Abstract
Since endophytes can affect metabolism of host plants, they are expected to be used to improve crop quality, especially for crops with organoleptic sensitive products such as wine grape. However, details of metabolic interactions between endophytes and host plants were less understood. In this work, we used high pressure liquid chromatography (HPLC) to analyze the metabolites of fruit flesh cells of grape treated with dual culture of different endophytic fungal strains (EFS). We observed that the dual-culture with different fungal strains show different metabolites composition in grape cells. In response to different EFS, quantities of detected metabolites in grape cells varied from 6 to 17 in this assay, and 1 to 11 novel metabolites were introduced into metabolome of grape cells. Dual-culture with fungal strains CS2, RH16 and RH5 introduced the highest quantities (10 or 11) of novel metabolites in grape cells. More importantly, the modification of metabolic profiles in grape cells via fungal endophytes appeared to be fungal strain/genus-specificity. Overall, this work revealed that introduction of specific metabolites in host plants may be one consequence during the process of endophytes-host metabolic interactions, which raise the possibility to shape grape qualities and characteristics using tool of fungal endophytes.
Introduction
Endophytes were intensively studied during the past decades for the great potential of novel valuable metabolites which have medicinal, agricultural and industrial applications [1–5]. However, despite numerous reports, there still no major breakthroughs in terms of commercial exploitation of any endophytes as a source of bioactive molecules [6]. The symbiosis of endophytes conferred growth promotion and environmental adaptability effects to host plants on the other hand, have achieved applications [7–9]. The fact that the endophytes had metabolic impacts on host plants, further suggests the possibility of regulating the biochemical status of host plants with fungal endophytes [10]. This may be of great interest to food crops which give organoleptic sensitive products such as wine, coffee and tea. Therefore, clarifying the mechanisms undergo endophytes-hosts metabolic interaction is fundamental for this purpose. Studies concerned the interactions between endophytes and host plants had been documented [9, 11, 12], while the metabolic contributions of these endophytes to their host plants is less covered so far. Due to the complexity and difficulty to investigate the metabolic interactions between endophytes and their host plants in vivo, we instead analyzed metabolome of in vitro plant cells and dual culture system.
Dual culture has been successfully used in studying the physiological and morphological interactions between fungal endophytes and plant cells [13–15]. Previously, fungal endophytes were classified into categories based on their infective and detrimental abilities to the grape cells in dual cultures [13], but how these fungal strains furtherly influence the metabolites of grape fruit cells are expected. This report, however studied the impacts of multiple endophytic fungal strains on the metabolite profiles during the dual culture and provided some clues for the mechanism of plant cell-endophytes metabolic interactions.
Materials and methods
Grape calli preparation
Cell line (CBL, kindly provided by Professor Serge Delrot, director of Research Lab. of grapevine physio-ecological and functional genomics, France) induced from flesh of young grape berries (Vitis vinifera, cultivar: Cabernet sauvignon) was used in this study. B5 solution with 3% sucrose, 0.2 mg/L cytokinin, 0.1 mg/L naphthylacetic acid (NAA) and 0.8% agar was prepared as callus medium for callus sub-culture and the following dual culture with fungus. Prepared grape calli for this experiment were in the logarithmic growth phase.
Preparation of endophytic fungal strains
Foliar endophytic fungal strains (EFS, Table 1) were isolated from grape variety ‘Cabernet sauvignon’ (Vitis vinifera) and another local variety, ‘Rose honey’ (V. Vinifera L.× V. labrusca L.) in local vineyards (Yunnan province, China) (Table 1). Vineyards located in subtropical climate area, within the latitude from N26 to N27, and altitude from 1400 to 2500 m. Endophytic fungi isolation followed the tissue patch method [16], with some modifications. Briefly, healthy leaves (grapevine) without any symptoms of disease were chosen for EFS isolation. Leaves were done surface sterilization with the procedure of 75% ethanol, 1 minute; 3% sodium hypochlorite, 20 minutes; and washed 3 times in sterilized water. The surface sterilized leaves were then cut into 0.5×0.5 cm patches and pasted on potato dextrose agar (PDA) medium (containing 50 mg/L of carbenicillin and streptomycin) in petri dishes. Plates were incubated at 25 °C and examined the emergence of fungi every day. The emerged fungi were transferred to PDA plates to obtain pure cultures. Prior to initial plating, several samples were imprinted onto media and these imprinted plates were monitored for the lack of fungal growth to ensure the effectiveness of the surface sterilization. Pure cultured endophytic fungal strains were identified using ITS DNA sequences (Ma et al., 2014). Before performing dual culture with grape cells, fungal strains were plate cultured on potato dextrose agar (PDA) medium in petri dishes for one week.
Table 1. Endophytic Fungal strains (EFS) used in the experiment.
| Strain ID | Species | Strain ID | Species |
|---|---|---|---|
| RH5 | Trichothecium sp. | RH38 | Epicoccum nigrum |
| RH6 | Alternaria alternaria | RH43 | Alternaria arborescens |
| RH7 | Epicoccum nigrum | RH44 | Alternaria arborescens |
| RH12 | Niqrospora sphaerica | RH45 | Epicoccum sp. |
| RH16 | Alternaria sp. | RH46 | Niqrospora sp. |
| RH24 | Alternaria arborescens | RH48 | Colletotrichum gloesporioides |
| RH28 | Alternaria alternaria | CS2 | Colletotrichum gloesporioides |
| RH31 | Alternaria alternaria | CS11 | Epicoccum nigrum |
| RH32 | Alternaria alternaria | CS13 | Fusarium oxysporum |
| RH34 | Trichothecium roseum | CS16 | Alternaria sp. |
| RH37 | Epicoccum nigrum |
Fungal strain ID with ‘RH’ represents the endophytic fungal strain was isolated originally from grape cultivar Rose honey (Vitis. Vinifera L.× V. labrusca L.), and with ‘CS’ means the fungal strain was isolated from another grape cultivar Cabernet sauvignon (V. vinifera L.).
Establishment of fungi-calli dual-culture system
Sterilized 30 mL callus medium was added to each sterilized petri dishes to generate solid culture plates. Dual cultures were performed as described by Huang et al. (2017). Calli without fungal inoculation were used as callus control. Every treatment and control contains more than 3 biological replicates.
Sample harvest and pre-treatment
Calli were harvested after 10 days dual-culture with fungi. All harvested grape calli were grounded into powder with liquid nitrogen and then freezing dried in a vacuum freeze dryer.
HPLC assay for methanol extracts of grape cells
Ten milligram of freezing dried grape callus powder were accurately weighed and extracted with 500 μL of 60% methanol (contains 0.1% of hydrochloric acid) for 2 hours in an ultrasonic cleaner. Extracts were centrifuged briefly at 10000 rpm and supernatants were then filtered with 0.45 um filter columns. Ten micro liter of extracts were loaded and metabolites in grape cells were separated by reverse C18 column on a HPLC instrument (Agilent, USA) with 30 °C of column temperature. Elution phase is acetonitrile: water: formic acid = 35: 65: 0.1, with the elution speed of 1 mL/min, and detected with a UV detector at 254, 263 and 280 nm, respectively. Samples were eluted with the gradient procedures as illustrated in supplementary table (S1 Table).
Data analysis
UV detector at 254 nm detected the most metabolites in this HPLC assay, and data acquired at this detection wavelength were used in this analysis. Metabolome between treatments were directly compared the chromatograms. Peak areas were used to compare the relative contents of certain detected compounds in HPLC, and reported as means ± standard deviation of all replicates. And the statistical differences among treatments were determined using one-way ANOVA followed by Tukey’s test (P<0.05) on SPSS16.0 (SPSS Inc., Chicago, IL, USA). The confirmation of certain metabolites in one treatment determined only when this compounds appeared in two third of the replicates. Treatments were done Squared Euclidean distance Hierarchical clustering using SPSS 16.0 software, based on the appearance (1) and absence (0) matrix of all detected metabolites in grape cells. Heat-map were generated (on excel 2013) according to the mean peak areas of the appeared metabolites.
Results
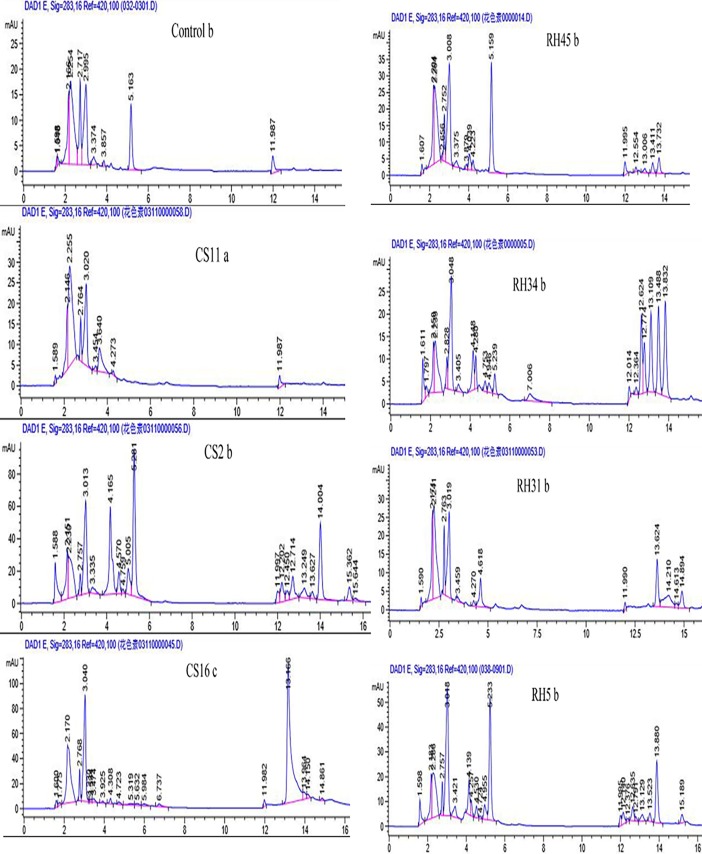
HPLC clearly displayed the differences of metabolite profiles in grape flesh cells after the dual-culture with fungal endophytes (Fig 1). Dual-culture with different endophytic fungal strains (EFS) led to the establishment of different metabolites patterns (Fig 1). In this HPLC assay, the detected metabolites mainly appeared within retention time from 2.0 to 16.0 minutes and obviously separated into two clusters, one appeared at retention times between 2.0 and 5.0 minutes, whereas the other appeared at retention within 12.0 and 14.0 minutes (Fig 1). In comparison with the control, dual-culture with EFS RH5, RH16, RH34 and others has robustly increased numbers of detected metabolites in grape cells (Fig 1, S1 Fig).
Fig 1. HPLC chromatograms of grape cell extracts after dual-cultured with different endophytic fungal strains (EFS).
Chromatograms were selectively displayed in the figure. Each chromatogram was marked the EFS which the grape cells dual-cultured, and the followed letter was the serial number of replicates. Chromatograms of all other detected samples can be found in supplementary materials (S1 Fig).
Clustering to all samples based on the presence (1) and absence (0) matrix of metabolites, biological replicates of one treatment tend to be clustered together with few exceptions (Fig 2), suggesting that metabolite change in grape cells by fungal endophytes are reproducible and EFS-specific. Exceptions were found in grape cells after co-cultured with three EFS: RH45, RH7 and RH32. Dual-culture with these EFS make the metabolites composition in grape cells varied among replicates (Fig 2).
Fig 2. A clustering (Squared Euclidean distance Hierarchical clustering using SPSS 16.0 software) to all replicates of the treatments based on the appearance (1) and absence (0) matrix of detected metabolites.
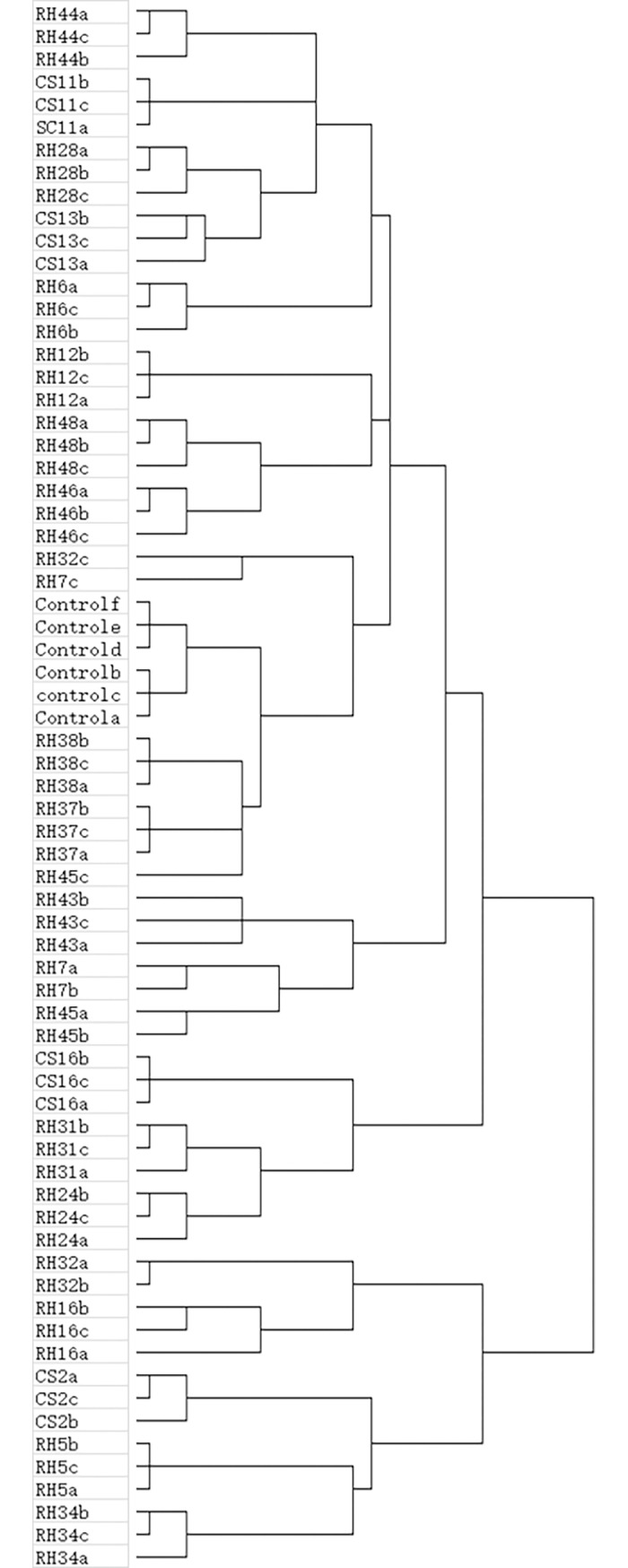
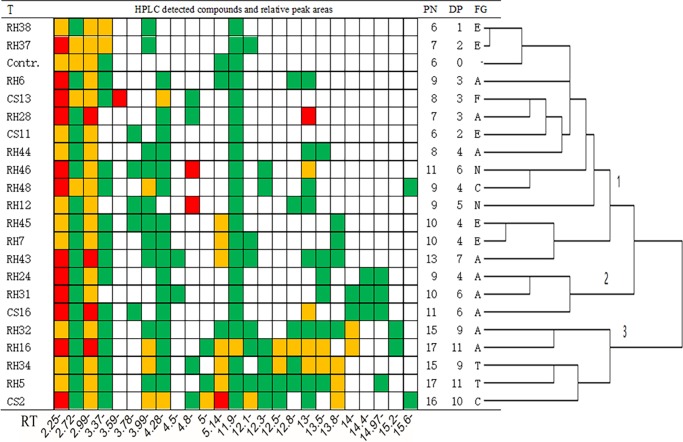
Fig 3 summarized the HPLC detected metabolite compositions, peak numbers and relative contents of these detected compounds in grape cells under different treatments, as well as the clustering of all those treatments in the experiment (Fig 3). In total 25 metabolites were detected in this assay and numbers of the detected metabolites in grape cells of different treatments varied from 6 to 17 (Fig 3). EFS CS2, RH16 and RH5 dual-cultured grape cells detected the highest quantities (16 or 17) of metabolites, whereas in EFS RH38, RH37, RH28, CS11 dual-cultured grape cells and controls detected less quantities (6 or 7) of metabolites (Fig 3). In comparison with control, 1 to 11 novel metabolites were produced in grape cells due to the existences of different EFS in the dual culture. Accordingly, dual culture with EFS CS2, RH16 and RH5 introduced the most quantities (10 or 11) of novel metabolites, whereas, dual culture with EFS RH38, RH37 and CS11 induced the least quantities (1 or 2) of novel metabolites into grape cells (Fig 3, S2 Table).
Fig 3. HPLC detected metabolomes and metabolites contents in grape cells, as well as the clustering to all treatments.
T: treatment (represent as endophytic fungal strain ID and the control (Contr)). HPLC detected compounds are marked as colored bricks, and different color represent the relative content (peak area) of the metabolites: 10 mAU*S ≤green <100 mAU*S; 100 ≤yellow < 500 mAU*S; red ≥ 500mAU*S. PN: peak numbers; DP: novel peak numbers when compared to the control; FG: genus of the EFS, E: Epicoccum; A: Alternaria; F: Fusarium; N: Niqrospora; C: Colletotrichum; T: Trichothecium. At the bottom of the figure displayed the retention time (RT) at which the metabolites appeared in this HPLC assay.
Apart from the metabolite at retention time of 3.59 minutes which only appeared in fungal strain CS13 (Fusarium sp.) dual-cultured grape cells, all other metabolites in this HPLC assay can be detected in two or more EFS treated grape cells (Fig 3). Notably, three specific metabolites at retention times of 4.5, 4.8 and 15.6 minutes were detected, and each of these metabolites was only present in two of the used EFS dual-cultured grape cells (Fig 3). Fungal strains RH31 and RH43 which introduced the metabolite at retention time of 4.5 minutes belong to same genus Alternaria. Co-culture with EFS RH48 and CS2 which from the genus Colletotrichum produce the metabolite at retention time of 15.6 minutes in grape cells. And additionally, EFS RH46 and RH12 initiated higher relative content of metabolite (peak areas>500 mAU*s) at retention time of 4.8 minutes, and these fungal strains also belong to the same genus Niqrospora (Fig 3, S2 Table).
Clustering analysis based on the metabolites patterns in grape cells, all EFS and control can be categorized into 3 groups. EFS RH37, RH38, RH6, CS13, RH28, CS11, RH44 were closely clustered with control (group 1), suggested the less metabolomics impacts of these EFS on grape cells (Fig 3). Except the control, fungal treatments in group 1 included all of the EFS from genus Epicoccum (5/5: five of five used EFS in this experiment), Niqrospora (2/2), Fusarium (1/1), and half of the EFS from genus Alternaria (4/9). Group 2 clustered with 3 EFS, and all these fungal strains belong to genus Alternaria. The left group contain 5 fungal strains, included two EFS from genus Trichothecium (2/2) and Alternaria (2/9), respectively, and another fungal strain CS2 from genus Colletotrichum (Fig 3). No obvious differences were observed in this experiment of the metabolic effects on grape cells between host and non-host isolated EFS (Fig 3).
Dual-culture with fungal endophytes not only modified the composition of metabolites, but also influenced the relative contents of the co-detected metabolites in the grape cells. Five metabolites at retention times of 2.25 (metabolite A+B), 2.72 (C), 2.99 (D), and 11.9 (E) minutes in this assay, were detected in all samples (Figs 1 and 3 and S2 Table). Relative contents (represent as peak areas mAU*S) of these metabolites and different significances among treatments were presented in Table 2. Relative content of these metabolites in grape cells were obviously varied due to the dual-cultured EFS, and some of these changes have reached the statistical significance (Table 2). For some examples, compared to the control, dual-culture with EFS RH16 and CS16 significantly promoted the content of metabolite C, whereas dual culture with another EFS CS11 suppressed the content of this metabolite (Table 2). Dual culture with EFS RH16 also significantly promoted the content of metabolite E in grape cells (Table 2).
Table 2. Peak areas (mAU*S) of co-detected metabolites in grape cells and the different significances.
| Compound | A+B (RT = 2.25) | C (RT = 2.73) | D (RT = 2.99) | E (RT = 11.9) |
|---|---|---|---|---|
| Control | 314.45±52.85a* | 131.33±54.54a | 165.89±23.93cd | 29.61±1.10b |
| 11RH6 | 967.31±699.53a | 75.14±33.32a | 259.95±87.09abcd | 17.39±7.79b |
| RH12 | 449.95±83.03a | 78.65±26.06a | 319.30±7.35abcd | 11.02±1.79b |
| RH28 | 868.14±204.53a | 78.24±8.00a | 576.35±190.93abc | 14.97±7.83b |
| RH32 | 375.67±133.15a | 55.19±20.49a | 308.81±25.05abcd | 35.06±27.29b |
| RH34 | 321.56±148.48a | 43.81±11.18a | 400.04±210.50abcd | 19.27±3.74b |
| RH37 | 592.72±643.11a | 206.92±238.69a | 273.37±242.95abcd | 28.01±3.27b |
| RH38 | 460.06±176.15a | 87.38±67.97a | 178.45±67.69cd | 21.05±1.89b |
| RH45 | 336.72±51.78a | 81.13±22.33a | 216.33±66.79abcd | 26.84±4.06b |
| RH46 | 646.90±25.72a | 85.54±32.38a | 390.41±55.22abcd | 15.27±5.28b |
| RH48 | 1176.38±402.78a | 128.10±43.79a | 363.04±36.75abcd | 42.00±18.22b |
| CS2 | 553.15±75.85a | 59.96±4.81a | 487.96±48.52abcd | 48.78±23.92b |
| CS11 | 342.38±66.18a | 54.78±13.84a | 137.29±24.08e | 15.84±3.98b |
| CS13 | 1100.10±570.09a | 220.35±134.67a | 361.06±246.69abcd | 30.71±10.51b |
| CS16 | 1203.52±456.04a | 70.66±18.90a | 621.24±36.69a | 26.20±0.92b |
| RH5 | 384.48±54.94a | 58.72±3.85a | 404.94±53.19abcd | 26.26±2.01b |
| RH7 | 400.32±13.41a | 97.15±11.74a | 307.72±71.02abcd | 15.65±3.15b |
| RH16 | 593.52±261.40a | 73.46±33.72a | 611.46±293.05ab | 105.13±70.65a |
| RH24 | 695.25±391.96a | 79.42±23.05a | 198.62±65.76bcd | 20.28±5.79b |
| RH31 | 926.19±465.05a | 81.08±12.26a | 258.50±91.48abcd | 10.78±1.03b |
| RH43 | 696.70±17.33a | 79.55±4.51a | 508.87±250.12abcd | 15.67±2.59b |
| RH44 | 247.98±58.41a | 52.14±8.83a | 197.52±29.39 23bcd | 26.60±19.12b |
Values were displayed as means of all replicates ± standard variations.
*Letters indicate the different significances of values within columns. And values followed with totally different letters are significantly different (P≤0.05).
Discussion
Metabolites in wine grape berries contribute most to wine qualities and characters [17]. How abiotic and biotic environmental factors such as temperature, water conditions, radiations, pathogens and others on wine grape chemistry and their resultant wines were intensively studied [18–22]. However, little is known how endophytes, one of biotic environmental factors, affect metabolism on grapevine and other plant. Endophytes had been proven to participate in the plant growth and stresses adaptability [7–9]. And all these changes in growth and adaptability for host plants were fundamentally the alteration of plant metabolisms. Re-inoculation of fungal endophytes to field grow grapevines had significant metabolic impacts on grape leaves and fruits, suggested the possible application of endophytes in grape quality regulation [10]. In fact, results in this field experiment integrated effects of exogenous fungal inoculation and the inoculation interfered endogenous endophytes communities. Our current report using dual culture system, which allowed to furtherly analyze the metabolic changes by certain EFS along on grape cells. Previous dual culture of these EFS with grape cells (shoot induced cells) had categorized fungal strains due to their specific interactions [13]. It will be of great interest if EFS also impose specific metabolic impacts on grape flesh cells.
Even not qualitatively identified all those detected metabolites in this HPLC assay, results of this assay were enough to confirm the significant effects of fungal endophytes on the metabolome of plant cells. Novel metabolites were produced as well as the relative contents of metabolites in grape cells were modified due to the existence of fungal endophytes (Fig 1, Table 2). Because of the symbiosis of fungal endophytes, similar circumstances may occur in natural growing plants in vivo. As expected, strain-specific fungal endophytes on grape cellular metabolome modification was observed (Fig 2, S1 Fig). Certain degrees of genus-specificity, however, was also detected in fungal endophyte-grape cell metabolic interactions (Fig 3). EFS belong to the same genus such as Epicoccum, Niqrospora and Alternaria tend to clustered together (Fig 3). And specific metabolites were only detected in grape cells dual-cultured with certain genus of fungal endophytes (Fig 3). Therefore, beside the specific morphological interactions[13], specific metabolic interactions also occur between fungal endophytes and grape cells. This will be important in guiding the selections of candidate EFS for purpose shaping the qualities and the characteristics of grape in viticulture.
Generally, mechanisms underlying the metabolic impacts of endophytes on host plant may employ pathways of: i) endophytes self-metabolizing; ii) endophytes and host co-metabolizing; and iii) Signaling [23]. The fact that novel metabolites were introduced into grape cell which has been confirmed in this work, implies the significant pathways of endophytes self-metabolizing and endophytes-host co-metabolizing during the endophyte-host metabolic interactions. But details on how certain novel metabolite produced and appeared in plant cells during the process of endophytes symbiosis need further researches.
In conclusion, the introduction of novel metabolites may be one mechanism that endophytes influence host plant’s metabolism. And these newly produced metabolites may employ pathways of endophytes self–metabolizing or endophytes and host co-metabolizing. The specific effects of endophytic fungal strains on grape cellular metabolites composition raise the possibility of purpose shaping of grape qualities and characters in viticulture by means of endophytes.
Supporting information
(PDF)
(PDF)
(PDF)
Acknowledgments
We greatly appreciate Professor Serge Delrot, director of Research Lab. of grapevine physio-ecological and functional genomics, in France, for his kind providing of grape cell line (CBL); and we thank Dr. Yunkun Dang for his earnest language revision for the manuscript.
Data Availability
Relevant data are contained within the paper and its supporting information files.
Funding Statement
Our research was financially supported totally by the National Natural Science Foundation of China (NSFC 31560538 and 31160070) and Yunnan University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Owen NL, Hundley N (2004) Endophytes–the chemical synthesizers inside plants. Science Progress 87: 79–99. doi: 10.3184/003685004783238553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao J, Zhou L, Wang J, Shan T, Zhong L, Liu X, et al. (2010) Endophytic fungi for producing bioactive compounds originally from their host plants. Curr Res, Technol Educ Trop Appl Microbiol Microbial Biotechnol 1: 567–576. [Google Scholar]
- 3.Chandra S (2012) Endophytic fungi: novel sources of anticancer lead molecules. Applied microbiology and biotechnology 95: 47–59. doi: 10.1007/s00253-012-4128-7 [DOI] [PubMed] [Google Scholar]
- 4.Nisa H, Kamili AN, Nawchoo IA, Shafi S, Shameem N, Bandh SA (2015) Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microbial Pathogenesis 82: 50–59. doi: 10.1016/j.micpath.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Kaul S, Gupta S, Ahmed M, Dhar MK (2012) Endophytic fungi from medicinal plants: a treasure hunt for bioactive metabolites. Phytochemistry Reviews 11: 487–505. doi: 10.1007/s11101-012-9260-6 [Google Scholar]
- 6.Venugopalan A, Srivastava S (2015) Endophytes as in vitro production platforms of high value plant secondary metabolites. Biotechnology Advances 33: 873–887. doi: 10.1016/j.biotechadv.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 7.Hubbard M, Germida J, Vujanovic V (2012) Fungal endophytes improve wheat seed germination under heat and drought stress. Botany 90: 137–149. org/10.1139/b11-091 [Google Scholar]
- 8.Waqas M, Khan AL, Kamran M, Hamayun M, Kang S-M, Kim Y-H, et al. (2012) Endophytic fungi produce gibberellins and indoleacetic acid and promotes host-plant growth during stress. Molecules 17: 10754–10773. doi: 10.3390/molecules170910754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres MS, White JF, Zhang X, Hinton DM, Bacon CW (2012) Endophyte-mediated adjustments in host morphology and physiology and effects on host fitness traits in grasses. Fungal Ecology 5: 322–330. org/10.1016/j.funeco.2011.05.006 [Google Scholar]
- 10.Yang M-Z, Ma M-D, Yuan M-Q, Huang Z-Y, Yang W-X, Zhang H-B, et al. (2016) Fungal Endophytes as a Metabolic Fine-Tuning Regulator for Wine Grape. PLoS ONE 11: e0163186 doi: 10.1371/journal.pone.0163186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unterseher M, Gazis R, Chaverri P, Guarniz CFG, Tenorio DHZ (2013) Endophytic fungi from Peruvian highland and lowland habitats form distinctive and host plant-specific assemblages. Biodiversity and conservation 22: 999–1016. doi: 10.1007/s10531-013-0464-x [Google Scholar]
- 12.Soliman SS, Trobacher CP, Tsao R, Greenwood JS, Raizada MN (2013) A fungal endophyte induces transcription of genes encoding a redundant fungicide pathway in its host plant. BMC plant biology 13: 93 doi: 10.1186/1471-2229-13-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L-h, Ao X-J, Shan h, Li h-X, Yang W-X, Zhang h-B, et al. (2017) In vitro specific interactions revealed the infective characteristics of fungal endophytes to grapevine. Vitis 56: 71–77. doi: 10.5073/vitis.2017.56.71–77 [Google Scholar]
- 14.Peters S, Draeger S, Aust HJ, Schulz B (1998) Interactions of dual cultures of endophytic fungi with host and nonhost plant calli. Mycologia 90: 360. [Google Scholar]
- 15.Sirrenberg A, Salzer P, Hager A (2010) Induction of mycorrhiza-like structures and defence reactions in dual cultures of spruce callus and ectomycorrhizal fungi. New Phytologist 130: 149–156. doi: 10.1111/j.1469-8137.1995.tb01825.x [Google Scholar]
- 16.Martin RC, Dombrowski JE (2015) Isolation and Identification of Fungal Endophytes from Grasses along the Oregon Coast. American Journal of Plant Sciences 6: 3216–3230. doi: 10.4236/ajps.2015.619313, [Google Scholar]
- 17.Zoecklein BW, Fugelsang KC, Gump BH, Nury FS (1995) Wine Analysis and Production. Food Quality & Preference 9: VII–VIII. [Google Scholar]
- 18.Deluc LG, Quilici DR, Decendit A, Grimplet J, Wheatley MD, Schlauch KA, et al. (2009) Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC genomics 10: 212 doi: 10.1186/1471-2164-10-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berli FJ, Bottini R (2013) UV-B and abscisic acid effects on grape berry maturation and quality. Journal of Berry Research 3: 1–14. doi: 10.3233/JBR-130047 [Google Scholar]
- 20.Martínez-Lüscher J, Torres N, Hilbert G, Richard T, Sánchez-Díaz M, Delrot S, et al. (2014) Ultraviolet-B radiation modifies the quantitative and qualitative profile of flavonoids and amino acids in grape berries. Phytochemistry 102: 106–114. doi: 10.1016/j.phytochem.2014.03.014 [DOI] [PubMed] [Google Scholar]
- 21.Casassa LF, Keller M, Harbertson JF (2015) Regulated Deficit Irrigation Alters Anthocyanins, Tannins and Sensory Properties of Cabernet Sauvignon Grapes and Wines. Molecules 20: 7820–7844. doi: 10.3390/molecules20057820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soubeyrand E, Basteau C, Hilbert G, van Leeuwen C, Delrot S, Gomès E (2014) Nitrogen supply affects anthocyanin biosynthetic and regulatory genes in grapevine cv. Cabernet-Sauvignon berries. Phytochemistry 103: 38–49. doi: 10.1016/j.phytochem.2014.03.024 [DOI] [PubMed] [Google Scholar]
- 23.Ludwig-Müller J (2015) Plants and endophytes: equal partners in secondary metabolite production? Biotechnology letters: 1–10. doi: 10.1007/s10529-015-1814-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
Relevant data are contained within the paper and its supporting information files.