Abstract
We report on the influence of pH and monovalent/divalent cations on the catalytic current response, internal electron transfer (IET), and structure of fructose dehydrogenase (FDH) by using amperometry, spectrophotometry, and circular dichroism (CD). Amperometric measurements were performed on graphite electrodes, onto which FDH was adsorbed and the effect on the response current to fructose was investigated when varying the pH and the concentrations of divalent/monovalent cations in the contacting buffer. In the presence of 10 mM CaCl2, a current increase of up to ≈ 240% was observed, probably due to an intra-complexation reaction between Ca2+ and the aspartate/glutamate residues found at the interface between the dehydrogenase domain and the cytochrome domain of FDH. Contrary to CaCl2, addition of MgCl2 did not show any particular influence, whereas addition of monovalent cations (Na+ or K+) led to a slight linear increase in the maximum response current. To complement the amperometric investigations, spectrophotometric assays were carried out under homogeneous conditions in the presence of a 1-electron non-proton-acceptor, cytochrome c, or a 2-electron-proton acceptor, 2,6-dichloroindophenol (DCIP), respectively. In the case of cytochrome c, it was possible to observe a remarkable increase in the absorbance up to 200% when 10 mM CaCl2 was added. However, by further increasing the concentration of CaCl2 up to 50 mM and 100 mM, a decrease in the absorbance with a slight inhibition effect was observed for the highest CaCl2 concentration. Addition of MgCl2 or of the monovalent cations shows, surprisingly, no effect on the electron transfer to the electron acceptor. Contrary to the case of cytochrome c, with DCIP none of the cations tested seem to affect the rate of catalysis. In order to correlate the results obtained by amperometric and spectrophotometric measurements, CD experiments have been performed showing a great structural change of FDH when increasing the concentration CaCl2 up to 50 mM, at which the enzyme molecules start to agglomerate, hindering the substrate access to the active site probably due to a chelation reaction occurring at the enzyme surface with the glutamate/aspartate residues.
Graphical Abstract.
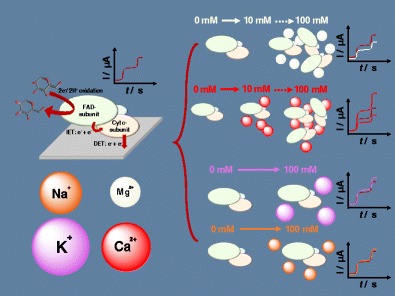
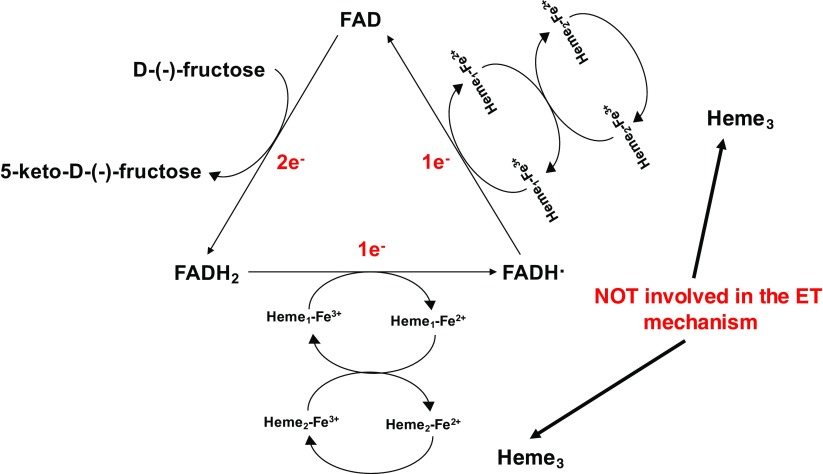
Fructose dehydrogenase (FDH) consists of three subunits, but only two are involved in the electron transfer process: (I) 2e−/2H+ fructose oxidation, (II) internal electron transfer (IET), (III) direct electron transfer (DET) through 2 heme c; FDH activity either in solution or when immobilized onto an electrode surface is enhanced about 2.5-fold by adding 10 mM CaCl2 to the buffer solution, whereas MgCl2 had an “inhibition” effect. Moreover, the additions of KCl or NaCl led to a slight current increase
Electronic supplementary material
The online version of this article (10.1007/s00216-018-0991-0) contains supplementary material, which is available to authorized users.
Keywords: Fructose dehydrogenase (FDH), Calcium chloride, Direct electron transfer (DET), Enzyme activity, Enzyme structure
Introduction
In the last 30 years, studies of direct electron transfer (DET) reactions between redox enzymes and electrodes have attracted much interest to understand the background reaction mechanism [1, 2] and thus gain sufficient basic knowledge to be able to establish the basis and to improve the performance of third generation biosensors and enzymatic fuel cells (EFCs) [3–6]. DET-based reactions have been explored for a number of redox proteins [7] such as cytochrome c (cyt c) [8–11], ferredoxin [12, 13], azurin [14, 15], and redox enzymes [16] such as peroxidases [17–23], hydrogenases [24], “blue” multi-copper oxidases (BMCO) [25–28], sulfite oxidase (SOx) [29, 30], alcohol PQQ dehydrogenase (ADH) [31, 32], cellobiose dehydrogenase (CDH) [33–37], D-fructose dehydrogenase (FDH) [38–41], etc. Several of the redox enzymes mentioned above, such as SOx, ADH, CDH, and FDH are composed of at least two different domains, where one domain is the catalytically active domain containing a bound cofactor such as FAD, PQQ, or MOCO, and a second domain, a cytochrome containing heme c or heme b, acting as an electron transfer domain or in other words “as a built in mediator” [31] connecting the redox enzyme to its natural electron acceptor, or when immobilized onto the surface of an electrode to the electrode if the enzyme is properly orientated on its surface. The electron transfer (ET) reaction between the catalytic and the cytochrome domain, the internal electron transfer (IET) reaction, can be very much dependent on pH, ion strength, and buffer constituents [42]. Studies to enable understanding what determines and limits the ET reactions are fundamental to increase the knowledge about protein structure, mechanisms of redox transformations of protein molecules, and metabolic processes involving redox transformations [43]. DET is the key issue to develop “reagentless” electrochemical biosensors [1–7, 44]. Among the flavocytochrome oxidoreductases, the DET mechanism for membrane bound FDH has not yet been elucidated [45] and therefore FDH has attracted a growing interest also regarding all factors that can influence the DET reaction (e.g., pH, cations, ionic strength, etc.) [46–48].
D-Fructose dehydrogenase (FDH; EC 1.1.99.11) from Gluconobacter japonicus NCBR 3260 is a heterotrimeric membrane-bound enzyme complex with a molecular mass of 146.4 kDa, consisting of three subunits/domains [45]: subunit I, which is the catalytic dehydrogenase domain (DHFDH) with a covalently bound flavin adenine dinucleotide (FAD) cofactor, where D-(–)-fructose is involved in a 2H+/2e- oxidation reaction to form 5-dehydro-D-(–)-fructose; subunit II, which is equivalent to the cytochrome domain (CYTFDH) acts as electron acceptor to subunit I and contains three heme c moieties covalently bound to the enzyme scaffold and two of them involved in the one-by-one ET pathway [45]; subunit III, which is not involved in the ET but rather plays a key role for the stability of the enzyme complex [38, 45, 49, 50]. FDH exhibits a strict substrate specificity to D-(–)-fructose and therefore has been used as bioelectrocatalyst for biosensor development both in direct and mediated ET (DET and MET) modes [51–54].
Unfortunately, the crystal structure of FDH is not yet available because of the difficulties involved with crystallization of a membrane bound protein with a high molecular weight (ca. 146 kDa). Despite these obvious difficulties, in the last few years much effort has been directed towards considering new crystallization methods [55]. The crystal structure would be of fundamental importance to clarify the ET mechanism of this enzyme with particular attention on the co-factor involved.
The suggested ET pathway for FDH when immobilised on the electrode surface and in the absence of any competing e- acceptors [38], is assumed to occur according to Scheme 1:
Oxidation of D-(–)-fructose to 5-keto-D-(–)-fructose involving 2e-/2H+ with the reduction of FAD to FADH2;
FADH2 is sequentially re-oxidized in two separate 1 ET steps. In the first FADH2 is partially re-oxidized to FADH⋅ through the IET pathway between the DHFDH and CYTFDH domains, whereby one of the three heme c (heme c1) is reduced. Next, the electron is transferred from heme c1 to a second heme c (heme c2) of the two hemes involved in the ET pathway and then to a final electron acceptor, which is the electrode when FDH is adsorbed onto the electrode surface;
FADH⋅ is finally re-oxidized to FAD by heme c1 and the electron is then transferred to heme c2 (which gives the second internal electron transfer (IET) step), which in turn is re-oxidized by the electrode whereby FDH is returned to its fully oxidized state.
Scheme 1.
Suggested ET mechanism of the oxidation reaction of FDH. D-(–)-fructose is oxidized to 5-keto-D-(–)-fructose releasing two electrons, which are transferred one by one initially through the FAD, followed by two heme c working as monoelectronic acceptors
The influence of ionic strength as well as various cationic and anionic species on enzymes has already been proven to modulate enzymatic activity or stability in different ways [56, 57]; e.g., CDH has two redox domains connected through a flexible linker [58–60], with an ET mechanism similar to FDH but with only one heme in its cytochrome domain as electron acceptor. Moreover, Sezer et al. showed an increase in terms of catalytic current of SOx by increasing the ionic strength of the buffer solution [61], while Feng et al. reported on the influence of the viscosity on the rate of the IET [62]. Nevertheless, each of these enzymes has its own and different dependency on pH and ionic strength because of the amino acids residues at the interface between the subunits connecting each other through linker regions.
The aim of this paper was to investigate whether there is any influence of pH and the concentration of various divalent/monovalent cations on the rate of the IET and the activity and the structure of FDH. First, FDH was immobilized onto graphite electrodes by a drop-casting procedure and amperometric measurements were carried out when shifting the pH in presence of various concentrations of divalent cations (CaCl2 and MgCl2) or monovalent cations (KCl and NaCl). Furthermore, the influence of pH on the rate of the IET was investigated in the presence of the CaCl2 concentration that yielded the highest current density for fructose. These results were compared with those obtained with the spectrophotometric assays carried out in the presence of 2,6-dichloroindophenol (DCIP, bielectronic acceptor) or of cytochrome c (cyt c, monoelectronic acceptor). Finally, circular dichroism (CD) measurements and homology modelling/docking studies were performed, confirming the hypothesis formulated on the IET of FDH.
Material and methods
Chemicals
D-(–)-fructose, calcium chloride (CaCl2), magnesium chloride hexahydrate (MgCl2 ⋅ 6 H2O), sodium chloride (NaCl), potassium chloride (KCl), cytochrome c from bovine heart (≥95% based on MW 12,327 Da, prepared using TCA), 2,6-dichloroindophenol (DCIP), sodium acetate (CH3COONa, NaAc), 3-(N-morpholino)propanesulfonic acid (MOPS), 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS), sodium dihydrogen phosphate (NaH2PO4), hydrochloric acid (HCl), sodium hydroxide (NaOH) were purchased from Sigma Aldrich (St. Louis, MO, USA). D-fructose dehydrogenase from Gluconobacter japonicus (FDH; EC 1.1.99.11) was purified from the culture supernatant of Gluconobacter japonicus NBRC 3260 obtained from the National Institute of Technology and Evaluation (Nishinomiya, Hyogo Prefecture, Japan) and solubilized in PBS buffer at pH 6 (50~500 mM) containing 0.1 mM 2-mercaptoethanol and 0.1% v/v Triton X-100 (volumetric activity measured with potassium ferricyanide at pH 4.5 = 420 ± 30 U mL−1, specific activity = 250 ± 30 U mg-1, protein concentration = 1.7 ± 0.2 mg mL−1) [49]. All solutions were prepared using Milli-Q water (R = 18.2 MΩ cm at 25 °C; TOC < 10 μg L−1, Millipore, Molsheim, France).
Electrochemical measurements
Graphite rods (Alfa Aesar GmbH and Co. KG, AGKSP grade, ultra “F” purity, and 3.05 mm diameter, Karlsruhe, Germany) were polished on wet emery paper (Turfbak Durite, P1200) and then carefully rinsed with Milli-Q water [56]. The enzyme-modified electrodes were prepared by allowing 5 μL of a FDH solution (volumetric activity measured with potassium ferricyanide at pH 4.5 and found to be 420 ± 30 U mL−1, specific activity = 250 ± 30 U mg-1, protein concentration = 1.7 ± 0.2 mg mL−1) to physically adsorb on the top of the graphite rod electrodes, overnight at 4 °C. The FDH modified graphite electrode, an Ag|AgCl (0.1 M KCl) reference electrode and a platinum wire counter electrode were fitted into a flow-through wall-jet electrochemical cell [63] connected to a flow injection analysis (FIA) system consisting of a peristaltic pump (Gilson, Villier-le-Bel, France) and a six-port valve electrical injector equipped with a 50 μL loop (injection volume 50 μL) (Rheodyne, Cotati, CA, USA) [63]. A potentiostat (Zäta Elektronik, Höör, Sweden) controlled the flow-through cell, and the response of the injected samples was shown on a strip chart recorder (Kipp and Zonen, Utrecht, The Netherlands) [64].
Spectrophotometric measurements
Two spectrophotometric assays were performed to measure the activity of FDH in solution as a function of both pH and the concentration of cations/ionic strength. In the first assay, the activity of the DHFDH domain was monitored measuring the time-dependent variation of the absorbance at λ = 520 nm (ε = 6.9 mM-1 cm-1) of a mixture containing 100 μL of 300 mM D-(–)-fructose, 780 μL of 50 mM NaAc buffer, 20 μL of enzyme solution, and 100 μL of 3 mM DCIP as two-electrons/two protons acceptor [56]. In the second assay, the activity of FDH and the internal electron transfer (IET) between the DHFDH and CYTFDH domains was monitored by using the one-electron acceptor cyt c, which communicates only with the CYTFDH domain due to steric hindrance because of its dimensions. The absorption is followed at 550 nm (ε = 19.6 mM−1 cm−1) of a mixture of 20 μL of a 1 mM cyt c solution, 100 μL of 300 mM D-(–)-fructose, 860 μL of 50 mM NaAc buffer pH 4.5, and 20 μL of the enzyme solution [65]. One unit of DCIP and cyt c activity was defined as the amount of FDH that reduces 1 μmol of DCIP or cyt c, respectively, per min under the applied conditions. All measured enzyme activities are the average of three measurements at 25 °C, whereas the average activity values in 50 mM NaAc buffer without additions of “extra” KCl, NaCl, MgCl2, or CaCl2 were set to 100% and the average activity values in the presence of divalent and monovalent cations in 50 mM NaAc buffer were related to those values. All measurements were carried out using a UV-Vis spectrophotometer 1800 (Shimadzu Europe GmbH, Duisburg, Germany).
Circular dichroism (CD) measurements
Changes in the secondary structure of FDH as a function of pH and addition of divalent cations to the solution were investigated through CD measurements using a CD spectrometer (J-815 Circular Dichroic Spectrometer, JASCO, Easton, MD, USA) [66]. These measurements were carried out in a 0.1 cm cuvette at 25 °C using a final protein concentration of 0.15 mg mL-1, diluted in 50 mM NaH2PO4 at pH 4.5 (unfortunately we could not use NaAc because the –COOH groups adsorb the light so we needed to use a non-adsorbing medium to obtain reliable data [67]). Then, the concentration of CaCl2 or MgCl2 was increased in the range of 0–100 mM to receive information about any changes in the secondary structure. The obtained spectra were further fitted with a method called β-structure selection (BeStSel) that takes into account the twist of β-structures for estimation of the secondary structure. This method can reliably distinguish parallel and anti-parallel β-sheets and accurately estimate the secondary structure for a broad range of proteins [68, 69]. Moreover, the secondary structure components applied by the method are characteristic to the protein fold, which in turn can be predicted to the level of topology in the CATH classification from a single CD spectrum.
Dynamic light scattering (DLS) measurements
The aggregation of FDH in presence of NaCl, KCl, CaCl2, MgCl2 at different concentrations, 0, 10, 50, and 100 mM was evaluated using Dynamic Light Scattering (Zetasizer Nano ZS90, Malvern Instruments Ltd, Malvern, UK) by considering polydispersion index (PDI) and diameter (nm).
Homology modeling and docking
The homology models of the individual DHFDH and CYTFDH domains of FDH (GenBank accession number A0A164AQ58_9SYNE) were generated using the Protein Homology/analogY Recognition Engine V 2.0 server (http://www.sbg.bio.ic.ac.uk/) [70] with the individually crystallized DH domains of FAD-glucose dehydrogenase (GDH) from Aspergillus flavus (PDB ID 4YNT [71]) and thiosulfate dehydrogenase (TSDBA) from Marichromatium purpuratum “as2 isolated” (PDB ID 5LO9 [72]) as templates. The FIREDOCK webserver (http://bioinfo3d.cs.tau.ac.il/FireDock/firedock.html) [73, 74] was used for protein–protein docking of the individual FDH domains. In total, 10 models from three clusters per enzyme were evaluated. The best model has been selected based on the lowest free energy [75]. Structures were visualized using the PyMOL Molecular Graphics System, ver. 1.4 (Schrödinger, New York, NY, USA).
Results and discussion
Influence of pH and divalent/monovalent cations on the direct electron transfer (DET) reaction of FDH: electrochemical study
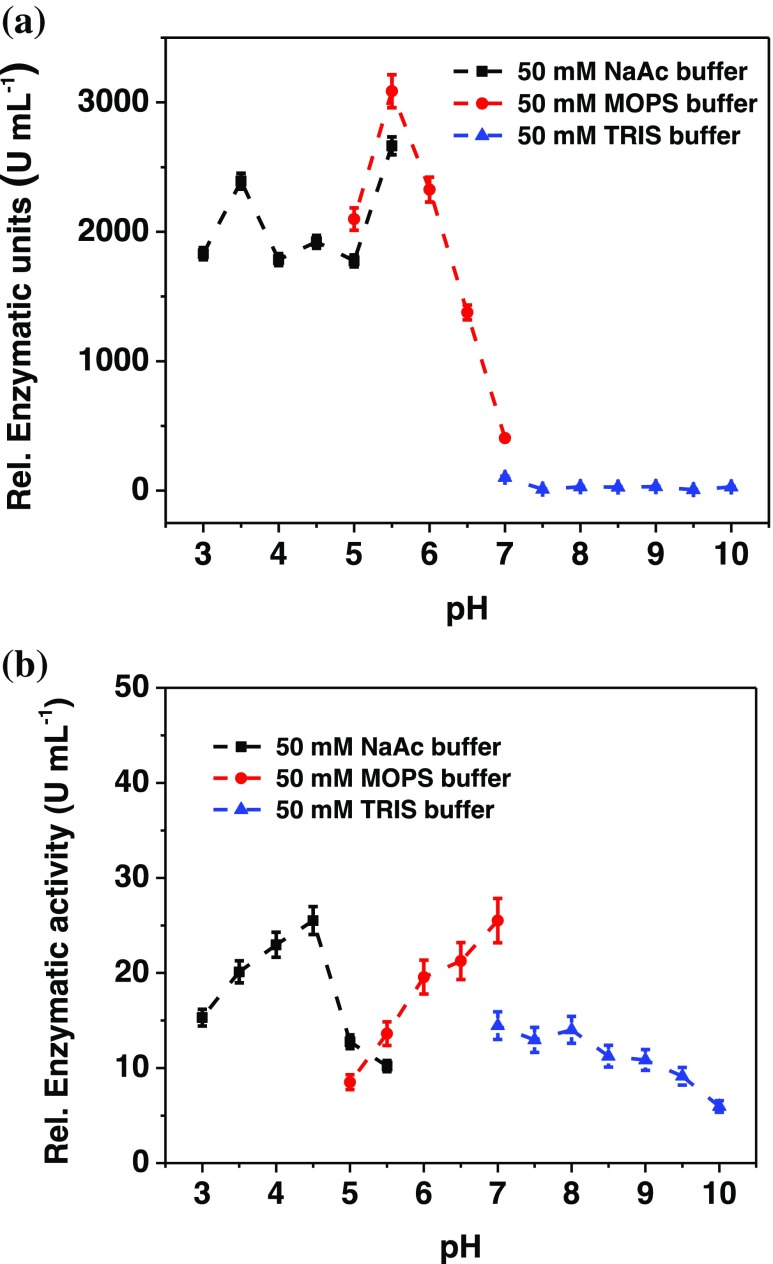
In order to study the influence of pH on the DET reaction of FDH with graphite electrodes, we performed amperometric measurements with graphite electrodes simply modified with adsorbed FDH. The measurements were performed with 5 mM D-(–)-fructose as substrate and by varying the pH in the running buffer between 3 and 7 using NaAc buffer between pH 3 and 5.5, MOPS buffer between pH 5 and 7. Figure 1a shows the current density (J, μA cm-2) dependence on pH. The optimum pH was found at 4.5. The response current rapidly decreased when increasing the pH above 4.5, thus assessing no DET activity starting at pH 7. These results are in good agreement with previous values reported in the literature [76].
Fig. 1.
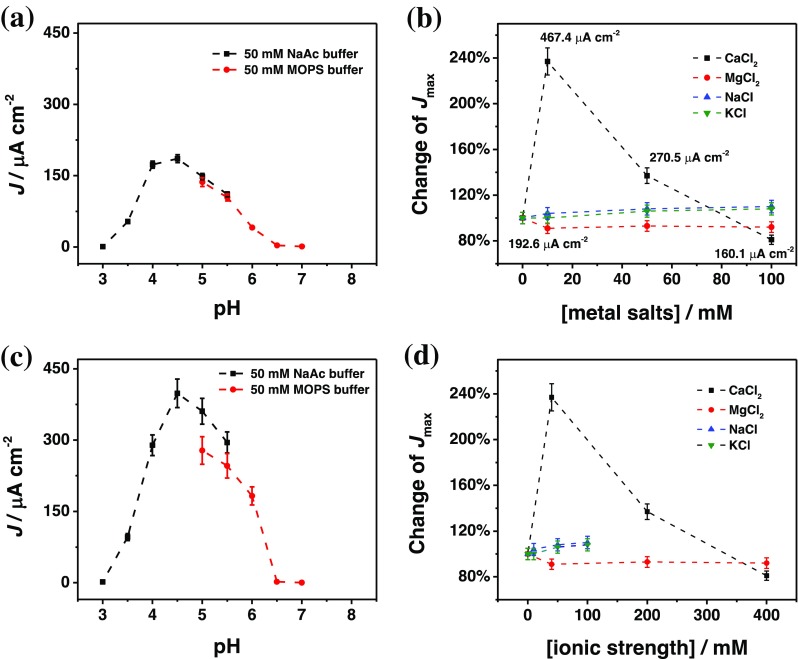
Dependence of the current density on pH (a) in the absence and (c) in the presence of 10 mM CaCl2 carried out in 50 mM NaAc buffer (pH 3–5.5, black), 50 mM MOPS buffer (pH 5–7, red), and 50 mM TRIS buffer (pH 7–10, blue) in the presence of 5 mM D-(–)-fructose; the relative maximal catalytic current densities (Jmax) on the concentration (b) and ionic strength (d) of CaCl2 (black), MgCl2 (red), NaCl (blue), and KCl (green) solutions (50 mM NaAc buffer pH 4.5) calculated from calibration curves performed in presence of different concentrations of D-(–)-fructose. Experimental conditions: applied potential (Eapp): +0.2 V versus Ag|AgClsat, flow rate: 0.5 mL min-1; injection volume 50 μL
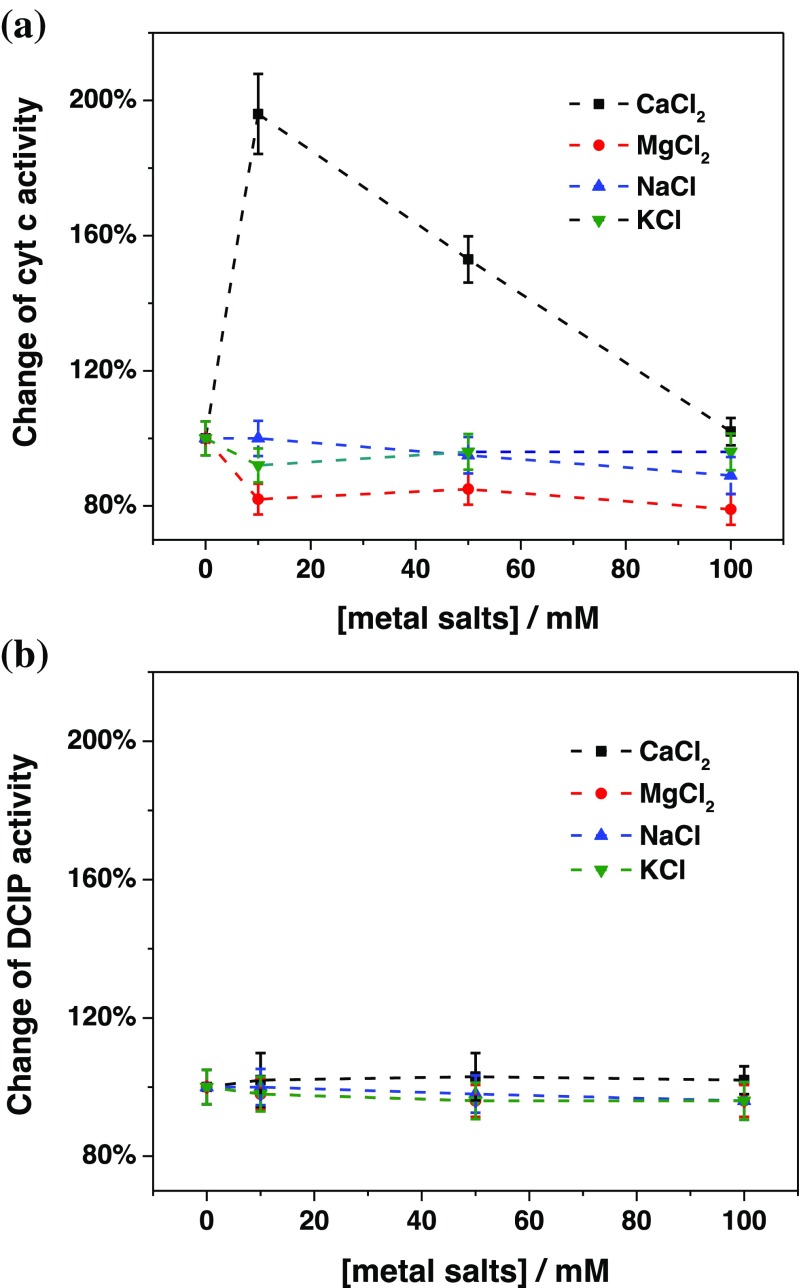
Next, the influence of divalent (CaCl2 and MgCl2) and monovalent (NaCl and KCl) cations was studied through registering calibration curves for D-(–)-fructose at pH 4.5 and varying the salt concentrations in the range of 0–100 mM. Figure 1b shows the dependence of maximum current, Jmax, on the concentration of CaCl2, MgCl2, NaCl, and KCl for FDH at pH 4.5. The Jmax values were obtained from the calibration curves and were set to 100% in the absence of extra added cations. All values obtained in the presence of the different cations are related to those values. Surprisingly, when increasing the concentration of CaCl2, a marked increase in Jmax was observed, with a maximum increase of 237%, corresponding to an average maximal current density of 467.4 ± 38.2 μA cm-2 for 10 mM CaCl2. However, at higher concentrations of CaCl2 than 10 mM, an unexpected Imax decrease was registered, yielding Jmax values of 140% and of 80%, at 50 mM 100 mM CaCl2, respectively. This particular trend for CaCl2 is probably due to the presence of side-chain carboxyl functions of surface-exposed aspartic acid (pKa = 3.86) and glutamic acid (pKa = 4.07) residues at the interface of the DHFDH and CYTFDH domains. These –COOH groups are ~90% deprotonated at pH 4.5, creating a strong electrostatic repulsion between the two domains [77]. It can be supposed that a possible complexation of Ca2+ with the carboxylic groups of the aspartic and glutamic acid residues takes place, resulting in a closer interaction and shortening the distance between the DHFDH and CYTFDH domains, thus increasing the rate of the IET [58]. On the other hand, the decrease in response signal when the concentration of Ca2+ is higher than 10 mM can probably be ascribed to a possible saturation of the aspartate/glutamate residues available at the interface between the DHFDH and CYTFDH domains [78]. Therefore, at concentrations higher than 10 mM, Ca2+ is now available for complexation of other aspartate/glutamate residues present on the surface of both subunits. Both the DHFDH and CYTFDH subunits show a partial positive charge at pH 4.5, which might now create a slight repulsion between the two subunits, which is, however, counter-balanced by the attractive interactions. A possible explanation for the inhibition effect observed at 100 mM CaCl2 could be the interaction with other enzyme molecules through a Ca2+-bridge [79].
Contrary to the effect by addition of CaCl2, addition of MgCl2 showed a 10% decrease in Jmax already at 10 mM MgCl2. This result could be ascribed to a lower affinity of Mg2+ compared with Ca2+ towards the carboxylic functions available at the interface of the DHFDH and CYTFDH domains. Mg2+ is expected mainly to be involved in the complexation reaction occurring between different enzyme molecules (Mg2+-bridge), which will partially hinder the substrate access to the active site of FDH and will therefore show a slight inhibition behavior evident already at low MgCl2 concentrations. Figure 1c shows that the enhanced effect on the catalytic current density by addition of CaCl2 occurs only in the pH range between 3.5 and 5.5 without any shift in the optimum pH of the enzyme activity, due to its presumed effect on the IET as described and discussed above.
In a series of previous investigations, the effect of ionic strength and addition of Ca2+ and Mg2+ on the catalytic performance of various CDHs was investigated both in solution and when adsorbed on graphite [56, 58, 80, 81]. The results obtained here for FDH are both similar and dissimilar with respect to those obtained for the various CDHs; e.g., for two ascomycete CDHs the addition of both Ca2+ and Mg2+ had a similar increasing effect on the catalytic efficiency and could even lead to a drastic shift in the pH optimum of the enzyme activity [80]. Thus, it seems as though further investigations on the background reasons for the effect by both ionic strength and by individual cations on the interactions between the various domains participating in the ET pathways in such multi-domain redox enzymes are necessary.
For the monovalent cations, further additions of both KCl and NaCl showed a slight linear increase in Jmax up to 100 mM, which is more evident in Fig. 1d, where the Jmax is plotted versus the ionic strength of the buffer. The concentration of the monovalent cations is directly related to the ionic strength, leading to a general increase in the catalytic current, as already reported in literature for many enzymes, whereas monovalent cations are usually not involved in any complexation reactions occurring at the protein surface [56].
Influence of pH and divalent/monovalent cations on the catalytic reaction of FDH: spectrophometric study
Two different spectrophotometric assays were performed to follow the catalytic reaction of FDH in solution; one with cyt c and one with DCIP, acting as mono- and bielectronic acceptors, respectively. DCIP may have a direct connection with subunit I (DHFDH), where the catalytic oxidation of D-fructose to 5-keto-D-fructose occurs [59], whereas cyt c may interact with both subunits, namely subunits I and II. Figure 2a shows the dependence of the activity of FDH with pH using DCIP as electron acceptor, and one can see that the highest activity is found between 3 and 6 corresponding to an activity of 3086 U mL-1 at pH 5.5. For pH values higher than 6, the activity rapidly decreases, indicating a progressive inactivation of DHFDH (subunit I). The dependence of the activity of FDH with pH when using cyt c is shown in Fig. 2b. In this case the activity exhibits two pH optima: one at 4.5 and one at 7, in agreement with previous results already reported in the literature [82]. These results could be ascribed to the different mechanisms of the interaction between FDH and cyt c: (1) ET may either proceed from the active site (FAD containing subunit I, DHFDH) via the heme groups (subunit II containing the three heme centers, CYTFDH) toward cyt c or (2) directly from the DHFDH to cyt c. A similar conclusion was reached when investigating the ET between FDH and cyt c by cyclic voltammetry at different scan rates [83, 84].
Fig. 2.
Dependence of the relative activities of (a) DCIP and (b) cyt c on pH carried out in 50 mM NaAc buffer (pH 3–5.5, black), 50 mM MOPS buffer (pH 5–7, red), and 50 mM TRIS buffer (pH 7–10, blue). Experimental conditions: [fructose]: 30 mM, [cyt c]: 100 μM, and [2,6-dichloroindophenol, DCIP]: 300 μM
As far as for the influence of mono- and divalent cations, the results of the spectrophotometric activity assays are shown in Fig. 3a and b. The results obtained with cyt c (Fig. 3a) are comparable with those obtained with the amperometric measurements (Fig. 1b) for immobilized FDH on graphite electrodes, as Ca2+ ions may affect the rate of the IET and therefore the rate of overall DET reaction. Figure 3b shows that there is no influence of either divalent or monovalent cations on the activity of FDH using DCIP as electron acceptor because DCIP is reduced directly by the FAD deeply buried within the DHFDH (subunit I) and is thus not affected by any change in the IET.
Fig. 3.
Dependence of the relative (a) cyt c and (b) DCIP activities on different concentrations of CaCl2 (black), MgCl2 (red), NaCl (blue), and KCl (green) in 50 mM NaAc buffer pH 4.5. Experimental conditions: [fructose]: 30 mM, [cyt c]: 100 μM, and [2,6-dichloroindophenol, DCIP]: 300 μM
Since the conversion rate of fructose at the FAD is not influenced much by the presence of different ions, the increase in current and cyt reduction found here may have arisen from an enhanced IET and/or a better interaction of the subunit II (heme subunit) with cyt c or the electrode.
The obtained results unequivocally confirm the influence of CaCl2 on the rate of the IET reaction, whereas there is no influence observed on the catalytic oxidation reaction of D-(–)-fructose at the DHFDH domain by CaCl2 or by the other cations tested.
Influence of CaCl2 on the secondary enzyme structure
In order to confirm the results obtained with the amperometric and spectrophotometric assays, CD measurements of FDH were performed increasing the concentration CaCl2 or MgCl2 in the range of 0–100 mM. Figure 4a–d show the CD spectra recorded for FDH in the presence of CaCl2, MgCl2, NaCl, and KCl. It is possible to observe that the secondary structure only to some extent shows minor changes up to a concentration of 10 mM, whereas at higher concentrations of CaCl2 the spectra reveal large changes that can be explained by agglomeration with other FDH molecules due to Ca2+-bridge interactions between enzyme molecules, as shown in Fig. 4a. Therefore, at low CaCl2 concentrations, it is possible to observe an intra-complexation caused by Ca2+ ions, which might increase the rate of the IET reaction, whereas at high CaCl2 concentrations inter-complexation reactions occur due to aggregation of enzyme molecules caused by the high CaCl2 concentration. Conversely, the CD spectra in the presence of low Mg2+ concentration already showed large changes, probably due to inter-complexation reactions occurring between different enzyme molecules, as depicted in Fig. 4b. Moreover, Na+ and K+ did not show any influence on the secondary enzyme structure, as reported in Fig. 4c and d, because they are not involved in any chelation reactions normally occurring with divalent cations [85]. Finally, formation of FDH multimers or aggregates was clearly proven by using dynamic light scattering (DLS) measurements as reported in Figs. S1–S4 and Table S1 in the Electronic Supplementary Material (ESM).
Fig. 4.
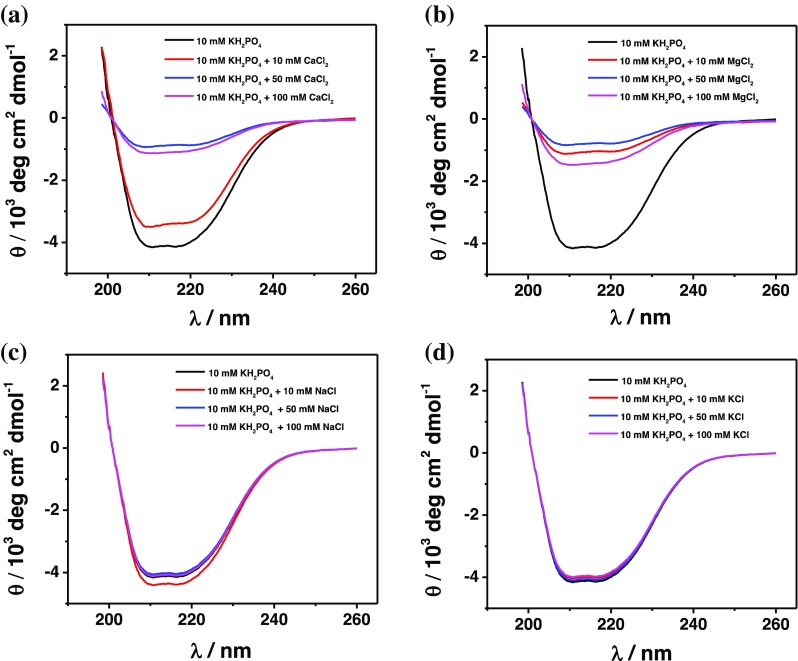
Circular dichroism (CD) spectra of FDH obtained in 10 mM KH2PO4 pH 4.5 at different concentrations of (a) CaCl2, (b) MgCl2, (c) NaCl, (d) KCl: 0 mM (black), 10 mM (red), 50 mM (purple), and 100 mM (blue)
Structural analysis and docking of FDH domains
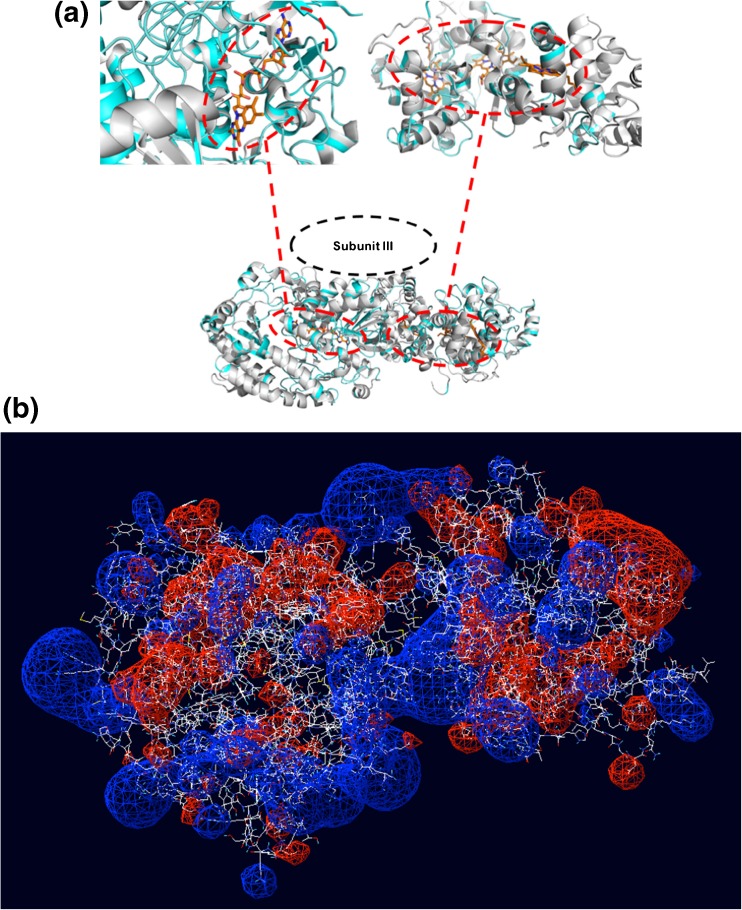
The already characterized FAD-glucose dehydrogenase from Aspergillus flavus (4YNT) and thiosulfate dehydrogenase (TSDBA) from Marichromatium purpuratum “as2 isolated” (5LO9) were used for modeling and docking studies of the DHFDH and CYTFDH domains, respectively. Figure 5a shows the homology models of the DHFDH and CYTFDH domains highlighting in the insets the position of the FAD cofactor (subunit I) and the three heme c (subunit II), respectively, while subunit III was not possible to model because of the absence of a similar structure. From previous studies [49] it was already stated that subunit III has no influence on the DET reaction but it is important for the overall stability of the enzymatic complex.
Fig. 5.
(a) Detailed representation of structural alignment between Subunit I of FDH (homology model, grey structure) and Aspergillus niger FAD-glucose dehydrogenase (PDB ID: 4ynt, light blue structure); and between Subunit II of FDH (homology model) and thiosulfate dehydrogenase (tsdba) from Marichromatium purpuratum “as2 isolated” form (PDB ID: 5lo9); (b) electrostatic map of Subunits I and II after docking obtained from the homology models
Moreover, the docking between the DHFDH and CYTFDH domains was needed to highlight those amino acids expected to be involved in the interaction between the two domains, viz., Asp, Glu, Lys, Arg, and Tyr. DHFDH exhibits five amino acid residues negatively charged at the interface, whereas CYTFDH contains three such amino acid residues. The occurrence of these residues could partially explain the reason for the highest rate of the IET reaction at pH 4.5, where the repulsion between the two domains is lowest, whereas at pH between 7 and 10 the repulsion between the two domains is high and will cause the low rate of the IET reaction. However, this model does not give any information about Ca2+-bridging interactions. Nevertheless, a concentration of 10 mM CaCl2 results in the upper limit in terms of an enhanced IET, probably because only a few possible matches between the negatively charged amino acid residues and Ca2+ ions are found at the interface between the DHFDH and the CYTFDH, while the number of negatively charged amino acids residues exposed on the other parts of the enzyme surface is much higher, confirming the possibility of other Ca2+-bridging interactions between different individual enzyme molecules allowing the aggregation process [86, 87].
Conclusions
This paper demonstrates the possibility to enhance the measurable activity of FDH either in solution or when immobilized onto an electrode surface about 2.5-fold by adding 10 mM CaCl2 to the buffer solution, whereas MgCl2 had no such effect. Additions of KCl or NaCl led to a slight linear increase in Jmax of about 10%. However, CaCl2 had an effect only at pH 4.5 because there was no shift in the optimum pH, in contrast to what was shown for ascomycete CDHs in previous papers [56, 58, 80, 81]. Moreover, the amperometric and spectrophotometric results were confirmed through conformational changes observed in the secondary structure of FDH, revealing unfolding of the protein occurring at high CaCl2 concentrations (50 and 100 mM) followed by aggregation.
By the homology models, it was assumed that Ca2+ at lower concentrations was chelated by the few amino acid residues negatively charged, shortening the distance between the DHFDH and CYTFDH domains, leading to an enhanced IET, while at higher concentrations of Ca2+, they give Ca2+-bridging interactions with other enzyme molecules.
These results show that studies of the mechanism of the IET between the DHFDH and CYTFDH domains are of great interest to shed further light into the physiological functions of FDH, as well as for the development of third generation biosensors and enzymatic fuel cells based on FDH.
Electronic supplementary material
(PDF 133 kb)
Acknowledgments
The authors thank the following agencies for financial support: The Swedish Research Council (Vetenskapsrådet project 2014-5908) and the European Commission (project “Bioenergy” FP7-PEOPLE-2013-ITN-607793). P.B. acknowledges a scholarship of the Erasmus+ Project Unipharma-Graduates, promoted by a Consortium of Italian Universities and coordinated by Sapienza University of Rome.
Biographies
Paolo Bollella
is a postdoc fellow at the Department of Chemistry and Drug Technologies, Sapienza University of Rome, Italy. In 2017, he obtained his PhD in Pharmaceutical Sciences with specialization in electroanalytical chemistry. His PhD work was focused on the green synthesis, characterization, and application of metal nanoparticles employed as transducers in new biosensors suitable in toxicological and pharmaceutical analysis.
Yuya Hibino
is a doctoral course student under the supervision of Professor Kenji Kano at the Division of Applied Life Sciences in Graduate School of Agriculture, Kyoto University, Japan. His work aims at the improvement of the performance of direct-electron transfer-type bioelectrocatalysis using protein engineering.
Kenji Kano
is Professor of Physical and Analytical Biochemistry, Division of Applied Life Sciences in Graduate School of Agriculture, Kyoto University, Japan. One of his recent research fields is bioelectrochemistry, including enzymatic biofuel cells, biosensors, and bioreactors. The other field concerns physical and analytical biochemistry on redox enzymes.
Lo Gorton
is Professor Emeritus in Analytical Chemistry, Lund University, Sweden. Currently he is also the President of the Bioelectrochemical Society (BES). His main interest is bioelectrochemical studies of direct and mediated electron transfer reactions between redox enzymes, biological membranes, living cells, and electrodes, and applications thereof in biosensors and biofuel cells.
Riccarda Antiochia
obtained her MSc degree in Chemistry in 1992 and in Pharmacy in 2009 and her PhD in Analytical Chemistry in 1996. She is currently Associate Professor at Sapienza University of Rome, Italy. Her fields of research are biosensors, studies on electrode kinetics, chemically modified electrodes, nanostructured electrode materials, liquid chromatography, and their use for determination and analysis of analytes of food and biomedical interest.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s00216-018-0991-0) contains supplementary material, which is available to authorized users.
Contributor Information
Lo Gorton, Email: Lo.Gorton@biochemistry.lu.se.
Riccarda Antiochia, Email: Riccarda.Antiochia@uniroma1.it.
References
- 1.Leger C, Bertrand P. Direct electrochemistry of redox enzymes as a tool for mechanistic studies. Chem Rev. 2008;108(7):2379–2438. doi: 10.1021/cr0680742. [DOI] [PubMed] [Google Scholar]
- 2.Frew JE, Hill HAO. Direct and indirect electron-transfer between electrodes and redox proteins. Eur J Biochem. 1988;172(2):261–269. doi: 10.1111/j.1432-1033.1988.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 3.Falk M, Blum Z, Shleev S. Direct electron transfer based enzymatic fuel cells. Electrochim Acta. 2012;82:191–202. doi: 10.1016/j.electacta.2011.12.133. [DOI] [Google Scholar]
- 4.Ghindilis AL, Atanasov P, Wilkins E. Enzyme-catalyzed direct electron transfer: Fundamentals and analytical applications. Electroanalysis. 1997;9(9):661–674. doi: 10.1002/elan.1140090902. [DOI] [Google Scholar]
- 5.Gorton L, Lindgren A, Larsson T, Munteanu F, Ruzgas T, Gazaryan I. Direct electron transfer between heme-containing enzymes and electrodes as basis for third generation biosensors. Anal Chim Acta. 1999;400(1):91–108. doi: 10.1016/S0003-2670(99)00610-8. [DOI] [Google Scholar]
- 6.Cracknell JA, Vincent KA, Armstrong FA. Enzymes as working or inspirational electrocatalysts for fuel cells and electrolysis. Chem Rev. 2008;108(7):2439–2461. doi: 10.1021/cr0680639. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong FA, Hill HAO, Walton NJ. Direct electrochemistry of redox proteins. Acc Chem Res. 1988;21(11):407–413. doi: 10.1021/ar00155a004. [DOI] [Google Scholar]
- 8.Wang L, Wang E. Direct electron transfer between cytochrome c and a gold nanoparticles modified electrode. Electrochem Commun. 2004;6(1):49–54. doi: 10.1016/j.elecom.2003.10.004. [DOI] [Google Scholar]
- 9.Ju H, Liu S, Ge B, Lisdat F, Scheller FW. Electrochemistry of cytochrome c immobilized on colloidal gold modified carbon paste electrodes and its electrocatalytic activity. Electroanalysis. 2002;14(2):141. doi: 10.1002/1521-4109(200201)14:2<141::AID-ELAN141>3.0.CO;2-Y. [DOI] [Google Scholar]
- 10.Eddowes MJ, Hill HAO. Electrochemistry of horse heart cytochrome c. J Am Chem Soc. 1979;101(16):4461–4464. doi: 10.1021/ja00510a003. [DOI] [Google Scholar]
- 11.Yeh P, Kuwana T. Reversible electrode-reaction of cytochrome c. Chem Lett. 1977;10:1145–1148. doi: 10.1246/cl.1977.1145. [DOI] [Google Scholar]
- 12.Yagati AK, Lee T, Min J, Choi J-W. Amperometric sensor for hydrogen peroxide based on direct electron transfer of spinach ferredoxin on Au electrode. Bioelectrochemistry. 2011;80(2):169–174. doi: 10.1016/j.bioelechem.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong FA, Heering HA, Hirst J. Reactions of complex metalloproteins studied by protein-film voltammetry. Chem Soc Rev. 1997;26(3):169–179. doi: 10.1039/cs9972600169. [DOI] [Google Scholar]
- 14.Lancaster KM, Farver O, Wherland S, Crane EJ, III, Richards JH, Pecht I, Gray HB. Electron transfer reactivity of type zero Pseudomonas aeruginosa azurin. J Am Chem Soc. 2011;133(13):4865–4873. doi: 10.1021/ja1093919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi QJ, Zhang JD, Andersen JET, Ulstrup J. Ordered assembly and controlled electron transfer of the blue copper protein azurin at gold (111) single-crystal substrates. J Phys Chem B. 2001;105(20):4669–4679. doi: 10.1021/jp0105589. [DOI] [Google Scholar]
- 16.Ferapontova EE, Shleev S, Ruzgas T, Stoica L, Christenson A, Tkac J, Yaropolov AI, Gorton L. Direct electrochemistry of proteins and enzymes. In: Palecek E, Scheller F, Wang J, editors. Electrochemistry of nucleic acids and proteins: towards electrochemical sensors for genomics and proteomics, Vol 1. Perspectives in bioanalysis. 2005. pp. 517–598.
- 17.Olloqui-Sariego JL, Zakharova GS, Poloznikov AA, Calvente JJ, Hushpulian DM, Gorton L, Andreu R. Interprotein coupling enhances the electrocatalytic efficiency of tobacco peroxidase immobilized at a graphite electrode. Anal Chem. 2015;87(21):10807–10814. doi: 10.1021/acs.analchem.5b01710. [DOI] [PubMed] [Google Scholar]
- 18.Li W-T, Wang M-H, Li Y-J, Sun Y, Li J-C. Linker-free layer-by-layer self-assembly of gold nanoparticle multilayer films for direct electron transfer of horseradish peroxidase and H2O2 detection. Electrochim Acta. 2011;56(20):6919–6924. doi: 10.1016/j.electacta.2011.06.023. [DOI] [Google Scholar]
- 19.Ruzgas T, Csöregi E, Emnéus J, Gorton L, MarkoVarga G. Peroxidase-modified electrodes: fundamentals and application. Anal Chim Acta. 1996;330(2/3):123–138. doi: 10.1016/0003-2670(96)00169-9. [DOI] [Google Scholar]
- 20.Christenson A, Dimcheva N, Ferapontova EE, Gorton L, Ruzgas T, Stoica L, Shleev S, Yaropolov AL, Haltrich D, Thorneley RNF, Aust SD. Direct electron transfer between ligninolytic redox enzymes and electrodes. Electroanalysis. 2004;16(13/14):1074–1092. doi: 10.1002/elan.200403004. [DOI] [Google Scholar]
- 21.Ferapontova EE. Direct peroxidase bioelectrocatalysis on a variety of electrode materials. Electroanalysis. 2004;16(13/14):1101–1112. doi: 10.1002/elan.200403003. [DOI] [Google Scholar]
- 22.Luis Olloqui-Sariego J, Zakharova GS, Poloznikov AA, Jose Calvente J, Hushpulian DM, Gorton L, Andreu R. Fenton-like Inactivation of tobacco peroxidase electrocatalysis at negative potentials. ACS Catal. 2016;6(11):7452–7457. doi: 10.1021/acscatal.6b01839. [DOI] [Google Scholar]
- 23.Gazaryan IG, Gorton L, Ruzgas T, Csöregi E, Schuhmann W, Lagrimini LM, Khushpul'yan DM, Tishkov VI. Tobacco peroxidase as a new reagent for amperometric biosensors. J Anal Chem. 2005;60(6):558–566. doi: 10.1007/s10809-005-0139-1. [DOI] [Google Scholar]
- 24.Vincent KA, Parkin A, Armstrong FA. Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem Rev. 2007;107(10):4366–4413. doi: 10.1021/cr050191u. [DOI] [PubMed] [Google Scholar]
- 25.Salaj-Kosla U, Pöller S, Schuhmann W, Shleev S, Magner E. Direct electron transfer of Trametes hirsuta laccase adsorbed at unmodified nanoporous gold electrodes. Bioelectrochemistry. 2013;91:15–20. doi: 10.1016/j.bioelechem.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Shleev S, Tkac J, Christenson A, Ruzgas T, Yaropolov AI, Whittaker JW, Gorton L. Direct electron transfer between copper-containing proteins and electrodes. Biosens Bioelectron. 2005;20(12):2517–2554. doi: 10.1016/j.bios.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Shleev S, Jarosz-Wilkolazka A, Khalunina A, Morozova O, Yaropolov A, Ruzgas T, Gorton L. Direct electron transfer reactions of laccases from different origins on carbon electrodes. Bioelectrochemistry. 2005;67(1):115–124. doi: 10.1016/j.bioelechem.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Pita M, Gutierrez-Sanchez C, Olea D, Velez M, Garcia-Diego C, Shleev S, Fernandez VM, De Lacey AL. High redox potential cathode based on laccase covalently attached to gold electrode. J Phys Chem C. 2011;115(27):13420–13428. doi: 10.1021/jp203643h. [DOI] [Google Scholar]
- 29.Ferapontova EE, Ruzgas T, Gorton L. Direct electron transfer of heme- and molybdopterin cofactor-containing chicken liver sulfite oxidase on alkanethiol-modified gold electrodes. Anal Chem. 2003;75(18):4841–4850. doi: 10.1021/ac0341923. [DOI] [PubMed] [Google Scholar]
- 30.Frasca S, Rojas O, Salewski J, Neumann B, Stiba K, Weidinger IM, Tiersch B, Leimkuehler S, Koetz J, Wollenberger U. Human sulfite oxidase electrochemistry on gold nanoparticles modified electrode. Bioelectrochemistry. 2012;87:33–41. doi: 10.1016/j.bioelechem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda T, Kobayashi D, Matsushita F, Sagara T, Niki K. Bioelectrocatalysis at electrodes coated with alcohol-dehydrogenase, a quinohemoprotein with heme-c serving as a built-in mediator. J Electroanal Chem. 1993;361(1/2):221–228. doi: 10.1016/0022-0728(93)87058-4. [DOI] [Google Scholar]
- 32.Ramanavicius A, Habermüller K, Csöregi E, Laurinavicius V, Schuhmann W. Polypyrrole entrapped quinohemoprotein alcohol dehydrogenase. Evidence for direct electron transfer via conducting-polymer chains. Anal Chem. 1999;71(16):3581–3586. doi: 10.1021/ac981201c. [DOI] [PubMed] [Google Scholar]
- 33.Tasca F, Gorton L, Harreither W, Haltrich D, Ludwig R, Nöll G. Highly efficient and versatile anodes for biofuel cells based on cellobiose dehydrogenase from Myriococcum thermophilum. J Phys Chem C. 2008;112(35):13668–13673. doi: 10.1021/jp805092m. [DOI] [Google Scholar]
- 34.Bollella P, Mazzei F, Favero G, Fusco G, Ludwig R, Gorton L, Antiochia R. Improved DET communication between cellobiose dehydrogenase and a gold electrode modified with a rigid self-assembled monolayer and green metal nanoparticles: the role of an ordered nanostructuration. Biosens Bioelectron. 2017;88(Suppl. C):196–203. doi: 10.1016/j.bios.2016.08.027. [DOI] [PubMed] [Google Scholar]
- 35.Bollella P, Ludwig R, Gorton L. Cellobiose dehydrogenase: insights on the nanostructuration of electrodes for improved development of biosensors and biofuel cells. Appl Mater Today. 2017;9:319–332. doi: 10.1016/j.apmt.2017.08.009. [DOI] [Google Scholar]
- 36.Ludwig R, Harreither W, Tasca F, Gorton L. Cellobiose dehydrogenase: A versatile catalyst for electrochemical applications. ChemPhysChem. 2010;11(13):2674–2697. doi: 10.1002/cphc.201000216. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig R, Ortiz R, Schulz C, Harreither W, Sygmund C, Gorton L. Cellobiose dehydrogenase modified electrodes: advances by materials science and biochemical engineering. Anal Bioanal Chem. 2013;405(11):3637–3658. doi: 10.1007/s00216-012-6627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai S, Yakushi T, Matsushita K, Kitazumi Y, Shirai O, Kano K. The electron transfer pathway in direct electrochemical communication of fructose dehydrogenase with electrodes. Electrochem Commun. 2014;38:28–31. doi: 10.1016/j.elecom.2013.10.024. [DOI] [Google Scholar]
- 39.Kamitaka Y, Tsujimura S, Kano K. High current density bioelectrolysis of D-fructose at fructose dehydrogenase-adsorbed and Ketjen black-modified electrodes without a mediator. Chem Lett. 2006;36(2):218–219. doi: 10.1246/cl.2007.218. [DOI] [Google Scholar]
- 40.Khan GF, Kobatake E, Shinohara H, Ikariyama Y, Aizawa M. Molecular interface for an activity controlled enzyme electrode and its application for the determination of fructose. Anal Chem. 1992;64(11):1254–1258. doi: 10.1021/ac00035a011. [DOI] [Google Scholar]
- 41.Khan GF, Shinohara H, Ikariyama Y, Aizawa M. Electrochemical behavior of monolayer quinoprotein adsorbed on the electrode surface. J Electroanal Chem. 1991;315(1/2):263–273. doi: 10.1016/0022-0728(91)80075-2. [DOI] [Google Scholar]
- 42.Page CC, Moser CC, Dutton PL. Mechanism for electron transfer within and between proteins. Curr Opin Chem Biol. 2003;7(5):551–556. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Murgida DH, Hildebrandt P. Redox and redox-coupled processes of heme proteins and enzymes at electrochemical interfaces. Phys Chem Chem Phys. 2005;7(22):3773–3784. doi: 10.1039/b507989f. [DOI] [PubMed] [Google Scholar]
- 44.Bollella P, Schulz C, Favero G, Mazzei F, Ludwig R, Gorton L, Antiochia R. Green synthesis and characterization of gold and silver nanoparticles and their application for development of a third generation lactose biosensor. Electroanalysis. 2017;29(1):77–86. doi: 10.1002/elan.201600476. [DOI] [Google Scholar]
- 45.Hibino Y, Kawai S, Kitazumi Y, Shirai O, Kano K. Construction of a protein-engineered variant of D-fructose dehydrogenase for direct electron transfer-type bioelectrocatalysis. Electrochem Commun. 2017;77:112–115. doi: 10.1016/j.elecom.2017.03.005. [DOI] [Google Scholar]
- 46.Sugimoto Y, Kawai S, Kitazumi Y, Shirai O, Kano K. Function of C-terminal hydrophobic region in fructose dehydrogenase. Electrochim Acta. 2015;176:976–981. doi: 10.1016/j.electacta.2015.07.142. [DOI] [Google Scholar]
- 47.Funabashi H, Murata K, Tsujimura S. Effect of pore size of MgO-templated carbon on the direct electrochemistry of D-fructose dehydrogenase. Electrochemistry (Tokyo, Jpn) 2015;83(5):372–375. doi: 10.5796/electrochemistry.83.372. [DOI] [Google Scholar]
- 48.Kizling M, Bilewicz R. Fructose dehydrogenase electron transfer pathway in bioelectrocatalytic reactions. ChemElectroChem, in press. 2017. doi:10.1002/celc.201700861
- 49.Kawai S, Goda-Tsutsumi M, Yakushi T, Kano K, Matsushita K. Heterologous overexpression and characterization of a flavoprotein-cytochrome c complex fructose dehydrogenase of Gluconobacter japonicus NBRC3260. Appl Environ Microbiol. 2013;79(5):1654–1660. doi: 10.1128/AEM.03152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsujimura S, Nishina A, Hamano Y, Kano K, Shiraishi S. Electrochemical reaction of fructose dehydrogenase on carbon cryogel electrodes with controlled pore sizes. Electrochem Commun. 2010;12(3):446–449. doi: 10.1016/j.elecom.2010.01.016. [DOI] [Google Scholar]
- 51.Tominaga M, Nomura S, Taniguchi I. D-Fructose detection based on the direct heterogeneous electron transfer reaction of fructose dehydrogenase adsorbed onto multi-walled carbon nanotubes synthesized on platinum electrode. Biosens Bioelectron. 2009;24(5):1184–1188. doi: 10.1016/j.bios.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Antiochia R, Lavagnini I, Magno F. Amperometric mediated carbon nanotube paste biosensor for fructose determination. Anal Lett. 2004;37(8):1657–1669. doi: 10.1081/AL-120037594. [DOI] [Google Scholar]
- 53.Antiochia R, Gorton L. A new osmium-polymer modified screen-printed graphene electrode for fructose detection. Sensors Actuators B. 2014;195:287–293. doi: 10.1016/j.snb.2014.01.050. [DOI] [Google Scholar]
- 54.Antiochia R, Vinci G, Gorton L. Rapid and direct determination of fructose in food: a new osmium-polymer mediated biosensor. Food Chem. 2013;140(4):742–747. doi: 10.1016/j.foodchem.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 55.Privé GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41(4):388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Schulz C, Ludwig R, Micheelsen PO, Silow M, Toscano MD, Gorton L. Enhancement of enzymatic activity and catalytic current of cellobiose dehydrogenase by calcium ions. Electrochem Commun. 2012;17(Suppl. C):71–74. doi: 10.1016/j.elecom.2012.01.031. [DOI] [Google Scholar]
- 57.Saboe PO, Conte E, Farell M, Bazan GC, Kumar M. Biomimetic and bioinspired approaches for wiring enzymes to electrode interfaces. Energy Environ Sci. 2017;10(1):14–42. doi: 10.1039/C6EE02801B. [DOI] [Google Scholar]
- 58.Kracher D, Zahma K, Schulz C, Sygmund C, Gorton L, Ludwig R. Inter-domain electron transfer in cellobiose dehydrogenase: modulation by pH and divalent cations. FEBS J. 2015;282(16):3136–3148. doi: 10.1111/febs.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamocky M, Ludwig R, Peterbauer C, Hallberg B, Divne C, Nicholls P, Haltrich D. Cellobiose dehydrogenase-a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr Protein Pept Sci. 2006;7(3):255–280. doi: 10.2174/138920306777452367. [DOI] [PubMed] [Google Scholar]
- 60.Tan T-C, Kracher D, Gandini R, Sygmund C, Kittl R, Haltrich D, Hallberg BM, Ludwig R, Divne C. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat Commun. 2015;6. doi:10.1038/ncomms8542. [DOI] [PMC free article] [PubMed]
- 61.Sezer M, Spricigo R, Utesch T, Millo D, Leimkuehler S, Mroginski MA, Wollenberger U, Hildebrandt P, Weidinger IM. Redox properties and catalytic activity of surface-bound human sulfite oxidase studied by a combined surface enhanced resonance Raman spectroscopic and electrochemical approach. Phys Chem Chem Phys. 2010;12(28):7894–7903. doi: 10.1039/b927226g. [DOI] [PubMed] [Google Scholar]
- 62.Feng C, Kedia RV, Hazzard JT, Hurley JK, Tollin G, Enemark JH. Effect of solution viscosity on intramolecular electron transfer in sulfite oxidase. Biochemistry. 2002;41(18):5816–5821. doi: 10.1021/bi016059f. [DOI] [PubMed] [Google Scholar]
- 63.Appelqvist R, Marko-Varga G, Gorton L, Torstensson A, Johansson G. Enzymatic determination of glucose in a flow system by catalytic oxidation of the nicotinamide coenzyme at a modified electrode. Anal Chim Acta. 1985;169:237–247. doi: 10.1016/S0003-2670(00)86226-1. [DOI] [Google Scholar]
- 64.Harreither W, Coman V, Ludwig R, Haltrich D, Gorton L. Investigation of graphite electrodes modified with cellobiose dehydrogenase from the ascomycete Myriococcum thermophilum. Electroanalysis. 2007;19(2/3):172–180. doi: 10.1002/elan.200603688. [DOI] [Google Scholar]
- 65.Baminger U, Subramaniam SS, Renganathan V, Haltrich D. Purification and characterization of cellobiose dehydrogenase from the plant pathogen Sclerotium (Athelia) rolfsii. Appl Environ Microbiol. 2001;67(4):1766–1774. doi: 10.1128/AEM.67.4.1766-1774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenfield NJ, Fasman GD. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 67.Johnson WC. Protein secondary structure and circular dichroism: a practical guide. Proteins: Struct Funct Bioinf. 1990;7(3):205–214. doi: 10.1002/prot.340070302. [DOI] [PubMed] [Google Scholar]
- 68.Micsonai A, Wien F, Kernya L, Lee Y-H, Goto Y, Réfrégiers M, Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc Natl Acad Sci. 2015;112(24):E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lees JG, Miles AJ, Wien F, Wallace B. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22(16):1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 70.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction, and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida H, Sakai G, Mori K, Kojima K, Kamitori S, Sode K. Structural analysis of fungus-derived FAD glucose dehydrogenase. Sci Rep. 2015;5:13498. doi: 10.1038/srep13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kurth JM, Brito JA, Reuter J, Flegler A, Koch T, Franke T, Klein E-M, Rowe SF, Butt JN, Denkmann K. Electron accepting units of the diheme cytochrome c TsdA, a bifunctional thiosulfate dehydrogenase/tetrathionate reductase. J Biol Chem. 2016;291(48):24804–24818. doi: 10.1074/jbc.M116.753863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins: Struct Funct Bioinf. 2007;69(1):139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 74.Duhovny D, Nussinov R, Wolfson HJ. Efficient unbound docking of rigid molecules. Lect Notes Comput Sci. 2002;2452:185–200. doi: 10.1007/3-540-45784-4_14. [DOI] [Google Scholar]
- 75.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Suppl. 2):W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia C, de Oliveira NG, Kubota L. New fructose biosensors utilizing a polypyrrole film and D-fructose 5-dehydrogenase immobilized by different processes. Anal Chim Acta. 1998;374(2):201–208. doi: 10.1016/S0003-2670(98)00259-1. [DOI] [Google Scholar]
- 77.Arthur EJ. “Exploring the solvent environment of biomolecular systems”. PhD thesis, The University of Michigan. 2015. https://deepblue.lib.umich.edu/handle/2027.42/116689. Accessed 13 Jan 2016
- 78.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. BBA-Prot Proteomics. 2006;1764(11):1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 79.Armstrong FA, Cox PA, Hill HAO, Lowe VJ, Oliver BN. Metal ions and complexes as modulators of protein-interfacial electron transport at graphite electrodes. J Electroanal Chem. 1987;217(2):331–366. doi: 10.1016/0022-0728(87)80228-0. [DOI] [Google Scholar]
- 80.Schulz C, Ludwig R, Gorton L. Polyethyleneimine as a promoter layer for the immobilization of cellobiose dehydrogenase from Myriococcum thermophilum on graphite electrodes. Anal Chem. 2014;86(9):4256–4263. doi: 10.1021/ac403957t. [DOI] [PubMed] [Google Scholar]
- 81.Kielb P, Sezer M, Katz S, Lopez F, Schulz C, Gorton L, Ludwig R, Wollenberger U, Zebger I, Weidinger IM. Spectroscopic observation of calcium-induced reorientation of cellobiose dehydrogenase immobilized on electrodes and its effect on electrocatalytic activity. ChemPhysChem. 2015;16(9):1960–1968. doi: 10.1002/cphc.201500112. [DOI] [PubMed] [Google Scholar]
- 82.Wettstein C, Kano K, Schäfer D, Wollenberger U, Lisdat F. Interaction of flavin-dependent fructose dehydrogenase with cytochrome c as basis for the construction of biomacromolecular architectures on electrodes. Anal Chem. 2016;88(12):6382–6389. doi: 10.1021/acs.analchem.6b00815. [DOI] [PubMed] [Google Scholar]
- 83.Ferapontova EE, Gorton L. Direct electrochemistry of heme multicofactor-containing enzymes on alkanethiol-modified gold electrodes. Bioelectrochemistry. 2005;66(1):55–63. doi: 10.1016/j.bioelechem.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 84.Sarauli D, Wettstein C, Peters K, Schulz B, Fattakhova-Rohlfing D, Lisdat F. Interaction of fructose dehydrogenase with a sulfonated polyaniline: application for enhanced bioelectrocatalysis. ACS Catal. 2015;5(4):2081–7. doi: 10.1021/acscatal.5b00136. [DOI] [Google Scholar]
- 85.Davies CW. The extent of dissociation of salts in water. Part VI. Some calcium salts of organic acids. J Chem Soc. 1938;(Resumed):277–81. 10.1039/JR9380000277.
- 86.Blondin G, Girerd JJ. Interplay of electron exchange and electron transfer in metal polynuclear complexes in proteins or chemical models. Chem Rev. 1990;90(8):1359–76. doi: 10.1021/cr00106a001. [DOI] [Google Scholar]
- 87.Bendall DS. Protein electron transfer. In: Bendall DS, editor. Interprotein electron transfer. BIOS Scientific Publishers Ltd, Oxford UK; 1996. pp. 43–68.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 133 kb)