SANT and SLIDE domains. Despite this unwrapping, the histone core remains largely unaltered. Although the Snf2-nucleosome structure does not induce unwrapping of DNA, it is only a fragment of the motor subunit and also lacks other accessory subunits of the SWI/SNF complex so likely presents an incomplete picture of interactions or DNA distortions within the nucleosome in the complex.
Extended Data Fig 6. Interaction of human Actin, Arp5 and Arp8 with human H2A-H2B dimers assessed by in vitro pulldown.
a, Actin and Actin-related proteins were all expressed with a C-terminal double-Strep tag and used as bait to capture untagged H2A-H2B dimers. The result supports the position of Arp5 in the reported structure. Assay products were visualised by SDS-PAGE and Coomassie staining. n=1. b, A comparison of Arp5-Ies6 and Arp5 nucleosome binding activity assayed by EMSA, demonstrating a lack of nucleosome binding activity by Arp5 at in vivo relevant concentrations in the absence of Ies6. Nucleosomes were labelled with AlexaFluor 488 (Thermo Fisher Scientific). Reaction species were visualised by fluorescent scan. n=1. c, Arp5-Ies6 and 0N100 nucleosome interaction measured by MST. d, Arp5-Ies6 and H2A-H2B interaction measured by MST. For gel source data, see Supplementary Figure 1. n=2 biologically independent experiments in all the graphs. Error bars represent SD from the mean values.
Extended Data Fig 7. INO80 SC1 is flexible in the INO80-nucleosome complex.
a, Individual particles (selected out of 775,804) with RUVBL1-RUVBL2 oriented similarly showing different orientations of SC1 (dashed lines). b, 2D class averages (~30 particles each) showing different orientations of SC1 relative to the RUVBL1-RUVBL2. c, Projections of the 3D reconstruction along the same angles of those in b, confirming the extra density as SC1. The white bar represents 100 Å.
Extended Data Fig. 8. INO80 is regulated by H3 tails.
a, Schematic diagram illustrating the histone tail truncations used in this study. b, Initial nucleosome sliding rates of human nucleosomes lacking different histone tails. Plots of raw data for each histone tail deletion, with Vmax obtained after fitting the data shown as a dotted line. Data are summarised in Figure 6a of the main text. n=2 biologically independent experiments in all the graphs. Error bars represent SD from the mean values.
Extended Data Fig. 9. INO80 is regulated by H3 tails.
a, ATPase data and Hill coefficients for data shown in Figure 6c of the main text. b, ATPase rates for mutations of the H3 tails. c, Nucleosomes carrying wildtype or mutated H3 tails show similar salt stability indicating that the mutations have not altered the stability of nucleosomes. n=2 biologically independent experiments in all the panels. Error bars represent SD from the mean values.
Extended Data Table 1. Electron microscopy data collection, image processing and model refinement statistics.
| INO80 nucleosome complex (EMDB-3954) (PDB 6ETX) |
|
|---|---|
| Data collection and processing | |
| Magnification | 129,000 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 80 |
| Defocus range (μm) | -2.0 to -4.0 |
| Pixel size (Å) | 1.13 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 775,804 |
| Final particle images (no.) | 26,416 |
| Map resolution (Å) | 4.8 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 4.0 to 8.0 |
| Refinement | |
| Initial model used (PDB code) | 2YQQ, 3LZ0, 5AV9, 5O9G, 50AF |
| Model resolution (Å) | 4.8 |
| Map sharpening B factor (Å2) | -100 |
| Refinement (Phenix) | |
| Map correlation coefficient (whole unit cell) | 0.88 |
| Map correlation coefficient (around atoms) | 0.74 |
| Model composition | |
| Non-hydrogen atoms | 38,759 |
| Protein residues | 4,753 |
| Nucleic acid residues | 300 |
| Ligands | 7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.447 |
| Validation | |
| MolProbity score | 1.58 (93rd percentile*(0Å - 99Å)) |
| Clashscore | 8.4 |
| Poor rotamers (%) | 1.5 |
| Ramachandran plot | |
| Favored (%) | 98.2 |
| Allowed (%) | 1.75 |
| Disallowed (%) | 0.02 |
Supplementary Material
Acknowledgments
We would like to thank Alistair Siebert and Yuriy Chaban at eBIC for assistance with data collection, and Chris Aylett for help in preparing the nucleosome model. EM access and support was provided by the UK national electron Bio-Imaging Centre (eBIC) (proposal EM14769), funded by the Wellcome Trust, MRC and BBSRC. The work was funded by the Wellcome Trust (DBW and XZ), Cancer Research UK (DBW) and an Imperial College President’s PhD Scholarship (RA).
Footnotes
Data Availability
All the data have been deposited in wwPDB with access codes EMDB 3954 (hINO80 core-nucleosome complex map), PDB 6ETX (protein coordinates).
Author contributions: DBW and XZ designed the studies. RA, RJA, MW and OW performed the cryoEM analysis, and built and refined the structural models. OW, EAM & LO prepared the samples. OW conducted the biochemical experiments. DBW and XZ wrote the manuscript with input from all the authors.
Competing Interests
The authors declare no competing financial interests.
References
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Dechassa ML, et al. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahma S, et al. INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat Commun. 2017;8:15616. doi: 10.1038/ncomms15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nodelman IM, et al. Interdomain Communication of the Chd1 Chromatin Remodeler across the DNA Gyres of the Nucleosome. Mol Cell. 2017;65:447–459 e446. doi: 10.1016/j.molcel.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnung L, Vos SM, Wigge C, Cramer P. Nucleosome-Chd1 structure and implications for chromatin remodelling. Nature. 2017 doi: 10.1038/nature24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, Li M, Xia X, Li X, Chen Z. Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature. 2017;544:440–445. doi: 10.1038/nature22036. [DOI] [PubMed] [Google Scholar]
- 7.Sundaramoorthy R, et al. Structural reorganization of the chromatin remodeling enzyme Chd1 upon engagement with nucleosomes. Elife. 2017;6 doi: 10.7554/eLife.22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 9.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willhoft O, Bythell-Douglas R, McCormack EA, Wigley DB. Synergy and antagonism in regulation of recombinant human INO80 chromatin remodeling complex. Nucleic Acids Res. 2016;44:8179–8188. doi: 10.1093/nar/gkw509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leonard JD, Narlikar GJ. A nucleotide-driven switch regulates flanking DNA length sensing by a dimeric chromatin remodeler. Mol Cell. 2015;57:850–859. doi: 10.1016/j.molcel.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udugama M, Sabri A, Bartholomew B. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol. 2011;31:662–673. doi: 10.1128/MCB.01035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu Y, et al. The Chd1 Chromatin Remodeler Shifts Nucleosomal DNA Bidirectionally as a Monomer. Mol Cell. 2017;68:76–88 e76. doi: 10.1016/j.molcel.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willhoft O, et al. Crosstalk within a functional INO80 complex dimer regulates nucleosome sliding. Elife. 2017;6 doi: 10.7554/eLife.25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Racki LR, et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aramayo RJ, et al. Cryo-EM structures of the human INO80 chromatin-remodeling complex. Nat Struct Mol Biol. 2018;25:37–44. doi: 10.1038/s41594-017-0003-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tosi A, et al. Structure and subunit topology of the INO80 chromatin remodeler and its nucleosome complex. Cell. 2013;154:1207–1219. doi: 10.1016/j.cell.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Conaway RC, Conaway JW. Multiple modes of regulation of the human Ino80 SNF2 ATPase by subunits of the INO80 chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2013;110:20497–20502. doi: 10.1073/pnas.1317092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe S, et al. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun. 2015;6:7108. doi: 10.1038/ncomms8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao W, et al. Assembly of the Arp5 (Actin-related Protein) Subunit Involved in Distinct INO80 Chromatin Remodeling Activities. J Biol Chem. 2015;290:25700–25709. doi: 10.1074/jbc.M115.674887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saravanan M, et al. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20883–20888. doi: 10.1073/pnas.1214735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerhold CB, et al. Structure of Actin-related protein 8 and its contribution to nucleosome binding. Nucleic Acids Res. 2012;40:11036–11046. doi: 10.1093/nar/gks842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 26.Ranjan A, et al. H2A histone-fold and DNA elements in nucleosome activate SWR1-mediated H2A.Z replacement in budding yeast. Elife. 2015;4:e06845. doi: 10.7554/eLife.06845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 28.Ranjan A, et al. Nucleosome-free region dominates histone acetylation in targeting SWR1 to promoters for H2A.Z replacement. Cell. 2013;154:1232–1245. doi: 10.1016/j.cell.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 30.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deindl S, et al. ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell. 2013;152:442–452. doi: 10.1016/j.cell.2012.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krietenstein N, et al. Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell. 2016;167:709–721 e712. doi: 10.1016/j.cell.2016.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012;492:280–284. doi: 10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan L, Wang L, Tian Y, Xia X, Chen Z. Structure and regulation of the chromatin remodeller ISWI. Nature. 2016;540:466–469. doi: 10.1038/nature20590. [DOI] [PubMed] [Google Scholar]
- 37.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 38.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 39.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 43.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.