Abstract
Colorectal cancer (CRC) is a disease involving a variety of genetic and environmental factors. Sirtuin-3 (Sirt3) is expressed at a low level in cancer tissues of CRC, but it is unclear how Sirt3 modulates colonic tumorigenesis. In this study, we found that gut microbiota play a central role in the resistance to CRC tumor formation in wild-type (WT) mice through APC (Adenomatous Polyposis Coli)-mutant mouse microbiota transfer via Wnt signaling. We also found that Sirt3-deficient mice were hypersusceptible to colonic inflammation and tumor development through altered intestinal integrity and p38 signaling, respectively. Furthermore, susceptibility to colorectal tumorigenesis was aggravated by initial commensal microbiota deletion via Wnt signaling. Mice with Sirt3-deficient microbiota transfer followed by chemically induced colon tumorigenesis had low Sirt3 expression compared to WT control microbiome transfer, mainly due to a decrease in Escherichia/Shigella, as well as an increase in Lactobacillus reuteri and Lactobacillus taiwanensis. Collectively, our data revealed that Sirt3 is an anti-inflammatory and tumor-suppressing gene that interacts with the gut microbiota during colon tumorigenesis.
Colorectal cancer: Beneficial bacteria prompt anti-tumor gene response
Boosting specific beneficial bacteria in the gut may enhance expression levels of a tumor-suppressing gene in colorectal cancer (CRC). Both genetic factors and the bacteria present in the gut play vital roles in CRC development. However, it is unclear exactly how genes interact with the bacteria to affect tumor growth. Man-tian Mi and co-workers at the Third Military Medical University in Chongqing, China, examined the role of a gene called Sirt-3 in CRC development. Mice lacking the Sirt-3 gene suffered severe chronic inflammation and developed tumors due to altered signalling pathways and reduced intestinal integrity. Further, the guts of the mice harboured more pathogenic bacteria than wild-type mice. The team also found lower levels of two key types of beneficial bacteria that would ordinarily prevent reduced Sirt-3 expression.
Introduction
Colorectal cancer (CRC) is a complex multifactorial disease with different incidences around the world1. Both genetic and environmental factors contribute to the pathogenesis of CRC, and a series of gene–environment interactions have been shown to contribute to disease susceptibility2,3. In recent years, with the development of high-throughput sequencing technology, the gut microbiome has emerged as a central environmental factor in CRC progression4–7. However, only a few CRC-related gene–environment interactions have been found, and it remains unclear how host genes interact with the gut microbiome.
Sirtuin-3 (Sirt3), an NAD+-dependent deacetylase, plays a role in metabolism regulation and energy balance. Recent studies have highlighted the importance of mitochondrial Sirt3 in the regulation of ROS8. Tissue-specific deletion of Sirt3 in murine models has shown that Sirt3 performs critical roles in complex diseases, such as metabolic syndrome and diabetes9,10. However, the function of Sirt3 in CRC is largely unknown. High expression of the gene encoding Sirt3 is negatively associated with lymph node metastasis and the tumor stage of CRC patients11. Sirt3 exerts a tumor-suppressive function in cancer cells12. Thus, further exploration of the biological function of Sirt3 in colon tumorigenesis could provide novel strategies for its treatment.
Sirtuins, such as Sirt1, have been linked to altered gut microbiota and susceptibility to colon tumorigenesis13,14. The human gut harbors trillions of microbes and plays an important role in the development and progression of CRC15. Various studies further support that there are multiple mechanisms by which gut microbiota contribute to CRC tumorigenesis, including the infection of direct enterotoxigenic bacterial pathogens, triggering of the T helper type-17 inflammatory response, carcinogenic Toll-like receptor signaling and production of microbial oncogenic metabolites16.
The aim of this study was to identify the tumor-suppressive function of Sirt3 using Sirt3-deleted murine models with colitis-induced tumorigenesis. Furthermore, we compared the gut microbiota of Sirt3-deficient and WT mice to explore the mechanisms of CRC pathogenesis. Finally, we removed or transferred gut microbiota to reveal host-microbiota interactions in CRC.
Materials and methods
Breeding and genotyping
Sirt3 knockout (SIRT3KO) mice (129/SvlmJ) were purchased from the Jackson Laboratory (Stock Number 012755, Bar Harbor, ME, USA), and male APC min/+ mice (Adenomatous Polyposis Coli,C57BL/6J) were obtained from the Model Animal Research Center of Nanjing University (Nanjing, China). Female WT mice (129/SvlmJ) were crossed with male SIRT3KO mice to generate the heterozygous females or hemizygous males, which were subsequently crossed to generate male Sirt3−/− mice and littermates of Sirt3+/+ mice. Age-matched male WT (129/SvlmJ or C57BL/6J) mice were used for fecal microbiota transplant by initial microbiota removal using an antibiotic cocktail for 3 weeks. The antibiotic treatment was administered via drinking water and consisted of a mixture of 0.2 g L−1 ampicillin, 0.1 g L−1 vancomycin, 0.2 g L−1 neomycin and 0.2 g L−1 metronidazole. The mice were maintained under specific pathogen-free (SPF) conditions in an animal experimental center at the Third Military Medical University (Chongqing, China). The protocol was approved by the Animal Care and Use Committee of the Third Military Medical University. Genotyping was done by routine PCR assays on tail DNA using an animal tissue DNA extraction kit and master mix (Tiangen, Beijing, China). The specific primers used to characterize the WT, knockout or mutant mice are listed in Supplemental Table S1.
Experimental design
In the initial experiment, male SIRT3KO or WT littermates (8 weeks old) were subjected to dextran sulfate sodium (DSS)-induced colitis by adding DSS (MW 36–40 kDa, MP Biologicals) to drinking water at a 2.5% weight volume for 6 days (Supplementary Figure 1). In the gene susceptibility cancer experiment, SIRT3KO mice and WT mice were exposed to an AOM/DSS colon tumor cycle and four DSS cycles, which is one additional cycle than previously described17. In the microbiota susceptibility cancer experiment, WT mice and WT mice colonized with 8-week-old male APC min/+ mice microbiota (multiple antibiotics for 3 weeks plus 2 week microbiota adjustment) were exposed to AOM/DSS colon tumor cycles. In the gut commensal microbiota cancer protection experiment, SIRT3KO mice and SIRT3KO mice receiving multiple antibiotics (from week 7 to week 10) were treated by AOM/DSS cycles as mentioned above. In the gene–microbiota interaction experiment, male C57BL/6J mice first received multiple antibiotics for 3 weeks, and the fecal microbiota of 10-week-old male SIRT3KO or WT littermate mice were then transferred for 2 weeks (once a week) according to a previous study18. Freshly obtained feces were diluted 1/100 (wt/vol) in sterile 0.01 M phosphate-buffered saline (PBS, pH 7.4), vortexed for 2 min, and centrifuged for 5 min at 400 × g. The upper suspensions were obtained and washed with PBS three times, and bacterial suspension was used for intragastric administration. Finally, microbiota-transferred C57BL/6J mice were exposed to AOM/DSS colon tumor cycles.
Quantification of cytokines
Colonic homogenates were obtained by a tissue homogenizer (IKA, Germany). Colonic myeloperoxidase (MPO) activity (Abcam, Inc.) and serum cytokine levels, including IL-1β (R&D Systems, Inc.), TNF-α (BioLegend, Inc.) and IL-6 (BioLegend, Inc.), were quantified by ELISA.
Western blot analysis
Proteins were extracted from colon tissues using tissue lysis buffer (Thermo Scientific) supplemented with a protease inhibitor cocktail (Roche Diagnostics). The samples were resolved in 10–15% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked in 5% nonfat milk for 1 h, and primary antibodies were incubated with the membranes separately under rotation overnight at 4 °C. The membranes were incubated with HRP-conjugated secondary antibody for 1 h, and the proteins were visualized using a chemoluminescence system (FUSION, France) with Millipore Immobilon ECL substrate (Millipore, Inc.). The primary antibodies were Caspase-3 (1:1000 dilution, Beyotime, China), β-catenin (1:1000 dilution, Beyotime), HIF-1α (1:1000, 3716, Beyotime, China), p38 (1:1000, Abcam), Claudin 8 (1:1000, Abcam), CLCN3 (1:1000, Abcam), Foxo3 (1:1000, Abcam), ERK (1:1000, Abcam), JNK (1:1000, Abcam), CLIC4 (1:1000, Abcam), CLCN4 (1:1000, Sigma), SLC26A2 (1:1000, Sigma), Sirt3 (1:1000, Cell Signaling), phospho-p53 (1:1000, Cell Signaling), GSK-3β (1:1000, Santa Cruz), Claudin15 (1:1000, Santa Cruz), Occludin (1:1000, Santa Cruz) and β-actin (1:1000, Bestbio, China).
Histological evaluation and immunofluorescence staining
Mouse colonic tissues were fixed in 10% neutral formalin followed by gradient dehydration. The tissues were then embedded in paraffin and sectioned at 5 μm thickness. The sectioned slides were stained with hematoxylin-eosin (HE) for microscopic assessment (Olympus, Tokyo, Japan). Histological score was evaluated blindly according to a previous research with minor modification19. Briefly, inflammation severity, ulceration and hyperplasia account for 30, 30, and 40%, respectively. Each field was divided into 5 grade levels (0, 1, 2, 3, and 4).
For immunofluorescence staining, colon sections were first treated with an antigen-retrieval solution (Beyotime) for 5 min followed by a wash with by phosphate-buffered saline (PBS) containing 0.1% Triton X-100 for 10 min. The sections were then blocked with 20% rabbit serum for 1 h. After incubation with primary antibody against Claudin15 or ERK at 4 °C overnight, the samples were incubated for 30 min with appropriate FITC- or Cy3-labelled secondary antibodies (Invitrogen, Thermo Scientific) at room temperature for 1 h. The nuclei were stained with DAPI at a concentration of 100 g mL−1. The slides were then examined with a Leica confocal microscope (Leica Inc., Germany), and images were analyzed with LAS Lite (Leica Inc., Germany).
Mouse fecal occult blood tests
Fecal occult blood tests were performed using freshly collected feces with a commercial kit (Nanjing Jiancheng Bioengineering Institute, Nanjing). Briefly, o-toluene solution and hydrogen peroxide were mixed with mouse feces, and the color changes were observed. The results were classified by the following 5 levels: solution turned blue within 3 min (0, negative), 2 min (1, weak positive), 30–60 s (2, positive), blue immediately (3), and dark blue immediately (4, strongly positive).
16S rRNA gene sequencing
DNA from stool and intestinal contents (QIAamp DNA Stool Mini Kit, Germany) was amplified using barcoded V3–V4 region primers targeting the bacterial 16S rRNA gene, sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA), analyzed using QIIME, and assessed by multiple parameters with STAMP software. For third-generation sequencing, purified fecal DNA was amplified using bacterial 16S rRNA full-length primers and sequenced using Pacbio RSII system (Pacific Biosciences, USA). Principal coordinates analysis (PCoA) was performed using R software.
Fluorescence in situ hybridization (FISH) with bacteria
Specific oligonucleotide probes were synthesized according to previous studies with the primers listed in Supplemental Table S220,21. Microbial FISH was performed as previously described22. Colon sections were fixed in 10% neutral formalin for 10 min, and they were then treated with 1 mg mL−1 lysozyme in 100 mM Tris–HCl (pH 7.2) for 10 min at room temperature. The sections were subsequently washed in PBS buffer (pH 7.2) and equilibrated in a FISH solution (900 mM NaCl, 20 mM Tris–HCl [pH 8.0], 0.01% sodium dodecyl sulfate, and 30% formamide). FISH was performed on slides overnight at 55 °C in a humidity chamber with 150 μL of FISH solution containing 4 μl of probe (50 ng/μl), reaching a final concentration of 4 ng μL−1. Finally, DAPI was added to the wash solution to a final concentration of 100 ng mL−1 for staining the nuclei.
Results
Sirt3 diminishes susceptibility to Colitis
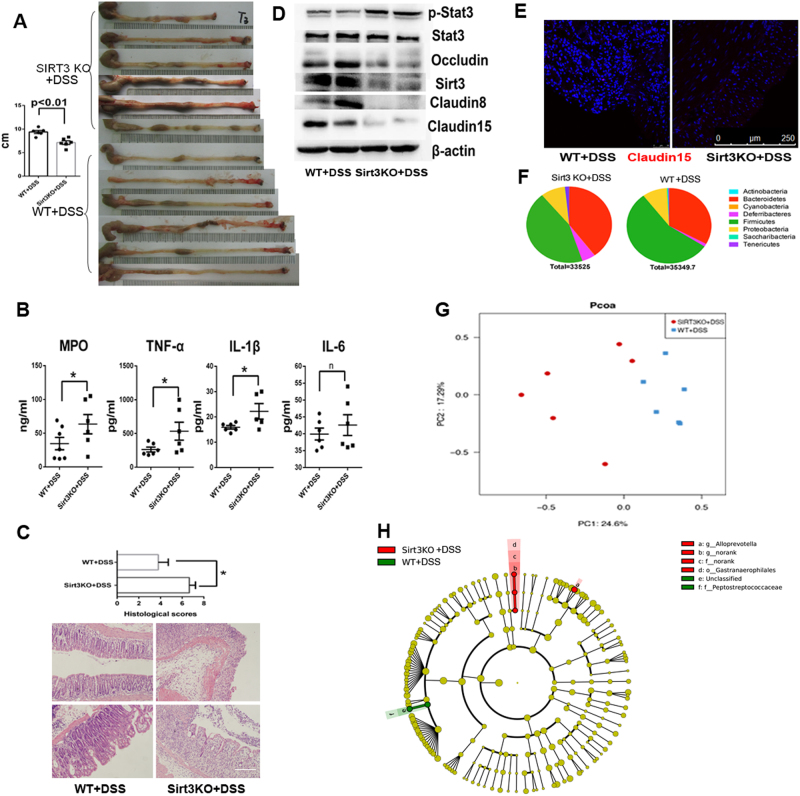
SIRT3KO mice subjected to DSS exhibited exacerbated colonic shortening (p < 0.01, Fig. 1a) and increased inflammatory cytokine levels compared to WT mice (MPO, IL-1β and TNF-α; p < 0.05, Fig. 1b). SIRT3KO + DSS mice displayed increased histological damage in the mucosal integrity of large intestine compared to WT mice (p < 0.05, Fig. 1c). The leaky gut phenotype is characterized by altered expression of tight junction components, including Occludin, Claudin8 and Claudin1523. Consistently, colons from SIRT3KO+DSS mice had decreased gene expression of Occludin, Claudin8 and Claudin15 compared to WT mice (Fig. 1d), demonstrating a deficit in intestinal barrier integrity. Consistent with the genotyping results, the colonic expression of Sirt3 significantly decreased in SIRT3KO+DSS mice compared to WT mice (Fig. 1d). Most notably, phosphorylated STAT3 was increased in SIRT3KO+DSS mice compared to WT mice (Fig. 1d). Immunofluorescence staining also demonstrated a low Claudin15 level in SIRT3KO+DSS colons (Fig. 1e). Additionally, SIRT3KO+DSS mice consistently exhibited increased levels of Bacteroides/Firmicutes compared to WT mice (Fig. 1f). PCoA analysis of 16S rRNA gene sequencing showed an alteration of the microbiome profiling in the colonic content in SIRT3KO+DSS mice (Fig. 1g). Furthermore, the microbial communities of Alloprevotella and an unclassified genus from the Gastranaerophilales order were significantly higher in SIRT3KO+DSS mice than in WT mice (P < 0.05, Fig. 1h). Collectively, these results identify a critical role for Sirt3 in actively regulating mucosal barrier function and gut homeostasis in mice.
Fig. 1. Sirt3 deficiency results in colitis aggravation.
a Gross pathology of SIRT3KO+DSS and WT+DSS mice colons on day 6 post-DSS. Note the extensive shortening in SIRT3KO mice (n = 6). b Inflammatory cytokine (MPO, IL-1β, TNF-α and IL-6) levels in SIRT3KO and WT mice on day 6 post-DSS (n = 6). Data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. c Western blots of intestinal epithelial proteins (Occuludin, Sirt3, Stat3, ph-Stat3, Claudin8 and Claudin15) from the colons of SIRT3KO or WT mice after treatment with DSS on day 6 post-treatment. d Histological scores and representative HE staining of distal colon sections obtained from SIRT3KO or WT mice after treatment with DSS. e Comparison of phylum-level proportional abundance from Illumina Miseq sequencing. f Featured altered microbial communities between SIRT3KO and WT mice after treatment with DSS (n = 6) from Illumina Miseq sequencing. The data were analyzed by the Mann–Whitney test. g Principal coordinates analysis (PCoA) of 16S rRNA sequences of gut microbiota obtained from SIRT3KO+DSS and WT+DSS mice (n = 6) from Illumina Miseq sequencing. PC1 and PC2 explain 24.60 and 17.29% of the variation, respectively. h Immunofluorescence staining of Claudin15 (red) and nuclei (blue) in the colon. Scale bars, 10 mm. n no significance
Resistance of WT mice to Colitis-associated cancer is diminished by APC min+/− mice microbiome transfer
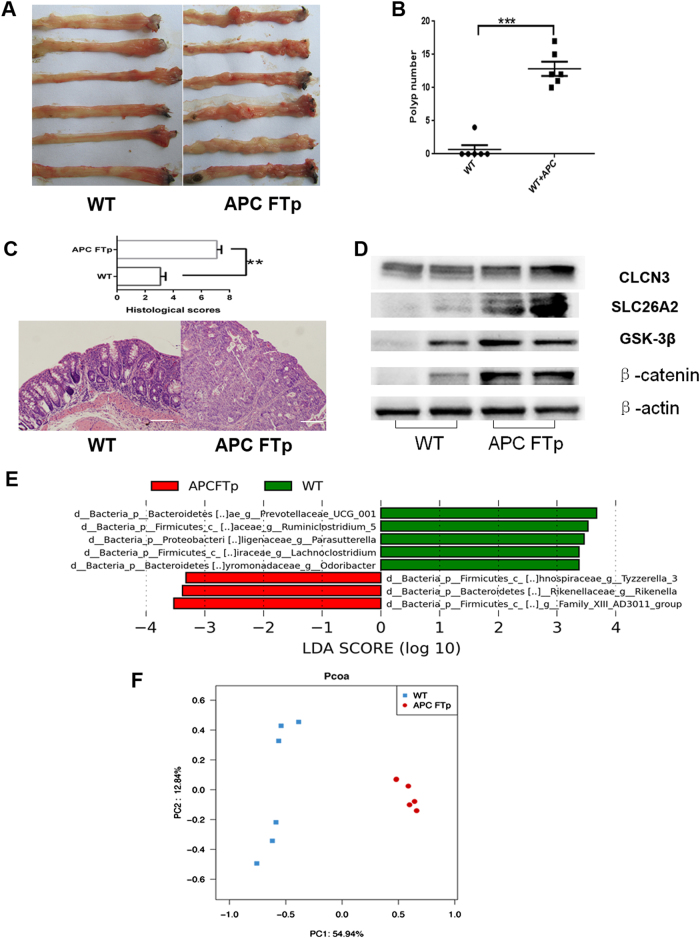
WT (129/SvlmJ) mice subjected to AOM/DSS tumorigenesis exhibited resistance to colonic tumor formation, while WT mice colonized with adult APC min+/− mice microbiota developed a significantly increased number of intestinal polyps (P < 0.001, Fig. 2a, b) and more histological tumor burden in the large intestine (P < 0.01, Fig. 2c). Chloride ion-dependent CLCN3 has been shown to prevent colon tumorigenesis in a previous study24. In the present study, there was no apparent difference of colonic CLCN3 expression in APC FTp mice and WT mice (Fig. 2d), but the expression of SLC26A2, which contributes to the intestinal exchange of chloride and sulfate, was increased in the colon of APC FTp mice and influenced the sulfate transport. Moreover, colons from APC FTp mice showed increased gene expression of GSK3β and β-catenin compared to WT mice (Fig. 2d), demonstrating an activation of the Wnt signaling pathway. With regard to the gut microbiome analysis, WT mice harbored high levels of Prevotellaceae UCG-001, Lachnoclostridium, Odoribacter, Ruminiclostridium and Parasuttella, while APC FTp mice harbored high levels of an unknown genus belonging to Family XIII-AD3011, Rikenella and Tyzzerella bacteria (Fig. 2e). Above the family level, APC FTp mice harbored increased levels of Peptococcaceae and Actinobacteria, as well as a decreased level of Gamma-proteobacteria (Fig. S2). There was also a significant difference in the microbiome profiling between APC FTp mice and WT mice (Fig. 2f). Collectively, these results indicated a key role for the gut microbiome in promoting susceptibility to colon cancer in mice.
Fig. 2. Microbiota modify the host genotype to colon cancer.
a Images of the distal colon from APC FTp and WT mice after AOM/DSS tumorigenesis. b Comparison of polyp numbers between APC FTp and WT mice (n = 6). Data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. c Histological scores and representative HE staining of distal colon sections obtained from APC FTp and WT mice after AOM/DSS tumorigenesis. d Western blot of intestinal CLCN3, SLC26A2, GSK3β and β-catenin expression from APC FTp or WT mice after AOM/DSS tumorigenesis. e Featured altered microbial communities between APC FTp and WT mice after AOM/DSS tumorigenesis (n = 6) from Illumina Miseq sequencing. Data were analyzed by the Mann–Whitney test. f PCoA of 16S rRNA sequences of gut microbiota obtained from the APC FTp and WT mice (n = 6) from Illumina Miseq sequencing. PC1 and PC2 explain 54.94 and 12.84% of the variation, respectively. n no significance
Sirt3 knockout mice are hypersusceptible to colonic tumor development
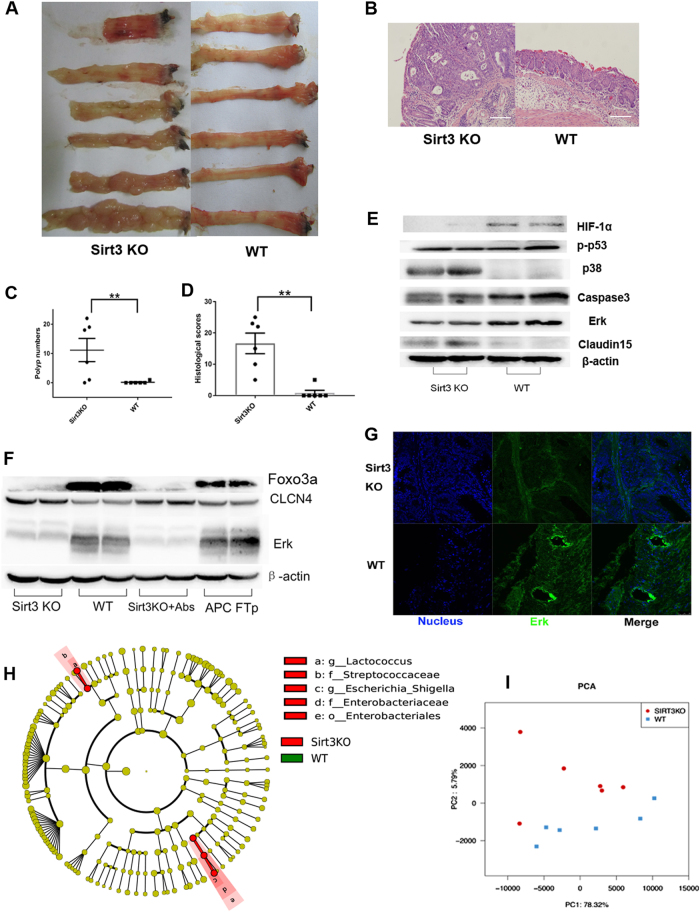
To evaluate the tumor-suppressive role of Sirt3, SIRT3KO and WT mice were subjected to AOM/DSS tumorigenesis (Fig. S3). As expected, SIRT3KO mice showed increased polyp numbers and an increased severity of colonic proliferation compared to WT mice (Fig. 3a–d), both of which suggest a tumor-suppressive role of Sirt3. Previous in vitro cancer cell studies have shown that Sirt3 regulates HIF-1α to suppress ROS and reduces intracellular NAD+ levels to repress Erk1/2 and upregulate p53 as well as activation of caspase-3 to induce cell death12. We investigated the expression of p-p53, HIF1-α, ERK, Caspase3 and p38 in the colon carcinogenesis of SIRT3KO and WT mice. SIRT3KO mice showed decreased expression of HIF1-α, ERK and Caspase3 but increased intestinal expression of p38 and Claudin15 compared to WT mice (Fig. 3e, f). We also found that SIRT3KO mice had lower liver ERK expression (Fig. 3g). CLCN4 has been implicated in the development of colon cancer, and higher CLCN4 expression has been observed in Sirt3-deficient mice (Fig. 3f)25. Consistently, the Sirt3 target protein, FOXO3a, was downregulated in Sirt3-deficient mice (Fig. 3f). Microbiome analysis revealed that Escherichia/Shigella and Lactococcus were more abundant in SIRT3KO samples than WT samples (P < 0.05, Fig. 3h). There was also a significant difference in the microbiome profiling between SIRT3KO and WT mice (Fig. 3i).
Fig. 3. Sirt3 deficiency results in increased susceptibility to colonic tumors.
a Images of distal colons from SIRT3KO and WT mice after AOM/DSS tumorigenesis. b Representative HE staining of distal colon sections obtained from SIRT3KO and WT mice after AOM/DSS tumorigenesis. c Comparison of polyp numbers between SIRT3KO and WT mice (n = 6). Data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. d Histological scores of distal colon sections from SIRT3KO and WT mice after AOM/DSS tumorigenesis (n = 6). e Western blots of intestinal HIF1-α, Erk, p-p53, p38, Claudin15 and caspase3 expression from SIRT3KO and WT mice after AOM/DSS tumorigenesis. f Western blots of liver CLCN4 and Erk expression from SIRT3KO and WT mice after AOM/DSS tumorigenesis. g Immunofluorescence staining of Erk (green) and nuclei (blue) in colons. Scale bars, 10 mm. h Featured altered microbial communities between SIRT3KO and WT mice after AOM/DSS tumorigenesis (n = 6) from Illumina Miseq sequencing. Data were analyzed by the Mann–Whitney test. i PCoA of 16S rRNA gene-sequencing analysis of gut microbes obtained from the indicated mice and treatments (n = 6) from Illumina Miseq sequencing. PC1 and PC2 explain 78.32 and 5.79% of the variation, respectively. n no significance
Presence of commensal microbiota reduces the incidence of colon tumors
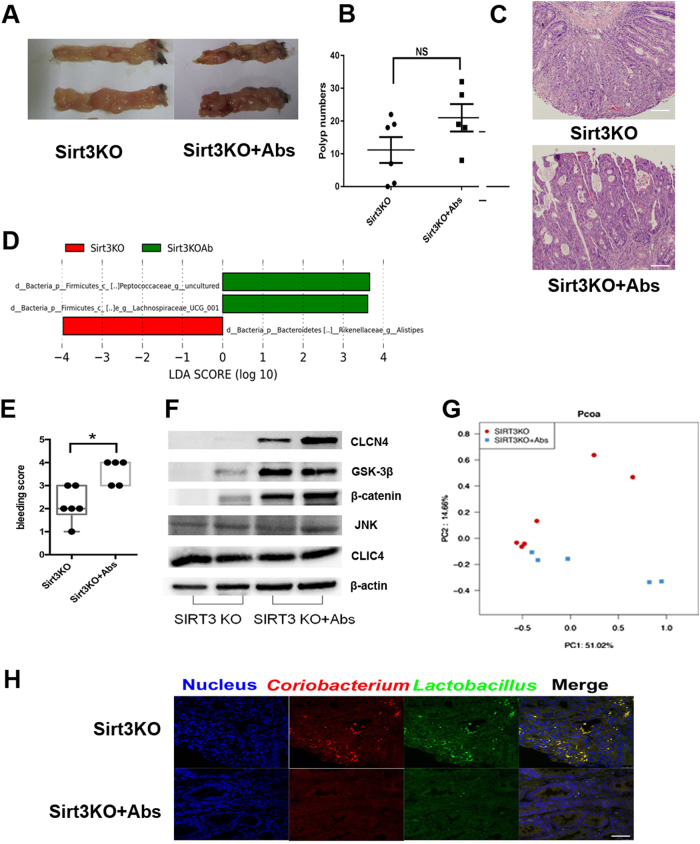
To determine if the presence of commensal microbiota influences AOM/DSS tumorigenesis, we pre-treated SIRT3KO mice (genotyping by Fig. S3) with a broad-spectrum antibiotic cocktail (vancomycin, neomycin, ampicillin and metronidazole) for initial microbiota removal and then subjected them to AOM/DSS cycles. As a result, the incidence of tumors increased, and mucosal bleeding was enhanced in antibiotic-treated mice (Fig. 4a–c). Evaluation of key bacterial taxa that discriminate SIRT3KO mice from antibiotic-treated mice revealed that the presence of commensal microbiota significantly altered the levels of several OTUs (Fig. 4d). Specifically, the genus Alistipes was more abundant in SIRT3KO mice, while an uncultured genus (belonging to the Peptococcaceae family) and genus Lachnospiraceae UCG-001 were more abundant in SIRT3KO+Abs mice (Fig. 4d). Intestinal bleeding was promoted in SIRT3KO+Abs mice as indicated by occult blood tests (Fig. 4e). Compared to SIRT3KO mice, SIRT3KO+Abs mice showed enhanced intestinal CLCN4, GSK3β and β-catenin expression but not JNK and CLIC4 expression (Fig. 4f). There was also a significant difference in the microbiome profiling between SIRT3KO and SIRT3KO+Abs mice (Fig. 4g). Additionally, SIRT3KO mice exhibited a gut tumor-probiotic effect with some beneficial bacterial taxa, such as Coriobacterium and Lactobacillus, which were enriched on tumor samples (Fig. 4h).
Fig. 4. Commensal microbiota protect against severe colon cancer in SIRT3KO mice.
a Images of distal colons from SIRT3KO and SIRT3KO+Abs mice after AOM/DSS tumorigenesis. b Comparison of polyp numbers between SIRT3KO and SIRT3KO+Abs mice (n = 5–6). The data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. c Representative HE staining of distal colon sections obtained from SIRT3KO and SIRT3KO+Abs mice after AOM/DSS tumorigenesis. d Featured altered microbial communities between SIRT3KO and SIRT3KO+Abs mice after AOM/DSS tumorigenesis (n = 5–6) from Illumina Miseq sequencing. Data were analyzed by the Mann–Whitney test. e Fecal occult blood tests of SIRT3KO and SIRT3KO+Abs mice (n = 5–6). f PCoA of 16S rRNA gene-sequencing analysis of gut microbes obtained from the SIRT3KO and SIRT3KO+Abs mice (n = 5–6) from Illumina Miseq sequencing. PC1 and PC2 explain 51.02 and 14.66% of the variation, respectively. g Western blots of intestinal CLCN4, JNK, CLIC4, GSK3β and β-catenin expression from SIRT3KO and SIRT3KO+Abs mice after AOM/DSS tumorigenesis. h FISH with Lactobacillus (green) and Coriobacterium (red) from distal colons of SIRT3KO and WT mice after AOM/DSS tumorigenesis. n no significance
Transmitted SIRT3 expression by the microbiome determines susceptibility to colon cancer
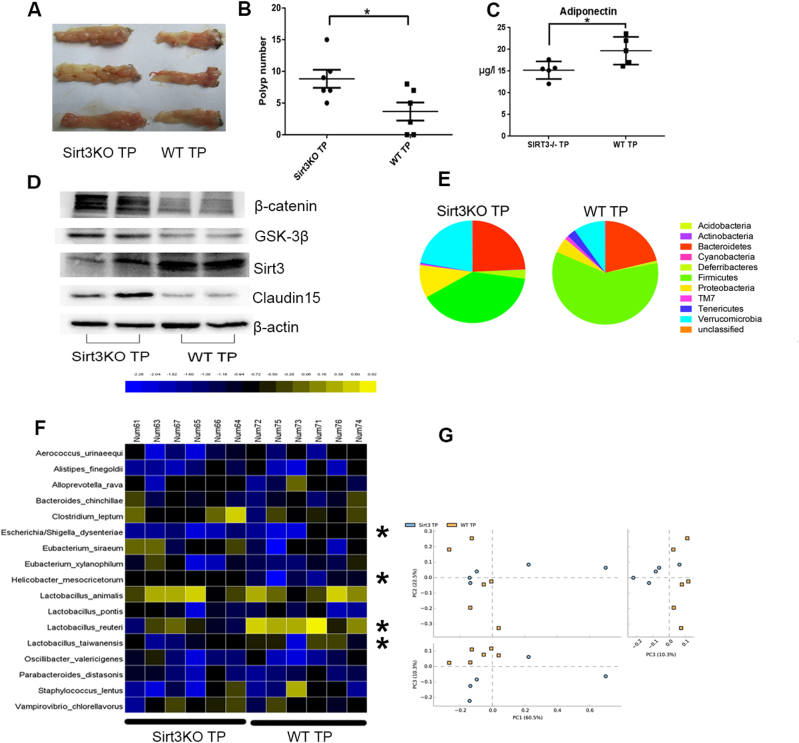
To identify the SIRT3-related gut microbes playing a key role in the development of colonic tumorigenesis, we transferred the gut microbiome of SIRT3KO mice into antibiotic-treated C57BL/6J mice in comparison with WT microbiome transfer. As expected, SIRT3KO TP mice were more susceptible to AOM/DSS tumorigenesis and developed more tumors than WT TP mice (P < 0.05, Fig. 5a, b). Moreover, the adiponectin level was significantly higher in WT TP mice than in SIRT3KO TP mice (P < 0.05, Fig. 5c). Microbiota transfer also impacted intestinal SIRT3, GSK3β, β-catenin and Claudin15 expression (Fig. 5d). Compared to WT TP mice, SIRT3KO TP mice consistently exhibited decreased levels of Firmicutes/Bacteroides and a higher proportion of Proteobacteria and Verrucomicrobia (Fig. 5e). Furthermore, an altered gut microbiome was observed with Escherichia/Shigella dysenteriae more abundant in SIRT3KO TP mice and Lactobacillus reuteri, Lactobacillus taiwanensis and Helicobacter mesocricetorum more abundant in WT TP mice (P < 0.05, Fig. 5f). These data suggested that Escherichia/S. dysenteriae, well-known infective pathogens, may play a role in colonic tumorigenesis but that Lactobacillus reuteri and Lactobacillus taiwanensis may exert a protective effect. H. mesocricetorum is a commensal rodent bacterium. The SIRT3KO TP mouse samples clustered distinctly from WT TP mice by PCoA (PC1 = 60.5% and PC3 = 10.3%), which also indicated different gut microbiome when Sirt3 was depleted (Fig. 5g). Finally, we assessed if Sirt3 activity was influenced by the gut microbiome, and we found that mice transplanted with SIRT3KO samples had low Sirt3 expression (Fig. 5c).
Fig. 5. High and low Sirt3 expression modified by microbiome transfer have different effects on colon tumorigenesis.
a Images of distal colons from SIRT3KO TP and WT TP mice after AOM/DSS tumorigenesis. b Comparison of polyp numbers between SIRT3KO TP and WT TP mice (n = 6). Data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. c Western blots of intestinal SIRT3, GSK3β, β-catenin and Claudin15 expression from SIRT3KO TP and WT TP mice after AOM/DSS tumorigenesis. d Comparison of serum adiponectin levels between SIRT3KO TP and WT TP mice (n = 5–6). Data are represented as the mean ± SEM. P < 0.05; **P < 0.01; ***P < 0.001; by unpaired Student’s t test. e Comparison of phylum-level proportional abundance of SIRT3KO TP and WT TP mice (n = 5–6) by Pacbio sequencing. f Heat map of bacterial species from feces that had a difference in abundance between SIRT3KOTP and WTTP mice (n = 6) by Pacbio sequencing. Data were analyzed by the Mann–Whitney test. g PCoA of SIRT3KO TP and WT TP mice (n = 6) by Pacbio sequencing. PC1 and PC3 explain 60.5 and 10.30% of the variation, respectively. n no significance
Discussion
It is well established that chronic inflammation of the colon can promote the incidence of carcinogenesis26,27. A clinical study also supported the idea that inflammation is an independent risk factor for colon neoplasia formation in IBD patients28. In this study, we aimed to uncover the potential role of Sirt3 in the regulation of intestinal barrier integrity and inflammation. In this study, mice lacking Sirt3 exhibited a higher inflammatory level, increased intestinal epithelial damage, and an altered gut microbiome compared to WT mice. Furthermore, Sirt3 deficiency induced altered expression of intestinal epithelial proteins, especially Claudin15. Previous research has shown that the expression of epithelial Claudin15 is positively associated with a significant deficit in intestinal barrier function23. We hypothesized that loss of Sirt3 would thus influence intestinal barrier integrity and increase susceptibility to colitis via Claudin15 and the STAT3 pathway.
Despite affecting the intestinal barrier integrity, Sirt3 is a multifunctional tumor suppressor that influences the expression of p38 and Caspase3 but not HIF1-α, ERK and p5312. A recent study demonstrated that p38 is required to maintain metabolism in CRC and that p38 inhibition can result in FOXO3 activation followed by caspase-8 and HER3 upregulation, thereby resulting in ERK1/2 over-activation in vivo29. To our knowledge, FOXO3, a direct target of Sirt3, can protect against oxidative damage30. These results indicate that the crosstalk between the ERK1/2 and p38 signaling plays a key role in Sirt3 susceptibility to CRC and that Sirt3 mainly inhibits intestinal p38 signaling as well as suppresses tumor growth and development. Here we identified an unexpected result for Sirt3 that differs from its known role in Claudin15 signaling. A possible explanation for the opposite expression of Claudin15 in colitis and colitis-associated colon cancer is that the colon of CRC is in a hyperproliferative state.
Previous research has found that Fusobacterium nucleatum-enriched intestinal cancer tissue with activated β-catenin signaling potentiates tumorigenesis in a mouse model31. Similar observations were made in our study. First, the transfer of the fecal microbiome of APC-mutant mice to WT mice eliminated resistance to AOM/DSS tumorigenesis with the activation of GSK3β/β-catenin. In contrast, high abundance of Rikenella might participate in sulfated mucin degradation, activate SLC26A2 expression and regulate intestinal sulfate transport with sulfated mucin desulfatase activity32. Hydrogen sulfide, a sulfate-reducing product, may induce DNA damage and play a role in colon carcinogenesis33. Moreover, the presence of commensal microbiota delayed the deterioration of gut carcinogenesis and suppressed β-catenin signaling. Previous research has reported that commensal microbiota induce a late onset of proinflammatory and protumorigenic responses after AOM/DSS34. The commensal microbiota were mainly characterized by a high abundance of Alistipes, which has been demonstrated as a potential probiotic at higher levels in tumor-bearing mice35. Additionally, a high abundance of Peptococcaceae has also been reported in tumor-bearing mice. Consistently, a recent study also reported that Peptostreptococcus anaerobius is detected with higher abundance in CRC and promotes mouse colorectal tumorigenesis by inducing intracellular cholesterol biosynthesis36.
Finally, transfer of Sirt3-deficient fecal microbiota to C57BL/6 mice exhibited aggravated inflammation-associated colon tumorigenesis due to a high abundance of potentially infective Escherichia/Shigella dysenteriae and low abundance of beneficial L. reuteri and L. taiwanensis. A previous study demonstrated that Escherichia/Shigella dysenteriae may promote p38 MAP kinase activation37. Lactobacillus spp., which exerts an inhibitory effect on Escherichia/Shigella in vitro and in vivo, has been demonstrated to have an anti-tumor effect, including apoptotic, antioxidant and immune-regulating functions38. A recent study revealed that L. reuteri and L. taiwanensis stimulate aryl hydrocarbon receptor (AHR) activity to reduce colitis and regulate gut microbiota39. In summary, the mechanism driving microbiome-linked colonic tumorigenesis may involve the p38 and GSK3β/β-catenin signaling pathways.
For the first time, we showed a Sirt3-microbiome interaction, of which C57BL/6 recipients colonized with the microbiome from SIRT3KO mice had decreased Sirt3 expression in comparison with WT microbiota transfer. The key microbes that prevent downregulation of Sirt3 expression during colorectal tumorigenesis are L. reuteri and L. taiwanensis, which may provide a precise therapeutic strategy for the prevention of Sirt3-deficient colon cancer by manipulation of the gut microbiome. Moreover, Sirt3 and commensal microbiota synergize to regulate the expression of CLCN4 in the development of colon cancer. CLCN4 has been shown to enhance colon cancer migration and metastasis. CLCN4 has been proposed as a risk factor for colon cancer, but its role in the early stages of colon cancer is unknown25. In summary, CLCN4 is critical for Sirt3-microbiome coordination with a tumor-suppressing effect, but further study is needed.
Considering the gut probiotic effect of SIRT3KO mice, it is possible that probiotic bacteria, such as Lactobacillus, may provide bacterial Sir2 (a gene was homologous to eukaryotic Sirt3) for deacetylase activity. A previous in vitro experiment demonstrated that bacterial Sir2 activity is elevated with co-culture of Lactobacillus acidophilus and Caco-2 cells40. Thus, Sirt3 may be crucial for a probiotic effect in gut tumorigenesis. In conclusion, Sirt3 is a novel colonic tumor suppressor that inhibits colonic tumorigenesis via an interaction with the gut microbiome.
Accession numbers
The original Miseq and Pacbio sequences of the 16S profiling have been deposited to a BioProject (PRJNA341959) in NCBI and are available under accession numbers SRS1736448–SRS1751469.
Electronic supplementary material
Acknowledgements
This study was supported by grants from the Chongqing Postdoctoral Research Project (Xm2014125).
Authors contributions:
Y.Z. and M.-t.M. designed the experiments; Y.Z., M.Z., and M.-t.C. performed the experiments: C.K., X.-l.W. and Y.Z. analyzed the data; C.K. and X.Z. contributed to the western blot experiment and bioinformatics analysis; B.W. and X.-l.W. contributed reagents and materials; Y.Z. and M.-t.M. wrote the paper; and H.-d.L. and S.-c.H. commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims inpublished maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s12276-017-0002-0.
References
- 1.Torre LA, et al. Global cancer statistics, 2012. Ca. Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kantor ED, et al. Gene-environment interaction involving recently identified colorectal cancer susceptibility loci. Cancer Epidemiol. Biomark. Prev. 2014;23:1824–1833. doi: 10.1158/1055-9965.EPI-14-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeller G, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 7.Kasai C, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: Terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol. Rep. 2016;35:325–333. doi: 10.3892/or.2015.4398. [DOI] [PubMed] [Google Scholar]
- 8.Bause AS, Haigis MC. SIRT3 regulation of mitochondrial oxidative stress. Exp. Gerontol. 2013;48:634–639. doi: 10.1016/j.exger.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Hirschey MD, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lantier L, et al. SIRT3 Is Crucial for maintaining skeletal muscle insulin action and protects against severe insulin resistance in high-fat-fed mice. Diabetes. 2015;64:3081–3092. doi: 10.2337/db14-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu C, Huang Z, Jiang H, Shi F. The Sirtuin 3 expression profile is associated with pathological and clinical outcomes in colon cancer patients. Biomed. Res Int. 2014;2014:871263. doi: 10.1155/2014/871263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014;5:e1047. doi: 10.1038/cddis.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakhan SE, Kirchgessner A. Gut microbiota and sirtuins in obesity-related inflammation and bowel dysfunction. J. Transl. Med. 2011;9:202. doi: 10.1186/1479-5876-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Sasso G, et al. Loss of Sirt1 function improves intestinal anti-bacterial defense and protects from colitis-induced colorectal cancer. PLoS ONE. 2014;9:e102495. doi: 10.1371/journal.pone.0102495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 16.Chen HM, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013;97:1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 17.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 18.Zheng P, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol. Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 19.Man SM, et al. Critical role for the DNA sensor aim2 in stem cell proliferation and cancer. Cell. 2015;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harmsen HJ, et al. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 2000;66:4523–4527. doi: 10.1128/AEM.66.10.4523-4527.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quevedo B, et al. Phylogenetic group- and species-specific oligonucleotide probes for single-cell detection of lactic acid bacteria in oral biofilms. BMC Microbiol. 2011;11:14. doi: 10.1186/1471-2180-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller S, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao EY, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Lactobacillus casei Zhang and vitamin K2 prevent intestinal tumorigenesis in mice via adiponectin-elevated different signaling pathways. Oncotarget. 2017;8:24719–24727. doi: 10.18632/oncotarget.15791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishiguro T, et al. Gene trapping identifies chloride channel 4 as a novel inducer of colon cancer cell migration, invasion and metastases. Br. J. Cancer. 2010;102:774–782. doi: 10.1038/sj.bjc.6605536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin. Gastroenterol. Hepatol. 2012;10:639–645. doi: 10.1016/j.cgh.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Beaugerie L, Itzkowitz SH. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015;372:1441–1452. doi: 10.1056/NEJMra1403718. [DOI] [PubMed] [Google Scholar]
- 28.Rutter M, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126:451–459. doi: 10.1053/j.gastro.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Chiacchiera F, et al. Blocking p38/ERK crosstalk affects colorectal cancer growth by inducing apoptosis in vitro and in preclinical mouse models. Cancer Lett. 2012;324:98–108. doi: 10.1016/j.canlet.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free. Radic. Biol. Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host. Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bomar L, Maltz M, Colston S, Graf J. Directed culturing of microorganisms using metatranscriptomics. MBio. 2011;2:e00012. doi: 10.1128/mBio.00012-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Y, et al. Gut microbiota protects against gastrointestinal tumorigenesis caused by epithelial injury. Cancer Res. 2013;73:7199–7210. doi: 10.1158/0008-5472.CAN-13-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, Leite-Moreira AF, Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev. Esp. Enferm. Dig. 2015;107:659–671. doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 36.Tsoi H, et al. Peptostreptococcus anaerobius induces intracellular cholesterol biosynthesis in colon cells to induce proliferation and causes dysplasia in mice. Gastroenterology. 2017;152:1419–1433. doi: 10.1053/j.gastro.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Lautz K, et al. NLRP10 enhances Shigella-induced pro-inflammatory responses. Cell. Microbiol. 2012;14:1568–1583. doi: 10.1111/j.1462-5822.2012.01822.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhong L, Zhang X, Covasa M. Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J. Gastroenterol. 2014;20:7878–7886. doi: 10.3748/wjg.v20.i24.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamas B, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016;22:598–605. doi: 10.1038/nm.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yanagihara S, Hirota T, Yamamoto N. Transcriptional response of Lactobacillus acidophilus L-92 after attachment to epithelial Caco-2 cells. J. Biosci. Bioeng. 2012;114:582–585. doi: 10.1016/j.jbiosc.2012.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.