Abstract
Mendelian adult-onset leukodystrophies are a spectrum of rare inherited progressive neurodegenerative disorders affecting the white matter of the central nervous system. Among these, cerebral autosomal dominant and recessive arteriopathy with subcortical infarcts and leukoencephalopathy, cerebroretinal vasculopathy, metachromatic leukodystrophy, hereditary diffuse leukoencephalopathy with spheroids, and vanishing white matter disease present with rapidly progressive dementia as dominant feature and are caused by mutations in NOTCH3, HTRA1, TREX1, ARSA, CSF1R, EIF2B1, EIF2B2, EIF2B3, EIF2B4, and EIF2B5, respectively. Given the rare incidence of these disorders and the lack of unequivocally diagnostic features, leukodystrophies are frequently misdiagnosed with common sporadic dementing diseases such as Alzheimer's disease (AD), raising the question of whether these overlapping phenotypes may be explained by shared genetic risk factors. To investigate this intriguing hypothesis, we have combined gene expression analysis (1) in 6 different AD mouse strains (APPPS1, HOTASTPM, HETASTPM, TPM, TAS10, and TAU) at 5 different developmental stages (embryo [E15], 2, 4, 8, and 18 months), (2) in APPPS1 primary cortical neurons under stress conditions (oxygen-glucose deprivation) and single-variant–based and single-gene–based (c-alpha test and sequence kernel association test (SKAT)) genetic screening in a cohort composed of 332 Caucasian late-onset AD patients and 676 Caucasian elderly controls. Csf1r was significantly overexpressed (log2FC > 1, adj. p-value < 0.05) in the cortex and hippocampus of aged HOTASTPM mice with extensive Aβ dense-core plaque pathology. We identified 3 likely pathogenic mutations in CSF1R TK domain (p.L868R, p.Q691H, and p.H703Y) in our discovery and validation cohort, composed of 465 AD and mild cognitive impairment (MCI) Caucasian patients from the United Kingdom. Moreover, NOTCH3 was a significant hit in the c-alpha test (adj p-value = 0.01). Adult-onset Mendelian leukodystrophy genes are not common factors implicated in AD. Nevertheless, our study suggests a potential pathogenic link between NOTCH3, CSF1R, and sporadic late-onset AD, which warrants further investigation.
Keywords: Alzheimer's disease, Mendelian leukodystrophies, CSF1R, NOTCH3
1. Introduction
Mendelian adult-onset leukodystrophies are a spectrum of rare chronic progressive disorders affecting the white matter of the central nervous system. Although a growing body of literature is reporting newly discovered forms, the most characterized adult-onset leukodystrophies are cerebral autosomal dominant and recessive arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL and CARASIL), cerebroretinal vasculopathy (CRV), metachromatic leukodystrophy (MLD), hereditary diffuse leukoencephalopathy with spheroids (HDLS) and vanishing white matter disease (VWM), caused by mutations in NOTCH3, HTRA1, TREX1, ARSA, CSF1R, EIF2B1, EIF2B2, EIF2B3, EIF2B4, EIF2B5, respectively (Joutel et al., 1996), (Hara et al., 2009), (Richards et al., 2007), (Fluharty et al., 1991), (Rademakers et al., 2011), (Scali et al., 2006). Given the rare incidence of these disorders (5/100,000 to only few cases reported), the lack of peculiar and distinctive (1) clinical features, generally represented by rapidly progressive dementia, behavioral changes, pyramidal and extrapyramidal signs and, less commonly, ischemic strokes and epileptic seizures; (2) magnetic resonance imaging (MRI) lesion patterns, normally characterized by T2-weighted periventricular and subcortical, patchy and later confluent white matter hyperintensities with prominent frontal involvement; and (3) neuropathological features, frequently a combination of diverse neurodegenerative hallmarks, these rare Mendelian disorders are most frequently underrecognized and misdiagnosed with common sporadic dementias such as Alzheimer's disease (AD). On the other hand, motor features like ataxia and spasticity may appear in the course of AD progression, particularly in the cases caused by or associated to PSEN1 mutations (Rossor et al., 2010) and AD patients may display MRI patterns and neuropathological features typical of adult-onset leukodystrophies (Smith et al., 2000, Marnane et al., 2016, Barber et al., 1999, Guerreiro et al., 2013), suggesting a potential common pathogenic ground.
In the past 10 years, next-generation sequencing (NGS) paved the way for groundbreaking discoveries in AD, showing that Mendelian rare disorders offer a unique window into the sporadic complex traits and, particularly, that rare alleles in TREM2, TYROBP, and NOTCH3, causative for adult-onset leukodystrophies, significantly influence the susceptibility for AD (Guerreiro et al., 2012, Guerreiro et al., 2013, Ma et al., 2015). Moreover, the sequencing of different mouse strains showed extensive similarities between mouse and human genome and validated the importance of using mouse models to illuminate the genetics of human diseases (Cheng et al., 2014, Yue et al., 2014). Nevertheless, NGS still presents 2 main challenges: (1) the huge amount of data generated is difficult to mine and (2) the investigation of rare coding variants requires several thousands of samples. Consequently, the need for experimental methods that accurately identify critical genes and strategies to empower association studies became priorities. Therefore, we have applied a combination of cortical and hippocampal gene expression analysis in 6 diverse AD mouse strains (APPPS1, HOTASTPM, HETASTPM, TPM, TAS10, and TAU) at 5 different developmental stages (embryo [E15], 2, 4, 8 and 18 months) to comprehensively study leukodystrophy gene expression pattern in relation to the progression of AD neuropathology and under stress conditions such as oxygen-glucose deprivation (OGD), which represents an in vitro model of ischemic stroke, a common feature in several adult-onset leukodystrophies and frequent comorbidity in AD. We then used exome and genome sequencing data in a cohort composed of 332 Caucasian late-onset AD (LOAD) patients and 676 Caucasian elderly controls to investigate rare coding variability in these main adult-onset Mendelian leukodystrophy genes. Among the studied genes, Csf1r was the only gene significantly overexpressed (log2FC>1, p-value < 0.05) in AD mouse models and its expression tightly correlated with the severity of dense-core plaque deposition. Moreover, we identified a total of 3 rare variants in CSF1R tyrosine kinase (TK) domain and TK flanking regions (p.L868R and p.D565N, p.G957R, respectively) present only in cases and very likely pathogenic. We then screened CSF1R in an independent cohort composed of 465 mild cognitive impairment (MCI) and AD cases, identifying 2 additional mutations in CSF1R TK domain (p.Q691H and p.H703Y). Finally, NOTCH3 was a significant hit in the gene-based analysis (adj p-value = 0.01), suggesting a potential role as a disease modifier. We conclude that rare coding variability in adult-onset Mendelian leukodystrophy genes is not a common risk factor for AD. However, CSF1R coding variants clustering in the TK domain and NOTCH3 may influence AD susceptibility.
2. Materials and methods
2.1. Adult-onset leukodystrophy gene selection
The selected genes are all Mendelian leukodystrophy causative genes with a core clinical hallmark represented by adult-onset subacute dementia with frontal predominance revealed by T2-weighted MRI (Supplementary Table S1). Moreover, all of these candidate genes present more than one of the following features: (1) previously reported misdiagnosis with AD (CADASIL, HDLS, CRV, MLD) (Guerreiro et al., 2012, Johannsen et al., 2001, Rademakers et al., 2011, Richards et al., 2007); (2) molecular interaction with other genes playing a key role in AD (NOTCH3 and CSF1R) (Otero et al., 2009, Thijs et al., 2003); (3) genes taking part to APP-amyloid beta (Aβ) metabolism (NOTCH3 and HTRA1) (Grau et al., 2005); (4) copresence of AD neuropathological hallmarks reported (NOTCH3 and CSF1R) (Baba et al., 2006, Paquet et al., 2010); and (5) most frequently mutated genes in adults with leukoencephalopathies (NOTCH3, EIF2B4, EIF2B5, and CSF1R) (Lynch et al., 2017).
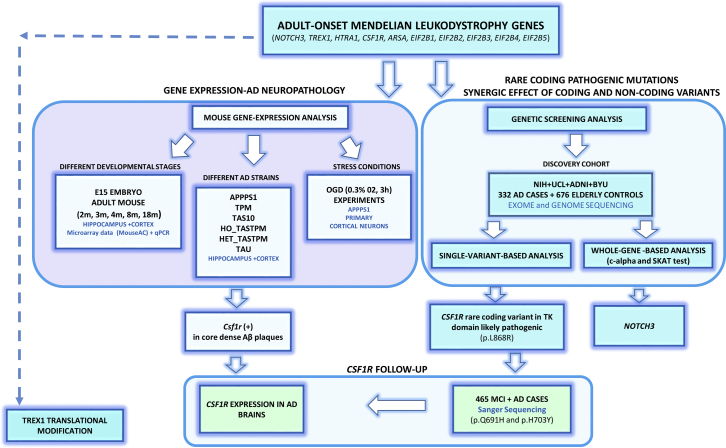
The pipeline followed in this study is described in Fig. 1.
Fig. 1.
Pipeline followed in the adult-onset leukodystrophy gene study. Abbreviations: AD, Alzheimer's disease; BYU, Brigham Young University; MCI, mild cognitive impairment; TK, tyrosine kinase; NIH, National Institutes of Health; UCL, University College London.
2.2. Gene expression analysis
We have used microarray data publicly available (MouseAC database [http://www.mouseac.org/]) (Matarin et al., 2015) and real-time polymerase chain reaction (RT-PCR) data to analyze Arsa, Csf1r, Eif2b1, Eif2b2, Eif2b3, Eif2b4, Eif2b5, Htra1, Notch3, and Trex1 gene expression (1) in the hippocampus and cortex of 6 different AD mouse strains (APPPS1, HOTASTPM, HETASTPM, TPM, TAS10, and TAU); (2) at 5 different time points (E15, 2, 4, 8, and 18 months), to comprehensively follow expression changes related to Aβ plaque density (HOTASTPM, HETASTPM, and TAS10), neurofibrillary tangles (TAU) and absence of pathology (E15 and TPM). Adult APPPS1 data for hippocampus were available only for 2 months of age, where no plaques were reported but only rare Aβ oligomers in the cortex and surrounding cortical vessels (Supplementary Fig. S1). Finally, considering that ischemic stroke is a common feature in several leukodystrophies and frequent comorbidity in AD, we used an in vitro model of ischemic stroke and performed OGD experiments in APPPS1 primary cortical neurons to test whether leukodystrophy gene expression pattern may have significantly differed between APPPS1 and wild-type (WT) mice under stress conditions.
2.3. Genetic screening
2.3.1. The discovery cohort
The discovery cohort was composed of 332 apparently sporadic AD cases and 676 elderly controls, neuropathologically and clinically confirmed, originating from the United Kingdom and North America. The mean age at disease onset was 71.66 years (range 41–94 years) for cases and the mean age of ascertainment was 78.15 years (range 60–102 years) for controls. Most of the cases (77%) were late-onset (>65 years at onset). Among the cases and controls, 42% and 51% were females, respectively; 58% and 47% of the cases and controls carried the APOE ε4 allele, respectively. The APOE ε4 allele was significantly associated to the disease status in the National Institutes of Health (NIH) and Alzheimer's Disease Neuroimaging Initiative (ADNI) series (p-value = 0.02 and 1.19 × 10E−9, respectively). This cohort has already been described elsewhere (Sassi et al., 2016). The threshold call rate for inclusion of the subject in analysis was 95%. On this cohort, we performed (1) gene-based analysis (SKAT and c-alpha tests) and (2) single-variant association analysis. Finally, we followed up, in an independent Caucasian data set, CSF1R, the only gene significantly overexpressed during AD most severe pathology (Fig. 1, Supplementary Table S2).
2.3.2. The follow-up data set
The follow-up data set was composed of 296 AD and 169 MCI late-onset cases (mean age at onset >75 years) from the United Kingdom (Supplementary Table S2). Written informed consent was obtained for each clinically assessed individual, and the study was approved by the appropriate institutional review boards. All samples had fully informed consent for retrieval and were authorized for ethically approved scientific investigation (UCLH Research Ethics Committee number 10/H0716/3, BYU IRB, Cardiff REC for Wales 08/MRE09/38+5, REC Reference 04/Q2404/130, National Research Ethics Service).
2.3.3. Exome and genome sequencing
DNA was extracted from blood or brain for cases and brain only for controls using standard protocols. Library preparation for NGS was based on Roche Nimblegen Inc. or TruSeq, Illumina protocols and has been described elsewhere (Sassi et al., 2016). Genome sequencing was performed in 199 controls, from the Cache County Study on Memory in Aging. All samples were sequenced with the use of Illumina HiSeq technology.
Sequence alignment and variant calling were performed against the reference human genome (UCSC hg19) and has been described in the Supplementary materials and methods.
Initial analysis excluded pathogenic mutations in APP, PSEN1, PSEN2, MAPT, GRN, and TREM2. All variants within the coding regions of the 10 adult-onset leukodystrophy candidate genes (ARSA [NM_000487]; CSF1R [NM_005211]; EIF2B1 [NM_001414]; EIF2B2 [NM_014239]; EIF2B3 [NM_001261418]; EIF2B4 [NM_001034116]; EIF2B5 [NM_003907]; HTRA1 [NM_002775]; NOTCH3 [NM_000435], and TREX1 [NM_016381] have been collected and analyzed, including 20.8 Megabase pairs of coding sequence.
2.3.4. Sanger sequencing
Mutations in CSF1R TK domain and flanking regions were validated with Sanger Sequencing. CSF1R was screened in an additional follow-up cohort composed of 296 AD and 169 MCI cases (Supplementary Materials and Methods).
2.4. Statistical analysis
In the single-variant analysis, allele frequencies were calculated for each low-frequency and rare coding variant in cases and controls, and Fisher's exact test on allelic association was performed. MouseAC data have been analyzed and false discovery rate (FDR) correction was applied.
The Supplementary data provide a more detailed description of the methods used (mouse and human gene expression analysis, OGD experiments, Sanger sequencing, statistical analysis, and bioinformatics).
3. Results
3.1. Gene expression analysis
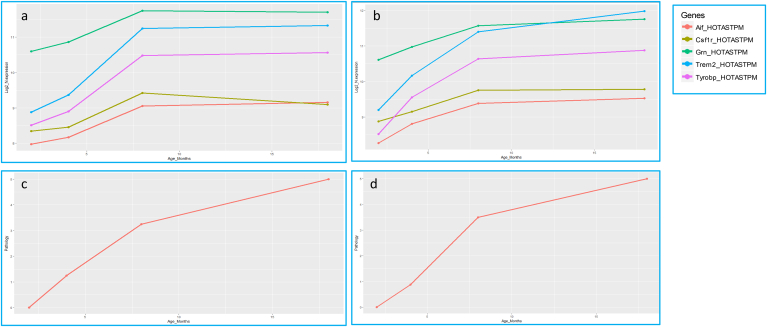
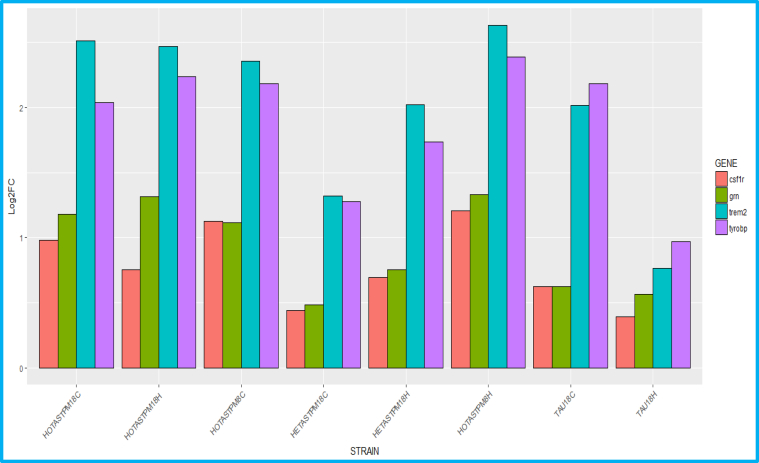
We do not report any significant differential expression in Arsa, Csf1r, Eif2b1, Eif2b2, Eif2b3, Eif2b4, Eif2b5, Htra1, Notch3, and Trex1 until the development of severe AD pathology, markedly pronounced in the most aggressive AD strain studied, HOTASTPM, homozygous for the Swedish mutation APP p.K670N/M671L and PSEN1 p.M146V, 8 months of age (Fig. 2A–D, Supplementary Tables S3 and S4). Here, Csf1r was up to 2 folds significantly overexpressed both in the hippocampus and cortex (log2FC = 1.2 and 1.1; adj p-value = 2.5e−07 and 8.7e−05, respectively) and presented a trend at 18 months both in the hippocampus and cortex (log2FC = 0.75 and 0.98; adj p-value = 2.7e−04 and 3e−04, respectively), in linear correlation with the most rapid and severe dense-core plaque deposition (0.8 dense-core plaque/month and 0.5 dense-core plaque/month between 4–8 months and 8–18 months of age, respectively) (http://www.mouseac.org/) (Fig. 2A–D, Supplementary Tables S3 and S4). Moreover, Csf1r overexpression positively correlated also with tau pathology, suggesting that Csf1r upregulation is not Aβ plaque specific. By contrast, Csf1r was downregulated when plaque deposition was minimal (HETTASTPM, TAS10, TPM, and TAU, 2m; TAS10 and TAU, 4m) (Supplementary Tables S3 and S4). Importantly, Csf1r upregulation relied on microglia infiltration and was coexpressed with other microglia markers such as Aif1, CD68, Trem2, Tyrobp, and Grn. Particularly, Csf1r and Grn displayed the same pattern of overexpression, which was between one-third to one-fourth of Tyrobp and Trem2 overall upregulation (Fig. 3, Supplementary Tables S3 and S4).
Fig. 2.
Log2-normalized expression of Csf1r, Grn, Trem2, Tyrobp, and Aif1 in HOTASTPM mice and related Aβ plaque pathology. (A–B) Log2-normalized expression of Csf1r, Grn, Trem2, Tyrobp and Aif1 in HOTASTPM mice (homozygous for the Swedish mutation APP p.K670N/M671L and PSEN1 p.M146V) at 4 different time points (2, 4, 8, and 18 months) in the hippocampus (A) and cortex (B) showing coexpression of the above genes. (C–D) Progression of AD pathology in hippocampus (C) and cortex (D), based on Aβ plaque density, in HOTASTPM mice at 4 different time points (2, 4, 8, and 18 months). Significant Csf1r, Grn, Trem2, Tyrobp, and Aif1 overexpression (Log2FC > 1, adj. p-value < 0.05) is detected at 8 months, in linear correlation with the most rapid and severe Aβ plaque deposition. Abbreviations: Aβ, amyloid beta; AD, Alzheimer's disease.
Raw data are taken from http://www.mouseac.org/
Fig. 3.
Log2FC of Csf1r, Grn, Trem2, and Tyrobp in different AD mouse strains during the most severe pathology, showing coexpression of Csf1r, Grn, Trem2, and Tyrobp. Particularly, Csf1r and Grn display the same overexpression pattern, which is, overall, 1/3 of Trem2 and Tyrobp upregulation. Abbreviations: C, cortex; H, hippocampus.
Raw data for this study are taken from http://www.mouseac.org/
3.2. Embryonal hippocampi and primary neuronal cortical cultures OGD experiments
We do not report any significant differential expression in the studied genes between APPPS1 and WT embryonal hippocampi and APPPS1 and WT primary cortical neurons after OGD experiments (Supplementary Table S5). This is likely due to the fact that most of the leukodystrophy genes are expressed on microglia, only moderately present in E15 hippocampi and in primary neuronal cortical cultures. In line with this observation, Csf1r and its ligands (Csf1 and Il34), Grn, Trem2, and Aif2 were significantly overexpressed in both APPPS1 and WT adult hippocampi compared to the embryonal ones (log2FC = 4, 2.45, 7.9, 1.5, 2.24, 3.4 and 4.2, 2.7, 7.9, 1.77, 3.1 and 3.8, respectively) (Supplementary Table S5). By contrast, Notch3 was up to 2 folds upregulated in both APPPS1 and WT embryonal hippocampi compared to adult hippocampi.
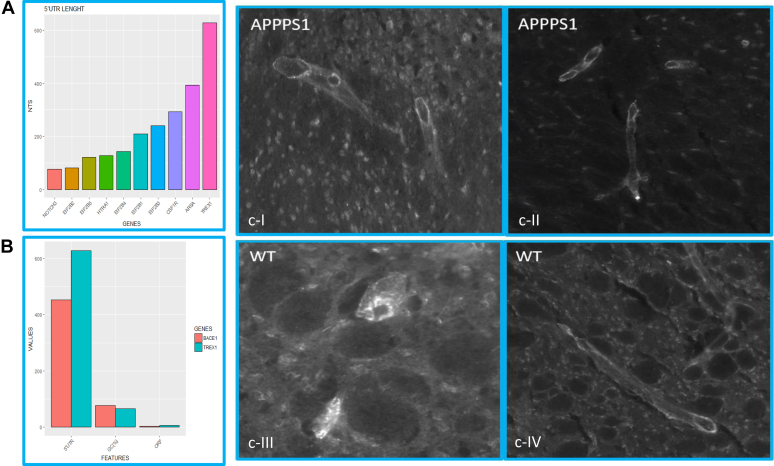
Moreover, we noticed that TREX1 5′UTR displays the typical features of many transcripts, like BACE1, that are translationally controlled by cellular stress (O'Connor et al., 2008): TREX1 5′UTR is indeed particularly long (628 nts), GC rich (65%), and predicted to contain 6 upstream open reading frames (http://www.ncbi.nlm.nih.gov/orffinder/) (Supplementary Fig. S2A, B), suggesting TREX1 transcript might be a target of translation control by one or more stress-activated pathway. Therefore, we have investigated TREX1 protein levels in both APPPS1 and WT adult brains, and we do not report any macroscopically significant difference (Supplementary Fig. S2CI-IV). This may be due to the fact that APPPS1 mice used for these experiments, being 2 months of age, did not display a severe pathology (Supplementary Fig. S1).
3.3. Genetic screening
The study population consisted of a total of 332 sporadic and mainly LOAD cases and 676 elderly controls of British and North American ancestry.
We do not report any pathogenic mutation in APP, PSEN1, and PSEN2 in our cohort. However, one of the controls was a heterozygous carrier of the protective variant APP p.A673T (minor allele frequency [MAF] 7 × 10E−4 in our cohort and MAF 5 × 10E−4 among the European non-Finnish, ExAC database, released on January 13, 2015).
We performed a single-variant and a single-gene association analysis in a predefined set of adult-onset Mendelian leukodystrophy genes (ARSA, CSF1R, EIF2B1, EIF2B2, EIF2B3, EIF2B4, EIF2B5, HTRA1, NOTCH3, and TREX1).
A total of 215 single-nucleotide variants have been identified. Among these, 77 (35.8%) were nonsynonymous, 59 (27.4%) were synonymous, and 13 (6%) UTR variants. Among the missense variants, 192 (95%) were very rare (MAF < 1%), 16 (7.9%) were low frequency (1% < MAF < 5%), and 12 (5.9%) were common (MAF > 5%). In addition, we report 4 novel coding variants (NOTCH3, p.A2146E, CSF1R p.G957R, and p.D565N and ARSA p.H425Y). Variant MAF and novel variants were based on ExAC database, European non-Finnish panel, and Exome Variant Server (EVS) European-American panel, released on March 14, 2016, or dbSNP 137 (Supplementary Table S6).
The overall variant frequency in our cohort was in line with the variant frequency reported in the American-European cohort in the Exome Variant server database (Supplementary Table S7).
3.3.1. Single-gene–based analysis
We carried out gene-wide analysis to combine the joint signal from multiple variants (coding variants and flanking UTRs) within a gene and to provide greater statistical power than that for single-marker tests. All the variants (nonsynonymous, synonymous, UTRs, and singletons) located within the studied genes and their exon-intron flanking regions were collapsed together, and their combined effect was studied.
NOTCH3 is the only significant hit in the c-alpha test (adj p-value = 0.01) (Supplementary Table S8a). The signal is driven by a common coding synonymous variant (p.P1521P) of moderate effect size (odds ratio = 1.755, confidence interval = 1.31–2.33), significant after Bonferroni correction (adj p-value = 0.02) (Supplementary Table S9). TREX1 is another hit in the c-alpha test, although nominally significant (adj p-value = 0.56), and the signal is mainly driven by a 5′UTR and synonymous (p.Y232Y) variants (Supplementary Tables S8a and S9). None of these variants were predicted to affect the splicing site (http://www.umd.be/HSF/) or a miRNA binding site (http://www.microrna.org/microrna/home.do).
3.3.2. Single-variant–based analysis
A total of 69 rare and low-frequency coding missense mutations were considered in the single-variant–based analysis in the studied genes. Among these, the majority (62.8%) were singletons (Supplementary Table S10).
Moreover, 41 missense variants (59.4%) were described as damaging variants by at least 2 of 3 in silico prediction softwares (SIFT, Polyphen and Mutation Taster).
The study possessed relatively low power to detect a significant association between cases and controls for low-frequency and rare variants, however, we analyzed these variants because we could not preclude the possibility that high-effect risk alleles were present.
EIF2B4 and CSF1R harbor the lowest and highest relative frequency of low-frequency and rare coding variants (mean = 1.27 and 5.13 low-frequency-rare variants per kb of coding sequence, respectively), with 81.25% of the rare and low-frequency coding variability in CSF1R clustering in the Ig-like domain (Supplementary Tables S11 and S10).
The main hits, although not significant, are rare variants with moderate to strong effect sizes (0.6 < odds ratio < 2.73) clustering to EIF2B4, NOTCH3, TREX1, and CSF1R (Supplementary Table S10).
None of the missense mutations leads to a premature stop codon.
3.3.3. Singletons in CSF1R TK domain
We report 2 heterozygous missense mutations in the CSF1R TK domain (exons 12–22, aa 582–910) in the discovery cohort (p.L868R and p.E694K), detected in one case and one control, respectively. Moreover, we found 4 likely pathogenic variants in the TK domain flanking regions (aa 538–581 and 911–972): CSF1R p.D565N, p.E916K, p.E920D, and p.G957R. We then screened CSF1R in an independent follow-up cohort of LOAD and MCI patients, and we identified 2 additional mutations in CSF1R TK domain (p.Q691H and p.H703Y) (Table 1).
Table 1.
Rare variants detected in CSF1R TK and TK flanking regions in the discovery and follow-up cohort (a)
| Position | rsID | cDNA | Aa change | Domain | ExAc | EVS | AD carriers (tot = 797) (%) | CTRLS carrier (tot = 676) | MT | p-value | Adj p-value | OR | CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr5:149433682 | Novel | c.2869G > C | p.G957R | TK flanking region | NA | NA | 1 (0.12) | 0 | Disease_causing | 1 | 1 | Inf | 0.021–Inf |
| chr5:149433888 | rs34030164 | c.2760G > C | p.E920D | TK flanking region | 3e-3 | 2.5e-3 | 2 (0.25) | 7 (1) | polymorphism | 0.089 | 1 | 0.24 | 0.024–1.26 |
| chr5:149433902 | rs142435467 | c.2746G > A | p.E916K | TK flanking region | 7e-4 | 9.3e-4 | 0 | 2 (0.29) | Disease_causing | 0.21 | 1 | 0 | 0–4.5 |
| chr5:149434851 | rs281860278 | c.2603T > G | p.L868R | TK | 8.9e-5 | NA | 1 (0.12) | 0 | Disease_causing | 1 | 1 | Inf | 0.021–Inf |
| chr5:149439287 | rs111943087 | c.2107C > T | p.H703Ya | TK | NA | NA | 1 (0.12) | 0 | Polymorphism | 1 | 1 | Inf | 0.021–Inf |
| chr5:149439315 | rs545858226 | c.2080G > A | p.E694K | TK | 1.4e-5 | NA | 0 | 1 (0.14) | Disease_causing | 0.45 | 1 | 0 | 0–33 |
| chr5:149439322 | Novel | c.2073G > C | p.Q691Ha | TK | 6e-5 | NA | 1 (0.12) | 0 | Polymorphism | 1 | 1 | Inf | 0.021–Inf |
| chr5:149441346 | Novel | c.1693G > A | p.D565N | TK flanking region | NA | NA | 1 (0.12) | 0 | Disease_causing | 1 | 1 | Inf | 0.021–Inf |
Key: AD, Alzheimer's disease; Aa, amino acid; tot, total; MT, Mutation Taster; Adj p-value, adjusted p-value, based on Bonferroni correction with 69 rare coding variants; OR, odds ratio; CI, confidence interval. NA, not available; Inf, infinity.
Variants detected in the follow-up cohort.
CSF1R TK mutation carriers (patients E, F, and H) presented a rather homogeneous phenotype (Table 2). All these carriers were LOAD cases displaying memory impairment at the onset. Behavioral and motor signs eventually appeared. In 2/3 patients, cardiovascular problems and strokes preceded the dementia. The neuropathology examination, available for patients H and I, showed aggressive and diffuse neurodegeneration (Braak 6 and CERAD C). Two of 3 carriers were heterozygous for APOE ε4 allele and do not report any familial history for dementia. By contrast, patient H was homozygous for APOE ε4 allele, had a family history for dementia (4 brothers) and plausibly the combination of these risk factors, likely coupled with a pre-existent cerebrovascular disorder, may explain the earlier age at onset compared to the other patients (64 years). Patient I carried a missense mutation in CSF1R TK flanking region (p.G957R) and displayed a different clinical picture, dominated by early-onset dementia (49 years) and language problems at the onset. Despite the small sample size, we do not report any association between age at onset, severity of the disease progression, and disease duration.
Table 2.
CSF1R TK and TK flanking region mutation carrier description
| Patient | CSF1R mutation | APOE | AO-AD | Family history of dementia | Disease duration | First symptom | Behavioral symptom | Motor symptom | Vascular risk factors | Misdiagnosis | CT/MRI | Neuropath |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient D | p.D565N | 34 | −92 | NA | NA | NA | NA | NA | NA | NA | NA | |
| Patient E | p.Q691H | 34 | 82–89 | Negative | 7y, rapid deterioration during the last 7 months | Memory problems | No | Symmetric patchy periventricular hyperintensities, mainly pronounced in the frontal lobes | ||||
| Patient F | p.H703Y | 34 | 79–82 | Negative | 3y | Memory problems | Irritability and anxiety | Intermittent mild rigidity, tremor and bradykinesia, mild left hemiparesis | Bilateral severe carotid artery stenosis, vertebrobasilar TIA | Vascular dementia | Hippocampal and temporal lobe atrophies, subcortical microbleeds (right basal ganglia), and small ischemic stroke (left pons), lacunar infarct right parietal lobe, centrum semiovale bilateral lesions | |
| Patient H | p.L868R | 44 | 64–75 | 4/4 siblings diagnosed with dementia | 11y | Short-term memory problems and dysphasia | Stroke at 65y | Vascular dementia | Severe microbleeding | Extensive Aβ and tau deposition (Braak VI and CERAD C), amyloid angiopathy and focal TDP-43 | ||
| Patient I | p.G957R | NA | 49–57 | Negative | 8y | Language problem | Aggression and paranoia later in the course of the disease | No | PNFA | Braak VI and CERAD C |
Key: Aβ, amyloid beta; AO-AD, age at onset-age at death; symp., symptoms; CT/MRI, computed tomography/magnetic resonance imaging; NA, not available; TIA, transient ischemic attack; PNFA, progressive nonfluent aphasia.
Detailed clinical description was available for 4 patients. The clinical, neuroimaging, and neuropathological features of the carriers are summarized in Table 2.
3.3.4. Patient H (p.L868R)
This male patient deceased at the age of 75 years. He was one of 5 siblings who all survived to old age, of whom 4 experienced memory problems or received a diagnosis of dementia or AD. The informant reported he experienced sudden decline following a stroke at 65 years. He had obvious short-term memory problems and dysphasia. At this stage, he was considered to have probable vascular dementia. Pathological examination of the brain concluded this patient had a high probability of AD. Neurofibrillary tangle stage was consistent with Braak stage VI, while plaque pathology met CERAD criteria for score C. In addition, there was evidence of amyloid angiopathy, focal TDP-43 positivity, and occasional glial inclusions.
3.3.5. Patient E (p.Q691H)
This patient deceased at the age of 89 years. She complained of memory problems at the age of 82 years, and 2 years later underwent an MRI scan, which showed symmetric patchy periventricular hyperintensities, mainly pronounced in the frontal lobe (Fig. 4A). Following annual visits involving neuropsychiatric testing, she received a diagnosis of AD at the age of 86 years. In the 3 years following her diagnosis, her symptoms were quite stable. She experienced a rapid deterioration in the last 7–8 months before her death.
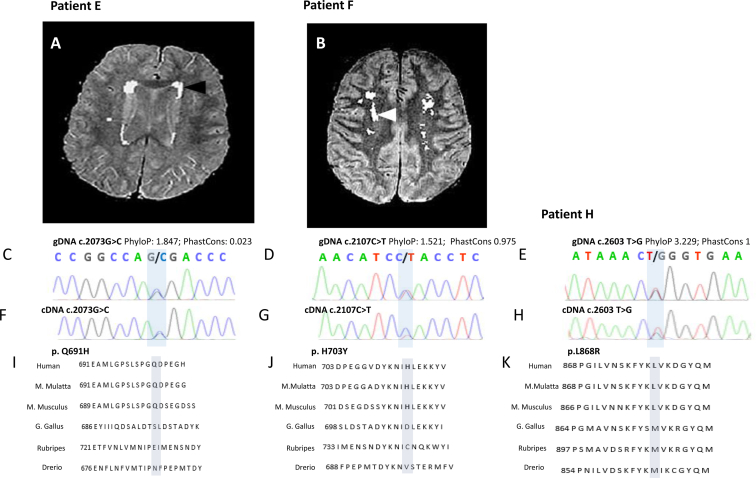
Fig. 4.
MRI scans, Sanger sequencing validation, and mutation domain conservation for patients E, F, and H. (A–B) Coronal T2-weighted MRI scan of patients E and F (for both taken 2y after onset of symptoms, aged 84y and 81y, respectively) showing symmetric patchy periventricular hyperintensities, mainly pronounced in the frontal lobes (A) and bilateral lesions localized to the centrum semiovale (B) (arrows). (C–E) Genomic DNA Sanger sequencing validation of CSF1R c.2073G>C, c.2107C>T, and c.2603 T>G mutations. (F–H) cDNA Sanger sequencing validation of c.2073G>C, c.2107C>T, and c.2603 T>G mutations. For patients F and H, cDNA sequence highlights a possible allelic imbalance, supporting the likely functional effect of gDNA c.2107C>T and gDNA c.2603 T>G mutations. (I–K) Conservation of p.Q691H, p. H703Y, and p.L868R in different species. PhastCons and PhyloP scores range between 0–1 and −14 to +6, respectively. For PhastCons, the closer to one, the more conserved; for PhyloP, conserved sites are assigned positive scores. Abbreviation: MRI, magnetic resonance imaging.
3.3.6. Patient F (p.H703Y)
This patient had a strong history of cardiovascular disease and reported memory symptoms, at the age of 79 years, followed by irritability and anxiety and 2 years later received a clinical diagnosis of probable AD. Computed tomography of the brain showed supratentorial atrophy, temporal lobe atrophy, and slight vascular changes. The patient also experienced intermittent motor symptoms that included mild rigidity, tremor, and slowness of movement. The MRI scans showed central and cortical atrophies and mild to moderate medial temporal lobe atrophy, as well as a small old hemorrhage, ischemic lesions, and bilateral lesions localized to the centrum semiovale (Fig. 4B).
Patient I's detailed description is in the Supplementary data.
3.4. Tissue expression of CSF1R
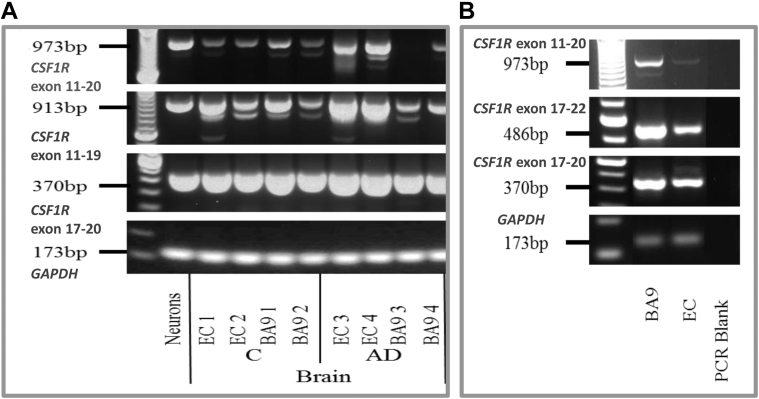
We followed up our findings checking CSF1R expression in LOAD and control brain samples. We selected the entorhinal cortex (EC) and BA9 preassociation cortex (BA9) because the brain regions primarily affected by AD spreading pathology (Khan et al., 2014). CSF1R was overexpressed in AD EC compared to AD BA9 preassociation cortex and control brains (Fig. 5A).
Fig. 5.
Tissue expression of CSF1R. (A) Relative CSF1R and GAPDH expression assessed by RT-PCR in postmortem brain RNA from normal control and AD individuals. (B) Expression of CSF1R transcripts and the house-keeping gene GAPDH in the EC and BA9 preassociation cortex (BA9) from patient H (c.2603T>G, p.L868R). RT-PCR for CSF1R exon 11 to 20, exon 11 to 19, exon 17 to 20, and exon 17 to 22 as indicated. Abbreviations: EC, entorhinal cortex; BA, Brodmann area; AD, Alzheimer’s disease; C, normal control; RT-PCR, Real Time PCR.
It was not possible to quantitatively compare levels of CSF1R in all the 3 CSF1R TK mutation carriers due to a lack of available brain tissue. However, cDNA Sanger Sequencing revealed a possible allelic imbalance, with the WT allele normally expressed and the mutated one only moderately both for patient F (p.H703Y) and patient H (p.L868R), suggesting a functional role of these mutations (Fig. 4G–H). RNA from the EC and the BA9 preassociation cortex was available for patient H and showed significantly lower expression of CSF1R (1) in the EC compared to BA9 preassociation cortex and (2) in patient H's EC compared to other AD patients and controls for all CSF1R primers tested (Fig. 5A–B).
4. Discussion
Mendelian adult-onset leukodystrophies clinically resemble common dementias such as AD, potentially implying they may be influenced by shared genetic risk factors.
To comprehensively investigate this hypothesis, we applied a combination of gene expression analysis in different AD mouse strains at diverse developmental stages (http://mouseac.org/) and single-variant–based and single-gene–based genetic screening in a cohort composed of 332 LOAD cases and 676 elderly controls from the United Kingdom and United States (Fig. 1). Divergent gene expression between AD and WT mouse strains was only detected in aged mice with severe dense-core plaque deposition (Fig. 2A–D, Supplementary Tables S3 and S4).
Csf1r was the only gene displaying a significant differential expression between AD and WT mouse strains. It was up to 2 folds significantly overexpressed both in the hippocampus and cortex in HOTASTPM mice aged 8 months (log2FC = 1.2 and 1.1; adj p-value = 2.5E−07 and 8.7E−05, respectively), and its overexpression linearly correlated with the rapidity of dense-core plaque deposition rather than with their overall load (Fig. 2A–D, Supplementary Tables S3 and S4). By contrast, Csf1r was downregulated when the pathology was minimal or absent (Supplementary Tables S3 and S4), suggesting that Csf1r upregulation was tightly driven by and consequential to dense-core plaque formation and, to a lesser extent, neurofibrillary tau tangles.
Importantly, Csf1r expression pattern relied on microglia infiltration as overexpression of Aif1 suggested (Fig. 2A–B, Supplementary Tables S3 and S4). Csf1r was coexpressed and shared the same expression pattern with Trem2, Tyrobp, and Grn, critical genes expressed on microglia, whose loss of function mutation in homozygosity is causative for adult-onset leukodystrophies such as polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL) (TREM2 and TYROBP) (Paloneva et al., 1993), and in heterozygosity causes frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP43) (GRN) (Baker et al., 2006) or is a significant risk factor for sporadic AD (TREM2) (Guerreiro et al., 2013), and whose overexpression is protective and limits AD neuropathology through a very effective clearance of Aβ plaques (GRN and TREM2) (Minami et al., 2014). Notably, in all the strains, Csf1r and Grn degree of expression correlated, and this was generally one-third of Tyrobp and Trem2 overall upregulation (Fig. 3, Supplementary Tables S3 and S4). This effect was not simply due to the aging process: we do not report significant differential expression in Csf1r, Grn, Trem2, and Tyrobp in the cortex and hippocampus of WT mice between 2 and 18 months of age (−0.04 < log2FC < 0.2 and 0.04 < log2FC < 0.4 in the hippocampus and cortex, respectively) (Supplementary Tables S3 and S4). Importantly, CSF1R, TREM2, and TYROBP have been already shown to cointeract (Otero et al., 2009) (https://string-db.org/). Here, we report GRN as an additional potential key player and its coexpression on microglia strengthens its synergic function. Therefore, this may imply that CSF1R, in concert with TREM2, TYROBP, and GRN, plays a key role in Aβ plaque removal, hypothesis supported by previous literature, reporting CSF1R overexpressed in AD patients particularly around senile plaques and taking part in Aβ removal (Akiyama et al., 1994) (Murphy et al., 2000) (Mizuno et al., 2011) (Boissonneault et al., 2009).
By contrast, no differential expression of any adult-onset leukodystrophy gene was observed in the E15 hippocampi and primary cortical neurons after OGD experiments between APPPS1 and WT strains, likely given the fact that these genes, although expressed in neurons, mainly exert a critical role on microglia (TREX1 and CSF1R), astrocytes (NOTCH3 and HTRA1), and endothelial cells (NOTCH3, HTRA1, TREX1, EIF2B2, EIF2B3, and EIF2B5) (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) that are minimally present in E15 hippocampi and primary cortical neuronal cultures.
In our discovery and validation cohorts, we detected 3 rare coding variants in the TK domain of CSF1R in 3 LOAD cases, one of these neuropathologically confirmed. Moreover, we report 2 cases harboring rare mutations in the TK flanking regions (aa 538–580 and aa 911–972, encoded by exon 12 and 22, respectively), where an additional causative mutation for HDLS has been recently described (c.1736G>A, p.R579Q) (Ghadiri et al., 2014). These variants are very likely pathogenic: they cluster to highly conserved domains among different species (average PhyloP and PhastCons scores = 2.2 and 0.6, respectively) (Fig. 4I–K) and have been detected only in cases.
Moreover, CSF1R p.L868R is a functional mutation as it was associated with a reduced expression of the mutated allele (Fig. 4H) and decreased CSF1R expression in the EC, a region primarily affected in AD and generally displaying CSF1R upregulation in AD patients (Fig. 5A and B). Importantly, a different amino acid change, clustering within the same codon (p.L868P) has been reported as de novo mutation, causative for HDLS (Rademakers et al., 2011). Remarkably, to date, any missense mutation in Mendelian gene domains harboring heterozygous causative mutations for autosomal dominant disorders such as familial AD and FTD (APP, PSEN1, PSEN2, and MAPT) has always been reported as pathogenic (Supplementary Table S12, http://www.molgen.ua.ac.be/ADMutations/). The only exception is represented by APP p.A673T, which is a very rare protective factor for AD (Jonsson et al., 2012). In addition, an intronic single-nucleotide polymorphism (SNP) in CSF1R, rs1010101, displayed a trend toward association (adj p-value = 2E−4), in a genome-wide association study performed in Caucasian LOAD patients (Wijsman et al., 2011), supporting CSF1R possible role in LOAD progression.
CSF1R mutation carriers presented a homogeneous phenotype, closely resembling HDLS. First, the symptom at onset was a memory deterioration followed by behavioral changes in 3/3 carriers. Second, T2-weighted MRI, available for 2 patients, showed symmetric patchy periventricular hyperintensities, mainly pronounced in the frontal lobe (patient E) and bilateral lesions localized to the centrum semiovale (patient F) (Fig. 4B), that represent common MRI findings in HDLS patients (Ghadiri et al., 2014), (Rademakers et al., 2011) (Boissé et al., 2010) (Mateen et al., 2010). Finally, senile plaques, amyloid angiopathy, and tau tangles have been reported also in the cortex and hippocampus of 2 familial and 1 sporadic HDLS patients (Baba et al., 2006) (Browne et al., 2003). Importantly, these 3 HDLS patients displaying AD neuropathological hallmarks presented an overall early age at onset (average 54 years [range 78–32 years]), developed Parkinsonism, atypical Parkinsonism, and motor impairment with increasing rigidity (Supplementary Table S13). By contrast, only patient F (20%) displayed intermittent mild rigidity, tremor, and bradykinesia, arguing for Parkinsonism, however within a neurological picture already dominated by cerebrovascular disorders, and patient H reported no sign of motor impairment besides 2 falls in few months. Nevertheless, although an earlier age at onset, Parkinsonism and distinctive motor features may be more common in HDLS patients presenting AD neuropathology than AD cases carrying CSF1R TK mutations; the average disease duration for both these HDLS and AD patients was 7 years. Therefore, in the absence of accurate differential diagnostic criteria, this combination of clinical, neuroimaging, and neuropathological features strikingly overlapping makes the definitive neurological diagnosis a real conundrum. The fact that potentially pathogenic mutations in the TK domain in heterozygosity may be detected either in databases and apparently healthy controls, may give rise to HDLS or may be rare risk factors for AD may be due to different factors modifying the mutation penetrance (Karle et al., 2013). Analogously to GRN missense mutations in AD and FTD, CSF1R mutation penetrance may be influenced by APOE genotype, aging, disease duration, or comorbidities such as cerebrovascular accidents, for which CSF1R has been already shown to play a critical protective role (Luo et al., 2013). In addition, it may be plausible that most HDLS patients may not display AD neuropathology due to the rapid progression of the disease (Baba et al., 2006).
Finally, NOTCH3 was a significant hit in the gene-based analysis (c-alpha test, adj p-value = 0.01). The signal was driven both by a common synonymous variant (p.P1521P) (Supplementary Table S9) that may influence gene expression (Sauna and Kimchi-Sarfaty, 2011) and 3 rare coding variants with large effect size (p.V1952M, p.V1183M, p.H170R, 2.73 <0R < 1.63), whose carrier frequency was between 2 and 3 times higher in cases compared to controls, although not significant (Supplementary Tables S6 and S10). Importantly, these rare variants (p.V1952M, p.V1183M, and p.H170R) have been already reported to be significantly associated with severity of white matter lesions in elderly with hypertension (Schmidt et al., 2011), suggesting a potential role as disease modifier in LOAD. In addition, we report a heterozygous pathogenic gain of cysteine mutation in NOTCH3 (p.R578C) detected in 1 control and already reported in a Korean patient with clinical suspicious CADASIL, implying that the penetrance of NOTCH3 mutations is variable (Kim et al., 2014).
Therefore, although canonical NOTCH3 mutations causative for CADASIL are highly stereotyped: (1) cluster in epidermal growth factor–like repeat domains, (2) in exons 3 and 4, and (3) consist in the gain or loss of cysteine; nevertheless, our study reports a possible synergetic effect of common and rare variants in NOTCH3 potentially influencing AD susceptibility through an increased risk for small vessel disease or white matter lesions. Our hypotheses are supported by a growing body of evidence showing that (1) NOTCH3 common variants (rs1043994, rs10404382, rs10423702, and rs1043997) are significantly associated with white matter lesions in elderly with hypertension (Schmidt et al., 2011) and (2) rare noncysteine mutations may be pathogenic as they have been reported in Korean and Japanese CADASIL patients, in a French case with small vessel disease, and have been associated to severe white matter lesions in elderly patients (Fouillade et al., 2008, Mizuno et al., 2008, Schmidt et al., 2011). Importantly, our findings add evidence to the pathogenic link between AD and CADASIL, displaying clinical shared features and rarely, as only few cases have been reported, neuropathological hallmarks characterized by Aβ plaques, amyloid angiopathy, and neurofibrillary tangles (Guerreiro et al., 2012, Gray et al., 1994, Paquet et al., 2010, Thijs et al., 2003). Biologically, presenilins cleaving both APP and NOTCH3 may bridge the gap between AD and CADASIL. However, whether NOTCH3 mutations or differential expression may accelerate a pre-existing AD or AD may contribute to CADASIL exacerbation remains to be elucidated.
In summary, adult-onset Mendelian leukodystrophy genes are not common factors in AD, therefore the genetic screening plays a pivotal role in the differential diagnosis. However, genetically diagnosed HDLS and CADASIL patients may display clinical, neuroimaging, and neuropathological features meeting the diagnostic criteria for AD, leaving the definitive diagnosis a significant challenge. Here, we report neuropathologically confirmed AD patients carrying likely pathogenic mutations in CSF1R TK domain and a potential association between AD and NOTCH3. Our study provides compelling evidence that HDLS, CADASIL, and AD may represent shades of the same disease spectrum. Moreover, we support previous studies, suggesting that CSF1R, in concert with TREM2, TYROBP, and GRN, may play a critical role in Aβ plaque clearance and therefore may represent a pivotal, although rare, genetic factor influencing AD susceptibility. Given the very rare frequency of CSF1R TK pathogenic mutations detected in the screened patients (0.3% LOAD carriers), our hypotheses should foster genetic screening in larger cohorts of both early-onset AD and LOAD cases and functional studies.
Disclosure statement
All the authors declare no competing financial or personal interests that can influence the presented work. However, MAN's participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging NIH, Bethesda, MD, USA, as a possible conflict of interest, and he also consults Illumina Inc, the Michael J. Fox Foundation, and the University of California Healthcare among others.
Acknowledgements
This study was supported by the Alzheimer's Research UK, the Medical Research Council (MRC), the Wellcome Trust/MRC Joint Call in Neurodegeneration Award (WT089698) to the UK Parkinson's Disease Consortium (whose members are from the University College London Institute of Neurology, the University of Sheffield, and the MRC Protein Phosphorylation Unit at the University of Dundee), grants (P50 AG016574, U01 AG006786, and R01 AG18023), the National Institute for Health Research Biomedical Research Unit in Dementia at University College London Hospitals, University College London; the Big Lottery (to KM); JB and RG's work is funded by fellowships from the Alzheimer's Society; Humboldt Fellowship (to CS) and the Intramural Research Programs of the National Institute on Aging and the National Institute of Neurological Disease and Stroke, National Institutes of Health (Department of Health and Human Services Project number, ZO1 AG000950-10). MAN's participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging NIH, Bethesda, MD, USA. The MRC London Neurodegenerative Diseases Brain Bank and the Manchester Brain Bank from Brains for Dementia Research are jointly funded from ARUK and AS. The Alzheimer's Research UK (ARUK) Consortium funded PP, DC, JJ, BMcG, ST, Queen's University Belfast, UK; RH, Royal Derby Hospital, UK; HK, University of Bonn, Germany; PGK, University of Bristol, UK; NMH, University of Leeds, UK; ERLCV, University of Newcastle, UK; DMM, SP-B, University of Manchester, UK; KB, JL, KM, University of Nottingham, UK; ADS, GW, DW, University of Oxford (OPTIMA), UK; CH, University of Southampton, UK.
Tissue samples were supplied by The London Neurodegenerative Diseases Brain Bank, which receives funding from the MRC and as part of the Brains for Dementia Research programme, jointly funded by Alzheimer's Research UK and Alzheimer's Society.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.neurobiolaging.2018.01.015.
Contributor Information
Celeste Sassi, Email: celeste.sassi@charite.de.
ARUK Consortium:
Peter Passmore, David Craig, Janet Johnston, Bernadette McGuinness, Stephen Todd, Reinhard Heun, Heike Kölsch, Patrick G. Kehoe, Emma R.L.C. Vardy, Nigel M. Hooper, David M. Mann, Stuart Pickering-Brown, Kristelle Brown, James Lowe, Kevin Morgan, A. David Smith, Gordon Wilcock, Donald Warden, and Clive Holmes
Appendix A. Supplementary data
Supplementary Fig. S1.
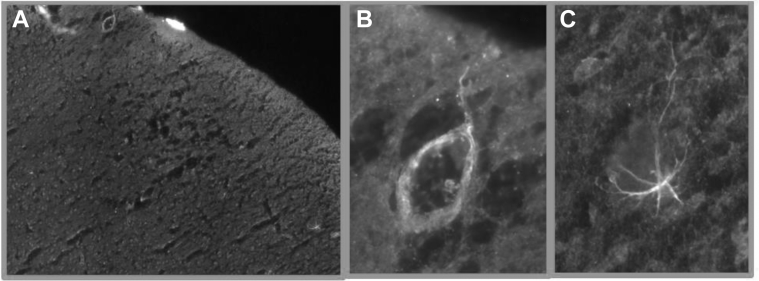
(A) Histological cortical section of APPPS1 mouse, 2 months of age, stained with Aβ oligomers (Ab126892). Rare Aβ oligomers are detected around small cortical vessels (B) and in the extracellular space, likely phagocytated by astrocytes (C), showing a very mild pathology and disease state. No Aβ plaques have been detected (data not shown).
Supplementary Fig. S2.
(A) 5′UTR length of the studied adult-onset leukodystrophy genes. TREX1 presents the longest 5′UTR region (628nts). (B) TREX1 displays analogous 5′UTR features of BACE1, known to be subjected to translational control: (1) 5′UTR length (628 and 453 nts, respectively); (2) GC content (65% and 77%, respectively), and (3) predicted open reading frames (ORFs) (6 and 3, respectively). (CI–IV) Cortical histological sections of APPPS1 and WT mice (aged 2 months), stained with TREX1 antibody (Abnova 68191), which mainly binds to endothelial cells and to a lesser extent neurons. No macroscopical difference has been detected between APPPS1 and WT mice. Abbreviation: WT, wild type.
References
- Akiyama H., Nishimura T., Kondo H., Ikeda K., Hayashi Y., McGeer P.L. Expression of the receptor for macrophage colony stimulating factor by brain microglia and its upregulation in brains of patients with Alzheimer’s disease and amyotrophic lateral sclerosis. Brain Res. 1994;639:171–174. doi: 10.1016/0006-8993(94)91779-5. [DOI] [PubMed] [Google Scholar]
- Baba Y., Ghetti B., Baker M.C., Uitti R.J., Hutton M.L., Yamaguchi K., Bird T., Lin W., DeLucia M.W., Dickson D.W., Wszolek Z.K. Hereditary diffuse leukoencephalopathy with spheroids: clinical, pathologic and genetic studies of a new kindred. Acta Neuropathol. (Berl.) 2006;111:300–311. doi: 10.1007/s00401-006-0046-z. [DOI] [PubMed] [Google Scholar]
- Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C.A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Barber R., Scheltens P., Gholkar A., Ballard C., McKeith I., Ince P., Perry R., O’Brien J. White matter lesions on magnetic resonance imaging in dementia with Lewy bodies, Alzheimer's disease, vascular dementia, and normal aging. J. Neurol. Neurosurg. Psychiatry. 1999;67:66–72. doi: 10.1136/jnnp.67.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissé L., Islam O., Woulfe J., Ludwin S.K., Brunet D.G. Neurological picture. Hereditary diffuse leukoencephalopathy with neuroaxonal spheroids: novel imaging findings. J. Neurol. Neurosurg. Psychiatry. 2010;81:313–314. doi: 10.1136/jnnp.2009.180224. [DOI] [PubMed] [Google Scholar]
- Boissonneault V., Filali M., Lessard M., Relton J., Wong G., Rivest S. Powerful beneficial effects of macrophage colony-stimulating factor on beta-amyloid deposition and cognitive impairment in Alzheimer’s disease. Brain J. Neurol. 2009;132:1078–1092. doi: 10.1093/brain/awn331. [DOI] [PubMed] [Google Scholar]
- Browne L., Sweeney B.J., Farrell M.A. Late-onset neuroaxonal leucoencephalopathy with spheroids and vascular amyloid. Eur. Neurol. 2003;50:85–90. doi: 10.1159/000072504. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Ma Z., Kim B.-H., Wu W., Cayting P., Boyle A.P., Sundaram V., Xing X., Dogan N., Li J., Euskirchen G., Lin S., Lin Y., Visel A., Kawli T., Yang X., Patacsil D., Keller C.A., Giardine B., Mouse ENCODE Consortium. Kundaje A., Wang T., Pennacchio L.A., Weng Z., Hardison R.C., Snyder M.P. Principles of regulatory information conservation between mouse and human. Nature. 2014;515:371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluharty A.L., Fluharty C.B., Bohne W., von Figura K., Gieselmann V. Two new arylsulfatase A (ARSA) mutations in a juvenile metachromatic leukodystrophy (MLD) patient. Am. J. Hum. Genet. 1991;49:1340–1350. [PMC free article] [PubMed] [Google Scholar]
- Fouillade C., Chabriat H., Riant F., Mine M., Arnoud M., Magy L., Bousser M.G., Tournier-Lasserve E., Joutel A. Activating NOTCH3 mutation in a patient with small-vessel-disease of the brain. Hum. Mutat. 2008;29:452. doi: 10.1002/humu.9527. [DOI] [PubMed] [Google Scholar]
- Ghadiri M., Buckland M.E., Sutton I.J., Al Jahdhami S., Flanagan S., Heard R., Barnett Y., Brennan J., Barnett M.H. Progressive neuropsychiatric symptoms and motor impairment. JAMA Neurol. 2014;71:794–798. doi: 10.1001/jamaneurol.2013.6308. [DOI] [PubMed] [Google Scholar]
- Grau S., Baldi A., Bussani R., Tian X., Stefanescu R., Przybylski M., Richards P., Jones S.A., Shridhar V., Clausen T., Ehrmann M. Implications of the serine protease HtrA1 in amyloid precursor protein processing. Proc. Natl. Acad. Sci. U. S. A. 2005;102:6021–6026. doi: 10.1073/pnas.0501823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray F., Robert F., Labrecque R., Chrétien F., Baudrimont M., Fallet-Bianco C., Mikol J., Vinters H.V. Autosomal dominant arteriopathic leuko-encephalopathy and Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 1994;20:22–30. doi: 10.1111/j.1365-2990.1994.tb00953.x. [DOI] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S., Hazrati L., Collinge J., Pocock J., Lashley T., Williams J., Lambert J.-C., Amouyel P., Goate A., Rademakers R., Morgan K., Powell J., St George-Hyslop P., Singleton A., Hardy J., Alzheimer genetic analysis group TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R.J., Lohmann E., Kinsella E., Brás J.M., Luu N., Gurunlian N., Dursun B., Bilgic B., Santana I., Hanagasi H., Gurvit H., Gibbs J.R., Oliveira C., Emre M., Singleton A. Exome sequencing reveals an unexpected genetic cause of disease: NOTCH3 mutation in a Turkish family with Alzheimer’s disease. Neurobiol. Aging. 2012;33:1008.e17–1008.e23. doi: 10.1016/j.neurobiolaging.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Shiga A., Fukutake T., Nozaki H., Miyashita A., Yokoseki A., Kawata H., Koyama A., Arima K., Takahashi T., Ikeda M., Shiota H., Tamura M., Shimoe Y., Hirayama M., Arisato T., Yanagawa S., Tanaka A., Nakano I., Ikeda S., Yoshida Y., Yamamoto T., Ikeuchi T., Kuwano R., Nishizawa M., Tsuji S., Onodera O. Association of HTRA1 mutations and familial ischemic cerebral small-vessel disease. N. Engl. J. Med. 2009;360:1729–1739. doi: 10.1056/NEJMoa0801560. [DOI] [PubMed] [Google Scholar]
- Johannsen P., Ehlers L., Hansen H.J. Dementia with impaired temporal glucose metabolism in late-onset metachromatic leukodystrophy. Dement. Geriatr. Cogn. Disord. 2001;12:85–88. doi: 10.1159/000051240. [DOI] [PubMed] [Google Scholar]
- Jonsson T., Atwal J.K., Steinberg S., Snaedal J., Jonsson P.V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., Hoyte K., Gustafson A., Liu Y., Lu Y., Bhangale T., Graham R.R., Huttenlocher J., Bjornsdottir G., Andreassen O.A., Jönsson E.G., Palotie A., Behrens T.W., Magnusson O.T., Kong A., Thorsteinsdottir U., Watts R.J., Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- Joutel A., Corpechot C., Ducros A., Vahedi K., Chabriat H., Mouton P., Alamowitch S., Domenga V., Cécillion M., Marechal E., Maciazek J., Vayssiere C., Cruaud C., Cabanis E.A., Ruchoux M.M., Weissenbach J., Bach J.F., Bousser M.G., Tournier-Lasserve E. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Karle K.N., Biskup S., Schüle R., Schweitzer K.J., Krüger R., Bauer P., Bender B., Nägele T., Schöls L. De novo mutations in hereditary diffuse leukoencephalopathy with axonal spheroids (HDLS) Neurology. 2013;81:2039–2044. doi: 10.1212/01.wnl.0000436945.01023.ac. [DOI] [PubMed] [Google Scholar]
- Khan U.A., Liu L., Provenzano F.A., Berman D.E., Profaci C.P., Sloan R., Mayeux R., Duff K.E., Small S.A. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer’s disease. Nat. Neurosci. 2014;17:304–311. doi: 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-E., Yoon C.W., Seo S.W., Ki C.-S., Kim Y.B., Kim J.-W., Bang O.Y., Lee K.H., Kim G.-M., Chung C.-S., Na D.L. Spectrum of NOTCH3 mutations in Korean patients with clinically suspicious cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Neurobiol. Aging. 2014;35:726.e1-6. doi: 10.1016/j.neurobiolaging.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Luo J., Elwood F., Britschgi M., Villeda S., Zhang H., Ding Z., Zhu L., Alabsi H., Getachew R., Narasimhan R., Wabl R., Fainberg N., James M.L., Wong G., Relton J., Gambhir S.S., Pollard J.W., Wyss-Coray T. Colony-stimulating factor 1 receptor (CSF1R) signaling in injured neurons facilitates protection and survival. J. Exp. Med. 2013;210:157–172. doi: 10.1084/jem.20120412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D.S., Rodrigues Brandão de Paiva A., Zhang W.J., Bugiardini E., Freua F., Tavares Lucato L., Macedo-Souza L.I., Lakshmanan R., Kinsella J.A., Merwick A., Rossor A.M., Bajaj N., Herron B., McMonagle P., Morrison P.J., Hughes D., Pittman A., Laurà M., Reilly M.M., Warren J.D., Mummery C.J., Schott J.M., Adams M., Fox N.C., Murphy E., Davagnanam I., Kok F., Chataway J., Houlden H. Clinical and genetic characterization of leukoencephalopathies in adults. Brain. 2017;140:1204–1211. doi: 10.1093/brain/awx045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Jiang T., Tan L., Yu J.-T. TYROBP in Alzheimer’s disease. Mol. Neurobiol. 2015;51:820–826. doi: 10.1007/s12035-014-8811-9. [DOI] [PubMed] [Google Scholar]
- Marnane M., Al-Jawadi O.O., Mortazavi S., Pogorzelec K.J., Wang B.W., Feldman H.H., Hsiung G.-Y.R., Alzheimer’s Disease Neuroimaging Initiative Periventricular hyperintensities are associated with elevated cerebral amyloid. Neurology. 2016;86:535–543. doi: 10.1212/WNL.0000000000002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarin M., Salih D.A., Yasvoina M., Cummings D.M., Guelfi S., Liu W., Nahaboo Solim M.A., Moens T.G., Paublete R.M., Ali S.S., Perona M., Desai R., Smith K.J., Latcham J., Fulleylove M., Richardson J.C., Hardy J., Edwards F.A. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Rep. 2015;10:633–644. doi: 10.1016/j.celrep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- Mateen F.J., Keegan B.M., Krecke K., Parisi J.E., Trenerry M.R., Pittock S.J. Sporadic leucodystrophy with neuroaxonal spheroids: persistence of DWI changes and neurocognitive profiles: a case study. J. Neurol. Neurosurg. Psychiatry. 2010;81:619–622. doi: 10.1136/jnnp.2008.169243. [DOI] [PubMed] [Google Scholar]
- Minami S.S., Min S.-W., Krabbe G., Wang C., Zhou Y., Asgarov R., Li Y., Martens L.H., Elia L.P., Ward M.E., Mucke L., Farese R.V., Gan L. Progranulin protects against amyloid β deposition and toxicity in Alzheimer’s disease mouse models. Nat. Med. 2014;20:1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Doi Y., Mizoguchi H., Jin S., Noda M., Sonobe Y., Takeuchi H., Suzumura A. Interleukin-34 selectively enhances the neuroprotective effects of microglia to attenuate oligomeric amyloid-β neurotoxicity. Am. J. Pathol. 2011;179:2016–2027. doi: 10.1016/j.ajpath.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Muranishi M., Torugun T., Tango H., Nagakane Y., Kudeken T., Kawase Y., Kawabe K., Oshima F., Yaoi T., Itoh K., Fushiki S., Nakagawa M. Two Japanese CADASIL families exhibiting Notch3 mutation R75P not involving cysteine residue. Intern. Med. Tokyo Jpn. 2008;47:2067–2072. doi: 10.2169/internalmedicine.47.1391. [DOI] [PubMed] [Google Scholar]
- Murphy G.M., Zhao F., Yang L., Cordell B. Expression of macrophage colony-stimulating factor receptor is increased in the AbetaPP(V717F) transgenic mouse model of Alzheimer’s disease. Am. J. Pathol. 2000;157:895–904. doi: 10.1016/s0002-9440(10)64603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T., Sadleir K.R., Maus E., Velliquette R.A., Zhao J., Cole S.L., Eimer W.A., Hitt B., Bembinster L.A., Lammich S., Lichtenthaler S.F., Hébert S.S., De Strooper B., Haass C., Bennett D.A., Vassar R. Phosphorylation of the translation initiation factor eIF2alpha increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60:988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero K., Turnbull I.R., Poliani P.L., Vermi W., Cerutti E., Aoshi T., Tassi I., Takai T., Stanley S.L., Miller M., Shaw A.S., Colonna M. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and beta-catenin. Nat. Immunol. 2009;10:734–743. doi: 10.1038/ni.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paloneva J., Autti T., Hakola P., Haltia M.J. Polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL) In: Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J., Bird T.D., Ledbetter N., Mefford H.C., Smith R.J., Stephens K., editors. GeneReviews(®) University of Washington, Seattle; Seattle, WA: 1993. [PubMed] [Google Scholar]
- Paquet C., Jouvent E., Mine M., Vital A., Hugon J., Chabriat H., Gray F. A cortical form of CADASIL with cerebral Aβ amyloidosis. Acta Neuropathol. (Berl.) 2010;120:813–820. doi: 10.1007/s00401-010-0758-y. [DOI] [PubMed] [Google Scholar]
- Rademakers R., Baker M., Nicholson A.M., Rutherford N.J., Finch N., Soto-Ortolaza A., Lash J., Wider C., Wojtas A., DeJesus-Hernandez M., Adamson J., Kouri N., Sundal C., Shuster E.A., Aasly J., MacKenzie J., Roeber S., Kretzschmar H.A., Boeve B.F., Knopman D.S., Petersen R.C., Cairns N.J., Ghetti B., Spina S., Garbern J., Tselis A.C., Uitti R., Das P., Van Gerpen J.A., Meschia J.F., Levy S., Broderick D.F., Graff-Radford N., Ross O.A., Miller B.B., Swerdlow R.H., Dickson D.W., Wszolek Z.K. Mutations in the colony stimulating factor 1 receptor (CSF1R) gene cause hereditary diffuse leukoencephalopathy with spheroids. Nat. Genet. 2011;44:200–205. doi: 10.1038/ng.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A., van den Maagdenberg A.M.J.M., Jen J.C., Kavanagh D., Bertram P., Spitzer D., Liszewski M.K., Barilla-Labarca M.-L., Terwindt G.M., Kasai Y., McLellan M., Grand M.G., Vanmolkot K.R.J., de Vries B., Wan J., Kane M.J., Mamsa H., Schäfer R., Stam A.H., Haan J., de Jong P.T.V.M., Storimans C.W., van Schooneveld M.J., Oosterhuis J.A., Gschwendter A., Dichgans M., Kotschet K.E., Hodgkinson S., Hardy T.A., Delatycki M.B., Hajj-Ali R.A., Kothari P.H., Nelson S.F., Frants R.R., Baloh R.W., Ferrari M.D., Atkinson J.P. C-terminal truncations in human 3’-5’ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Rossor M.N., Fox N.C., Mummery C.J., Schott J.M., Warren J.D. The diagnosis of young-onset dementia. Lancet Neurol. 2010;9:793–806. doi: 10.1016/S1474-4422(10)70159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi C., Nalls M.A., Ridge P.G., Gibbs J.R., Ding J., Lupton M.K., Troakes C., Lunnon K., Al-Sarraj S., Brown K.S., Medway C., Clement N., Lord J., Turton J., Bras J., Almeida M.R., ARUK Consortium. Holstege H., Louwersheimer E., van der Flier W.M., Scheltens P., Van Swieten J.C., Santana I., Oliveira C., Morgan K., Powell J.F., Kauwe J.S., Cruchaga C., Goate A.M., Singleton A.B., Guerreiro R., Hardy J. ABCA7 p.G215S as potential protective factor for Alzheimer’s disease. Neurobiol. Aging. 2016;46:235.e1-9. doi: 10.1016/j.neurobiolaging.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna Z.E., Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Scali O., Di Perri C., Federico A. The spectrum of mutations for the diagnosis of vanishing white matter disease. Neurol. Sci. 2006;27:271–277. doi: 10.1007/s10072-006-0683-y. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Zeginigg M., Wiltgen M., Freudenberger P., Petrovic K., Cavalieri M., Gider P., Enzinger C., Fornage M., Debette S., Rotter J.I., Ikram M.A., Launer L.J., Schmidt R., CHARGE consortium Neurology working group Genetic variants of the NOTCH3 gene in the elderly and magnetic resonance imaging correlates of age-related cerebral small vessel disease. Brain J. Neurol. 2011;134:3384–3397. doi: 10.1093/brain/awr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.D., Snowdon D.A., Wang H., Markesbery W.R. White matter volumes and periventricular white matter hyperintensities in aging and dementia. Neurology. 2000;54:838–842. doi: 10.1212/wnl.54.4.838. [DOI] [PubMed] [Google Scholar]
- Thijs V., Robberecht W., De Vos R., Sciot R. Coexistence of CADASIL and Alzheimer’s disease. J. Neurol. Neurosurg. Psychiatry. 2003;74:790–792. doi: 10.1136/jnnp.74.6.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman E.M., Pankratz N.D., Choi Y., Rothstein J.H., Faber K.M., Cheng R., Lee J.H., Bird T.D., Bennett D.A., Diaz-Arrastia R., Goate A.M., Farlow M., Ghetti B., Sweet R.A., Foroud T.M., Mayeux R., NIA-LOAD/NCRAD Family Study Group Genome-wide association of familial late-onset Alzheimer’s disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. Plos Genet. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F., Cheng Y., Breschi A., Vierstra J., Wu W., Ryba T., Sandstrom R., Ma Z., Davis C., Pope B.D., Shen Y., Pervouchine D.D., Djebali S., Thurman R.E., Kaul R., Rynes E., Kirilusha A., Marinov G.K., Williams B.A., Trout D., Amrhein H., Fisher-Aylor K., Antoshechkin I., DeSalvo G., See L.-H., Fastuca M., Drenkow J., Zaleski C., Dobin A., Prieto P., Lagarde J., Bussotti G., Tanzer A., Denas O., Li K., Bender M.A., Zhang M., Byron R., Groudine M.T., McCleary D., Pham L., Ye Z., Kuan S., Edsall L., Wu Y.-C., Rasmussen M.D., Bansal M.S., Kellis M., Keller C.A., Morrissey C.S., Mishra T., Jain D., Dogan N., Harris R.S., Cayting P., Kawli T., Boyle A.P., Euskirchen G., Kundaje A., Lin S., Lin Y., Jansen C., Malladi V.S., Cline M.S., Erickson D.T., Kirkup V.M., Learned K., Sloan C.A., Rosenbloom K.R., Lacerda de Sousa B., Beal K., Pignatelli M., Flicek P., Lian J., Kahveci T., Lee D., Kent W.J., Ramalho Santos M., Herrero J., Notredame C., Johnson A., Vong S., Lee K., Bates D., Neri F., Diegel M., Canfield T., Sabo P.J., Wilken M.S., Reh T.A., Giste E., Shafer A., Kutyavin T., Haugen E., Dunn D., Reynolds A.P., Neph S., Humbert R., Hansen R.S., De Bruijn M., Selleri L., Rudensky A., Josefowicz S., Samstein R., Eichler E.E., Orkin S.H., Levasseur D., Papayannopoulou T., Chang K.-H., Skoultchi A., Gosh S., Disteche C., Treuting P., Wang Y., Weiss M.J., Blobel G.A., Cao X., Zhong S., Wang T., Good P.J., Lowdon R.F., Adams L.B., Zhou X.-Q., Pazin M.J., Feingold E.A., Wold B., Taylor J., Mortazavi A., Weissman S.M., Stamatoyannopoulos J.A., Snyder M.P., Guigo R., Gingeras T.R., Gilbert D.M., Hardison R.C., Beer M.A., Ren B., Mouse ENCODE Consortium A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.