Abstract
The realization of a synthetic biology approach to microbial (1R,2S,5R)-(−)-menthol (1) production relies on the identification of a gene encoding an isopulegone isomerase (IPGI), the only enzyme in the Mentha piperita biosynthetic pathway as yet unidentified. We demonstrate that Δ5-3-ketosteroid isomerase (KSI) from Pseudomonas putida can act as an IPGI, producing (R)-(+)-pulegone ((R)-2) from (+)-cis-isopulegone (3). Using a robotics-driven semirational design strategy, we identified a key KSI variant encoding four active site mutations, which confer a 4.3-fold increase in activity over the wild-type enzyme. This was assisted by the generation of crystal structures of four KSI variants, combined with molecular modeling of 3 binding to identify key active site residue targets. The KSI variant was demonstrated to function efficiently within cascade biocatalytic reactions with downstream Mentha enzymes pulegone reductase and (−)-menthone:(−)-menthol reductase to generate 1 from 3. This study introduces the use of a recombinant IPGI, engineered to function efficiently within a biosynthetic pathway for the production of 1 in microorganisms.
Keywords: biosynthetic (−)-menthol production, ketosteroid isomerase, isopulegone isomerase, enzyme engineering, robotics
Introduction
Limonene and its related derivatives are the most abundant, naturally sourced monoterpenoids known.1 These natural products are commonly used in the perfume, fragrance, and flavor industries.2,3 For example, (R)-limonene ((R)-4) is the major essential oil constituent of orange peels and is often found in cleaning products.4 In contrast Mentha piperita (peppermint) generates (S)-limonene ((S)-4), which is subsequently converted into menthol isomers (Scheme 1A).2,5 The most commercially useful isomer is 1, known by its characteristic cooling anesthetic effects and aroma.6 It also has antibacterial, anticancer, and anti-inflammatory activities, making it a valuable natural product.7−10
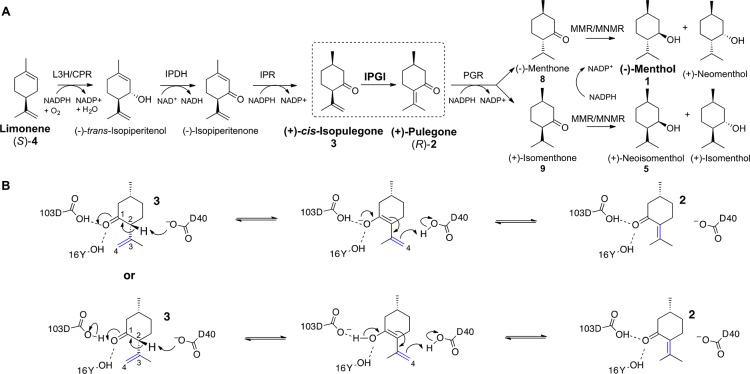
Scheme 1. (A) Biosynthesis of 1 and (+)-Neoisomenthol (5) from (S)-4 and (B) Proposed KSI Catalytic Mechanism with 3, on the Basis of the Known Mechanism of the Enzyme towards Δ5-3-Ketosteroids14.
Abbreviations: L3H = (−)-limonene-3-hydroxylase; IPDH = (−)-trans-isopiperitenol dehydrogenase; IPR = (−)-isopiperitenone reductase; IPGI = (+)-cis-isopulegone isomerase; PGR = (+)-pulegone reductase; MMR = (−)-menthone:(−)-menthol reductase; MNMR = (−)-menthone:(−)-neomenthol reductase.
Commercially, 1 is utilized both as a pure compound and as a component of the essential oil of peppermint, with an estimated 30000 tons consumed annually.11 While the majority of 1 is extracted from Mentha canadensis, a large proportion is generated synthetically (US $300 million annually).12 The two major synthetic routes to 1 are the Haarmann–Reimer route from m-cresol and the Takasago synthesis utilizing β-pinene.13 However, the flavor and fragrance industries and/or consumers may demand that these compounds be supplied from natural sources, particularly when they are used as food additives.
Naturally sourced peppermint oil can vary in both relative composition of its monoterpenoid constituents and overall yields due to fluctuating environmental conditions. This leads to price volatility and increased arable land competition for more profitable crops, such as biofuel biomass production. Optimization of peppermint oil production through conventional breeding methods is difficult, as it is a sterile hybrid plant.2,15 Additionally, purification of 1 from essential oil requires expensive and low-yielding steam-distillation and filtration processes to generate a usable product.16,17 Therefore, alternative clean biosynthetic routes to these compounds are a commercially attractive prospect to generate 1 with a “natural” label.
One alternative “natural” route is to introduce the biosynthetic pathway of 1 into cost-effective and even food-compatible microorganisms.18,19 This general synthetic biology approach generates biological factories, where rapid fine chemicals production can be achieved sustainably using nonpetroleum renewable feedstocks, with a reduced land burden and higher product consistency and purity.20−22 Such an approach has proved commercially successful in the semisynthetic industrial-scale production (ca. 35 tons per annum) by Sanofi of artemisinin, a major active ingredient in modern malarial treatments. Similarly, the semisynthetic production of Taxol has been demonstrated, where microorganism biofactories generated the precursor taxadiene using introduced pathways.20
We recently described the incorporation of the M. piperita based biosynthetic pathway from (R)-2 to 1 into Escherichia coli.23 An in vitro protocol was established where 1 was successfully generated, utilizing E. coli cell extracts as the biocatalyst source. However, a more cost-effective route would be an in vivo approach, where the full pathways from simple carbon sources to 1 would be incorporated and the microorganisms would be grown on waste feedstock. The six enzymatic steps from (S)-4 to 1 in Mentha sp. have been well characterized,5,24−30 and the native enzymes for five of these reactions have been sequenced and functionally expressed in E. coli (Scheme 1A).4,5,23,28,30,31 The remaining enzyme is isopulegone isomerase (IPGI), which catalyzes the double bond migrating isomerization (C3–C4 to C2–C3) of 3 to (R)-2 (Scheme 1B).26 Early studies with impure native Mentha IPGI identified a 54 kDa protein complex2,24 exhibiting cofactor-free isomerization, utilizing a mechanism similar to bacterial Δ5-3-ketosteroid isomerases (KSI).26 Therefore, the IPGI mechanism likely involves deprotonation of the C2α hydrogen of 3 by an enzymatic proton acceptor (D40 in KSI), to form an enolate intermediate (Scheme 1B), stabilized by hydrogen-bonding residues (Y16 and D103 in KSI). A proton-donating residue then transfers its proton to C4, leading to double bond migration to C2–C3 and re-formation of the carbonyl group. As no significant ketosteroid isomerase-like proteins have been annotated in plant genomes, the protein sequence of native IPGI is likely to remain elusive until the completion of the full annotation of the Mentha longifolia genome sequence.32
To complete the construction of a pathway from (S)-4 to 1 in E. coli, we investigated a well-studied bacterial Δ5-3-ketosteroid isomerase from Pseudomonas putida(14,33,34) and found that it exhibited low activity toward 3. To improve the IPGI activity, we performed three rounds of automated mutagenesis to improve yields of 2. The crystal structures of four KSI variants were generated, to assist the structure-guided mutagenesis approach to improve the binding of 3 to the enzyme. Molecular dynamics simulations and DFT modeling were applied to the most successful variant, to investigate the mode of increase in activity. The performance of the optimized KSI variant within both in vitro and in vivo biotransformation cascades from 3 to 1 was demonstrated. Therefore, the identification and optimization of a substitute IPGI is a critical step in the future generation of a potentially industrial E. coli strain capable of producing “natural” 1.
Experimental Section
Design and Synthesis of KSI Variants
The wild-type gene sequence encoding the Δ5-3-ketosteroid isomerase from P. putida (UniProt ID: P07445) was designed using the GeneGenie online tool, containing a C-terminal His6-tag.35 Mutagenic oligonucleotides for the first-generation libraries encoding ambiguous codons for 16 residues were designed optimized for E. coli expression using CodonGenie (Tables S1 and S2).36 Eight overlapping DNA oligonucleotides (up to 80 nt in length) were used to assemble the full length genes using the SpeedyGenes gene synthesis method (section S2.5 in the Supporting Information).37,38 Purified genes were ligated into linearized pET21b (Novagen) to generate a C-terminally His6-tagged enzyme by In-Fusion cloning (Clontech), according to the manufacturer’s protocol. Cloning products were transformed into T7 Express competent E. coli cells and grown on LB agar plates containing 100 μg/mL ampicillin, followed by incubation overnight at 37 °C.
The second- and third-generation variants were constructed using asymmetric PCR mutagenesis,39,40 followed by ligation into linearized pET21b by In-Fusion cloning. These mutations were designed as double or triple variants of the first-generation residues identified as providing hits (V88I, L99X, and D103S). The final variant was a combination of four individual positive mutations (V88I/L99V/V101A/D103S).
Gene Synthesis of Mentha Biocatalysts
The protein sequences for the following enzymes from M. piperita were obtained from UniProt (http://www.uniprot.org): pulegone reductase (MpPGR; Q6WAU0) and (−)-menthone:(−)-menthol reductase (MMR; Q5CAF4). The respective gene sequences were designed and synthesized and subcloned into pET21b by GeneArt (ThermoFisher), incorporating a C-terminal His6-tag and applying codon optimization techniques of rare codon removal for optimal expression in E. coli. Each construct was transformed into E. coli T7 Express and grown on LB agar plates containing 100 μg/mL ampicillin, followed by incubation overnight at 37 °C.
Robotics-Driven Production and Purification of Variant Libraries
Robotic colony picking and auto induction media inoculation (Formedium, 100 μg/mL ampicillin) of first-generation libraries were performed in deep-well plates (Hamilton Robotics), including replica plating into LB (Formedium, 100 μg/mL ampicillin) for glycerol stock generation. Plates were covered with a Breathseal sealer (Greiner Bio-One) and incubated at 30 °C and 1000 rpm for 24 h. Each variant underwent robotic cell lysis, extraction, and purification using Ni-NTA resin (Qiagen; 50 μL per variant) prior to enzymatic assays (see section S3.1 in the Supporting Information for details). Enzyme concentrations were determined using the Bradford assay (Bio-Rad) following the manufacturer’s protocol.
KSI Variant Biotransformation Screen
Substrate 3 (85% purity) was synthesized by Dr. Aisling Miller (University of Manchester), using a combination of chemical and enzymatic transformations.41 Reactions (200 μL) were performed in 50 mM Tris pH 7.0 containing 3 (1 mM) and KSI extract (189 μL) and incubated at 30 °C for 24 h (180 rpm). Samples were prepared for GCMS analysis by solvent extraction with 180 μL of ethyl acetate containing 0.01% sec-butylbenzene as the internal standard and dried with anhydrous MgSO4. GC-MS achiral quantitative analysis was conducted on a 7890B GC coupled to a 5975 series MSD quadrupole mass spectrometer and equipped with a 7693 autosampler (Agilent Technologies). Chiral product analysis was performed by analyzing reactions by GC using an Agilent Technologies 7890A GC system with an FID detector and a Chirasil-DEX-CB column (Agilent; 25 m, 0.32 mm, 0.25 μm). Further chromatography details can be found in section S3.2 in the Supporting Information.
Yields of (R)-2 were corrected for the initial concentration present in substrate 3 (15%). Due to limitations in the quantity of in-house synthesized 3, three colonies per plate were pooled together to test for an increase in yield of (R)-2. Pooled samples exhibiting an increased (R)-2 yield in comparison to wild-type KSI were rescreened individually, as above, to identify the well(s) responsible for the improved yield. Quantification was performed by comparing the peak areas to the calibration curves of authentic standards. Vendor binary files were converted to open mzXML data format42 using ProteoWizard msConvert.43 Automated peak profiling and quantification was conducted using in-house scripts written in R.
Comparative Activity of the Variants
First-generation KSI variant hits and later mutants generated by site-specific mutagenesis were cultured from glycerol stocks in LB medium containing 100 μg/mL ampicillin as above. The KSI-encoding plasmids were extracted and purified (Macherey-Nagel) and underwent Sanger sequencing to identify the mutation present. Larger scale cultures (500 mL) were generated for each variant in the same antibiotic-selective LB medium and incubated at 37 °C (180 rpm) to an OD600nm value of 0.6–0.8, followed by induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were incubated overnight at 25 °C (180 rpm) and the cells harvested by centrifugation (3000g for 5 min). Each variant underwent extraction (4 mL of lysis buffer) and purification (0.5 mL Ni-NTA resin) using the same protocol and buffers as in the initial screen (section S3.1 in the Supporting Information). Biotransformations were performed with 1 mM 3 as above (10 μM KSI), and the variants that did not display improved yield of (R)-2 above wild type (false positives) were discarded.
Steady-State Reactions with (+)-cis-Isopulegone
Continuous steady-state reactions with 3 were performed with selected purified variants to investigate the effect of the mutation(s) on the reaction rate. Reaction mixtures (100 μL) were composed of 50 mM Tris pH 7.0 containing 1 mM 3 within UV-Star microplates (Greiner Bio-One) and covered with a ClearVue sealing sheet. Product formation was detected by monitoring the absorbance at 260 nm for 1 h at 20 °C using a CLARIOstar microplate reader (BMG Labtech). Concentrations were determined by comparison to a standard curve (Figure S1), and data were corrected for both 3 and (R)-2 loss over 1 h (control substrate and product data).
Cascade Biocatalysis
The Mentha enzymes MpPGR and MMR were produced (500 mL of culture; 20 °C) and purified according to the method for the KSI variants (section S3.1 in the Supporting Information). Reactions (200 μL) were performed in 50 mM Tris pH 7.0 containing 10 μM KSI, 1 μM MpPGR, 0.3 μM MMR, 1 mM 3, and a cofactor recycling system (10 U Sigma glucose dehydrogenase, 10 μM NADP+, and 15 mM d-glucose) to supply MpPGR and MMR with NADPH.23 Reaction mixtures were incubated at 30 °C for 24 h (180 rpm), and the extracts were analyzed by GC-MS as before.
Pathway Assembly and in Vitro Activity
The genes encoding KSI variant V88I/L99V/V101A/D103S, MpPGR, and MMR and their respective ribosomal binding sequences (RBS) were amplified by PCR and ligated into pET21b using In-Fusion cloning to create an expression construct under the control of one T7 promoter. The RBS of MMR was modified by PCR mutagenesis to generate three constructs with lower predicted translation initiation rates (Table S3). Each construct was transformed into competent E. coli NEB5α cells and underwent culture growth, plasmid purification, and DNA sequencing as before.
The plasmids were transformed into competent E. coli NiCo21 (DE3) expression cells and cultured on LB agar containing 100 μg/mL ampicillin. Single colonies were selected and grown in 200 mL of phosphate buffered Terrific Broth pH 7.0 containing 0.4% glycerol and 100 μg/mL ampicillin, followed by induction with 0.1 mM IPTG. The cultures were incubated overnight at 20 °C and harvested by centrifugation (3000g). Soluble cell extracts were generated using lysis buffer (3 mL pH 7.0), sonication, and centrifugation as above. Cascade biocatalysis reactions were set up as before, except the purified enzymes were replaced with 50 μL of cell extract.
Protein Crystallization and Structure Solution
Purified KSI variants D103S, V88I/L99V, V88I/L99V/D103S and V88I/L99V/V101A/D103S (10 mg/mL) were crystallized using a Mosquito robot (TTP Labtech) and the following crystallization solutions: (i) 10–25% PEG3350 with 0.2 M MgCl2, (ii) 10–25% PEG3350 with 0.2 M ammonium acetate (pH 4.6), and (iii) 10–25% PEG3350 with 0.2 M ammonium acetate (pH 5.5). Crystals were flash-frozen in 30% glycerol and data collected at the Diamond light source (beamlines IO4 and IO3). Reflections were merged and scaled with Xia2.44 All mutant structures were solved by molecular replacement using Phaser-MR (CCP4)45 and wild-type KSI (PDB code: 1OPY) as a search model. Initial model building was done with AutoBuild (PHENIX)46 followed by iterative cycles of manual model building and refinement in COOT47 and Phenix.refine,48 respectively. Mutations could be clearly identified in Fo – Fc difference maps and were modeled accordingly. The final data collection and refinement statistics are found in Table S4. The atomic coordinates and structure factors (6F4Y, 6F50, 6F53, and 6F54) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Molecular Modeling
DFT modeling was performed using Gaussian09 revision D.01.49 An active site model was created from the crystal structure of KSI with bound androstenedione (6; PDB ID 1OHS) including the side chains and α-carbons of the 15 first-shell residues shown in Figure S4, with substrate 3 built in using the coordinates of the corresponding ligand 6 atoms as the starting point. Energy minimization was carried out using the B3LYP hybrid functional and 6-31G(d) basis sets50 with the D3 version of Grimme’s dispersion.51 Since the active site is solvent exposed, a continuum solvent model was used to mimic the shielding effect of water. Molecular dynamics simulations were carried out using Gromacs 4.6.1 with the Gromos 53A6 force field and periodic boundary conditions.52,53 For WT KSI, the crystal structure of KSI with bound 6 (PDB: 1OHS) was used, with substrate 3 built in using the coordinates of the corresponding ligand 6 atoms as the starting point. For KSI variant V88I/L99V/V101A/D103S, ligand 3 was placed in the active site of the crystal structure (PDB: 1OPY) by aligning the protein to the wild-type KSI model. The energy minimization parameters are described in detail in section S6 in the Supporting Information.
Results and Discussion
Identification of KSI as a (+)-cis-Isopulegone Isomerase
Prior studies with the partially purified native M. piperita isomerase suggested its mechanism resembled that of the bacterial Δ5-3-ketosteroid isomerase family (EC 5.3.3.1).26 These enzymes catalyze the isomerization of Δ5-3-ketosteroids, such as 5(10)estrene-3,17-dione, to the equivalent Δ4-3-ketosteroid by intramolecular transfer of the C4β proton to the C6β position.54 Prior studies showed that KSI from P. putida reacts with the truncated substrate 3-cyclohexen-1-one, forming the more stable 2-cyclohexen-1-one.55 Therefore, we postulated that KSI might react with the structurally related compound 3 to generate (R)-2 using a similar mechanism (Scheme 1B).
A C-terminally His6-tagged version of wild-type KSI from P. putida was generated, and biotransformations with 3 showed that 2 was formed (35% yield). Chiral GC analysis confirmed that the product generated was (R)-2, the enantiomeric form required for 1 biosynthesis (Table S5).2 An additional minor quantity (<1% yield) of the side product (−)-trans-isopulegone (7) was also detected. As the (R)-2 yields were judged to be too low for biotechnological applications, we employed a directed evolution strategy to engineer KSI for improved synthesis of (R)-2.
Automated Directed Evolution of KSI
Comparative studies of KSI with single- and multiple-ring substrates showed that, while the specificity (kcat/Km) is 27000-fold higher for steroid substrates, the reaction rates were within 2-fold.55 This suggests that remote binding interactions with steroid substrates have a marked effect on the strength of substrate binding, and single-ring substrates such as 3 may have poor binding affinity. Therefore, our initial KSI mutagenesis strategy was to increase binding of 3 by targeting every amino acid known or predicted to interact with steroid substrates. An examination of the cocrystal structure of wild-type KSI bound to the reaction intermediate analogue equilenin (PDB 1OH0)34 showed that 16 residues were located within 5 Å of the ligand. Given the overall apolar nature of the KSI active site and 3, maintaining this environment would likely maximize our search for optimized mutations while minimizing the size of the variant libraries and subsequent screening effort. Reducing the number of screening assays was critical, given the limited availability of the in-house-synthesized substrate 3.
Twelve of these residues were targeted for individual limited randomizing mutagenesis, where the general physicochemical properties of the amino acids were conserved (Table S1). Residues Y16 and Y57 are both known to partake in H bonding with the substrate carbonyl moiety; therefore, these residues were mutated combinatorially, using an ambiguous codon (WVC) that encodes other residues capable of H bonding. Similarly, residues V88 and L99 were selected for combinatorial mutations using an ambiguous codon (DTK) for hydrophobic residues, given their close proximity to the substrate analogue.34 Variant sequences were created by gene synthesis using ambiguous codons (Table S2) and constructed using the SpeedyGenes protocols.37,38 In total, 158 variants were created and screened for improvement in (R)-2 production using automated robotic culturing and protein purification protocols (summarized in Figure S2). Comparative biotransformations were performed with 3, with quantitation and product identification by GC-MS, to identify variants with activity higher than that of wild-type KSI.
Four enzyme variants were identified as having minor increases in (R)-2 yield (1.4–1.7 fold), three of which contained mutations of residue L99 (L99I, L99V, and V88I/L99V; Table 1). Interestingly, both L99I and L99V involve mutations to a smaller side chain, suggesting that these changes may affect 3 binding by reducing steric hindrance. The fourth improved variant D103S also contained a smaller side chain; however, there was also a change from a negatively charged moiety to a polar functionality. As a serine hydroxyl group can participate in H bonding, this mutation might maintain the native enzyme hydrogen-bonding interaction with the substrate carbonyl group.
Table 1. Biotransformations of Wild-Type and Variant KSI with 3a.
| round | variant | activityb | rel activity (fold) |
|---|---|---|---|
| wild type | 13.88 ± 0.08 | 1.0 | |
| 1 | D103S | 20.09 ± 1.16 | 1.4 |
| L99I | 23.58 ± 0.10 | 1.7 | |
| L99V | 18.46 ± 0.48 | 1.3 | |
| V88I/L99V | 19.39 ± 0.57 | 1.4 | |
| 2 | V88I/L99V/D103S | 26.47 ± 1.59 | 1.9 |
| L99V/D103S | 25.51 ± 0.83 | 1.8 | |
| L99I/D103S | 25.91 ± 0.58 | 1.9 | |
| 3 | V88I/L99V/V101A/D103S | 63.10 ± 0.61 | 4.5 |
Duplicate reactions (200 μL) were performed in 50 mM Tris pH 7.0 containing 1 mM 3 and 10 mM KSI. After a 24 h incubation at 30 °C (180 rpm), reaction mixtures were extracted with 180 μL of ethyl acetate containing 0.01% sec-butylbenzene and dried with anhydrous MgSO4. Products were analyzed by GC-MS using a DB-WAX column.
Activity is expressed as μM (R)-2 produced per μM KSI in 24 h.
Second Round: Compiling Mutations
To investigate the combinatorial effect of the initial screen variants, the L99X-containing single and double mutants were each combined with D103S using asymmetric PCR mutagenesis.39,40 Each variant was purified and underwent biotransformations to determine any improvement in (R)-2 yield by GC-MS (Table 1). Unfortunately, these double and triple variants showed only moderate improvements in (R)-2 yield in comparison to wild-type enzyme (1.8–1.9-fold).
Third Round: Structure-Guided Mutation
To ensure the mutations in the active site and surrounding residues did not affect the overall fold of KSI, we solved the X-ray crystal structures of the four key variants D103S, V88I/L99V, V88I/L99V/D103S, and V88I/L99V/V101A/D103S (Table S4). Each of these structures showed backbone conformations virtually identical with the cocrystal structure of wild-type KSI bound to the reaction intermediate analogue equilenin (Figure S3),34 with RMSD values of 0.427, 0.418, 0.443, and 0.357, respectively. All of the mutations could be clearly identified in Fo – Fc difference maps and were incorporated without any major displacement of the backbone atoms or surrounding residues. We did note minor rearrangements of backbone atoms within the loop region 90–98 (results not shown). This loop is exposed to the bulk solvent and shows above average B factors in comparison to surrounding residues, implying the observed differences are due to inherent flexibility, rather than any mutational changes to neighboring residues.
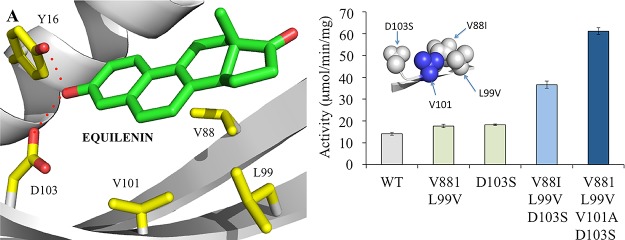
Inspecting the spatial arrangement of these mutations showed that residue V101 was positioned directly between L99V and D103S, highlighting it as another potential target for mutagenesis (Figure 1B, inset). Given that most of the existing variants resulted in a shortening of the amino acid side chain, the triple variant was modified to incorporate the mutation V101A. Biotransformations were performed on the KSI V88I/L99V/V101A/D103S variant, which showed a more dramatic increase in (R)-2 production over wild-type enzyme (4.5-fold; Table 1). Therefore, the modification of V101 to an alanine residue has resulted in over a 2-fold increase in (R)-2 formation over the triple variant.
Figure 1.
Location of key active site mutations implicated in improving KSI activity toward 3. (A) Residues located in the equilenin-binding region of wild type KSI from P. putida (PDB: 1OH0).34 The residues and equilenin are shown as atom colored sticks with yellow and green carbons, respectively. Interactions are shown as red dotted lines. The backbone is shown as a gray cartoon. (B) Comparative steady-state activity of wild-type and variant KSI enzymes. Reaction mixtures (100 μL) were composed of 50 mM Tris pH 7.0 containing 1 mM 3. The absorbance was monitored at 260 nm for 1 h at 20 °C. Inset: location of the variant residues in KSI V881/L99V/D103S. The backbone and mutations are shown as gray ribbons and balls, respectively (blue balls for V101).
To investigate the effect of the mutations further, the specific activity of the variants with substrate 3 was determined under pseudo-steady-state conditions (1 h reactions). This method takes advantage of the enolization of 3 to (R)-2, which results in the development of a peak around 260 nm (Figure S1). First-generation variants showed minor rate increases (around 1.3-fold), while a near 2-fold improvement was seen with the triple variant V88I/L99V/D103S (Figure 1B). The addition of mutation V101A generated a 4.3-fold increase in specific activity in comparison to the WT (61.17 μmol/(min mg) in comparison to 14.16 μmol/(min mg); Figure 1B). Therefore, the increases in the overall reaction rate mirror the increased product yields detected in the 24 h biotransformations.
Modeling and Molecular Dynamics Simulations for KSI Variants
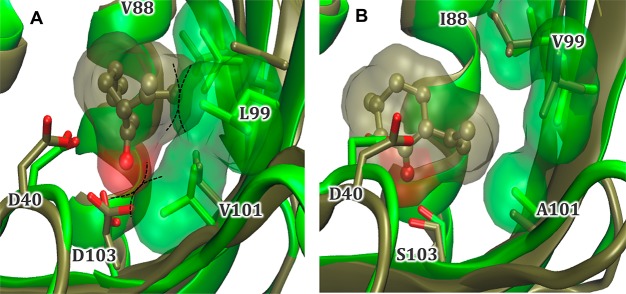
We performed comparative DFT modeling and MD simulations between wild-type KSI and variant V88I/L99V/V101A/D103S to investigate the origin(s) of activity rate changes. The DFT optimized model for wild type KSI suggests that the substrate 3 carbonyl group hydrogen bonds with D103 (<2.0 Å), but the orientation is nonideal for catalysis due to a less favorable interaction distance for the proton abstraction step (Figure 2, Figure S4, and Scheme 1B). Additionally, residue L99 has reoriented outward from the active site, causing a twist in the β-hairpin from residue 88 to 99 (Figure S2).
Figure 2.
Overlay of energy-minimized structure (green) and representative structure from MD simulations (brown) for (A) wild-type KSI and (B) variant V88I/L99V/V101A/D103S modeled with 3 in the active site. The solvent-accessible surface areas for the substrate and residues 88, 99, and 101 from the representative and minimized structures, respectively, are shown as transparent surfaces. The dotted lines illustrate potential steric clashes between the substrate and enzyme. The MD simulations were performed using Gaussian09 revision D.01.49
MD simulations of variant KSI with 3 restrained in a reactive conformation suggests that the decrease in bulk of variant residues I88, V99, and A101 allows the substrate to comfortably adopt this position (Figure 2B). The position of 3 looks more ideal, with a D40–substrate hydrogen–oxygen distance of 2.04 Å in the DFT model. Therefore, the decrease in bulk of variant residues I88, V99, and A101 has allowed this reorienting of the substrate, while still maintaining a (slightly longer) hydrogen-bonding interaction with residue S103 (D103 in wild type). MD simulations with the variant model showed no repositioning of the hairpin loop from 88 to 99 was necessary, as 3 fit more easily within the active site. Overall, molecular simulations suggest that these four mutations have improved binding of 3 in a functional conformation within the active site. Therefore, the V88I/L99V/V101A/D103S variant was selected for further studies to see if it can be used in cascade reactions for the biosynthetic production of 1 from 3.
One-pot (−)-Menthol Production with Purified Enzymes
Initial cascade reactions were performed with purified enzymes to eliminate any interference from competing E. coli enzymes present in whole cells and cell extracts (Table 2). Interestingly, reactions with KSI variant V88I/L99V/V101A/D103S showed the formation of ∼10% byproducts, such as trans-isopulegone (7). This suggests that the presence of the four mutations has altered the range of possible binding conformations of 3, leading to additional isomer formation. The pathway from 3 to 1 also requires the presence of two Mentha enzymes, namely pulegone reductase (MpPGR)29 and (−)-menthone:(−)-menthol reductase (MMR), to generate (−)-menthone (8) and 1, respectively (Scheme 1A).2 Initial biocatalytic reactions of purified MpPGR with (R)-2 showed it produced near-equivalent yields of 8 and (+)-isomenthone (9) (ratio 49:51; Table 2). This differs slightly from previous studies of a non-His6-tagged recombinant form of MpPGR, which showed an 8:9 ratio of 55:45.29 However, this enzyme showed a significantly higher yield of 8 than had been seen with the homologous enzyme from Nicotiana tabacum (NtDBR) performed under the same reaction conditions.56 Similarly, purified MMR-His6 demonstrated activity toward 8 similar to that in previous studies,5 showing primarily 1 production (Table 2).
Table 2. Biotransformations of Individual Enzymes and Cascading Reactions with Mentha Monoterpenoidsa.
| substrate
remaining/product yield (μM) |
||||||
|---|---|---|---|---|---|---|
| biocatalyst | 3 | (R)-2 | 8 | 9 | 3 | 5 |
| Individual Enzymes | ||||||
| KSI WT | 361.7 ± 11.5 | 210.4 ± 6.0 | N/A | N/A | N/A | N/A |
| KSI variantb,c | 40.7 ± 0.7 | 436.3 ± 10.8 | N/A | N/A | N/A | N/A |
| MpPGR | N/A | 10.0 ± 1.1 | 153.3 ± 5.8 | 160.1 ± 4.5 | N/A | N/A |
| MMR | N/A | N/A | 8.0 ± 2.0 | N/A | 280.7 ± 10.3 | N/A |
| Cascading Reactions | ||||||
| cascadec | 16.7 ± 1.2 | 10.3 ± 0.1 | 25.2 ± 0.9 | 16.6 ± 0.5 | 63.4 ± 3.8 | 11.1 ± 0.6 |
| cell extractc | 17.1 ± 0.1 | 11.5 ± 0.5 | 60.6 ± 1.9 | 34.8 ± 0.3 | 52.8 ± 0.9 | 18.6 ± 0.8 |
| whole cellsc | 32.5 ± 6.7 | 1.0 ± 2.1 | 44.7 ± 10.3 | 98.6 ± 2.4 | 158.8 ± 0.4 | 19.6 ± 0.1 |
Duplicate reactions (200 μL) were performed in 50 mM Tris pH 7.0 containing 1 mM monoterpenoid substrate, enzyme(s), and cofactor recycling system (10 U Sigma glucose dehydrogenase, 10 μM NADP+, and 15 mM d-glucose). After a 24 h incubation at 30 °C (180 rpm), reaction mixtures were extracted with 180 μL of ethyl acetate containing 0.01% sec-butylbenzene and dried with anhydrous MgSO4. Products were analyzed by GC-MS using a DB-WAX column (Figure S7). Reaction mixtures with individual enzymes had enzyme concentrations of 10 μM. The in vitro cascading reaction had enzyme concentrations of 10, 2, and 0.3 μM for KSI variant, MpPGR, and MMR, respectively. Control biotransformations of each compound in the absence of enzymes (data not shown) showed losses occurred over 24 h due to nonenzymatic degradation. Therefore, the product yields are likely an underestimation of the true yields. The cell extract and whole cell slurry volumes in the cascading reactions were 50 μL. Substrates are shown in boldface. N/A = not applicable.
KSI variant = V88I/L99V/V101A/D103S.
Additional products were detected that were not quantified (e.g., 7 and (+)-neoiso-isopulegol (10)).
Initial cascade biocatalytic reactions of purified KSI wild-type and variant V88I/L99V/V101A/D103S with MpPGR and MMR with 3 showed that the major product was an isomer of isopulegol (results not shown). Chiral GC analysis of the reaction mixture showed that the main isomer formed was (+)-neoiso-isopulegol (10), with a minor proportion of (−)-isopulegol generated (Figure S6).57 However, product 1 was also detected, with 5.1-fold higher yields obtained for reactions containing V88I/L99V/V101A/D103S variant KSI (46 μM vs 9 μM).
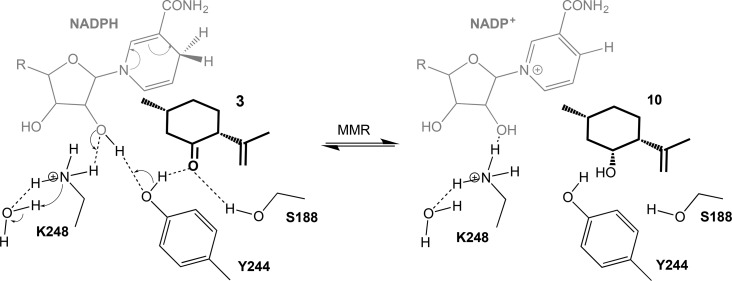
Subsequent reactions with each individually purified biocatalyst showed that this side product was obtained by the action of MMR on 3, which differs from earlier studies.5 However, this apparent disparity is likely due to the 144-fold reduction in the reaction time of the earlier study in comparison to this work (6–9 min vs 24 h).5 It is likely that the mechanism of action of MMR with 3 proceeds in a manner similar to that for the short chain dehydrogenase/reductase (SDR) ketoreduction mechanism described previously for MMR and MNMR with substrates 8 and 9 (Scheme 2).58 Unfortunately, the formation of 10 by MMR appeared to dominate over the reaction with 8, likely due to the lack of the latter substrate at the start of the reaction.
Scheme 2. Proposed Mechanism of Action of the NADPH-Dependent MMR-Catalyzed Reduction of 3 to 10.
This is adapted from the proposed SDR ketoreductase mechanism of MNMR with 8.58
Subsequent cascade reactions were performed with a 30-fold molar excess of KSI over MMR, to allow KSI activity with 3 to predominate. Using this approach, we dramatically increased 1 titers (63.4 μM; 14.1% conversion; Table 2) relative to 10, producing a near 1:1 ratio of products. In addition, a surprising yet advantageous observation was that relatively lower levels of 9 (and subsequently 5) than expected were produced for MMR, with a combined ratio of 76:24 ((8 + 1):(9 + 5)). Therefore, these studies have demonstrated that KSI variant V88I/L99V/V101A/D103S can be utilized in cascade reactions as the first step in generating 1 from 3.
Cascading Production of 3 Using Cell-Free Extracts and Whole Cells
We have previously reported the biosynthesis of menthol isomers from (R)-2, using E. coli cell extracts expressing the enzymes NtDBR and MMR.23 To further extend this approach, we developed a single expression construct in pET21b containing KSI variant V88I/L99V/V101A/D103S, MpPGR, and MMR, with each gene under the control of one T7 promoter (Table S3). This enabled the IPTG-dependent coexpression of all three genes in E. coli. Biotransformations were performed using cell extracts with 3 in the presence of a cofactor recycling system, the latter to supply MpPGR and MMR with NADPH. As expected, the major product obtained was 10, due to the promiscuous activity of MMR (results not shown).
To mimic the effect of increasing the KSI:MMR concentration ratio in vivo, we modified the RBS sequence upstream of MMR to three alternative ones, each with predicted translation initiation rates (TIR) less than the existing rate of 4100 (1000, 500, and 200; Table S3).59,60 Of the three constructs, only that containing an RBS with a predicted TIR of 500 showed any MMR activity, leading to the production of both 1 and 10 (Table 2). Cell extracts of this construct yielded 53 μM of 1, similar to that obtained with the purified enzyme one-pot biocatalytic reactions. The higher levels of 8 and 9 detected reflects the lower in vivo concentration of MMR within this construct. Reactions were also conducted with whole cells, producing nearly 3-fold higher levels of 1 in comparison to cell extract biotransformations (159 μM, 27% total yield; Table 2) with proportionally less 10.
These studies have demonstrated that these three enzymes have great potential in an overall strategy for in vivo production of 1 in E. coli. Ideally KSI V88I/L99V/V101A/D103S would be incorporated into an E. coli construct containing all the remaining enzymes required for 1 production from (S)-4 (Scheme 1A). This would enable one-pot biotransformations or in vivo fermentations to be performed using the inexpensive and renewable starting material (S)-4. Ultimately, the inclusion of an (S)-4 production pathway would enable E. coli to produce 1 in vivo from simple sugars.61 This would avoid the addition of high concentrations of toxic monoterpenoid precursors and eliminate the need for exogenous cofactor recycling systems. Additionally, the low in vivo concentration of 3 would likely minimize pathway hijacking to form 10, increasing the overall yields of 1.
Concluding Remarks
The implementation of a synthetic biology approach to microbial 1 production has been hampered by the absence of the sequence for the native Mentha pathway enzyme IPGI and the lack of identification of plant proteins exhibiting homology to bacterial Δ5-3-ketosteroid isomerases known to catalyze this reaction on steroid substrates. We took the approach of engineering an existing bacterial Δ5-3-ketosteroid isomerase to improve its activity toward 3, using a structure-modeling-guided, semirational design to identify target residues. Our implementation of a robotics platform to ensure rapid and reproducible automated gene assembly, enzyme purification, and microscale biotransformation screening enabled the identification of potential variant biocatalysts with limited substrate availability. The incorporation of four targeted mutations into KSI enabled a 4.3-fold increase in IPGI activity, generating a biotechnologically useful enzyme with significant in vivo activity. Further engineering studies could potentially improve its functionality, thereby driving the cascade reactions toward more efficient 1 production.
This is the first report of a non-Mentha enzyme with IPGI activity, thereby identifying the “missing link” to complete the enzymatic route from (S)-4 to 1. This will enable for the first time the full construction of a biosynthetic pathway for microbial production of 1. The demonstration of successful 1 production from 3 by engineered E. coli adds to a growing number of studies highlighting the potential usefulness of microbial hosts in monoterpenoid production. It is anticipated that integrating our existing KSI-MpPGR-MMR construct into a larger chassis will enable the production of menthol isomers from inexpensive precursors, providing a sustainable and renewable production platform to meet the growing global demand for natural menthol products.
Acknowledgments
We thank Colin Levy for protein crystallization and data collection and the wider SYNBIOCHEM team for their contributions. The authors acknowledge the assistance given by IT Services and the use of the Computational Shared Facility at The University of Manchester. The UK Catalysis Hub (http://www.ukcatalysishub.co.uk) is kindly thanked for resources and support. Funding was provided by the Biotechnology and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council (EPSRC) under grants BB/J015512/1 and BB/M017702/1.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscatal.7b04115.
Additional experimental details, materials used, additional tables and figures, results, GC/MS analyses, and discussion of computational and crystallographic studies (PDF)
The authors declare the following competing financial interest(s): A.C., H.S.T., and N.S.S. are named inventors on a submitted patent describing the work in this paper.
Supplementary Material
References
- Duetz W. A.; Bouwmeester H.; van Beilen J. B.; Witholt B. Appl. Microbiol. Biotechnol. 2003, 61, 269–277. 10.1007/s00253-003-1221-y. [DOI] [PubMed] [Google Scholar]
- Croteau R. B.; Davis E. M.; Ringer K. L.; Wildung M. R. Naturwissenschaften 2005, 92, 562–577. 10.1007/s00114-005-0055-0. [DOI] [PubMed] [Google Scholar]
- Leavell M. D.; McPhee D. J.; Paddon C. J. Curr. Opin. Biotechnol. 2016, 37, 114–119. 10.1016/j.copbio.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Alonso-Gutierrez J.; Chan R.; Batth T. S.; Adams P. D.; Keasling J. D.; Petzold C. J.; Lee T. S. Metab. Eng. 2013, 19, 33–41. 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Davis E. M.; Ringer K. L.; McConkey M. E.; Croteau R. Plant Physiol. 2005, 137, 873–881. 10.1104/pp.104.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt E. E.; Betts B. A.; Kotey F. O.; Humbert D. J.; Griffith T. N.; Kelly E. W.; Veneskey K. C.; Gill N.; Rowan K. C.; Jenkins A.; Hall A. C. Eur. J. Pharmacol. 2008, 590, 120–126. 10.1016/j.ejphar.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Freires I. A.; Denny C.; Benso B.; de Alencar S. M.; Rosalen P. L. Molecules 2015, 20, 7329–7358. 10.3390/molecules20047329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens U. R.; Stöber M.; Vetter H. Eur. J. Med. Res. 1998, 3, 539–545. [PubMed] [Google Scholar]
- Kim S.-H.; Lee S.; Piccolo S. R.; Allen-Brady K.; Park E.-J.; Chun J. N.; Kim T. W.; Cho N.-H.; Kim I.-G.; So I.; Jeon J.-H. Biochem. Biophys. Res. Commun. 2012, 422, 436–441. 10.1016/j.bbrc.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Li Q.; Wang X.; Yang Z.; Wang B.; Li S. Oncology 2009, 77, 335–341. 10.1159/000264627. [DOI] [PubMed] [Google Scholar]
- Kamatou G. P. P.; Vermaak I.; Viljoen A. M.; Lawrence B. M. Phytochemistry 2013, 96, 15–25. 10.1016/j.phytochem.2013.08.005. [DOI] [PubMed] [Google Scholar]
- Croteau R. B.; Davis E. M.; Ringer K. L.; Wildung M. R. Naturwissenschaften 2005, 92, 562–577. 10.1007/s00114-005-0055-0. [DOI] [PubMed] [Google Scholar]
- Sell C.A fragrant introduction to terpenoid chemistry; Royal Society of Chemistry: Cambridge, UK, 2003; pp 43–64. [Google Scholar]
- Pollack R. M. Bioorg. Chem. 2004, 32, 341–353. 10.1016/j.bioorg.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Lange B. M.; Mahmoud S. S.; Wildung M. R.; Turner G. W.; Davis E. M.; Lange I.; Baker R. C.; Boydston R. A.; Croteau R. B. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16944–16949. 10.1073/pnas.1111558108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B. M.; Mahmoud S. S.; Wildung M. R.; Turner G. W.; Davis E. M.; Lange I.; Baker R. C.; Boydston R. A.; Croteau R. B. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 16944–16949. 10.1073/pnas.1111558108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence B. M. In Mint: The genus Mentha; Hardman R., Ed.; CRC Press: Boca Raton, FL, USA, 2007; pp 1–547. [Google Scholar]
- Chemler J. A.; Koffas M. A. Curr. Opin. Biotechnol. 2008, 19, 597–605. 10.1016/j.copbio.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Mitchell W. Curr. Opin. Chem. Biol. 2011, 15, 505–515. 10.1016/j.cbpa.2011.05.017. [DOI] [PubMed] [Google Scholar]
- Ajikumar P. K.; Xiao W. H.; Tyo K. E. J.; Wang Y.; Simeon F.; Leonard E.; Mucha O.; Phon T. H.; Pfeifer B.; Stephanopoulos G. Science 2010, 330, 70–74. 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. C.; Ellington A. D. ACS Synth. Biol. 2015, 4, 1053–1055. 10.1021/acssynbio.5b00190. [DOI] [PubMed] [Google Scholar]
- Winter J. M.; Tang Y. Curr. Opin. Biotechnol. 2012, 23, 736–743. 10.1016/j.copbio.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toogood H. S.; Cheallaigh A. N.; Tait S.; Mansell D. J.; Jervis A.; Lygidakis A.; Humphreys L.; Takano E.; Gardiner J. M.; Scrutton N. S. ACS Synth. Biol. 2015, 4, 1112–1123. 10.1021/acssynbio.5b00092. [DOI] [PubMed] [Google Scholar]
- Croteau R.; Venkatachalam K. V. Arch. Biochem. Biophys. 1986, 249, 306–315. 10.1016/0003-9861(86)90007-X. [DOI] [PubMed] [Google Scholar]
- Crowell P. L.; Gould M. N. Crit. Rev. Oncog. 1994, 5, 1–22. 10.1615/CritRevOncog.v5.i1.10. [DOI] [PubMed] [Google Scholar]
- Kjonaas R. B.; Venkatachalam K. V.; Croteau R. Arch. Biochem. Biophys. 1985, 238, 49–60. 10.1016/0003-9861(85)90139-0. [DOI] [PubMed] [Google Scholar]
- Lupien S.; Karp F.; Wildung M.; Croteau R. Arch. Biochem. Biophys. 1999, 368, 181–192. 10.1006/abbi.1999.1298. [DOI] [PubMed] [Google Scholar]
- Ringer K. L.; Davis E. M.; Croteau R. Plant Physiol. 2005, 137, 863–872. 10.1104/pp.104.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer K. L.; McConkey M. E.; Davis E. M.; Rushing G. W.; Croteau R. Arch. Biochem. Biophys. 2003, 418, 80–92. 10.1016/S0003-9861(03)00390-4. [DOI] [PubMed] [Google Scholar]
- Wüst M.; Little D. B.; Schalk M.; Croteau R. Arch. Biochem. Biophys. 2001, 387, 125–136. 10.1006/abbi.2000.2248. [DOI] [PubMed] [Google Scholar]
- Duetz W. A.; Bouwmeester H.; Beilen J. B. v.; Witholt B. Appl. Microbiol. Biotechnol. 2003, 61, 269–277. 10.1007/s00253-003-1221-y. [DOI] [PubMed] [Google Scholar]
- Vining K. J.; Johnson S. R.; Ahkami A.; Lange I.; Parrish A. N.; Trapp S. C.; Vroteau R. B.; Straub S. C. K.; Pandelova I.; Lange B. M. Mol. Plant 2017, 10, 323–339. 10.1016/j.molp.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Ha N.-C.; Choi G.; Choi K. Y.; Oh B.-H. Curr. Opin. Struct. Biol. 2001, 11, 674–678. 10.1016/S0959-440X(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Kim S. W.; Cha S.-S.; Cho H.-S.; Kim J.-S.; Ha N.-C.; Cho M.-J.; Joo S.; Kim K. K.; Choi K. Y.; Oh B.-H. Biochemistry 1997, 36, 14030–14036. 10.1021/bi971546+. [DOI] [PubMed] [Google Scholar]
- Swainston N.; Currin A.; Day P. J.; Kell D. B. Nucleic Acids Res. 2014, 42, W395–W400. 10.1093/nar/gku336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swainston N.; Currin A.; Green L.; Breitling R.; Day P. J.; Kell D. B. PeerJ. Comput. Sci. 2017, 3, e120. 10.7717/peerj-cs.120. [DOI] [Google Scholar]
- Currin A.; Swainston N.; Day P. J.; Kell D. B. Protein Eng., Des. Sel. 2014, 27, 273–280. 10.1093/protein/gzu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currin A.; Swainston N.; Day P. J.; Kell D. B. Methods Mol. Biol. (N. Y., NY, U. S.) 2017, 1472, 63–78. 10.1007/978-1-4939-6343-0_5. [DOI] [PubMed] [Google Scholar]
- Perrin S.; Gilliland G. Nucleic Acids Res. 1990, 18, 7433–7438. 10.1093/nar/18.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S. H.; Madison E. L. Nucleic Acids Res. 1997, 25, 3371–3372. 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ní Cheallaigh A.; Mansell D.; Toogood H.; Tait S.; Lygidakis A.; Scrutton N.; Gardiner J.. Submitted for publication.
- Pedrioli P. G. A.; Eng J. K.; Hubley R.; Vogelzang M.; Deutsch E. W.; Raught B.; Pratt B.; Nilsson E.; Angeletti R. H.; Apweiler R.; Cheung K.; Costello C. E.; Hermjakob H.; Huang S.; Julian R. K.; Kapp E.; McComb M. E.; Oliver S. G.; Omenn G.; Paton N. W.; Simpson R.; Smith R.; Taylor C. F.; Zhu W.; Aebersold R. Nat. Biotechnol. 2004, 22, 1459–1466. 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- Chambers M. C.; Maclean B.; Burke R.; Amodei D.; Ruderman D. L.; Neumann S.; Gatto L.; Fischer B.; Pratt B.; Egertson J.; Hoff K.; Kessner D.; Tasman N.; Shulman N.; Frewen B.; Baker T. A.; Brusniak M.-Y.; Paulse C.; Creasy D.; Flashner L.; Kani K.; Moulding C.; Seymour S. L.; Nuwaysir L. M.; Lefebvre B.; Kuhlmann F.; Roark J.; Rainer P.; Detlev S.; Hemenway T.; Huhmer A.; Langridge J.; Connolly B.; Chadick T.; Holly K.; Eckels J.; Deutsch E. W.; Moritz R. L.; Katz J. E.; Agus D. B.; MacCoss M.; Tabb D. L.; Mallick P. Nat. Biotechnol. 2012, 30, 918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G.; Lobley C. M.; Prince S. M. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1260–73. 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P. V.; Grosse-Kunstleve R. W.; Echols N.; Headd J. J.; Moriarty N. W.; Mustyakimov M.; Terwilliger T. C.; Urzhumtsev A.; Zwart P. H.; Adams P. D. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Petersson G. A.; Nakatsuji H.; Li X.; Caricato M.; Marenich A.; Bloino J.; Janesko B. G.; Gomperts R.; Mennucci B.; Hratchian H. P.; Ortiz J. V.; Izmaylov A. F.; Sonnenberg J. L.; Williams-Young D.; Ding F.; Lipparini F.; Egidi F.; Goings J.; Peng B.; Petrone A.; Henderson T.; Ranasinghe D.; Zakrzewski V. G.; Gao J.; Rega N.; Zheng G.; Liang W.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Throssell K.; Montgomery J. A. Jr.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Keith T.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Lyengar S. S.; Tomasi J.; Cossi M.; Millam J. M.; Klene M.; Adamo C.; Cammi R.; Ochterski J. W.; Martin R. L.; Morokuma K.; Farkas O.; Foresman J. B.; Fox D. J.. Gaussian 09 D.01; Gaussian, Inc.: Wallingford, CT, 2016.
- Becke A. D. J. Chem. Phys. 1993, 98, 5648–5652. 10.1063/1.464913. [DOI] [Google Scholar]
- Grimme S.; Antony J.; Ehrlich S.; Krieg H. J. Chem. Phys. 2010, 132, 154104. 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- Oostenbrink C.; Villa A.; Mark A. E.; Van Gunsteren W. F. J. Comput. Chem. 2004, 25, 1656–1676. 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Ha N. C.; Choi G.; Choi K. Y.; Oh B. H. Curr. Opin. Struct. Biol. 2001, 11, 674–678. 10.1016/S0959-440X(01)00268-8. [DOI] [PubMed] [Google Scholar]
- Schwans J. P.; Kraut D. A.; Herschlag D. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 14271–14275. 10.1073/pnas.0901032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansell D. J.; Toogood H. S.; Waller J.; Hughes J. M. X.; Levy C. W.; Gardiner J. M.; Scrutton N. S. ACS Catal. 2013, 3, 370–379. 10.1021/cs300709m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedenburg G.; Jendrossek D.; Breuer M.; Juhl B.; Pleiss J.; Seitz M.; Klebensberger J.; Hauer B. Appl. Environ. Microbiol. 2012, 78, 1055–1062. 10.1128/AEM.07059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygidakis A.; Karuppiah V.; Hoeven R.; Ní Cheallaigh A.; Leys D.; Gardiner J. M.; Toogood H. S.; Scrutton N. S. Angew. Chem., Int. Ed. 2016, 55, 9596–9600. 10.1002/anie.201603785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H. M.; Mirsky E. A.; Voigt C. A. Nat. Biotechnol. 2009, 27, 946–950. 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis H. M. Methods Enzymol. 2011, 498, 19–42. 10.1016/B978-0-12-385120-8.00002-4. [DOI] [PubMed] [Google Scholar]
- Alonso-Gutierrez J.; Chan R.; Batth T. S.; Adams P. D.; Keasling J. D.; Petzold C. J.; Lee T. S. Metab. Eng. 2013, 19, 33–41. 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.