Abstract
Circular RNAs (circRNAs) play important roles in the initiation and development of different diseases. Here, we detected their role in intervertebral disc (IVD) degeneration. An Arraystar human circular RNA microarray assay was used to detect circRNAs in normal and degenerated human IVD nucleus pulposus (NP) tissues. The role of circ-4099 in IVDD and its mechanism were evaluated by qRT-PCR and gain-of-function/loss-of-function studies. Interaction networks for competing endogenous RNAs (ceRNAs), miRNAs, and miRNA target gene were detected by bioinformatics analysis, RNA immunoprecipitation and luciferase assay. Expression of seventy-two circRNAs were increased by more than twofold in degenerated NP tissues. qRT-PCR showed that the expression of circ-4099 in NP tissues was consistent with that of the array screening. Over-expression of circ-4099 increased the expression of Collagen II and Aggrecan and decreased the secretion of the pro-inflammatory factors IL-1β, TNF-α, and PGE2. TNF-α treatment increased circ-4099 expression in NP cells. NF-κB/MAPK inhibitors or shRNAs abolished the inductive effects of TNF-α on circ-4099 expression. We further demonstrated that circ-4099 was able to function as a “sponge” by competitively binding miR-616-5p, which reversed the suppression of Sox9 by miR-616-5p. We used DNA pull-down and spectrometry experiments to show that TNF-α can promote circ-4099 transcription through upregulation of GRP78. We provide the first evidence that shows circRNAs are differentially expressed in degenerated and normal NP tissues. Circ-4099 may play a role in a protective mechanism and be part of a compensatory response that maintains the synthesis and secretion of the extracellular matrix in NP cells and might be a protective factor in IVD degeneration as well as restore NP cell function.
Spinal disease: Circular RNA linked to disc degeneration
A circular RNA molecule helps protect against degenerative disc disease. Hua Wang and coworkers from Sun Yat-Sen University in Guangzhou, China, examined whether circular RNAs, regulatory molecules that take the form of closed RNA loops, contribute to intervertebral disc degeneration, a condition in which connective tissue in the spine breaks down over time, causing back pain and weakness. They found 72 circular RNAs that were either significantly over- or under-expressed in the inner core tissue of intervertebral discs from patients with this condition. They showed that one of these circular RNAs, circ-4099, increased the production of key cartilage proteins. This RNA also blocked the activity of another non-circular regulatory RNA that normally inhibits a molecular pathway needed for proper cartilage formation. Enhancing the activity of this protective molecule could help treat degenerative disc disease.
Introduction
Low back pain (LBP) is one of the most common and costly health problems, affecting ~80% of the population during their life time, with total costs exceeding $100 billion per year1,2. Intervertebral disc (IVD) degeneration-related disorder is the major contributor of LBP and associated disabilities3. The hallmark of IVD degeneration is decreased cell density4 and extracellular matrix (ECM)5. Nucleus pulposus (NP) cells that produce proteoglycan aggrecan and collagens to form complex ECM play a key role in maintaining disc health6. IVD degeneration results in increased production of degradative enzymes2 together with decreased synthesis of ECM7. These catabolic processes are thought to be mediated by a number of cytokines, including TNF-α and IL-1β8. Thus, TNF-α and IL-1β are the most selected reagents to induce IVD degeneration cellular models in vitro8. Although many factors have been demonstrated to participate in the IVD degeneration process, the exact pathophysiology regarding IVD degeneration requires further investigation.
Circular RNAs (circRNAs), which were discovered more than 20 years ago, are a novel type of widely expressed non-coding RNA (ncRNA) that are different from linear RNAs (miRNAs are also linear) and represent an understudied class of ncRNAs9. circRNAs form a covalently closed continuous loop, are conserved across species, and usually show tissue-specific or developmental stage-specific expression10. circRNAs are more stable than linear RNAs because of their higher nuclease stability, which gives them great advantages as novel biomarkers in clinical applications11. circRNAs have been shown to function as natural miRNA sponge transcripts that compete for binding with endogenous RNAs in diverse species12. The interactions between circRNAs and miRNAs suggest the potential importance of circRNAs in disease regulation. Hansen et al13. showed a highly expressed circRNA in human and mouse brain tissue and the degeneration process. Li et al11. demonstrated that cir-ITCH expression was low in esophageal cancer compared to peri-tumoral tissue, which indicated that cir-ITCH may have an inhibitory effect on esophageal cancer. Hansen et al12. demonstrated that ciRS-7 probably serves as a crucial factor that is significantly engaged in neuronal function and is responsible for neurological disorders and brain tumor development. Ghosal et al14. revealed circRNA SNPs related to a wide range of diseases, including esophageal, gastric, and prostate cancer, multiple sclerosis, and neurodegenerative diseases, such as Parkinson’s disease, Alzheimer’s disease, among others. However, the roles of circRNAs in IVD degeneration remain a largely uncharted territory.
In this study, we performed circRNA expression profiling analysis in degenerated compared with non-degenerated human IVD NP tissues using an Arraystar human circRNA microarray. This array is the first commercially available circRNA microarray (Arraystar, Inc., USA) designed for the global profiling of human circRNAs. Our data also rely on published paired-end sequencing data and splice sites15,16.
Materials and Methods
Collection and grading of human NP Tissues
Degenerated IVD tissue samples were collected as surgical waste from patients undergoing spinal surgery in our hospital. Control non-degenerated samples were obtained from patients suffering from vertebral fracture or scoliosis (Supplemental Table 1). Samples were collected according to the approval of the local ethics committee, and informed consent was obtained from every patient or family members before collection. Assessment of IVD degeneration was performed according to the description by Pfirrmann et al17. Samples were divided into two groups, six non-degenerated (grade I/II) and six degenerated (grade IV/V) samples.
Isolation of NP cells and treatment
Human NP cells were isolated from non-degenerated IVD tissues from spine fracture patients undergoing spinal surgery using a previously described method18. NP cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) and 10% fetal bovine serum (FBS) supplemented with antibiotics. To investigate the effect of cytokines, cells were treated with TNF-α (50 ng/ml, Peprotech, NJ) for 24 h.
Array data production
Total RNA from NP tissues was extracted using the Trizol reagent (Invitrogen) and then purified using a NucleoSpin® RNA clean-up Kit (Macherey-Nagel). Extracted total RNA was quantified with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The sample preparation and microarray hybridization were performed by KangChen, Inc., based on Arraystar’s standard protocols. Briefly, total RNA was digested with Rnase R (Epicentre, Inc.) to remove linear RNAs and enrich for circRNAs. Then, the enriched circRNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled cRNAs were hybridized onto the Arraystar Human circRNA Array (8 × 15K, Arraystar). After washing the slides, the arrays were scanned by the Agilent Scanner G2505C.
Differential expression analysis
The Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed. Differentially expressed circRNAs with statistical significance between the two groups were identified through Volcano Plot filtering. Differentially expressed circRNAs between two samples were identified through fold change filtering. Hierarchical Clustering was performed to show the distinguishable circRNA expression patterns among samples.
miRNA prediction and co-expression networks
circRNA/miRNA interactions were predicted with Arraystar’s homemade miRNA target prediction software (Rockville, USA), whose principles are based on the TargetScan and miRanda prediction algorithms. The differentially expressed circRNAs within the comparisons were annotated in detail with the circRNA/miRNA interaction information. To further elucidate correlations between the circRNAs and miRNAs, potential circRNA–miRNA–mRNA interaction analysis was conducted by Cytoscape. The size of each node represents the number of putative miRNAs functionally connected to each circRNA.
Plasmid construction
We constructed the circRNA over-expression plasmid used in this study. The construct method was previously published12. The resulting construct (pcDNA3.1-cir-DENN5A) was verified by direct sequencing with the cir-DENN5A -F (CGG AAT TCT GAA ATA TGC TAT CTT AC AGC ATT ATG CCA GTA CATA CAG GCT) and cir-DENN5A –R (CGG GAT CCT CAA GAA AAA ATA TAT TCA CGC TGT GAG TAG AGC AGG ATT TGA) primers.
The full-length sequences of DENN5A and the Sox9 gene were amplified using PCR methods. The reporter promoter contains cir-DENN5A, Sox9-wild type and Sox9-mutation and was constructed with a previous method; the resulting construct was verified by direct sequencing with the following primers: (1) cir-DENN5A -F (AAA CTC GAG CAT TAT GCC AGT ACA TAC AGG CTT CTAA) and cir-DENN5A -R (AAA GCG GCC GCG CTG TGA GTA GAG CAG GAT TTG AA), (2) Sox9-WT -F (AAA ACT CGA GGG AGG CCT CCC ACG AAG) and Sox9-WT -R (AAA AGC GGC CGC TTT CTT TTT AAT GCA ATG TAT ATT TAT TGT AAA CA), and (3) Sox9-MT -F (CCA TTA CCA AAA ACT CGG GAT TTA TAC ATA TTT TTA GAT AAA ATT AAA TGC TC) and Sox9-MT -R (ATC CCG AGT TTT TGG TAA TGA ATC ATA CAC AGT ACA TAC TAA AAA TAT T).
Site-directed mutagenesis
The Sox9-W reporter plasmid was used to mutate the miR-616-5p site (TTTTGAG to AAAACTC). Mutants were generated with the QuickChange II XL site-directed mutagenesis kit (Stratagene, Wilmington, DE, USA) using forward and reverse primer pairs containing the desired mutation following the manufacturer’s instructions. The mutations were verified by sequencing.
RNA isolation and qRT-PCR
Following treatment, total RNA was extracted from NP tissues or NP cells using the Trizol reagent (Invitrogen) as described above. Two micrograms of total DNA-free RNA was used to synthesize cDNA with the SuperScript III cDNA synthesis kit (Invitrogen). Reactions were set up in triplicate in 96-well plates using 1 µl cDNA with PrimeScriptTM RT-PCR Kit (Takara) to which gene-specific forward and reverse PCR primers were added. Each set of samples included a template-free control.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was completed using the Taq qPCR Master Mix (Promega) on an Mx3005P real-time PCR System (Stratagene, La Jolla, CA) following the manufacturer’s instructions. Divergent rather than more commonly used convergent circ-4099primers were designed and synthesized by Sangon Biotech. β-actin was used to normalize mRNA expression. Melting curves were analyzed to verify the specificity. The following gene-specific primer pairs were used: (1) Human-β-actin, F 5′-GCA TGG GTC AGA AGG ATT CCT-3′ and R 5′-TCG TCC CAG TTG GTG ACG AT-3′; (2) Homo Circ-4099, F3 5′-CCA GAG ACC AAG TAC CAA TGAG-3′ and R 5′-GTA CTG GCA TAA TGG CTG TGA-3′; (3) p65, F 5′-ACA ACA ACC CCT TCC AAG AAGA-3′ and R 5′-AGA GTT TCG GTT CAC TCG GC-3′; (4) IKKβ, F 5′-ATC CAG ATC ATG AGA AGG CTG AC-3′ and R 5′-AGC AAG GTG AGG ATG GCAC-3′; (5) COLL2, F 5′-GCC AAA GGT GAA CAA GGA GA-3′ and R 5′-TCG AGC ACC TTT AGG ACC AT-3′; (6) Aggrecan, F 5′-GTC GTG GTA AAA GGC ATC GT-3′ and R 5′-TGC CTC TTG GAA GGT GAA CT-3′; (7) MMP-3, F 5′-TTC TCG AAT CAC GGA GGA AGC CAA-3′ and R5′-ACG ACG CCT TCC ATG GAT CTT CTT-3′; (8) ADAMTS5, F 5′-GTC CAA ATG CAC TTC AGC CAC GAT-3′ and R 5′-AAT GTC AAG TTG CAC TGC TGG GTG-3′; and (9) DENND5A (linear + circRNA), -F 5′-TCT ACC ACA TGC ACA ATG CT -3′ and -R 5′-GTC CCG GCT AAT GTC ATA GG -3′.
Protein extraction and western blotting
NP cells were placed on ice immediately after treatment, washed with ice-cold PBS, and harvested in mammalian protein extraction reagent buffer (Pierce), as reported earlier by Risbud et al19. Protein concentrations were measured using a BCA Protein Assay Kit (Pierce, USA). Total cellular proteins were resolved on 8–12% SDS-polyacrylamide gels and transferred via electroblotting to PVDF membranes (Millipore). The membranes were blocked in 5% non-fat dry milk with TBST and incubated overnight at 4 °C with antibodies against Aggrecan (ACAN), Collagen II, Sox9, p65, IKKβ, GRP94, and GRP78 from Abcam. After washing with TBST, the membranes were incubated with anti-rabbit or anti-Rat HRP-conjugated secondary antibodies and detected with ECL Plus (Millipore, USA) reagent. The results were then quantified with a multigauge densitometry system (Fujifilm, Tokyo, Japan).
Transient transfections and luciferase assays
NP cells were transfected with the reporter plasmids described above using Lipofectamine 2000. Cells were co-transfected with miRNAs according to the manufacturer’s instructions. Each group included three replicates and triplicate independent experiments were performed. In the luciferase screening assay, we used one internal control (RL reporter) and two negative controls (miRNA control and luciferase reporter lacking the circDENN5A 3′-UTR). Each miRNA or NC-RNA was co-transfected with RL reporter and FL reporter with or without the cir-DENN5A 3′-UTR. For comparison, the FL activity was first normalized with RL activity. The effect of each miRNA on luciferase reporter with the circDENN5A 3′-UTR was then normalized with that of luciferase reporter without the circDENN5A 3′-UTR. Finally, the fold change was calculated for each miRNA compared with NC.
Lentiviral particle production and viral transduction
HEK 293T cells were seeded into 10 cm plates in DMEM with 10% FBS 2 days before transfection. Cells were transfected with 2.5 μg of sh-control, sh-p65, or sh-IKKβ plasmids along with 1.875 μg psPAX2 and 0.625 μg pMD2.G. After 16 h, the transfection media was removed and replaced by DMEM with 5% FBS and penicillin/streptomycin. Lentiviral particles were harvested at 48 h post-transfection. NP cells were plated in DMEM with 5% FBS. Cells in 10 cm plates were transduced with 5 ml of viral particle-conditioned media with 6 μg/ml polybrene. After 24 h, the conditioned media was replaced by DMEM with 5% FBS. Cells were collected for RNA or protein extraction after treatment.
RNA FISH
hNP cells cultured on a glass coverslip were fixed in 4% paraformaldehyde for 30 min and permeabilized with 0.1 % Triton X-100 for 30 min. The cells were washed three times with PBS and treated with pre-hybridization buffer (2× saline-sodium citrate) containing 10 % formamide. Hybridization was performed using a similar buffer, but with the addition of competitor RNA (tRNA) and competitor protein (BSA) to reduce background. The FITC-labeled circ-4099 and miR-616-5p probe.
RNA-binding protein immunoprecipitation assay
RNA-binding protein immunoprecipitation (RIP) experiments were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). The Ago-RIP assay was conducted in hNP cells stably expressing Flag-AGO2 or Flag-GFP. We first constructed the lentivirus vectors harboring Flag-Ago2 or Flag-GFP and established two stable cell lines after lentivirus transduction in hNP cells (denoted as Flag-GFP and Flag-Ago2 cells). Approximately 107 cells were pelleted and re-suspended in an equal pellet volume of RIP Lysis Buffer (~100 ml) plus protease and RNase inhibitors. The cell lysates (100 ml) were incubated with 5 mg of control mouse IgG-coated or antibody against Flag peptide-coated (Sigma-Aldrich) beads with rotation at 4 °C overnight. After treating with proteinase K, the immunoprecipitated RNAs were extracted with the RNeasy MinElute Cleanup Kit (Qiagen) and reverse transcribed using the Prime-Script RT Master Mix (TaKaRa). The abundance of miR-616-5p and Circ-4099 was detected by qRT-PCR assay.
Biotin-coupled miRNA capture
The biotin-coupled miRNA pull-down assay was performed as described previously. Briefly, 3′-end biotinylated miR-616 mimic or control RNA (RiboBio) were transfected into hNP cells at a final concentration of 20 nM for 1 day. The biotin-coupled RNA complexes were pulled down by incubating the cell lysates with streptavidin-coated magnetic beads (Life Technologies). The abundance of circDENN5A in the bound fractions was evaluated by qRT-PCR analysis.
Biotin-promoter pull-down assay and mass spectrometry
Double-stranded circDENN5A promoter was synthesized by PCR and labeled with biotin-14-dCTP according to the manufacturer’s instructions (19518-018, Invitrogen, Grand Island, NY, USA). Biotin-labeled DNA was diluted in 10 mM Tris-HCl, 10 mM MgCl2, 25 mM NaCl, and 10 % glycerol and incubated with nuclear protein extract at 4 °C for 12 h. In total, 100 μl streptavidin-linked magnetic beads (88816, Thermo Scientific, Waltham, MA USA) were used to pull down the biotinylated DNA at room temperature for 2 h. The bead-DNA-protein complexes were then washed with 1× binding and washing buffer (5 mM Tris-HCl, 1 M NaCl, 0.5 mM EDTA, and 0.005 % Tween 20 four times. The proteins were precipitated and diluted in 100 μl protein lysis buffer. One-shot mass spectrometry analyses were then performed to analyze the purified nuclear proteins.
Results
circRNAs are differentially expressed in degenerated and non-degenerated NP tissues
Given the high abundance of circRNAs in other tissues, we aimed to evaluate the role of circRNAs in the IVD degeneration process. We analyzed human circRNA expression in degenerated versus non-degenerated IVD NP tissues(Table 1). Among the 5397 circRNAs, 21 were downregulated and 51 upregulated by more than twofold in degenerated compared with non-degenerated NP tissues (P < 0.05) (Table 2) (Fig. 1a, b). Hierarchical clustering analysis indicated distinguishable circRNA expression profiles between the samples and homogeneity within groups (Fig. 1c). Circ-4099, circ-68610, circ-2069, and circ-92328 as well as circ-4099 were used for further analysis.
Table 1.
Selected patient characteristics
| Gender | Age | Diagnosis | Operation level | IVDD classification | |
|---|---|---|---|---|---|
| 1 | M | 30 | L3 burst fracture | L2–4 | I |
| 2 | F | 32 | L2 burst fracture | L1–3 | II |
| 3 | M | 35 | L2 burst fracture | L1–3 | I |
| 4 | M | 38 | L2 burst fracture | L1–3 | II |
| 5 | F | 28 | Scoliosis | L4–5 | II |
| 6 | F | 26 | L3 burst fracture | L2–4 | I |
| 7 | M | 48 | Intervertebral disc herniation (L5/S1) | L5–S1 | IV |
| 8 | M | 52 | Intervertebral disc herniation (L5/S1) | L5–S1 | IV |
| 9 | M | 50 | Lumbar spondylolisthesis (L5) | L5–S1 | V |
| 10 | F | 49 | Lumbar spondylolisthesis (L5) | L5–S1 | V |
| 11 | F | 51 | Lumbar spondylolisthesis (L4) | L4–5 | V |
| 12 | M | 48 | Lumbar spondylolisthesis (L5) | L5–S1 | V |
Table 2.
Top 10 differentially expressed circRNAs (degenerated vs. non-degenerated)
| circRNA | Regulation | Type | FC (abs) | Alias | FDR | P-value |
|---|---|---|---|---|---|---|
| hsa_circRNA_100759 | Up | Exonic | 6.0436796 | circ_0004099 | 0.001985008 | 2.80285E-05 |
| hsa_circRNA_103555 | Up | Exonic | 5.5301653 | circ_0068610 | 0.083401104 | 0.025051229 |
| hsa_circRNA_102113 | Up | Exonic | 5.3087065 | circ_0002069 | 0.023900268 | 0.002867487 |
| hsa_circRNA_101353 | Up | Exonic | 4.5647371 | circ_0031897 | 0.070983991 | 0.018858289 |
| hsa_circRNA_104574 | Up | Exonic | 4.0113787 | circ_0083756 | 0.070942531 | 0.018794621 |
| hsa_circRNA_103029 | Up | Exonic | 3.8610047 | circ_0059955 | 0.016096515 | 0.001457627 |
| hsa_circRNA_100719 | Up | Exonic | 3.6146979 | circ_0020390 | 0.015623803 | 0.001380433 |
| hsa_circRNA_104135 | Up | Exonic | 3.5263885 | circ_0007874 | 0.098387237 | 0.032936772 |
| hsa_circRNA_400101 | Up | Intronic | 3.5095173 | circ_0092328 | 0.053147953 | 0.011768583 |
| hsa_circRNA_103541 | Up | Exonic | 3.5020867 | circ_0068462 | 0.046516597 | 0.009053098 |
| hsa_circRNA_103225 | Down | Exonic | 3.105779 | circ_0063331 | 0.045605649 | 0.00875982 |
| hsa_circRNA_104194 | Down | Exonic | 2.8688053 | circ_0004712 | 0.028477579 | 0.00389463 |
| hsa_circRNA_101004 | Down | Exonic | 2.6683336 | circ_0000375 | 0.112304139 | 0.040901878 |
| hsa_circRNA_000596 | Down | Antisense | 2.6262258 | circ_0000661 | 0.047669869 | 0.009363936 |
| hsa_circRNA_101412 | Down | Exonic | 2.5754826 | circ_0005252 | 0.041823488 | 0.007759216 |
| hsa_circRNA_104040 | Down | Exonic | 2.54998 | circ_0075410 | 0.098387237 | 0.032945284 |
| hsa_circRNA_400027 | Down | Intronic | 2.5112 | circ_0092367 | 0.06739006 | 0.017092378 |
| hsa_circRNA_103818 | Down | Exonic | 2.4968942 | circ_0072279 | 0.107230858 | 0.037801246 |
| hsa_circRNA_103639 | Down | Exonic | 2.3998995 | circ_0007646 | 0.070058343 | 0.018376124 |
| hsa_circRNA_100024 | Down | Exonic | 2.2937773 | circ_0009135 | 0.106912695 | 0.03757954 |
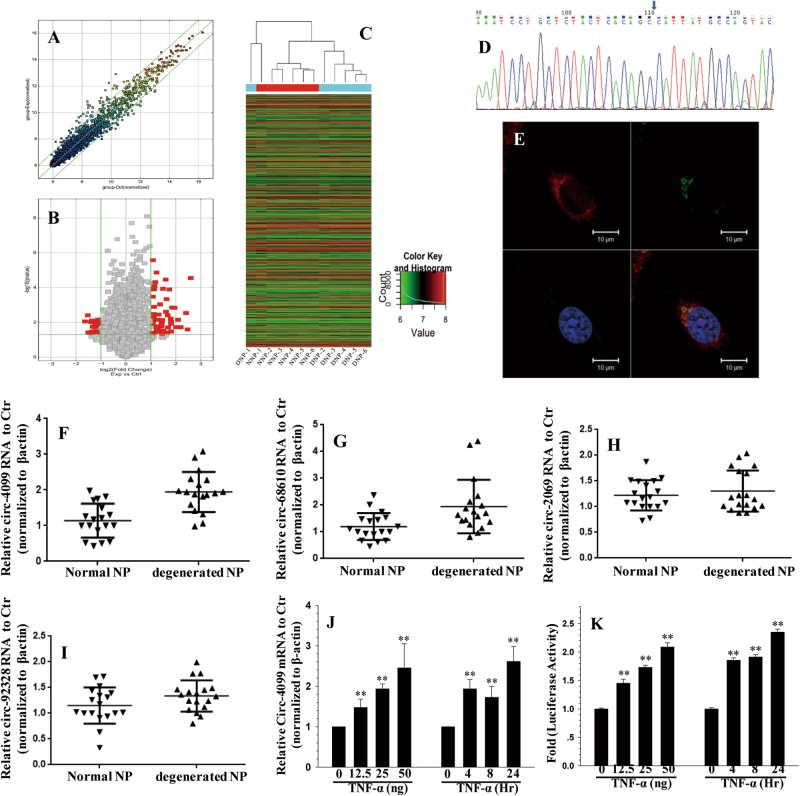
Fig. 1. circRNA profile in IVDD.
a Scatter plot of circRNA expression. The circRNAs above the top green line and below the bottom green line indicated more than a twofold change between the degenerated and normal IVD NP tissues. b Volcano Plot of the differentially expressed circRNAs. The red points in the plot represent differentially expressed circRNAs with statistical significance. c Hierarchical clustering shows the distinguishable circRNA expression profile between the two groups and homogeneity within the groups. d The sequence of the amplified Circ-4099 product was completely consistent with CircBase. e Co-localization between circ-4099 (red) and miR-616-5p (green) was observed by RNA in situ hybridization in hNP cells after co-transfection with circ-4099-expressing and miR-616-5p-expressing vectors. The nuclei were stained with DAPI (blue). Scale bar = 10 µm. f The expression levels of circ-4099 are significantly higher in degenerated NP tissues than in non-degenerated NP tissues. g–i The expression levels of circ-68610/2069/92328 are higher in degenerated NP tissues than in non-degenerated NP tissues. j qRT-PCR analysis shows that TNF-α upregulates the expression of circ-4099 in a dose-dependent and time-dependent manner. For time-dependent studies, 50 ng/ml TNF-α was used. k hNP cells were transfected with the circ-4099-Luc promoter and the induction in luciferase activity was measured following TNF-α treatment (P < 0.001)
circRNA amplification
To validate the efficacy and specificity of the divergent primers to detect the four circRNAs, we performed real-time RT-PCR experiments. A single-peak melting curve indicated that there were no primer dimers or non-specific amplification. The sequence of the amplified circ-4099 product was completely consistent with CircBase (Fig. 1d, other sequence data not shown). These results showed that we could reliably detect and quantify circ-4099 in NP tissues or cells by qRT-PCR.
The localization of circ-4099 in NP cells
We next investigated the localization of this circRNA in NP cells. Fluorescence in situ hybridization (FISH) against circ-4099 demonstrated that the circular form of DENND5A preferentially localized in the cytoplasm (Fig. 1e). Taken together, our results show that circ-4099 is an abundant and stable circRNA expressed in different human cells.
Increased Circ-4099 expression in degenerated NP tissues
To validate the circRNA microarray results, we selected circ-4099, circ-68610, circ-2069, and circ-92328, which exhibited significant changes in expression among the differentially expressed circRNAs and performed qPCR to analyze the changes in expression. The data confirmed that circ-4099 was upregulated in degenerated NP tissues with 2.5-fold change by qPCR (P < 0.001; Fig. 1f). The expression of circ-68610 (Fig. 1g), circ-2069 (Fig. 1h), and circ-92328 (Fig. 1i) were also increased in degenerated NP tissues, but were not as consistent as circ-4099, so circ-4099 was used for further analysis.
TNF-α induced the expression of circ-4099 in the NP cells
The cytokine TNF-α was used to establish a cellular model of IVD degeneration in vitro8. To test the premise that cytokines involved with IVD degeneration control circ-4099, NP cells were treated with TNF-α and the expression of circ-4099 was analyzed. qRT-PCR analysis showed that TNF-α upregulated circ-4099 expression in a dose-dependent and time-dependent manner. For time-dependent studies, 50 ng/ml TNF-α was used. (Fig. 1j)
To further investigate whether cytokines enhance circ-4099 activity in human NP cells, we transfected cells with the circ-4099-Luc wild-type reporter and measured their activity following TNF-α treatment. The results of these experiments showed that TNF-α increased the activity of the circ-4099-Luc reporter in a dose-dependent and time-dependent manner. (Fig. 1k)
TNF-α promotes circ-4099 expression through the MAPK and NF-κB pathways
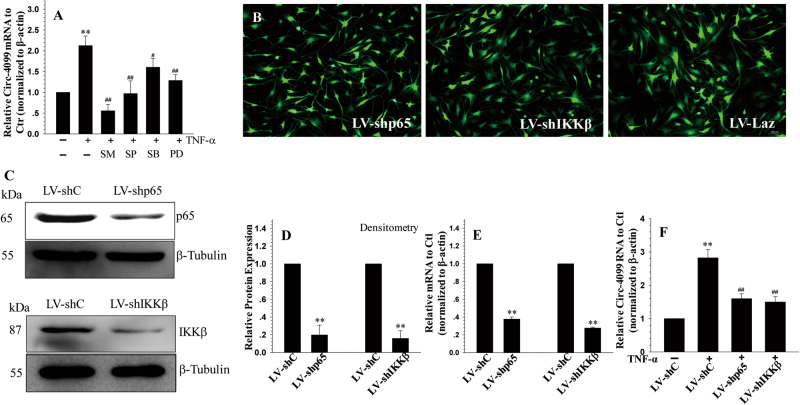
As the MAPK and NF-κB pathways are the main pathways that induce TNF-α, we determined whether the NF-κB and MAPK signaling pathways control the cytokine-dependent induction of circ-4099 expression. NP cells were treated with pathway-specific inhibitors (SM7368 (10 μΜ), SB203580 (10 μΜ), PD98059 (50 μΜ), or SP60025 (10 μΜ) (Calbiochem)) 1 h prior to exposure to TNF-α. When induced by TNF-α, pretreatment with inhibitors resulted in the suppressed expression of circ-4099, with more suppression observed in the NF-κB inhibitor group. (Fig. 2a)
Fig. 2. MAPK and NF-κB regulation of circ-4099 expression.
a qRT-PCR analysis shows that the TNF-α-dependent induction of circ-4099 in NP cells is blocked by MAPK and NF-kB pathway inhibitors. b Immunofluorescence detection of GFP in human NP cells transduced with lentivirus (LV) co-expressing GFP and either sh-p65 or sh-Ikkβ showed high transduction efficiency. c Western blot, d corresponding densitometry, and e RT-PCR analyses of cells transduced with LV-shLaz, LV-shp65, and LV-shIkkβ. Expression of p65 and Ikkβ is significantly blocked by the respective shRNAs compared with the control. f qRT-PCR analysis of circ-4099 expression in cells infected with LV-shC, LV-shp65, and LV-shIkkβ following TNF-α treatment. Note that the TNF-α-dependent induction of circ-4099 is significantly blocked by suppression of NF-κB pathway components
To further elucidate the role of the NF-κB signaling pathway in the regulation of circ-4099 expression, we silenced the expression of individual NF-κB signaling components and transduced human NP cells with lentivirus co-expressing GFP and p65-shRNA or IKKβ-shRNA. Figure 2b shows that there was robust GFP expression in the virally transduced cells, indicating a high level of transduction efficiency and transgenic expression. There was a significant decrease in p65 and IKKβ protein (Fig. 2c, d) and mRNA (Fig. 2e) expression in cells transduced with sh-p65 and sh-IKKβ, respectively, when compared to cells transduced with the control shRNA. Importantly, suppression of NF-κB signaling components significantly decreased the inductive effects of TNF-α on circ-4099 expression (Fig. 2f).
The effects of circ-4099 on ECM degradation and inflammatory cytokine secretion in IVD
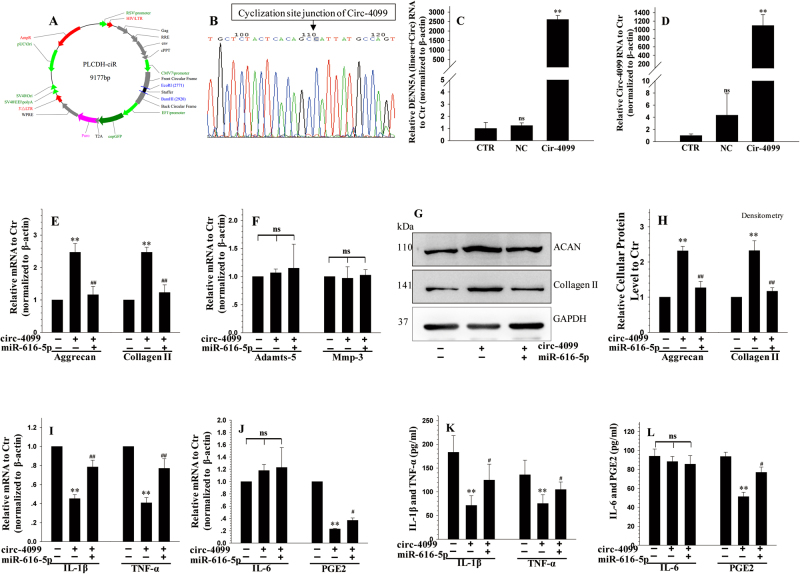
To analyze the effects of circ-4099 on ECM degradation, we examined the effects of circ-4099 over-expression in IVD NP cells. First, we designed a circ-4099 over-expression plasmid with the circ-4099 F (CGG AAT TCT GAA ATA TGC TAT CTT ACA GCA TTA TGC CAG TAC ATA CAG GCT) and circ-4099 R (CGG GAT CCT CAA GAA AAA ATA TAT TCA CGC TGT GAG TAG AGC AGG ATT TGA) primers (Fig. 3a, b). Then, hNP cells were transfected with circ-4099, NC (PLCDH-ciR) or negative control. Total RNA was collected from the transfected cells and qPCR was used to detect the expression of circ-4099 and DENND5A (linear + circRNA). The results showed that circ-4099 over-expression can extremely enhance the expression of circ-4099; the qPCR products were also used to confirm the circ-4099 sequences. These data confirmed the validity and efficiency of the circ-4099 over-expression plasmid. (Fig. 3c, d)
Fig. 3. The effects of Circ-4099 on the expression of extracellular matrix and inflammatory cytokines in hNP.
a Plasmid map of the Circ-4099 over-expression plasmid. b The qPCR products were used to confirm the Circ-4099 sequence. c, d hNP cells were transfected with Circ-4099, NC (PLCDH-ciR) or negative control, and total RNA was collected. qPCR was used to detect the expression of Circ-4099 and its linear RNA (DENND5A) (linear + circRNA). The results showed that the Circ-4099 over-expression plasmid can extremely enhance Circ-4099 expression. These data confirm the validity and efficiency of the Circ-4099 over-expression plasmid. e, f qRT-PCR showed that Circ-4099 over-expression led to increased Collagen II and Aggrecan mRNA expression, which was suppressed by treatment with miR-616-5p mimics, while MMP3 and ADAMTS5 showed few significant differences in regulation. g, h Western blotting also showed that he Collagen II and Aggrecan protein expression levels were significantly increased by circ-4099 over-expression treatment. We next tested the effects of Circ-4099 on expression of inflammatory cytokines in hNP. i, j The IL-1β, TNF-α, and PGE2 mRNA levels were significantly suppressed in hNP cells transduced with Circ-4099 over-expression plasmid, which was inhibited by treatment with miR-616-5p mimics. k, l IL-1β, TNF-α, IL-6, and PGE2 cytokine release into medium was detected by ELISA, which showed similar results
Next, we examined whether circ-D over-expression could impact the expression of Collagen II and Aggrecan, two main ECM molecules important for NP tissues. As a consequence of circ-D over-expression, Collagen II, and Aggrecan mRNA expression increased, while MMP3 and ADAMTS5 showed few changes in expression. (Fig. 3e, f)
We further performed western blot to examine the expression of Collagen II and Aggrecan at the protein level. WB and densitometry analysis with three replicates showed that the Collagen II and Aggrecan protein levels were significantly increased by circ-4099 over-expression and that this effect was reversed by co-transfection with miR-616-5p mimics in NP cells, which indicated that miR-616-5p may function as a circ-4099 sponge. (Fig. 3g,h) These results showed that the increased circ-4099 activities restored functional ECM synthesis in NP cells. However, the regulation of circ-4099 in IVDD and its target gene remain unknown.
We next tested whether circ-4099 can regulate pro-inflammatory genes, including IL-1β, TNF-α, IL-6, and PGE2, in hNP cells. Circ-4099 over-expression significantly suppressed IL-1β, TNF-α, and PGE2 mRNA levels in hNP cells, but not IL-6 (Fig. 3i, j). Additionally, miR-616-5p mimics can reverse the suppression effects of circ-4099 on pro-inflammatory genes. The release of IL-1β, TNF-α, IL-6, and PGE2 into medium was detected by ELISA, which showed similar results (Fig. 3k, l). As it has been shown that pro-inflammatory cytokines play a vital role in the pathogenic processes underlying painful IVD degenerative diseases, circ-4099 may regulate disc degeneration and LBP processes.
Annotation of circRNA–miRNA interactions and target genes
We used gene co-expression networks to predict the circRNA targets. Previous studies have demonstrated that circRNAs were correlated with miRNAs and play a crucial role in fine tuning the levels of miRNA that mediate gene expression regulation by sequestering the miRNAs. To explore how circRNAs might participate in regulating IVD degeneration, we attempted to predict circRNA–microRNA–mRNA interactions using the TargetScan20 and miRanda21 software. The differentially expressed circRNAs within the comparisons were annotated in detail with the circRNA–miRNA–mRNA interaction information. There were a total of 5 miRNAs and more than 100 mRNAs in the network. In the co-expression networks, each gene corresponds to a node and three genes are connected by a string, indicating a tight correlation between the genes and a potential regulatory relationship. Among the mRNAs, Sox9, which can positively regulate the expression of chondrocyte-specific genes, including aggrecan and collagen II22, was used for further analysis. (S Figs. 1–2).
Circ-4099 serves as a sponge for miR-616-5p
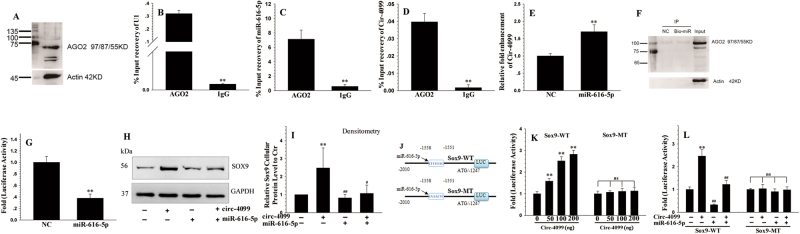
Given that circRNAs have been shown to act as miRNA sponges and that circ-4099 is abundant and stable in the cytoplasm, we next investigated the ability of circ-4099 to bind to miR-616-5p. We performed RNA-binding protein immunoprecipitation by qPCR23 to determine whether AGO2 was associated with miR-616-5p transcripts, which can bind circ-4099. Lysates from hNP cells were immunoprecipitated with anti-AGO2 antibody (0.1, 1 μg) or control IgG antibody (0.1, 1 μg) and subjected to qRT-PCR to quantify the expression of miR-616-5p and circ-4099. The results showed that the expression of AGO2 protein is high in hNP cells (Fig. 4a, b) and that endogenous miR-616-5p pulled down from Flag-AGO2 cells was specifically enriched by qRT-PCR analysis (Fig. 4c). Circ-4099 was also detected in these RNA–protein complexes (Fig. 4d), which suggested that circ-4099 could bind to miR-616-5p transcripts and be indirectly captured by anti-AGO2. We further performed RNA–RNA pull-down qPCR tests using a biotin-coupled miR-616-5p mimic to detect miR-616-5p-related RNAs and proteins. We observed an approximately twofold enrichment of circ-4099 in the miR-616-5p-captured fraction compared with the negative control (Fig. 4e), which demonstrated the binding of circ-4099 to miR-616-5p. WB showed significantly higher AGO2 protein levels in the miR-616-5p-captured fraction compared with the negative control (Fig. 4f). The co-localization experiments (FISH) showed that there is overlapped localization between circ-4099 and miR-616-5p, which supported circ-4099 interacting with miR-616-5p (Fig. 1d). We further constructed a circ-4099 fragment and inserted it immediately downstream of the luciferase reporter gene (LUC-circ-4099). We hypothesized that circ-4099-associated miRNAs may potentially inhibit the luciferase activity, presumably via the miRNA-mediated activation of deadenylation and subsequent exonucleolytic degradation. Supporting this hypothesis, we observed that miR-616-5p mimic treatment decreased LUC-circ-4099 plasmid luciferase activity (Fig. 4g). These results suggested that Circ-4099 may serve as a binding platform for AGO2 and miRNAs.
Fig. 4. Circ-4099 and miR-616-5p binding by RIP and RNA–RNA pull-down assays.
a The expression of AGO2 protein can be detected by WB in hNP cells. b RIP assay showed that AGO2 was associated with U1, which acts as a positive control. c, d RIP assay showed that the anti-Ago2 antibody efficiently captured miR-616-5p transcripts; circ-4099 was also detected in these RNA–protein complexes, which shows that it may bind to miR-616-5p transcripts and be indirectly captured by anti-Ago2. e RNA–RNA pull-down qRT-PCR showed an approximately twofold enrichment of circ-4099 in the miR-616-5p-captured fraction compared with the negative control. f WB showed significantly higher AGO2 protein in the miR-616-5p-captured fraction compared with the negative control. g miR-616-5p mimic treatment caused decreased luciferase activity with the LUC-circ-4099 plasmid. h Western blot and i corresponding densitometry analyses showed that Sox9 mRNA expression can be upregulated by circ-4099 over-expression, which can be suppressed by treatment with miR-616-5p mimics. j schematic of constructs Sox9-WT and Sox9-WT containing mutation in miR-616-5p sites generated by site-directed mutagenesis. k Co-transfection with circ-4099 results in a dose-dependent increase in Sox9-WT activity, but no significant induction at the Sox9-MT promoter. l miR-616-5p mimics blocked the inductive effects of circ-4099 on the wild-type Sox9-WT promoter, while no significant responses was observed with the Sox9-MT promoter
Circ-4099 is targets Sox9 through miR-616-5p as a ceRNA
It has been reported that circRNAs function as miRNA “sponges” that naturally sequester and competitively suppress miRNA activity12. We assumed that circ-4099 functions as a decoy to regulate Sox9 expression through the same mechanism. According to the network above, there were five miRNA-binding sites in circ-4099, including miR-508-5p, miR-561-5p, miR-616-5p, miR-328-5p, and miR-761 (S Fig. 1). The sequences in the circ-4099 3′-UTR matched these miRNAs. We identified miR-616-5p as circ-4099 and Sox9 targets through informatics analysis. The results showed that miR-616-5p also matched the Sox9 3′-UTR. To analyze the effects of circ-4099 on Sox9 expression, we examined the effects of circ-4099 over-expression in IVD NP cells. qPCR showed that Sox9 mRNA expression can be upregulated by circ-4099 over-expression, which can be suppressed by treatment with miR-616-5p mimics. (Fig. 4h,i) We inferred that circ-4099-induced Sox9 regulation by functioning as a ceRNA (competing endogenous RNAs). Furthermore, a luciferase reporter was constructed to determine whether miR-616-5p can directly target the Sox9 3′-UTR(Fig. 4j). Co-transfection with circ-4099 resulted in a dose-dependent increase in Sox9 activity (Fig. 4k), while miR-616-5p mimics blocked the inductive effects of circ-4099 on the wild-type Sox9 promoter (Fig. 4l). To confirm that Sox9 promoter activity is responsive to miR-616-5p, we evaluated the roles of the conserved miR-616-5p-binding sites in controlling Sox9 promoter activity. Using a Sox9-W fragment as a template, we generated a mutant reporter that contained a mutation in the miR-616-5p-binding sites (Fig. 4j) and examined the effects of circ-4099 and miR-616-5p mimics on its activity. In contrast to the wild-type promoter, the single site mutants were less responsive to circ-4099 or miR-616-5p mimic treatment (Fig. 4l).
Transcription factors responsible for circ-4099 expression
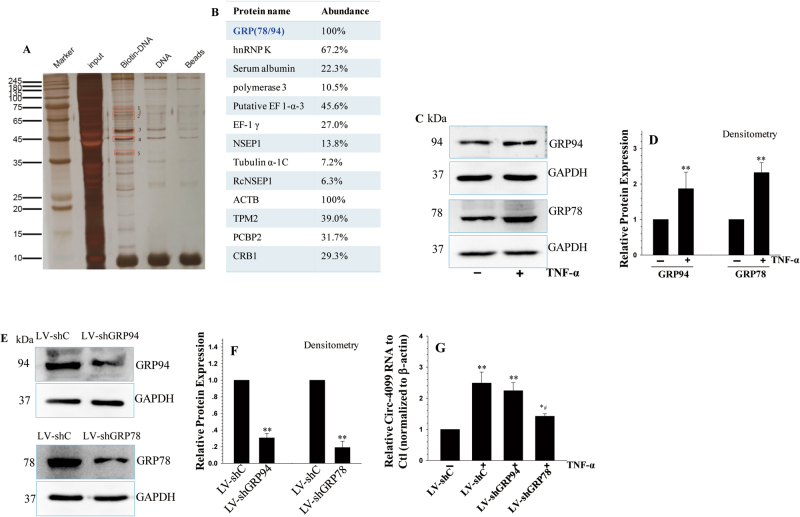
To investigate the transcription factors that are responsible for circ-4099 expression, we performed a DNA pull-down experiment in nuclear extracts using the biotin-labeled circ-4099 promoter. Nuclear proteins from hNP cells were incubated and pulled down by a biotin-labeled circ-4099 promoter (Fig. 5a). Spectrometry analyses were then performed to analyze the purified nuclear proteins. Among these proteins, we found that glucose-regulated protein had been reported to be related to IVDD (Fig. 5b). Thus, GRP78/94 was used for further analysis. Using WB analysis, we found that GRP78 protein was increased in the nucleus following TNF-α treatment (Fig. 5c, d); in contrast, no NF-κB protein was among the pulled down proteins. WB also showed no bands for p65 or IKK-β among the pulled down proteins when the nuclear extracts were incubated with anti-p65 and anti-IKKβ antibodies, respectively (data not shown). This means there may be an indirect contact between the NF-κB pathway and circ-4099. After GRP78/94 were individually silenced (Fig. 5e, f), only GRP78 dysregulation obviously affected circ-4099 expression under TNF-α treatment (Fig. 5g).
Fig. 5. Transcription factors responsible for circ-4099 expression.
a Biotin-labeled circ-4099 promoter was mixed with nuclear extracts from hNP cells. b The eluted proteins in the red box were analyzed using mass spectrometry methods. c, d The GRP78/94 protein levels in the nuclei of hNP cells treated with TNF-α for 24 h were analyzed by WB. e, f The GRP78/94 protein levels in hNP cells transfected with shRNA vectors for 48 h were analyzed by WB. g circ-4099 expression was analyzed by qRT-PCR in hNP cells transfected with the sh-GRP78 RNA vector for 48 h and then treated with TNF-α for 24 h
Discussion
circRNAs are a new enigmatic class of RNAs with unknown cell-specific functions, and the role of circRNAs in IVD remains largely unknown. To systematically explore the role of circRNAs in human IVD degeneration, we analyzed the differential expression of circRNAs in non-degenerated and degenerated IVD NP tissues. We detected thousands of well-expressed stable circRNAs, which often showed tissue-specific or developmental stage-specific expression. Arraystar human circRNA microarray analysis revealed high and specific expression of circRNAs in degenerated IVD NP tissues, with 51 circRNAs upregulated and 21 circRNAs downregulated by more than twofold in degenerated NP tissues, suggesting that circRNAs may play important roles in the initiation and development of IVD degeneration. Through gain-of-function and loss-of-function studies, we found that circ-4099 is important in the ECM degradation process in IVD degeneration. A series of experiments illustrated the correlation between circ-4099 and miR-616-5p, demonstrating that circ-4099 plays an important role as an miRNA sponge in the IVDD development process.
Circ-4099 is the most upregulated circRNA among all of the detected circRNAs, displaying a sixfold upregulation in degenerated NP tissues compared with normal NP tissues. Circ-4099 is located on exonic chromosome 11 and is aligned in a sense orientation to the known protein-coding gene DENND5A. DENND5A contains DENN domains and interacts with Rab6, Rab11, and Rab39 as well as small GTPases that are indispensable for intracellular membrane trafficking24. DENND5A is a cancer driver candidate gene and functions in epithelial cell polarity as indicated by bioinformatics analysis; it participates in both intracellular trafficking and cell cycle progression25. In this study, we found that a circular form of DENN5A is constantly expressed in human NP tissues and was increased expression during IVD degeneration. This suggested that Circ-4099 may play a role in the IVD degeneration process. We designed primers, and head-to-tail splicing was assayed by real-time RT-PCR after reverse transcription with divergent primers and Sanger sequencing. The predicted circ-4099 head-to-tail junctions were validated to show their high accuracy based on our predictions.
Previous studies have indicated that circRNAs may play important roles in the initiation and development of different diseases, especially aging and degeneration-related diseases. Westholm et al26. revealed that circRNAs substantially increase during CNS aging and that circRNAs represent a class of aging biomarkers in Drosophila. Liu et al27. identified a new circRNA involved in cartilage injury and OA development and progression, which suggests that circRNA-CER could be used as a potential target in OA therapy. The NP and cartilage share a common developmental lineage at the molecular level, which is the main functional component in IVD and consists of small chondrocyte-like cells with typical chondrocyte-like morphology19. To further document the involvement of circ-4099 in IVD degeneration, we detected the expression of circ-4099 in an IVDD cellular model created with TNF-α8. The results showed that TNF-α can induce the expression of circ-4099, which is consistent with previous data. Gain-of-function/loss-of-function studies were used to detect the exact role of circ-4099 during NP cell matrix synthesis and degradation, which is crucial for normal disc function and results in spine mechanical stability. The results showed that circ-4099 regulates the synthesis of Collagen II and Aggrecan, but not MMP3 and ADAMTS5. We also found that Sox9 is a target gene of circ-4099 and that Sox9 can positively regulate the expression of chondrocyte-specific genes, including aggrecan and collagen II22. In this way, circ-4099 may achieve regulation of NP cells through Sox9.
Previous studies have demonstrated that the most activated cytokine pathways are the NF-κB and MAPK signaling pathways. To test whether a similar pattern is observed in the regulation of circRNAs in NP cells, we co-treated NP cells with NF-κB or MAPK pathway-specific inhibitors. We first used NF-κB pathway-specific inhibitors, which led to significant suppression of the induction of circ-4099 expression by TNF-α to almost the basal level. To detect the exact NF-κB pathway sub-unit involved in this regulation, we performed lentiviral p65/IKKβ shRNA studies. The results showed that inhibition of TNF-α-dependent circ-4099 expression was followed by suppression of p65 and IKKβ signaling, highlighting the importance of the NF-κB pathway in controlling circ-4099 expression. Moreover, we observed that blocking the NF-κB pathway does not maintain the basal circ-4099 expression levels, which suggested that there might be a NF-κB-independent mechanism controlling baseline transcription. To further explore this regulatory system, we treated NP cells with MAPK pathway-specific inhibitors. The results showed that pretreatment with p38/JNK/ERK inhibitors partially counteracted the inductive effects of TNF-α, and there was differential sensitivity of circ-4099 expression to MAPK isoforms. Based on these findings, it is reasonable to assume that cytokines regulate the expression of circ-4099 by controlling the activity of specific MAPK isoforms and through NF-κB signaling in NP cells. Thus, the regulation of circ-4099 by cytokines is more complex than was originally thought.
It has been reported that circRNAs can function as miRNA sponges; however, since most circRNAs show low abundance and have short lengths, many circRNAs may not serve as miRNA sponges12,28. In this study, we found that circ-4099 harbors miRNA-binding sites for miR-508-5p, miR-561-5p, miR-616-5p, miR-328-5p, and miR-761. Although many miR target genes were predicted by miRNA software, only a few of them have been experimentally demonstrated to be direct targets in IVD degeneration. Sox9 is a miR-616-5p target gene related to IVD degeneration. Several studies have reported that miR-616 might play important roles in regulating cell proliferation, cell growth and migration, chondrogenic differentiation of human adipose-derived stem cells, and chondrogenesis29,30. Previous studies have demonstrated that decreased Sox9 expression in NP cells is associated with IVD degeneration31. Sox9 is a master regulator of NP cell differentiation, and Sox9 over-expression can enhance the expression of typical matrix protein-coding genes (Col2, Aggrecan, Versican, etc.)32,33. Sox9 over-expression can also reverse the upregulation of MMP-3, MMP-13, ADAMTS-5 induced by IL-1β34, suggesting that Sox9 could be a potential therapeutic target. The pivotal role of Sox9 in ECM degradation has been reported to be regulated by microRNAs, such as MicroRNA-30a, which promotes ECM degradation in articular cartilage via downregulation of Sox935; miR-101, which promotes condylar cartilage cell apoptosis by inhibiting Sox936; and MicroRNA-145, which regulates MSC chondrogenic differentiation by targeting Sox937. As the circRNA mechanism is complicated and many other molecules participate in the IVD degradation process, in this study, we only focused on miR-616-5p, which targets both circ-4099 and Sox9. To verify this speculation, the binding effects of circ-4099 and miR-616-5p were demonstrated by FISH, RIP for AGO2, a RNA–RNA pull-down-qPCR assay and a luciferase reporter gene assay. Gain-of-function and loss-of-function studies were performed to demonstrate that circ-4099-regulated Sox9 expression.
Using DNA pull-down and spectrometry experiments, we found that TNF-α can promote circ-4099 transcription through GRP78 upregulation. GRP78 is known to be expressed in early embryonic development38 and protects against cell death by increasing the protein-folding capacity in the ER or redistributes to mitochondria to form complexes with BIK39. Our previous study indicated that the increased GRP78 in degenerative discs can be considered to be a negative-feedback mechanism to reset the stress sensors in the ER, which leads to increased ER protein-folding capacity, maintenance of disc cell integrity and survival4. The exact role and dedicated mechanism require further analysis.
In our study, we constructed a network that included circRNAs and mRNAs: a circRNA–miRNA–mRNA network. The network indicated the potential association between circRNAs and their target genes, and provided an important reference for studying the interactions between other differentially expressed circRNAs and their potential targets.
Collectively, this is the first systematic analysis of circRNA expression and its roles in IVD NP tissues. Our data indicate that 72 circRNAs were either over-expressed or under-expressed by more than twofold in IVDD, with Circ-4099 increased by approximately sixfold, which may have biological consequences on the progression of IVD degeneration. Over-expression of circ-4099 increased the expression of collagen II and Aggrecan. TNF-α promoted Circ-4099 expression through the MAPK and NF-κB pathways. We further confirmed that circ-4099 functions as a “sponge” by competitively binding miR-616-5p, which reverses the suppression of Sox9 by miR-616-5p, and participates in ECM synthesis in NP cells.
A previous study showed that increased Sox9 can positively regulate the expression of chondrocyte-specific genes, including aggrecan and collagen II22. These results indicated that circ-4099 may be a protective gene. Thus, controlling circ-4099 activities during disc disease may restore NP cell functions and delay or reverse the IVDD process. Further studies should focus on the exact molecular mechanisms and pathophysiological roles of circ-4099 in IVDD as a potential new therapeutic strategy for IVDD treatment.
Electronic supplementary material
Supplement Construction of reporter plasmid
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81401840 and 81371984), Sun Yat-sen University Starting Funds for Young Teachers (Grant No.16ykpy31), and the Guangdong Province Medical research fund project (No. 2016A030313259).
Author contributions
W.H. contributed to the study design and microarray analysis, identified specific circRNAs, and wrote the manuscript. H.P.H. and W.J.R. contributed to the cell regulation experiments and the RNA FISH analysis. P.H.H. collected the clinical samples and performed the RNA–protein interaction analysis. L.J., L.Z.M., and L.H. performed the in vitro analysis experiments. J.W.Y. and Z.Z.M. contributed to the study design, collected the data and performed the data analysis. All the authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Hua Wang and Peiheng He.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s12276-018-0056-7.
Contributor Information
Hua Wang, Phone: +0086(20)-87331675, Email: wangxucheng@gmail.com.
Weiying Jiang, Phone: +0086(20)-87331928, Email: Jiangwy@mail.sysu.edu.cn.
Zhaomin Zheng, Phone: +0086(20)-87331675, Email: zhengzm1@163.com.
References
- 1.van Dieen JH, Kuijer PP, Burdorf A, Marras WS, Adams MA. Non-specific low back pain. Lancet. 2012;379:1874. doi: 10.1016/S0140-6736(12)60803-4. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, et al. Tumor necrosis factor-alpha- and interleukin-1beta-dependent matrix metalloproteinase-3 expression in nucleus pulposus cells requires cooperative signaling via syndecan 4 and mitogen-activated protein kinase-NF-kappaB axis: implications in inflammatory disc disease. Am. J. Pathol. 2014;184:2560–2572. doi: 10.1016/j.ajpath.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livshits G, et al. Lumbar disc degeneration and genetic factors are the main risk factors for low back pain in women: the UK Twin Spine Study. Ann. Rheum. Dis. 2011;70:1740–1745. doi: 10.1136/ard.2010.137836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, et al. Role of death receptor, mitochondrial and endoplasmic reticulum pathways in different stages of degenerative human lumbar disc. Apoptosis. 2011;16:990–1003. doi: 10.1007/s10495-011-0644-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang SZ, Rui YF, Lu J, Wang C. Cell and molecular biology of intervertebral disc degeneration: current understanding and implications for potential therapeutic strategies. Cell Prolif. 2014;47:381–390. doi: 10.1111/cpr.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts S, Evans H, Trivedi J, Menage J. Histology and pathology of the human intervertebral disc. J. Bone Jt. Surg. Am. 2006;88(Suppl 2):10–14. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 7.Sun F, Qu JN, Zhang YG. Animal models of disc degeneration and major genetic strategies. Pain Physician. 2013;16:E267–E275. [PubMed] [Google Scholar]
- 8.Wang H, et al. Inflammatory cytokines induce NOTCH signaling in nucleus pulposus cells: implications in intervertebral disc degeneration. J. Biol. Chem. 2013;288:16761–16774. doi: 10.1074/jbc.M112.446633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Li F, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XO, et al. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 18.Risbud MV, et al. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell Biochem. 2006;98:152–159. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 19.Hunter CJ, Matyas JR, Duncan NA. Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: a species comparison. J. Anat. 2004;205:357–362. doi: 10.1111/j.0021-8782.2004.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 22.Li P, et al. Estrogen enhances matrix synthesis in nucleus pulposus cell through the estrogen receptor beta-p38 MAPK pathway. Cell Physiol. Biochem. 2016;39:2216–2226. doi: 10.1159/000447915. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J. Biol. Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Cancer driver candidate genes AVL9, DENND5A and NUPL1 contribute to MDCK cystogenesis. Oncoscience. 2014;1:854–865. doi: 10.18632/oncoscience.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westholm JO, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, et al. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Sci. Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybak-Wolf A, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Zhang D, et al. MicroRNA-616 promotes the migration, invasion and epithelial-mesenchymal transition of HCC by targeting PTEN. Oncol. Rep. 2016;35:366–374. doi: 10.3892/or.2015.4334. [DOI] [PubMed] [Google Scholar]
- 30.Ma S, et al. MicroRNA-616 induces androgen-independent growth of prostate cancer cells by suppressing expression of tissue factor pathway inhibitor TFPI-2. Cancer Res. 2011;71:583–592. doi: 10.1158/0008-5472.CAN-10-2587. [DOI] [PubMed] [Google Scholar]
- 31.Gruber HE, Norton HJ, Ingram JA, Hanley EN., Jr. The SOX9 transcription factor in the human disc: decreased immunolocalization with age and disc degeneration. Spine. 2005;30:625–630. doi: 10.1097/01.brs.0000155420.01444.c6. [DOI] [PubMed] [Google Scholar]
- 32.Lee YJ, Kong MH, Song KY, Lee KH, Heo SH. The relation between Sox9, TGF-beta1, and proteoglycan in human intervertebral disc cells. J. Korean Neurosurg. Soc. 2008;43:149–154. doi: 10.3340/jkns.2008.43.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun W, et al. Sox9 gene transfer enhanced regenerative effect of bone marrow mesenchymal stem cells on the degenerated intervertebral disc in a rabbit model. PLoS ONE. 2014;9:e93570. doi: 10.1371/journal.pone.0093570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H, et al. Lentiviral vector-mediated over-expression of Sox9 protected chondrocytes from IL-1beta induced degeneration and apoptosis. Int J. Clin. Exp. Pathol. 2015;8:10038–10049. [PMC free article] [PubMed] [Google Scholar]
- 35.Chang T, et al. MicroRNA-30a promotes extracellular matrix degradation in articular cartilage via downregulation of Sox9. Cell Prolif. 2016;49:207–218. doi: 10.1111/cpr.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Wang ZX, Wang ZS, Li QF. Effect of microRNA-101 on apoptosis of rabbit condylar cartilage cells by inhibiting target gene SOX9. Asian Pac. J. Trop. Med. 2015;8:502–505. doi: 10.1016/j.apjtm.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Yang B, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS ONE. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun FC, et al. Localization of GRP78 to mitochondria under the unfolded protein response. Biochem J. 2006;396:31–39. doi: 10.1042/BJ20051916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang YH, Zhao CQ, Jiang LS, Dai LY. Cyclic stretch-induced apoptosis in rat annulus fibrosus cells is mediated in part by endoplasmic reticulum stress through nitric oxide production. Eur. Spine J. 2011;20:1233–1243. doi: 10.1007/s00586-011-1718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Construction of reporter plasmid