Abstract
The brain harbors billions of neurons that form distinct neural circuits with exquisite specificity. Specific patterns of connectivity between distinct neuronal cell types permit the transfer and computation of information. The molecular correlates that give rise to synaptic specificity are incompletely understood. Recent studies indicate that cell-surface molecules are important determinants of cell type identity and suggest that these are essential players in the specification of synaptic connectivity. Leucine-rich repeat (LRR)-containing adhesion molecules in particular have emerged as key organizers of excitatory and inhibitory synapses. Here, we discuss emerging evidence that LRR proteins regulate the assembly of specific connectivity patterns across neural circuits, and contribute to the diverse structural and functional properties of synapses, two key features that are critical for the proper formation and function of neural circuits.
Brain circuits: Regulating synaptic connections
Further analysis of synaptic proteins will provide insights into the functioning of neural circuits and associated brain disorders. The brain houses numerous highly specialized neuron types, which transfer and process information via a complex network of synaptic connections. Every neuron develops its own distinctive synapses with specific functions, but exactly how this is achieved is not clear. Joris de Wit and Anna Schroeder at the VIB Center for Brain and Disease Research in Leuven, Belgium, reviewed recent research into the leucine-rich repeat-containing (LRR) proteins, which are thought to be major organizers of synaptic connectivity and key regulators of healthy neural circuit development. Further investigations into the functionality of LRR proteins in the brain will not only improve understanding of neural circuitry but also provide insights into synaptic impairments in brain disorders like schizophrenia.
Molecular correlates of synaptic specificity and diversity
A rich diversity of neuronal cell types is found in the brain, with each neuron participating in its own specific network of connections. Synaptic connections allow for information to not only be conveyed from one cell type to another, but also to be differentially processed. This ability is made possible because synapses between different types of neurons have distinct structural and functional properties, as well as synaptic plasticity dynamics1–4. Synapse type-specific properties include, but are not limited to, neurotransmitter type, dendritic spine shape, short- and long-term plasticity (LTP) properties, pre-synaptic release probability, and post-synaptic receptor composition.
The molecular logic that underlies the specificity and diversity of synaptic connectivity is poorly understood. Recent single-cell sequencing studies reveal that only a few key gene classes determine cell identity, including transcription factors and cell-surface proteins5–10. These studies have also found that many proteins that confer particular synaptic properties, such as ion channels, are differentially expressed in discrete neuronal cell types. Cell type-specific repertoires of cell-surface and synaptic proteins provide the building blocks for the synapses a neuron makes and receives, but how they instruct specific connectivity and diversity of synapses is largely unknown. What has become clear is that these building blocks do not distribute evenly across the neuron and all of its synapses. The differential expression of synaptic proteins, including vesicular transporters, receptor subunits, and scaffolding proteins, at particular synaptic connections, is well-documented11. Thus, cell type-specific expression of cell-surface and synaptic proteins might contribute to defining the specificity, and the particular properties, of the synaptic connections that neuron makes. However, the mechanisms by which synaptic proteins are differentially localized to specific connections, or how they orchestrate the diverse properties of synapses, are largely unknown.
Synaptic adhesion proteins are a class of cell-surface proteins with an essential role in shaping neural connectivity. These molecules organize synaptic development by forming select trans-synaptic complexes and recruiting diverse synaptic proteins. One family of synaptic adhesion molecules, the leucine-rich repeat (LRR)-containing proteins, has emerged as key organizers of both excitatory and inhibitory synapses12–16. These proteins are so named because of the LRR structural motifs they each contain, which array in tandem and curve to form a horseshoe-shaped structure, the LRR domain17–19. The concave side of the curved motif stack, which constitutes a continuous β-sheet, makes the LRR domain an excellent protein-binding motif20. Variation in the length and number of LRR repeats, as well as in secondary structures on the convex side, affects the domain curvature, permitting its interaction with a diversity of ligands. Through interactions with various synaptic proteins mediated by their extracellular LRR domains and cytoplasmic tails, LRR-containing proteins exert their roles as synaptic organizers. In addition to these molecular properties, LRR proteins display cell type-specific expression patterns. Recent single-cell sequencing studies of retinal bipolar neurons and cortical GABAergic interneurons revealed differential, cell type-specific expression patterns of LRR proteins8,10, suggesting that different neuronal cell types express their own repertoires of LRR proteins. Together, these various attributes make LRR-containing synaptic adhesion molecules candidates for regulating synaptic specificity and diversity in neural circuits.
In this review, we discuss the role of different LRR-containing adhesion molecules, which for simplification purposes we will refer to here as LRR proteins, in synaptic connectivity and diversity. We focus on the mammalian nervous system and refer the reader to several excellent reviews that address the issue of synaptic specificity in invertebrates17,21–25. We first provide an overview of the major synaptogenic LRRs that have been identified in recent years. We consider the evidence that indicates a role for LRR proteins in the development of specific synaptic connectivity in a variety of neural circuits, focusing on those in cortex, hippocampus, and retina. We then discuss emerging evidence that suggests that LRR proteins might contribute to the specification of synaptic diversity in mammalian neural circuits. Finally, we briefly consider the role of LRR proteins in brain disorders, as several genes encoding LRR-containing synaptic adhesion molecules have been shown to have associations with neuropsychiatric disease, including autism and schizophrenia4,12,26.
Synaptogenic LRR proteins and their synaptic ligands
The main focus of this review will be on five major LRR protein subfamilies with synapse formation-promoting, or synaptogenic, capacity in cultured neurons. Here, we first provide a brief overview of these subfamilies and describe what is currently known regarding the extracellular synaptic binding partners of these adhesion molecules.
NGLs
The netrin-G ligand (NGL) protein family comprises three members, NGL1–3, also known as the LRRC4 genes27–29. NGLs localize to the post-synaptic membrane and form trans-synaptic complexes with their pre-synaptic binding partners. NGL-1 binds lipid-anchored netrin-G1, and NGL-2 binds netrin-G2. NGL-3, on the other hand, does not bind netrin-Gs, but binds all leukocyte common antigen-related (LAR) family receptor protein tyrosine phosphatases (RPTPs), including LAR, PTPδ, and PTPσ29. When expressed on the surface of heterologous cells, all NGLs induce pre-synaptic differentiation in contacting axons from co-cultured neurons28,29. The fact that netrin-Gs lack a cytoplasmic domain suggests the existence of an interacting co-receptor that would allow NGL-1 and NGL-2 to induce pre-synaptic differentiation. Indeed, NGL-1 binding to netrin-G1 induces recruitment of pre-synaptic LAR to the complex30. Netrin-G2 must operate via another, as-of-yet unidentified, transmembrane co-receptor.
LRRTMs
The LRR transmembrane neuronal protein (LRRTM) family31,32 comprises four members, LRRTM1–4. LRRTMs are post-synaptic proteins that form trans-synaptic complexes with pre-synaptic neurexins33,34. LRRTM1, LRRTM2, and LRRTM3 interact with neurexins in a splice site-dependent manner, as they bind neurexins specifically lacking an insert at splice site 434–36. LRRTM4 differs in that it binds neurexins with or without a splice site 4 insert, and preferentially binds heparan sulfate proteoglycans, most prominently glypicans, in trans35,37–39. LRRTM1, LRRTM2, LRRTM4, and to a lesser degree, LRRTM3, are all capable of inducing pre-synaptic differentiation in contacting axons31,33,34. Since glycosylphosphatidylinositol (GPI)-anchored glypicans lack a cytoplasmic domain, the LRRTM4-triggered recruitment of the pre-synaptic co-receptor PTPσ is required to induce pre-synaptic differentiation in culture37.
Slitrks
Slit and NTRK-like family members (Slitrk) proteins comprise a larger LRR protein subfamily, with six members, Slitrk1–640. Post-synaptic Slitrks engage in trans-synaptic interactions with PTPδ and PTPσ from the LAR-RPTP family, but not with LAR itself41,42. All Slitrk family members are capable of inducing pre-synaptic differentiation41,43. While Slitrk1, −2, −4, −5, and −6 have primarily been implicated in the regulation of excitatory synapse development in vitro, Slitrk3 is an inhibitory synapse-specific LRR protein41,43.
SALMs
The family of synaptic adhesion-like molecules (SALMs), also known as LRR and fibronectin type III domain-containing proteins (LRFNs), comprises five members, SALM1-544–46. The molecular interactions that SALM proteins form both in cis and in trans are complex and are yet to be fully understood. SALM1-3 form homo- and heteromeric complexes in cis47, and SALM4 inhibits SALM3 function in cis48. SALM4 and −5 participate in homophilic adhesion in trans48, and both SALM3 and SALM5 bind to LAR-RPTPs49,50. Only SALM3 and SALM5 have synaptogenic activity in co-cultures of heterologous cells and neurons51.
FLRTs
The fibronectin LRR transmembrane (FLRT) protein family comprises three members, FLRT1–352. FLRTs bind latrophilins (LPHN) 1–3 in trans, with promiscuity between isoforms53. However, FLRTs also engage in other important molecular interactions, as will be discussed later in this review. Unlike LRRTMs, Slitrks, NGLs, and SALMs, FLRTs expressed in heterologous cells do not induce pre-synaptic differentiation in contacting axons. However, FLRT3 knockdown (KD) in cultured hippocampal neurons decreases excitatory synapse density53.
Control of synaptic protein composition by LRR proteins
In addition to their interactions with binding partners across the synaptic cleft, LRR proteins also recruit specific components of the synaptic proteome, largely via their cytoplasmic tails. A common theme has emerged of LRR proteins interacting with, or influencing the localization of, post-synaptic scaffolding proteins, as well as glutamate receptor subunits.
Artificial LRR protein clustering assays allow for the analysis of LRR protein-mediated post-synaptic protein recruitment. Aggregation of either NGL-2 or NGL-3 on the surface of dendrites induces NMDA glutamate receptor (NMDAR) clustering, while NGL-3 also induces AMPA glutamate receptor (AMPAR) clustering28,29. Clustering of LRRTM1 or LRRTM2 on the dendrites of hippocampal neurons induces co-clustering of NMDAR subunit GluN1 and AMPAR subunit GluA1, as well as PSD-95 and SynGAP, a PSD-95-associated post-synaptic density (PSD) protein31. SALM2 clustering on cultured neuron dendrites induces co-clustering of PSD-95 as well as the PSD protein GKAP46. SALM3 aggregation on dendrites also induces clustering of PSD-9551. LRR proteins are, therefore, not only able to mediate pre-synaptic differentiation, but also post-synaptic differentiation.
Studies that have investigated the consequences of LRR protein knockout (KO) also provide insight into how these molecules influence synaptic protein composition. SALM1 KO increases expression of NMDAR subunit GluN2A, but decreases expression of PSD-95 in all hippocampal regions54. GluA1 expression was also found to be reduced in SALM1 KO hippocampus, suggesting a role of SALM1 in enhancing synaptic incorporation of AMPARs in cooperation with PSD-95. On striatal neurons, Slitrk5 localizes to dendritic spines, where it regulates the expression of glutamate receptors55. In Slitrk5 KO mice, expression of NMDAR subunits GluN2A and GluN2B, as well as AMPAR subunits GluA1 and GluA2, are decreased in striatum, while PSD-95 amounts are unchanged. LRRTM4 KO results in a reduction in the expression of PSD-95 family proteins in dentate gyrus (DG) synaptosome fractions, while basal levels of GluA1 and GluA2 are unaffected39.
In conclusion, LRR proteins organize synapses via interactions with binding partners across the synaptic cleft, as well as by interacting with essential synaptic proteins. In the next section, we discuss the roles of LRR proteins in shaping synaptic connectivity in neural circuits.
Role of LRR proteins in circuit development
LRR proteins have been implicated in all steps of circuit formation, from neuronal migration and neurite outgrowth, to the formation and functional assembly of synaptic contacts. Here, we focus on three main brain regions—cortex, hippocampus, and retina—to highlight the diverse roles of LRR proteins in the control of neural circuit development.
Cortical development: cell migration and neurite outgrowth
FLRT proteins play an important role in cortical neuron migration. FLRT2 and FLRT3 ectodomains are shed from neurons during cortical development56. By binding to Unc5-receptors on cortical neuron axons or somata, FLRTs act as repulsive guidance cues. KO of FLRT2, or of its receptor, Unc5D, results in an accelerated radial migration of cortical neurons56. FLRT1/3 double KO also affects cortical neuron migration, but with a more dramatic effect, as this results in the unusual formation of macroscopic sulci during embryogenesis in what would normally be a smooth mouse neocortex57. Reduced intercellular adhesion following loss of FLRT1 and FLRT3 allows for a larger proportion of immature neurons to reach the upper cortical plate, as it permits accelerated radial migration of neurons as well as enhanced neuron clustering along the tangential axis. Interestingly, endogenous FLRT1/3 expression levels are lower in human neocortex compared to mouse neocortex57, suggesting that FLRT-regulated intercellular adhesion might be involved in species-specific differences in cortical folding.
LRR proteins regulate neurite outgrowth in the thalamocortical system. NGL-1 is highly expressed in embryonic striatum and cortex, which both receive innervation from thalamocortical axons (TCAs)27. Binding of NGL-1 to netrin-G1 on developing TCAs stimulates neurite growth toward their target regions. Thalamic axon growth is blocked upon application of soluble NGL-1 or upon treatment with phosphatidylinositol phospholipase C, which cleaves GPI-anchored surface proteins, including netrin-Gs. FLRTs also contribute to the correct formation of thalamocortical connectivity. FLRT3, which is highly expressed in rostral TCAs, determines the Netrin-1 responsiveness of the Robo1 receptor, also expressed in TCAs58. In the presence of Slit1, Robo1 and FLRT3 receptors induce Netrin-1 attraction by upregulating surface DCC receptors. Intermediate TCAs that normally lack FLRT3 are insensitive to Netrin-1. This Netrin-1 sensitivity adjustment, governed by FLRT3 expression, allows rostral and intermediate TCAs to target different regions of cortex. Consequently, loss of FLRT3 from TCAs that normally express it results in axon guidance defects58.
Hippocampal circuits: development of synaptic connectivity
The hippocampus is an attractive system for the investigation of the role of LRR proteins in regulating synaptic connectivity because of the accessible laminar organization of synaptic afferents in this brain region. Multiple studies have focused on the Schaffer collateral (SC) synapses that form in CA1 stratum radiatum (SR), where CA3 axons terminate on the proximal apical dendrites of CA1 pyramidal neurons. Several LRR proteins regulate the development of SC-CA1 synapses. NGL-2 localizes to the dendritic segments of CA1 pyramidal neurons that receive SC input, while the pre-synaptic binding partner of NGL-2, netrin-G2, localizes to SC axons59. NGL-2 KD or KO selectively reduces the density of spines in the CA1 dendritic segment receiving SC input, while the density of spines on the distal tuft of CA1 pyramidal neurons is unaffected60 (Fig. 1), demonstrating that the NGL-2—netrin-G2 complex regulates synapse development in an input-specific manner. Unlike DeNardo et al.60, Matsukawa et al.61 found no differences in CA1 pyramidal neuron spine densities in either netrin-G2 KO or NGL-2 KO mice, neither in juveniles nor adults, possibly due to differences in genetic backgrounds of the mice used in these studies.
Fig. 1. LRR proteins regulate synapse development in CA1 pyramidal neurons.
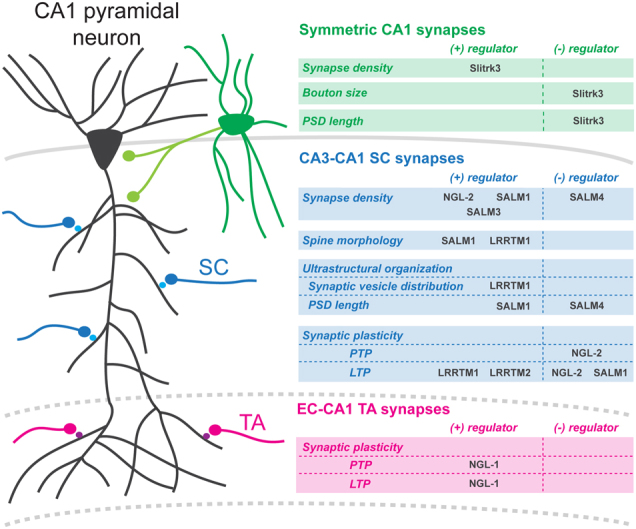
Symmetric CA1 synapses. Slitrk3 regulates density, pre-synaptic bouton size, and ultrastructural organization of inhibitory synapses formed on CA1 pyramidal cell bodies and in CA1 stratum radiatum (SR), presumably by local GABAergic interneurons (green). CA3-CA1 SC synapses. Different LRR proteins regulate synapse density on the proximal apical dendritic segment, where Schaffer collateral (SC) axons synapse onto CA1 neurons (black). NGL-2, SALM1, and SALM3 positively regulate dendritic spine density, whereas SALM4 negatively regulates SC spine density. LRRTM1, SALM1, and SALM4 influence synapse structure or ultrastructure. LRRTM1 loss results in longer, possibly less mature, spines, as well as a dispersion of vesicles in the pre-synaptic bouton. SALM1 loss results in odd-looking spines with thin protrusions emerging from the spine heads, as well as a decrease in post-synaptic density (PSD) length and an increase in the proportion of perforated synapses. SALM4 loss results in an increase in PSD length. LRRTM1/2, NGL-2, and SALM1 affect short- or long-term synaptic plasticity. Loss of LRRTM1/2 results in a long-term potentiation (LTP) block. Loss of SALM1 results in enhanced LTP. Loss of netrin-G2, the pre-synaptic binding partner of NGL-2, also results in enhanced LTP as well as facilitation of post-tetanic potentiation (PTP). EC-CA1 TA synapses. At temporoammonic (TA) synapses, formed on distal CA1 dendrites by incoming afferents from layer III (LIII) entorhinal cortex (EC) neurons, NGL-1 KO results in attenuation of PTP. Loss of the pre-synaptic binding partner of NGL-1, netrin-G1, at TA synapses also results in attenuated LTP
LRRTM1 KO results in a small decrease in SC-CA1 synapse density in electron microscopic (EM) analysis, although the same study found no change in spine density using Golgi staining of CA1 dendrites62. Similarly, triple KD of LRRTM1, LRRTM2, and neuroligin-3 (NLGN3) in a NLGN1 KO background had no significant effect on dendritic spine density in CA1 SR compared to NLGN1 KO mice63, suggesting that the role of LRRTMs in regulating SC-CA1 synapse density is relatively minor.
Multiple members of the SALM family regulate synapse density in CA1 neurons. SALM1 KO mildly reduces dendritic spine number in CA1 SR, based on Golgi staining of CA1 dendrites (Fig. 1)54. SALM5 KD in CA1 neurons in organotypic hippocampal slice cultures strongly decreases the frequency, but not the amplitude, of miniature excitatory post-synaptic currents (mEPSCs), suggesting a reduction in excitatory synapse density following loss of SALM549. SALM3 KO also decreases mEPSC frequency, and in addition reduces PSD density as measured by EM analysis50, indicating that loss of SALM3 reduces excitatory synapse density. SALM4 KO, on the other hand, increases mEPSC frequency and PSD density in CA1 neurons, indicating increased excitatory synapse number (Fig. 1)48. Interestingly, this increase occurs because SALM4 normally cis-inhibits SALM3, which has synaptogenic activity in culture51, resulting in a suppression of excitatory synapse development48. Consistent with these observations from cell culture, double KO of SALM3 and SALM4 normalizes excitatory synapse number in CA1 pyramidal neurons48.
LRR proteins also contribute to the development of inhibitory synaptic connectivity in CA1 pyramidal neurons. Slitrk3 KO mice display a reduced density of inhibitory synapses, in CA1 SR, as well as in the CA1 cell body layer (Fig. 1)43. Slitrk3 physically interacts with neuroligin-2 (NLGN2), a major organizer of inhibitory synapses64. NLGN2 KO selectively impairs perisomatic inhibitory synapses in CA1 neurons65. Targeted disruption of the Slitrk3–NLGN-2 interaction, by engineering a mutant mouse that lacks the Slitrk3 LRR critical for the interaction with NLGN2, reduces inhibitory synapse density in CA1 SR and in stratum pyramidale64, demonstrating that Slitrk3 and NLGN2 interact to synergistically promote inhibitory synapse development in CA1 neurons.
Another hippocampal circuit that has been utilized for the study of LRR proteins in regulating synaptic connectivity is found in the DG, where perforant path (PP) afferents from entorhinal cortex synapse on the middle and distal portion of DG granule cell dendrites. FLRT3 KD in DG granule cells mildly reduces spine density in the middle dendritic segment receiving PP input (Fig. 2)53. LRRTM3 KO also results in a modest decrease in dendritic spine density in the same dendritic segment of DG granule cells35. LRRTM4 KO mice show a similar reduction in spine density on the outer segment of DG granule cell dendrites39 (Fig. 2), indicating a contribution of LRRTM3 and LRRTM4 in regulating the density of PP-DG granule cell synapses.
Fig. 2. LRR proteins regulate synapse development in dentate gyrus granule cells.
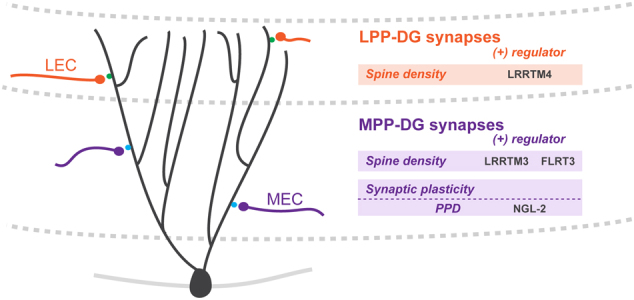
LPP-DG synapses. Lateral perforant path (LPP) axons from layer II (LII) lateral EC (LEC) synapse on distal dentate gyrus (DG) granule cell dendrites in the outer molecular layer (OML). Loss of LRRTM4 results in a reduction in spine density at LPP-DG synapses. MPP-DG synapses. Medial perforant path (MPP) synapses form between incoming LII medial EC (MEC) afferents and DG granule cell dendrites in the middle molecular layer (MML). At MPP-DG synapses, loss of FLRT3 or LRRTM3 results in a reduction in spine density. Loss of NGL-2, or its pre-synaptic binding partner, netrin-G2, results in an increase in the paired-pulse ratio (PPR) that corresponds to an attenuated paired-pulse depression (PPD)
Retinal circuits: photoreceptor synapse formation and laminar targeting
LRR proteins are important regulators of pathway-specific laminar organization in retinal circuits. In retinal horizontal cells (HCs), NGL-2 localizes to the tips of axons66, rather than to dendrites as in hippocampal neurons (Fig. 3a). HC axons synapse onto rod photoreceptors (PRs) in the outer plexiform layer (OPL), which express netrin-G2. HC axons from NGL-2 KO mice overshoot their laminar target, the OPL, and form fewer synapses with rod PRs. Consequently, rod-mediated signaling in the outer retina, as recorded by in vivo electroretinograms, is impaired. Connections between HC dendrites and cone PRs are unaffected in NGL-2 KO mice, and laminar targeting of rod PR-contacting dendrites of another cell type, bipolar cells (BCs), to rod PRs is also intact in the absence of NGL-266, showing a rod PR pathway-specific role for NGL-2 in HCs in regulating connectivity of the first retinal synapse.
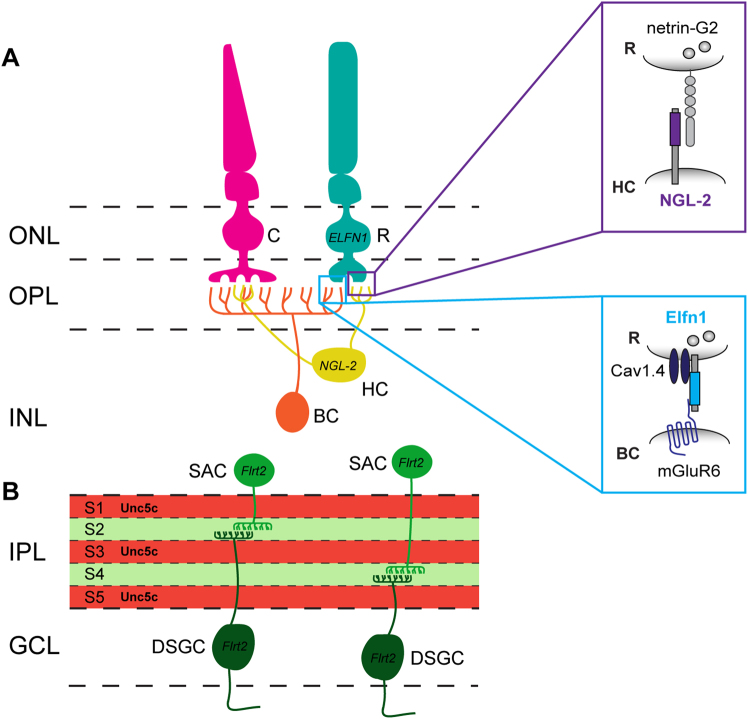
Fig. 3. LRR proteins mediate functional wiring of retinal synapses.
a Synaptic wiring in the outer plexiform layer. Photoreceptors (PR) form triad synapses with horizontal cells (HC) and bipolar cells (BC) in the outer plexiform layer (OPL). NGL-2 localizes to HC axon tips, which synapse onto netrin-G2-expressing rod PRs. NGL-2 KO causes HC axons to overshoot their laminar target, the OPL, and form fewer synapses with rod PRs, while HC dendrite—cone PR synapses that do not contain NGL-2 are unaffected. Elfn1 is expressed by rod PRs, which synapse onto ON-BCs in the OPL. Neighboring cone PR synapses formed on ON-BCs do not express Elfn1. Synaptic targeting of Elfn1 is mediated by α2δ4, an auxiliary subunit of the Cav1.4 channel. Once at the synapse, Elfn1 bridges a trans-synaptic complex between pre-synaptic, glutamate release-directing Cav1.4, and post-synaptic, glutamate-sensing mGluR6. Elfn1 KO in rod PRs disrupts the formation of rod PR synapses on ON-BCs, while neighboring cone PR synapses on ON-BCs are unaffected. b Laminar targeting in the inner plexiform layer. FLRT2 may mediate sublaminar targeting in the inner plexiform layer (IPL). FLRT2 protein is highly expressed in sublaminae (S) 2 and 4 of the IPL, where the dendrites of both starburst amacrine cells (SACs) and direction-selective ganglion cells (DSGCs) arborize. Unc5c is expressed in a complementary pattern in S1/3/5 of the IPL. FLRT2-expressing neurons are repelled by Unc5c in vitro, suggesting that Unc5c might restrict FLRT2-positive SAC and DSGC dendrites to S2/4 of the developing IPL. ONL outer nuclear layer, OPL outer plexiform layer, INL inner nuclear layer, IPL inner plexiform layer, GCL ganglion cell layer
Extracellular LRR fibronectin type III domain-containing protein 1 (Elfn1) also plays a critical role in the formation of PR synapses (Fig. 3a). Elfn1 is selectively expressed by rod PRs, is absent from cone PRs, and localizes to rod PR axon terminals, where it forms a trans-synaptic complex with mGluR6 on ON-BC dendrites67. Expression and synaptic targeting of Elfn1 is mediated by α2δ4, an extracellular auxiliary subunit of Cav1.4, which is a pre-synaptic voltage-gated Ca2+ channel68. Once synaptically localized, Elfn1 aligns glutamate release-directing Cav1.4 with the post-synaptic, glutamate-sensing mGluR6 receptor (Fig. 3a). Elfn1 KO specifically disrupts the formation of rod synapses on ON-BCs, while neighboring cone PR synapses on ON-BCs are unaffected. Consequently, synaptic transmission between rod PRs and ON-BCs is selectively impaired67, indicating that Elfn1 plays a critical role in rod PR synaptogenesis and function.
FLRTs may play a role in mediating laminar targeting in the inner plexiform layer (IPL) of the retina (Fig. 3b). FLRTs, as well as the Unc5 proteins they bind, exhibit differential, laminar-restricted expression patterns in the developing IPL, and elicit repulsion or attraction in subpopulations of retinal neurons in ex vivo stripe assays69. FLRT2 and Unc5C display striking, alternating laminar patterns in the IPL, suggesting that repulsive interactions between these proteins may contribute to laminar organization. FLRT2 is expressed by both starburst amacrine cells (SACs) and ON–OFF direction-selective ganglion cells (ooDSGC), synaptic partners whose neurites co-stratify in the same IPL laminae. Neurites from both cell types are repelled by Unc5C in vitro69, which is prominently expressed in the neighboring laminae, suggesting that repulsive interactions between FLRT2-expressing SAC or ooDSGC neurites and neighboring Unc5C-expressing neurites causes laminar restriction of SAC and ooDSGC neurites in the IPL.
Role of LRR proteins in synaptic diversity
In contrast to the multiple studies that have addressed the development of specific synaptic connectivity following manipulation of LRR proteins in various neural circuits, little is known about the role LRR proteins might play in the regulation of synapse type-specific structural and functional properties. However, their specific expression patterns in the brain, and their ability to interact in trans across the synaptic cleft, as well as recruit specific proteins to the synapse, makes them interesting candidates for regulating synaptic diversity.
Regulation of synaptic shape and ultrastructural organization
Few studies have described an effect of LRR protein loss on synaptic shape, but in SALM1 KO CA1 neurons, dendritic spines look oddly shaped, with thin protrusions rising from the spine heads (Fig. 1)54. Moreover, analysis of synaptic ultrastructure in CA1 SR using EM reveals an increase in the ratio of perforated to total asymmetrical synapses, but a reduced PSD length54. SALM4 KO, on the other hand, increases PSD length48, suggesting that SALMs maintain normal spine morphology and synaptic ultrastructure. LRRTM1 KO also affects synapse morphology in CA1 neurons, as spines appear to be longer, and possibly less mature62, and EM ultrastructural analysis suggests a more dispersed distribution of synaptic vesicles following loss of LRRTM162. With regard to inhibitory synapses in CA1, loss of Slitrk3 increases pre-synaptic bouton size and PSD length at symmetric perisomatic synapses on CA1 pyramidal neurons (Fig. 1)64.
Regulation of pre-synaptic release properties
Arguably, the best example of an LRR protein that instructs specific synaptic functional properties is Elfn1, which controls pre-synaptic release probability in a target cell-dependent manner. The axons of CA1 pyramidal neurons form synapses onto two types of interneurons: somatostatin (SOM)-positive and parvalbumin (PV)-positive cells. These synapses are facilitating or depressing, depending on the target cell: CA1-SOM synapses are facilitating and have a low release probability, whereas CA1-PV synapses are depressing and have a high release probability. Elfn1 is selectively expressed in SOM cells and localizes to the excitatory post-synaptic compartment of these cells where it binds to the pre-synaptic mGluR7 receptor on CA1 axons to instruct a low release probability70,71. Loss of Elfn1 increases release probability in CA1-SOM synapses and, conversely, Elfn1 overexpression in PV cells decreases release probability at CA1-PV synapses70,71.
Regulation of short- and long-term plasticity properties
In addition to influencing properties of basal synaptic transmission, LRRs have been shown to act as regulators of synaptic efficacy by influencing short- or long-term synaptic plasticity. The best example for a role of LRR proteins in regulating synapse type-specific plasticity properties comes from work on NGLs/netrin-Gs in CA1 and DG neurons. Loss of netrin-G1 impairs post-tetanic potentiation (PTP), a form of pre-synaptic plasticity, at temporoammonic (TA) synapses formed on distal CA1 dendrites (Fig. 1)61. This defect is phenocopied in NGL-1 KO mice, indicating that the netrin-G1–NGL-1 interaction is required for PTP at this synapse. Loss of netrin-G2, on the other hand, facilitates PTP at SC-CA1 synapses61 (Fig. 1), demonstrating that netrin-G - NGL interactions differentially influence short-term plasticity at specific hippocampal synapses. This can also be seen at PP-DG synapses in DG granule cells, where loss of netrin-G1 from synapses in the outer molecular layer does not affect synaptic facilitation in response to paired-pulse stimuli, but loss of either netrin-G2 or NGL-2 from synapses in the middle molecular layer results in an attenuated paired-pulse depression (Fig. 2).
NGL protein interactions also differentially affect LTP at different CA1 inputs. LTP at TA-CA1 synapses in netrin-G1 KO mice is attenuated, whereas LTP at SC-CA1 synapses in netrin-G2 KO mice is facilitated (Fig. 1)61. Furthermore, LTP is not affected at SC-CA1 synapses in netrin-G1 KO and at SC-CA1 synapses in netrin-G2 KO, again demonstrating an input-specific regulation of synaptic plasticity properties by NGL proteins and their pre-synaptic binding partners.
The most dramatic effect of LRR proteins on LTP is observed following double KD (DKD) of LRRTM1 and LRRTM2 in CA1 pyramidal neurons. LRRTM1/LRRTM2 DKD blocks LTP at SC-CA1 synapses in neonatal hippocampus, and severely impairs it in young adult hippocampus (Fig. 1)72. Analysis of surface GluA1 levels following chemical LTP induction in cultured neurons reveals that LRRTM DKD does not impair the initial NMDAR-triggered delivery of AMPARs to the plasma membrane. However, newly inserted AMPARs are not maintained at the synaptic surface, resulting in a greater proportion of extrasynaptic surface AMPARs, at the expense of synaptic ones. However, overall surface GluA1 content is decreased following induction of LTP, indicating that extrasynaptic AMPARs may also be more unstable in the absence of LRRTMs72. Finally, SALMs have recently been implicated in LTP, as SALM1 KO results in enhanced LTP at SC-CA1 synapses (Fig. 1)54.
LRR proteins in behavior and neurological disease
Many genes encoding LRR proteins are linked to neurological disorders, including autism, schizophrenia, Tourette’s, obsessive compulsive disorder (OCD), and even Alzheimer’s disease12,26,73,74. Resolving the molecular mechanisms of LRR proteins will therefore allow us to determine not only how they contribute to synaptic diversity in healthy circuits, but also how loss or alteration of LRR proteins results in synaptic dysfunction as well as behavioral irregularities.
While many LRR protein genes exhibit cell type-specific expression patterns, they are still often expressed in multiple cell types across brain regions. This suggests that most LRR proteins operate in different circuits, implying that their respective contributions to circuit dysfunction in cases of disease are both divergent from one another and complex. This is emphasized by the fact that loss of particular LRR proteins is known to manifest in distinctive behavioral phenotypes. For instance, Slitrk5 KO mice display OCD-like behavior, as they present with severe facial lesions associated with excessive grooming55. In relation to this, it has also been shown that rare functional mutations in SLITRK5 contribute to the genetic risk for OCD in humans75. SALM1 KO mice, on the other hand, exhibit autism-like behavioral abnormalities, such as social withdrawal and increased stereotyped activities54. Functionally defective SALM1 mutations have also been found in human autism patients54. LRRTM1 is an emerging candidate gene for schizophrenia76,77. LRRTM1-deficient mice show behavioral impairments that partly resemble the signs of schizophrenia, such as social discrimination and spatial memory deficits62. These diverse examples indicate the importance of understanding how LRR proteins operate in neural circuits throughout the brain.
Conclusion
LRR proteins have emerged as major regulators of neural circuit development. A wealth of studies has shown that these proteins regulate every step in the development of precise connectivity: from cell migration and neurite outgrowth, to targeting and synapse development. In addition, LRR proteins regulate synapse type-specific features, such as synaptic morphology, ultrastructural organization, pre-synaptic release probability, and short-term plasticity and LTP properties, suggesting that these proteins also contribute to the diversity of synapses. Recent insight suggests that distinct neuronal cell types express different combinations of LRR proteins8. A major challenge will be to decipher how cell type-specific repertoires of LRR proteins regulate the specificity of connectivity, as well as the structural and functional diversity of a neuron’s synapses. Determining how different LRR proteins collaborate in this process, even with other cell-adhesion molecules at the synapse, is another challenge. Addressing these challenges will require an extensive analysis of LRR protein function in neural circuits in vivo. As many LRR proteins have been associated with brain disorders, this effort will not only permit a deeper understanding of the molecular mechanisms that specify synaptic connectivity and diversity in neural circuits, but will also help us understand how specific synaptic impairments can manifest in various neurological disorders.
Acknowledgements
A.S. is supported by an FWO PhD fellowship. Work in the authors’ lab on the role of LRR proteins in synaptic connectivity is supported by a European Research Council Starting grant (#311083), an FWO Odysseus grant, KU Leuven Methusalem grant, FWO Project grant G094016N, and ERANET NEURON 2015.
Conflict of interest
This manuscript is commissioned by Dr. Eunjoon Kim and Jaewon Ko for a 2018 special issue on synapse assembly, neural circuit development, and brain disorders.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- 2.Sanes JR, Yamagata M. Many paths to synaptic specificity. Annu. Rev. Cell Dev. Biol. 2009;25:161–195. doi: 10.1146/annurev.cellbio.24.110707.175402. [DOI] [PubMed] [Google Scholar]
- 3.Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu. Rev. Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Südhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Földy C, et al. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl Acad. Sci. USA. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuccillo MV, et al. Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TM, et al. An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. Elife. 2016;5:e22757. doi: 10.7554/eLife.22757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul A, et al. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 2017;171:522–539. doi: 10.1016/j.cell.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, et al. Classifying Drosophila olfactory projection neuron subtypes by single-cell RNA sequencing. Cell. 2017;171:1206–1220. doi: 10.1016/j.cell.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shekhar K, et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Rourke NA, Weiler NC, Micheva KD, Smith SJ. Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat. Rev. Neurosci. 2012;13:365–379. doi: 10.1038/nrn3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 2014;37:539–550. doi: 10.1016/j.tins.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jang S, Lee H, Kim E. Synaptic adhesion molecules and excitatory synaptic transmission. Curr. Opin. Neurobiol. 2017;45:45–50. doi: 10.1016/j.conb.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Ko J, Kim E. Leucine-rich repeat proteins of synapses. J. Neurosci. Res. 2007;85:2824–2832. doi: 10.1002/jnr.21306. [DOI] [PubMed] [Google Scholar]
- 15.Nam J, Mah W, Kim E. The SALM/Lrfn family of leucine-rich repeat-containing cell adhesion molecules. Semin. Cell Dev. Biol. 2011;22:492–498. doi: 10.1016/j.semcdb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 16.Woo J, Kwon S, Kim E. The NGL family of leucine-rich repeat-containing synaptic adhesion molecules. Mol. Cell. Neurosci. 2009;42:1–10. doi: 10.1016/j.mcn.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 17.de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu. Rev. Cell Dev. Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 18.Kajava AV. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- 19.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 20.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001;11:725–732. doi: 10.1016/S0959-440X(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 21.Colón-Ramos DA. Synapse formation in developing neural circuits. Curr. Top. Dev. Biol. 2009;87:53–79. doi: 10.1016/S0070-2153(09)01202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong W, Luo L. Genetic control of wiring specificity in the fly olfactory system. Genetics. 2014;196:17–29. doi: 10.1534/genetics.113.154336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu B, Wang KH, Nose A. Molecular mechanisms underlying neural circuit formation. Curr. Opin. Neurobiol. 2009;19:162–167. doi: 10.1016/j.conb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margeta MA, Shen K, Grill B. Building a synapse: lessons on synaptic specificity and presynaptic assembly from the nematode C. elegans. Curr. Opin. Neurobiol. 2008;18:69–76. doi: 10.1016/j.conb.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanes JR, Zipursky SL. Review design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proenca CC, Gao KP, Shmelkov SV, Rafii S, Lee FS. Slitrks as emerging candidate genes involved in neuropsychiatric disorders. Trends Neurosci. 2011;34:143–153. doi: 10.1016/j.tins.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin JC, Ho W, Gurney A, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat. Neurosci. 2003;6:1270–1276. doi: 10.1038/nn1148. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, et al. NGL family PSD-95-interacting adhesion molecules regulate excitatory synapse formation. Nat. Neurosci. 2006;9:1294–1301. doi: 10.1038/nn1763. [DOI] [PubMed] [Google Scholar]
- 29.Woo J, et al. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009;12:428–437. doi: 10.1038/nn.2279. [DOI] [PubMed] [Google Scholar]
- 30.Song YS, Lee H, Prosselkov P, Itohara S, Kim E. Trans-induced cis interaction in the tripartite NGL-1, netrin-G1 and LAR adhesion complex promotes development of excitatory synapses. J. Cell Sci. 2013;126:4926–4938. doi: 10.1242/jcs.129718. [DOI] [PubMed] [Google Scholar]
- 31.Linhoff MW, et al. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laurén J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/S0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 33.de Wit J, et al. LRRTM2 interacts with neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Um JW, et al. LRRTM3 regulates excitatory synapse development through alternative splicing and neurexin binding. Cell Rep. 2016;14:808–822. doi: 10.1016/j.celrep.2015.12.081. [DOI] [PubMed] [Google Scholar]
- 36.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 2010;30:7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko JS, et al. PTPσ functions as a presynaptic receptor for the glypican-4/LRRTM4 complex and is essential for excitatory synaptic transmission. Proc. Natl Acad. Sci. USA. 2015;112:1874–1879. doi: 10.1073/pnas.1410138112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Wit J, et al. Unbiased discovery of glypican as a receptor for LRRTM4 in regulating excitatory synapse development. Neuron. 2013;79:696–711. doi: 10.1016/j.neuron.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddiqui TJ, et al. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron. 2013;79:680–695. doi: 10.1016/j.neuron.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Aruga J, Mikoshiba K. Identification and characterization of Slitrk, a novel neuronal transmembrane protein family controlling neurite outgrowth. Mol. Cell. Neurosci. 2003;24:117–129. doi: 10.1016/S1044-7431(03)00129-5. [DOI] [PubMed] [Google Scholar]
- 41.Yim YS, et al. Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc. Natl Acad. Sci. USA. 2013;110:4057–4062. doi: 10.1073/pnas.1209881110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Um JW, et al. Structural basis for LAR-RPTP/Slitrk complex-mediated synaptic adhesion. Nat. Commun. 2014;5:5423. doi: 10.1038/ncomms6423. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi H, et al. Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat. Neurosci. 2012;15:389–398. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. A novel family of adhesion-like molecules that interacts with the NMDA receptor. J. Neurosci. 2006;26:2174–2183. doi: 10.1523/JNEUROSCI.3799-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Ko J, et al. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 47.Seabold GK, et al. The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J. Biol. Chem. 2008;283:8395–8405. doi: 10.1074/jbc.M709456200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lie E, et al. SALM4 suppresses excitatory synapse development by cis-inhibiting trans-synaptic SALM3–LAR adhesion. Nat. Commun. 2016;7:12328. doi: 10.1038/ncomms12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi Y, et al. SALM5 trans-synaptically interacts with LAR-RPTPs in a splicing-dependent manner to regulate synapse development. Sci. Rep. 2016;6:26676. doi: 10.1038/srep26676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, et al. Splicing-dependent trans-synaptic SALM3-LAR-RPTP interactions regulate excitatory synapse development and locomotion. Cell Rep. 2015;12:1618–1630. doi: 10.1016/j.celrep.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mah W, et al. Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J. Neurosci. 2010;30:5559–5568. doi: 10.1523/JNEUROSCI.4839-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lacy SE, Bönnemann CG, Buzney EA, Kunkel LM. Identification of FLRT1, FLRT2, and FLRT3: a novel family of transmembrane leucine-rich repeat proteins. Genomics. 1999;62:417–426. doi: 10.1006/geno.1999.6033. [DOI] [PubMed] [Google Scholar]
- 53.O’Sullivan ML, et al. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morimura N, et al. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Nat. Commun. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 55.Shmelkov SV, et al. Slitrk5 deficiency impairs corticostriatal circuitry and leads to obsessive-compulsive–like behaviors in mice. Nat. Med. 2010;16:598–602. doi: 10.1038/nm.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamagishi S, et al. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5‐positive neurons. EMBO J. 2011;30:2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.del Toro D, et al. Regulation of cerebral cortex folding by controlling neuronal migration via FLRT adhesion molecules. Cell. 2017;169:621–635. doi: 10.1016/j.cell.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 58.Levya-Díaz E, et al. FLRT3 is a Robo1-interacting protein that determines Netrin-1 attraction in developing axons. Curr. Biol. 2014;24:494–508. doi: 10.1016/j.cub.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 59.Nishimura-Akiyoshi S, Niimi K, Nakashiba T, Itohara S. Axonal netrin-Gs transneuronally determine lamina-specific subdendritic segments. Proc. Natl Acad. Sci. USA. 2007;104:14801–14806. doi: 10.1073/pnas.0706919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DeNardo LA, de Wit J, Otto-Hitt S, Ghosh A. NGL-2 regulates input-specific synapse development in CA1 pyramidal neurons. Neuron. 2012;76:762–775. doi: 10.1016/j.neuron.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsukawa H, et al. Netrin-G/NGL complexes encode functional synaptic diversification. J. Neurosci. 2014;34:15779–15792. doi: 10.1523/JNEUROSCI.1141-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takashima N, et al. Impaired cognitive function and altered hippocampal synapse morphology in mice lacking LRRTM1, a gene associated with schizophrenia. PLoS ONE. 2011;6:e22716. doi: 10.1371/journal.pone.0022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soler-llavina GJ, Fuccillo MV, Ko J, Malenka RC. The neurexin ligands, neuroligins and leucine-rich repeat transmembrane proteins, perform convergent and divergent synaptic functions in vivo. Proc. Natl Acad. Sci. USA. 2011;108:16502–16509. doi: 10.1073/pnas.1114028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li J, et al. Molecular dissection of neuroligin 2 and Slitrk3 reveals an essential framework for GABAergic synapse development. Neuron. 2017;96:808–826. doi: 10.1016/j.neuron.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulopoulos A, et al. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through Gephyrin and Collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 66.Soto F, Watkins KL, Johnson RE, Schottler F, Kerschensteiner D. NGL-2 regulates pathway-specific neurite growth and lamination, synapse formation, and signal transmission in the retina. J. Neurosci. 2013;33:11949–11959. doi: 10.1523/JNEUROSCI.1521-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao Y, et al. Mechanism for selective synaptic wiring of rod photoreceptors into the retinal circuitry and its role in vision. Neuron. 2015;87:1248–1260. doi: 10.1016/j.neuron.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, et al. The auxiliary calcium channel subunit α2δ4 is required for axonal elaboration, synaptic transmission, and wiring of rod photoreceptors. Neuron. 2017;93:1359–1374. doi: 10.1016/j.neuron.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visser JJ, et al. An extracellular biochemical screen reveals that FLRTs and Unc5s mediate neuronal subtype recognition in the retina. Elife. 2015;4:e08149. doi: 10.7554/eLife.08149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sylwestrak EL, Ghosh A. Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science. 2012;338:536–540. doi: 10.1126/science.1222482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tomioka NH, et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat. Commun. 2014;5:4501. doi: 10.1038/ncomms5501. [DOI] [PubMed] [Google Scholar]
- 72.Soler-llavina GJ, et al. Leucine-rich repeat transmembrane proteins are essential for maintenance of long-term potentiation. Neuron. 2013;79:439–446. doi: 10.1016/j.neuron.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katayama K, et al. Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol. Psychiatry. 2010;15:177–184. doi: 10.1038/mp.2008.97. [DOI] [PubMed] [Google Scholar]
- 74.Majercak J, et al. LRRTM3 promotes processing of amyloid-precursor protein by BACE1 and is a positional candidate gene for late-onset Alzheimer’s disease. Proc. Natl Acad. Sci. USA. 2006;103:17967–17972. doi: 10.1073/pnas.0605461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song M, et al. Rare synaptogenesis-impairing mutations in SLITRK5 are associated with obsessive compulsive disorder. PLoS ONE. 2017;12:e0169994. doi: 10.1371/journal.pone.0169994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ludwig KU, et al. Supporting evidence for LRRTM1 imprinting effects in schizophrenia. Mol. Psychiatry. 2009;14:743–745. doi: 10.1038/mp.2009.28. [DOI] [PubMed] [Google Scholar]