Abstract
Apoptosis signal-regulating kinase 1 (ASK1) is a critical cellular stress sensor that senses diverse reactive chemotypes and integrates these chemical signals into a single biological pathway response. It is unknown whether ASK1 senses all stressors in the same way, or if unique stress-specific mechanisms detect distinct chemotypes. In order to answer this question, we treated ASK1-expressing cells with two distinct stress activators, H2O2 and 4-hydroxy-2-nonenal (HNE), and monitored the phosphorylation state of ASK1. Phosphorylation is an important regulator for the activity of ASK1 and we hypothesized that these two chemically distinct molecules may produce differences in the phosphorylation state of ASK1. Shotgun mass spectrometry and manual validation identified 12 distinct ASK1 phosphosites. Targeted parallel reaction monitoring assays were used to track the phosphorylation dynamics of each confirmed site in response to treatment. Eleven phosphosites exhibited dynamic response to one or both treatments. Six of these sites were identified in both H2O2 and HNE treated cells and four of these exhibited a consistent response between the two molecules. The results confirm that different chemotypes produce distinct phosphorylation patterns in concert with activation of a common MAPK pathway.
Introduction
Apoptosis signal-regulating kinase 1 (ASK1) is a mitogen activated protein kinase kinase kinase that acts as a critical sensor for cell stress. Activation of ASK1 results in downstream activation of P38 and JNK and subsequent stress response and apoptosis.1, 2 ASK1 is activated by diverse stress signals including, but not limited to, reactive oxygen species, cytokines, endotoxins, calcium influx, and electrophilic agents,3-18 although it has been most notably studied for its role in transducing cellular responses to oxidative stress.3-5, 19 An unanswered question in the field is how ASK1 senses these chemotypically diverse molecules and actuates a common downstream response.
ASK1 is thought to transduce stress signals through two mechanisms. ASK1 activation coincides with remodeling of ASK1 multi-protein complexes, which regulate kinase activation.2, 20-23 ASK1 also undergoes dynamic post-translational modifications (PTMs), particularly phosphorylation, which may activate or deactivate ASK1 signaling.21, 24-27
Phosphorylation of Thr-838 activates ASK1. This modification occurs either by ASK1 autophosphorylation or through the action of other kinases.21, 28 In contrast, phosphorylation of the serine residues at positions Ser-83 and Ser-966 have the opposite effect, inactivating ASK1 through the AKT pro-survival pathway (Ser-83)25 and through the formation of an inactive complex including ASK1 and 14-3-3 protein (Ser-966).27 Two other phosphorylation events (Tyr-718 and Ser-1033) have been linked to inhibition of ASK1 activity, but involve unknown mechanisms.
The significance of active PTMs in ASK1 has been evaluated by examining the effects of individual stressors on site-specific ASK1 mutants. This approach has limited throughput and also requires the generation of specific phosphoantibodies for each site to monitor phosphorylation dynamics. Studies of ASK1 regulation through phosphorylation are further complicated by the large size of the protein (150 kDa) and by the limited structural information about this protein, which has not yet been crystallized. For example, 231 serines, threonines and tyrosines within the ASK1 protein all are potential phosphorylation sites. Thirty phosphorylation sites have been previously reported, but only five have been linked to activity, predominantly under conditions of treatment with TNFα19, 24 and H2O2.27, 29 It is not known whether these five sites (and potentially others) serve as master phospho-regulators of ASK1 function for all stress signals or if dynamic phosphorylation at these sites is specific to oxidative stress only. Indeed, the broader question of whether chemically distinct activators of ASK1 signaling produce distinct sets of dynamic phosphorylations has never been addressed.
In order to test the generalizability of the phospho-regulation of ASK1, we chose to map phosphorylation sites in response to treatment of cells with either H2O2 or 4-hydroxy-2-nonenal (HNE). HNE is a prototypical lipid electrophile that is generated endogenously in cells under conditions of oxidative stress, but which acts primarily through covalent adduction of cysteine thiol residues, as opposed to the reversible oxidation of protein thiols by H2O2.30-33 HNE has been previously shown to activate ASK1 and the downstream pathway to a similar degree as H2O2,8, 20 but whether ASK1 is regulated by the same mechanisms in response to these chemically diverse signals is not known.
Because of the differences in chemical mechanisms between H2O2 and HNE, we hypothesized that different dynamic phosphorylation patterns on ASK1 would be observed for each stressor, but which ultimately activate the same stress pathway. To test this hypothesis, we used both data-dependent shotgun liquid chromatography-tandem mass spectrometry (LC-MS/MS) and parallel reaction monitoring (PRM) methods to map and quantify sites of phosphorylation on ASK1 in response to treatment of an ASK1-expressing cell line with H2O2 and HNE. We report here the interplay between oxidative and electrophilic activation of ASK1 and the phosphorylative response of the protein to these stressors.
Experimental Procedures
DNA construct
The tandem-tagged construct for ASK1 (ASK1-TAG) was generated in a pcDNA3.1 plasmid (Life Technologies, Grand Island, NY) by Genscript (Piscataway, NJ). The ASK1 sequence previously described1 was modified to exchange the HA tag for a tandem HA-FLAG tag and is available from Addgene (#69726).
Antibodies and reagents
All ASK1 protein IPs were performed with EZview red anti-FLAG beads (Sigma-Aldrich, St. Louis, MO). Western blots were performed with the following primary antibodies: FLAG (8146S), beta-actin (4967S), SAPK/JNK (9252S), phospho-SAPK/JNK (9255S), P-38 (9212S), phospho-P-38 (9215), ASK1 phosphoThr-838 (3765S), phosphoSer-83 (3761s), phosphoSer-967 (3764S) from Cell Signaling Technology (Danvers, MA), and ASK1 (SC-5294; Santa Cruz Biotechnology, Dallas, TX). Secondary antibodies were obtained from Life Technologies (Grand Island, NY): anti-mouse 680 (A21058) and anti-rabbit 680 (A21109).
Cell lines and cell culture
Stable Transfection
HEK-293 cells (#CRL-1573;ATCC, Manassas, VA) were grown in DMEM (12430; Life Technologies) supplemented with 10% fetal bovine serum (Atlas Biologicals, Fort Collins, CO) at 37°C in a humidified atmosphere with 5% CO2. The ASK1 expressing cell line was generated from HEK-293 cells by transfecting the ASK1-TAG plasmid into cells using Lipofectamine 2000 (Life Technologies) following the manufacturer's instructions. Twenty-four hr following transfection, cells were split 1:10 and after an additional 24 hr were switched into selection medium consisting of the HEK-293 medium described above supplemented with 1.5 mg/mL Geneticin (Life Technologies) and maintained in this medium for two weeks to select for cells expressing the plasmid of interest. Stock cell cultures were maintained thereafter in HEK-293 media supplemented with 1mg/mL Geneticin. Prior to harvesting, cells were plated in 15 cm dishes in HEK-293 media supplemented with 0.5 mg/mL Geneticin.
HNE and H2O2 stimulation
All cells were treated with 20 μM MG-132 (Sigma-Aldrich) in serum-free media for two hr at 37 °C prior to treatment with stressors. Following proteasomal inhibition, cells were treated with ethanol (HNE vehicle, final medium concentration 0.08% v/v), 10 μM HNE, 50 μM HNE, PBS (H2O2vehicle) 500 μM H2O2, or 5000 μM H2O2. The treatments were prepared in serum-free media and placed on the cells for one hr at 37°C. Cells then were harvested with a cell scraper in the treatment medium and immediately centrifuged at 100 × g for five min at 4°C. The cell pellets then were washed twice with ice cold phosphate buffered saline (PBS) and stored at -80°C until use.
Cell viability assays
Cells were plated in 96-well plates at a density of 10,000 cells/well and allowed to grow for 24 hr. Medium was then removed and replaced with 100 μL/well of serum-free DMEM containing the specified concentration of HNE or H2O2 and the cells were incubated at 37°C for 24 hr. To measure cell viability, 10 μL/well of WST reagent (Sigma, 11644807001) was added and the cells were incubated for 1 hr. Absorbance readings then were taken at 450 nm and 650 nm with a Spectramax M4 spectrophotometer (Molecular Devices, Sunnyvale, CA) according to the manufacturer's protocol.
Immunoprecipitation and western blotting
Frozen cell pellets were lysed on ice in 500 μL of NETN buffer (50 mM HEPES pH 7.5, 150 mM NaCl, and 1% Igepal supplemented with 10 μL/mL of HALT protease and phosphatase inhibitor (Life Technologies), per 15 cm plate for 30 min with occasional inversion. Lysates then were clarified by centrifugation at 10,000 × g for 10 min at 4°C. Protein in the clarified lysates was measured with the BCA assay (Life Technologies). Protein concentrations were then adjusted to 2 mg/mL. Next, 5 mg of total protein was added in a final volume of 2.5 mL to antibody beads pre-washed with NETN. For each IP, a 50 μL slurry of antibody-bead conjugate was washed twice with 1 mL of NETN prior to incubation with protein lysates. After addition of the protein lysate, the beads were rotated at 4°C for 1 hr to allow for capture of the target protein. Following incubation, the beads were pelleted by centrifugation at 100 × g and the supernatant was discarded. The beads were then washed twice with 1 mL of NETN buffer and the bound protein was resuspended in 80 μL of lithium dodecyl sulfate (LDS) sample buffer diluted 1:1 with NETN and supplemented with 50 mM dithiothreitol (DTT). The samples were then frozen at -80°C until use.
For western blot analysis, cell lysis and IP were carried out as described above. Gels for sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) were loaded with either 20 μg of input protein or 2 μL of IP and run for 50 min at a constant 180 V. Proteins then were transferred to polyvinylidene fluoride (PVDF) membranes (Life Technologies) using the BioRad (Hercules, California) wet transfer system operated at a constant 300 mA current for 90 min at 4°C. Membranes were blocked in a 1:1 mixture of Tris-buffered saline containing 0.05% Tween-20 (TBST) and blocking buffer (Cat. # MB-070; Rockland, Limerick, PA) for one hr at room temperature while rocking. Primary antibodies were diluted (1:1000 for tag and protein antibodies, and 1:500 for phospho antibodies) in the same buffer used for blocking, added to the membrane, and incubated at 4°C overnight with rocking. After incubation, the membranes were washed three times with blocking buffer, incubated with the appropriate secondary antibody diluted 1:10,000 in the same buffer used for the primary antibody, and allowed to rock for 30 min at room temperature. The membranes then were washed three times with TBST, visualized using a LiCor Odyssey (Lincoln, Nebraska) instrument, and analyzed using the Image Studio Lite software (LiCor).
Preparation of peptides for MS analyses
Bead-bound ASK1 protein was eluted in LDS buffer and heated at 95°C for 10 min prior to SDS-PAGE on a 10% Bis-Tris gel (Life Technologies). The gels were loaded with 35 μL of each sample elution and then were run at a constant 180 V for three min, after which the run was paused. Each sample lane was then re-loaded with the remaining 35 μL of each sample and the gel runs were resumed for an additional 47 min. Gels were then stained with Simply Blue safe stain (Life Technologies) for one min in a microwave oven at maximum power and then allowed to destain in distilled water for two-three hr. Each sample lane was cut as a single band and diced into ∼1 mm cubes. The gel pieces were placed into individual Eppendorf tubes and washed twice with 200 μL of 100 mM ammonium bicarbonate (AmBic). Samples then were reduced with 5 mM dithiothreitol in AmBic for 30 min at 60°C while shaking at 1000 rpm and then alkylated with 10 mM iodoacetamide in AmBic for 20 min in the dark at room temperature. Excess blue dye was then removed with three 200 μL washes in 50 mM AmBic:acetonitrile (1:1, v/v) and the gel pieces were dehydrated in 100% acetonitrile. The gel pieces were rehydrated in 200 μL of 25 mM AmBic with 200 ng of Trypsin Gold™ (Promega, Madison, WI) per sample and placed in a 37°C incubator for 16 hr. Peptides were extracted from the SDS gel with three 20 min incubations in 200 μL of 60% acetonitrile / 40% water containing 1% formic acid; each extract was evaporated to dryness in vacuo. The peptides were redissolved in 30% acetonitrile / 70% water containing 0.1% formic acid and stored at -80°C until use.
Mass spectrometry
Shotgun LC-MS/MS
Shotgun analyses of the single gel fraction samples were carried out on a Q Exactive Plus mass spectrometer (ThermoFisher Scientific, San Jose, CA) equipped with a Proxeon nLC1000 LC (ThermoFisher Scientific) and a Nanoflex source (ThermoFisher Scientific). Peptide mixtures were evaporated in vacuo and resuspended in 2% acetonitrile / 98% water containing 0.1% formic acid and loaded onto an 20 cm long column with a 75 μm internal diameter (Cat. # PF360-75-10-N-5; New Objective, Woburn, MA) packed with 3 μm particle size and 120 Å pore size ReproSil-Pur C18-AQ resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and separated over a 180 min gradient with a mobile phase containing aqueous acetonitrile and 0.1% formic acid programmed from 2-5% acetonitrile over 5 min, then 5 to 25% acetonitrile over 150 min, then 25-90% acetonitrile over 5 min, followed by 20 min at 90% acetonitrile, all at a flow rate of 300 nL/min. A single MS1 scan from m/z 300-1800 at 70,000 resolution with an automatic gain control (AGC) value of 3e6 and max injection time of 64 msec was recorded as profile data. A top 10 method was used, whereby the 10 most intense precursors were automatically chosen for MS2 analysis and a dynamic exclusion window of 20 sec was employed. For each MS2 scan, a resolution of 17,500, an AGC value of 1e5, a max injection time of 175 msec, a 1.4 m/z isolation window, and a normalized collision energy of 27 was used and centroid data were recorded.
Parallel reaction monitoring (PRM) targeted MS
PRM assays were performed on the same Q Exactive Plus instrument and LC system using the same gradient described above. The PRM method consisted of an MS1 scan at 17,500 resolution with an AGC value of 3e6, max injection time of 30 msec, and scan range from m/z 380-1500 recorded as profile data. This was followed by 10 targeted MS2 scans at a resolution of 17,500 and with an AGC value of 1e5, a max injection time of 175 msec, a 0.5 m/z isolation window, a fixed first mass of 150 m/z, normalized collision energy of 27, and recorded as profile data. The targeted-MS2 methods were controlled with a timed inclusion list containing the target precursor m/z value, charge, and a retention time window that was determined from shotgun analyses.
Data analysis
Shotgun datafiles were converted to mzml format using Proteowizard version 3.0.5211.34 The mzml files were searched using MS-GF+ version 951735 against the human Refseq version 54 database (Sep 25, 2012; 34,590 entries). Following the initial search, the mzml files were researched against a subset FASTA database generated from the initial search results, which consisted of 277 entries in the H2O2 experimental set and 315 entries in HNE experimental set; both initial search results corresponded to a 1% peptide FDR. A semi-tryptic search was employed with a maximum of four missed cleavages allowed. A fixed carbimidomethyl modification on cysteine, a variable oxidation on methionine, and a pyro-glu on glutamine were allowed with a maximum of 3 dynamic modifications per peptide with a precursor ion tolerance of 10 ppm and a fragment ion tolerance of 20 ppm. For phosphorylation studies, an additional variable modification of 79.966331 Da on serine, threonine, or tyrosine was employed. A target-decoy search was employed using a reverse sequence database to allow calculation of FDR for peptide-spectrum matches36. Protein-level FDR was calculated by dividing the number of reverse sequence proteins identified by the total number of proteins identified, multiplying by two and converting to a percentage. All search result files were parsimoniously assembled in IDPicker 3 version 3.1.643.0.37
PRM runs were designed and analyzed using Skyline38 and the three most intense transitions per peptide were used for quantitation. To normalize the data between independent runs, the integrated peak area for each peptide was divided by the total quantifiable signal (TQS) observed in each run. The TQS value corresponds to the sum of the peak areas corresponding to all transitions from all the peptides targeted in each run. This normalization corrected for both instrumental variance and slight sample-to-sample discrepancies that occurred during immunoprecipitation.
All phosphorylation site assignments were confirmed by manual spectrum annotation as described.39 For each peptide, precursor purity was confirmed by MS1, major peaks were unambiguously assigned and direct evidence of site modifications were required (i.e., b- and/or y-ion fragments containing the modification). The precursor purity was confirmed by examining the MS1 scan immediately prior to peptide isolation to determine if any m/z signal other than from the targeted peptide fell within the isolation window employed for the run (1.4 m/z for shotgun or 0.5 m/z for PRM). If any other signal was present at more than 10% of the intensity of the targeted precursor, this spectrum was not considered for further evaluation. Additionally, the selected peptide peak was required to have a mass error in the MS1 scan no larger than 10 ppm. Spectra that passed this filter then were manually annotated using mMass version 5.5.040, 41, which allowed for automated spectral annotations of b- and y-ions, internal fragment ions, neutral loss ions for H2O, NH3, CO, and H3PO4, as well as annotations of PTM-specific mass-shifted ions. We required that the assigned peaks explain at least 60% of the total ion current for each spectrum to be considered acceptable. Furthermore, for peptides with phosphorylated threonine or serine residues present, we required the presence of direct evidence of the site modification in the form of a phosphoric acid neutral loss peak (from either the precursor or a fragment ion) or specific mass shifted ions for the spectrum to be accepted. We then divided the peptides that passed all of these filters into two categories: peptides with and without ambiguous site localizations. Peptides with ambiguous PTM site localizations were those where more than one modifiable residue was present in the sequence such that the PTM could not be definitively assigned to one residue (e.g., a peptide with two sequential serine residues). These ambiguous PTM sites were counted as single phosphosites because they represented a single phosphorylation event in the sequence. Unambiguous PTM assignments were those spectra where no other site-assignment of the PTM could reasonably explain the MS/MS data.
Experimental Design and Statistical Rationale
Treatment of ASK1-TAG cells by HNE and H2O2 were each performed in biological quadruplicate (i.e., in four separate cultures of cells). PRM analyses of phosphorylated peptides in HNE treated cells were performed in triplicate, all other MS assays for phosphopeptide detection and quantitation were performed in quadruplicate. The HNE adduct site mapping experiments were performed in biological triplicate. Statistical comparisons of the relative amount of each phosphorylated peptide were performed using the Student's t-test as pair-wise comparisons of each treatment concentration to the zero level treatment. Replicate numbers were chosen to be sufficient to evaluate reproducibility and provide enough data points for the t-test.
Results
Activation of the ASK1 MAPK signaling pathway by HNE and H2O2 in HEK-293 cells
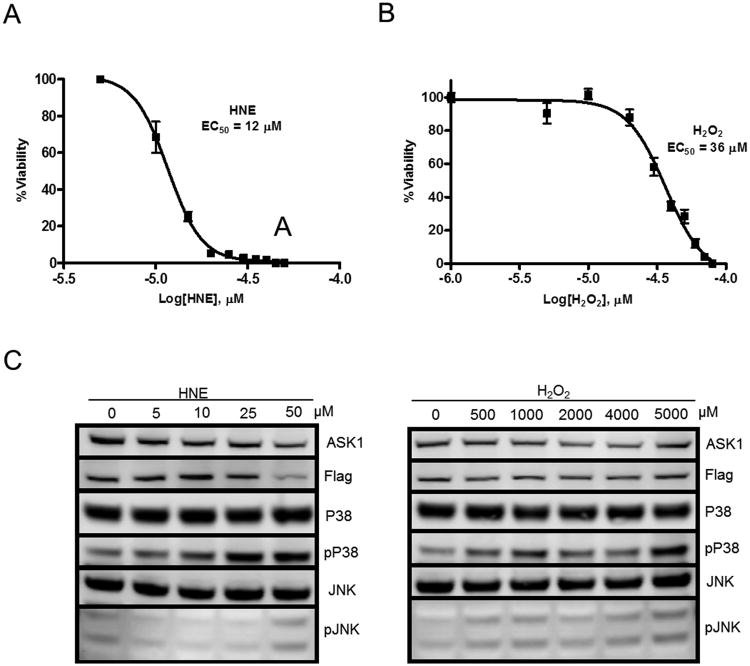
In order to investigate the differential stress sensing ability of ASK1, we first stably expressed a tandem tagged version of the ASK1 protein (ASK1-TAG) in HEK-293 cells. We then characterized the response of these cells to HNE and H2O2 treatment using WST cell survival assays and Western blot-based MAPK pathway activation assays. In the 24 hr WST assay, the treated cells were found to have an EC50 concentration for cytotoxicity of approximately 10 μM for HNE and 36 μM for H2O2 (Figure 1 A & B). Despite the low EC50 value obtained for the H2O2 cells, this concentration in a one hr exposure produced minimal stress activation of the ASK1 MAPK pathway (data not shown). Thus, we conducted a dose-escalation experiment with higher concentrations of H2O2 to find a one hr treatment level at which the downstream pathway was activated similarly to treatment with HNE at 10 μM and 50 μM (Figure 1C). Based on the results of these pathway activation Western analyses, we used 0, 10, and 50 μM HNE and 0, 500, and 5000 μM H2O2 for all further studies.
Figure 1. ASK1-TAG cell characterization.
Twenty-four hr WST-based cell death assays were performed with (A) HNE and (B) H2O2 to determine the EC50 concentration for each compound in the cell line. (C) Western blot showing activation of the ASK1 MAPK pathway after one hr treatment with each compound at varying concentrations
Identification and quantitation of ASK1 phosphopeptides
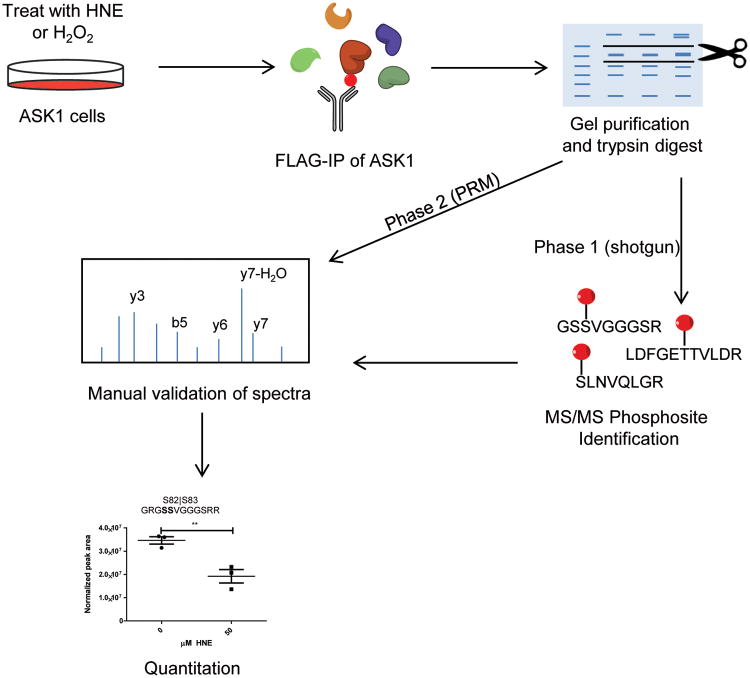
In order to identify phosphorylated ASK1 peptides, the ASK1-cells were first pretreated with MG-132 to inhibit proteasomal protein degradation and then treated with either HNE or H2O2 at the concentrations described above. Following treatment, cells were harvested and lysed, and the tagged ASK1 protein was immunoprecipitated with anti-FLAG beads. The ASK1 was recovered and further purified by 1-dimensional SDS-PAGE and analyzed by MS as shown in Figure 2. A one hour treatment was used for phosphosite mapping studies to capture initial responses to HNE or H2O2, rather than subsequent adaptations to the onset of toxicity. Moreover, longer timecourse studies were precluded by the toxicity of MG132.
Figure 2. Workflow for identification, validation, and quantitation of phosphorylated peptides on ASK1.
Phase 1 consisted of a series of discovery experiments to generate a list of putative phosphorylation sites on ASK1. Phase 2 consisted of a targeted reanalysis of the same samples via a PRM method followed by quantitation of validated phosphorylation sites.
We employed a two-step approach to identifying phosphorylated ASK1 peptides. The first step was a data-dependent shotgun analysis of the immunopurified samples in order to identify potential sites of phosphorylation (Figure 2, phase 1). We next targeted all of the identified sites by reanalyzing the same samples with a scheduled PRM method to quantify each peptide (Figure 2, phase 2). We then performed manual annotation of the putative phosphopeptide spectra obtained in the PRM analysis to confirm each phosphorylation. PRM data for these validated sites were used to quantify each phosphopeptide under the treatment conditions compared to the control (0 μM) level.
Table 1 lists all of the phosphopeptides and phosphosites that were identified in the shotgun and PRM analyses and that passed our manual validation criteria. For some of these phosphopeptides, the exact site of phosphorylation could not be unambiguously localized from the MS/MS data. For purposes of this study, phosphorylation events that were localized to either of two residues within a peptide are referred to hearafter as phosphosites, together with phosphorylation events that were unambiguously localized. In the HNE treated samples, an initial list of 25 putative phosphopeptides from the shotgun data was reduced to 14 manually validated phosphopeptides. In the H2O2 treated samples, an initial list of 23 putative phosphopeptides was reduced to 11 manually validated phosphopeptides. These phosphopeptides supported identification of eight distinct phosphosites in H2O2-treated cells and eight phosphosites in HNE-treated cells; five of these were present in both HNE and H2O2 treated samples. Of the 12 identified phosphosites, seven have not been previously reported. Annotated MS/MS spectra for each of these sites are provided in Figure S1.
Table 1.
Manually validated ASK1 phosphorylation sites identified by shotgun and PRM MS.
| Status1 | A.A. site | Stimulation | Identified sequences |
|---|---|---|---|
| Known | S82/83 | Both | GSSVGGGSRR |
| Known | S82/83 | Both | GRGSSVGGGSR |
| Known | S82/83 | Both | GRGSSVGGGSRR |
| Known | S82/83 | H2O2 | GSSVGGGSR |
| Novel | T140/141 | HNE | LDFGETTVLDR |
| Novel | S268/269 | H2O2 | VAQASSSQYFR |
| Novel | Y355 | H2O2 | FHYAFALNR |
| Known | T947/S952 | HNE | KKKTQPKLSALSAGSNEYLR |
| Known | T947/S952 | Both | KKTQPKLSALSAGSNEYLR |
| Novel | S955 | Both | LSALSAGSNEYLR |
| Known | S958 | Both | LSALSAGSNEYLR |
| S955/S958 | HNE | TQPKLSALSAGSNEYLR | |
| S955/S958 | HNE | KTQPKLSALSAGSNEYLR | |
| Novel | T1000/S1004 | HNE | TRAKSCGERDVKGIR |
| Novel | T1000/S1004 | HNE | TRAKSCGERDVK |
| Novel | S1004 | Both | AKSCGERDVK |
| Known | T1059 | H2O2 | ILTEDQDKIVR |
| Novel | S1240 | HNE | SLNVQLGR |
Known=previously reported site, Novel=unreported phosphorylation site
Incomplete digestion consistently yielded multiple peptides representing four phosphorylated sequences (Table 1). The sequence containing Ser82/83 was represented by four distinct peptides. The sequence containing Thr947 and Ser952 was represented by two distinct peptides. The sequence containing Ser955 and Ser958 was represented by four distinct peptides and the sequence representing Thr1000 and Ser1004 was represented by four distinct peptides.
Of the five known ASK1 dynamic phosphorylation sites, only one (Ser-83) was detected in these MS experiments. Of the other four sites, Tyr-71824, 42 has been studied in the context of interferon gamma and TNFα stress and Ser-103326 was examined in the context of serum withdrawal and have not been observed in studies of H2O2 stress. Ser-966 previously was reported to be dynamic in response to oxidative stress, but was not observed here, possibly due to the fact that the tryptic peptide would have been long (28 amino acids) and may not have ionized well. Even if it had been detected, it would have contained 8 serine residues, which would have made site localization difficult.
Thr-838 is the only known activating site on ASK1 and is expected to be observed as dynamically phosphorylated in response to both H2O2 and HNE. However the tryptic peptide containing this site was not detected in the shotgun or PRM runs, is also long (24 amino acids), and contains four threonine residues. PRM analyses failed to detect a synthetic standard for the phosphorylated Thr-838 peptide, except at very high concentrations (data not shown), which indicates that this peptide may be poorly ionized.
Comparison of MS methodology with antibody-based detection
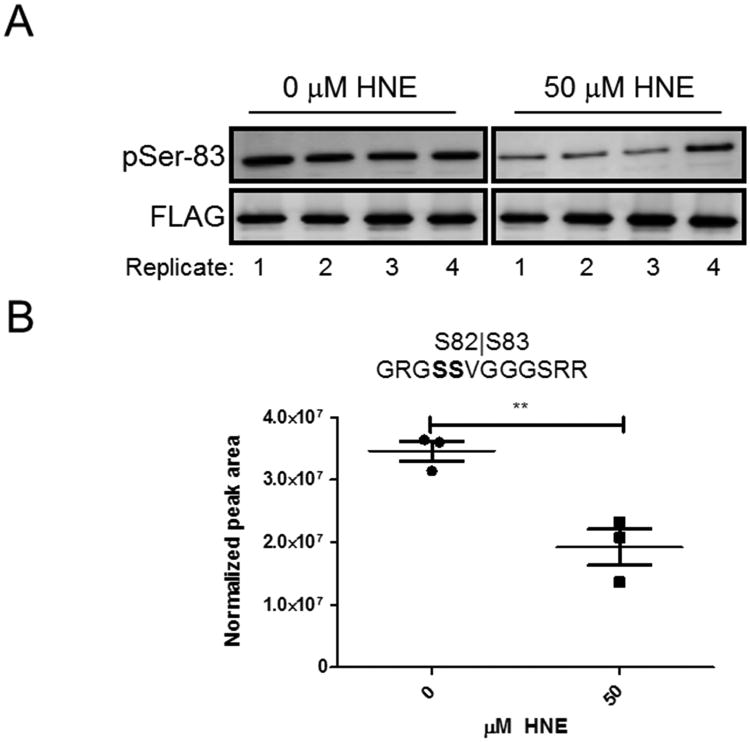
We compared Western blot based results for a well-characterized phosphorylation in ASK1 (Ser-83) to our PRM results (Figure 3). In response to increasing HNE, both the Western blot and the PRM show a decrease in the amount of phosphorylation at Ser-83 (compare Figure 3B with Figure 3D). Because the peptide that contains Ser-83 also contains a serine at position 82, we were not able to differentiate between the two potential phosphorylation sites with the MS/MS data. However, the assignment to Ser-83 is supported by previous literature evidence, which includes mutational analyses.25, 43
Figure 3. Comparison of Western blot and PRM for pSer-83 detection.
(A) Western blot of ASK1-pSer-83 and the FLAG tag in ASK1-TAG cells with or without HNE treatment. (B) Quantitation of the pSer-83 containing peptide in PRM assays at both concentration points.
Dynamics of phosphorylation sites in HNE and H2O2 treated samples
In ASK1-TAG cells treated with three concentrations of HNE, we quantified 13 phosphopeptides covering eight different phosphorylation sites (Figure S2). Of these six phosphosites, five exhibited a significant dynamic response to HNE treatment at one or both treatment levels compared to untreated samples (Table 2). Ser955/Ser958 displayed a 7-fold phosphorylation increase at 10 μM HNE, but no significant phosphorylation change at 50 μM HNE. Small changes (<2-fold) on several other phosphosites were not HNE concentration-dependent, except for phosphorylation at Thr947/Ser952.
Table 2.
ASK1 phosphorylation dynamics in response to increasing concentration levels of HNE and H2O2.
| Peak area ratio HNE1 | Peak area ratio H2O21 | ||||
|---|---|---|---|---|---|
|
| |||||
| Module | Peptide Modified Sequence2 | 0 to 10μM | 0 to 50μM | 0 to 500μM | 0 to 5000μM |
|
| |||||
| S82 GRGSSVGGGSR | 0.7 | 4.9** | |||
| S82 GSSVGGGSRR | 0.7 | 6.5*** | |||
| S82 GSSVGGGSR | 0.9 | 2.4 | |||
| A | S83 GRGSSVGGGSRR | 0.7 | 3.6**** | ||
| S82|S83 GRGSSVGGGSR | 0.9 | 0.7 | |||
| S82|S83 GRGSSVGGGSRR | 0.6 | 0.6** | |||
| S82|S83 GSSVGGGSRR | 0.9 | 0.6 | |||
|
| |||||
| B | YT140 LDFGETTVLDR | 2.1* | 2.5 | ||
|
| |||||
| C | S269 VAQASSSQYFR | 1.3 | 1.7 | ||
|
| |||||
| D | Y355 FHYAFALNR | 0.2* | 0.4* | ||
|
| |||||
| T947|S952 KKTQPKLSALSAGSNEYLR | 0.7* | 0.1**** | |||
| E | T947|S952 KKKTQPKLSALSAGSNEYLR | 1.0 | 0.2** | ||
| S952KKKTQPKLSALSAGSNEYLR | 1.1 | 1.9 | |||
|
| |||||
| S955 LSALSAGSNEYLR | 1.5** | 2.8*** | |||
| S958 LSALSAGSNEYLR | 1.6* | 2.3** | |||
| F | S955|S958 KTQPKLSALSAGSNEYLR | 7.0*** | 1.1 | ||
| S955|S958 LSALSAGSNEYLR | 0.9 | 0.7 | |||
| S955|S958 TQPKLSALSAGSNEYLR | 2.4** | 0.7 | |||
|
| |||||
| T1000|S1004 TRAKSC[+57]GERDVK | 0.2* | 1.8 | |||
| G | T1000|S1004 TRAKSC[+57]GERDVKGIR | 2.0** | 1.4 | ||
| S1004 AKSC[+57]GERDVK | 0.5* | 0.8* | 0.6 | 12.8**** | |
|
| |||||
| H | S1059 ILTEDQDKIVR | 2.8 | 2.9* | ||
|
| |||||
| I | S1240 SLNVQLGR | 1.0 | 0.8 | 0.9 | 0.7** |
Peak areas for the indicated peptides were measured by PRM. Peak area ratio values of 1 indicates there was no difference detected between treatments, values >1 indicate an increase, and values<1 indicate a decrease in phosphopeptide peak area after treatment. Significance is included as follows
p≤0.05,
p≤0.01,
p≤0.001. and
p≤0.0001, Student's T-test.
Phosphorylated residue is highlighted in bold.
Treatment of the cells with H2O2, evoked more significant, concentration-dependent phosphorylation changes. We quantified 12 phosphopeptides covering nine different phosphorylation sites (Figure S3). Of these nine phosphosites, seven displayed significant concentration-based dynamic responses to at least one of the treatment levels compared to the zero treatment level (Table 2). The changes produced by H2O2 were distinct in magnitude and direction from those produced by HNE. For example, Ser82/Ser83 phosphorylation increased up to 6.5-fold, as measured by 3 peptides, whereas phosphorylation of this site was marginally decreased by HNE. Similarly, whereas HNE decreased phosphorylation at Ser1004, 5 mM H2O2 increased phosphorylation at the same site by 12.8-fold (Table 2).
Antibody-based analysis of phosphorylation dynamics
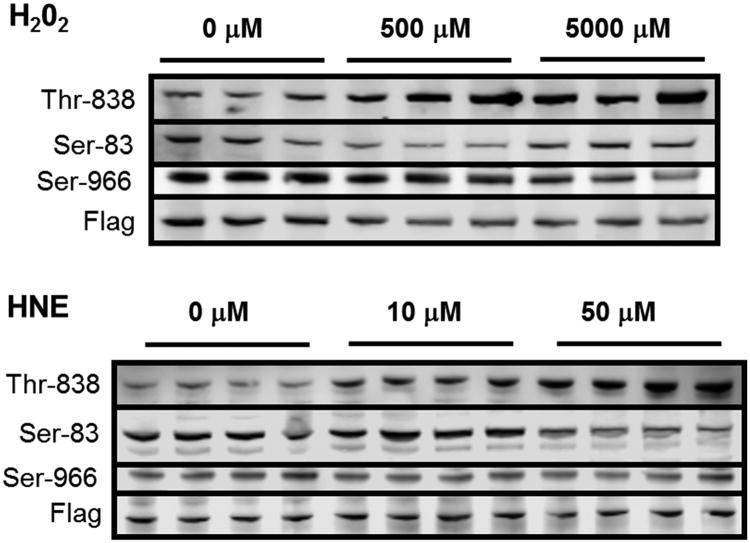
Of the previously reported dynamic phosphorylation sites, we only were able to observe Ser-83 with our MS method. Several of the other phosphorylation sites were on tryptic peptides that are too long or short for successful MS analysis. To analyze these sites, we performed Western blots with the three commercially available ASK1 anti-phosphoantibodies (Figure 4). In addition to Ser-83, we were able to measure the dynamics of phosphorylated Thr-838 and Ser-966. Both of these residues exhibited dynamic changes consistent with previous literature – Thr-838 phosphorylation increased with increasing H2O2 and HNE treatment and Ser-966 phosphorylation decreased with increasing H2O2 and HNE treatment.
Figure 4. Western blot data for ASK1 activation.
Phosphorylation of Thr-838, Ser-83, and Ser-966 on ASK1 was examined with increasing concentration of (A) H2O2 and (B) HNE after a one hr treatment.
Discussion
Of the 231 serine, threonine, and tyrosine residues which could be potentially phosphorylated in ASK1, only five have been shown to regulate its kinase activity. Four sites (Ser-83, Tyr-718, Ser-966, and Ser-1033)24-27, 42 have been reported to negatively regulate ASK1, whereas one site (Thr-838)21, 28 has been reported to enhance activity. All five dynamic sites were identified through mutagenesis experiments and have been studied only in the context of perturbation with the broad-spectrum oxidant H2O2 and the cytokine TNFα. However, ASK1 is responsible for the transduction of stress signals from many other chemotypes including electrophiles and other reactive oxidant species. Here we demonstrate that both HNE and H2O2 result in enhanced phosphorylation of Thr-838, but that these chemicals produce other distinct, largely non-overlapping phosphorylation changes at other sites (Table 2) These data suggest new hypotheses for the chemotype-selectivity of ASK1-mediated stress signal transduction.
To interpret the differential phosphorylation dynamics of ASK1 sites in response to HNE and H2O2, we have assigned the sites to 9 modules (A-I) as shown in Table 2. The modules contain distinct phosphorylation sites and some modules include multiple sites that are not distinguishable from the MS/MS spectra. Modules B-D contain phosphosites only detected in H2O2-treated cells, whereas module H contains a phosphosite only detected in HNE treated cells. The remaining modules contain phosphosites detected in both H2O2- and HNE-treated cells. However, the dynamics of the H2O2- and HNE-responsive sites were frequently distinct.
Module A contains Ser-82 and Ser-83; the latter is well known as a dynamically modified site in ASK1. Previous reports described a decreased Ser-83 phosphorylation upon treatment with oxidants and subsequent activation of ASK1.25, 44 HNE treatment also decreased Ser-83 phosphorylation, whereas H2O2 (5 mM) increased pSer-83. This is counter to what has been previously reported, but previous studies typically only examined phosphorylation of this residue after H2O2 treatments at lower concentrations.44-46 It is therefore possible that this residue is dephosphorylated at lower oxidant concentrations and then increasingly phosphorylated at higher oxidant levels. Importantly, AKT1 is the kinase known to phosphorylate ASK1 at Ser-83 and H2O2 has been shown to stimulate AKT1 activity in a time- and dose-dependent manner.47-49 Thus, there may be a switch point at which the activity of AKT1 outcompetes the activity of the phosphatase that dephosphorylates Ser-83.
Module E contains Thr-947 and Ser-952, whose phosphorylation was indistinguishable in HNE-treated cells, but which were strongly decreased by HNE. In contrast, phosphorylation of Ser-952 was detectable, but not significantly increased by H2O2. Module F contains Ser-955 and Ser-958, which both exhibited increased phosphorylation upon treatment with both stressors. HNE-induced phosphorylation was increased only at 10 μM, whereas H2O2-induced phosphorylation was highest at 5 mM.
Interestingly, Ser-955 and 958 are both located relatively close to a known 14-3-3 docking site (residues 963-968) in ASK127, 50 and could potentially influence 14-3-3 binding. It was previously shown that dephosphorylation of Ser-966 correlated with decreased 14-3-3 binding and increased ASK1 activity.27 Here we observed that increased concentrations of H2O2 decreased pSer-966, as detected by Western blotting, but no decrease in this site was observed in response to HNE treatment. Our recently published study20 reported that in ASK1 expressing cells, the 14-3-3 association with ASK1 either increased or remained constant in comparison to the control in response to HNE treatment. This is consistent with our current observation that phosphorylation of Ser-966 (14-3-3 docking site) remains unchanged during HNE stimulation but decreases with H2O2 treatment. ASK1 thus may respond to various stimuli with different alterations of phosphorylation or protein-protein interactions.
Module G contains Thr-1000 and Ser-1004 and also was responsive to both stressors. In the H2O2 treated cells, Ser-1004 underwent a dramatic, concentration-dependent increase in phosphorylation. However, in the HNE treated cells, Ser-1004 phosphorylation either decreased or increased, depending on which peptide was measured, which may indicate differential phosphorylation of Thr-1000 and Ser-1004. This difference in response patterns again suggests that ASK1 senses chemically distinct stressors by different mechanisms.
Our data demonstrate that chemically distinct activators both induce phosphorylation of Thr-838, which is the signature activating phosphorylation for ASK1. Nevertheless, HNE and H2O2 produce distinct patterns of altered ASK1 phosphorylation. Both stressors alter multiple phosphorylations in modules A, E, F, G and I, but many of these changes differ either in direction (increase versus decrease) or in stressor concentration dependence. Although H2O2 and HNE produce well-characterized protein modifications, we do not yet have any information how these alter ASK1 phosphorylation. Our attempts to detect HNE adducts on ASK1 from treated cells yielded insufficient quantities of modified peptides for reliable analysis (data not shown). The presence of a core group of phosphorylated residues in ASK1 that are dynamically altered by two dissimilar chemotypes could start to explain how a single sensor protein is able to transduce so many different chemical signals into a single biological pathway response. It is possible that many or all of the stressors that activate ASK1 will exert fine control over the phosphorylation of Thr-838, Ser-955, and Ser-958 residues to control the activity of ASK1. Future studies with additional stress activators of ASK1 will be necessary to test this hypothesis.
Supplementary Material
Acknowledgments
Funding Information: This work was supported by National Institutes of Health Grant R01ES022936 (to D. C. L. and B.M.). J. D. F. and C. M. B. were supported by the Training Program in Environmental Toxicology T32ES007028. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- AGC
automatic gain control
- AmBic
ammonium bicarbonate
- AKT1
Protein Kinase B
- ASK1
Apoptosis Signal-regulating Kinase 1
- DTT
dithiothreitol
- EC50
half maximal effective concentration
- FBS
fetal bovine serum
- HNE
4-hydroxy-2-nonenal
- IP
immunoprecipitation
- IRP
internal reference peptide
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LDS
lithium dodecyl sulfate
- LRP
labelled reference peptide
- MAPK
mitogen-activated protein kinase
- MG132
cell-permeable protease inhibitor
- MS
mass spectrometry
- NETN
50mM hepes, 150mM NaCl, 1% igepal lysis buffer pH 7.5
- PBS
phosphate buffered saline
- PRM
parallel reaction monitoring
- PTM
post-translational modification
- PVDF
polyvinylidene fluoride
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide electrophoresis
- TBST
tris-buffered saline plus 0.05% Tween-20
- TNFα
tumor necrosis factor alpha
- TQS
total quantifiable signal
Footnotes
Supporting information: The complete list of proteins identified during this study and its respective spectra counts can be find in the supplemental documents 150107_IDpicker_Shotgun_MS_H2O2_set.xls and 150728_IDpicker_Shotgun_MS_HNE_set.xls. The shotgun RAW files can be found at at ftp://MSV000079606@massive.ucsd.edu and the PRM skyline files can be downloaded at https://panoramaweb.org/labkey/Liebler_ASK_PTM.url. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 2.Shiizaki S, Naguro I, Ichijo H. Activation mechanisms of ASK1 in response to various stresses and its significance in intracellular signaling. Advances in biological regulation. 2013;53:135–144. doi: 10.1016/j.jbior.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Noguchi T, Naguro I, Ichijo H. Apoptosis signal-regulating kinase 1 in stress and immune response. Annual review of pharmacology and toxicology. 2008;48:199–225. doi: 10.1146/annurev.pharmtox.48.113006.094606. [DOI] [PubMed] [Google Scholar]
- 4.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. The EMBO journal. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuzawa A, Saegusa K, Noguchi T, Sadamitsu C, Nishitoh H, Nagai S, Koyasu S, Matsumoto K, Takeda K, Ichijo H. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nature immunology. 2005;6:587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 6.Takeda K, Matsuzawa A, Nishitoh H, Tobiume K, Kishida S, Ninomiya-Tsuji J, Matsumoto K, Ichijo H. Involvement of ASK1 in Ca2+-induced p38 MAP kinase activation. EMBO reports. 2004;5:161–166. doi: 10.1038/sj.embor.7400072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & development. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soh Y, Jeong KS, Lee IJ, Bae MA, Kim YC, Song BJ. Selective activation of the c-Jun N-terminal protein kinase pathway during 4-hydroxynonenal-induced apoptosis of PC12 cells. Molecular pharmacology. 2000;58:535–541. doi: 10.1124/mol.58.3.535. [DOI] [PubMed] [Google Scholar]
- 9.Lee KW, Zhao X, Im JY, Grosso H, Jang WH, Chan TW, Sonsalla PK, German DC, Ichijo H, Junn E, Mouradian MM. Apoptosis signal-regulating kinase 1 mediates MPTP toxicity and regulates glial activation. PLoS One. 2012;7:e29935. doi: 10.1371/journal.pone.0029935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim PL, Liu J, Go ML, Boelsterli UA. The mitochondrial superoxide/thioredoxin-2/Ask1 signaling pathway is critically involved in troglitazone-induced cell injury to human hepatocytes. Toxicol Sci. 2008;101:341–349. doi: 10.1093/toxsci/kfm273. [DOI] [PubMed] [Google Scholar]
- 11.Usuki F, Fujita E, Sasagawa N. Methylmercury activates ASK1/JNK signaling pathways, leading to apoptosis due to both mitochondria- and endoplasmic reticulum (ER)-generated processes in myogenic cell lines. Neurotoxicology. 2008;29:22–30. doi: 10.1016/j.neuro.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa H, Maeda S, Hikiba Y, Ohmae T, Shibata W, Yanai A, Sakamoto K, Ogura K, Noguchi T, Karin M, Ichijo H, Omata M. Deletion of apoptosis signal-regulating kinase 1 attenuates acetaminophen-induced liver injury by inhibiting c-Jun N-terminal kinase activation. Gastroenterology. 2008;135:1311–1321. doi: 10.1053/j.gastro.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Shinkai Y, Iwamoto N, Miura T, Ishii T, Cho AK, Kumagai Y. Redox cycling of 1,2-naphthoquinone by thioredoxin1 through Cys32 and Cys35 causes inhibition of its catalytic activity and activation of ASK1/p38 signaling. Chem Res Toxicol. 2012;25:1222–1230. doi: 10.1021/tx300069r. [DOI] [PubMed] [Google Scholar]
- 14.Kuo CT, Chen BC, Yu CC, Weng CM, Hsu MJ, Chen CC, Chen MC, Teng CM, Pan SL, Bien MY, Shih CH, Lin CH. Apoptosis signal-regulating kinase 1 mediates denbinobin-induced apoptosis in human lung adenocarcinoma cells. J Biomed Sci. 2009;16:43. doi: 10.1186/1423-0127-16-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon MJ, Jeong KS, Choi EJ, Lee BH. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD)-induced activation of mitogen-activated protein kinase signaling pathway in Jurkat T cells. Pharmacol Toxicol. 2003;93:186–190. doi: 10.1034/j.1600-0773.2003.930406.x. [DOI] [PubMed] [Google Scholar]
- 16.Pramanik KC, Srivastava SK. Apoptosis signal-regulating kinase 1-thioredoxin complex dissociation by capsaicin causes pancreatic tumor growth suppression by inducing apoptosis. Antioxid Redox Signal. 2012;17:1417–1432. doi: 10.1089/ars.2011.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang M, Shen X. Critical role of ASK1 in the 6-hydroxydopamine-induced apoptosis in human neuroblastoma SH-SY5Y cells. J Neurochem. 2006;97:234–244. doi: 10.1111/j.1471-4159.2006.03730.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang W, Tiffany-Castiglioni E, Koh HC, Son IH. Paraquat activates the IRE1/ASK1/JNK cascade associated with apoptosis in human neuroblastoma SH-SY5Y cells. Toxicol Lett. 2009;191:203–210. doi: 10.1016/j.toxlet.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 19.Gotoh Y, Cooper JA. Reactive oxygen species- and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-alpha signal transduction. J Biol Chem. 1998;273:17477–17482. doi: 10.1074/jbc.273.28.17477. [DOI] [PubMed] [Google Scholar]
- 20.Federspiel JD, Codreanu SG, Palubinsky AM, Winland AJ, Betanzos CM, McLaughlin B, Liebler DC. Assembly Dynamics and Stoichiometry of the Apoptosis Signal-regulating Kinase (ASK) Signalosome in Response to Electrophile Stress. Mol Cell Proteomics. 2016;15:1947–1961. doi: 10.1074/mcp.M115.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tobiume K, Saitoh M, Ichijo H. Activation of apoptosis signal-regulating kinase 1 by the stress-induced activating phosphorylation of pre-formed oligomer. J Cell Physiol. 2002;191:95–104. doi: 10.1002/jcp.10080. [DOI] [PubMed] [Google Scholar]
- 22.Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27:8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katagiri K, Matsuzawa A, Ichijo H. Regulation of apoptosis signal-regulating kinase 1 in redox signaling. Methods Enzymol. 2010;474:277–288. doi: 10.1016/S0076-6879(10)74016-7. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Zhang W, Zhang R, Zhang H, Min W. SOCS1 inhibits tumor necrosis factor-induced activation of ASK1-JNK inflammatory signaling by mediating ASK1 degradation. J Biol Chem. 2006;281:5559–5566. doi: 10.1074/jbc.M512338200. [DOI] [PubMed] [Google Scholar]
- 25.Kim AH, Khursigara G, Sun X, Franke TF, Chao MV. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol Cell Biol. 2001;21:893–901. doi: 10.1128/MCB.21.3.893-901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujii K, Goldman EH, Park HR, Zhang L, Chen J, Fu H. Negative control of apoptosis signal-regulating kinase 1 through phosphorylation of Ser-1034. Oncogene. 2004;23:5099–5104. doi: 10.1038/sj.onc.1207668. [DOI] [PubMed] [Google Scholar]
- 27.Goldman EH, Chen L, Fu H. Activation of apoptosis signal-regulating kinase 1 by reactive oxygen species through dephosphorylation at serine 967 and 14-3-3 dissociation. J Biol Chem. 2004;279:10442–10449. doi: 10.1074/jbc.M311129200. [DOI] [PubMed] [Google Scholar]
- 28.Takeda K, Shimozono R, Noguchi T, Umeda T, Morimoto Y, Naguro I, Tobiume K, Saitoh M, Matsuzawa A, Ichijo H. Apoptosis signal-regulating kinase (ASK) 2 functions as a mitogen-activated protein kinase kinase kinase in a heteromeric complex with ASK1. J Biol Chem. 2007;282:7522–7531. doi: 10.1074/jbc.M607177200. [DOI] [PubMed] [Google Scholar]
- 29.Morita K, Saitoh M, Tobiume K, Matsuura H, Enomoto S, Nishitoh H, Ichijo H. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. The EMBO journal. 2001;20:6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benedetti A, Comporti M, Esterbauer H. Identification of 4-hydroxynonenal as a cytotoxic product originating from the peroxidation of liver microsomal lipids. Biochim Biophys Acta. 1980;620:281–296. doi: 10.1016/0005-2760(80)90209-x. [DOI] [PubMed] [Google Scholar]
- 31.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fritz KS, Kellersberger KA, Gomez JD, Petersen DR. 4-HNE adduct stability characterized by collision-induced dissociation and electron transfer dissociation mass spectrometry. Chem Res Toxicol. 2012;25:965–970. doi: 10.1021/tx300100w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 34.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nature communications. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 37.Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. Journal of proteome research. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichols AM, White FM. Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. Methods Mol Biol. 2009;492:143–160. doi: 10.1007/978-1-59745-493-3_8. [DOI] [PubMed] [Google Scholar]
- 40.Strohalm M, Kavan D, Novak P, Volny M, Havlicek V. mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Analytical chemistry. 2010;82:4648–4651. doi: 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- 41.Niedermeyer TH, Strohalm M. mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One. 2012;7:e44913. doi: 10.1371/journal.pone.0044913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Min W, He Y, Qin L, Zhang H, Bennett AM, Chen H. JAK2 and SHP2 reciprocally regulate tyrosine phosphorylation and stability of proapoptotic protein ASK1. J Biol Chem. 2009;284:13481–13488. doi: 10.1074/jbc.M809740200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan ZQ, Feldman RI, Sussman GE, Coppola D, Nicosia SV, Cheng JQ. AKT2 inhibition of cisplatin-induced JNK/p38 and Bax activation by phosphorylation of ASK1: implication of AKT2 in chemoresistance. J Biol Chem. 2003;278:23432–23440. doi: 10.1074/jbc.M302674200. [DOI] [PubMed] [Google Scholar]
- 44.Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009;28:4261–4271. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang R, Luo D, Miao R, Bai L, Ge Q, Sessa WC, Min W. Hsp90-Akt phosphorylates ASK1 and inhibits ASK1-mediated apoptosis. Oncogene. 2005;24:3954–3963. doi: 10.1038/sj.onc.1208548. [DOI] [PubMed] [Google Scholar]
- 46.Seong HA, Jung H, Ichijo H, Ha H. Reciprocal negative regulation of PDK1 and ASK1 signaling by direct interaction and phosphorylation. J Biol Chem. 2010;285:2397–2414. doi: 10.1074/jbc.M109.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanc A, Pandey NR, Srivastava AK. Distinct roles of Ca2+, calmodulin, and protein kinase C in H2O2-induced activation of ERK1/2, p38 MAPK, and protein kinase B signaling in vascular smooth muscle cells. Antioxid Redox Signal. 2004;6:353–366. doi: 10.1089/152308604322899422. [DOI] [PubMed] [Google Scholar]
- 48.Cui XL, Ding Y, Alexander LD, Bao C, Al-Khalili OK, Simonson M, Eaton DC, Douglas JG. Oxidative signaling in renal epithelium: Critical role of cytosolic phospholipase A2 and p38(SAPK) Free Radic Biol Med. 2006;41:213–221. doi: 10.1016/j.freeradbiomed.2006.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw M, Cohen P, Alessi DR. The activation of protein kinase B by H2O2 or heat shock is mediated by phosphoinositide 3-kinase and not by mitogen-activated protein kinase-activated protein kinase-2. Biochem J. 1998;336(Pt 1):241–246. doi: 10.1042/bj3360241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.