Abstract
Kinesin-1 is a nanoscale molecular motor that walks towards the fast growing (plus) ends of microtubules, hauling molecular cargo to specific reaction sites in cells. Kinesin-driven transport is central to the self-organisation of eukaryotic cells and shows great promise as a tool for nano-engineering1. Recent work hints that kinesin may also play a role in modulating the stability of its microtubule track, both in vitro2,3 and in vivo4, but results are conflicting5–7 and mechanisms are unclear. Here we report a new dimension to the kinesin-microtubule interaction, whereby strong-binding state (ATP-bound and apo) kinesin-1 motor domains inhibit the shrinkage of GDP-microtubules by up to 2 orders of magnitude and expand their lattice spacing by ~1.6%. Our data reveal an unexpected mechanism by which the mechanochemical cycles of kinesin and tubulin interlock, allowing motile kinesins to influence the structure, stability and mechanics of their microtubule track.
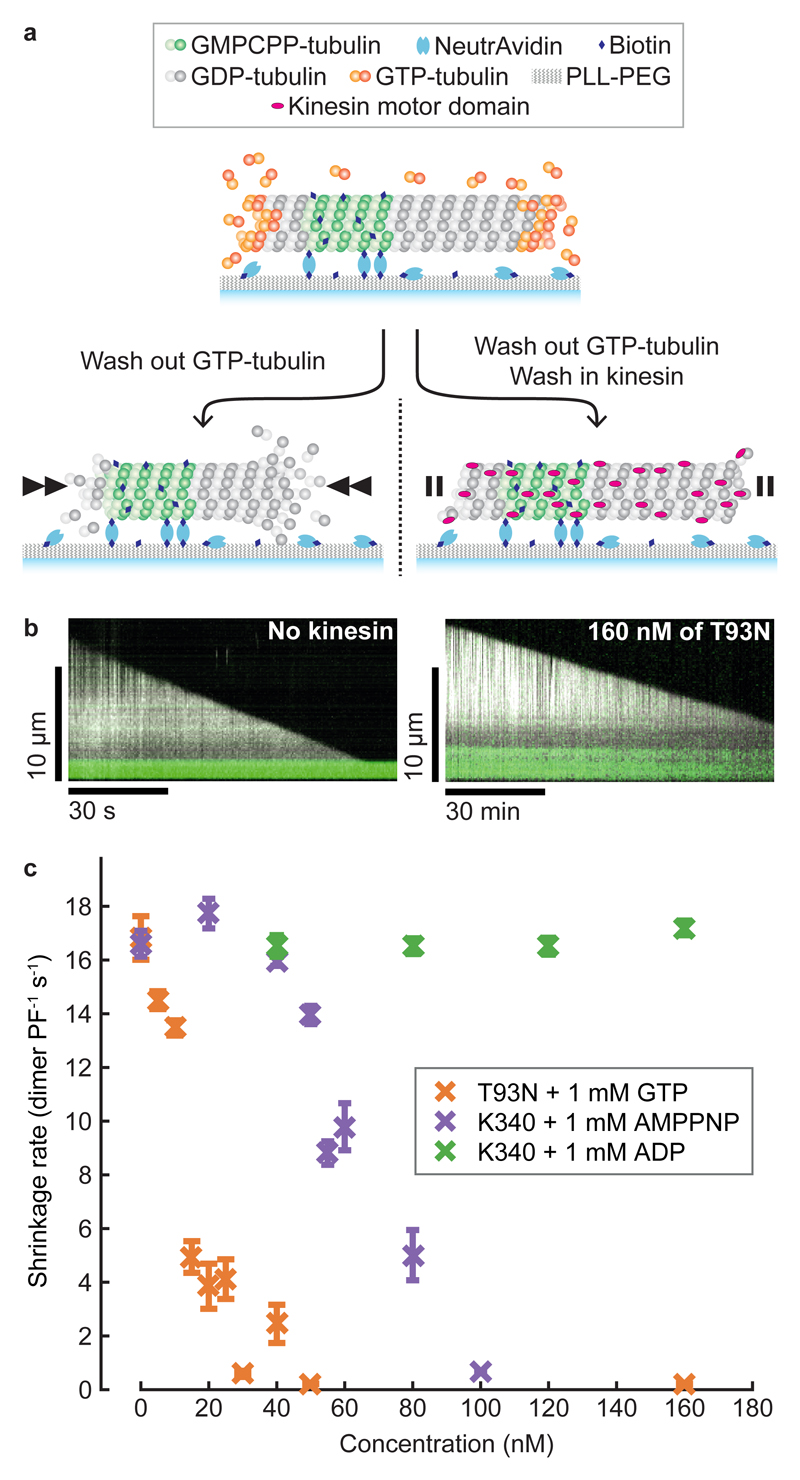
As kinesin molecules walk along microtubules, their motor domains cycle through a series of nucleotide-specific conformations. We tested whether these different nucleotide states of kinesin-1 motor domains influence microtubule stability. First, we attached fluorescent microtubule ‘seeds’ to the inside of a flow chamber via biotin-NeutrAvidin linkages and flowed in GTP-tubulin, causing dynamic microtubules to grow from the seeds (Fig. 1a). We then initiated microtubule depolymerisation by washing out tubulin, whilst simultaneously flowing in monomeric kinesin-1 motor domains. We first tested the effect of a kinesin motor domain mutant (T93N) that enriches the apo state of the motor8. The apo state of kinesin is a strong-binding state, meaning that it binds tightly and stereospecifically to microtubules9. We found that T93N reduced microtubule shrinkage to 1% of the control rate (Fig. 1b,c). We then used wild-type kinesin motor domains supplemented with either AMPPNP, a non-hydrolysable ATP analogue that causes a strong-binding state, or ADP, which causes a weak-binding state, and compared their effects. Wild-type kinesin is immotile in AMPPNP and ADP. We found that AMPPNP-wild-type kinesin inhibited microtubule shrinkage similarly to T93N, whilst ADP-wild-type kinesin had no detectable effect (Fig. 1c). We conclude that the strong-binding states of kinesin powerfully inhibit the shrinkage of GDP-microtubules.
Figure 1. Strong-binding state kinesins inhibit GDP-microtubule shrinkage.
a, Schematic representation of a tubulin depletion assay. Dynamic microtubules shrink rapidly when GTP-tubulin is depleted (left) unless bound to strong-binding state kinesins (right). b, Representative kymographs of microtubules shrinking in the absence (left) and presence (right) of T93N. Note the different time scales. Dynamic microtubules are shown in white (dark-field) and fluorescent seeds in green (epi-fluorescence). c, Shrinkage rates of microtubules in the presence of kinesins and nucleotides. GTP was included with T93N only. Error bars are mean ± SEM. We analysed a total of n=317, 225, 168 microtubules for the T93N-GTP, wild-type-AMPPNP and wild-type-ADP conditions, respectively.
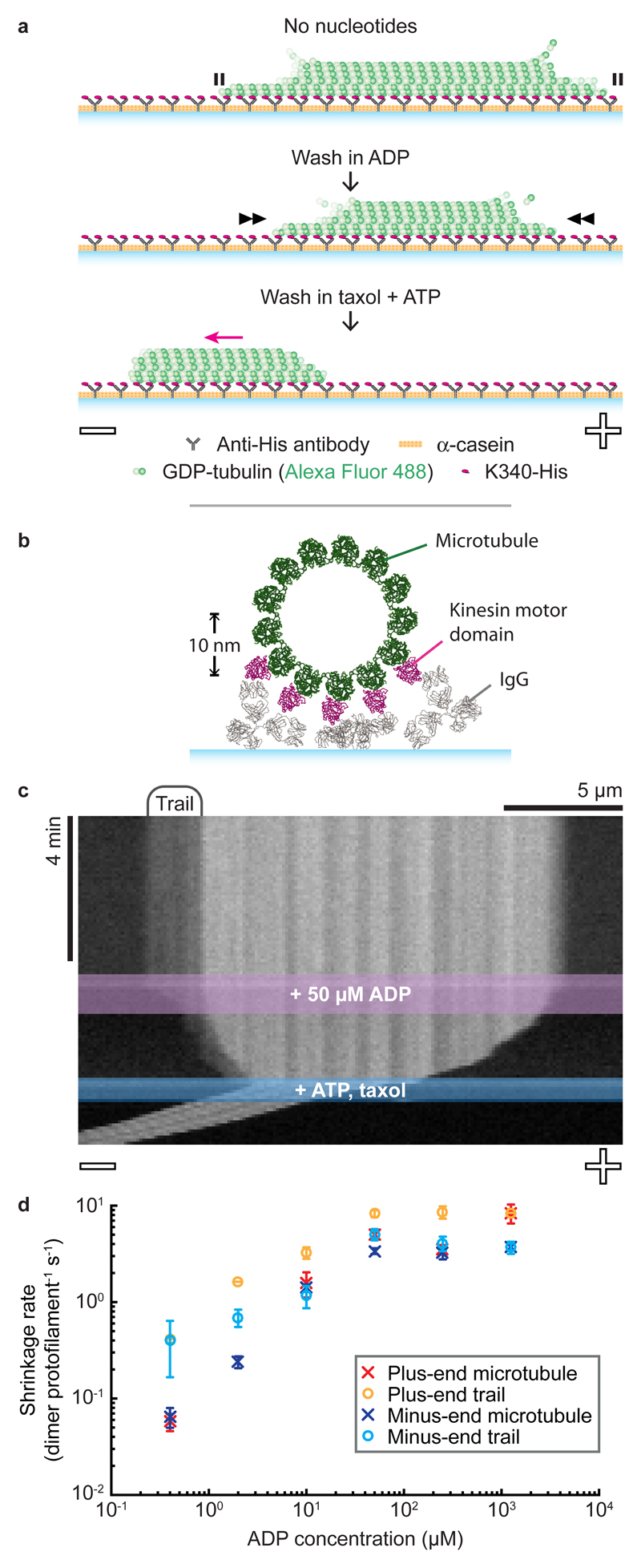
Next, we bound GDP-microtubules to a kinesin-coated coverslip in a flow chamber, triggered depolymerisation by washing out residual GTP-tubulin, and again observed microtubule shrinkage (Fig. 2a). Geometric constraints suggest that in this arrangement at most 5 protofilaments can bind to the kinesin surface (Fig. 2b). Despite this, entire microtubules were stabilised (Fig. 2c). We then flowed solutions through the channel in 2 steps (Fig. 2a). First, ADP was flowed in, reducing the fraction of kinesins in a strong-binding state and thereby increasing the microtubule shrinkage rate (Fig. 2c, Supplementary Movie 1). By titrating the ADP concentration, we found that microtubule shrinkage rates could be fine-tuned over 2 orders of magnitude (Fig. 2d, Supplementary Table 1). Comparing the inhibition of microtubule shrinkage by kinesin in solution (maximally 0.21 ± 0.02 (25) dimer protofilament-1 s-1 (mean ± SEM (n))) with that of the kinesin surface (0.06 ± 0.01 (6) dimer protofilament-1 s-1 in the presence of 400 nM ADP) shows that surface immobilisation enhances the stabilising effect of kinesin, despite kinesin binding being restricted to only a subset of protofilaments.
Figure 2. Microtubules are stabilised when kinesins bind to one side of the lattice.
a, Schematic of a kinesin-clamp assay. Microtubule polarity is labelled -/+. b, Cross-sectional view of a kinesin-clamp assay, showing IgG (PDB:1IGY) and kinesin-bound microtubule (PDB:4UXT) structures to provide scale. c, Representative kymograph of a microtubule in a kinesin-clamp. d, Average shrinkage rates of microtubules and their trails. Error bars are mean ± SEM, reflecting inter-microtubule variability. n-values are given in Supplementary Table 1.
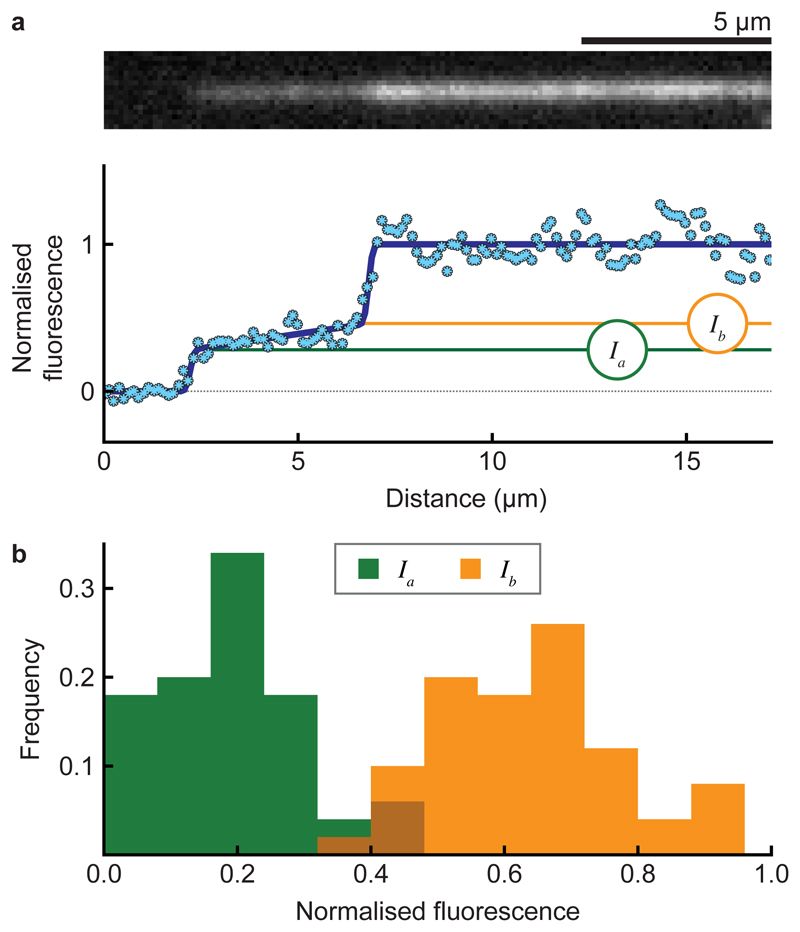
Frequently, faint fluorescent trails were visible on the kinesin-coated surface in the wake of retreating microtubule tips. These shrank endwise upon addition of ADP, suggesting that their tubulin is still assembled into protofilaments (Fig. 2c, Supplementary Movie 1). Trails were tapered, and fluorescence intensity analysis (Fig. 3a, Supplementary Methods and Supplementary Fig. 1-2) indicated that at their tips they contain 2-3 protofilaments (Fig. 3b). On average, trails can shrink faster than their microtubule stem because they appear transiently, typically forming, lengthening and retracting multiple times during the shrinkage of each surface-attached microtubule (Supplementary Fig. 3). As a final step in these experiments, we flowed in a buffer containing taxol and ATP, triggering kinesin-driven sliding to reveal the microtubule polarity.
Figure 3. Subsets of protofilaments are stabilised by a kinesin-coated surface.
a, Model fit to the intensity profile of a microtubule tip (bottom) with the associated fluorescence image (top). (Ia) is the intensity at the tip and (Ib) at the base of the trail. b, Histogram of (Ia) and (Ib) values for microtubules (n = 50), normalised to the intensities of their parent microtubules in the no-nucleotide phase of the experiment. Mean ± SD is 0.19 ± 0.11 and 0.64 ± 0.13 for Ia and Ib, respectively.
Why does a kinesin-coated surface stabilise microtubules but also cause them to split? Taxol-stabilised microtubules have recently been shown to split on a kinesin-coated surface10. However, ATP-driven kinesin stepping was essential to this process, which is not the case for our taxol-free GDP-microtubules. Several strands of evidence suggest that kinesin binding can change the lattice conformation and mechanics of microtubules. A kinesin-coated surface has been reported to reduce the Young’s modulus of taxol-stabilised microtubules11. Structural changes have also been reported following kinesin binding to taxol-stabilised12 and GMPCPP-bound microtubules13. Furthermore, the longitudinal compaction of the microtubule lattice that accompanies GTP hydrolysis is reduced in kinesin-bound microtubules14, suggesting that kinesin influences the longitudinal spacing between tubulin subunits in the microtubule lattice. We therefore hypothesised that kinesin binding modifies the axial spacing between GDP-tubulin subunits in the microtubule lattice. Binding kinesins to one side of a microtubule, as in our surface assay, would then change the lattice spacing on that side but not the other, causing shear stress that could facilitate splitting.
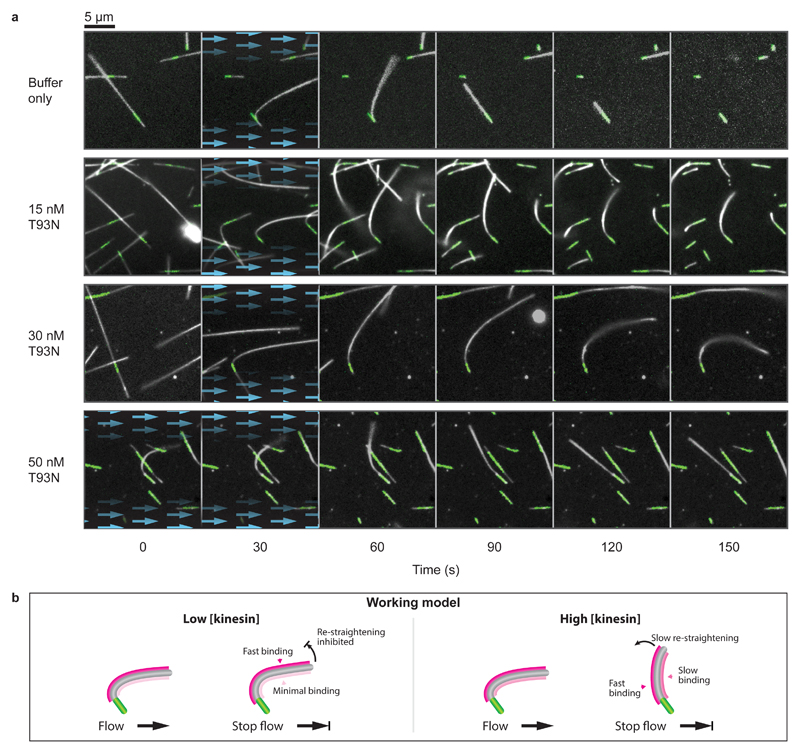
To test the idea that kinesin binding stabilises a distinct conformation of the microtubule lattice, we used hydrodynamic flow to bend tethered dynamic microtubules, thereby expanding the microtubule lattice on the convex side and compacting it on the concave side (Fig. 4, Supplementary Movie 2). We supplemented T93N into the flow and observed the mechanical response of microtubules upon stopping the flow. In the absence of kinesins, the microtubules quickly recoiled to a straight conformation and rapidly depolymerised. Remarkably, low concentrations of T93N (15-30 nM) blocked this recoil, effectively locking the GDP-microtubules in a curved conformation as well as inhibiting their shrinkage. These data suggest that indeed strong-binding state kinesins preferentially bind and stabilise a distinct longitudinal lattice spacing of GDP-microtubules.
Figure 4. Nucleotide-free motor domains can bend-lock microtubules.
a, Time-lapse images of microtubule bending experiments for a range of kinesin concentrations. Blue arrows highlight the presence and direction of fluid flow. Dynamic microtubules appear white (dark-field) and fluorescent seeds are marked in green (epi-fluorescence). Each condition was tested twice on independent occasions. Microtubules shown here have been selected for having similar orientations. A more extensive selection is given in Supplementary Movie 2, which shows a complete range of orientations and lengths. b, Working model. We propose that kinesin binds preferentially to the stretched (convex) side of the microtubule and also stabilises this expanded region of the microtubule lattice. At high kinesin concentrations, the convex side of the microtubule would quickly saturate. Binding would also occur slowly on the concave side, causing this side to expand, progressively re-straightening the microtubule.
We noticed that in the presence of higher concentrations of kinesin (≥ 50 nM), the curved microtubules tended slowly to re-straighten. To explain this observation, we speculate that strong-binding state kinesins bind preferentially but not exclusively to one side of curved microtubules. At high kinesin concentrations, the favoured side of the microtubule would then quickly become fully occupied, while binding would continue more slowly on the unfavoured side, ultimately driving the microtubule back into a straight conformation (Fig. 4b). Kinesins have previously been reported to bind preferentially to GTP-microtubules, which have an expanded lattice spacing compared to GDP-microtubules15. In light of this, we postulated that strong-binding state kinesins drive an increase in the lattice spacing of GDP-microtubules.
In order to directly test this point, we grew dynamic microtubules as in the previous experiments but this time we capped their exposed tips with non-biotinylated fluorescent GMPCPP-tubulin, producing stable GDP-microtubules that were tethered to the surface only at one end (Fig. 5a). The GDP-microtubules and their stabilised caps were fluorescently labelled in different colours and imaged using TIRF microscopy. We used pressure-driven microfluidics to align the microtubules in a constant hydrodynamic flow containing either wild-type apo-kinesin motor domains or 1 mM ADP. When kinesin was introduced, the GDP-bound segment of the microtubule lengthened as predicted (Fig. 5b, Supplementary Movie 3).
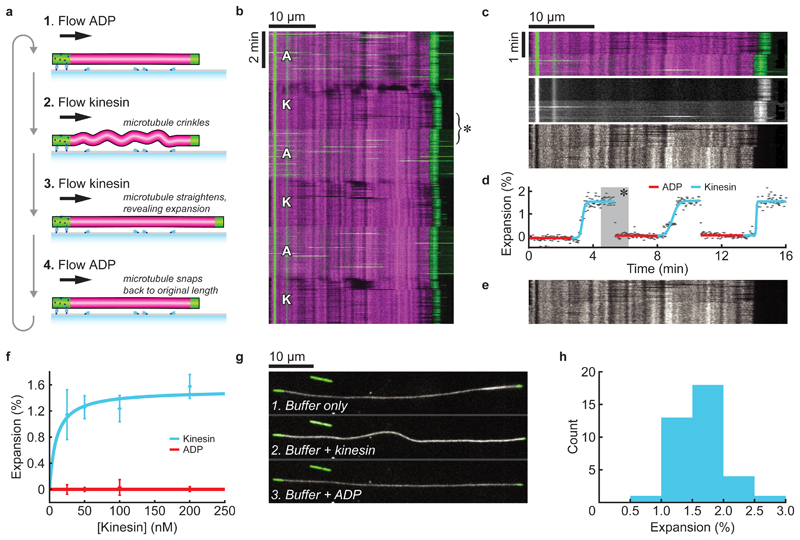
Figure 5. Kinesin increases the lattice spacing of GDP-microtubules.
a, Fluorescently labelled GDP-microtubules (rhodamine; magenta) were grown from surface-bound GMPCPP-tubulin seeds (Alexa Fluor 488; green) and then capped with GMPCPP-tubulin. Buffer was flowed through the channel at a constant rate and while switching between ADP and kinesin containing solutions. Microtubules briefly crinkled when kinesin was added at high concentrations, before straightening to reveal they had lengthened. Addition of ADP caused immediate recoil of the microtubules to their original lengths. b, Representative kymograph of a microtubule changing length as the flow switched between 1 mM ADP (A) and 200 nM kinesin (K) containing buffers. A constant volumetric flow rate was maintained throughout the experiment (74.3 ± 0.3 µl/min; mean ± SD, 480 time points). c, Magnified region from panel b (marked *), highlighting the compaction observed when switching from kinesin-containing to ADP-containing solutions. Top: merge. Middle: Green imaging channel (GMPCPP-tubulin and beads). Buffer exchange is visible due to the high background provided by the fluorescent beads (after ~50 sec). The untethered (rightmost) tip retracts in <4 s (2 frames) upon switching to ADP. Bottom: Magenta imaging channel (GDP-tubulin). An abrupt rift can be seen in the fiducial markings on the GDP-microtubule when ADP is added. The effect becomes increasingly pronounced further away from the tethered end, consistent with the microtubule lattice uniformly compacting. d, Kinesin-driven expansion of the microtubule shown in b (* again marks the region in c). Each data point corresponds to the inverse scaling factor that best maps the given GDP-microtubule profile (row) to match the average profile when ADP was present. The ADP points serve as an internal control for this measurement technique. See Supplementary Fig. 5 for further detail on this method. e, Transformed image from c, generated by scaling each profile using the inverse of the fitted values in d. Such images were used to visually check for errors. See Supplementary Animation 1 for a full-image transformation. f, microtubule expansion measured over a range of kinesin concentrations. A standard binding curve fitted to the mean values is shown, which saturates at 1.52%. Error bars show standard deviation. n-values are listed in Supplementary Table 2. g, Sequential images of a complementary experiment where GDP-microtubules (shown in white, dark-field) were encouraged into the focal plane using methylcellulose and fluid flow was used only intermittently for exchanging solutions. When kinesin was added, the microtubule bowed to accommodate expansion of its lattice between sparse points that were loosely stitched to the surface. The microtubule straightened and shortened upon addition of ADP. h, Expansion of surface-stitched GDP-microtubules, given by the relative contour lengths of GDP-microtubules in the presence and absence of kinesin. Mean ± SD is 1.64 ± 0.39 (n=37 microtubules).
TIRF microscopy visualises an optical section ~100 nm deep and microtubules remained visible throughout our experiment, indicating that the flow constrained them within this 100 nm thick section. Strikingly, microtubules briefly crinkled upon switching from ADP to kinesin, causing them to dip in and out of the TIRF illumination (Supplementary Movie 4). This transient crinkling can also be seen in our initial microtubule bending experiments (≥ 50 nM in Supplementary Movie 2). The crinkles progressively straightened (within 10-30 s) to reveal that the microtubule had expanded. Switching to ADP caused microtubules to quickly recoil to their original length (<4 sec, 2 frames in our data) as the kinesin unbound (Fig. 5c, d). Quantification reveals that apo-kinesin binding to GDP-microtubules increases their length by uniformly expanding the microtubule lattice along its axis (Fig. 5c-e; Supplementary Animation 1). The kinesin-induced lattice expansion appears to be fully reversible and the expand-and-recoil cycle can be repeated multiple times (Fig. 5d). We measured the extent of lattice expansion over a range of kinesin concentrations. The increase in lattice spacing saturated at 1.5% (Fig. 5f, Supplementary Table 2).
Occasional point-interactions between the microtubules and the surface occurred during these experiments, but these interactions were typically transient and the microtubules visibly quivered in the flow (Supplementary Movies 3 and 4). Under these conditions, kinesin is prohibited from stepping by the absence of ATP and GTP in the imaging buffers. We can therefore rule out the possibility of kinesin generating forces by stepping along the microtubule. Since any bending or crinkling produced by kinesin binding will reduce the apparent length of a microtubule in a kymograph, it is possible that our measurements slightly underestimate the full extent of kinesin-induced lattice expansion.
We additionally performed experiments where we used methylcellulose rather than flow to encourage the microtubules to remain in focus, introducing a flow only intermittently to exchange buffers. Under these conditions microtubules tended to become ‘stitched’ to the coverslip at sparse interaction sites. Introducing apo-kinesin then caused the microtubules to bow locally between these tethering points (Fig. 5g, Supplementary Movie 5), emphasising the expansion of the lattice. We used 200 nM of apo-kinesin in these experiments, a concentration sufficient to maximise the lattice expansion effect of kinesin binding (Fig. 5f). By measuring the change in contour length between the two stabilised microtubule caps, we confirmed that the bowing of the microtubule between tethering points accommodates a 1.6% expansion of the GDP-microtubule lattice (Fig. 5h). Microtubules became more densely ‘stitched’ to the surface if multiple cycles were performed but this did not influence the measured expansion (Supplementary Fig. 4). When ADP was flowed through the channel, the microtubules again recoiled to their original lengths (Fig. 5g).
Our data show that strong-binding state kinesin stabilises the GDP-lattice of dynamic microtubules, and concomitantly increases their axial lattice spacing by 1.6%. Kinesins are known to bind to the intra-dimer interface of αβ-tubulin, away from the inter-dimer contacts of the microtubule lattice13,14,16. This suggests to us that kinesin binding allosterically modifies the conformation of GDP-tubulin, giving it properties more similar to GTP-tubulin. Assuming no twisting of the microtubule occurs, a 1.6% axial expansion equates to approximately 1.3 Å per 80 Å tubulin dimer, similar to the 1.7 Å difference observed by cryo-EM between GMPCPP-microtubule-kinesin and GDP-microtubule-kinesin structures16. Strong-binding of kinesin has previously been reported to alter the structure of both taxol-GDP-microtubules12 and GMPCPP-microtubules13. Moreover, a long-range, ATP-dependent, cooperative effect has been described whereby the first few kinesins that bind facilitate subsequent binding events in the same region of the microtubule, again suggestive of a kinesin-induced conformational change17.
We envisage that the ability of strong-binding state kinesin to stabilise GDP-microtubules by inducing a conformational change in their tubulin subunits provides at least a partial mechanistic explanation for the surface-bound depolymerisation trails and the bend-locking phenomenon reported here. Thus, a microtubule landing on and binding to a kinesin-coated surface would likely become stretched on its surface-bound side. This stretching would create shear stress in the lattice and potentially contribute towards splitting the microtubule to form the trails observed in our kinesin-clamp experiments. Similarly, for the microtubule bend-locking, expanding the longitudinal microtubule lattice spacing by 1.6% exclusively on one side of the microtubule would be more than sufficient to account for the observed kinesin-stabilisation of curvature. Indeed, full occupancy on one side with zero occupancy on the other would produce a radius of curvature of 1.6 µm, assuming a microtubule has a diameter of 25 nm, far tighter than we observe in any of our post-flow data.
We have worked with kinesin-1, the best-studied kinesin, but it is possible that the mechanism we report here is common to other kinesins. Kif14 is a slow kinesin that binds to microtubules in a rigor-like conformation and inhibits their shrinkage18. Kinesin-5 is reported to stabilise protofilament assemblies during microtubule growth19, potentially due to its strong-binding state stabilising the polymer. Kip220 also dwells at microtubule ends and increases microtubule stability.
Our work reveals a specific action of strong-binding state kinesins in stabilising the GDP-lattice of dynamic microtubules. Microtubule-activated ADP release creates a strong (apo) state and this process is affected by the tubulin and kinesin species21, by post-translational modifications22 and by the nucleotide state23 of the microtubule. Importantly, the residence time of kinesin in the strong-binding states is also influenced by mechanical force24. Such forces will arise in vivo wherever kinesins do mechanical work, for example at kinetochores, in microtubule bundles25, at cortical attachment sites26, and during the transport of vesicles against a resistance27. It will be important now to understand the role of these various effects in determining how kinesin motility may feed back on microtubule dynamics.
In conclusion, our data reveal a novel mechanism that allows kinesin-1 to feed back on the structure and stability of its microtubule track. Recent advances in the remote control of kinesin motility, such as photo-switchable fuels28, suggest the potential for precise spatial control of these effects.
Methods
Proteins and biochemical reagents
Tubulin was purified from pig brains as previously2 with additional steps as follows. Tubulin was polymerised in 50 mM PIPES, 1.2 mM MgSO4, 1 mM EGTA, 1mM GTP, 1 mM dithiothreitol (DTT) and 186 mg ml-1 glutamic acid and incubated for 60 min at 37 °C microtubules were centrifuged in a TLA 100.3 rotor at 85,000 rpm for 20 min at 35 °C resuspended in K-PEM with 1 mM GTP, 1 mM MgSO4 and 1 mM DTT, cooled to 4 °C and centrifuged in a TLA 100.3 rotor at 85,000 rpm for 20 min. The supernatant was run through a Hiprep 26/10 desalting column into K-PEM buffer (100 mM PIPES, 1 mM MgSO4, 2 mM EGTA (Fisher); adjusted to pH 6.9 with KOH) and 20 µM GTP. Tubulin concentrations were determined using E280 = 105,838 M-1 cm-1.
X-rhodamine labelled tubulin was purchased from Cytoskeleton Inc. (USA). Alexa Fluor 488 (Molecular Probes) labelled tubulin was prepared using standard protocols, as previously2.
Kinesin was purified as previously29. Kinesin concentrations were determined using E280 = 15,300 M-1 cm-1.
Nucleotides were from Jena Biosciences (Germany). Other reagents were from Sigma (UK) unless stated otherwise.
Bead-mPEG crosslinking
0.5 µm yellow-green carboxylated FluoSpheres (Thermofisher) were diluted to 1% solids and activated using 10 mg/ml 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDAC) in pH 6 MES buffer and mixed gently at room temperature for 30 min. Beads were then centrifuged, resuspended in 10 mg/ml of methoxypolyethylene glycol amine 750 (mPEG-amine) in pH 7.4 PBS and mixed gently at room temperature for 2 h. Adding 90 mM glycine quenched the reaction. After 30 min the beads were centrifuged and resuspended in 0.1% Tween20 in K-PEM 5 times and stored at 4 °C.
Flow chamber assembly (for manual flow-through)
Flow chambers were assembled from 22x22 mm no. 1.5 glass coverslips (Menzel, Germany) and 76x26 mm 1-1.2 mm thickness glass slides (Menzel, Germany). Double-sided Scotch tape was sandwiched between the glass surfaces to form a 2 mm wide channel. The periphery of the chamber was further secured using nail polish excluding the channel ends, which were left open. Solutions were drawn through the channel by using grade 1 Whatman filter paper.
Microfluidics
Microfluidic flow chambers were assembled by stacking the following: a 50x22mm no. 1.5 glass coverslip (Menzel, Germany), cleaned using the protocol from the tubulin depletion assays, or a 50x22mm cyclo-olefin polymer coverslip (188 µm thick; ZF14-188, Zeon) cut using a Silhouette Portrait plotter cutter; double-sided adhesive tape (ArCare 90445, kindly provided by Adhesives Research), with a Y-shaped channel cut using a plotter cutter (two 8x0.75 mm inlets joining a 22x1.5 mm channel); a 40x22mm COP coverslip with portholes. Chip-to-tubing connectors were assembled using ring magnets (8.16 mm OD x 3.5 mm ID, First4magnets) pressed into a 3D-printed ABS magnet holder, which was later used to align the magnets with the portholes in the COP coverslip, then cast in polydimethylsiloxane (PDMS; Sylgard 184, Dow Corning) to fill the ring magnets and produce a 0.8 mm thick cushion that forms the interface between the magnets and the microfluidic chip. A 1.25 mm biopsy punch was used to bore holes through the PDMS-magnet core. 127 µm ID x 1.59 mm OD polyether ether ketone (PEEK) tubing (Upchurch) was inserted into the holes, forming a tight seal. The flow chamber was placed on a custom ferromagnetic stainless steel (430) microscope stage, which sealed the connections by attracting the magnets and held the sample in position. Flow was driven using an MFCS-EZ, inlets were controlled using an L-switch, and the outlet flow was monitored using an M flow unit (Fluigent). A Matlab (Mathworks) function was written to fully integrate control of the microfluidics with the microscope.
Tubulin depletion and microtubule bending assay
Coverslips were sonicated (600 W bath, Ultrawave) in 3% Neutracon detergent (Decon Laboratories, UK) for 30 min at 60 °C before undergoing extensive wash-sonication cycles in ultrapure water (18.2 MΩ). A flow chamber was assembled, filled with 0.2 mg ml-1 PLL-PEG-biotin (SuSoS, Switzerland) and incubated for 30 min. It was washed with K-PEM before adding 1 mg ml-1 NeutrAvidin (Thermo Fisher Scientific) for 5 min and washing again. Microtubule seeds (polymerised using 26 µM of 15% labelled Alexa488-tubulin and 1 mM GMPCPP in K-PEM at 37 °C for 25 min) were pelleted, diluted to ~ 60 nM, injected into the chamber and incubated for 5 min. After washing the chamber, dynamic microtubule extensions were grown by flowing through with 15 µM tubulin, 1 mM GTP, GOC oxygen scavenger (4.5 mg ml-1 glucose, 0.2 mg ml-1 glucose oxidase, 35 µg ml-1 catalase, 0.5% (v/v) β-mercaptoethanol), 1 mg ml-1, 1 mg ml-1 BSA and 0.1% (v/v) Tween20 in K-PEM. Microtubules were grown for > 15 min at 25 °C prior to imaging with epifluorescence and dark-field illumination. Tubulin was depleted by flowing through pre-warmed (25 °C) K-PEM, supplemented with kinesin and nucleotides as described in the main text. Microtubule bending was achieved by rapidly drawing solutions through the channel using Whatman filter paper.
Kinesin-clamp assay
Fluorescence controls (colour-segmented stabilised microtubules) were prepared by incubating 5 µM 30% labelled Alexa Fluor 288 tubulin and 0.2 mM Guanosine-5’-[(α,β)-methyleno]triphosphate (GMPCPP) in K-PEM at 37 °C for 60 min and pelleted in an airfuge (Beckman Coulter) at 25 psi for 10 min. The supernatant was discarded and the pellet resuspended in pre-warmed 5 µM 30% labelled X-rhodamine tubulin and 0.2 mM GMPCPP in K-PEM. Microtubules were left to anneal at room temperature then diluted 50-fold before use.
Coverslips were sonicated (600 W bath) at room temperature in a 1:1 solution of methanol and HCl for 30 min; then sonicated for 4×5 min in ultrapure water, 60 min in 0.2 M KOH and 5×5 min in ultrapure water. They were spun dry using a Spin Clean (Technical video), incubated at 100 °C for 30 min and plasma-cleaned (PLASMA clean 4, ILMVAC) for 5 min. Coverslips were then silanised by immersing in 0.05% dimethyldichlorosilane in trichloroethylene for 60 min, washed in methanol, sonicated for 5×5 min in methanol and spun dry.
A flow chamber was assembled using a silanised coverslip and filled with 0.1 mg ml-1 anti-6xHistidine antibodies (372900) for 10 min. The chamber was then flushed with 0.5 mg ml-1 α-casein and incubated for 5 min, then with 75 nM K340 for 10 min, and washed with 10 chamber volumes of K-PEM. Stabilised segmented microtubules were then introduced. Unbound microtubules were washed out immediately with K-PEM. Dynamic microtubules, polymerised by incubating 50 µM 30% labelled Alexa Fluor 488 tubulin (same stock as used for fluorescence controls) and 1 mM GTP in K-PEM for 45 min at 37 °C were diluted 20-fold in warm (37 °C) K-PEM and immediately flowed through the chamber ahead of 10 chamber volumes of warm K-PEM. The sample was imaged using epifluorescence, and ADP in K-PEM was introduced at the desired concentration. Once microtubules had shortened sufficiently, 10 µM taxol and 200 µM ATP in K-PEM was flowed in.
Microtubule expansion assay with microfluidics
microtubules were attached to the glass surface as in the tubulin depletion assay or by using COP coverslips, which were plasma-treated (air, 3 min), immersed in 3% (3-Aminopropyl)triethoxysilane (APTES) in ethanol for 2 hours, placed in an ultrasonic bath in ethanol for 5 min (2 times), then washed extensively with water before drying with N2 gas. COP flow chambers were filled with 1 mM BS(PEG)5 (PEGylated bis(sulfosuccinimidyl)suberate) in PBS for 30 min, washed thoroughly with PBS, incubated with 0.1 mg/ml anti-biotin antibodies for 5 min and then 50 mM Tris buffer pH 8 for 30 min. Dynamic microtubules were polymerised following the same initial steps as the tubulin depletion assay, except 20 µM of 10% x-rhodamine-labelled tubulin was used to grow dynamic extensions, which were left for 1 h to polymerise. Buffer exchanges were performed using the microfluidic pump. Microtubules were capped by incubating with 6 µM of 15 % Alexa-488 tubulin + 1 mM GMPCPP for 20 min before switching to 1% Tween20 for 5 min. Two pressure lines were calibrated to achieve the same user-defined flow rate immediately before imaging with TIRF microscopy. K-PEM with 1X GOC, 1% Tween20, 0.02% methylcellulose (1500 cP) and 1 mg/ml casein or 2 mg/ml BSA was flowed through with either 1 mM ADP + 0.003-0.01% mPEG-beads or 200 nM K340, alternating every 80x2 s frames, cycling 3 times. Images shown were background-subtracted in the green imaging channel by subtracting the median and applying a rolling ball (r=200) to each frame using Fiji.
Surface-stitched microtubule expansion assay
Coverslips were incubated in 1 M HCl at 50 °C for 12-15 hours, rinsed with ultrapure water twice, sonicated in ultrapure water for 30 min, sonicated in ethanol for 30 min, rinsed in ethanol and dried by spinning or using nitrogen gas. Microtubules were polymerised as in the microtubule expansion assay with microfluidics, except the microtubule seeds and caps were labelled to 30% with Alexa488 and dynamic extensions were grown using unlabelled tubulin. Microtubules were capped for 10 min before washing with 100 µl of 0.1% Tween in K-PEM. Hereafter, each buffer contained GOC, 0.1% Tween20 and 0.02% methylcellulose (4000 cP) in K-PEM. The chamber was washed with 30 µl of buffer before imaging. During imaging, 40 µl of 200 nM K340 was flowed through by hand, followed later by 40 µl of 1 mM ADP. Flowing through 100 µl of buffer depleted the ADP, after which a new field of view could be imaged and the kinesin and ADP flows repeated. We imaged no more than five times in a single flow chamber.
Dark-field/epifluorescence microscopy
Images were captured by an EM-CCD camera (iXonEM+DU-897E, Andor) fitted to a Nikon E800 microscope with a Plan Fluor 100x NA 0.5-1.3 variable iris objective. A custom-built enclosure with an air heater (Air-Therm ATX, World Precision Instruments) was used to keep samples at 25 °C. Dark-field illumination was achieved using a 100 W mercury lamp connected to the microscope via a fibre optic light scrambler (Technical video), cold mirror, 500-568 nm band-pass filter (Nikon) and a dark-field condenser (Nikon). A stabilised mercury lamp (X-cite exacte, Lumen Dynamics) provided illumination for epifluorescence, connected to the microscope with a light pipe. Motorised filter wheels (Ludl Electronic Products) housed the fluorescence excitation and emission filters: 485/20 and 536/40 for Alexa-488 and 586/20 and 628/32 for X-rhodamine (Chroma). Combined dark-field and fluorescence imaging was achieved using an FF505/606-Di01-25x36 dichroic mirror (Semrock) and electronic shutters to switch between illumination modes. The shutters, filter wheels and camera were controlled using Metamorph software (Molecular Devices).
TIRF microscopy
Images were captured by an EM-CCD camera (iXon3 888, Andor) fitted to a Warwick Open-Source Microscope (WOSM; wosmic.org), which was equipped with 473 nm (Cobolt) and 561 nm (Obis) laser lines, a Di01-R488/561 dichroic mirror (Semrock), a Nikon 100x NA 1.49 TIRF objective, and ZET473NF and ZET561NF emission filters (Chroma). Acquisition was triggered using a Matlab script, which used Micromanager for capturing images, launched WOSM macros for all other microscope functionality, and controlled and monitored the microfluidics. Pixels were 130 nm. Data were acquired at 23 °C.
Analysis of microtubule shrinkage rates
Data were analysed in Matlab (Mathworks). Kymographs were generated by averaging the cross-section of a straight-line ROI over 11 pixels (rotated by using the imrotate function first) for each frame of an image stack. Shrinkage rates were measured by manually tracing kymographs using the impoly function and calculating the slope. Time and distance calibration was automated using the image metadata. Rates in this paper assume a conversion of 125 dimer protofilament-1 = 1 µM. Fluorescence analysis of kinesin-clamp data is presented in Supplementary Methods. Plotting and statistical tests were also carried out using Matlab.
Analysis of microtubule expansion assay with microfluidics
Microtubules were carefully selected for analysis, rejecting any that did not conform to their original trajectory at the end of an ADP-kinesin cycle, which was assessed by visual inspection. Kymographs were generated using the Fiji plugin KymoResliceWide with a 15 pixel cross-section. Subsequent analysis was performed using Matlab. Expansion of the microtubule was measured for each time point as shown in Supplementary Fig. 5. These measurements were then analysed for each ADP or kinesin interval: the mean expansion value was taken for each ADP interval, whereas a logistic curve was fitted (using bisquare robust fitting) to each ADP-kinesin transition to estimate the kinesin-driven expansion (as shown in Fig. 5d).
Analysis of surface-stitched microtubule expansion assay
Coordinates of microtubules were extracted from dark-field images using the Fiji plugin JFilament30, mapped onto the fluorescence channel and used to generate a 5-pixel-wide linescan. Fluorescence profiles of the cap and seed were each fitted with a Gaussian error function in Matlab. The length of the GDP-microtubule is given by the distance between the point of inflection on each curve. The microtubule length change was then assessed by taking the mean length of the manually identified plateaus, as shown in Supplementary Fig. 4b. Points deviating by greater than 5% from the median in these intervals were discarded before fitting. The average length of kinesin-free GDP-microtubules analysed for this paper was 41 µm.
Supplementary Material
Supplementary information is available in the online version of the paper.
Acknowledgements
The authors thank D. R. Drummond and N. Sheppard for assistance with protein purification, and T. A. McHugh for commenting on the manuscript. This research was funded by the Biotechnology and Biological Sciences Research Council (grant number BB-G530233-1) via the Systems Biology Doctoral Training Centre, University of Warwick; and the Wellcome Trust (grant number 103895/Z/14/Z).
Footnotes
Author contributions
D. R. P. and R. A. C. designed experiments. N. J. B. provided mathematical insight. D. R. P. designed the analyses, collected and analysed the data, developed the microfluidics interface, and produced the manuscript and figures. All authors contributed towards the discussion and interpretation of results, and editing the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Reprints and permission information is available online at www.nature.com/reprints.
Data availability statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Bachand GD, Spoerke ED, Stevens MJ. Microtubule-based nanomaterials: Exploiting nature's dynamic biopolymers. Biotechnol Bioeng. 2015;112:1065–1073. doi: 10.1002/bit.25569. [DOI] [PubMed] [Google Scholar]
- 2.Katsuki M, Drummond DR, Cross RA. Ectopic A-lattice seams destabilize microtubules. Nat Commun. 2014;5:3094. doi: 10.1038/ncomms4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- 4.Marceiller J, Drechou A, Durand G, Perez F, Poüs C. Kinesin is involved in protecting nascent microtubules from disassembly after recovery from nocodazole treatment. Exp Cell Res. 2005;304:483–492. doi: 10.1016/j.yexcr.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Kowalski RJ, Williams RC. Unambiguous Classification of Microtubule-Ends In Vitro: Dynamic Properties of the Plus- and Minus-Ends. Cell Motil Cytoskeleton. 1993;26:282–290. doi: 10.1002/cm.970260403. [DOI] [PubMed] [Google Scholar]
- 6.Daire V, et al. Kinesin-1 regulates microtubule dynamics via a c-Jun N-terminal kinase-dependent mechanism. J Biol Chem. 2009;284:31992–32001. doi: 10.1074/jbc.M109.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura Arimura Fukata TNY, Watanabe H, Iwamatsu A, Kaibuchi K. Tubulin and CRMP-2 complex is transported via Kinesin-1. J Neurochem. 2005;93:1371–1382. doi: 10.1111/j.1471-4159.2005.03063.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross RA. The kinetic mechanism of kinesin. Trends Biochem Sci. 2004;29:301–309. doi: 10.1016/j.tibs.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 10.VanDelinder V, Adams PG, Bachand GD. Mechanical splitting of microtubules into protofilament bundles by surface-bound kinesin-1. Sci Rep. 2016;6:39408. doi: 10.1038/srep39408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabir AMR, et al. Biomolecular Motor Modulates Mechanical Property of Microtubule. Biomacromolecules. 2014;15:1797–1805. doi: 10.1021/bm5001789. [DOI] [PubMed] [Google Scholar]
- 12.Krebs A, Goldie KN, Hoenger A. Complex formation with kinesin motor domains affects the structure of microtubules. J Mol Biol. 2004;335:139–153. doi: 10.1016/j.jmb.2003.10.039. [DOI] [PubMed] [Google Scholar]
- 13.Morikawa M, et al. X-ray and Cryo-EM structures reveal mutual conformational changes of Kinesin and GTP-state microtubules upon binding. EMBO J. 2015;34:1270–1286. doi: 10.15252/embj.201490588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alushin GM, et al. High-Resolution Microtubule Structures Reveal the Structural Transitions in αβ-Tubulin upon GTP Hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakata T, Niwa S, Okada Y, Perez F, Hirokawa N. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J Cell Biol. 2011;194:245–255. doi: 10.1083/jcb.201104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:1–11. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muto EE, Sakai HH, Kaseda KK. Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J Cell Biol. 2005;168:691–696. doi: 10.1083/jcb.200409035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arora K, et al. KIF14 binds tightly to microtubules and adopts a rigor-like conformation. J Mol Biol. 2014;426:2997–3015. doi: 10.1016/j.jmb.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Hancock WO. Kinesin-5 is a microtubule polymerase. Nat Commun. 2015;6:8160. doi: 10.1038/ncomms9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hibbel A, et al. Kinesin Kip2 enhances microtubule growth in vitro through length-dependent feedback on polymerization and catastrophe. eLife. 2015;4:e10542. doi: 10.7554/eLife.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alonso MC, et al. An ATP Gate Controls Tubulin Binding by the Tethered Head of Kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vale RD, Coppin CM, Malik F, Kull FJ, Milligan RA. Tubulin GTP hydrolysis influences the structure, mechanical properties, and kinesin-driven transport of microtubules. J Biol Chem. 1994;269:23769–23775. [PubMed] [Google Scholar]
- 24.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 25.Cross RA, McAinsh A. Prime movers: the mechanochemistry of mitotic kinesins. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- 26.Hendricks AG, et al. Dynein Tethers and Stabilizes Dynamic Microtubule Plus Ends. Curr Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blehm BH, Schroer TA, Trybus KM, Chemla YR, Selvin PR. In vivo optical trapping indicates kinesin's stall force is reduced by dynein during intracellular transport. Proc Natl Acad Sci USA. 2013;110:3381–3386. doi: 10.1073/pnas.1219961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perur N, Yahara M, Kamei T, Tamaoki N. A non-nucleoside triphosphate for powering kinesin-microtubule motility with photo-tunable velocity. ChemComm. 2013;49:9935–9937. doi: 10.1039/c3cc45933k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.