Abstract
Ethylene responses in Arabidopsis are mediated by a small family of receptors, including the ETR1 gene product. Specific mutations in the N-terminal ethylene-binding domain of any family member lead to dominant ethylene insensitivity. To investigate the mechanism of ethylene insensitivity, we examined the effects of mutations on the ethylene-binding activity of the ETR1 protein expressed in yeast. The etr1-1 and etr1-4 mutations completely eliminated ethylene binding, while the etr1-3 mutation severely reduced binding. Additional site-directed mutations that disrupted ethylene binding in yeast also conferred dominant ethylene insensitivity when the mutated genes were transferred into wild-type Arabidopsis plants. By contrast, the etr1-2 mutation did not disrupt ethylene binding in yeast. These results indicate that dominant ethylene insensitivity may be conferred by mutations that disrupt ethylene binding or that uncouple ethylene binding from signal output by the receptor. Increased dosage of wild-type alleles in triploid lines led to the partial recovery of ethylene sensitivity, indicating that dominant ethylene insensitivity may involve either interactions between wild-type and mutant receptors or competition between mutant and wild-type receptors for downstream effectors.
Genetic studies in Arabidopsis have provided evidence that ethylene perception in plants is mediated by a family of receptors, including the ETR1 gene product. The ETR1 gene encodes a protein with homology to the two-component His kinase regulators that control a variety of signaling cascades in prokaryotic systems and some eukaryotic systems (Chang et al., 1993). While ETR1 was the first ethylene receptor to be identified in plants (Bleecker et al., 1988), additional screens for ethylene-insensitive seedlings and cloning by sequence similarity indicate that additional genes mediate ethylene sensitivity in Arabidopsis (Hua et al., 1995, 1998; Sakai et al., 1998). The ERS1, ETR2, EIN4, and ERS2 genes all show a high degree of sequence similarity to ETR1 and appear to comprise a small family of ethylene receptors. Dominant point mutations that confer ethylene insensitivity in planta have been isolated in the ETR1, ETR2, and EIN4 genes, and all of these mutations are located within the putative transmembrane domains in the N-termini of these genes. Similar mutations introduced into ERS1 and ERS2 also confer dominant insensitivity when transformed into Arabidopsis plants (Hua et al., 1995, 1998). These studies indicate that a single mutation in any one of these five genes is sufficient to render plants insensitive to ethylene throughout the plant.
Subsequent biochemical experiments have confirmed that the ETR1 gene encodes an ethylene receptor. The N-terminal hydrophobic domain of the ETR1 protein binds ethylene with high affinity when expressed in yeast (Schaller and Bleecker, 1995). The ethylene-binding (sensor) domain of ETR1 consists of three putative membrane-spanning subdomains that are modeled as alpha helices (Rodriguez et al., 1999). Notably, the etr1-1 mutation in subdomain 2 abolishes ethylene binding by the yeast-expressed protein (Schaller and Bleecker, 1995). Biochemical studies demonstrated that a copper ion in the N-terminal hydrophobic domain of ETR1 is required for ethylene binding, and that the etr1-1 mutation abolishes the capacity of the receptor to coordinate this ion (Rodriguez et al., 1999).
Genetic evidence indicates that the ETR1 receptor family signals through the Raf-like kinase CTR1. Loss-of-function mutations in CTR1 show a constitutive triple-response phenotype, indicating that CTR1 acts as a negative regulator of ethylene-response pathways (Kieber et al., 1993). Recently, Hua and Meyerowitz (1998) demonstrated that combining loss-of-function mutants in three or more members of the ETR1 family also results in plants with a constitutive ethylene-response phenotype. These results favor a model for receptor signaling in which the ETR1 receptor family acts in conjunction with CTR1 to suppress response pathways in the absence of ethylene. Ethylene binding would convert receptors to a non-signaling state, resulting in derepression of the response pathway.
Based on the concept of ethylene as a negative regulator of the ETR1 receptor family, we hypothesized that dominant insensitivity to ethylene could result from mutations that disrupt ethylene-binding activity and lock receptors in a signaling state. The finding that the etr1-1 mutation disrupts ethylene binding is consistent with this hypothesis (Schaller and Bleecker, 1995). We investigated whether this relationship could be extended to the other mutant alleles, some of which do not completely abolish ethylene sensitivity in planta (Guzman and Ecker, 1990; Chen and Bleecker, 1995). To further explore the relationship between ethylene binding and dominant insensitivity, we tested whether novel mutations in ETR1 that abolished ethylene binding in yeast could confer ethylene insensitivity to plants transformed with these mutant genes.
MATERIALS AND METHODS
Plant Materials
The etr1-1, etr1-2 (Bleecker et al., 1988; Chen and Bleecker, 1995), etr1-3 (Guzman and Ecker, 1990), etr1-4 (Chang et al., 1993), etr2-1 (Sakai et al., 1998), and ein4-3 mutants (Hua et al., 1998) have been previously described. To reduce the probability of mutations at additional loci, mutant lines were back-crossed to Arabidopsis ecotype Columbia (wild type) at least three times before physiological experiments. Heterozygous diploid plants were obtained by crossing homozygous mutants and wild-type plants (ecotype Columbia). Triploid plants were made by crossing homozygous mutant or wild-type diploid plants to wild-type tetraploid plants (ecotype Bensheim), and the resulting F1 seeds were used in the studies described.
Growth Conditions and Ethylene Treatments
Seedling growth-response assays were carried out as described by Chen and Bleecker (1995). For the experiment shown in Figure 4, flow-through chambers were used with an ethylene concentration of 35 μL L−1. Ethylene concentrations were determined by GC using a column of Carboxen 1000 (45/60-mesh size, Supelco, Bellefonte, PA), with ethylene as the calibration standard.
Figure 4.
Analysis of ethylene binding and ethylene responses in C65S and H69A mutants. A, Ethylene binding by C65S and H69A proteins expressed in yeast. Dark bars represent the amount of [14C]ethylene bound (disintegrations per minute per gram of yeast) by transgenic yeast incubated with 0.07 μL L−1 [14C]ethylene. Samples were run in triplicate and sd bars are shown. White bars represent the amount of [14C]ethylene bound (disintegrations per minute per gram of yeast) by single samples of transgenic yeast incubated with 0.07 μL L−1 [14C]ethylene and 1,000 μL L−1 [12C]ethylene (background binding). WT, Wild type. B, Analysis of the triple response in dark-grown seedlings. Transgenic lines are shown for plants transformed with the wild-type ETR1 gene (WT), the ETR1 gene containing a C65S mutation (C65S), and the ETR1 gene containing an H69A mutation (H69A). For comparison, wild-type (ethylene-sensitive) and etr1-1 mutant (ethylene-insensitive) seedlings were grown under the same conditions. Seeds were grown for 4 d in 35 μL L−1 ethylene, and two independent transgenic lines are shown for each construct used. C, Expression level of ETR1 protein. Expression levels of ETR1 protein were determined for the transgenic lines from B. Membranes were isolated from etiolated seedlings grown in air for 4 d. Membrane protein (15 μg) was subjected to SDS-PAGE, and ETR1 was visualized by western blot.
Measurement of Seedling-Growth Response
To measure root and hypocotyl length, seedlings were grown on vertical plates and photographed with slide film or scanned directly using a Vista-S6E scanner (UMAX, Fremont, CA). Slides were scanned and saved as TIFF files, and measurements were made using the NIH Image program (version 1.62, National Institutes of Health, Bethesda, MD). Ethylene dose-response data were fitted to the Hill equation, used to describe relationships of plant hormone concentrations and responses (Weyers et al., 1987). Dose-response curves were generated using COPLOT (CoHort, Berkeley, CA) and MacCurve Fit (version 1.3, Macintosh) and were based on non-linear least squares regression.
Construction of Site-Directed Mutants for Yeast and Transgenic Plant Expression
Site-directed mutations for yeast expression were introduced into a full-length ETR1 cDNA clone (cETR1-5.2) subcloned into the pALTERII vector. Mutations were introduced using the Altered Sites mutagenesis system (Promega) according to the manufacturer's instructions, and mutations were confirmed by dideoxy sequencing (Sanger et al., 1977). Constructs were transformed using the lithium acetate method (Schiestl and Gietz, 1989) into Saccharomyces cerevisiae strain LRB520 (MATαhis3leu2trp1 ura3-52yck2-1::HIS3) and expressed in yeast as previously described (Schaller and Bleecker, 1995). For wild-type ETR1 expression in Arabidopsis, a 7.3-kb genomic fragment subcloned into the EcoRI site of pBlueScript (Chang et al., 1993) was removed by digestion with BamHI and SalI and cloned into pBIN19 (Bevan, 1984). For mutant gene expression in Arabidopsis the ETR1 genomic clone was subcloned into pALTERII. Site-directed mutants of the genomic ETR1 fragment were confirmed by dideoxy sequencing, excised from the PALTERII vector, and subcloned into pBIN19 using BamHI and SalI.
Ethylene Binding to Transgenic Yeast
Ethylene-binding experiments were performed as previously described (Schaller and Bleecker, 1995) using a modification of the method originally described by Sisler (1979). Yeast cells were grown to mid-log phase at 30°C, harvested by centrifugation at 1,500g for 5 min, washed with water, and collected in 1-g aliquots by vacuum filtration on glass fiber discs (Whatman). Yeast samples (1 g) were sealed in jelly jars containing an injection port in the lid. Samples were incubated in the presence of [14C]ethylene with or without 1,000 μL L−1 [12C]ethylene. [14C]ethylene (specific activity = 56.9 mCi/mmol) was used as the mercuric perchlorate complex, and trapped ethylene was released by the addition of 1 mL of saturated LiCl. After incubation for 4 h at 22°C, samples were removed from the jars, aired for 6 min, then transferred to individual jars with 0.3 mL of mercuric perchlorate in a scintillation vial. These jars were heated to 65°C for 90 min, and then allowed to stand for 24 h to trap the ethylene released from the samples. Trapped [14C]ethylene was quantified by liquid scintillation counting.
Protein Isolation
For membrane isolation, yeast cells were grown according to standard procedures (Ausubel et al., 1994) and isolated by centrifugation for 5 min at 5,000 rpm in a centrifuge (model RC-5B Superspeed, Sorvall), washed with water, and respun at 3,100 rpm for 5 min (AccuSpin FR, Beckman). Yeast cells (25 g) were resuspended in 45 mL of membrane extraction buffer (100 mm NaCl, 50 mm Tris-HCl, pH 6.4, 1% [v/v] DMSO, and 1 mm PMSF), and cells were disrupted by two 1-min treatments with a bead beater (BioSpec Products, Bartlesville, OK) separated by a 30-s interval. Cell debris were pelleted at 10,000 rpm for 10 min and discarded. Membranes were isolated from the supernatant by ultracentrifugation at 30,000 rpm (SW41 Ti rotor, Beckman) for 30 min (model L8–70, Beckman) and resuspended in membrane resuspension buffer (10 mm MES [2-(N-morpholino)-ethanesulfonic acid], pH 6.0, 10% [w/v] Suc, and 1% [v/v] DMSO) at a concentration of 5 g/mL (membranes from 5 g of yeast cells/mL of buffer), and frozen at −80°C.
The TCA precipitation method was used for the isolation of total yeast proteins. Yeast cultures (25 mg) used in the ethylene-binding assays were incubated with cold TCA for 15 min on ice. Yeast were spun down for 5 min at top speed in a microcentrifuge, and the acid supernatant was removed. The cells were then resuspended in 100 μL of SDS-PAGE loading buffer (125 mm Tris [pH 6.8], 20% [v/v] glycerol, 4% [w/v] SDS, and 0.01% [w/v] bromphenol blue), and incubated for 1 h at 37°C before SDS-PAGE analysis (Laemmli, 1970).
For isolation of Arabidopsis membranes, etiolated seedlings (1 g) were homogenized at 4°C in extraction buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 10 mm EDTA, and 20% [v/v] glycerol) containing the protease inhibitors PMSF (1 mm), leupeptin (10 μg/mL), and pepstatin (1 μg/mL). The homogenate was strained through Miracloth (Calbiochem) and centrifuged at 8,000g for 15 min. The supernatant was centrifuged at 100,000g for 30 min, and the membrane pellet was resuspended in 10 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, and 10% (v/v) glycerol with protease inhibitors.
Protein concentration was determined by the Lowry method (Lowry et al., 1951) after addition of samples to 0.4% (w/v) deoxycholate to extract protein from membranes. BSA was used as a standard.
Western Blotting and Protein Quantification
For western blotting, proteins were fractionated via SDS-PAGE using 8% (w/v) gels, transferred to PVDF membranes (Millipore), and incubated for 1 h with the HRR antibody (Schaller et al., 1995) at a 1:2,500 dilution. Immunodecorated proteins were visualized with a chemiluminescent system according to the manufacturer's instructions (Kirkegaard and Perry, Gaithersburg, MD). Exposed film was then scanned, saved as TIFF files in Photoshop (version 4.0, Adobe Systems, Mountain View, CA), and the immunodetectable protein bands were quantified densitometrically using imaging software.
Arabidopsis Transformation and Growth
Constructs in the pBIN19 vector were introduced into Agrobacterium tumefaciens (strain GV3101) and used to transform Arabidopsis ecotype Columbia by vacuum infiltration (Bechtold et al., 1993). Seeds were plated onto agar plates, and transformed plants were selected based on resistance to kanamycin (50 μg/mL) and/or insensitivity to ethylene. Plants were selfed and homozygous lines identified in subsequent generations. For growth of Arabidopsis plants in pots, a 3:1 mixture of Metromix 360 (Scotts-Sierra Horticultural Products, Marysville, OH) to perlite was used. Plants were maintained in an environmental growth chamber at 22°C with a 16-h daylength.
RESULTS
Effects of Mutations in the ETR1 Gene on Seedling-Growth Response
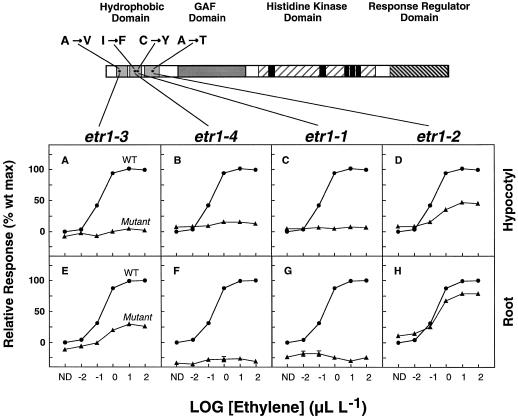
There are four known dominant mutations in the ETR1 gene: etr1-1 (Bleecker et al., 1988), etr1-2 (Chen and Bleecker, 1995), etr1-3 (formerly ein1-1; Guzman and Ecker, 1990), and etr1-4 (Chang et al., 1993). All were isolated using screens for seedlings that were insensitive to ethylene-mediated growth inhibition (Bleecker et al., 1988). The mutations are clustered in the three predicted transmembrane domains of the protein, and all are point mutations that result in the following amino acid substitutions: Ala-31 to Val (etr1-3), Ile-62 to Phe (etr1-4), Cys-65 to Tyr (etr1-1), and Ala-102 to Thr (etr1-2) (Fig. 1).
Figure 1.
Ethylene dose-response curves of seedling growth for the etr1 alleles. A schematic of ETR1 indicates the protein's predicted domains. The three predicted transmembrane subdomains within the N-terminal hydrophobic domain are shaded light gray. The GAF domain (Aravind and Ponting, 1997) is shaded dark gray. The His kinase domain is indicated by diagonal lines; black boxes within this area represent the five consensus motifs (H, N, G1, F, and G2) found in bacterial His kinases. The response regulator domain is indicated by diagonal bars. The point mutations that constitute the four dominant etr1 alleles are indicated. Single-letter abbreviations for amino acids are designated. Ethylene dose-response curves of seedling growth for the etr1 alleles are shown for hypocotyl (A–D) and root (E–H). Dose-response curves are shown for wild type (•, WT) and mutants (▴), including etr1-1 (C and G), etr1-2 (D and H), etr1-3 (A and E), and etr1-4 (B and F). Mutant and wild-type dark-grown seedlings were incubated for 3 d in air or a range of ethylene concentrations (0–1,000 μL L−1) (log values shown on the x axis). Hypocotyl and root length values are reported as a percentage of the wild-type maximum (wt max) and represent the means ± se of 50 measurements. Error bars are not shown if smaller than the smallest displayed. ND, No detectable ethylene.
To determine the effects of these four mutations on ethylene sensitivity in planta, we analyzed the ethylene dose response for hypocotyl and root growth in mutant and wild-type etiolated seedlings. Seedlings were grown in the presence of a range of ethylene concentrations (0–1,000 μL L−1). Hypocotyl and root lengths were measured after 3 d of growth in the dark, and dose-response relationships were compared with isogenic wild-type seedlings.
Plants homozygous for the etr1-1 (Fig. 1, C and G) and etr1-4 alleles (Fig. 1, B and F) showed no responsiveness to applied ethylene in either roots or hypocotyls. The dose-response curves for these mutants appear as negative responses compared with wild-type seedlings, because wild-type seedlings grown in air showed a slight response to endogenous ethylene that the mutants did not. In the etr1-3 line, the maximum ethylene response of seedling growth was 10% of the wild type in the hypocotyl (Fig. 1A) and 23% of wild type in the root (Fig. 1E). In the etr1-2 line, the mutant seedlings had a maximum response of 42% of wild type in the hypocotyl (Fig. 1D) and 60% in the root (Fig. 1H).
These results indicate that the etr1-1 and etr1-4 alleles completely eliminate seedling-growth responses over a range of ethylene concentrations, and confirm previous observations that the etr1-2 and etr1-3 alleles are only partially effective in eliminating seedling-growth responses (Guzman and Ecker, 1990; Chen and Bleecker, 1995). Furthermore, for the etr1-2 and etr1-3 alleles, the partial ethylene sensitivity was more apparent in the root than in the hypocotyl.
Effects of Mutations on the Ethylene-Binding Activity of the ETR1 Receptor
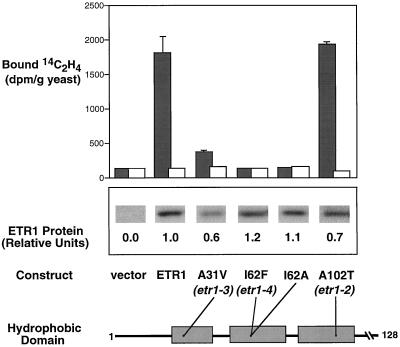
Based on the result that the etr1-1 mutation completely abolished ethylene binding by the yeast-expressed protein (Schaller and Bleecker, 1995), we examined the effects of the other existing mutations on the ethylene-binding capacity of mutant receptors expressed in yeast. Site-directed mutations were introduced into the ETR1 cDNA, corresponding to the etr1-2, etr1-3, and etr1-4 alleles. Each mutant protein was expressed in yeast and assayed for ethylene binding. Protein expression levels for the mutants were quantified on western blots probed with an anti-ETR1 antibody (HRR) (Schaller et al., 1995), and all strains produced immunodetectable protein at levels of at least 60% relative to yeast expressing the ETR1 protein.
Yeast expressing the etr1-4 protein showed no detectable ethylene binding, even though the level of immunodetectable protein was slightly higher in the etr1-4-expressing yeast strain than in the wild-type ETR1-expressing strain (Fig. 2). A more conservative substitution at this residue (I62A) also completely eliminated ethylene binding, indicating that the disruption of binding by the etr1-4 mutation does not result simply from substituting a bulky side chain (I62F) at this position.
Figure 2.
Analysis of ethylene binding by mutant ETR1 proteins. A schematic of the amino acids 1 to 128 of the ETR1 protein is shown, and relative positions of amino acid changes (•) in the three hydrophobic domains are included. For site-directed mutations, single-letter abbreviations are noted for amino acids. Dark bars represent the amount of [14C]ethylene bound (disintegrations per minute per gram of yeast) by transgenic yeast incubated with 0.07 μL L−1 [14C]ethylene. Samples were run in triplicate and sd bars are shown. White bars represent the amount of [14C]ethylene bound (disintegrations per minute per gram of yeast) by single samples of transgenic yeast incubated with 0.07 μL L−1 [14C]ethylene and 1,000 μL L−1 [12C]ethylene (background binding). Total yeast proteins were isolated and analyzed by western-blot analysis. The expression level of each of the mutants was determined by densitometric quantification of western blots, and is reported as expression level relative to wild-type ETR1 protein.
When the etr1-3 protein containing the A31V mutation was expressed in yeast, a greatly reduced level of ethylene binding was detected. In this case, the level of immunodetectable protein was also reduced to 60% of wild type. However, even when corrected for expression level, the etr1-3 protein showed only 23% of the binding activity measured in yeast expressing wild-type ETR1. By contrast, protein expressed from the construct containing the etr1-2 mutation displayed high levels of ethylene-binding activity. When corrected for the level of immunodetectable protein, the etr1-2 protein showed 50% more binding activity than the wild-type protein.
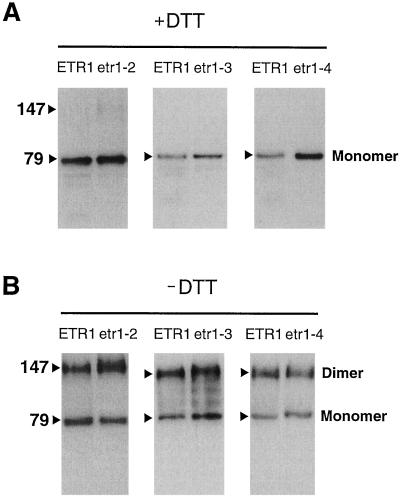
Western-blot analysis indicated that each of the yeast-expressed mutant proteins were membrane associated and formed disulfide-linked dimers, similar to the wild-type ETR1 protein (Schaller et al., 1995) (Fig. 3). Therefore, the effects of the site-directed mutations on ethylene binding did not appear to result from gross disruptions of the protein structure or membrane association.
Figure 3.
Western-blot analysis indicates mutant ETR1 proteins form disulfide-linked dimers. Membranes isolated from yeast expressing each of the mutant ETR1 proteins were incubated in the presence (+) or absence (−) of 100 mm DTT for 1 h at 37°C, and separated by SDS-PAGE. Western-blot analysis comparing wild-type ETR1 protein to the mutant proteins was carried out using an anti-ETR1 antibody (HRR). In the presence of reducing agent (A), the proteins migrate as a 79-kD monomer, while in the absence of reducing agent (B), a 147-kD dimer is also detected.
Taken together, these data indicate that not all of the dominant mutations in ETR1 abolish ethylene binding, and the ethylene-insensitive phenotype observed in the etr1 mutant lines may be caused by defects in ethylene binding, signal transduction, or both.
Two Novel Site-Directed Mutations in ETR1 Abolish Ethylene Binding in Yeast and Confer Dominant Insensitivity to Transgenic Arabidopsis Plants
The finding that the etr1-1, etr1-3, and etr1-4 mutations reduce or eliminate the ability of ETR1 to bind ethylene raises the possibility that the dominant ethylene insensitivity exhibited by the mutant plants results from the disruption of the receptor's ability to bind ethylene (Schaller and Bleecker, 1995). We therefore hypothesized that any mutation in ETR1 that eliminates ethylene binding by ETR1 expressed in yeast might also cause dominant ethylene insensitivity when transformed back into Arabidopsis. To test this, two site-directed mutations that have not been identified from screens for ethylene-insensitive seedlings (C65S and H69A) were introduced in ETR1 (Schaller and Bleecker, 1995; Rodriguez et al., 1999). We then analyzed the effects of these mutations on ethylene binding by the yeast-expressed protein (Rodriguez et al., 1999) and on ethylene sensitivity in planta.
Cys-65 and His-69 are likely candidates to coordinate the copper ion that mediates ethylene binding to the ETR1 receptor (Schaller and Bleecker, 1995; Rodriguez et al., 1999). We hypothesized that if metal coordination is required for ethylene binding, mutations in residues critical for metal coordination would abolish ethylene binding. Consistent with our hypothesis, the C65S and H69A mutations completely eliminate ethylene binding by the yeast-expressed protein (Rodriguez et al., 1999) (Fig. 4A).
To determine the effects of the C65S and H69A mutations on ethylene sensitivity in planta, these mutations were also made in a 7.3-kb genomic DNA fragment containing the ETR1 gene (Chang et al., 1993), cloned into a plant transformation vector (Bevan, 1984), and transformed into wild-type Arabidopsis (Bechtold et al., 1993). Transgenic lines that segregated for kanamycin resistance as single loci were identified and used for further analysis of ethylene responses.
Seedlings transformed with the mutant ETR1 genomic clones were clearly insensitive to ethylene (Fig. 4B). Dark-grown transgenic seedlings gassed with ethylene lacked the triple-response phenotype, which includes a shortened hypocotyl and root, radial thickening of the hypocotyl, and an exaggerated apical hook. Instead, the transgenic seedlings showed the etiolated morphology typical of air-grown plants, and mimicked the appearance of ethylene-insensitive etr1-1 mutant plants. Transgenic plants grown in the light for 2 weeks also showed no responsiveness to ethylene, and lacked such morphological changes as a shortened hypocotyl and root and compressed leaves (data not shown). Because ethylene insensitivity in these transgenic plants occurs in a genetic background containing wild-type ETR1, the ethylene insensitivity conferred by the C65S and H69A mutations is dominant. In contrast, plants transformed with the wild-type ETR1 genomic fragment showed normal ethylene responses (Fig. 4B).
The expression level of ETR1 protein in transgenic plants was determined by performing western-blot analysis with membranes isolated from etiolated seedlings (Fig. 4C). All of the transgenic lines had higher levels of ETR1 protein compared with the level in wild-type seedlings, which is consistent with effective transgene expression. The variability in protein levels may be due to positional effects upon transgene expression. The higher levels of immunodetectable ETR1 present in the transgenic lines cannot account for the ethylene insensitivity observed in the lines transformed with the site-directed mutations, as the lines transformed with wild-type ETR1 and the C65S construct show similar levels of immunodetectable protein. These results indicate a strong correlation between dominant ethylene insensitivity in planta and a lack of ethylene binding by the ETR1 receptor.
Effects of Gene Copy Number on Mutant Phenotypes of the Dominant Ethylene-Insensitive Lines
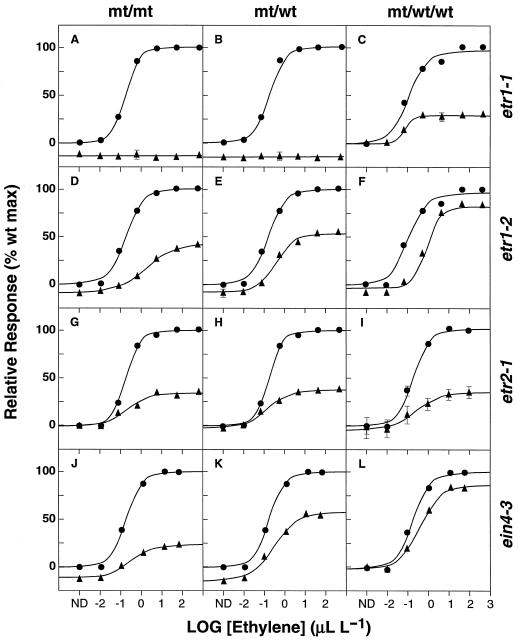
To determine the degree of dominance of mutations in four ethylene-insensitive lines, we examined the relationship between the seedling-response phenotype and mutant gene copy number. Lines that were heterozygous for the etr1-1 allele showed no response to applied ethylene and were indistinguishable from homozygous mutants (Fig. 5, A and B). To further increase wild-type over mutant gene copy number, we crossed a homozygous etr1-1 diploid line to a wild-type tetraploid line (ecotype Bensheim), providing us with triploid F1 progeny with two wild-type and one mutant allele of ETR1. Interestingly, we found that when the ratio of wild-type to mutant gene number was increased in a triploid background, the effects of the etr1-1 mutation were partially attenuated (Fig. 5C). The mutant/wild-type/wild-type lines displayed 27% of the maximum seedling response shown by the wild-type/wild-type/wild-type controls, indicating that partial responsiveness to applied ethylene was restored by increasing the wild-type allele copy number. The possibility that the partial recovery of responsiveness in the triploid line was due to genetic background effects of the ecotype Bensheim parent line was ruled out by additional experiments with Bensheim/Columbia heterozygote diploid lines (Chen and Bleecker, 1995, and Q.G. Chen and A.B. Bleecker, unpublished data).
Figure 5.
Effects of gene dosage on the etr1, etr2, and ein4 dominant mutant phenotypes. Dose-response curves for seedling-growth response in hypocotyl tissues are shown for wild-type (•) and mutants (▴) including etr1-1 (A–C), etr1-2 (D–F), etr2-1(G–I), and ein4-3 (J–L). Genotypes of mutants are indicated as mt/mt for homozygous diploid mutants, mt/wt for heterozygous diploid mutants, and mt/wt/wt for heterozygous triploid mutants. Values represent the means ± se of 20 measurements. Error bars are not shown if smaller than the symbol. ND, No detectable ethylene.
Restoration of ethylene responsiveness by increasing the wild-type allele dosage was even more apparent in the case of the weaker etr1-2 allele (Fig. 5, D–F). The percent wild-type response increased from 42% in the homozygous etr1-2 mutant (Fig. 5D) to 55% in the heterozygote (Fig. 5E). This result does demonstrate the dominance of the mutant over the wild-type allele for even a weak etr1 mutant, but also shows that the dominance is incomplete. Increasing the ratio of wild-type to mutant copies in the mutant/wild-type/wild-type triploid line increased responsiveness to 84% of that obtained in the wild-type/wild-type/wild-type control, demonstrating that the effects of this mutant allele can be almost completely overcome by increasing the wild-type allele copy number (Fig. 5F).
We included two additional lines in our dose-response analysis that contain dominant mutations in the putative ethylene receptors ETR2 and EIN4 to compare the extent of their dominance to the etr1 lines. The etr2-1 mutation results in the conversion of Pro-66 to Leu (Sakai et al., 1998), while the ein4-3 mutation results in the conversion of Thr-117 to Met (Hua et al., 1998). Like etr1-2 mutants, both etr2-1 and ein4-3 mutants showed some sensitivity to ethylene, even in the homozygous mutant lines. The ein4-3 mutation also showed a gene dosage effect similar to etr1-2 in that dominance was incomplete (Fig. 5, J–L). Interestingly, the etr2-1 allele was more completely dominant than the other alleles tested. Mutants showed about 35% of wild-type responsiveness in the presence of zero, one, or two copies of the wild-type allele (Fig. 5, G–I). Similar results were also obtained with the more sensitive root-growth assay (Q.G. Chen and A.B. Bleecker, unpublished results).
DISCUSSION
The genetic evidence from combined loss-of-function mutations in members of the ETR1 receptor family supports an inverse-agonist model for receptor signaling in which unbound receptors repress the ethylene-response pathway (Hua and Meyerowitz, 1998). Consistent with this model, we hypothesized that mutations in the ETR1-like genes that cause dominant ethylene insensitivity in planta could work by disrupting the ethylene-binding activity of the ethylene sensor domain. This in turn could lock the receptors in a constitutively active signaling state in both air and ethylene. The yeast expression system allowed us to examine the specific effects of mutations in the ethylene sensor domain on ethylene binding.
Two of the four mutations tested in this study resulted in mutant proteins unable to bind ethylene. Specifically, the etr1-1 (Schaller and Bleecker, 1995) and etr1-4 mutations, which result in strongly ethylene-insensitive plants, completely abolished ethylene binding by the receptor. This relationship may also hold for the other members of the ETR1-like family, because the amino acid substitution equivalent to etr1-4 causes dominant insensitivity when it occurs in EIN4 or is introduced in ERS1 (Hua et al., 1995) and ERS2 (Hua et al., 1998).
We also determined that novel site-directed mutations that would abolish ethylene binding and in turn confer dominant insensitivity to transgenic plants could be introduced into ETR1. Both the C65S and H69A mutations, located within the second hydrophobic subdomain of ETR1, completely abolished ethylene binding in yeast (Schaller and Bleecker, 1995; Rodriguez et al., 1999) and conferred dominant ethylene insensitivity in planta. These two mutations not only strengthen the link between ethylene binding and dominant insensitivity, but illustrate the possibility of modulating ethylene sensitivity in plants by selective mutation of the ETR1 gene.
In addition, we have found that there may be a quantitative relationship between ethylene binding and insensitivity: etr1-3 plants retained some sensitivity to ethylene, while the mutant form of the protein bound a very small but detectable amount of ethylene. One possible explanation for the reduced level of binding we detected in the etr1-3 protein is that the mutant etr1-3 receptors may have a reduced affinity for ethylene. This could result in an incomplete derepression of the ethylene transduction pathway, producing plants that exhibit partial insensitivity to ethylene.
A second hypothesis to account for how dominant mutations result in receptors constitutively signaling even in the presence of ethylene is that signal propagation, rather than signal perception, is altered. Our studies with the etr1-2 protein are consistent with this hypothesis: ethylene-binding assays carried out with yeast expressing the etr1-2 protein indicated that this mutant protein was able to bind ethylene in excess of wild-type binding levels. While we cannot rule out the possibility that the etr1-2 mutation disrupts ethylene binding in planta (e.g. through forming heterodimer complexes), our data suggest that the etr1-2 mutant protein is altered in its ability to transduce, rather than to perceive, the ethylene signal. Furthermore, we must also consider the possibility that the dominant mutations that do affect ethylene binding may also affect signal output. For example, if the mutations that disrupt ethylene binding also lock the receptor in a hyperactive signaling state, an explanation would be provided for the observation that similar mutations in any one of five receptor isoforms leads to ethylene insensitivity.
A more complete understanding of how mutations in the ETR1 family lead to dominant ethylene insensitivity requires more knowledge about the mechanism used by the receptors to transmit signals to downstream effectors. A reasonable assumption is that ethylene binding to the sensor domain of a receptor induces a conformational change that is propagated to the transmitter domain. Transduction of the signal could occur via modulation of His kinase activity in the transmitter domain, as it does in many bacterial two-component regulators (Parkinson, 1993). His kinase activity has been demonstrated for the ETR1 kinase domain expressed in yeast (Gamble et al., 1998). Alternatively, conformational changes resulting from ethylene binding could be transmitted directly to downstream effectors via protein/protein interactions. In support of this possibility, Clark et al. (1998) found that the transmitter domains of ETR1 and ERS1 interact with the regulatory domain of the CTR1 kinase in yeast two-hybrid and in vitro pull-down assays.
With either of the above mechanisms, mutations anywhere in the receptor that lock the transmitter domain in the appropriate signaling state would lead to dominant insensitivity to ethylene. In this regard, the etr1-2 mutation is located in the third hydrophobic subdomain, between the residues that are known to be required for ethylene binding (Schaller and Bleecker, 1995) and the cytoplasmic transmitter domain. Ethylene-induced conformational changes would likely be propagated through the third hydrophobic subdomain. This could make the third hydrophobic subdomain particularly susceptible to mutations that lock the transmitter in a particular signaling mode. It will be of interest to make additional mutations in this domain of ETR1 and examine their effects on ethylene binding by the receptor and ethylene responses in transgenic plants. It will also be interesting to examine the effects of the ein4-3 mutation on ethylene binding, as this mutation is also located in the third hydrophobic subdomain of EIN4 (Hua et al., 1998).
While genetic studies are consistent with the hypothesis that mutations isolated thus far in the ETR1 family achieve dominance through a gain-of-function mechanism (Hua and Meyerowitz, 1998), these experiments did not clarify whether mutant receptor isoforms act through interactions with other wild-type receptor isoforms or independently. The insensitivity of the etr2-1 mutant allele to increased wild-type allele dosage in the triploid line experiments (Fig. 5I) is consistent with the latter possibility (assuming that the Bensheim background has a functional ETR2 allele). However, in the case of the etr1-1, etr1-2, and ein4-3 alleles, increasing the ratio of wild-type allele dosage reduced the level of ethylene insensitivity in the triploid lines. This recovery of sensitivity by an increased ratio of wild-type to mutant alleles could result if the dominant insensitivity caused by these alleles is mediated through interactions of wild-type and mutant receptor isoforms. These could take the form of receptor heterodimers and/or higher order multimeric complexes of receptor clusters. Interestingly, the latter possibility has been suggested for the ETR1-related bacterial chemoreceptors, based on models in which alterations in one receptor can change the function of receptors held in close proximity by non-covalent interactions (Liu et al., 1997).
Alternatively, this gene dosage effect could result from increased competition by wild-type receptor isoforms for downstream effectors (e.g. CTR1). Either of these models could account for the reduction in ethylene insensitivity with increased wild-type gene dosage.
ACKNOWLEDGMENTS
We thank C. Chang for the tetraploid Bensheim lines. A.E.H. thanks T.A. Richmond for computer assistance and K. Elliot for assistance with illustrations. We thank Anita Klein, Wayne Fagerberg, and members of the Bleecker lab for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–9603679 to G.E.S. and grant no. MCB-9513463 to A.B.B.), the HATCH project (grant no. 386 to G.E.S.), the Department of Energy (grant no. DE–FG02–91ER20029 to A.B.B.), and the Department of Energy-National Science Foundation-U.S. Department of Agriculture Collaborative Research in Plant Biology Program (grant no. BIR92–20331 to support A.E.H.).
LITERATURE CITED
- Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Strohl K, eds (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Ser III. 1993;316:1194–1199. [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsisthaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Bleecker AB. Analysis of ethylene signal transduction kinetics associated with seedling-growth responses and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C. Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA. 1998;95:5401–5406. doi: 10.1073/pnas.95.9.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by ArabidopsisERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz EM. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Levit M, Lurz R, Surette MG, Stock JB. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Parkinson JS. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenberg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB. The ethylene response mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Geitz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sisler EC. Measurement of ethylene binding in plant tissue. Plant Physiol. 1979;64:538–542. doi: 10.1104/pp.64.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers JDB, Paterson NW, Brook A. Towards a quantitative definition of plant hormone sensitivity. Plant Cell Environ. 1987;14:1–12. doi: 10.1111/j.1365-3040.1987.tb02073.x. [DOI] [PubMed] [Google Scholar]