Abstract
Transcriptomics, the high-throughput characterization of RNAs, has been instrumental in defining pathogenic signatures in human autoimmunity and autoinflammation. It enabled the identification of new therapeutic targets in IFN-, IL-1- and IL-17-mediated diseases. Applied to immunomonitoring, transcriptomics is starting to unravel diagnostic and prognostic signatures that stratify patients, track molecular changes associated with disease activity, define personalized treatment strategies, and generally inform clinical practice. Herein, we review the use of transcriptomics to define mechanistic, diagnostic, and predictive signatures in human autoimmunity and autoinflammation. We discuss some of the analytical approaches applied to extract biological knowledge from high-dimensional data sets. Finally, we touch upon emerging applications of transcriptomics to study eQTLs, B and T cell repertoire diversity, and isoform usage.
Keywords: autoimmunity, autoinflammation, transcriptomics, mechanisms, patient stratification, therapeutic targets
INTRODUCTION
Human autoimmune diseases result from complex interactions between genetic and environmental factors that build up over time until the appearance of clinical symptoms. Their common denominator is the loss of tolerance to self-antigens, which can be organ specific, as in type 1 diabetes (T1D), or widely expressed, as in systemic lupus erythematosus (SLE), leading to the appearance of autoantibodies and/or autoreactive T cells. While underlying adaptive immunity alterations have been considered for decades, the pathogenic role of innate immunity in many of these diseases is also becoming evident. Recently, the term autoinflammation was coined to describe a group of diseases characterized by predominant activation of the innate immune system in the absence of detectable B or T cell autoreactivity. Autoinflammatory diseases manifest with fevers and multiorgan inflammation, as is the case in familial cryopyrinopathies (1). Within this spectrum, there are diseases with both autoimmune and autoinflammatory features, such as psoriasis, a disease featuring innate immunity dysregulation driven by autoreactive Th17 cells (2). Traditionally, all of these diseases have been treated with nonspecific anti-inflammatory and immunosuppressive agents that exhibit significant morbidity.
The breakthrough discovery of tumor necrosis factor alpha (TNF-α) as a pathogenic hub in rheumatoid arthritis (RA) (3, 4) heralded a new era of targeted therapeutics. Recent scientific and technological advances have led to the discovery of associated genetic risk factors and dysregulated immune pathways, enabling the development of targeted treatments that are successfully replacing nonspecific immunosuppression. These include monoclonal antibodies and fusion proteins targeting cytokines and their receptors, as well as small molecules targeting intracellular signaling pathways. IL-1 blockade has, for instance, been approved to treat a wide variety of diseases ranging from familial cryopyrinopathies to more common, sporadic inflammatory arthritides, such as systemic onset juvenile idiopathic arthritis (sJIA) and gout (5). Targeting various cytokines, including TNF-α, IL-12/23, and more recently IL-17, has shown great success in psoriasis (6–8). RA can also be controlled in many patients by targeting TNF-α, IL-6, or Janus kinases, and by antibody-mediated B cell depletion.
While therapies are becoming available for many autoimmune and autoinflammatory diseases, patients display significant heterogeneity in response to treatment. Indeed, 30–40% of RA, psoriasis, and inflammatory bowel disease (IBD) patients do not respond to TNF-α blockade, even though the clinical presentation of nonresponders is identical to that of patients achieving complete remission (9). Monoclonal antibodies targeting B cells in SLE showed promise in phase II but failed in several phase III clinical trials (10), even though there are numerous reports of successful outcomes in noncontrolled studies (11). This suggests that similar clinical profiles can result from distinct pathogenic mechanisms. Conversely, a drug targeting a single pathogenic hub can successfully treat distinct clinical phenotypes. As there are currently no transcriptional biomarkers that can predict response to a particular drug, there is a need to better characterize disease pathogenesis at the molecular level and on an individual basis. This could lead to identification of therapeutic targets, successful patient stratification in clinical trials, and personalized treatment strategies.
For nearly two decades, transcriptomics has enabled the simultaneous measurement of thousands of RNA transcripts in accessible tissues, including blood, urine, synovial fluid, saliva, and biopsy specimens using high-throughput technologies such as genome-wide DNA microarrays (12), as well as focused assays such as PCR and Nanostring. Previous studies in blood leukocytes and tissues demonstrated the applicability of microarrays to characterize molecular networks involved in cancer (13–18), autoimmunity (19, 20), infection (21–26), and vaccination (27–32). The recent advent of next-generation sequencing (NGS) (33) enabled comprehensive characterization of noncoding RNAs, including micro-RNAs (miRNAs) and long noncoding RNAs (lncRNAs) (34), and qualitative and quantitative measurements of isoforms through long-read sequencing (35, 36). The transcriptome is now routinely assessed in combination with whole-genome (WGS) or whole-exome (WES) sequencing for expression quantitative trait loci (eQTL) analyses, and with chromatin accessibility (ATAC-seq) (37) to further define genetic and epigenetic regulators of transcription. Targeted sequencing enables the characterization of T and B cell repertoires (38, 39). Finally, high-throughput single-cell sequencing is transforming the analysis of heterogeneous cell compartments and the identification of new populations (40).
In the next sections, we discuss how transcriptomics has informed us on the pathogenesis of human autoimmune and autoinflammatory diseases. We touch upon emerging applications of this technology to study eQTLs, repertoire diversity, and isoform usage. We focus on systemic diseases and summarize some of the lessons we learned from monitoring the transcriptome of children with SLE.
TRANSCRIPTOMICS TO UNRAVEL MECHANISMS OF INFLAMMATORY DISEASES
Transcriptional profiling of patients with human autoimmune diseases started at the turn of the 21st century, following the footsteps of cancer studies. Thus far, these studies have proven instrumental in revealing a role for interferons (IFNs) in a large number of autoimmune and autoinflammatory diseases, helping expand the novel field of interferonopathies (41, 42). Gene expression profiling also identified therapeutic targets in diseases driven by innate immunity activation that respond to IL-1 blockade (43). In psoriasis, the skin transcriptome enabled the identification of some of the most successful therapeutic targets to date (44). Whereas NGS is now taking gene expression profiling to a new level, the studies that we summarize under this section mostly used hybridization-based platforms.
Interferonopathies
The term interferonopathy refers to diseases where elevated type I IFN levels and/or biological activity is found in biological specimens. These patients characteristically share a prominent blood IFN signature that can be used as a complementary biomarker.
IFN families
IFNs belong to three families of cytokines originally discovered through their antiviral properties (45–47). Their pathogenic role in autoimmunity was first inferred from trials in humans. Use of recombinant IFN-α to treat hepatitis C and various malignancies was soon followed by reports of organ-specific and systemic autoimmunity (48).
The human type I IFN family includes 13 IFN-α subtypes together with IFN-β, IFN-ε, IFN-κ, and IFN-Ω. These cytokines have antiviral, antiproliferative, and immunomodulatory effects. Signaling through the canonical type I IFN receptor (IFNAR) activates the cytoplasmic tyrosine kinases JAK1 and TYK2 and leads to the phosphorylation of STAT1 and STAT2, which together with interferon regulatory factor (IRF) 9 form the ISGF3 transcription factor complex. This complex translocates to the nucleus and binds to response elements in the promoter regions of hundreds of IFN-stimulated genes (ISGs) (49). Type I IFNs display broad biological effects that can trigger autoimmunity through the activation of B cells, T cells, and dendritic cells (DCs) (42). In vitro, IFN-α enhances the differentiation of activated B cells into immunoglobulin-secreting plasma cells (50) and upregulates the expression of BAFF and APRIL on monocyte-derived DCs (moDCs) (51), promoting B cell survival. It also induces differentiation of monocytes intomoDCs (52), and upregulation of T cell costimulatory molecules such as MHC class II, CD80, and CD86. In mice, IFN-α-primed naive CD8+ T cells undergo proliferation and acquire effector functions (53), and type I IFN stimulates moDCs to induce differentiation of naive T cells into helper T cells (54).
Type II IFN (IFN-γ) is mainly produced by natural killer (NK) and T cells. IFN-γ signals through its unique receptor (IFNGR), which is expressed by most cell types. Activation of IFNGR leads to phosphorylation of STAT1 homodimers and subsequent expression of genes containing IFN-γ-activated sites (GASs), many of which are also induced by type I IFN (47). Type III IFN is the most recently discovered IFN family and includes IFN-λ1 (IL-29), IFN-λ2 (IL-28A), IFN-λ3 (IL-28B), and IFN-λ4 (55). Type III IFNs are produced by a variety of cell populations, including plasmacytoid DCs (pDCs), regulatory T cells, macrophages, and hepatocytes. Receptor ligation triggers signaling through the JAK/STAT cascade and induces the ISGF3 transcriptional complex, which elicits a gene signature overlapping with that of type I IFN (47).
Cellular sensors and cells involved in type I IFN production
Type I IFNs, as well as other proinflammatory cytokines, are secreted after activation of pattern-recognition receptors (PRRs) on immune cells with pathogen-derived nucleic acids (56). Although sensing of pathogen-derived nucleic acids is fundamental to mount antimicrobial immune responses, inappropriate detection of self–nucleic acids can lead to autoimmunity. PRRs that recognize nucleic acids include endosomal members of the Toll-like receptor family (TLR3, 7–9) and cytosolic sensors for DNA and RNA. The latter include retinoic acid–inducible gene 1 (RIG-I)-like receptors (RLRs) recognizing cytosolic dsRNA and 5′pppRNA, and AIM2-like receptors (ALRs) recognizing cytosolic DNA. Cyclic GMP-AMP (cGAMP) synthase (cGAS) was recently identified as a cytosolic PRR that detects DNA and leads to the production of cGAMP. This molecule binds to and activates the stimulator of IFN genes (STING), which is located in the mitochondria-associated membrane of the endoplasmic reticulum. In addition to STING, the mitochondrial antiviral-signaling protein (MAVS) is also found in this membrane-rich region, where it interacts with the RLRs RIG-I and MDA5 to further propagate innate immune signaling (57, 58).
Activation of STING and/or MAVS leads to phosphorylation and translocation to the nucleus of IRF3 and IRF7 and to activation of the NF-κB pathway. Once in the nucleus, IRF3, IRF7, and NF-κB subunits bind to specific binding sites in the IFN-β promoter as well as to the promoters of a subset of ISGs, including IRFs and PRRs (59). In humans, STING is ubiquitously expressed in hemopoietic and nonhemopoietic cells such as endothelial cells. It is characteristically absent in neutrophils, which rely on a unique repertoire of cytosolic DNA receptors, such as SOX2 (60).
Sensing of nucleic acids through endosomal receptors and IFN production is a characteristic of the natural IFN-producing cells or pDCs, which secrete high amounts of these cytokines in response to viruses or self-antigens following ligation to TLR7 or TLR9 (61, 62). Viruses and endogenous nucleic acids can also enter pDCs through Fc receptors when bound by antibodies. Whether engagement of TLR7 or TLR9 results in the production of type I IFN or proinflammatory cytokines depends on the compartment in which these TLRs encounter their ligands (63). TLR9-mediated recognition of large DNA-containing immune complexes is dependent, however, on a distinct convergence of phagocytic and autophagic pathways involving autophagy-related proteins but not the conventional autophagic preinitiation complex (64). Overall, production of type I IFN is tightly regulated by several surface receptors in pDCs (65).
The IFN signature
Transcripts induced by members of the different IFN families represent the IFN signature. This signature was originally defined in vitro by challenging mixed populations or individual cell types with recombinant IFN cytokines, mainly IFN-α2, IFN-β, IFN-γ, and members of the IFN-λ family, using microarray platforms. Such treatment leads to transcriptional stimulation of more than 5,000 genes in humans, as reported in the Interferome database (66). However, these approaches only captured changes in mRNA and not alternatively spliced isoforms or novel miRNAs and lncRNAs (47).
The signatures induced by members of the three IFN families overlap significantly. In fact, most components of the type I IFN signature are also induced by type II IFN, making the signature a broad marker of IFN activity (67). Indeed, under certain conditions, the type I and type II IFN pathways can reinforce each other, as small amounts of type II IFN enhance signaling through TLRs leading to type I IFN secretion (68), and type I IFN signaling induces the expression of the IFN-γR, thus cross-amplifying downstream pathways.
Among the numerous approaches that have been developed to characterize the IFN signature, we described a modular analytical framework that reduces the dimension of blood transcriptional data in order to facilitate functional interpretation, enable comparative analyses across multiple data sets and diseases, and improve robustness of biomarker signatures (Figure 1). This approach is based on modules that correspond to sets of genes coexpressed across multiple diseases and built via an unbiased data-driven approach. Mapping changes in gene expression at the module level generates disease-specific transcriptional fingerprints and provides a stable framework for the visualization and functional interpretation of microarray data (69). These transcriptional modules were used for the selection of biomarkers in patients with a diverse array of inflammatory diseases (70). Following functional interpretation, identified modules were linked to the two components driving differential gene expression in blood: changes in cell population frequency (e.g., B cell, cytotoxic cell modules), and cell-intrinsic transcript abundance (e.g., inflammation, IFN). Using a repertoire of 260 whole blood–derived modules, we could associate three of them with the IFN pathway (M1.2, M3.4, and M5.12). These modules were strongly upregulated in many autoimmune and infectious diseases (70–72) (Figure 2).
Figure 1.
A module framework to biologically interpret the blood transcriptome. (a) Reference whole-blood data sets are selected to represent a large spectrum of immune conditions, including autoimmunity, infectious diseases, cancer, and immunodeficiency. (b) Sets of coexpressed genes are extracted (69). (c) Modules are functionally annotated using a combination of knowledge-based (e.g., pathway enrichment analysis) and data-driven (e.g., hierarchical clustering, module enrichment in isolated leukocyte populations) methods. (d ) Module fingerprints can be derived for disease groups or for individual samples and mapped as module grids. The color represents the directionality of the module (red, up; blue, down) when compared to the reference control, and the intensity of the color represents the proportion of module probes that pass the significance threshold. (e) The modules can also be used as regular gene sets for gene set enrichment analyses (e.g., GSEA, QuSAGE, Q-Gen). Modified from Reference 72 with permission.
Figure 2.
The interferon (IFN) signature in systemic lupus erythematosus (SLE). (a) Unsupervised hierarchical clustering of the data reveals a prominent type I IFN signature in ~85% of SLE samples. (b) The module fingerprint of SLE, normalized to a group of reference healthy controls, reduces data dimensionality and enables rapid biological interpretation, revealing overexpression of the three IFN modules. (c) Gene-level network displaying the connectivity between IFN-inducible transcripts (circles) that positively correlate with disease activity. Modules are represented as squares. Nodes are colored according to the normalized expression of the transcripts in the high disease activity sample group. Significant transcripts were selected with a linear mixed model that compared high and low disease activity samples while accounting for patient ethnicity and treatment. Modified from Reference 72 with permission.
The Immunological Genome Project (ImmGen) recently performed a thorough dissection of the transcriptional response to IFN across many murine hematopoietic cell types together with detailed dynamic and chromatin analyses in B lymphocytes. Statistical integration of these data, together with other large data sets related to the IFN response across genetic variation in humans and mice, enabled the construction of a combined regulatory network for IFN. Predictions from this network were validated by genetic perturbation in both species. This regulatory map might help dissect the association between the IFN signature and disease (73).
Monogenic interferonopathies
The term interferonopathy was first proposed by Yanick Crow in 2011 (41) to define a group of Mendelian disorders associated with upregulation of type I IFN. Initially, these disorders included variants of Aicardi-Goutières syndrome (AGS), spondy-loenchondrodysplasia (SPENCD), and familial cases of SLE due to complement deficiency. Adoption of NGS rapidly expanded the spectrum of these type I IFN–mediated disorders, with the discovery of mutations in genes encoding a wide range of molecules involved in positive and negative regulation of IFN production, including nucleases, nucleic acid sensors, and signaling molecules (Table 1) (74).
Table 1.
Interferon- and IL-1-mediated autoimmune and autoinflammatory diseases
| Disease | Clinical featuresa | Causal mutationsb | Signat.Ref. | |
|---|---|---|---|---|
| Interferon | ||||
| Monogenic | Aicardi-Goutières syndrome | Inflammatory encephalopathy, cerebral calcifications, CLE symptoms | TREX1, SAMHD1, RNASEH2A-C, ADAR1, IFIH1, ISG15, USP18 | 239–241 |
| Familial chilblain lupus | CLE symptoms | TREX1, SAMHD1 | 228 | |
| CANDLE | Myositis, lipodystrophy, neutrophilic dermatitis | PSMB8 | 242 | |
| Adenosine deaminase 2 deficiency | Fever, hypertension, livedo reticularis, early onset stroke | CECR1 | 243 | |
| SAVI | Vasculitis/gangrene, interstitial lung disease | TMEM173 | 244 | |
| Singleton-Merten syndrome | Dental dysplasia, aortic calcifications, skeletal abnormalities, glaucoma, psoriasis | DDX58 | 245 | |
| Complement deficiencies | CLE and SLE-like symptoms, recurrent infections | C1q, C1r, C1s, C4 | ND | |
| Spondyloenchondrodysplasia | Skeletal abnormalities, SLE-like symptoms | ACP5/TRAP | 246 | |
| DNAse1L3 deficiency | Pediatric-onset SLE, lupus nephritis, hypocomplementemic urticarial vasculitis syndrome | Dnase1L3 | ND | |
| Common variable immunodeficiency | Infections, autoimmune manifestations | > 25 genes | 247c | |
| Complex | SLE | Rash, arthritis, lymphopenia, nephritis, serositis, seizure, psychosis | 19, 20, 72, 78, 79 | |
| Dermatomyositis, juvenile dermatomyositis, polymyositis | Muscle inflammation and proximal weakness | 92 | ||
| Sjögren syndrome | Inflammation of lacrimal and salivary glands | 90 | ||
| Rheumatoid arthritis (patient subset) | Inflammation of joints | 96–98 | ||
| Systemic sclerosis (patient subset) | Skin, kidney, lung inflammation/fibrosis | 101, 102 | ||
| Type 1 diabetes (preclinical) | Destruction of insulin-producing beta cells of the pancreas | 104 | ||
| Thyroiditis | Thyroid inflammation | NDd | ||
| IL-1f | ||||
| Monogenic | Cryopyrin-associated periodic syndromes | Fever, rash, headaches, arthritis | NLRP3 | 120 |
| Familial Mediterranean fever | Fever, rash, serositis, arthritis, amyloidosis | MEFV | ND | |
| Hyper-IgD syndrome | Fever, rash, headaches, arthritis | MVK | ND | |
| NLRC4 deficiency | Fever, rash, MAS ± enterocolitis | NLRC4 | 119 | |
| Deficiency of IL-1 receptor antagonist | Fever, severe skin and bone inflammation | IL1RN | ND | |
| Familial systemic onset juvenile arthritis | Fever, rash, polyarthritis | LACC1 | ND | |
| Pyogenic arthritis, pyoderma gangrenosum, and acne syndrome | Pyogenic sterile arthritis, pyoderma gangrenosum, acne | PSTPIP1 | 248 | |
| Majeed syndrome | Fever, pustular skin lesions, aseptic osteomyelitis, dyserythropoietic anemia | LPIN2 | ND | |
| Complex | Idiopathic recurrent pericarditis | Recurrent pericarditis | Sporadice | ND |
| Systemic juvenile idiopathic arthritis/Still’s disease (adults) | Fever, rash, arthritis, serositis, MAS | 43, 113, 114 | ||
| Type 1 diabetes (postdiagnosis) | Destruction of insulin-producing beta cells of the pancreas | 124 | ||
| Kawasaki disease | Fever, rash, mucositis, vasculitis, coronary artery aneurysms | 121–123 | ||
| Gout | Episodic inflammatory arthritis | ND | ||
| Schnitzler syndrome | Urticaria, fever, bone/joint pain, bone lesions | ND | ||
| Behçet disease | Fever, recurrent ulcers, vasculopathy | ND | ||
| Periodic fever, aphthous stomatitis, pharyngitis, and cervical adenitis syndrome | Fever, recurrent ulcers | ND | ||
| Henoch-Schönlein purpura | Purpura, abdominal pain, nephritis | ND | ||
| Sweet syndrome (neutrophilic dermatosis) | Fever, neutrophilic skin inflammation | ND | ||
| Synovitis, acne, pustulosis, hyperostosis, and osteitis syndrome | Synovitis, acne, pustulosis, hyperostosis, osteitis | ND | ||
| Hidradenitis suppurativa | Inflammation of sweat glands | ND | ||
| Autoimmune inner ear disease | Sensorineural hearing loss | ND | ||
Nonexhaustive list of symptoms.
Causal mutations are only known for monogenic diseases. For complex diseases, no single known mutation causes them.
Monogenic causes have been identified in ~30% of patients. An IFN signature is detected in the blood of patients with inflammatory manifestations.
Most common autoimmune disease induced by administration of recombinant IFN.
IRP is mainly sporadic, but rare familial cases have been identified and linked to mutations in TNFRSF1A and MEFV.
Clinical response to IL-1 inhibition.
Abbreviations: CANDLE, chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature; CLE, cutaneous lupus erythematosus; MAS, macrophage activation syndrome; ND, not defined; SAVI, STING-associated vasculopathy with onset in infancy; SLE, systemic lupus erythematosus.
Most patients with monogenic interferonopathies display elevated type I IFN protein and/or biological activity in serum and/or cerebrospinal fluid. These patients characteristically share a prominent blood IFN signature, which can be used as a complementary diagnostic biomarker as well as to assess response to emergent medications targeting the mutated pathways. Functional dissection of these pathways is proving fundamental to understanding how nucleic acid sensing and signaling regulate type I IFN production. We have learned, in addition, that the range of phenotypes associated with mutations within these pathways is much broader than previously realized (Table 1). In addition, patients’ relatives occasionally carry the mutation and display blood IFN signatures in the absence of clinical symptoms, adding a level of complexity that needs to be addressed (74).
Sporadic interferonopathies
While advances in genetics paved the way to understanding monogenic interferonopathies, transcriptomics was crucial to linking IFN with a number of sporadic autoimmune and inflammatory diseases, such as SLE.
SLE
SLE can be considered the prototype sporadic interferonopathy. Serum from SLE patients displays increased IFN-α activity (75, 76) and induces the differentiation of healthy monocytes into mature DCs in an IFN-α-dependent fashion (77). ISGs are significantly enriched in patient peripheral blood mononuclear cells (PBMCs) (19, 20) and whole blood (72, 78). Many longitudinal studies also revealed an association between this signature and SLE disease activity (72, 79–81). In addition, the signature is silenced following high-dose intravenous steroid therapy, which controls disease flares (19, 82).
After more than a decade of monitoring the SLE transcriptome, however, the specific contribution of different IFN families and family members to both the IFN signature and overall SLE pathogenesis is not yet fully resolved. Administration of IFN-γ in humans can induce an SLE-like disease, and both murine and in vitro human studies support a pathogenic role for this cytokine in SLE, especially in the context of lupus nephritis (83, 84). There are limited data suggesting dysregulation of the type III IFN pathway in SLE. Serum IFN-λ2 is elevated in SLE patients compared with healthy donors, and serum IFN-λ1 levels were found associated with disease activity, anti-dsDNA antibodies, glomerulonephritis, and arthritis (85). Both type II and III IFNs could contribute to the IFN signature and therefore explain either partial response or lack of response to type I IFN blockade in SLE.
Indeed, using our modular approach to longitudinally follow up SLE patients, we observed an IFN signature in 85% of children and 87% of adults (72, 78). Each of the three described IFN modules displayed a distinct activation threshold (M1.2<M3.4<M5.12), and a similar gradient was observed within clinically quiescent patients in the adult group, for whom moderate-to-strong modular scores were associated with higher anti-dsDNA titers and lymphopenia. Longitudinal analyses revealed both stable (M1.2) and variable (M3.4 and M5.12) components over time in individual patients. Comparing these modular IFN signatures to other previously established IFN scores (80, 86), the latter correlated strongly with IFN modules, but they were not redundant. Importantly, the limited set of genes selected for IFN scores by other groups belong mostly to M1.2 (the least variable module) and to a lesser extent M3.4, but very few belong to M5.12 (the more variable IFN module). In the modular study, all patients with strong IFN signatures had both IFN-α- and IFN-γ-positive scores, as described by Kirou et al. (81), which is in agreement with the possible implication of type II IFN in the activity of modules M3.4 and M5.12. The modular IFN score was also more sensitive to longitudinal changes than reductionist approaches using smaller gene sets (78).
Which IFNs contribute to the SLE signature and disease pathogenesis remains an open question. In addition, the cellular origin, sensors, and endogenous ligands that give rise to these patient signatures have not been fully characterized. Of note, most monogenic interferonopathies linked to dysregulated cytosolic nucleic acid sensing or proteasome dysfunction do not trigger immune complex–mediated systemic autoimmunity. However, those due to mutations in genes linked to IFN production through endosomal pathways and pDCs, such as TRAP (87) and C1q (88), cause symptoms that overlap with SLE, therefore supporting amore prominent role of TLRs in systemic forms of the disease.
The definite proof that IFNs play a role in SLE pathogenesis would come from clinical trials. Several anti-type I IFN therapies have been evaluated so far. The anti-IFN-α monoclonal antibodies sifalimumab and rontalizumab have completed phase II trials with mixed results in terms of reduction of clinical disease activity measures and suppression of the IFN gene signature. Anifrolumab, an anti-IFNAR monoclonal antibody that antagonizes all type I IFNs, has progressed to phase III. An alternate therapy targeting type I IFN is an IFN-α kinoid (IFN-K) vaccine composed of IFN-α2b coupled to a carrier protein that induces polyclonal IFN-α neutralizing antibodies. Results of a phase I/IIa trial showed induction of anti-IFN-α antibodies and decreased expression of both IFN-induced and B cell activation–associated gene transcripts, without significant adverse events (89). A phase I clinical trial of an anti-IFN-γ monoclonal antibody, AMG811, showed that it reduced IFN-γ-related gene expression. Currently, there are no drugs that specifically target type III IFNs in clinical trials, but depletion of pDCs, which is being tested, would block both type I and type III IFN production by these cells (85). The jury is therefore still out.
Other sporadic diseases with IFN signature
In addition to SLE, type I IFN has been associated with the pathogenesis of various rheumatic diseases. In patients with Sjögren syndrome, where exocrine glands are progressively destroyed by lymphocyte infiltrates, an IFN signature was detected both in blood (90) and in minor salivary glands (91). Inflammatory myopathies, including dermatomyositis, juvenile dermatomyositis, and polymyositis, also exhibited enriched IFN-inducible signatures in blood as well as in affected muscle and skin (92). The blood IFN signature correlated with disease activity and was abolished by immunosuppressive treatments (93, 94). The dermatomyositis skin signature is very similar to that of cutaneous lupus and herpes simplex-2 infection based on an enrichment of ISGs (95). A blood IFN signature is also present in subgroups of RA patients (96–98).
Importantly, IFN might be critical in early disease stages in a number of conditions. Thus, a type I IFN signature was identified in the skin lesions but not in the blood of patients with psoriasis (99). pDCs infiltrate the skin early during development of psoriasis, and can initiate psoriasis in an IFN-α-dependent manner in a mouse xenograft model (100). The blood of a subset of patients with systemic sclerosis also exhibits an IFN signature (101, 102), which is especially prominent during the earliest disease phases, even before overt skin fibrosis (103). Finally, transient increased expression of type I IFN signature is found during preclinical T1D and is a risk factor for autoimmunity in children with genetic predisposition to the disease (104).
The variety of clinical phenotypes associated with IFN signatures supports the idea that IFN plays an essential pathogenic role but additional factors determine the clinical outcome. How different nucleic acid ligands, sensors, and cell types/tissues contribute to IFN production and disease manifestations remains unknown, however, in the vast majority of nonfamilial interferonopathies.
IL-1-Mediated Diseases
IL-1β is an important mediator of innate and adaptive immunity (105, 106). IL-1β is translated as a precursor that requires cleavage by caspase 1, an enzyme that is activated in the cytosol by inflammasomes (107). As described above for monogenic interferonopathies, recent advances in genetics have provided unprecedented insight into mutations that cause monogenic autoinflammatory diseases. Cytokine blockade has proven effective in a subset of these conditions, particularly those associated with increased IL-1 activity (108). These include syndromes where the production of IL-1 is increased by mutation of innate immune sensors such as NLRP3 or NLRC4, upstream signaling molecules such as PSTPIP1, downstream signaling molecules or receptor antagonists such as IL-1Ra (109). In addition to rare monogenic diseases, IL-1β is involved in the pathogenesis of sporadic inflammatory and autoimmune disorders (106) (Table 1). Thus, the availability of therapies specifically targeting IL-1 has enabled the recognition of a pathogenic role for this cytokine in the underlying inflammation of many common diseases, including gout, cardiovascular disease, type 2 diabetes, the metabolic syndrome, and even cancer (110).
The IL-1 family of receptors contains a domain highly homologous to the cytoplasmic domains of all TLRs. Consequently, inflammation due to TLR ligands and IL-1 family members triggers similar downstream signaling and overlapping transcriptional profiles (111). Using a functional genomics approach, we uncovered the central role of IL-1β in the pathogenesis of sJIA, a sporadic and aggressive form of juvenile arthritis that does not respond to anti-TNF-α therapy (112). Analysis of transcriptional patterns from active patient PBMCs yielded a signature overlapping those of infectious and various inflammatory diseases, whereas a signature linked to erythropoiesis was more specific to the disease (113, 114). However, transcriptional changes elicited by culturing healthy PBMCs with patient serum included increased IL-1β and IL-1β-inducible genes such as IL1R1 and IL1R2 (43). Subsequent treatment of a small group of sJIA patients refractory to conventional treatments with the IL-1 receptor antagonist anakinra yielded complete and lasting remission in 72% of patients. Recent randomized clinical trials of anakinra; the anti-IL-1β monoclonal antibody canakinumab; or rilonacept, a dimeric fusion protein of IL-1R1 and its accessory protein that neutralizes IL-1, confirmed the efficacy of IL-1β blockade in this disease (115–117). Of note, blockade of IL-6, which is induced by IL-1β, is also effective in sJIA (118).
The sJIA signature partially overlaps with signatures found in monogenic inflammasome-mediated diseases. Upregulation of NLRC4, IL1R2, TLR5, and S100 family members, for example, is found in sJIA as well as in patients with monoallelic missense mutations in NLRC4 (119). A similar signature was described in patients with cryopyrinopathies, including upregulation of transcription of members of the IL-1 family; neutrophil-related chemokines and their receptors; markers of neutrophilic inflammation, such as S100A12; and potassium channel-encoding genes, such as KCNJ15 (120). Despite this signature overlap, no mutations in inflammasome-related genes have been found so far in sJIA patients.
An IL-1 signature has also been identified in PBMCs from patients with Kawasaki disease (KD), a sporadic, self-limited vasculitis that affects coronary arteries in young children (121–123). IL-1 blockade in KD is currently being tested in clinical trials. We and others reported an IL-1 signature in PBMCs of patients with T1D (124), and plasma from patients with recent-onset T1D induced an IL-1 signature in healthy PBMCs (125). However, canakinumab and anakinra did not prove effective in two randomized clinical trials, suggesting that targeting systemic inflammation is not sufficient to halt this organ-specific autoimmune disease once clinical symptoms manifest (126).
There is an expanding list of monogenic and sporadic inflammatory diseases where targeting IL-1 has been reported successful in clinical trails and/or case reports, but no transcriptomic analyses have been conducted (Table 1). Availability of such studies, especially prior to and after successful IL-1 blockade, might enable the identification of discrete transcriptional changes that could inform on disease-specific pathogenic mechanisms.
Multiple Cytokines, One Disease: The Case of Psoriasis
Psoriasis, one of the most common chronic inflammatory diseases in humans, is an example of dysregulated cooperation between innate and adaptive immune responses. It is also a great example of how translational research, and in particular transcriptomics, can uncover pathogenic mechanisms and novel therapeutic targets.
Early research, as well as trials with T cell–targeted therapies, set the stage for the role of adaptive immunity in this disease. Increased expression of IL-12 p40 subunit mRNA in lesions from patients led to the classification of psoriasis as a Th1 disease. Indeed, ustekinumab, an antibody against IL-12 p40, was developed to target IL-12 and inhibit Th1 cells. This therapy led to significant improvement in 70% of patients (127) and seemed to confirm the pathogenic role of the IL-12/Th1 axis. IL-23, however, shares the p40 subunit with IL-12, and psoriatic skin displayed tenfold upregulation of both IL-12 p40 and IL-23 p19 mRNAs (128). In experimental settings, the IL-23/Th17 axis was subsequently described as mediating skin changes compatible with those of psoriasis (129).
The success of blocking the IL-23/IL-17 axis in psoriasis is unprecedented. Antibodies against IL-17A (ixekizumab and secukinumab), the IL-17 receptor A subunit (brodalumab), or IL-23p19 (guselkumab) reverse clinical, histologic, and molecular features of disease in 80% to 90% of patients. Furthermore, IL-17 blockade with high doses of brodalumab reverses expression of almost all disease-specific signatures (130), suggesting that IL-17A or IL-17F acts upstream of any inflammatory cascade leading to disease. To date, at least six biologic antagonists of the IL-23/Th17 axis are undergoing phase II and phase III trials in psoriasis (44). Finally, transcriptional profiling of the skin of patients enrolled in a trial with TNF-α blockers, which the US Food and Drug Administration approved for psoriasis in 2006, demonstrated that clinical response was linked to suppression of the IL-17 axis (131).
Monogenic cases of psoriasis vulgaris, pityriasis rubra pilaris, and pustular psoriasis have been attributed to mutations in the CARD14 gene. These mutations, which lead to CARD14 gain of function, activate NF-κB and upregulate a subset of psoriasis-associated genes in keratinocytes, including chemokine (C-C motif ) ligand 20 (CCL20) and IL-8. It is thought that after a triggering event such as epidermal injury, these raremutations in CARD14 induce inflammatory cell recruitment by keratinocytes, including Th17 cells (132). Mutations in the IL36RN gene (which encodes the IL-36Ra protein) have also been described in familial systemic pustular psoriasis (133). IL-36 family members are induced upon IL-17A stimulation of keratinocytes, leading to IL-23 gene expression in DCs and keratinocytes, thus amplifying and sustaining the chronic inflammatory state.
While mutations in the above-described genes are not found in the more common sporadic forms of psoriasis (134), their functional elucidation has shed fundamental light on how end organ–specific dysfunction might set the stage for systemic inflammation in this disease.
APPLICATION OF TRANSCRIPTOMICS IN THE CLINIC
Many autoimmune diseases exhibit similar clinical presentations, dynamic disease course, and heterogeneous responses to treatment. Although the use of transcriptomics in the current clinical management of autoimmunity remains limited, efforts are under way to identify gene signatures that can (a) diagnose disease, (b) assess disease prognosis, (c) act as surrogate scores of clinical disease activity, and (d) monitor or predict response to treatment (Figure 3).
Figure 3.
Five applications of transcriptomics for the clinic. ❶ Disease diagnosis: Disease-specific signatures can help assign patients to disease A, B, or C. ❷ Disease molecular stratification: Patients with the same clinical disease may exhibit distinct transcriptional signatures, suggesting distinct underlying mechanisms of pathogenesis. The level of molecular heterogeneity can vary from disease to disease. ❸ Disease prognosis: A transcriptional signature can help predict the course of disease, including onset, flare, or prolonged remission. ❹ Treatment response monitoring: Transcriptional fingerprints obtained at regular intervals after initiation of treatment can help segregate responders from nonresponders and characterize the mechanisms of response to treatment. ❺ Treatment response prediction: A signature obtained ideally before initiation of treatment can predict the long-term response to therapy and help customize therapeutic regimen accordingly.
Disease Diagnosis
Fever, arthritis, arthralgia, fatigue, anemia, leukopenia and antinuclear antibodies are common manifestations in several autoimmune diseases and viral infections. Accurate diagnosis of an autoimmune disease can be challenging in the absence of specific symptoms, which is frequent around the time of disease onset. Traditional diagnostic platforms in autoimmunity have focused on the measurement of autoantibodies (135), although multiplex assays are gradually being developed (136). Transcriptomics has helped define common and specific signatures across diseases.
In sJIA, a 12-gene PBMC signature could accurately distinguish it from other febrile illnesses (113). In IBD, colonic gene signatures revealed significant differences between ulcerative colitis and Crohn disease (137), and a 12-gene PBMC signature enriched for immunoglobulins could distinguish these two diseases (138). Similarly, a synovial tissue signature could distinguish between RA and osteoarthritis (139). Several meta-analyses were conducted to identify core and specific gene signatures across autoimmune diseases (140, 141). A functional genomics approach identified genes specifically modulated in healthy PBMCs challenged with plasma from T1D patients when compared to plasma from several infectious inflammatory conditions, including influenza and bacterial pneumonia (142).
None of these signatures has made it yet to the clinic. While they might be important to reveal disease mechanisms, these studies are so far not designed and powered to uncover true diagnostic biomarkers. The identification of the latter requires comparisons across a larger spectrum of diseases and inclusion of larger numbers of patients, combined with appropriate classification methods that validate their sensitivity and specificity in independent cohorts.
Disease Prognosis
Biomarkers that can predict clinical outcome, such as disease development in predisposed individuals, flares, or progression to specific tissue/organ involvement, are of great interest to clinicians. Their identification necessitates prospective studies in large cohorts over long follow-up periods, as exemplified by a study of US Armed Forces personnel that queried a biobank of more than 30 million serum samples to discover that the appearance and diversification of autoantibodies preceded the clinical onset of SLE (143).
Several prospective studies have been conducted to identify early biomarkers of T1D development. The DIPP (Type I Diabetes Prediction and Prevention) project is a large initiative established in Finland in 1994 to identify and immunomonitor children at risk of T1D to unravel the pathomechanisms of T1D development. The contribution of environmental factors such as infection and diet has also been incorporated into these studies. One hypothesis is that onset of disease can, in genetically predisposed individuals, be triggered by a diabetogenic enterovirus (144). The Diabetes Autoimmunity Study in the Young (DAISY) was designed to assess whether enteroviral infections could predict progression to T1D in children seropositive for islet autoantibodies (145). The BABYDIET study sought to assess whether delaying the introduction of gluten in infants at risk of islet autoimmunity could reduce the incidence of T1D (146). Blood transcriptional profiling of subjects from these cohorts identified signatures associated with disease development. In the DAISY cohort, antibody-positive children exhibited a five-gene lymphocyte signature that could predict the rate of progression to clinical T1D (147). In both the BABYDIET and DIPP cohorts, a blood IFN response preceded seroconversion (104, 148). The role of type I IFNs, and possibly of an early viral challenge, in T1D pathology is supported by a recent study of antibody affinity in individuals with central immune tolerance defects due to AIRE deficiency. In this monogenic autoimmune polyendocrine syndrome, most patients develop antibodies against type I IFN, but only those whose autoantibodies display low neutralizing capacity progress to T1D (149).
Transcriptomics studies have also been performed in isolated blood T cells to identify signatures that would predict disease course (e.g., frequency of flares). A CD8+ T cell gene signature could predict long-term prognosis in patients with SLE and antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) (150), or separate clinically coherent IBD patients into two groups with subsequently different disease courses (151). In both studies, genes associated with poor prognosis were involved in IL-7 receptor and T cell receptor signaling. Furthermore, a signature of CD8+ T cell exhaustion predicted better prognosis across several autoimmune diseases, including T1D, AAV, SLE, and idiopathic pulmonary fibrosis (152). These studies suggest that purified cell subsets could be a source of transcriptional prognostic biomarkers that otherwise would be undetectable in heterogeneous tissues such as whole blood. The advent of single-cell sequencing technologies could further unveil leukocyte subset heterogeneity and help identify specific biomarkers to forecast disease course.
Clinical Score Surrogates and Disease Activity Assessment
There is significant value in defining objective molecular scores that could act as surrogates of clinical scores, which are nonexhaustive, sometimes depend on subjective evaluation by a physician, and can be difficult to record in routine clinical practice. In SLE, attempts to correlate the SLE disease activity index (SLEDAI) with a blood IFN signature have yielded controversial results (79–81, 86). This could be due to inappropriate study design, as few longitudinal studies have measured the full extent of the signature, possibly introducing biases during the knowledge-driven data-reduction process.
We developed a multivariate transcriptional U score combining expression of genes from modules that correlated positively (IFN response, neutrophils) or negatively (ribosomal proteins, T cells)with SLEDAI(79). This score correlated better with SLEDAI than its individual components and could be used to longitudinally monitor disease progression in an independent cohort of untreated patients. Recently, we expanded the study in a longitudinal cohort of 158 pediatric SLE patients followed up for up to four years (72). Close to 1,000 blood samples were collected and their transcriptomes profiled by microarray. In parallel, extensive clinical and laboratory data were recorded, including detailed SLEDAI information. Linear mixed models were used to assess the influence of disease activity, patient ethnicity, treatment, SLEDAI components, and nephritis class on the transcriptome. While IFN and neutrophil signatures exhibited strong correlation with the SLEDAI, a plasma cell signature was the most robust correlate of disease activity in two independent patient groups and was enriched in African American patients, who usually present with more severe forms of SLE. Importantly, the large sample size enabled us to account for ethnicity and treatment, which affect clinical presentation and blood signatures.
Because the SLEDAI is a weighted composite score of 24 clinical parameters, which can be broadly classified as serological, musculoskeletal, renal, or neurologic, in principle a specific score obtained from different combinations of symptoms could be associated with different blood signatures. The inclusion of a SLEDAI component category in the model revealed that the neutrophil signature was associated with the development of lupus nephritis, one of the more severe complications of the disease. Conversely, IFN and plasma cell signatures were enriched across all disease presentations. This suggests that the complex makeup of the clinical score should be recorded and considered when interpreting molecular profiles from patients with systemic autoimmune diseases. It also brings up the significance of specific signatures, such as the neutrophil signature, in lupus nephritis. Indeed, this signature was originally attributed to the presence of low-density neutrophils in SLE PBMCs (19). In fact, the signature reflects activation of both low- and high-density neutrophils, which become a remarkable source of endogenous interferogenic DNA by releasing oxidized mitochondrial DNA (153). Why this blood signature is linked to nephritis remains to be elucidated. The transcriptome of laser-captured glomeruli from nephritic patients revealed the presence of a myeloid signature corresponding to neutrophils, monocytes, and DCs. In contrast to findings from blood, glomeruli that displayed increased type I IFN signature were associated with milder pathological features, especially lower fibrosis. Whether this reflects a role of IFN in early disease stages, as fibrosis is a feature of long-standing disease, remains to be elucidated. Yet, as in the blood, an increased B cell/plasma cell signature paralleled disease activity (154).
Biomarkers that can predict changes in disease activity are very relevant to clinical practice. Their identification will require frequent and standardized sample collection during periods of flares and remissions to capture molecular profiles within a short window of flare onset. Such studies will be facilitated by on-going efforts in sample volume reduction (e.g., finger prick sampling) (27), technology miniaturization (e.g., microfluidics) (155), standardization of analytical techniques, and collaboration across clinical and research institutions to maximize sample size.
Monitoring Response to Treatment
Patients with autoimmune diseases display significant heterogeneity in response to treatment: 30% of RA and 40% of ulcerative colitis patients do not respond to anti-TNF-α therapy (156, 157), and 30% of sJIA patients do not respond to IL-1 blockade (115). Although both anti-IFN-α and anti-CD20 therapies show efficacy in subsets of SLE patients, the development of a universal targeted mode of therapy has mostly failed in phase III clinical trials (10, 158), perhaps reflecting the molecular heterogeneity of the disease.
Transcriptomics has been applied to monitor systemic responses to several targeted biologics. IL-1 blockade was followed by rapid and sustained downregulation of IL-1 signaling and proinflammatory genes in the blood of patients with sJIA (113, 115, 159), neonatal-onset multisystem inflammatory disease (NOMID) (160), and Blau syndrome–related uveitis (161). In several phase I SLE trials, a dose-dependent inhibition of IFN-inducible genes was observed in the blood of patients treated with anti-IFN-α monoclonal antibodies (162–165). In systemic sclerosis, the IFN signature of both blood and skin was suppressed by an anti-IFNAR monoclonal antibody (166), and a wound signature in lesional skin biopsies decreased following treatment with the tyrosine kinase inhibitor imatinib (167). In psoriatic skin, TNF-α, IL-17A, and IL-23 blockade downregulated many proinflammatory genes, including several IL-17-inducible ones (168, 169). Anti-TNF-α therapy also effectively downregulated proinflammatory genes in the blood of patients with intravenous immunoglobulin–resistant KD (170) and patients with anti-TNF-α-responsive RA (98). In multiple sclerosis (MS), monitoring of PBMC responses to IFN-β therapy highlighted an early and long-lasting increase in IFN-inducible genes (171). Furthermore, PBMCs from nonresponders did not exhibit the signature (172), supporting the value of the IFN signature as a biomarker of response to therapy in this setting.
In addition to confirming the modulatory effects of biologics on their targeted pathways, these immunomonitoring studies have yielded some important insight regarding immune pathway interactions. For example, both IL-1 and TNF-α blockade in sJIA resulted in increased blood IFN-inducible transcripts, confirming in vivo the antagonism between IL-1 or TNF-α and IFN signaling (173, 174). Similarly, the overlap between genes suppressed by anti-TNF-α and anti-IL-17A therapies in psoriasis patients highlighted the synergy elicited by these two cytokines (168).
Predicting Response to Treatment
While transcriptional monitoring during treatment can help uncover molecular networks modulated by therapeutic agents and separate responders from nonresponders a posteriori, whether these signatures can improve clinical decision making before treatment is not established. Because of the cost and side effects of current biological therapies, and the need to rapidly intervene to limit long-term disability in many autoimmune diseases, biomarkers that can match patients with the optimum therapy are actively sought out.
Most treatment prediction studies have focused on responses to anti-TNF-α and anti-CD20 therapy in RA (175). Two independent studies could predict responsiveness to therapy with anti-TNF-α alone (176) or in combination with methotrexate (177) using patient PBMC transcriptional profiles. There was no overlap, however, between the few transcripts that passed statistical tests in these two studies. In synovial tissue, a baseline myeloid signature was associated with good responses to this therapy (178). An elevated baseline IFN response in neutrophils and PBMCs predicted better responses to anti-TNF-α (179) and anti-IL-6 (180) therapies, respectively. However, an increased blood IFN signature and a failure to downregulate key inflammatory genes (e.g., IL8, IL1B, NFKBIA, CCL4) after TNF-α blockade were both associated with poor long-term clinical outcome (98, 181, 182). On the other hand, a high baseline IFN response was associated with poor clinical outcome following anti-CD20 therapy (183–185). In addition, nonresponders to anti-CD20 therapy exhibited an increased expression of IgJ mRNA, a marker of antibody-secreting plasmablasts (186). These combined observations first suggest that RA patients who display high baseline IFN response may benefit from anti-TNF-α therapy, while those who do notmay respond better to anti-CD20 therapy. Second, given the immunostimulatory effect of type I IFN on B cells (187), patients displaying a high plasmablast signaturemay benefit from increased dosage of anti-CD20 therapy, IFN blockade or eventually plasma cell–specific therapies.
Altogether, an association between major transcriptional networks at baseline and long-term therapeutic outcomes has not been firmly established in the autoimmunity field. Integration with additional genomic information [e.g., single-nucleotide polymorphism (SNP) analysis] together with monitoring larger patient cohorts would be necessary to identify robust predictive biomarkers of response to treatment.
Patient Stratification
The heterogeneity of patient responses to targeted therapies observed in many diseases has prompted the search for genomic biomarkers that could stratify patients to improve the chances of success by customizing therapeutic interventions.
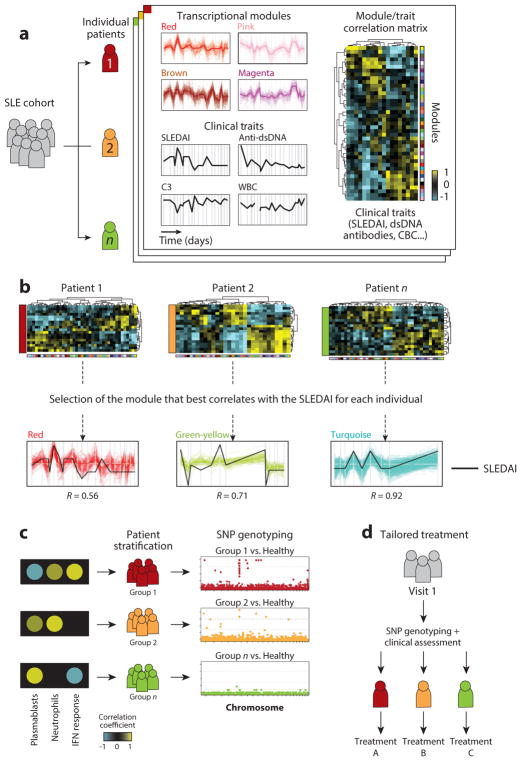
In our recent SLE study, we used longitudinally collected data to develop personalized transcriptional immunoprofiles (72). Blood signatures that best correlated with clinical disease activity over time were subsequently used to stratify patients based on the major molecular pathways driving these signatures (Figure 4). This approach revealed significant heterogeneity in SLE molecular classes. An important lesson from this study is that in order to identify meaningful immune networks, information must be recorded at multiple time points and reflect different clinical disease activity levels, rather than the presence or absence of a signature at a single time point being considered (188). Overall, our findings support that a priori binning of SLE patients into molecular subclasses might help clinical trial design and improve success rate.
Figure 4.
Stratification of SLE patients through individual immunoprofiling. (a) Modules of coexpressed genes over time are identified and a module/trait correlation matrix is generated for each patient. (b) The module that best correlates with a clinical trait of interest, such as SLEDAI, is selected for each individual. (c) Patients are stratified according to the immune network that best correlates with the trait of interest. Patients are genotyped and SNP analysis is conducted for each patient group. (d ) Definition of tailored treatment based on genotype and early clinical assessment of SLE patients. Abbreviations: CBC, complete blood count; C3, complement component 3; SLE, systemic lupus erythematosus; SLEDAI, SLE disease activity index;WBC, white blood cell count. Modified from Reference 72 with permission.
These observations will need to be validated in clinical settings, through clinical trials that incorporate patient molecular profiles in their design. Reducing time, cost, and complexity of molecular stratification will also be paramount to its application in the clinic. Along this line, we identified a subset of transcripts that could be used in cost-effective targeted assays such as Nanostring or multiplex PCR to stratify SLE patients. We also investigated genetic associations of stratification through a combination of traditional SNP enrichment and eQTL analyses. These preliminary results, which highlighted numerous genes in pathways associated with SLE pathogenesis, will require validation in larger cohorts.
Altogether, these studies are paving the way to personalized approaches to therapy that could reduce the use of nonspecific agents to prevent treatment toxicity and potentially dangerous broad immunosuppression. Larger studies incorporating longitudinal sampling and thorough collection of patient clinical data should be designed to validate the clinical applications of this approach.
EMERGING APPLICATIONS OF TRANSCRIPTOMICS IN AUTOIMMUNITY
Repertoire Analysis
The adaptive immune system is composed of a vast repertoire of B and T cells with distinct antigen specificities that provide long-term protection against a broad array of pathogens. In autoimmune diseases, the diversification processes at the origin of this repertoire yield autoreactive B and T cell clones that are central to disease pathogenesis (189). Understanding when, where, and how these autoreactive clones arise as well as their pathogenic role in disease is critical. Targeted high-throughput sequencing of the variable regions of the B and T cell receptors can be applied to characterize repertoire diversity and clonal expansion in various immunological states, including autoimmunity (38, 39, 190, 191).
In SLE, targeted RNA-Seq combined with flow cytometry and proteomics enabled the characterization of the diversity of antibody-secreting cells in the blood of patients experiencing disease-associated flares. These studies revealed the existence of highly autoreactive clones derived from a distinct subset of newly activated naive cells that could persist in circulation for months (192) and suggest that B cell–depleting treatments could be refined to target specifically this small subset, possibly preventing recurring flares and broader depletion of B cell compartments.
In MS, the B cell repertoire was sequenced in paired tissue samples from the central nervous system (CNS) and draining cervical lymph nodes (CLNs) to establish whether CNS-invading antigen-specific B cells could mature in secondary lymphoid compartments (193). Clonally expanded B cells were found in both compartments, founding members of clones could be found in the CLNs, and antigen-specific B cells could traffic freely across the tissue barrier after maturation outside the CNS. This study contributes to explain the immunomodulatory mechanisms of current MS therapies that deplete circulating B cells via anti-CD20 therapy or inhibit lymphocyte migration to the CNS via anti-VLA-4 therapy (194).
In RA, both B and T cell repertoires have been interrogated. A DNA barcoding method combined with recombinant antibody expression and enzyme-linked immunosorbent assay (ELISA) enabled the identification of new target antigens of autoreactive antibodies, including α-enolase, citrullinated fibrinogen, and citrullinated histone H2B (195). An analysis of the B cell repertoire in blood and synovial fluid from patients with early or established RA revealed that the synovium was enriched for expanded autoreactive B and plasma cell clones especially in the initial phase of the disease, suggesting that early intervention at the site of inflammation may be most beneficial (196). A similar approach combining TCR sequencing and single-cell transcriptomics identified transcriptional shifts within the most expanded memory CD4+ T cell clones in both blood and synovium and revealed increased senescence and altered transcription of homing genes in synovial clones (197).
Overall, repertoire analysis is further unraveling the diversity of autoantigens and the expansion and migration dynamics of autoreactive clones in various tissues. Perhaps the greatest challenge will be to expand these studies to tissues that are difficult to sample to obtain a more comprehensive picture of pathogenic B and T cell receptor diversity.
eQTLs
Genome-wide association studies (GWAS) have identified hundreds of genetic variants associated with autoimmune diseases in large patient cohorts, most of which are located in noncoding regions of the genome (198). To define true causal variants that directly affect disease pathogenesis, integrative approaches are combining genetic information with transcriptomics, epigenetics, protein-interaction networks, and functional annotations. These strategies, for example, have enabled the identification of eQTLs, which are genomic loci that influence the expression levels of mRNAs. eQTLs act either in cis or in trans if they map close to or are distant from the gene affected, respectively. They can be identified by combining SNP genotyping, by SNP array or whole-exome/genome sequencing, with expression profiling, originally by microarray and more recently by RNA-Seq, usually captured at a single time point during active disease (199).
eQTL analyses have been conducted in SLE (200, 201), T1D (200), RA (201–203), IBD (201), celiac disease (201), MS (201), Sjögren syndrome (204), ankylosing spondylitis (205), and AAV (206), among others. A large eQTL meta-analysis in more than 8,000 peripheral blood samples identified both cis- and trans-eQTLs associated with disease processes in SLE and T1D (200). Variants mapped to noncoding enhancer regions across six autoimmune diseases led to the development of a multiple enhancer variant hypothesis, where small contributions of several SNPs in linkage disequilibrium at the same noncoding loci can influence multiple enhancers of either the same gene or genes encoding proteins functionally related and enriched in common pathways. This hypothesis might explain how common traits contribute to complex diseases (201).
eQTLs have also been identified in specific cell subsets (207). cis-eQTL mapping in five primary leukocyte populations from patients with IBD or AAV and from healthy controls revealed that specific eQTL associations can only be detected in patients with active inflammation and disappear following treatment (206). Applying a joint modeling approach between cell types, as opposed to analyzing eQTLs in each population separately, showed that about half of detected eQTLs were shared between populations in both disease groups. Combining these results with previous GWAS further identified new eQTLs at 34 IBD-associated loci. Another approach mapped eQTLs to population-specific enhancer regions in RA, identifying different patterns of SNP–enhancer gene connections between monocytes and B and T cells (203). Mapping eQTLs to discrete leukocyte populations in active and remission phases of autoimmunity may therefore help uncover disease-, disease activity–, and population-specific eQTLs, in addition to highlighting leukocyte subsets involved in pathogenesis.
Several groups have developed integrative strategies to improve the identification of causal variants from GWAS. The Probabilistic Identification of Causal SNPs (PICS) algorithm integrated genetic, transcriptomic, and epigenetic measurements to identify causal variants associated with 21 autoimmune diseases across various immune cell subsets (208). Candidate risk genes were prioritized in RA by combining functional annotations, epigenetic chromatin marks, cis-eQTLs, knockout mouse phenotypes, cancer and primary immunodeficiency databases, and molecular pathway enrichment analyses. Some of the genes identified using this approach are the targets of approved therapies for RA, thus providing empirical evidence that integrative genetic analyses can lead to drug discovery (202). Recently, a massive parallel reporter assay was applied to >30,000 variants to pinpoint causal cis-eQTLs. Among >800 variants altering allele expression, CRISPR/Cas9 gene editing validated a risk allele for ankylosing spondylitis that controls expression of the proinflammatory prostaglandin EP4 receptor (205).
eQTLs have also been applied to disease prognosis. A noncoding polymorphism in the regulatory transcription factor FOXO3A was associated with milder disease activity but not with diagnosis of Crohn disease and RA (209). Mechanistically, isolated monocytes from minor allele homozygotes challenged with various TLR ligands displayed increased FOXO3 transcription and produced less TNF-α and more IL-10 in response to lipopolysaccharide, in a TGF-β-dependent manner. Importantly, this study demonstrated that polymorphisms associated with disease prognosis can be distinct from those associated with disease susceptibility. These observations should encourage the continuous expansion of patient databases to provide numbers large enough to enable eQTL studies both across and within diseases.
Isoforms
Alternative splicing (AS) is an important mechanism of biological diversity in eukaryotes, as it allows multiple mRNA isoforms to be transcribed from a single gene, leading to translation of multiple proteins (210–214). The human genome consists of ~23,000 genes; however, >95% of multi-exonic pre-mRNAs are alternatively spliced to generate >100,000 isoforms. The alternate mRNA isoforms that are translated into proteins can have opposing functions, such as pro- versus antiapoptotic activities. There are five AS modes, exon skipping being the most common in mammals. An understanding of the importance of AS in the development of autoimmune diseases is starting to emerge. A spliced variant of cytotoxic T-lymphocyte antigen 4 (CTLA-4) is linked to human autoimmunity, most notably T1D (215). Although very few studies have focused on the pathophysiological role of isoforms in autoimmunity, it is likely that many other mRNA isoforms are linked to immune functions and diseases. Previous reports focusing on the correlation of immune cell activation and isoform expression have used either exon microarrays or RNA-Seq, which are known to provide minimal information about detection of novel full-length isoforms.
CTLA-4 is critical for negative regulation of T cell proliferation. Several genetically associated polymorphisms have been identified within the CTLA-4 gene. Furthermore CTLA-4 knockout animals as well as treatment of patients with anti-CTLA4 support its role in autoimmunity (215). Two isoforms of CTLA-4 are produced by AS in human cells: a full-length transmembrane isoform (flCTLA-4), and a soluble isoform (sCTLA-4) in which exon 3 is skipped. Resting T cells express sCTLA-4 but downregulate its expression upon activation, whereas they rapidly increase expression of flCTLA-4. Variations in CTLA-4 gene splicing are involved in a number of autoimmune diseases, such as T1D (215) and myasthenia gravis (216). Thus, a spectrum of CTLA-4 isoforms has been detected in patients with myasthenia gravis, and when the levels of sCTLA-4 are reduced by RNAi, the potency of regulatory T cells significantly decreases.
In SLE, susceptibility loci that affect splicing have also been identified (217). For example, three SNPs in the B cell scaffold protein with ankyrin repeats (BANK1) gene confer susceptibility to SLE. One of these SNPs affects relative splicing efficiency and upregulates production of a novel BANK1 isoform that lacks exon 2 (218), which encodes a putative inositol 1,4,5-triphosphate receptor (IP3R)-binding domain that might attenuate BANK1-mediated signaling.
Regulation of the T cell receptor ζ chain (TCRζ) pre-mRNA by AS further illustrates the potential role of AS in SLE (219–221). Two isoforms lead to reduced expression of the TCR/CD3 complex and decreased production of IL-2 upon stimulation. Reduced availability of IL-2 results in a decrease in regulatory T cells and ensuing autoimmunity.
IRF5, which regulates type I IFN expression and mediates TLR signaling, is associated with SLE susceptibility (222–226). In mice, loss of Irf5 protects from lupus-like disease. In humans, IRF5 is constitutively expressed in pDCs, moDCs, and activated B cells and exists as multiple alternatively spliced transcripts. Distinct groups of IRF5 SNPs and genetic variants form haplotypes that confer risk for, or protection from, SLE development. Genetic association studies have begun to reveal the genetic risk conferred by these risk haplotypes. Both IRF5 expression and AS are significantly elevated in PBMCs of SLE patients. Standard molecular cloning techniques as well as RNA-Seq for de novo junction discovery have revealed that SLE patients express a unique IRF5 transcript signature, and an IRF5-SLE risk haplotype defines the top four most abundant IRF5 transcripts expressed in SLE patients (225).
Overall, deciphering the connections between AS splicing and autoimmunity holds promise for a better understanding of both processes.
CONCLUSIONS
Transcriptomics has been essential to unravel pathogenic mechanisms of several human autoimmune and autoinflammatory diseases. It is increasingly used to monitor disease course and response to treatment and is starting to yield diagnostic and predictive biomarkers. In combination with genetic data, it helps in pinpointing true pathogenic variants among the many identified by GWAS. It is providing a wealth of information about noncoding RNAs and their potential role as biomarkers and/or pathogenic players in these diseases (227, 228). Rapidly improving sequencing technologies, including long-read platforms such as single-molecule real-time (SMRT) sequencing (229) and droplet-based sequencing (230), are enabling the characterization of isoform usage in disease, the rapid and cost-effective sequencing of thousands of single cells at a time and even the analysis of single-cell transcriptomes under in vivo conditions (231).
Without doubt, the application of NGS to characterize the microbiome is already yielding fundamental information relevant to the development of autoimmunity and autoinflammation (232–235). These studies will be the object of much more research.
Despite these developments, direct applications to the clinic have been hindered by a relative scarcity of studies adequately designed and powered for predictive biomarker detection. The field of autoimmunity research will benefit from emulating some of the efforts in cancer research, in particular the establishment of large consortia, centralized tissue biobanks, and databanks, such as the ones provided by The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov) and the International Cancer Genome Consortium (ICGC; https://dcc.icgc.org) (236). The recent announcement for the Human Functional Genomics Project (237), which aims at understanding the interindividual variation of human immune responses in health and disease, should contribute to providing the basis for stratifying patients for personalized treatment strategies.
Finally, new approaches for detecting, measuring, and combining a wide range of biomedical information, including molecular, genomic, cellular, clinical, behavioral, physiological, and environmental parameters, as promoted by the Precision Medicine Initiative (238), will undoubtedly help in understanding and developing rational treatment approaches not only for cancer, but also for autoimmune and autoinflammatory diseases.
Acknowledgments
We acknowledge support from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (P50 AR054083-01 to V.P.), the National Institute of Allergy and Infectious Diseases (U19 AI089987 to J.B. and V.P. and U19 AIO82715 to V.P.), and the Baylor-Scott & White Health Care Research Foundation.
Footnotes
DISCLOSURE STATEMENT
J.B. is a member of the Board of Directors and the Chairman of the Scientific Advisory Board of Neovacs (Paris, France), a biotechnology company focused on the use of vaccines eliciting anticytokine responses. V.P. has consulted with Neovacs and Astra Zeneca for the development of anti-IFN therapies. V.P. and J.B. hold a patent (#8221748) on compositions and methods for the treatment of systemic onset juvenile idiopathic arthritis with IL-1 antagonists.
LITERATURE CITED
- 1.Stoffels M, Kastner DL. Old dogs, new tricks: monogenic autoinflammatory disease unleashed. Annu Rev Genom Hum Genet. 2016;17:245–72. doi: 10.1146/annurev-genom-090413-025334. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa A, Siewert K, Stohr J, Besgen P, Kim SM, et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J Exp Med. 2015;212:2203–12. doi: 10.1084/jem.20151093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maini RN, Brennan FM, Williams R, Chu CQ, Cope AP, et al. TNF-alpha in rheumatoid arthritis and prospects of anti-TNF therapy. Clin Exp Rheumatol. 1993;11(Suppl 8):S173–75. [PubMed] [Google Scholar]
- 4.Elliott MJ, Maini RN, Feldmann M, Long-Fox A, Charles P, et al. Treatment of rheumatoid arthritis with chimeric monoclonal antibodies to tumor necrosis factor α. Arthritis Rheum. 1993;36:1681–90. doi: 10.1002/art.1780361206. [DOI] [PubMed] [Google Scholar]
- 5.Dinarello CA. An expanding role for interleukin-1 blockade from gout to cancer. Mol Med. 2014;20(Suppl 1):S43–58. doi: 10.2119/molmed.2014.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366:1181–89. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 7.Leonardi C, Matheson R, Zachariae C, Cameron G, Li L, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–99. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 8.Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, et al. Secukinumab in plaque psoriasis—results of two phase 3 trials. N Engl J Med. 2014;371:326–38. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- 9.Siebert S, Tsoukas A, Robertson J, McInnes I. Cytokines as therapeutic targets in rheumatoid arthritis and other inflammatory diseases. Pharmacol Rev. 2015;67:280–309. doi: 10.1124/pr.114.009639. [DOI] [PubMed] [Google Scholar]
- 10.Kalunian KC, Merrill JT, Maciuca R, McBride JM, Townsend MJ, et al. A phase II study of the efficacy and safety of rontalizumab (rhuMAb interferon-alpha) in patients with systemic lupus erythematosus (ROSE) Ann Rheum Dis. 2016;75:196–202. doi: 10.1136/annrheumdis-2014-206090. [DOI] [PubMed] [Google Scholar]
- 11.Aguiar R, Araujo C, Martins-Coelho G, Isenberg D. Use of rituximab in systemic lupus erythematosus: a single center experience over 14 years. Arthritis Care Res. 2017;69:257–62. doi: 10.1002/acr.22921. [DOI] [PubMed] [Google Scholar]
- 12.Alizadeh A, Eisen M, Botstein D, Brown PO, Staudt LM. Probing lymphocyte biology by genomic-scale gene expression analysis. J Clin Immunol. 1998;18:373–79. doi: 10.1023/a:1023293621057. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 14.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–37. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 15.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 16.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Res. Netw. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Netw. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. PNAS. 2003;100:2610–15. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–77. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–66. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, et al. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–37. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 25.Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, et al. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. PNAS. 2002;99:972–77. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–17. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obermoser G, Presnell S, Domico K, Xu H, Wang Y, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–44. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banchereau R, Baldwin N, Cepika AM, Athale S, Xue Y, et al. Transcriptional specialization of human dendritic cell subsets in response to microbial vaccines. Nat Commun. 2014;5:5283. doi: 10.1038/ncomms6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cech TR, Steitz JA. The noncoding RNA revolution—trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Au KF, Sebastiano V, Afshar PT, Durruthy JD, Lee L, et al. Characterization of the human ESC transcriptome by hybrid sequencing. PNAS. 2013;110:E4821–30. doi: 10.1073/pnas.1320101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharon D, Tilgner H, Grubert F, Snyder M. A single-molecule long-read survey of the human transcriptome. Nat Biotechnol. 2013;31:1009–14. doi: 10.1038/nbt.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–18. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol. 2014;32:158–68. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodsworth DJ, Castellarin M, Holt RA. Sequence analysis of T-cell repertoires in health and disease. Genome Med. 2013;5:98. doi: 10.1186/gm502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–20. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- 42.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–92. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 43.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005;201:1479–86. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda S, Krueger JG, Guttman-Yassky E. The translational revolution and use of biologics in patients with inflammatory skin diseases. J Allergy Clin Immunol. 2015;135:324–36. doi: 10.1016/j.jaci.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Isaacs A, Lindenmann J. Virus interference. I The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–67. [PubMed] [Google Scholar]
- 46.McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart TA. Neutralizing interferon alpha as a therapeutic approach to autoimmune diseases. Cytokine Growth Factor Rev. 2003;14:139–54. doi: 10.1016/s1359-6101(02)00088-6. [DOI] [PubMed] [Google Scholar]
- 49.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–34. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 51.Joo H, Coquery C, Xue Y, Gayet I, Dillon SR, et al. Serum from patients with SLE instructs monocytes to promote IgG and IgA plasmablast differentiation. J Exp Med. 2012;209:1335–48. doi: 10.1084/jem.20111644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santini SM, Di Pucchio T, Lapenta C, Parlato S, Logozzi M, Belardelli F. A new type I IFN-mediated pathway for the rapid differentiation of monocytes into highly active dendritic cells. Stem Cells. 2003;21:357–62. doi: 10.1634/stemcells.21-3-357. [DOI] [PubMed] [Google Scholar]
- 53.Le Bon A, Thompson C, Kamphuis E, Durand V, Rossmann C, et al. Cutting edge: enhancement of antibody responses through direct stimulation of B and T cells by type I IFN. J Immunol. 2006;176:2074–78. doi: 10.4049/jimmunol.176.4.2074. [DOI] [PubMed] [Google Scholar]
- 54.Cucak H, Yrlid U, Reizis B, Kalinke U, Johansson-Lindbom B. Type I interferon signaling in dendritic cells stimulates the development of lymph-node-resident T follicular helper cells. Immunity. 2009;31:491–501. doi: 10.1016/j.immuni.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Wack A, Terczynska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802–9. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 57.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–88. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 58.Roers A, Hiller B, Hornung V. Recognition of endogenous nucleic acids by the innate immune system. Immunity. 2016;44:739–54. doi: 10.1016/j.immuni.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 59.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia P, Wang S, Ye B, Du Y, Huang G, et al. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nat Immunol. 2015;16:366–75. doi: 10.1038/ni.3117. [DOI] [PubMed] [Google Scholar]
- 61.Vallin H, Blomberg S, Alm GV, Cederblad B, Ronnblom L. Patients with systemic lupus erythematosus (SLE) have a circulating inducer of interferon-alpha (IFN-α) production acting on leucocytes resembling immature dendritic cells. Clin Exp Immunol. 1999;115:196–202. doi: 10.1046/j.1365-2249.1999.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Investig. 2005;115:407–17. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 64.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–97. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol. 2015;15:471–85. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rusinova I, Forster S, Yu S, Kannan A, Masse M, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–46. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall JC, Casciola-Rosen L, Berger AE, Kapsogeorgou EK, Cheadle C, et al. Precise probes of type II interferon activity define the origin of interferon signatures in target tissues in rheumatic diseases. PNAS. 2012;109:17609–14. doi: 10.1073/pnas.1209724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Negishi H, Osawa T, Ogami K, Ouyang X, Sakaguchi S, et al. A critical link between Toll-like receptor 3 and type II interferon signaling pathways in antiviral innate immunity. PNAS. 2008;105:20446–51. doi: 10.1073/pnas.0810372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–80. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pascual V, Chaussabel D, Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–71. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis. 2015;212:213–22. doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Banchereau R, Hong S, Cantarel B, Baldwin N, Baisch J, et al. Personalized immunomonitoring uncovers molecular networks that stratify lupus patients. Cell. 2016;165:551–65. doi: 10.1016/j.cell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mostafavi S, Yoshida H, Moodley D, LeBoite H, Rothamel K, et al. Parsing the interferon transcriptional network and its disease associations. Cell. 2016;164:564–78. doi: 10.1016/j.cell.2015.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crow YJ, Manel N. Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol. 2015;15:429–40. doi: 10.1038/nri3850. [DOI] [PubMed] [Google Scholar]
- 75.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301:5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 76.Preble OT, Black RJ, Friedman RM, Klippel JH, Vilcek J. Systemic lupus erythematosus: presence in human serum of an unusual acid-labile leukocyte interferon. Science. 1982;216:429–31. doi: 10.1126/science.6176024. [DOI] [PubMed] [Google Scholar]
- 77.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–43. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 78.Chiche L, Jourde-Chiche N, Whalen E, Presnell S, Gersuk V, et al. Modular transcriptional repertoire analyses of adults with systemic lupus erythematosus reveal distinct type I and type II interferon signatures. Arthritis Rheumatol. 2014;66:1583–95. doi: 10.1002/art.38628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009;18:980–89. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-α pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 82.Guiducci C, Gong M, Xu Z, Gill M, Chaussabel D, et al. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–41. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yokoyama H, Takabatake T, Takaeda M, Wada T, Naito T, et al. Up-regulated MHC-class II expression and gamma-IFN and soluble IL-2R in lupus nephritis. Kidney Int. 1992;42:755–63. doi: 10.1038/ki.1992.344. [DOI] [PubMed] [Google Scholar]
- 84.Haas C, Ryffel B, Le Hir M. IFN-gamma is essential for the development of autoimmune glomerulonephritis in MRL/Ipr mice. J Immunol. 1997;158:5484–91. [PubMed] [Google Scholar]
- 85.Oon S, Wilson NJ, Wicks I. Targeted therapeutics in SLE: emerging strategies to modulate the interferon pathway. Clin Transl Immunol. 2016;5:e79. doi: 10.1038/cti.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Higgs BW, Liu Z, White B, Zhu W, White WI, et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis. 2011;70:2029–36. doi: 10.1136/ard.2011.150326. [DOI] [PubMed] [Google Scholar]
- 87.An J, Briggs TA, Dumax-Vorzet A, Alarcon-Riquelme ME, Belot A, et al. Tartrate-resistant acid phosphatase deficiency in the predisposition to systemic lupus erythematosus. Arthritis Rheumatol. 2017;69:131–42. doi: 10.1002/art.39810. [DOI] [PubMed] [Google Scholar]
- 88.Santer DM, Hall BE, George TC, Tangsombatvisit S, Liu CL, et al. C1q deficiency leads to the defective suppression of IFN-alpha in response to nucleoprotein containing immune complexes. J Immunol. 2010;185:4738–49. doi: 10.4049/jimmunol.1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lauwerys BR, Hachulla E, Spertini F, Lazaro E, Jorgensen C, et al. Down-regulation of interferon signature in systemic lupus erythematosus patients by active immunization with interferon alpha-kinoid. Arthritis Rheum. 2013;65:447–56. doi: 10.1002/art.37785. [DOI] [PubMed] [Google Scholar]
- 90.Emamian ES, Leon JM, Lessard CJ, Grandits M, Baechler EC, et al. Peripheral blood gene expression profiling in Sjögren’s syndrome. Genes Immun. 2009;10:285–96. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren’s syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–44. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 92.Greenberg SA. Type 1 interferons and myositis. Arthritis Res Ther. 2010;12(Suppl 1):S4. doi: 10.1186/ar2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007;13:59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walsh RJ, Kong SW, Yao Y, Jallal B, Kiener PA, et al. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum. 2007;56:3784–92. doi: 10.1002/art.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wong D, Kea B, Pesich R, Higgs BW, Zhu W, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PLOS ONE. 2012;7:e29161. doi: 10.1371/journal.pone.0029161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66:1008–14. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reynier F, Petit F, Paye M, Turrel-Davin F, Imbert PE, et al. Importance of correlation between gene expression levels: application to the type I interferon signature in rheumatoid arthritis. PLOS ONE. 2011;6:e24828. doi: 10.1371/journal.pone.0024828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sekiguchi N, Kawauchi S, Furuya T, Inaba N, Matsuda K, et al. Messenger ribonucleic acid expression profile in peripheral blood cells from RA patients following treatment with an anti-TNF-α monoclonal antibody, infliximab. Rheumatology. 2008;47:780–88. doi: 10.1093/rheumatology/ken083. [DOI] [PubMed] [Google Scholar]
- 99.Bowcock AM, Shannon W, Du F, Duncan J, Cao K, et al. Insights into psoriasis and other inflammatory diseases from large-scale gene expression studies. Hum Mol Genet. 2001;10:1793–805. doi: 10.1093/hmg/10.17.1793. [DOI] [PubMed] [Google Scholar]
- 100.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med. 2005;202:135–43. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology. 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 102.Bos CL, van Baarsen LG, Timmer TC, Overbeek MJ, Basoski NM, et al. Molecular subtypes of systemic sclerosis in association with anti-centromere antibodies and digital ulcers. Genes Immun. 2009;10:210–18. doi: 10.1038/gene.2008.98. [DOI] [PubMed] [Google Scholar]
- 103.Brkic Z, van Bon L, Cossu M, van Helden-Meeuwsen CG, Vonk MC, et al. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann Rheum Dis. 2016;75:1567–73. doi: 10.1136/annrheumdis-2015-207392. [DOI] [PubMed] [Google Scholar]
- 104.Ferreira RC, Guo H, Coulson RM, Smyth DJ, Pekalski ML, et al. Atype I interferon transcriptional signature precedes autoimmunity in children genetically at risk for type 1 diabetes. Diabetes. 2014;63:2538–50. doi: 10.2337/db13-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arbore G, West EE, Spolski R, Robertson AA, Klos A, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science. 2016;352:aad1210. doi: 10.1126/science.aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–18. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sharma D, Kanneganti TD. The cell biology of inflammasomes: mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213:617–29. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jesus AA, Goldbach-Mansky R. IL-1 blockade in autoinflammatory syndromes. Annu Rev Med. 2014;65:223–44. doi: 10.1146/annurev-med-061512-150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moghaddas F, Masters SL. Monogenic autoinflammatory diseases: cytokinopathies. Cytokine. 2015;74:237–46. doi: 10.1016/j.cyto.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 110.Cavalli G, Dinarello CA. Treating rheumatological diseases and co-morbidities with interleukin-1 blocking therapies. Rheumatology. 2015;54:2134–44. doi: 10.1093/rheumatology/kev269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alsina L, Israelsson E, Altman MC, Dang KK, Ghandil P, et al. A narrow repertoire of transcriptional modules responsive to pyogenic bacteria is impaired in patients carrying loss-of-function mutations in MYD88 or IRAK4. Nat Immunol. 2014;15:1134–42. doi: 10.1038/ni.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bruck N, Schnabel A, Hedrich CM. Current understanding of the pathophysiology of systemic juvenile idiopathic arthritis (sJIA) and target-directed therapeutic approaches. Clin Immunol. 2015;159:72–83. doi: 10.1016/j.clim.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 113.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204:2131–44. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, et al. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum. 2007;56:3793–804. doi: 10.1002/art.22981. [DOI] [PubMed] [Google Scholar]
- 115.Quartier P, Allantaz F, Cimaz R, Pillet P, Messiaen C, et al. A multicentre, randomised, double-blind, placebo-controlled trial with the interleukin-1 receptor antagonist anakinra in patients with systemic-onset juvenile idiopathic arthritis (ANAJIS trial) Ann Rheum Dis. 2011;70:747–54. doi: 10.1136/ard.2010.134254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ruperto N, Brunner HI, Quartier P, Constantin T, Wulffraat N, et al. Two randomized trials of canakinumab in systemic juvenile idiopathic arthritis. N Engl J Med. 2012;367:2396–406. doi: 10.1056/NEJMoa1205099. [DOI] [PubMed] [Google Scholar]
- 117.Ilowite NT, Prather K, Lokhnygina Y, Schanberg LE, Elder M, et al. Randomized, double-blind, placebo-controlled trial of the efficacy and safety of rilonacept in the treatment of systemic juvenile idiopathic arthritis. Arthritis Rheumatol. 2014;66:2570–79. doi: 10.1002/art.38699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.De Benedetti F, Brunner HI, Ruperto N, Kenwright A, Wright S, et al. Randomized trial of tocilizumab in systemic juvenile idiopathic arthritis. N Engl J Med. 2012;367:2385–95. doi: 10.1056/NEJMoa1112802. [DOI] [PubMed] [Google Scholar]
- 119.Canna SW, de Jesus AA, Gouni S, Brooks SR, Marrero B, et al. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat Genet. 2014;46:1140–46. doi: 10.1038/ng.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Balow JE, Jr, Ryan JG, Chae JJ, Booty MG, Bulua A, et al. Microarray-based gene expression profiling in patients with cryopyrin-associated periodic syndromes defines a disease-related signature and IL-1-responsive transcripts. Ann Rheum Dis. 2013;72:1064–70. doi: 10.1136/annrheumdis-2012-202082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hoang LT, Shimizu C, Ling L, Naim AN, Khor CC, et al. Global gene expression profiling identifies new therapeutic targets in acute Kawasaki disease. Genome Med. 2014;6:541. doi: 10.1186/s13073-014-0102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Popper SJ, Shimizu C, Shike H, Kanegaye JT, Newburger JW, et al. Gene-expression patterns reveal underlying biological processes in Kawasaki disease. Genome Biol. 2007;8:R261. doi: 10.1186/gb-2007-8-12-r261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fury W, Tremoulet AH, Watson VE, Best BM, Shimizu C, et al. Transcript abundance patterns in Kawasaki disease patients with intravenous immunoglobulin resistance. Hum Immunol. 2010;71:865–73. doi: 10.1016/j.humimm.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92:3705–11. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 125.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a molecular signature in human type 1 diabetes mellitus using serum and functional genomics. J Immunol. 2008;180:1929–37. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 126.Moran A, Bundy B, Becker DJ, DiMeglio LA, Gitelman SE, et al. Interleukin-1 antagonism in type 1 diabetes of recent onset: Two multicentre, randomised, double-blind, placebo-controlled trials. Lancet. 2013;381:1905–15. doi: 10.1016/S0140-6736(13)60023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leonardi CL, Kimball AB, Papp KA, Yeilding N, Guzzo C, et al. Efficacy and safety of ustekinumab, a human interleukin-12/23 monoclonal antibody, in patients with psoriasis: 76-week results from a randomised, double-blind, placebo-controlled trial (PHOENIX 1) Lancet. 2008;371:1665–74. doi: 10.1016/S0140-6736(08)60725-4. [DOI] [PubMed] [Google Scholar]
- 128.Lee E, Trepicchio WL, Oestreicher JL, Pittman D, Wang F, et al. Increased expression of interleukin 23 p19 and p40 in lesional skin of patients with psoriasis vulgaris. J Exp Med. 2004;199:125–30. doi: 10.1084/jem.20030451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, et al. Interleukin-22, a TH17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–51. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 130.Russell CB, Rand H, Bigler J, Kerkof K, Timour M, et al. Gene expression profiles normalized in psoriatic skin by treatment with brodalumab, a human anti-IL-17 receptor monoclonal antibody. J Immunol. 2014;192:3828–36. doi: 10.4049/jimmunol.1301737. [DOI] [PubMed] [Google Scholar]
- 131.Zaba LC, Suarez-Farinas M, Fuentes-Duculan J, Nograles KE, Guttman-Yassky E, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol. 2009;124:1022–30. e395. doi: 10.1016/j.jaci.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jordan CT, Cao L, Roberson ED, Pierson KC, Yang CF, et al. PSORS2 is due to mutations in CARD14. Am J Hum Genet. 2012;90:784–95. doi: 10.1016/j.ajhg.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marrakchi S, Guigue P, Renshaw BR, Puel A, Pei XY, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N Engl J Med. 2011;365:620–28. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 134.Berki DM, Liu L, Choon SE, Burden AD, Griffiths CE, et al. Activating CARD14 mutations are associated with generalized pustular psoriasis but rarely account for familial recurrence in psoriasis vulgaris. J Investig Dermatol. 2015;135:2964–70. doi: 10.1038/jid.2015.288. [DOI] [PubMed] [Google Scholar]
- 135.Tozzoli R. Recent advances in diagnostic technologies and their impact in autoimmune diseases. Autoimmun Rev. 2007;6:334–40. doi: 10.1016/j.autrev.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 136.Fritzler MJ. Advances and applications of multiplexed diagnostic technologies in autoimmune diseases. Lupus. 2006;15:422–27. doi: 10.1191/0961203306lu2327oa. [DOI] [PubMed] [Google Scholar]
- 137.Costello CM, Mah N, Hasler R, Rosenstiel P, Waetzig GH, et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLOS Med. 2005;2:e199. doi: 10.1371/journal.pmed.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, et al. Molecular classification of Crohn’s disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8:51–61. doi: 10.2353/jmoldx.2006.050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ungethuem U, Haeupl T, Witt H, Koczan D, Krenn V, et al. Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis. Physiol Genom. 2010;42A:267–82. doi: 10.1152/physiolgenomics.00004.2010. [DOI] [PubMed] [Google Scholar]
- 140.Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes Immun. 2013;14:67–82. doi: 10.1038/gene.2012.55. [DOI] [PubMed] [Google Scholar]
- 141.Liu H, Liu J, Toups M, Soos T, Arendt C. Gene signature-based mapping of immunological systems and diseases. BMC Bioinform. 2016;17:171. doi: 10.1186/s12859-016-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Levy H, Wang X, Kaldunski M, Jia S, Kramer J, et al. Transcriptional signatures as a disease-specific and predictive inflammatory biomarker for type 1 diabetes. Genes Immun. 2012;13:593–604. doi: 10.1038/gene.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 144.Rodriguez-Calvo T, von Herrath MG. Enterovirus infection and type 1 diabetes: closing in on a link? Diabetes. 2015;64:1503–5. doi: 10.2337/db14-1931. [DOI] [PubMed] [Google Scholar]
- 145.Stene LC, Oikarinen S, Hyoty H, Barriga KJ, Norris JM, et al. Enterovirus infection and progression from islet autoimmunity to type 1 diabetes: the Diabetes and Autoimmunity Study in the Young (DAISY) Diabetes. 2010;59:3174–80. doi: 10.2337/db10-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hummel S, Pfluger M, Hummel M, Bonifacio E, Ziegler AG. Primary dietary intervention study to reduce the risk of islet autoimmunity in children at increased risk for type 1 diabetes: the BABYDIET study. Diabetes Care. 2011;34:1301–5. doi: 10.2337/dc10-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jin Y, Sharma A, Bai S, Davis C, Liu H, et al. Risk of type 1 diabetes progression in islet autoantibody-positive children can be further stratified using expression patterns of multiple genes implicated in peripheral blood lymphocyte activation and function. Diabetes. 2014;63:2506–15. doi: 10.2337/db13-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kallionpaa H, Elo LL, Laajala E, Mykkanen J, Ricano-Ponce I, et al. Innate immune activity is detected prior to seroconversion in children with HLA-conferred type 1 diabetes susceptibility. Diabetes. 2014;63:2402–14. doi: 10.2337/db13-1775. [DOI] [PubMed] [Google Scholar]
- 149.Meyer S, Woodward M, Hertel C, Vlaicu P, Haque Y, et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–95. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McKinney EF, Lyons PA, Carr EJ, Hollis JL, Jayne DR, et al. A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med. 2010;16:586–91. doi: 10.1038/nm.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee JC, Lyons PA, McKinney EF, Sowerby JM, Carr EJ, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Investig. 2011;121:4170–79. doi: 10.1172/JCI59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–16. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Caielli S, Athale S, Domic B, Murat E, Chandra M, et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Peterson KS, Huang JF, Zhu J, D’Agati V, Liu X, et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Investig. 2004;113:1722–33. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–89. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 156.Rubbert-Roth A, Finckh A. Treatment options in patients with rheumatoid arthritis failing initial TNF inhibitor therapy: a critical review. Arthritis Res Ther. 2009;11(Suppl 1):S1. doi: 10.1186/ar2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Roda G, Jharap B, Neeraj N, Colombel JF. Loss of response to anti-TNFs: definition, epidemiology, and management. Clin Transl Gastroenterol. 2016;7:e135. doi: 10.1038/ctg.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Merrill JT, Neuwelt CM, Wallace DJ, Shanahan JC, Latinis KM, et al. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: The randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 2010;62:222–33. doi: 10.1002/art.27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nirmala N, Brachat A, Feist E, Blank N, Specker C, et al. Gene-expression analysis of adult-onset Still’s disease and systemic juvenile idiopathic arthritis is consistent with a continuum of a single disease entity. Pediatr Rheumatol Online J. 2015;13:50. doi: 10.1186/s12969-015-0047-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, et al. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1βinhibition. N Engl J Med. 2006;355:581–92. doi: 10.1056/NEJMoa055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Simonini G, Xu Z, Caputo R, De Libero C, Pagnini I, et al. Clinical and transcriptional response to the long-acting interleukin-1 blocker canakinumab in Blau syndrome-related uveitis. Arthritis Rheum. 2013;65:513–18. doi: 10.1002/art.37776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yao Y, Richman L, Higgs BW, Morehouse CA, de los Reyes M, et al. Neutralization of interferon-α/β-inducible genes and downstream effect in a phase I trial of an anti-interferon-αmonoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785–96. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 163.Merrill JT, Wallace DJ, Petri M, Kirou KA, Yao Y, et al. Safety profile and clinical activity of sifalimumab, a fully human anti-interferon alpha monoclonal antibody, in systemic lupus erythematosus: a phase I, multicentre, double-blind randomised study. Ann Rheum Dis. 2011;70:1905–13. doi: 10.1136/ard.2010.144485. [DOI] [PubMed] [Google Scholar]
- 164.Petri M, Wallace DJ, Spindler A, Chindalore V, Kalunian K, et al. Sifalimumab, a human anti-interferon-alpha monoclonal antibody, in systemic lupus erythematosus: a phase I randomized, controlled, dose-escalation study. Arthritis Rheum. 2013;65:1011–21. doi: 10.1002/art.37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McBride JM, Jiang J, Abbas AR, Morimoto A, Li J, et al. Safety and pharmacodynamics of rontalizumab in patients with systemic lupus erythematosus: results of a phase I, placebo-controlled, double-blind, dose-escalation study. Arthritis Rheum. 2012;64:3666–76. doi: 10.1002/art.34632. [DOI] [PubMed] [Google Scholar]
- 166.Wang B, Higgs BW, Chang L, Vainshtein I, Liu Z, et al. Pharmacogenomics and translational simulations to bridge indications for an anti-interferon-alpha receptor antibody. Clin Pharmacol Ther. 2013;93:483–92. doi: 10.1038/clpt.2013.35. [DOI] [PubMed] [Google Scholar]
- 167.Chung L, Fiorentino DF, Benbarak MJ, Adler AS, Mariano MM, et al. Molecular framework for response to imatinib mesylate in systemic sclerosis. Arthritis Rheum. 2009;60:584–91. doi: 10.1002/art.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, Gudjonsson JE. Early tissue responses in psoriasis to the antitumour necrosis factor-alpha biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br J Dermatol. 2014;171:97–107. doi: 10.1111/bjd.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sofen H, Smith S, Matheson RT, Leonardi CL, Calderon C, et al. Guselkumab (an IL-23-specific mAb) demonstrates clinical and molecular response in patients with moderate-to-severe psoriasis. J Allergy Clin Immunol. 2014;133:1032–40. doi: 10.1016/j.jaci.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 170.Ogihara Y, Ogata S, Nomoto K, Ebato T, Sato K, et al. Transcriptional regulation by infliximab therapy in Kawasaki disease patients with immunoglobulin resistance. Pediatr Res. 2014;76:287–93. doi: 10.1038/pr.2014.92. [DOI] [PubMed] [Google Scholar]
- 171.Serrano-Fernandez P, Moller S, Goertsches R, Fiedler H, Koczan D, et al. Time course transcriptomics of IFNB1b drug therapy in multiple sclerosis. Autoimmunity. 2010;43:172–78. doi: 10.3109/08916930903219040. [DOI] [PubMed] [Google Scholar]
- 172.Sturzebecher S, Wandinger KP, Rosenwald A, Sathyamoorthy M, Tzou A, et al. Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain. 2003;126:1419–29. doi: 10.1093/brain/awg147. [DOI] [PubMed] [Google Scholar]
- 173.Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–23. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 174.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. PNAS. 2005;102:3372–77. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Burska AN, Roget K, Blits M, Soto Gomez L, van de Loo F, et al. Gene expression analysis in RA: towards personalized medicine. Pharmacogenomics J. 2014;14:93–106. doi: 10.1038/tpj.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lequerre T, Gauthier-Jauneau AC, Bansard C, Derambure C, Hiron M, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Oliveira RD, Fontana V, Junta CM, Marques MM, Macedo C, et al. Differential gene expression profiles may differentiate responder and nonresponder patients with rheumatoid arthritis for methotrexate (MTX) monotherapy and MTX plus tumor necrosis factor inhibitor combined therapy. J Rheumatol. 2012;39:1524–32. doi: 10.3899/jrheum.120092. [DOI] [PubMed] [Google Scholar]
- 178.Dennis G, Jr, Holweg CT, Kummerfeld SK, Choy DF, Setiadi AF, et al. Synovial phenotypes in rheumatoid arthritis correlate with response to biologic therapeutics. Arthritis Res Ther. 2014;16:R90. doi: 10.1186/ar4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wright HL, Thomas HB, Moots RJ, Edwards SW. Interferon gene expression signature in rheumatoid arthritis neutrophils correlates with a good response to TNFi therapy. Rheumatology. 2015;54:188–93. doi: 10.1093/rheumatology/keu299. [DOI] [PubMed] [Google Scholar]
- 180.Sanayama Y, Ikeda K, Saito Y, Kagami S, Yamagata M, et al. Prediction of therapeutic responses to tocilizumab in patients with rheumatoid arthritis: biomarkers identified by analysis of gene expression in peripheral blood mononuclear cells using genome-wide DNA microarray. Arthritis Rheumatol. 2014;66:1421–31. doi: 10.1002/art.38400. [DOI] [PubMed] [Google Scholar]
- 181.van Baarsen LG, Wijbrandts CA, Rustenburg F, Cantaert T, van der Pouw Kraan TC, et al. Regulation of IFN response gene activity during infliximab treatment in rheumatoid arthritis is associated with clinical response to treatment. Arthritis Res Ther. 2010;12:R11. doi: 10.1186/ar2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Koczan D, Drynda S, Hecker M, Drynda A, Guthke R, et al. Molecular discrimination of responders and nonresponders to anti-TNF alpha therapy in rheumatoid arthritis by etanercept. Arthritis Res Ther. 2008;10:R50. doi: 10.1186/ar2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Thurlings RM, Boumans M, Tekstra J, van Roon JA, Vos K, et al. Relationship between the type I interferon signature and the response to rituximab in rheumatoid arthritis patients. Arthritis Rheum. 2010;62:3607–14. doi: 10.1002/art.27702. [DOI] [PubMed] [Google Scholar]
- 184.Raterman HG, Vosslamber S, deRidder S, Nurmohamed MT, Lems WF, et al. The interferon type I signature towards prediction of non-response to rituximab in rheumatoid arthritis patients. Arthritis Res Ther. 2012;14:R95. doi: 10.1186/ar3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sellam J, Marion-Thore S, Dumont F, Jacques S, Garchon HJ, et al. Use of whole-blood transcriptomic profiling to highlight several pathophysiologic pathways associated with response to rituximab in patients with rheumatoid arthritis: data from a randomized, controlled, open-label trial. Arthritis Rheumatol. 2014;66:2015–25. doi: 10.1002/art.38671. [DOI] [PubMed] [Google Scholar]
- 186.Owczarczyk K, Lal P, Abbas AR, Wolslegel K, Holweg CT, et al. A plasmablast biomarker for nonresponse to antibody therapy to CD20 in rheumatoid arthritis. Sci Transl Med. 2011;3:101ra92. doi: 10.1126/scitranslmed.3002432. [DOI] [PubMed] [Google Scholar]
- 187.Kiefer K, Oropallo MA, Cancro MP, Marshak-Rothstein A. Role of type I interferons in the activation of autoreactive B cells. Immunol Cell Biol. 2012;90:498–504. doi: 10.1038/icb.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jourde-Chiche N, Chiche L, Chaussabel D. Introducing a new dimension to molecular disease classifications. Trends Mol Med. 2016;22:451–53. doi: 10.1016/j.molmed.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 189.Rosenblum MD, Remedios KA, Abbas AK. Mechanisms of human autoimmunity. J Clin Investig. 2015;125:2228–33. doi: 10.1172/JCI78088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Calis JJ, Rosenberg BR. Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol. 2014;35:581–90. doi: 10.1016/j.it.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Robinson WH. Sequencing the functional antibody repertoire–diagnostic and therapeutic discovery. Nat Rev Rheumatol. 2015;11:171–82. doi: 10.1038/nrrheum.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tipton CM, Fucile CF, Darce J, Chida A, Ichikawa T, et al. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat Immunol. 2015;16:755–65. doi: 10.1038/ni.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Stern JN, Yaari G, Vander Heiden JA, Church G, Donahue WF, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med. 2014;6:248ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Planas R, Jelcic I, Schippling S, Martin R, Sospedra M. Natalizumab treatment perturbs memory- and marginal zone-like B-cell homing in secondary lymphoid organs in multiple sclerosis. Eur J Immunol. 2012;42:790–98. doi: 10.1002/eji.201142108. [DOI] [PubMed] [Google Scholar]
- 195.Tan YC, Kongpachith S, Blum LK, Ju CH, Lahey LJ, et al. Barcode-enabled sequencing of plasmablast antibody repertoires in rheumatoid arthritis. Arthritis Rheumatol. 2014;66:2706–15. doi: 10.1002/art.38754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Doorenspleet ME, Klarenbeek PL, de Hair MJ, van Schaik BD, Esveldt RE, et al. Rheumatoid arthritis synovial tissue harbours dominant B-cell and plasma-cell clones associated with autoreactivity. Ann Rheum Dis. 2014;73:756–62. doi: 10.1136/annrheumdis-2012-202861. [DOI] [PubMed] [Google Scholar]
- 197.Ishigaki K, Shoda H, Kochi Y, Yasui T, Kadono Y, et al. Quantitative and qualitative characterization of expanded CD4+ T cell clones in rheumatoid arthritis patients. Sci Rep. 2015;5:12937. doi: 10.1038/srep12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 199.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 200.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Okada Y, Wu D, Trynka G, Raj T, Terao C, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Walsh AM, Whitaker JW, Huang CC, Cherkas Y, Lamberth SL, et al. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol. 2016;17:79. doi: 10.1186/s13059-016-0948-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nat Genet. 2013;45:1284–92. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tewhey R, Kotliar D, Park DS, Liu B, Winnicki S, et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell. 2016;165:1519–29. doi: 10.1016/j.cell.2016.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Peters JE, Lyons PA, Lee JC, Richard AC, Fortune MD, et al. Insight into genotype-phenotype associations through eQTL mapping in multiple cell types in health and immune-mediated disease. PLOS Genet. 2016;12:e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–10. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lynch KW. Consequences of regulated pre-mRNA splicing in the immune system. Nat Rev Immunol. 2004;4:931–40. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- 211.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 212.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–69. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Martinez NM, Lynch KW. Control of alternative splicing in immune responses: Many regulators, many predictions, much still to learn. Immunol Rev. 2013;253:216–36. doi: 10.1111/imr.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ueda H, Howson JM, Esposito L, Heward J, Snook H, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 216.Gu M, Kakoulidou M, Giscombe R, Pirskanen R, Lefvert AK, et al. Identification of CTLA-4 isoforms produced by alternative splicing and their association with myasthenia gravis. Clin Immunol. 2008;128:374–81. doi: 10.1016/j.clim.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 217.Sestak AL, Nath SK, Sawalha AH, Harley JB. Current status of lupus genetics. Arthritis Res Ther. 2007;9:210. doi: 10.1186/ar2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–16. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 219.Tsuzaka K, Fukuhara I, Setoyama Y, Yoshimoto K, Suzuki K, et al. TCRζ mRNA with an alternatively spliced 3′-untranslated region detected in systemic lupus erythematosus patients leads to the down-regulation of TCRζ and TCR/CD3 complex. J Immunol. 2003;171:2496–503. doi: 10.4049/jimmunol.171.5.2496. [DOI] [PubMed] [Google Scholar]
- 220.Tsuzaka K, Setoyama Y, Yoshimoto K, Shiraishi K, Suzuki K, et al. A splice variant of the TCRζ mRNA lacking exon 7 leads to the down-regulation of TCRζ, the TCR/CD3 complex, and IL-2 production in systemic lupus erythematosus T cells. J Immunol. 2005;174:3518–25. doi: 10.4049/jimmunol.174.6.3518. [DOI] [PubMed] [Google Scholar]
- 221.Moulton VR, Grammatikos AP, Fitzgerald LM, Tsokos GC. Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription. PNAS. 2013;110:1845–50. doi: 10.1073/pnas.1214207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Feng D, Sangster-Guity N, Stone R, Korczeniewska J, Mancl ME, et al. Differential requirement of histone acetylase and deacetylase activities for IRF5-mediated proinflammatory cytokine expression. J Immunol. 2010;185:6003–12. doi: 10.4049/jimmunol.1000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mancl ME, Hu G, Sangster-Guity N, Olshalsky SL, Hoops K, et al. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J Biol Chem. 2005;280:21078–90. doi: 10.1074/jbc.M500543200. [DOI] [PubMed] [Google Scholar]
- 224.Stone RC, Feng D, Deng J, Singh S, Yang L, et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis Rheum. 2012;64:788–98. doi: 10.1002/art.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Stone RC, Du P, Feng D, Dhawan K, Ronnblom L, et al. RNA-Seq for enrichment and analysis of IRF5 transcript expression in SLE. PLOS ONE. 2013;8:e54487. doi: 10.1371/journal.pone.0054487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Richez C, Yasuda K, Bonegio RG, Watkins AA, Aprahamian T, et al. IFN regulatory factor 5 is required for disease development in the FcγRIIB−/−Yaa and FcγRIIB−/− mouse models of systemic lupus erythematosus. J Immunol. 2010;184:796–806. doi: 10.4049/jimmunol.0901748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Alevizos I, Illei GG. microRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6:391–98. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ravenscroft JC, Suri M, Rice GI, Szynkiewicz M, Crow YJ. Autosomal dominant inheritance of a heterozygous mutation in SAMHD1 causing familial chilblain lupus. Am J Med Genet A. 2011;155A:235–37. doi: 10.1002/ajmg.a.33778. [DOI] [PubMed] [Google Scholar]
- 229.Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–14. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Barnett CP, Todd EJ, Ong R, Davis MR, Atkinson V, et al. Distal arthrogryposis type5Dwith novel clinical features and compound heterozygous mutations in ECEL1. Am J Med Genet A. 2014;164A:1846–49. doi: 10.1002/ajmg.a.36342. [DOI] [PubMed] [Google Scholar]
- 232.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–73. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Investig. 2014;124:3617–33. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–53. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8:75. doi: 10.1186/s13073-016-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Int. Cancer Genome Consort. Hudson TJ, Anderson W, Artez A, Barker AD, et al. International network of cancer genome projects. Nature. 2010;464:993–98. doi: 10.1038/nature08987. Corrigendum. 2010. Nature 465:966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Netea MG, Joosten LA, Li Y, Kumar V, Oosting M, et al. Understanding human immune function using the resources from the Human Functional Genomics Project. Nat Med. 2016;22:831–33. doi: 10.1038/nm.4140. [DOI] [PubMed] [Google Scholar]
- 238.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–95. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Crow YJ, Chase DS, Lowenstein Schmidt J, Szynkiewicz M, Forte GM, et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A. 2015;167A:296–312. doi: 10.1002/ajmg.a.36887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, et al. Human intracellular ISG15 prevents interferon-α/βover-amplification and auto-inflammation. Nature. 2015;517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Meuwissen ME, Schot R, Buta S, Oudesluijs G, Tinschert S, et al. Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TORCH syndrome. J Exp Med. 2016;213:1163–74. doi: 10.1084/jem.20151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liu Y, Ramot Y, Torrelo A, Paller AS, Si N, et al. Mutations in proteasome subunit beta type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 2012;64:895–907. doi: 10.1002/art.33368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou Q, Yang D, Ombrello AK, Zavialov AV, Toro C, et al. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med. 2014;370:911–20. doi: 10.1056/NEJMoa1307361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–18. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Jang MA, Kim EK, Now H, Nguyen NT, Kim WJ, et al. Mutations in DDX58, which encodes RIG-I, cause atypical Singleton-Merten syndrome. Am J Hum Genet. 2015;96:266–74. doi: 10.1016/j.ajhg.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Briggs TA, Rice GI, Daly S, Urquhart J, Gornall H, et al. Tartrate-resistant acid phosphatase deficiency causes a bone dysplasia with autoimmunity and a type I interferon expression signature. Nat Genet. 2011;43:127–31. doi: 10.1038/ng.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Park J, Munagala I, Xu H, Blankenship D, Maffucci P, et al. Interferon signature in the blood in inflammatory common variable immune deficiency. PLOS ONE. 2013;8:e74893. doi: 10.1371/journal.pone.0074893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Smith EJ, Allantaz F, Bennett L, Zhang D, Gao X, et al. Clinical, molecular, and genetic characteristics of PAPA syndrome: a review. Curr Genom. 2010;11:519–27. doi: 10.2174/138920210793175921. [DOI] [PMC free article] [PubMed] [Google Scholar]