Abstract
Renal cell carcinoma (RCC) is one of the 10 most common cancers worldwide, and to date, a strong systemic therapy has not been developed to treat RCC, even with the remarkable modern advances in molecular medicine mostly due to our incomplete understanding of its tumorigenesis. There is a dire unmet need to understand the etiology and progression of RCC, especially the most common subtype, clear cell RCC (ccRCC), and to develop new treatments for RCC. Genetically engineered mouse (GEM) models are able to mimic the initiation, progression, and metastasis of cancer, thus providing valuable insights into tumorigenesis and serving as perfect preclinical platforms for drug testing and biomarker discovery. Despite substantial advances in the molecular investigation of ccRCC and monumental efforts that have been performed to try to establish autochthonous animal models of ccRCC, this goal has not been achieved until recently. Here we present a review of the most exciting progress relevant to GEM models of ccRCC.
Kidney cancer: developing useful mouse models
Genetically engineered mouse models allow more detailed understanding of kidney cancers, say researchers in China. Current treatments for clear cell renal cell carcinoma (ccRCC), a common form of kidney cancer, are limited, and mouse models can help by revealing how tumors are initiated and progress. Zhigang Ji and Weibin Hou at the Peking Union Medical College Hospital in Beijing reviewed recent progress in ccRCC mouse models. In contrast to the longtime stagnation of only being able to produce ccRCC-like cell structures and cystic lesions, researchers recently have successfully produced true ccRCCs in mice by deactivating tumor suppressor genes, overexpressing oncogenic genes, or enhancing signaling pathways, in various combinations. Hou and Ji suggest that these exciting ccRCC mouse models could be especially useful for the investigation of the emerging immunotherapy for ccRCC.
Introduction
Renal cell carcinoma (RCC) is one of the 10 most frequently diagnosed cancers worldwide and accounts for approximately 3% of adult malignancies1,2. Clear cell RCC (ccRCC) is the most common subtype, accounting for approximately 75% of cases, and is characterized by a high lipid content, which gives rise to its name3. Our accumulating knowledge on the biology of RCC4,5 has led to the development of targeted therapy blocking the VHL-hypoxia-inducible factor (HIF)-vascular endothelial growth factor cascade or mTOR pathway in ccRCC6, leading to an improved clinical outcome7. However, a subset of patients is intrinsically refractory to targeted therapies, and most patients manifest tumor regression only during the first few months, followed by a short interval of an efficacy plateau. Drug resistance occurs almost inevitably after a median of 6–15 months of treatment, leading to cancer progression and, eventually, death8. Beginning in 2015, we have seen a rebirth of immunotherapy, with immune checkpoint inhibitors showing great promise in advanced RCC patients who are refractory to other treatments9. Last year, preclinical studies of a HIF2a antagonist for ccRCC showed bright prospects10,11. The prominent role of these emerging treatments in the management of metastatic RCC still needs to be established by ongoing trials, while potential challenges underlying these treatments and the opportunities to perform precision medicine based on all of these available therapies urgently call for a better preclinical animal model.
Genetic engineered animal models have served as a powerful tool to explore tumorigenesis and serve as preclinical models for drug testing, which is normally difficult or impossible to pursue clinically12. Although genetic engineered mouse (GEM) models of various cancers have been successfully established13, autochthonous cancer models of ccRCC that recapitulate its cardinal molecular and cellular features have been actively pursued but have failed, even though ccRCC is among one of the few human cancers known to evolve from a specific gene mutation4,5. Fortunately, the past few years have witnessed extraordinary success at this daunting task. In this review, we systematically describe the currently available animal models for the study of kidney cancer and discuss the in vivo role of the involved candidate genes and their cooperation in ccRCC tumorigenesis as revealed by these models. Since animal models are not only indispensable for investigating the pathogenesis of cancer in vivo but also powerful tools for preclinical investigations in cancer treatment and prevention, we will also discuss the potential utility of these various newly developed animal models of ccRCC.
Conditional inactivation of Vhl in kidney failed to produce RCC
Modern sequencing technology has shown a prevalence for biallelical inactivation of VHL in 92% of sporadic ccRCCs14; additionally, the VHL mutation is supposed to occur at the earliest stage of tumorigensis15. The role of VHL as a tumor suppressor gene (TSG) in cancer has been established by the fact that reintroducing wide-type VHL into VHL-null RCC cell lines perturbs their capacity to form tumors in immune-compromised mice in vivo16,17. However, transforming kidney cells by inactivation of VHL has basically been in vain. Homozygous germline inactivation of the Vhl gene in mice led to embryonic lethality18. A mosaic deletion19 or a heterozygous disruption20,21 of Vhl. however, does not predispose mice to RCC, in contrast to the situation in humans22. Further efforts have focused on the use of conditional inactivation of TSG or activation of oncogenes by the Cre-Lox system in adult renal tissue and cells of interest at the desired time. This conditional model better mimics the somatic mutation of sporadic cancer and is able to control spatiotemporal mutations of the genes.
Generally believed to be the cellular origin of ccRCC, proximal tubular epithelial cells have been the first choices as conditional targeting cells in the kidney. Rankin and colleagues developed the first conditional knockout mouse model using the phosphoenolpyruvate carboxykinase-Cre system to inactivate VHL in renal proximal tubule cells. They observed cellular proliferation and lipid accumulation and, most importantly, macroscopic renal cyst development at a frequency of 18% in mice older than 1 year, but no RCC development was observed before the mouses arrived at an age of 25 months23. Although VHL-associated renal cysts are usually considered to be preneoplastic lesions, this model is far from being a model that can be used to study RCC.
In addition to the mouse model, Noonan and colleagues attempted to develop a zebrafish larvae model. The proximal pronephric tubule of the vhl−/− zebrafish kidney has a clear cell histology, with tubule dilation, cilia disorganization, glycogen accumulation, and aberrant cell proliferation, which makes it a potential model of early-stage RCC. Interestingly, their zebrafish model showed promising results for the newly emerging HIF2a inhibitor treatment24.
Because accumulated evidence has indicated that ccRCC can at least partly originate from non-proximal tubules, such as the distal convoluted tubules and collecting ducts3, and the proliferative advantage due to loss of VHL function seems to preferentially be conferred to distal rather than proximal tubular cells25, some researchers have attempted to target non-proximal tubule cells of the kidney.
Frew et al.26 performed a conditional knockout of Vhl in distal tubules and collecting ducts using the Ksp1.3-Cre system (also expressed in proximal tubules) and found no abnormalities other than hydronephrosis. Recently, Pritchett and colleagues generated a mouse model using Hoxb7-Cre to knockout Vhl in kidney cells of the collecting ducts and a subset of distal tubules. Although these mice exhibited highly penetrant tubule abnormalities, including dilation, hyperplasia, presence of clear cells and cysts, and a disrupted structure, coupled with interstitial fibrosis and inflammation, no development of ccRCC has been induced27. Other groups have also tested inactivation of Vhl in mice using the Pax8-Cre system, which targets the complete tubular system28, or THP-Cre system, which targets the medullary thick ascending loop of Henle and the early distal tubule29, but no RCC or even renal cysts have been induced.
These results enabled us to arrive at the conclusion that VHL loss alone is insufficient for tumorigenesis in the kidney. Haase has provided several potential explanations for the inability of VHL inactivation to produce RCC in animals. For instance, he believes a specific gain-of-function mutation in VHL may be a requirement, the genetic background of mice may play a role, or VHL-associated renal lesions may only arise from a specific, rare type of kidney cell that is usually not targeted30. However, a more generally accepted reason is that additional mutations in other TSGs or oncogenes are required to form specific combinations of genetic alterations, together with the loss of VHL function, to initiate tumorigenesis3. In accordance with this hypothesis, researchers have combined the deletion of VHL with the deletion of other TSGs and/or activation of oncogenes in an attempt to provoke ccRCC development.
Combination of Vhl inactivation with other TSGs or oncogenes
The failure to produce ccRCC in mice by inactivating Vhl alone, together with the facts that ccRCC as well as renal cysts arise at a much lower frequency in relation to the total number of sites of VHL loss of function in the kidneys of VHL patients25, support the idea of combining additional genetic alterations with inactivation of VHL to recapitulate the tumorigenesis of ccRCC. Accumulating molecular investigations in ccRCC have revealed recurrent altered genes or pathways besides VHL inactivation. These genes and pathways, either by enhancing the activities of signaling pathways that are already involved in VHL molecular networks or interfering with other signaling pathways that may function cooperatively with the VHL pathway and serve as oncogenes or TSGs, are good candidates to be included ccRCC models.
Inactivation of Vhl and Pten
As one of the most frequently disturbed TSGs in sporadic cancers, PTEN negatively regulates the PI3K/AKT pathway, which plays a crucial role in the biological processes of cell metabolism, survival, and proliferation, among others31. The involvement of PTEN and the PI3K/AKT pathway in ccRCC was recognized nearly 20 years ago32,33 and has been reconfirmed by recent comprehensive genomic studies that indicate that approximately one-fifth of patients harbor mutations in genes involved in this pathway14,34. Moreover, the PI3K/AKT axis, cooperating with VHL, also plays a role in maintaining the primary cilium35, the reduced frequency of which is a feature of renal cystic diseases and ccRCC36,37.
Based on these facts, Frew and colleagues performed conditional co-deletion of Vhl and Pten in mouse kidney epithelial cells using the Ksp1.3-Cre system. One-hundred percent of these genetically engineered kidneys developed cysts that were lined with less ciliated and clear cell epithelial cells within 2 months of age, with 8% of the cysts presenting papillary projections (defined as atypical cystic lesions). Since all mice were sacrificed within 3–6 months of birth to avoid renal failure, no overt tumors were observed in this study. However, the combinatorial deletion of Vhl and Pten elicited cyst formation with a short latency, encouraging further exploration in this direction26.
Inactivation of Vhl and Trp53
The TP53 gene is another important TSG. Its product, p53, serves as a guardian of the genome and protects cells from DNA damage and transformation38. TP53 is regarded as a cornerstone in tumorigenesis due to its common alteration in diverse types of human cancer and involvement in approximately half of human cancers39,40. Unlike the PTEN gene, mutations of which usually only abrogate its tumor-suppressing function, mutations of TP53 usually confer it with oncogenic functions41. Although TP53 mutations only present in a small subset of ccRCC patients (2.08%)34, p53 pathway-related genes, including MDM2, CHEK2, ATM, and CDKN2A, are functionally mutated in approximately one-fourth of patients14. Moreover, TP53 mutations are much more prevalent in ccRCC with sarcomatoid dedifferentiation42 and in metastatic subclones of RCC43, indicating the pivotal role of TP53 alterations in cancer aggressiveness.
To investigate the role of TP53 mutations in RCC tumorigenesis and progression, Albers and colleagues performed conditional co-inactivation of Vhl and Trp53 in the mouse kidney epithelium using the Ksp1.3-Cre system. They found that three-fourths of mice with the Vhl and Trp53 double deletion gave rise to renal cysts after 1 year; one-eighth of these was atypical lesions. Additionally, a total of 16 neoplasms were also observed containing cells that showed altered proliferation as well as cytoplasmic clearing. Further molecular investigation of these neoplastic lesions revealed elevated target of rapamycin complex 1 (mTORC1) activity and MYC expression. These findings collectively demonstrate that inactivation of Vhl and Trp53 together induces development of renal cysts and ultimately causes progression to dysplastic and malignant lesions, the cells of which recapitulate some of the cellular and molecular features that are characteristic of human ccRCC44.
Inactivation of Vhl and Kif3a
The primary cilium is a microtubule-based organelle that is found in several cell types. In the kidney, it predominantly functions as a luminal flow sensor and is also involved in regulating tubular cell proliferation45. Our realization of the importance of the primary cilium in renal cytogenesis and tumorigenesis, and the fact that at least one subset of ccRCC develops in a cyst-dependent manner, have driven scientists to investigate the effects of Vhl inactivation together with specifically ablating primary cilia in kidney. To achieve this, the kinesin family member 3A (Kif3a) gene, which encodes a protein subunit of the kinesin-II microtubule motor complex that is essential for cilia formation, was conditionally deleted with Vhl in mouse kidney epithelial cells using the Ksp1.3-Cre system. The combined loss of Vhl and Kif3a caused renal cysts in all of the experimental participants, with a shortened latency of cyst initiation as well as increased total cystic burden and frequency of atypical cyst development46.
Inactivation of Vhl, Kif3a, and Trp53
Inspired by the facts that ablation of the primary cilium by deleting Kif3a accelerates renal cyst formation in Vhl-mutated kidney and that double inactivation of Vhl and Trp53 induces malignant lesions at a low frequency and long latency, a combination of mutations of Vhl, Trp53, and Kif3a in the mouse kidney was developed to investigate whether ablation of the primary cilium could accelerate tumor formation. The results showed that the triple-mutated kidney developed more cysts and neoplasms compared with double-mutated (Kif3a/Trp53) kidneys. However, this result is not perfect due to the inability of the triple mutation to cause ccRCC. It is strange that although the researchers tried to emphasize the tumor suppressor role of the primary cilium, they never compared these triple-mutant mice with Vhl/Trp53-mutated mice. This was disappointing because there is only one variable, the Kif3a mutation, that results in primary cilium deletion between the two mouse models, and comparing these two mouse models directly should be easier to draw a convincing conclusion47.
Inactivation of Vhl, Trp53, and Rb1
Recently, an exciting study from Harlander and colleagues showed success in developing an autochthonous mouse model of ccRCC by combining deletions of Vhl, Trp53, and Rb1 in renal epithelial cells48. More than 80% of the triple-mutant mice developed tumors within 25–61 weeks of somatic mutation, with an average of five tumors per mouse and a gender bias of more tumors in male mice, which is similar to the situation in human. Pathological investigation of these tumors revealed classic clear cell tumor cells, intratumoral hemorrhage, and necrosis, with pushing instead of infiltrative tumor margins. Immunohistochemical staining demonstrated nuclear accumulation of HIF1α and HIF2α as well as strong immunoactivity of CAIX and tissue markers indicating proximal tubule origin. Clinical diagnostic markers of ccRCC, such as PAX8 and pan-cytokeratin, were present in all cases. Pathway activation studies in these tumors confirmed strong mTORC1 activation and little MAPK pathway activity. An analysis of RNA sequencing between mouse tumors and human ccRCC showed a strong correlation in their global transcriptional profiles. RNA-sequencing analysis also confirmed the proximal tubule origin of ccRCC in these mice models. Genomic sequencing confirmed the mutation of Vhl, Trp53, and Rb1, and demonstrated a similarity in the classes of mutations, mutational load, and inter- and intratumoral heterogeneity between the modeled ccRCC and human ccRCC. Collectively, this triple-mutant mouse model recapitulates many of the pathological and molecular features of human ccRCC. Based on these facts, this mouse model has been tested in preclinical therapeutic studies, with individual ccRCC showing different sensitivity to sunitinib and everolimus treatment, suggesting the potential significant usefulness of this model for interrogating mechanisms of drug resistance, in tracking cancer evolution, and in identifying therapeutic biomarkers. Unfortunately, this autochthonous mouse model of ccRCC is not without limitations. Although the pathways regulated by these genes are frequently altered in ccRCC via different mechanisms14,34, simultaneous inactivation of Vhl, Trp53, and Rb1 is not common in human ccRCC. On the other hand, the frequently mutated TSGs in human ccRCC, including PBRM1, BAP1, SETD2, or JARID1C/KDM5C, are not found to be altered in the modeled ccRCCs. At a minimum, this triple-mutant ccRCC model may reflect a subset of human ccRCCs.
Inactivation of Vhl and Bap1
The advent of next-generation sequencing technology has led to the identification of recurrent mutations that were previously unknown in human sporadic ccRCCs. In addition to the well-known VHL inactivation, the most prominent mutations are chromatin-remodeling genes located on chromosome 3p together with VHL, including PBRM149, BAP150, and SETD251. This distinguishing genetic feature of ccRCC has not only illuminated cancer biology to a further degree but also offered us a compelling rationale to assume that these genetic alterations might function cooperatively with VHL inactivation to cause ccRCC. This allows us to “open the curtain” on our ability to manipulate chromatin-remodeling genes together with Vhl in a GEM model.
BRCA1-associated protein-1 (BAP1) is a nuclear-localized histone deubiquitinase that binds to the BRCA1 RING finger domain and functions as a TSG in various cancers52,53. Loss of BAP1 in sporadic ccRCC has been reported to be as high as 14% and defines a high-grade subset of ccRCC50. To investigate the effects of the combined inactivation of the Vhl and Bap1 in the mouse kidney, Wang and colleagues developed a mouse model using the Six2-Cre system to specifically induce gene deletion in nephron progenitor cells. However, mice deficient in Bap1 were obviously moribund and died before 1 month of age. Then, mice deficient in Vhl and one allele of Bap1 were developed. Although initially morphologically normal, these mice displayed increasingly extensive abnormalities in histology starting at 1 month of age. Most importantly, lesions spanning from premalignant cysts to cystic and solid RCC have been identified, resembling the manifests of kidneys from patients with VHL syndrome. Although the tumors were only 0.25–1.8 mm diameter and organized in a micropapillary or solid cystic form, the tumor cell cytoplasm was generally eosinophilic and clear. Further molecular investigation revealed positive staining for CAIX, which is a HIF-targeted protein, and elevated staining for phosphor-S6, which is a marker of mTORC1 activation. These results indicated that this mouse model deficient in Vhl and one allele of Bap1 was able to reproduce some of the features of human ccRCC. Loss of 3p in humans can simultaneously cause both VHL and BAP1 deletion, but this would not occur in mouse due to the location of these two genes on different chromosomes. This study also partly explained why a GEM model of ccRCC is lacking54.
Based on their partial success in producing a mouse model of ccRCC deficient in Vhl and one allele of Bap1, the same group further investigated the double deletion of Bap1 and Vhl in mouse kidney using different Cre drivers. Six2-Cre-induced Vhl deletion and loss of heterozygosity of Bap1 in mice led to mice dying shortly after birth, and kidney tumors that were too small; Sglt2-Cre or Villin-Cre failed to induce tumors. The investigators finally succeeded using the PAX8-Cre system, which is a nephric lineage transcription factor that targets the complete tubular system. These mice, which had a biallelic deficiency of Vhl and Bap1, lived to approximately 3 months of age. By that time, all mice presented with small RCCs and cystic lesions in kidneys, and the morphological features of their tumor cells were similar to those of the Six2-Cre mice. On the other hand, mice deficient in Vhl and one allele of Bap1 had a median survival of 14.5 months, displaying cystic lesions and tumors at 10 and 11 months of age, respectively. Both of these Pax8-Cre mouse models showed increased expression of HIF target genes and increased activation of mTORC1, which is consistent with the clinical features of human ccRCC with BAP1 deficiency. Another interesting and unexpected finding of this study is that the Bowman’s capsule, rather than the proximal tubule, as was previously thought, has been proposed to be the cell of origin of ccRCC based on their results55.
Inactivation of Vhl and Pbrm1
Large-scale genomic-sequencing projects identified PBRM1, the product of which is a subunit of the SWI/SNF chromatin-remodeling complex, which is also located at chromosome 3p close to VHL, as the second most commonly altered gene after VHL in sporadic ccRCC49. Several other studies have confirmed this conclusion and revealed the prevalence of this somatic gene mutation in ccRCC to be 40–55%14,50,56. However, our knowledge of PBRM1 in cancer biology is limited.
Nargund and colleagues pioneered a project to investigate the role of Pbrm1 in the tumorigenesis of ccRCC by deletion of both Vhl and Pbrm1 to produce an autochthonous ccRCC mouse model. They used the Ksp-Cre system to induce a conditional deletion of Vhl and Pbrm1 in mouse kidney epithelial cells starting from the embryonic stage. Mice deficient in both Vhl and Pbrm1 developed preneoplastic polycystic kidney diseases at 6–9 months of age with a 30% incidence, and histologic examination displayed a 50% tumor incidence at 10 months of age. These tumors showed key features of human ccRCC, including a clear cytoplasm, positive staining for CAIX, and hyperactive mTORC1 signaling. The origin of these tumors from kidney proximal tubules, the downregulated OXPHOS genes, and the gene expression profile all suggested that the induced kidney tumor resembled ccRCC rather than other types of RCC, such as papillary and chromophobe RCC57.
Another group developed a RCC GEM model based on Pax8-Cre deletion of Vhl and Pbrm1. Their results showed that approximately 85% of double-deleted mice presented bilateral, multiple, large, and homogeneous tumors at the age of 9 months and 100% at the age of 13 months. The tumor size increased along with age, until almost completely replacing the whole kidney at 16 months of age. These tumors exhibited obviously pushing borders and typical CAIX staining55.
Both of the studies confirmed the tumor suppressor role of PBRM1 and that loss of VHL and PBRM1 in the kidney predisposes mice to ccRCC that recapitulates key features of human ccRCC, which is potentially useful for further research. However, the two models have contradictory conclusions in terms of the mTORC1 activation status: one is hyperactive and the other is hypoactive. Additionally, the model using the Pax8-Cre system had a higher penetrance. These differences might reflect a different cell of origin of ccRCC in these two models.
Interestingly, Gu and colleagues studied the simultaneous deletion of Vhl/Bap1 and Vhl/Pbrm1 in mice and found that Bap1 and Pbrm1 not only drives ccRCC formation but also plays an important role in determining the tumor grade55. This result is compatible with the fact that mutations of BAP1 and PBRM1 have been repeatedly confirmed to be associated with the tumor grade, aggressiveness, and patient survival in human ccRCC cohorts50,58. This group further investigated the pathway underlying the tumor grade differences in PBRM1- and BAP1-deficient ccRCC. The activating status of mTORC1, which is overactivated in BAP1-deficient but not PBRM1-deficient ccRCC, leads to a high-grade and aggressive tumor. Deletion of one copy of TSC1 to active mTORC1 drives to low-grade tumors in PBRM1-deficient ccRCC to high-grade tumors55. This conclusion is important in clinical practice.
Constitutive activation of Hif1α
In addition to inactivation of TSGs, activation of oncogenes plays an equally important role in tumorigenesis. The most important molecular change in ccRCC is the inactivation of VHL, the product of which is a component of an E3 ubiquitin ligase complex that targets the a-subunits of the HIFs for ubiquitination and proteolytic degradation59. In most ccRCC, the loss of VHL function leads to stabilization and constitutive activation of HIFs, which account for up to 95% of ccRCCs14. Since the activation of HIFs substantially contributes to the critical phenotypes of ccRCC, including angiogenesis, invasiveness, and lipid storage60, its role in tumorigenesis of ccRCC has been investigated61. The supplementary role of HIFs as oncogenes in ccRCC has been clearly shown by the fact that co-deletion of the Hif1a/Hif2a or Arnt genes can rescue cystic phenotypes in Vhl mutant kidneys and cystic and neoplastic phenotypes in Vhl/Trp53 mutant kidneys62. However, the direct and independent oncogenic role of HIFs in the tumorigenesis of ccRCC was described by Fu and colleagues, who developed a GEM model with constitutively active Hif1α or Hif2α specifically expressed in renal proximal tubules using the type 1γ-glutamyl transpeptidase Cre system. Their results showed that Hif1α but not Hif2α activation drives the formation of “clear” cells and renal cysts, as well as other pathological and molecular features of early-stage human ccRCC. However, no bona fide ccRCC has been observed in these mice61. This study emphasized the oncogenic role of HIF1α but not HIF2α in ccRCC tumorigenesis, which is in contrast with the prevalent opinion regarding HIF1α as a TSG and HIF2α as an oncogene in ccRCC63. The insufficiency of inducing bona fide ccRCC in this GEM model indicates that further study is required to clarify the oncogenic role of HIF1α in the tumorigenesis of ccRCC.
Inactivation of Vhl and activation of Notch1
Notch signaling is an evolutionarily conserved pathway of cell–cell communication and cell fate determination, with its biological function being critically context-dependent in cancer64,65. In ccRCC, NOTCH1 expression is significantly increased compared with adjacent non-neoplastic tissues; further investigation of this pathway in ccRCC has revealed its oncogenic role in promoting tumor proliferation and migration by downregulating PTEN expression and increasing the activity of the PI3K/AKT pathway66–68.
Johansson and colleagues performed an in vivo study to determine whether activating Notch signaling could induce tumor formation in renal proximal tubular epithelial cells. They established a conditional mouse model with a concurrent deletion of the Vhl gene and ectopic expression of constitutively active Notch1 (NICD1) using the Kap2-iCre system to target proximal tubular cells. Though only one neoplastic lesion was found in 12 mice, there were extensive numbers of dysplastic cells that had a clear cytoplasm with lipid accumulation, which mimics an important hallmark of ccRCC in human69. A further in vivo study supported the role of activating Notch signaling in inducing cytoplasmic lipid accumulation in ccRCC, and inhibition of Notch signaling was able to reverse this process69.
Another group developed a similar mouse model and also showed dysplastic changes in these tubular cells with deleted Vhl and activated Notch. They further performed a comprehensive molecular transcription analysis to compare the molecular changes of Notch-driven tubular cells70 with those of human ccRCC, revealing that a considerable proportion of dysregulated genes in human ccRCC were driven by NOTCH pathway activation. However, the above results seem to indicate that Notch activation together with Vhl deletion is not sufficient to cause ccRCC, at least in proximal tubular cells.
Vhl and Cdkn2a(Ink4a/Arf) deletion with Myc activation
The MYC gene, which is mapped to chromosome 8q24, is one of the most commonly amplified oncogenes in various human cancers, including ccRCC. Overactivation of MYC confers a selective growth advantage to cells and orchestrates many hallmark features of cancer71. MYC activation has been found to be the top-ranking molecular pathway that is dysregulated in ccRCC when using adjacent normal tissues as controls72. Increased levels of MYC gene expression have been found in most RCC72 and could result from different genetic and epigenetic mechanisms, including focal amplification of 8q24 harboring MYC73 and regulation of MYC expression by single-nucleotide polymorphisms74.
However, unless in a permissive context, MYC overexpression alone is incapable of inducing cellular transformation or initiating tumorigenesis71. Due to the crucial role of MYC activation in ccRCC, as well as the high prevalence of VHL deletion in ccRCC, it is easy to propose a hypothesis that VHL deletion might serve as the permissive context for MYC in ccRCC development. Bailey and colleagues provided solid evidence for this hypothesis by developing a mouse model of RCC with a conditional deletion of Vhl and overexpression of Myc in renal tubular cells. RCC developed at a high incidence (67%) with modest clear cell changes. However, they did not stop there. In addition to Vhl deletion and MYC overexpression, inactivation of the Cdkn2a gene was added to the mouse model. The Cdkn2a gene, the mutations of which are implicated in 31% of ccRCC34, encodes the CDK4/6 inhibitor p16(ink4a), which is involved in cellular senescence by regulating the RB pathway, as well as a linked protein Arf that regulates the p53 pathway75. The new mouse model had a 100% penetrance of tumor formation, with its histological manifestations and gene expression profiles having a striking resemblance to human ccRCC. Additionally, Cdkn2a(Ink4a/Arf) deletion promoted macroscopic metastases of ccRCC to the liver in one-third of mice76. This aggressive feature of ccRCC has never been recapitulated in any other autochthonous ccRCC mouse models.
Conclusions
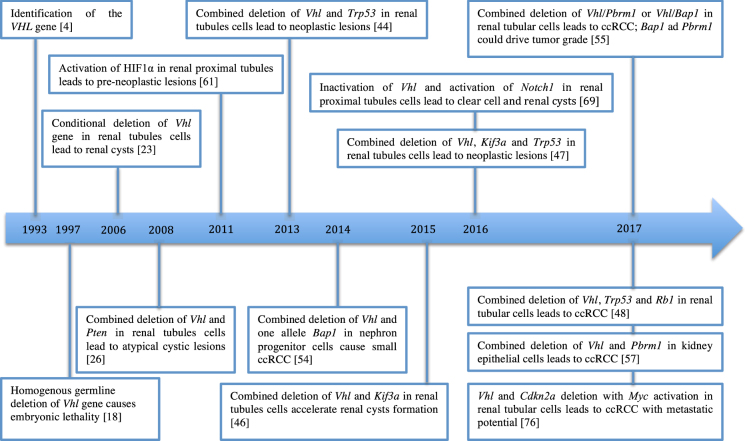
The high versatility of the genetic tools that are currently available makes the study of GEM models attractive and vital. GEM cancer models can mimic most of the genetic aberrations found in human cancers; thus, they are valuable tools to define the causal role of genetic events in cancer. Here more than 10 different important models were introduced (as shown in Fig. 1). Most of these models only reproduce the precursor lesions or few of the important phenotypes of ccRCC, showing the difficultly in successfully developing an autochthonous ccRCC model. The good news is that this year, five models were able to fully reproduce a bona fide ccRCC in adult mice (Table 1).
Fig. 1.
Timeline. Key events in the method of generating GEM models of ccRCC
Table 1.
Comparison of the key features of the five established GEM models of ccRCC
| GEM models | Cre-Lox system | Penetrance (age) | Tumor diameter | Pathological features | Pathway activation | Tumor aggressiveness |
|---|---|---|---|---|---|---|
| Vhl −/− Pbrm −/− 57 | Ksp-Cre | 50% (≥10 months) | (Typical tumors) 2 mm | Clear cytoplasm with lipid and glycogen; positive CAIX, LTL, CD31, Ki-67; negative THP, CD45, cleaved caspase-3 | Overexpression of HIF1 and JAK-STAT pathway genes; reduced expression of oxidative phosphorylation genes; hyperactive mTORCI signaling. | No local invasion; no metastases; transplanted tumor is metastatic |
| Vhl −/− Pbrm −/− 55 | Pax8-Cre | 85.7% (9–13 months)100% (>13 months) | 0.5–12 mm | Moderately clear cell with lipid and glycogen, pushing borders, small nested architectures, no necrotic regions, low-grade; positive CAIX, CD10, Ki-67, CD31, vimentin | Upregulation of HIF1 and HIF2 targeted genes; weak or no activation of mTORC1 pathway in most tumors | Not available |
| Vhl –/− Bap1 +/− 55 | Pax8-Cre | 100% (11 months) | 0.7–2.4 mm | Cystic masses, cells with pleomorphism, eosinophilic or clear cytoplasm with lipid and glycogen, high-grade; positive CAIX, CD10, Ki-67, CD31, vimentin | Upregulation of HIF1 and HIF2 targeted genes; overactivation of mTORC1 pathway | Focal lymphovascular invasion |
| Vhl − /− Trp53 -/- Rb1 -/- 48 | KSP1.3-Cre | 82% (25–61 weeks) | (Typical tumors) 5 mm | Grade 3/4 tumors in acinar, solid or pseudopapillary pattern, clear cytoplasm; necrotic regions and intratumor hemorrhage, pushing margins; positive CAIX, CD10, CD31, vWF, nuclear HIF1α/HIF2α, PAX8, pan-cytokeratin, aquaporin1; negative THP, NCC, aquaporin2 | Upregulation of HIF1 and HIF2 targeted genes; overactivation of mTORC1 pathway; no activation of RAS-MAPK pathway | No invasion; no metastases |
| Vhl−/− Cdkn2a−/− Myc activation76 | Ksp-Cre | 100% (N/A, expected 20–30 weeks) | (Most tumors) over 3 mm | Solid or tightly packed tubule-papillary tumor, high-grade, clear cytoplasm, necrotic regions | Upregulation of genes related to epithelial mesenchymal transition process | 1/3 of mice have metastases in liver |
CAIX carbonic anhydrase IX, LTL lotus tetragonolobus lectin, THP Tamm-Horsfall protein, vWF von Willebrand factor, MAPK mitogen activated protein kinase
These five established ccRCC models were derived from completely different combinations of genetic alterations, but all included Vhl inactivation, emphasizing the critical role of VHL in ccRCC tumorigenesis even though VHL inactivation alone cannot initiate ccRCC. TP53, RB1, MYC, and CDKN2A, which cooperate with VHL in ccRCC tumorigenesis, are among the most frequently altered genes across human cancer40,77,78, indicating that ccRCC initiation could share common pathways with other cancers. The BAP1 and PBRM1 genes represent ccRCC-specific TSGs that are not common in other cancers, indicating a relatively specific method of tumorigenesis of ccRCC. This means that ccRCC could arise via different mechanisms, which is also exemplified by cyst-dependent and cyst-independent pathways46, and the term ccRCC may describe a group of distinct and heterogeneous diseases. This inter-model heterogeneity mimics the intertumoral heterogeneity of ccRCC. Although none of these mice could recapitulate every aspect of ccRCC, every model might potentially represent a subset of ccRCC, serving different purposes.
Another recurrent mutated TSG on chromosome 3p is SET domain-containing 2 (SETD2), which encodes a histone methyltransferase and mutated in ccRCC at a rate of 10–20%14,34,56. Knocking down SETD2 in renal primary tubular epithelial cells in vitro has revealed that SETD2 facilitates a malignant transformation of cells to ccRCC by increasing cell proliferation and bypassing the senescence barrier79. Generation of a GEM ccRCC model deficient in Setd2 and Vhl would be promising.
Hopefully, these abovementioned new GEM models of ccRCC will be able to be widely used to investigate the biological mechanisms underlying tumor progression, lead to the discovery of new tumor biomarkers, and to test new drugs, which are currently dominated by in vitro studies and murine xenograft models derived from RCC cell lines or human ccRCC explants. These GEM models are more advantageous, especially for the research on the currently emerging immunotherapy of RCC, which cannot be done well on in vitro cell lines or immune-compromised xenograft mouse models. An autochthonous mouse model would offer an excellent opportunity to investigate the therapeutic efficacy and find relevant biomarkers in a genetically controlled setting in the context of a non-compromised tumor microenvironment and immune reaction. A mouse model with metastatic potential would provide the perfect tool for performing molecular investigations of metastatic mechanisms.
Several attempts to generate ccRCC mouse models were close to success, but failed. They are capable of recapitulating some of the cardinal hallmarks of ccRCC, such as a clear cell histology and preneoplastic cystic lesions, shedding some light on the mystery of ccRCC, but they do not lead to a bona fide ccRCC. One important explanation for these failures is that conditional knockout mice with a complete loss of proteins cannot adequately model gene mutations that produce mutant or truncated proteins that have residual activities. A good example of this is about TP53 mutations: patient-relevant Trp53 hotspot mutations in mice are much more oncogenic than Trp53 knockouts80,81. A similar story applies to Brca1 mutations and knockout in mice82,83. The revolutionary technology CRISPR/Cas9 could efficiently introduce defined mutations and might be a good choice for introducing specific mutations into GEM models13. Other ways to improve the outcome might be controlling different gene alterations in chronological order rather than simultaneously, for example, deleting Vhl first and then other genes after an interval. This suggestion is based on an observation that the tumor phenotype is directly shaped by the order of acquisition of genetic alterations84 and that the order of genetic mutations influences the clonal evolution of cancer85. Altering genes in order rather than simultaneously might better resemble the natural process of tumorigenesis, although this manipulation would increase the difficulty of experiments.
Acknowledgements
W.H. is now supported by a fellowship from the China Scholarship Council (201506210081) to study in Heidelberg University, Germany (2015–2018).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hsieh JJ, et al. Renal cell carcinoma. Nat. Rev. Dis. Prim. 2017;3:17009. doi: 10.1038/nrdp.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Frew IJ, Moch H. A clearer view of the molecular complexity of clear cell renal cell carcinoma. Annu. Rev. Pathol. 2015;10:263–289. doi: 10.1146/annurev-pathol-012414-040306. [DOI] [PubMed] [Google Scholar]
- 4.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 5.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 6.Clark PE. The role of VHL in clear-cell renal cell carcinoma and its relation to targeted therapy. Kidney Int. 2009;76:939–945. doi: 10.1038/ki.2009.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal-cell carcinoma. N. Engl. J. Med. 2017;376:354–366. doi: 10.1056/NEJMra1601333. [DOI] [PubMed] [Google Scholar]
- 8.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10:992–1000. doi: 10.1016/S1470-2045(09)70240-2. [DOI] [PubMed] [Google Scholar]
- 9.Motzer RJ, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1803–1813. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho H, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat. Rev. Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 13.Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017;9:137–153. doi: 10.15252/emmm.201606857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 15.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliopoulos O, Kibel A, Gray S, Kaelin WG., Jr. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1995;1:822–826. doi: 10.1038/nm0895-822. [DOI] [PubMed] [Google Scholar]
- 17.Gnarra JR, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl Acad. Sci. USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnarra JR, et al. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl Acad. Sci. USA. 1997;94:9102–9107. doi: 10.1073/pnas.94.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma W, et al. Hepatic vascular tumors, angiectasis in multiple organs, and impaired spermatogenesis in mice with conditional inactivation of the VHL gene. Cancer Res. 2003;63:5320–5328. [PubMed] [Google Scholar]
- 20.Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc. Natl Acad. Sci. USA. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleymenova E, et al. Susceptibility to vascular neoplasms but no increased susceptibility to renal carcinogenesis in Vhl knockout mice. Carcinogenesis. 2004;25:309–315. doi: 10.1093/carcin/bgh017. [DOI] [PubMed] [Google Scholar]
- 22.Lonser RR, et al. von Hippel-Lindau disease. Lancet. 2003;361:2059–2067. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 23.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66:2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noonan HR, et al. Loss of vhl in the zebrafish pronephros recapitulates early stages of human clear cell renal cell carcinoma. Dis. Model Mech. 2016;9:873–884. doi: 10.1242/dmm.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandriota SJ, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/S1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 26.Frew IJ, et al. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J. 2008;27:1747–1757. doi: 10.1038/emboj.2008.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchett TL, Bader HL, Henderson J, Hsu T. Conditional inactivation of the mouse von Hippel-Lindau tumor suppressor gene results in wide-spread hyperplastic, inflammatory and fibrotic lesions in the kidney. Oncogene. 2015;34:2631–2639. doi: 10.1038/onc.2014.197. [DOI] [PubMed] [Google Scholar]
- 28.Schietke RE, et al. Renal tubular HIF-2alpha expression requires VHL inactivation and causes fibrosis and cysts. PLoS ONE. 2012;7:e31034. doi: 10.1371/journal.pone.0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schley G, et al. Hypoxia-inducible transcription factors stabilization in the thick ascending limb protects against ischemic acute kidney injury. J. Am. Soc. Nephrol. 2011;22:2004–2015. doi: 10.1681/ASN.2010121249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haase VH. The VHL tumor suppressor in development and disease: functional studies in mice by conditional gene targeting. Semin. Cell Dev. Biol. 2005;16:564–574. doi: 10.1016/j.semcdb.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alimov A, et al. Somatic mutation and homozygous deletion of PTEN/MMAC1 gene of 10q23 in renal cell carcinoma. Anticancer Res. 1999;19(5B):3841–3846. [PubMed] [Google Scholar]
- 33.Velickovic M, Delahunt B, McIver B, Grebe SK. Intragenic PTEN/MMAC1 loss of heterozygosity in conventional (clear-cell) renal cell carcinoma is associated with poor patient prognosis. Mod. Pathol. 2002;15:479–485. doi: 10.1038/modpathol.3880551. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thoma CR, et al. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat. Cell Biol. 2007;9:588–595. doi: 10.1038/ncb1579. [DOI] [PubMed] [Google Scholar]
- 36.Schraml P, et al. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Mod. Pathol. 2009;22:31–36. doi: 10.1038/modpathol.2008.132. [DOI] [PubMed] [Google Scholar]
- 37.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol. Renal Physiol. 2005;289:F1159–F1169. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 38.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 39.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 40.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 42.Malouf GG, et al. Genomic characterization of renal cell carcinoma with sarcomatoid dedifferentiation pinpoints recurrent genomic alterations. Eur. Urol. 2016;70:348–357. doi: 10.1016/j.eururo.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 43.Bousquet G, et al. Tracking sub-clonal TP53 mutated tumor cells in human metastatic renal cell carcinoma. Oncotarget. 2015;6:19279–19289. doi: 10.18632/oncotarget.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Albers J, et al. Combined mutation of Vhl and Trp53 causes renal cysts and tumours in mice. EMBO Mol. Med. 2013;5:949–964. doi: 10.1002/emmm.201202231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoder BK. Role of primary cilia in the pathogenesis of polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:1381–1388. doi: 10.1681/ASN.2006111215. [DOI] [PubMed] [Google Scholar]
- 46.Lehmann H, Vicari D, Wild PJ, Frew IJ. Combined deletion of Vhl and Kif3a accelerates renal cyst formation. J. Am. Soc. Nephrol. 2015;26:2778–2788. doi: 10.1681/ASN.2014090875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guinot A, Lehmann H, Wild PJ, Frew IJ. Combined deletion of Vhl, Trp53 and Kif3a causes cystic and neoplastic renal lesions. J. Pathol. 2016;239:365–373. doi: 10.1002/path.4736. [DOI] [PubMed] [Google Scholar]
- 48.Harlander S, et al. Combined mutation in Vhl, Trp53 and Rb1 causes clear cell renal cell carcinoma in mice. Nat. Med. 2017;23:869–877. doi: 10.1038/nm.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duns G, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 52.Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 53.Ventii KH, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang SS, et al. Bap1 is essential for kidney function and cooperates with Vhl in renal tumorigenesis. Proc. Natl Acad. Sci. USA. 2014;111:16538–16543. doi: 10.1073/pnas.1414789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu YF, et al. Modeling renal cell carcinoma in mice: Bap1 and Pbrm1 inactivation drive tumor grade. Cancer Discov. 2017;7:900–917. doi: 10.1158/2159-8290.CD-17-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scelo G, et al. Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun. 2014;5:5135. doi: 10.1038/ncomms6135. [DOI] [PubMed] [Google Scholar]
- 57.Nargund AM, et al. The SWI/SNF protein PBRM1 restrains VHL-loss-driven clear cell renal cell carcinoma. Cell Rep. 2017;18:2893–2906. doi: 10.1016/j.celrep.2017.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kapur P, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 60.Lee KE, Simon MC. SnapShot: hypoxia-inducible factors. Cell. 2015;163:1288–1288. doi: 10.1016/j.cell.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Fu L, Wang G, Shevchuk MM, Nanus DM, Gudas LJ. Generation of a mouse model of Von Hippel-Lindau kidney disease leading to renal cancers by expression of a constitutively active mutant of HIF1alpha. Cancer Res. 2011;71:6848–6856. doi: 10.1158/0008-5472.CAN-11-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schonenberger D, et al. Formation of renal cysts and tumors in Vhl/Trp53-deficient mice requires HIF1alpha and HIF2alpha. Cancer Res. 2016;76:2025–2036. doi: 10.1158/0008-5472.CAN-15-1859. [DOI] [PubMed] [Google Scholar]
- 63.Schodel J, et al. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur. Urol. 2016;69:646–657. doi: 10.1016/j.eururo.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Capaccione KM, Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34:1420–1430. doi: 10.1093/carcin/bgt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S, et al. NOTCH1 functions as an oncogene by regulating the PTEN/PI3K/AKT pathway in clear cell renal cell carcinoma. Urol. Oncol. 2013;31:938–948. doi: 10.1016/j.urolonc.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 67.Ai Q, et al. High-level expression of Notch1 increased the risk of metastasis in T1 stage clear cell renal cell carcinoma. PLoS ONE. 2012;7:e35022. doi: 10.1371/journal.pone.0035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sjolund J, et al. Suppression of renal cell carcinoma growth by inhibition of Notch signaling in vitro and in vivo. J. Clin. Invest. 2008;118:217–228. doi: 10.1172/JCI32086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johansson E, et al. Simultaneous targeted activation of Notch1 and Vhl-disruption in the kidney proximal epithelial tubular cells in mice. Sci. Rep. 2016;6:30739. doi: 10.1038/srep30739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bielesz B, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J. Clin. Invest. 2010;120:4040–4054. doi: 10.1172/JCI43025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014;4:a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao M, Shuin T, Misaki H, Kubota Y. Enhanced expression of c-myc and epidermal growth factor receptor (C-erbB-1) genes in primary human renal cancer. Cancer Res. 1988;48:6753–6757. [PubMed] [Google Scholar]
- 73.Yamaguchi S, et al. The allelic loss of chromosome 3p25 with c-myc gain is related to the development of clear-cell renal cell carcinoma. Clin. Genet. 2003;63:184–191. doi: 10.1034/j.1399-0004.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 74.Grampp S, et al. Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nat. Commun. 2016;7:13183. doi: 10.1038/ncomms13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J. Cancer. 2012;130:1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey ST, et al. MYC activation cooperates with Vhl and Ink4a/Arf loss to induce clear cell renal cell carcinoma. Nat. Commun. 2017;8:15770. doi: 10.1038/ncomms15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cousin S, et al. Clinical impact of extensive molecular profiling in advanced cancer patients. J. Hematol. Oncol. 2017;10:45. doi: 10.1186/s13045-017-0411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li J, et al. Functional studies on primary tubular epithelial cells indicate a tumor suppressor role of SETD2 in clear cell renal cell carcinoma. Neoplasia. 2016;18:339–346. doi: 10.1016/j.neo.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 81.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 82.Drost R, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 83.Drost R, et al. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J. Clin. Invest. 2016;126:2903–2918. doi: 10.1172/JCI70196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herbet M, Salomon A, Feige JJ, Thomas M. Acquisition order of Ras and p53 gene alterations defines distinct adrenocortical tumor phenotypes. PLoS Genet. 2012;8:e1002700. doi: 10.1371/journal.pgen.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kent DG, Green AR. Order matters: the order of somatic mutations influences cancer evolution. Cold Spring Harb. Perspect. Med. 2017;7:a027060. doi: 10.1101/cshperspect.a027060. [DOI] [PMC free article] [PubMed] [Google Scholar]